Summary
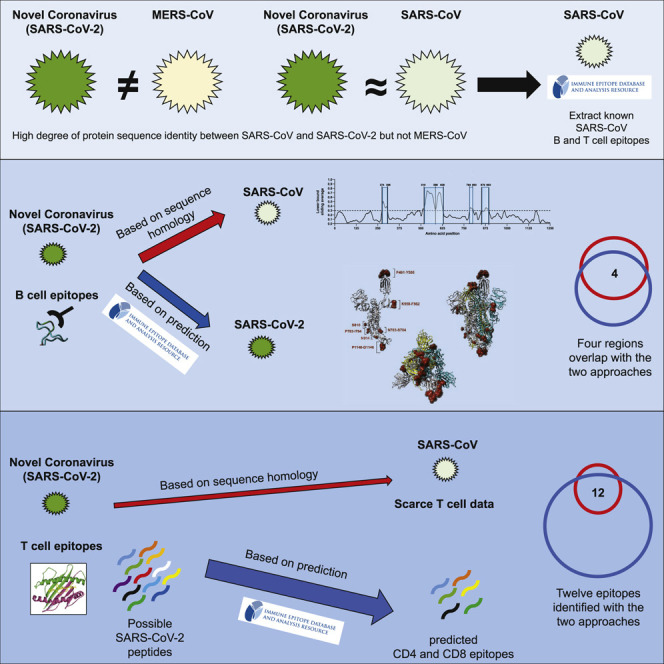
Effective countermeasures against the recent emergence and rapid expansion of the 2019 novel coronavirus (SARS-CoV-2) require the development of data and tools to understand and monitor its spread and immune responses to it. However, little information is available about the targets of immune responses to SARS-CoV-2. We used the Immune Epitope Database and Analysis Resource (IEDB) to catalog available data related to other coronaviruses. This includes SARS-CoV, which has high sequence similarity to SARS-CoV-2 and is the best-characterized coronavirus in terms of epitope responses. We identified multiple specific regions in SARS-CoV-2 that have high homology to the SARS-CoV virus. Parallel bioinformatic predictions identified a priori potential B and T cell epitopes for SARS-CoV-2. The independent identification of the same regions using two approaches reflects the high probability that these regions are promising targets for immune recognition of SARS-CoV-2. These predictions can facilitate effective vaccine design against this virus of high priority.
Keywords: SARS-CoV, COVID-19, SARS-CoV-2, coronavirus, T cell epitope, B cell epitope, infectious disease, sequence conservation
Graphical Abstract
Highlights
-
•
Ten experimentally defined regions within SARS-CoV have high homology with SARS-CoV-2
-
•
Parallel bioinformatics predicted potential B and T cell epitopes for SARS-CoV-2
-
•
Independent approaches identified the same immunodominant regions
-
•
The conserved immune regions have implications for vaccine design against multiple CoVs
Grifoni et al. identify potential targets for immune responses to the 2019 novel coronavirus (SARS-CoV-2) by sequence homology with closely related SARS-CoV and by a priori epitope prediction using bioinformatics approaches. This analysis provides essential information for understanding human immune responses to this virus and for evaluating diagnostic and vaccine candidates.
Introduction
On December 31, 2019, the Chinese Center for Disease Control (China CDC) reported a cluster of severe pneumonia cases of unknown etiology in the city of Wuhan in the Hubei province of China. Shortly thereafter, public health professionals identified the likely causative agent to be a novel Betacoronavirus (SARS-CoV-2). The current outbreak, COVID-19, has 81,109 confirmed cases worldwide with 2,718 deaths, as of February 26, 2020, according to the World Health Organization (WHO) in collaboration with the China CDC and public health centers in other countries. Although the majority of cases have occurred in China, a small number have been confirmed in 24 other countries, including Japan, Thailand, South Korea, Singapore, Vietnam, India, the United States, Canada, Germany, France, Italy, and the United Arab Emirates. These numbers are changing rapidly. For up-to-date information about COVID-19, see the WHO website at https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
The Immune Epitope Database and Analysis Resource (IEDB) is a repository of epitope-related information curated from the scientific literature in the context of infectious disease, allergy, and autoimmunity (Vita et al., 2019). The IEDB also provides bioinformatic tools and algorithms that allow for the analysis of epitope data and prediction of potential epitopes from novel sequences. The Virus Pathogen Resource (ViPR) is a complementary repository of information about human pathogenic viruses that integrates genome, gene, and protein sequence information with data about immune epitopes, protein structures, and host responses to virus infections (Pickett et al., 2012).
Limited information is currently available on which parts of the SARS-CoV-2 sequence are recognized by human immune responses. Such knowledge is of immediate relevance and would assist vaccine design and facilitate the evaluation of vaccine candidate immunogenicity, as well as monitoring of the potential consequences of mutational events and epitope escape as the virus is transmitted through human populations.
Although no epitope data are yet available for SARS-CoV-2, there is a significant body of information about epitopes for coronaviruses in general, and in particular for Betacoronaviruses like SARS-CoV and MERS-CoV, which cause respiratory disease in humans (de Wit et al., 2016, Song et al., 2019). Here, we used the IEDB and ViPR resources to compile known epitope sites from other coronaviruses, map corresponding regions in the SARS-CoV-2 sequences, and predict likely epitopes. We also used validated bioinformatic tools to predict B and T cell epitopes that are likely to be recognized in humans and to assess the conservation of these epitopes across different coronavirus species.
Results
A Wealth of Data Related to Coronaviruses Is Available in the IEDB
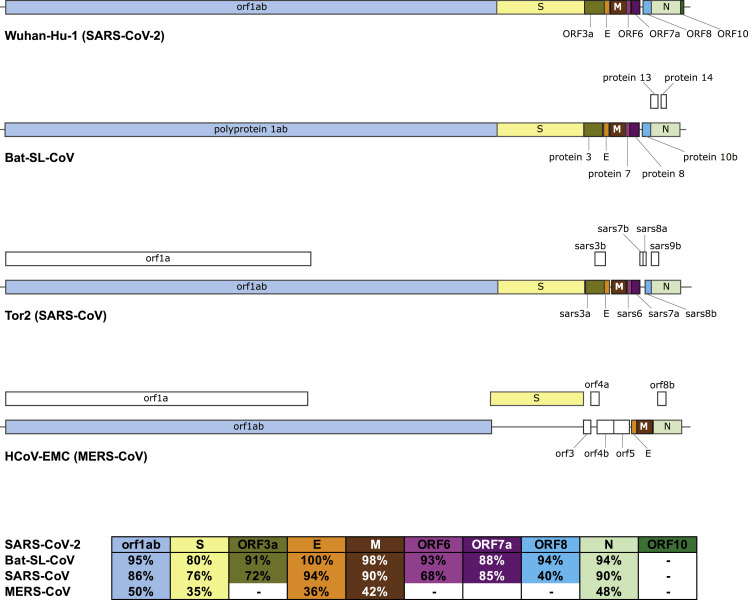
Coronaviruses belong to the family Coronaviradae, order Nidovirales, and can be further subdivided into four main genera (Alpha-, Beta-, Gamma-, and Deltacoronaviruses). Several Alpha- and Betacoronaviruses cause mild respiratory infections and common cold symptoms in humans, whereas others are zoonotic and infect birds, pigs, bats, and other animals. In addition to SARS-CoV-2, two other coronaviruses, SARS-CoV and MERS-CoV, caused large disease outbreaks that had high (10%–30%) lethality rates and widespread societal impact upon emergence (Figure 1 ) (de Wit et al., 2016, Song et al., 2019).
Figure 1.
Comparison of SARS-CoV-2 (Wuhan-Hu-1) Genome Structure with Its Closest Bat Relative (bat-SL-CoVZXC21), Tor2 SARS-CoV, and HCoV-EMC MERS-CoV
Above: Coding sequence (CDS) regions corresponding to homologous proteins between the four viruses are filled with the same color in the genome schematic to indicate homology; regions with no homology to the predicted SARS-CoV-2 proteins are colored white. Below: Table of pairwise protein similarities (expressed as % identity) between SARS-CoV-2 and the other three viruses.
The immune response to SARS-CoV-2 in humans awaits characterization, but human immune responses against other coronaviruses have been investigated. As of January 27, 2020, the IEDB has curated 581 linear, and 81 as discontinuous, B cell epitopes that have been reported in the peer-reviewed literature. In addition, 320 peptides have been reported as T cell epitopes (Table 1 ). The vast majority of these epitopes are derived from Betacoronavirues, and more specifically from SARS-CoV, which alone accounts for over 60% of them. In terms of the host in which the various B and T cell epitopes were recognized (Table 2 ), most epitopes (either B or T) were defined in humans or murine systems. Notably, all but 2 of the 417 B and T cell epitopes described in humans are from Betacoronaviruses, with 398 of them coming from SARS-CoV.
Table 1.
IEDB Inventory of Coronavirus B and T Cell Epitopes
| Epitope set | Type | Coronavirus |
Total | ||||
|---|---|---|---|---|---|---|---|
| Alpha | Beta |
Gamma | |||||
| SARS-CoV | MERS-CoV | Other | |||||
| B cell | Conformational | 18 | 27 | 23 | 2 | 11 | 81 |
| Linear | 81 | 405 | 5 | 60 | 30 | 581 | |
| T cell | 61 | 164 | 25 | 54 | 16 | 320 | |
Table 2.
IEDB Inventory of Coronavirus B and T Cell Epitopes
SARS-CoV-2 Similarity to Other Betacoronaviruses
Comparison of a consensus SARS-CoV-2 protein sequence to sequences for SARS-CoV, MERS-CoV and bat-SL-CoVZXC21 revealed a high degree of similarity (expressed as % identity) between SARS-CoV-2, bat-SL-CoVZXC21, and SARS-CoV, but a more limited similarity with MERS-CoV (Figure 1). This is in agreement with a recent paper published on February 7, 2020 that shows the highest similarity between SARS-CoV-2 and SARS or SARS-like CoVs (Wu et al., 2020). Further, SARS-Cov is the closest related virus to SARS-CoV-2 for which a significant number of epitopes have been defined in humans (and other species) and that also causes human disease with lethal outcomes. Accordingly, in the following analyses, we focused on comparing known SARS-CoV epitope sequences to the SARS-CoV-2 sequence.
We first assessed the distribution of SARS-CoV-derived epitopes as a function of the protein of origin (Table 3 ). In the context of B cell responses, most of the 12 antigens in the SARS-CoV proteome are associated with epitopes, with the greatest number derived from spike glycoprotein, nucleoprotein, and membrane protein (Table 3). The paucity of B cell epitopes associated with the other proteins is likely because, on average, B cell epitope screening studies to date have probed regions constituting less than 20% of each respective sequence, including <1% of the Orf 1ab polyprotein. By comparison, the complete span of the spike glycoprotein, nucleoprotein, and membrane protein sequences have been probed at least to some extent in B cell assays.
Table 3.
IEDB Inventory of Coronavirus B and T Cell Epitopes
| SARS-CoV Proteins | B Cell | T Cell |
|---|---|---|
| Spike glycoprotein | 279 | 48 |
| Nucleoprotein | 113 | 33 |
| Membrane protein | 20 | 4 |
| Replicase polyprotein 1ab | 8 | 9 |
| Protein 3a | 2 | 7 |
| Envelope small membrane protein | 2 | 0 |
| Non-structural protein 3b | 2 | 0 |
| Protein 7a | 2 | 0 |
| Protein 9b | 2 | 0 |
| Non-structural protein 6 | 1 | 0 |
| Protein non-structural 8a | 1 | 0 |
T cell epitope total includes epitopes recognized in humans and/or transgenic mice.
A similar situation was observed in the case of T cell epitopes. Here, we only considered epitopes whose recognition is restricted by human leukocyte antigen (HLA) major histocompatibility complex (MHC), because MHC polymorphism typically results in different epitopes being recognized in humans and mice.
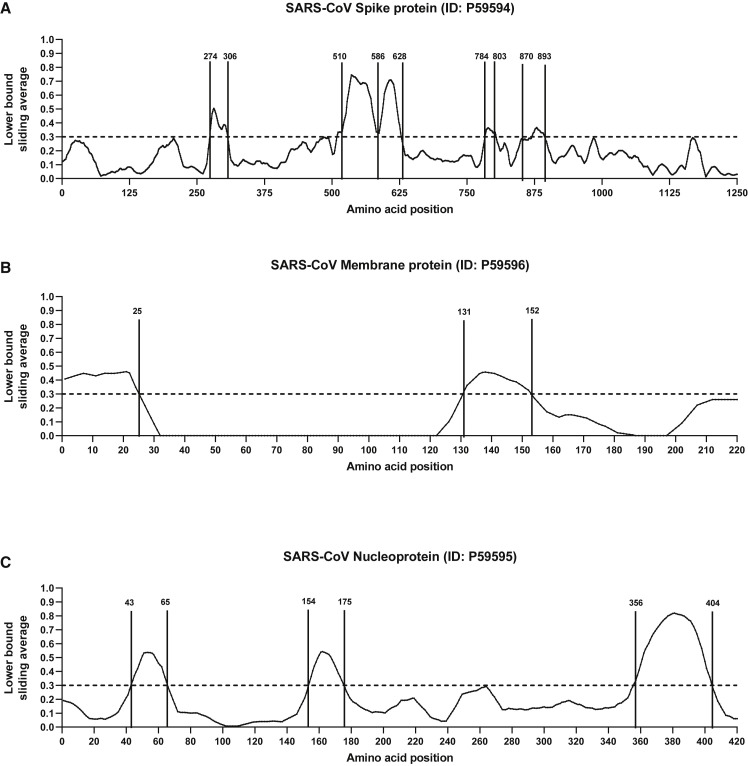
Defining Immunodominant Regions within the SARS-CoV Genome
B cell epitopes derived from SARS-CoV were mapped back to a SARS-CoV reference sequence using the IEDB’s Immunobrowser tool (Dhanda et al., 2018). This tool combines all records available along a reference sequence and produces a response factor (RF) score that accounts for the positivity rate (how frequently a residue was found in a positive epitope) and the number of records (how many independent assays are reported). Dominant regions were identified considering residues stretches where the RF score was ≥0.3.
Analyses of the spike glycoprotein, membrane protein, and nucleoproteins are shown in Figure 2 . In the case of the spike glycoprotein (Figure 2A), we identify five regions of potential interest (residues 274–306, 510–586, 587–628, 784–803, and 870–893), all representing regions associated with high immune response rates. Three of these immunodominant regions are located in the S1 subunit in the CTD2 and CTD3 (C-terminal domain), whereas the other two are in the HR1 domain of the S2 subunit.
Figure 2.
B Cell Immunodominant Regions Based on SARS-Specific Epitope Mapping
RF score for each amino acid position was calculated (see STAR Methods) and plotted over the SARS-CoV consensus sequence of spike glycoprotein (A), membrane protein (B), and nucleoprotein (C).
Next, we aligned the SARS-CoV B cell epitope region sequences to the SARS-CoV-2 sequence to calculate the percentage identity between each of the SARS-CoV-dominant regions and SARS-CoV-2 (Table 4 ). Of the 10 regions identified, 6 had 90% or more identity with SARS-CoV-2, 2 were between 80%–89% identical, and 2 had lower but still appreciable homology (69% and 78%).
Table 4.
Dominant SARS-CoV B Cell Epitope Regions
| SARS-CoV |
SARS-CoV-2 |
||||
|---|---|---|---|---|---|
| Sequence | Max RF | Sequence | Proteina | Mapped Start–End | Identity (%) |
| DAVDCSQNPLAELKCSVKSFEIDKGIYQTSNF | 0.504 | DAVDCALDPLSETKCTLKSFTVEKGIYQTSN | S | 287–317 | 69 |
| VCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTDSVRDPKTSEILDISPCSFGGVSVIT | 0.745 | VCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVI | S | 524–598 | 80 |
| GTNASSEVAVLYQDVNCTDVSTAIHADQLTPAWRIYSTGNN | 0.709 | GTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGS | S | 601–640 | 78 |
| FSQILPDPLKPTKRSFIED | 0.365 | FSQILPDPSKPSKRSFIE | S | 802–819 | 89 |
| FGAGAALQIPFAMQMAYRFNGIG | 0.367 | FGAGAALQIPFAMQMAYRFNGI | S | 888–909 | 100 |
| MADNGTITVEELKQLLEQWNLVIG | 0.460 | MADSNGTITVEELKKLLEQWNLVI | M | 1–24 | 92 |
| PLMESELVIGAVIIRGHLRMA | 0.457 | PLLESELVIGAVILRGHLRI | M | 132–151 | 90 |
| PQGLPNNTASWFTALTQHGKEE | 0.537 | RPQGLPNNTASWFTALTQHGK | N | 42–62 | 95 |
| NNAATVLQLPQGTTLPKGFYA | 0.543 | NNNAATVLQLPQGTTLPKGF | N | 153–172 | 95 |
| KHIDAYKTFPPTEPKKDKKKKTDEAQPLPQRQKKQPTVTLLPAADMDD | 0.82 | NKHIDAYKTFPPTEPKKDKKKKTDEAQPLPQRQKKQPTVTLLPAADM | N | 355–401 | 90 |
S, surface glycoprotein; M, membrane protein; N, nucleocapsid phosphoprotein
In a similar analysis, T cell epitopes were also found to be predominantly associated with spike glycoprotein and nucleoprotein (Table 3). Table 5 shows a listing of the most dominant SARS-CoV individual epitopes identified to date in humans. We also aligned the SARS-CoV T cell epitope sequences and calculated for each epitope the percentage identity to SARS-CoV-2. For each T cell epitope, Table 5 shows the antigen of origin, the epitope sequence, the homologous SARS-CoV-2 sequence, and the corresponding percentage of sequence identity. Overall, the nucleocapsid phosphoprotein and membrane-derived epitopes were most conserved (8/10 and 2/3, respectively, had ≥ 85% identity with SARS-CoV-2). The Orf1ab and surface glycoprotein epitopes were moderately conserved (3/7 and 10/23, respectively, had ≥85% identity with SARS-CoV-2), and Orf 3a epitopes were the least conserved.
Table 5.
Dominant SARS-CoV T Cell Epitopes
| SARS |
SARS-CoV-2 |
|||||
|---|---|---|---|---|---|---|
| Sequence | RF Score | HLA Restrictiona | Sequence | Protein | Mapped Start–End | Identity (%) |
| VRGWVFGSTMNNKSQSVI | 0.15 | DRB1∗04:01 | IRGWIFGTTLDSKTQSLL | S | 101–118 | 50 |
| CTFEYISDAFSLD | 0.21 | DRB1∗04:01 | CTFEYVSQPFLMD | S | 166–178 | 62 |
| DAFSLDVSEKSGN | 0.62 | DRB1∗04:01 | QPFLMDLEGKQGN | S | 173–185 | 38 |
| TNFRAILTAFSPAQDIW | 0.32 | DRB1∗04:01 | TRFQTLLALHRSYLTPGDSSSGW | S | 236–258 | 17 |
| KSFEIDKGIYQTSNFRVV | 0.40 | DRB1∗04:01, DRB1∗07:01 | KSFTVEKGIYQTSNFRVQ | S | 304–321 | 78 |
| STFFSTFKCYGVSATKL | 0.50 | DRB1∗07:01, DR8 | SASFSTFKCYGVSPTKL | S | 371–387 | 82 |
| KLPDDFMGCV | 0.55 | A∗02:01 | KLPDDFTGCV | S | 424–433 | 90 |
| NIDATSTGNYNYKYRYLR | 0.29 | Class II | NLDSKVGGNYNYLYRLFR | S | 440–457 | 56 |
| YLRHGKLRPFERDISNVP | 0.16 | DRB1∗04:01 | YLYRLFRKSNLKPFERDI | S | 451–468 | 58 |
| RPFERDISNVPFS | 0.36 | DRB1∗04:01 | KPFERDISTEIYQ | S | 462–474 | 54 |
| KSIVAYTMSLGADSSIAY | 0.15 | DRB1∗04:01, DRB1∗07:01 | QSIIAYTMSLGAENSVAY | S | 690–707 | 72 |
| SIVAYTMSL | 0.29 | A∗02:01 | SIIAYTMSL | S | 691–699 | 89 |
| TECANLLLQYGSFCTQL | 0.50 | DR8 | TECSNLLLQYGSFCTQL | S | 747–763 | 94 |
| VKQMYKTPTLKYFGGFNF | 0.20 | DRB1∗04:01 | VKQIYKTPPIKDFGGFNF | S | 785–802 | 78 |
| ESLTTTSTALGKLQDVV | 0.42 | DRB1∗04:01 | DSLSSTASALGKLQDVV | S | 936–952 | 71 |
| ALNTLVKQL | 0.29 | A∗02:01 | ALNTLVKQL | S | 958–966 | 100 |
| VLNDILSRL | 0.29 | A∗02:01 | VLNDILSRL | S | 976–984 | 100 |
| LITGRLQSL | 0.42 | A∗02:01 | LITGRLQSL | S | 996–1004 | 100 |
| QLIRAAEIRASANLAATK | 0.20 | DRB1∗04:01 | QLIRAAEIRASANLAATK | S | 1011–1028 | 100 |
| SWFITQRNFFSPQII | 0.60 | DRB1∗04:01 | HWFVTQRNFYEPQII | S | 1101–1115 | 73 |
| RLNEVAKNL | 0.42 | A∗02:01 | RLNEVAKNL | S | 1185–1193 | 100 |
| NLNESLIDL | 0.29 | A∗02:01 | NLNESLIDL | S | 1192–1200 | 100 |
| FIAGLIAIV | 0.80 | A∗02:01 | FIAGLIAIV | S | 1220–1228 | 100 |
| RFFTLGSITAQPVKI | 0.18 | B∗58:01 | RIFTIGTVTLKQGEI | Orf 3a | 6–20 | 40 |
| SITAQPVKI | 0.29 | B∗58:01 | TVTLKQGEI | Orf 3a | 12–20 | 22 |
| TLACFVLAAV | 0.59 | A∗02:01 | TLACFVLAAV | M | 61–70 | 100 |
| GLMWLSYFV | 0.59 | A∗02:01 | GLMWLSYFI | M | 89–97 | 89 |
| HLRMAGHSL | 0.40 | Class I | HLRIAGHHL | M | 148–156 | 78 |
| ALNTPKDHI | 0.29 | A∗02:01 | ALNTPKDHI | N | 138–146 | 100 |
| LQLPQGTTL | 0.29 | A∗02:01 | LQLPQGTTL | N | 159–167 | 100 |
| GETALALLLL | 0.38 | B∗40:01 | GDAALALLLL | N | 215–224 | 80 |
| LALLLLDRL | 0.29 | A∗02:01 | LALLLLDRL | N | 219–227 | 100 |
| LLLDRLNQL | 0.42 | A∗02:01 | LLLDRLNQL | N | 222–230 | 100 |
| RLNQLESKV | 0.42 | A∗02:01 | RLNQLESKM | N | 226–234 | 89 |
| TKQYNVTQAF | 0.29 | Class I | TKAYNVTQAF | N | 265–274 | 90 |
| GMSRIGMEV | 0.42 | A∗02:01 | GMSRIGMEV | N | 316–324 | 100 |
| MEVTPSGTWL | 0.42 | B∗40:01 | MEVTPSGTWL | N | 322–331 | 100 |
| QFKDNVILL | 0.50 | A∗24:02 | NFKDQVILL | N | 345–353 | 78 |
| CLDAGINYV | 0.42 | A∗02:01 | CLEASFNYL | Orf 1ab | 2139–2147 | 56 |
| WLMWFIISI | 0.42 | A∗02:01 | WLMWLIINL | Orf 1ab | 2292–2300 | 67 |
| ILLLDQVLV | 0.42 | A∗02:01 | ILLLDQALV | Orf 1ab | 2498–2506 | 89 |
| LLCVLAALV | 0.42 | A∗02:01 | SACVLAAEC | Orf 1ab | 2840–2848 | 56 |
| ALSGVFCGV | 0.42 | A∗02:01 | SLPGVFCGV | Orf 1ab | 2942–2950 | 78 |
| TLMNVITLV | 0.42 | A∗02:01 | TLMNVLTLV | Orf 1ab | 3639–3647 | 89 |
| SMWALVISV | 0.42 | A∗02:01 | SMWALIISV | Orf 1ab | 3661-3669 | 89 |
S, surface glycoprotein; M, membrane protein; N, nucleocapsid phosphoprotein.
Restrictions defined only in HLA-transgenic mice are indicated by the italicized font.
Prediction of SARS-CoV-2 B Cell Epitopes
To define potential B cell epitopes by an alternative method, we used the predictive tools provided with the IEDB. B cell epitope predictions were carried out using the SARS-CoV-2 surface glycoprotein, nucleocapsid phosphoprotein, and membrane glycoprotein sequences, which, as described above, were found to be the main protein targets for B cell responses to other coronaviruses. In parallel, we performed predictions for linear B cell epitopes with Bepipred 2.0 (Jespersen et al., 2017) and for conformational epitopes with Discotope 2.0 (Kringelum et al., 2012). Both prediction algorithms are available on the IEDB B cell prediction tool page (http://tools.iedb.org/main/bcell/). A full list of B cell epitope prediction results per amino acid position per protein is provided in Table S1.
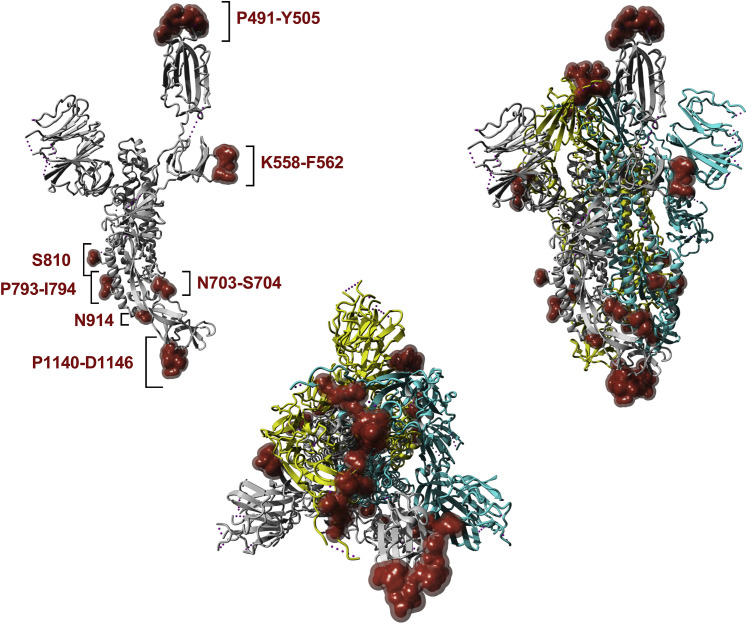
Using Bepipred 2.0 and a cutoff of ≥0.55 (corresponding to a specificity cutoff of 80%) (Jespersen et al., 2017), the surface glycoprotein had the highest number of predicted B cell epitopes, followed by membrane glycoprotein and nucleocapsid phosphoprotein (Table S2). To predict and map conformational B cell epitopes, we used the recently submitted SARS-CoV-2 spike glycoprotein structure (PDB: 6VSB). A list of surface glycoprotein amino acid positions having a high probability of being included in predicted B cell epitopes, based on analysis with the Discotope 2.0 algorithm, is shown in Table S1 (cutoff of ≥−2.5, corresponding to 80% specificity). We then localized the relevant amino acid positions onto the model structure, which allowed the identification of seven predicted epitope residue/regions (491–505, 558–562, 703–704, 793–794, 810, 914, and 1140–1146) in the surface glycoprotein (Figure 3 ).
Figure 3.
SARS-CoV-2 Spike Glycoprotein (PDB: 6VSB)
The calculated surface of the top 13 amino acid residues predicted to be B cell epitopes based on ranking performed with Discotope 2.0 are shown in red. The monomer is shown in the upper left. The upper right and lower center present the trimer in two different orientations. 3D-rendering was performed using YASARA (Krieger and Vriend, 2014).
Prediction of SARS-CoV-2 T Cell Epitopes
To predict CD4 T cell epitopes, we used the method described by Paul and co-authors (Paul et al., 2015a), as implemented in the Tepitool resource in IEDB (Paul et al., 2016). This approach was designed and validated to predict dominant epitopes independently of ethnicity and HLA polymorphism, taking advantage of the extensive cross-reactivity and repertoire overlap between different HLA class II loci and allelic variants. Here, we selected peptides that have a median consensus percentile ≤20, a threshold associated with epitope panels responsible for about 50% of target-specific responses. Using this threshold, we identified 241 candidates in the SARS-CoV-2 sequence (see Table S3).
In previous experiments, we showed that pools based on similar peptide numbers can be generated by sequential lyophilization (Carrasco Pro et al., 2015). These peptide pools (or megapools) incorporate predicted or experimentally validated epitopes and allow measurement of magnitude and characterization of the phenotype of human T cell responses in infectious disease indications such as Bordetella pertussis, Mycobacteria tuberculosis, Dengue, and Zika viruses (Carrasco Pro et al., 2015, da Silva Antunes et al., 2018, Grifoni et al., 2017, Grifoni et al., 2018). The SARS-CoV-2 CD4 megapool covers all 10 predicted proteins, with the number of potential epitopes proportional to the size of each protein (Table S4).
In parallel, we also sought to define likely CD8 epitopes. Here, a different approach was required because the overlap between different HLA class I allelic variants and loci is more limited to specific groups of alleles, or supertypes (Sidney et al., 2008). Following a previously validated approach (Weiskopf et al., 2013), we assembled a set of the 12 most prominent HLA class I alleles that have been shown to allow broad coverage of the general population, as described in the STAR Methods (see also Table S5). We then performed HLA class I binding predictions using the Net MHC pan 4.0 EL algorithm (Jurtz et al., 2017) available at the IEDB. For each allele, we selected the top 1% scoring peptides in the SARS-CoV-2 sequence, as ranked based on prediction. After eliminating redundancies and nested peptides, we obtained a final “in silico” megapool of 628 unique predicted epitopes. Table S6 lists those unique predicted epitopes per protein, indicating for each their respective HLA restriction(s).
Correspondence between the Epitopes Identified by the Two Different Approaches
The epitopes identified by homology to the experimentally defined SARS-CoV epitopes shown in Tables 4 and 5 were next compared with the epitopes identified by epitope predictions shown in Tables S2, S3, and S6. The epitopes independently identified in both approaches are presumed to be the most valuable leads.
We first compared B cell immunodominant regions identified in SARS-CoV and mapped to the homologous SARS-CoV-2 proteins (Table 4), with the predicted linear (Table S2) and conformational (Table S1) B cell epitopes. Out of the five B cell immunodominant regions from the SARS spike glycoprotein that were mapped to SARS-CoV-2, three regions overlapped with those identified by BebiPred 2.0, and two overlapped with regions predicted by Discotope 2.0 (Figure 3; Table S1). No overlap was observed for the five regions of SARS-CoV membrane protein and nucleoprotein that mapped to SARS-CoV-2 and those predicted by BebiPred 2.0. As stated above, no Discotope 2.0 prediction was available for those two proteins.
The prediction analysis performed with Discotope 2.0 based on the SARS-CoV-2 spike glycoprotein PDB structure independently confirms two of the likely epitope regions defined on the basis of SARS-CoV data. Specifically, one dominant epitope corresponds to the 524–598 epitope from Table 5, which overlaps with the 558–562 predicted epitope, and the 802–819 region is also predicted (cf., the predicted 810 residue is in the middle of this region). Finally, the 888–909 region is narrowly missed, because residue 914, which is predicted, is right outside of the epitope.
When we compared the SARS-CoV T cell epitopes that mapped to SARS-CoV-2 (Table 5) with the predicted CD4 and CD8 T cell epitopes (Tables S3 and S6, respectively), we found that 12 of 17 SARS-CoV-2 T cell epitopes with high sequence identity (≥90%) to the SARS-CoV were independently identified by the two methods. Another 7 of 16 epitopes with moderate sequence identity (70%–89%), and 6 of 12 epitopes with low sequence identity (<70%) were also identified by both methods. The lack of absolute correspondence is not surprising, given that the experimental data are derived from a skewed set of HLA restrictions (largely HLA A∗02:01) and that our HLA class I prediction strategy targeted a more limited set of alleles selected to represent the most frequent worldwide variants; at the same time, the class II predictions are expected to cover 50% of the class II responses (Paul et al., 2015b).
Discussion
The present study identifies likely targets of the human immune response to SARS-CoV-2, encompassing both the B and T cell arms of the adaptive immune response. This is of relevance in the face of the ever growing medical and societal urgency surrounding COVID-19, especially given the current scarcity of experimental data regarding any corresponding immune response. The approach we followed is based on establishing several lines of evidence that clearly pinpoint SARS-CoV as a relevant model to extrapolate likely targets of responses to SARS-CoV-2, the virus associated with COVID-19.
The first line of evidence pertains to the fact that of coronaviruses known to infect humans, SARS-CoV is the most similar in phylogenetic terms to SARS-CoV-2. The second line of evidence is that SARS-CoV-2 is the most (and highly) similar to SARS-CoV at the level of sequence identity. Third, when we critically reviewed the knowledge related to the precise epitopes recognized by adaptive responses in the context of coronaviruses in aggregate, it was apparent that all but 2 of the 417 B and T cell epitopes described in humans to date are from Betacoronaviruses, with 398 of them coming from SARS-CoV.
Our analysis showed that certain SARS-CoV regions were dominant for B cell responses and that those regions were well conserved in terms of sequence with SARS-CoV-2. Five regions contain epitopes recognized by neutralizing antibodies in SARS convalescent sera (Guo et al., 2004, Shichijo et al., 2004). Among those, of particular interest is the 587–628 region nesting the 604−625 peptide, which was identified in a SARS convalescent patient and found to have the capacity to elicit antibodies that efficiently prevent infection in non-human primates (Hu et al., 2005, Wang et al., 2016).
Two regions were identified from membrane protein (1–25 and 131–152) (Figure 2B), and three regions were identified for nucleoprotein (43–65, 154–175, and 356–404) (Figure 2C). The two regions in the membrane protein have been shown to elicit marked IgM and IgG responses and a broad spectrum of recognition, highlighting them as potential diagnostic candidates (Chow et al., 2006, Wang et al., 2003). Of the three regions identified in the nucleoprotein, 156–175 has shown strong reactivity against SARS patient sera and immunogenicity in multiple species, including mice, monkeys, and humans (Liu et al., 2006).
Because of the overall high level of sequence similarity of SARS-CoV and SARS-CoV-2, we infer that the regions dominant in SARS-CoV have a high likelihood to also be dominant in SARS-CoV-2, even if the actual sequences are different. This hypothesis is in agreement with the recent cryoelectron microscopy (cryoEM) structure of the spike glycoprotein of SARS-CoV-2, showing a high resemblance in the overall structure with the SARS-CoV spike protein (Wrapp et al., 2020). In the same study, however, the authors do not observe cross-recognition of SARS-CoV monoclonal antibodies with the SARS-CoV-2. Indeed, they observed no reactivity with SARS-CoV antibodies that recognize the SARS-CoV-2 spike receptor binding domain (RBD), despite the fact that SARS-CoV-2 retains the same capability to bind the ACE2 receptor of SARS-CoV (Wrapp et al., 2020). This suggests that the B cell prediction performed on the RBD domain will require further studies.
We also analyzed the SARS-CoV T cell epitopes. In these cases, epitopic regions and individual epitopes were more widely dispersed throughout the respective proteins, which made the identification of discrete, dominant epitopic regions more difficult. This outcome is not unexpected given that T cells recognize short peptides generated from cellular processing of viral antigens that can be derived from any segment of the protein.
It is generally expected that CD8 T cell epitopes will be derived from both structural and nonstructural proteins (Tian et al., 2019), because both types of proteins are endogenously processed by infected cells. In the case of class II epitopes, structural proteins would be of particular interest, as they are most likely to provide help by cognate interaction (Sette et al., 2008). When examining the homologous regions of SARS-CoV, it has been found that the likely T cell epitopes are positive in assays such as ELISPOT, intra-cellular staining (ICS), and multimer/tetramer staining (see, e.g., Cheung et al., 2007, Cheung et al., 2008, Kohyama et al., 2009, Tsao et al., 2006, Yang et al., 2009).
We also sought to address potential SARS-CoV-2 epitopes by a completely different method, namely utilizing the epitope predictions hosted by the IEDB (Dhanda et al., 2019, Vita et al., 2019). For B cell epitopes, we used methods that predict linear epitopes (Jespersen et al., 2017), and in the case of the spike glycoprotein where a reliable structure recently became available (Wrapp et al., 2020), the Discotope 2.0 (Kringelum et al., 2012) method that also predicts epitopes based on protein conformation and residue exposure. The Discotope prediction independently confirmed two of the likely epitope regions defined on the basis of SARS-CoV data.
In the case of T cell epitopes, we utilized predictive algorithms (Jurtz et al., 2017, Paul et al., 2016) to map hundreds of potential human epitopes to account for HLA polymorphism and for the fact that T cell epitopes are typically derived from both structural and non-structural proteins and not limited to exposed regions. Here, as an independent validation of the predictions, we asked whether the predictions effectively identified the relatively few epitopes identified experimentally in SARS-CoV, restricted by human HLA, and conserved in SARS-CoV-2. Indeed, we found that 12 of 17 SARS-CoV-2 T cell epitopes with high sequence identity (≥90%) to the SARS-CoV were independently identified by the epitope predictions based on SARS-CoV-2 sequences.
In conclusion, the use of available information related to SARS-CoV epitopes in conjunction with bioinformatic predictions points to specific regions of SARS-CoV-2 that have a high likelihood of being recognized by human immune responses. The observation that many B and T cell epitopes are highly conserved between SARS-CoV-2 and SARS-CoV is important. Vaccination strategies designed to target the immune response toward these conserved epitope regions could generate immunity that is not only cross-protective across Betacoronaviruses but also relatively resistant to ongoing virus evolution.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| SARS-CoV-2 spike glycoprotein 3D-structure | Wrapp et al., 2020 | PDB ID: 6VSB |
| Wuhan-Hu-1 RNA isolate | NCBI nuccore database | GenBank:MN908947 |
| ORF10 protein | NCBI protein database | NCBI: YP_009725255.1 |
| Nucleocapsid phosphoprotein | NCBI protein database | NCBI: YP_009724397.2 |
| ORF8 protein | NCBI protein database | NCBI: YP_009724396.1 |
| ORF7a protein | NCBI protein database | NCBI: YP_009724395.1 |
| ORF6 protein | NCBI protein database | NCBI: YP_009724394.1 |
| membrane glycoprotein | NCBI protein database | NCBI: YP_009724393.1 |
| envelope protein | NCBI protein database | NCBI: YP_009724392.1 |
| ORF3a protein | NCBI protein database | NCBI: YP_009724391.1 |
| surface glycoprotein | NCBI protein database | NCBI: YP_009724390.1 |
| orf1ab polyprotein | NCBI protein database | NCBI: YP_009724389.1 |
| Software and Algorithms | ||
| YASARA | Krieger and Vriend, 2014 | http://www.yasara.org |
| IEDB | Vita et al., 2019 | https://www.iedb.org |
| BebiPred 2.0 | Jespersen et al., 2017 | http://tools.iedb.org/bcell/ |
| Discotope 2.0 | Kringelum et al., 2012 | http://tools.iedb.org/bcell/ |
| NetMHCpan EL 4.0 | Jurtz et al., 2017 | http://tools.iedb.org/mhci/ |
| Tepitool | Paul et al., 2016 | http://tools.iedb.org/tepitool/ |
Lead Contact and Materials Availability
Please contact A.S. (alex@lji.org) for aliquots of synthesized sets of peptides identified in this study. There are restrictions to the availability of the peptide reagents due to cost and limited quantity.
Method Details
IEDB Analysis of Coronavirus T and B Epitopes
T and B cell epitopes for coronaviruses were identified by searching the IEDB at the end of January 2020. Queries were performed broadly for coronaviruses (taxonomy ID no. 11118), selecting positive assays in T cell, B cell and/or ligand contexts. Characteristics of each unique epitope (i.e., species, protein of provenance, positive assay type(s), MHC restriction) were tabulated, as well as the total number of donors tested and corresponding total number of donors with positive responses in B or T cell assays, and as a function of host. Finally, T or B cell assay specific response frequency scores (RF) were calculated broadly (i.e., any host), or for specific contexts (e.g., T cell assays in humans). Specifically, RF = [(r – sqrt(r)]/t, where r is the total number of responding donors and t is the total number of donors tested (Carrasco Pro et al., 2015)).
SARS-CoV (tax ID no. 694009) sequence epitope density was visualized with the IEDB Immunobrowser tool (Dhanda et al., 2018). To identity contiguous dominant regions, RF scores for each residue were recalculated to represent a sliding 10 residue window.
Comparison of Coronavirus Sequences to SARS-CoV-2
All full-length protein sequences from SARS-CoV and MERS-CoV were retrieved from ViPR (https://www.viprbrc.org/brc/home.spg?decorator=corona) on 31 January 2020. In order to exclude sequences of experimental strains, sequences from “unknown,” mouse, and monkey hosts were excluded from analysis. Remaining sequences were aligned using the MUSCLE algorithm in ViPR. Sequences causing poor alignments in a preliminary analysis were removed before computing the final alignment. The consensus protein sequences of each virus group were determined from the final alignments using the Sequence Variation Analysis tool in ViPR. Protein sequences from natural virus isolates with sequences identical to the SARS-CoV and MERS-CoV consensus were selected for use in epitope sequence analysis.
Determination of SARS-CoV-2 Sequence Conservation
Each Wuhan-Hu-1 (GeneBank: MN908947) protein sequence was compared against the consensus protein sequences from SARS-CoV and MERS-CoV and the protein sequences from closest bat relative (bat-SL-CoVZXC21) using the BLAST algorithm (ViPR; https://www.viprbrc.org/brc/blast.spg?method=ShowCleanInputPage&decorator=corona) to compute the pairwise identity between Wuhan-Hu-1 proteins and their comparison target.
SARS-CoV-2 B Cell Epitope Prediction
Linear B cell epitope predictions were carried out on three different coronavirus proteins: surface glycoprotein (S), nucleocapsid phosphoprotein (N) and membrane glycoprotein (M) (NCBI: YP_009724390.1, YP_009724397.2 and YP_009724393.1, respectively) as the homologous versions of these proteins are the primary targets of B cell immune responses for SARS-CoV. We used the BebiPred 2.0(Jespersen et al., 2017) algorithm embedded in the B cell prediction analysis tool available in IEDB (Zhang et al., 2008). For each protein, the epitope probability score for each amino acid and the probability of exposure was retrieved. Potential B cell epitopes were predicted using a cutoff of 0.55 (corresponding to specificity greater than 0.81 and sensitivity below 0.3) and considering sequences having more than 7 amino acid residues. Structure-based antibody prediction was performed by using Discotope 2.0 (Kringelum et al., 2012), available in IEDB (Zhang et al., 2008) and a positivity cutoff greater than −2.5 was applied (corresponding to specificity greater than or equal to 0.80 and sensitivity below 0.39), using the SARS-CoV-2 spike glycoprotein structure (PDB ID: 6VSB).
SARS-CoV-2 T Cell Epitope Prediction
Epitope prediction was carried out using the ten proteins predicted for the reference SARS-CoV-2 isolate, Wuhan-Hu-1. The corresponding protein accession identification numbers are: NCBI: YP_009725255.1 (Orf 10), NCBI: YP_009724397.2 (N), NCBI: YP_009724396.1 (Orf 8), NCBI: YP_009724395.1 (Orf 7a), NCBI: YP_009724394.1 (Orf 6), NCBI: YP_009724393.1 (M), NCBI: YP_009724392.1 (Envelope protein, E), NCBI: YP_009724391.1 (Orf 3a), NCBI: YP_009724390.1 (S), and NCBI: YP_009724389.1 (Orf 1ab).
For CD4 T cell epitope prediction, we applied a previously described algorithm that was developed to predict dominant HLA class II epitopes, using a median consensus percentile of prediction cutoff ≤ 20 as recommended (Paul et al., 2015b). For CD8 T cell epitope prediction, we selected the 12 most frequent HLA class I alleles in the worldwide population (Middleton et al., 2003, Paul et al., 2013), using a phenotypic frequency cutoff ≥ 6%. The specific alleles included were: HLA-A∗01:01, HLA-A∗02:01, HLA-A∗03:01, HLA-A∗11:01, HLA-A∗23:01, HLA-A∗24:02, HLA-B∗07:02, HLA-B∗08:01, HLA-B∗35:01, HLA-B∗40:01, HLA-B∗44:02, HLA-B∗44:03. The SARS-CoV-2 protein sequences were run against this set of alleles using the NetMHCpan EL 4.0 algorithm and a size range of 8-14mers (Jurtz et al., 2017). For each HLA class I allele analyzed, we selected the top 1% epitopes ranked based on prediction score. To generate a final set for synthesis, duplicate peptides (i.e., those selected for multiple alleles) were reduced to a single occurrence, and nested peptides were ensconced within longer sequences, up to 14 residues in length, before assigning the multiple corresponding HLA restrictions for each region.
Quantification and Statistical Analysis
No statistical analyses were utilized in the present theoretical study, based on data in the published literature and publicly available databases. Calculations of % identity and response factor scores were performed as described in the Method Details, above.
Data and Code Availability
All data presented and analyzed in the present study was retrieved from the IEDB and PDB, as described above. The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and Supplemental Materials. Text files of data downloaded from the IEDB are available from the corresponding author on request.
Acknowledgments
We thank Erica Ollmann Saphire, Sharon Schendel, and Mitchell Kronenberg for critical reading of the manuscript and numerous helpful suggestions. We also thank Jason McLellan and Barney Graham for early access to the PDB structure of the SARS-2 spike glycoprotein. Support for the work included funding provided through NIH-NIAID contracts 75N9301900065 (A.S.) and 75N93019C00001 (A.S. and B.P.). Additional support was provided by NIH-NIAID contract 75N93019C00076 (Y.Z. and R.H.S.).
Author Contributions
A.G., J.S., and Y.Z. performed analyses and wrote the paper; R.H.S. wrote the paper and provided direction for viral analyses; B.P. conceived the project, wrote the paper, and provided expertise to bioinformatic components; and A.S. conceived the project, wrote the paper, and provided overall direction for the study.
Declaration of Interests
La Jolla Institute for Immunology (LJI) has filed a patent application regarding this manuscript.
Published: March 16, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2020.03.002.
Supplemental Information
References
- Carrasco Pro S., Sidney J., Paul S., Lindestam Arlehamn C., Weiskopf D., Peters B., Sette A. Automatic Generation of Validated Specific Epitope Sets. J. Immunol. Res. 2015;2015:763461. doi: 10.1155/2015/763461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung Y.K., Cheng S.C., Sin F.W., Chan K.T., Xie Y. Induction of T-cell response by a DNA vaccine encoding a novel HLA-A∗0201 severe acute respiratory syndrome coronavirus epitope. Vaccine. 2007;25:6070–6077. doi: 10.1016/j.vaccine.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung Y.K., Cheng S.C., Sin F.W., Chan K.T., Xie Y. Investigation of immunogenic T-cell epitopes in SARS virus nucleocapsid protein and their role in the prevention and treatment of SARS infection. Hong Kong Med. J. 2008;14(Suppl 4):27–30. [PubMed] [Google Scholar]
- Chow S.C., Ho C.Y., Tam T.T., Wu C., Cheung T., Chan P.K., Ng M.H., Hui P.K., Ng H.K., Au D.M., Lo A.W. Specific epitopes of the structural and hypothetical proteins elicit variable humoral responses in SARS patients. J. Clin. Pathol. 2006;59:468–476. doi: 10.1136/jcp.2005.029868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R., Paul S., Sidney J., Weiskopf D., Dan J.M., Phillips E., Mallal S., Crotty S., Sette A., Lindestam Arlehamn C.S. Correction: Definition of Human Epitopes Recognized in Tetanus Toxoid and Development of an Assay Strategy to Detect Ex Vivo Tetanus CD4+ T Cell Responses. PLoS One. 2018;13:e0193382. doi: 10.1371/journal.pone.0193382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda S.K., Mahajan S., Paul S., Yan Z., Kim H., Jespersen M.C., Jurtz V., Andreatta M., Greenbaum J.A., Marcatili P., Sette A., Nielsen M., Peters B. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019;47:W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda S.K., Vita R., Ha B., Grifoni A., Peters B., Sette A. ImmunomeBrowser: a tool to aggregate and visualize complex and heterogeneous epitopes in reference proteins. Bioinformatics. 2018;34:3931–3933. doi: 10.1093/bioinformatics/bty463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Angelo M.A., Lopez B., O’Rourke P.H., Sidney J., Cerpas C., Balmaseda A., Silveira C.G.T., Maestri A., Costa P.R. Global Assessment of Dengue Virus-Specific CD4+ T Cell Responses in Dengue-Endemic Areas. Front. Immunol. 2017;8:1309. doi: 10.3389/fimmu.2017.01309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Costa-Ramos P., Pham J., Tian Y., Rosales S.L., Seumois G., Sidney J., de Silva A.D., Premkumar L., Collins M.H. Cutting Edge: Transcriptional Profiling Reveals Multifunctional and Cytotoxic Antiviral Responses of Zika Virus-Specific CD8+ T Cells. J. Immunol. 2018;201:3487–3491. doi: 10.4049/jimmunol.1801090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.P., Petric M., Campbell W., McGeer P.L. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology. 2004;324:251–256. doi: 10.1016/j.virol.2004.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Li L., Kao R.Y., Kou B., Wang Z., Zhang L., Zhang H., Hao Z., Tsui W.H., Ni A. Screening and identification of linear B-cell epitopes and entry-blocking peptide of severe acute respiratory syndrome (SARS)-associated coronavirus using synthetic overlapping peptide library. J. Comb. Chem. 2005;7:648–656. doi: 10.1021/cc0500607. [DOI] [PubMed] [Google Scholar]
- Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45(W1):W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohyama S., Ohno S., Suda T., Taneichi M., Yokoyama S., Mori M., Kobayashi A., Hayashi H., Uchida T., Matsui M. Efficient induction of cytotoxic T lymphocytes specific for severe acute respiratory syndrome (SARS)-associated coronavirus by immunization with surface-linked liposomal peptides derived from a non-structural polyprotein 1a. Antiviral Res. 2009;84:168–177. doi: 10.1016/j.antiviral.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E., Vriend G. YASARA View - molecular graphics for all devices - from smartphones to workstations. Bionformatics. 2014;30:2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kringelum J.V., Lundegaard C., Lund O., Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput. Biol. 2012;8:e1002829. doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.J., Leng C.H., Lien S.P., Chi H.Y., Huang C.Y., Lin C.L., Lian W.C., Chen C.J., Hsieh S.L., Chong P. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24:3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D., Menchaca L., Rood H., Komerofsky R. New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens. 2003;61:403–407. doi: 10.1034/j.1399-0039.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- Paul S., Weiskopf D., Angelo M.A., Sidney J., Peters B., Sette A. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J. Immunol. 2013;191:5831–5839. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Dillon M.B.C., Arlehamn C.S.L., Huang H., Davis M.M., McKinney D.M., Scriba T.J., Sidney J., Peters B., Sette A. A population response analysis approach to assign class II HLA-epitope restrictions. J. Immunol. 2015;194:6164–6176. doi: 10.4049/jimmunol.1403074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Lindestam Arlehamn C.S., Scriba T.J., Dillon M.B., Oseroff C., Hinz D., McKinney D.M., Carrasco Pro S., Sidney J., Peters B., Sette A. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J. Immunol. Methods. 2015;422:28–34. doi: 10.1016/j.jim.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Sidney J., Sette A., Peters B. TepiTool: A Pipeline for Computational Prediction of T Cell Epitope Candidates. Curr. Protoc. Immunol. 2016;114:18.19.1–18.19.24. doi: 10.1002/cpim.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Moutaftsi M., Moyron-Quiroz J., McCausland M.M., Davies D.H., Johnston R.J., Peters B., Rafii-El-Idrissi Benhnia M., Hoffmann J., Su H.P. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008;28:847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichijo S., Keicho N., Long H.T., Quy T., Phi N.C., Ha L.D., Ban V.V., Itoyama S., Hu C.J., Komatsu N. Assessment of synthetic peptides of severe acute respiratory syndrome coronavirus recognized by long-lasting immunity. Tissue Antigens. 2004;64:600–607. doi: 10.1111/j.1399-0039.2004.00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Peters B., Frahm N., Brander C., Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses. 2019;11 doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Grifoni A., Sette A., Weiskopf D. Human T Cell Response to Dengue Virus Infection. Front. Immunol. 2019;10:2125. doi: 10.3389/fimmu.2019.02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao Y.P., Lin J.Y., Jan J.T., Leng C.H., Chu C.C., Yang Y.C., Chen S.L. HLA-A∗0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem. Biophys. Res. Commun. 2006;344:63–71. doi: 10.1016/j.bbrc.2006.03.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wen J., Li J., Yin J., Zhu Q., Wang H., Yang Y., Qin E., You B., Li W. Assessment of immunoreactive synthetic peptides from the structural proteins of severe acute respiratory syndrome coronavirus. Clin. Chem. 2003;49:1989–1996. doi: 10.1373/clinchem.2003.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T., Zhu H., Liu J., Xu Y., Xie J. Immunodominant SARS Coronavirus Epitopes in Humans Elicited both Enhancing and Neutralizing Effects on Infection in Non-human Primates. ACS Infect. Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Angelo M.A., de Azeredo E.L., Sidney J., Greenbaum J.A., Fernando A.N., Broadwater A., Kolla R.V., De Silva A.D., de Silva A.M. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA. 2013;110:E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., James E., Roti M., Huston L., Gebe J.A., Kwok W.W. Searching immunodominant epitopes prior to epidemic: HLA class II-restricted SARS-CoV spike protein epitopes in unexposed individuals. Int. Immunol. 2009;21:63–71. doi: 10.1093/intimm/dxn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Wang P., Kim Y., Haste-Andersen P., Beaver J., Bourne P.E., Bui H.H., Buus S., Frankild S., Greenbaum J. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513-8. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented and analyzed in the present study was retrieved from the IEDB and PDB, as described above. The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and Supplemental Materials. Text files of data downloaded from the IEDB are available from the corresponding author on request.