Abstract
Immune signatures measured at baseline and immediately prior to vaccination may predict the immune response to vaccination. Such pre-vaccine assessment might allow not only population-based, but also more personalized vaccination strategies (‘precision vaccination’). If baseline immune signatures are predictive, the underlying mechanism they reflect may also determine vaccination outcome. Thus, baseline signatures might contribute to identifying interventional targets to be modulated prior to vaccination in order to improve vaccination responses. This concept has the potential to transform vaccination strategies and usher in a new approach to improve global health.
Highlights
Extensive baseline variability in immune responses (e.g., antibody titers) among individuals in given populations is increasingly being appreciated as a major contributor to vaccine response heterogeneity.
The concept of ‘baseline may predict outcome’ has recently been reported for human influenza virus, yellow fever virus, and hepatitis B virus, as well as malaria vaccination. This concept might also apply to other vaccines.
The ability to predict who might respond to immunization (and to what extent) might offer avenues for optimization of current vaccination strategies.
We posit that this simple concept might be useful and significant for vaccine design: if ‘baseline determines outcome, then altering baseline prior to vaccination could alter outcome’.
This approach could potentially lead to tailored (precision) vaccines ensuring that the majority, or all individuals vaccinated, respond by eliciting a protective immune response (i.e., devoid of non-responder individuals). Presumably, this approach might also allow the administration of fewer vaccine doses, potentially arriving at one vaccine dose only.
Capturing ‘Baseline’ via Systems Immunology
Vaccines preventing infections or diseases are among the most effective life-saving medical interventions in history [1]. In the past, vaccine design was largely empirical. However, this approach has thus far mostly failed to tackle complex infections such as HIV, Mycobacterium tuberculosis (TB), and Plasmodium sp. (malaria), as well as cancers and other noncommunicable diseases. This failure has been attributed to the lack of insight into the underlying mechanisms of how vaccines induce protection (i.e., the rules of immunity) [2,3]. Recent technological advances, including highly multiplexed immune profiling and data-driven computational modeling, have raised the prospect of identifying these rules more globally. Application of systems biology to vaccines [or ‘systems vaccinology’ (see Glossary)] involves assessing the molecular and cellular state of the immune system before and after vaccination in a comprehensive and unbiased multiomic manner (‘omics’). This is then used to develop data-driven models to predict post-vaccination, pathogen-specific immune responses (e.g., antigen-specific antibody titers); through these, the goal is to identify key molecular immune parameters that correlate with, and potentially shape, vaccine responses. This approach has already led to new insights. For instance, correlation between the early post-vaccination host response and outcome (e.g., antibody responses) has raised the hypothesis that the microbiome may be involved in vaccination responses. Antibiotic-induced shifts in the microbiome can influence responses to influenza virus vaccination in mice and potentially in humans [4., 5., 6.]. This unbiased systems approach is increasingly being applied to vaccine design and testing [7]. This has also led to an increasing appreciation of the extensive baseline and response variability in many immune parameters among individuals within a population [8]. Given the pervasive population heterogeneity, being able to predict who might respond to a given vaccine is necessary. Moreover, understanding how immune status prior to vaccination shapes vaccination responses is important. This has recently been thought to be possible for human influenza virus, hepatitis B virus (HBV), and malaria vaccination [9., 10., 11., 12., 13., 14.,41]. Specifically, the aim would be to assess whether a subject’s immune status prior to vaccination allows a predictive response (i.e., the concept of ‘baseline predicts outcome’).
An important assumption and implication embodied in this hypothesis is that ‘if baseline determines outcome, then altering baseline before vaccination might potentially alter outcome’. There is evidence, albeit indirect and preliminary, supporting this hypothesis. The purpose of this Opinion is to highlight the potential of this paradigm. If one can reshape baseline immune status to optimize vaccine responsiveness, it might allow the design of vaccination strategies that can lead to a more effective, safe, and protective immune response (i.e., eliminating non-responder vaccinees). It may also enable strategies that allow the administration of fewer vaccine doses (ideally only one dose) (Box 1 ). Furthermore, many licensed interventions (e.g., drugs, adjuvants, biologics) known to have immunomodulatory functions might be potentially repurposed to modify baseline status in a targeted fashion. In addition, tools have advanced to allow testing this paradigm in humans, including single-cell large-scale immune profiling and computational modeling; these approaches may contribute to determining, for example, which immune baseline modulators to administer in a tailored fashion and for what types of vaccines.
Box 1. Relevance to Precision Health.
The concept that ‘baseline may predict outcome’ lends itself to precision public health (i.e., at the population level). This is already done with influenza virus vaccines every year, where different vaccines against influenza virus and/or different numbers of doses of the same vaccine are administered depending on the age of the individual and their prior vaccine status [61]. For example, a 3-year-old child might receive one dose (if previously vaccinated) or two doses (if vaccine naïve) of either a quadrivalent intranasal live-attenuated vaccine or an inactivated vaccine, whereas an individual aged >65 years might receive a single dose of standard quadrivalent inactivated vaccine, a high-dose vaccine, or a formulation that includes the MF59 adjuvant.
Alt-text: Box 1
Here, we review evidence supporting the notion that baseline immune status can predict and potentially impact vaccine responses. We hypothesize that iterative application of population-based systems vaccinology studies following the administration of immune modulators and vaccines will help decipher how baseline immune status might impact vaccine outcome. Identification of these predictive parameters and rules is an important step towards reaching the ultimate goal: the ability to predict a vaccine’s outcome prior to its administration, along with tailored vaccine designs protective for all.
Baseline Might Predict Outcome for Influenza Virus Vaccination
One hundred years ago, the 1918 influenza virus pandemic emerged [15]. A new influenza A/H1N1 virus had infected one-third of the world’s population and killed between 50 and 100 million people within 1–2 years [16]. Put in perspective, the pandemic killed more people in 12 months than HIV-1/AIDS has in the last 35 years. Today, influenza virus infections still loom as one of our greatest public health threats [16]. While great advances have been made in understanding the influenza virus, progress has been slow in understanding how the human immune system prevents and controls influenza virus infection and how to develop better vaccines. As a result, our current influenza virus vaccines are only moderately effective in blunting annual epidemics and would offer limited, or no, protection against a global pandemic due to novel influenza viral strains [16].
The response to influenza virus vaccination can be highly variable across a given population, leaving some individuals without protection even when the seasonal vaccine matches the circulating influenza viral strains. The change in antibody titers against influenza virus following vaccination is a reasonable correlate of protection (CoP) and is associated with ‘intrinsic’ factors such as age [9,10], sex [17], pre-existing antibody titers [9,10], and prior vaccination history [18]. However, much of the variance in antibody responses to influenza virus vaccination remains unexplained [10]. Based on studies in twins, genetics seems to contribute only marginally to this heterogeneity in vaccination outcomes in adults, suggesting environmental factors as key drivers [19,20]. While early systems-biology analyses of influenza virus vaccination have identified antibody response predictors, these have been based on post-vaccination parameters, such as the magnitude of plasmablast increases on day 7, and changes in blood host-derived transcripts on days 1–3 after vaccination [21,22]. These observations raised questions on whether baseline determinants of these post-vaccination parameters might exist, ultimately shaping the quality and quantity of antibody responses following vaccination [21,22]. In searching for this ‘root’ predictor, both seasonal and the pandemic H1N1 (pH1N1) viral strains were considered in a framework transforming multimodal data sets and natural population variation into computational models designed to predict serological responses to influenza virus vaccination [10]. Using baseline data alone, independent of age, sex, initial serology, and the antigen specificity of the T and B cell populations for vaccine antigens, high versus low human responders could be robustly predicted [10]. The essential parameters contributing to this prediction comprised the frequency of a few circulating immune cell subsets (especially B and T cell subpopulations), and these were among the most temporally stable within subjects over a period of 2 months. However, some of these immune cell subpopulation parameters also had the highest between-subject variation and thus were likely to be predictive because they delineated distinct baseline immune states among individuals [10]. Such baseline biomarkers, if robustly validated, are particularly attractive because they might potentially provide relevant information on the timing at which measurement should occur [8].
Additional evidence for the existence of baseline predictors for influenza virus vaccine responses came from additional studies where peripheral-blood naïve helper CD4+ T cell counts and class-switched memory B cell frequencies at the time of vaccination correlated with antibody responses to the annual trivalent inactivated influenza vaccine (TIV) and also following pH1N1 vaccination [23., 24., 25.]. Recently, multiple influenza virus vaccination cohorts spanning distinct annual seasons [from the Human Immunology Project Consortium (HIPC) and the National Institutes of Health (NIH) Center for Human Immunology (CHI)] and geographic locations (located in North America) were leveraged to identify baseline transcriptional predictive signatures of antibody responses to influenza virus vaccination [9]. This analysis revealed baseline predictive signatures for younger (<35 years of age) individuals [9], similar to what was observed in the previously identified baseline predictive signatures [10]. Together, the signatures emerging from multiple omics approaches (e.g., cell population analyses plus transcriptomics) could potentially capture the inherent biological state of individuals during vaccination more comprehensively. For influenza virus, pre-vaccination host-specific predictive signatures exist beyond those that reflect intrinsic variables such as age and sex and these predictors are likely to reflect cumulative effects of the environment in addition to, potentially, past exposure to influenza virus. However, pre-existing frequencies of antigen-specific B or CD4+ T cells in blood are often not associated with antibody responses to influenza virus vaccination [10,26,27], suggesting that the known baseline predictive signatures may be independent of antigen-specific immunity. Further assessment of these signatures and the search for additional predictors in diverse populations will be essential to help guide the development of next-generation vaccines that can provide persistent immunity against influenza viral strains, especially under pandemic scenarios (Box 2 ).
Box 2. Age-Dependent Baseline Predictors.
In the HIPC-CHI multicohort meta-analysis of influenza virus vaccination discussed, a putative baseline predictor in older individuals was found (although not validated in an independent cohort) and the signature was distinct from that in young individuals [9]. Specifically, baseline parameters that positively correlated with antibody responses in the young (<35 years of age) tended to negatively correlate with antibody responses in older individuals (>65 years of age). While the underlying mechanisms remain unclear, this observation (which emerged from data across several vaccination seasons) suggested that young and older adults might indeed share baseline predictive parameters, but their effects on vaccination outcomes could be age dependent.
Alt-text: Box 2
Baseline Might Predict Outcome for HBV Vaccination
Approximately 30% of the world’s population (i.e., about 2 billion persons) have serological evidence of HBV infection [28]. Each year, approximately 620 000 HBV-infected persons die from chronic liver disease [29]. The World Health Organization (WHO) recommends universal neonatal/infant HBV immunization as well as vaccination of adults at risk for HBV infection [30]. All current HBV vaccines comprise hepatitis B surface antigen (HBs) and employ a two- to four-dose vaccination schedule [31]. The serological CoP against chronic HBV infection is one of the best defined for any vaccine, constituting a titer of ≥10 mIU/ml anti-HBs antibody (defined by the WHO Anti-HBs Reference Preparation) (18,22). There is a direct correlation between quantitative antibody titers and protection [28]. Some individuals achieve an anti-HBs response of ≥10 mIU/ml after just one or two doses of vaccine [28]; specifically, 30–50% after one dose and 50–75% after two doses [32]. Unfortunately, persistent non-responders, 5–10% of the vaccinated population, stay unprotected even after a completed vaccination schedule [29,33]. Similar to influenza virus vaccination, cell population and transcriptional parameters measured at baseline (i.e., prior to vaccination) might predict HBV vaccine response outcome. Specifically, in adults aged ≥65 years, higher expression of inflammatory response transcripts and increased frequencies of proinflammatory innate immune cells in peripheral blood correlate with lower anti-HBs antibody responses [11]. This was also the case for younger adults aged ~20–50 years, where non-responders showed a more activated state of immunity before vaccination than responders; this was determined from the increased expression of the granulin precursor gene (GRN) and higher granulocyte numbers in peripheral blood in non-responding young individuals relative to responders [12,13]. Thus, it might be possible to define, at baseline, predictive signatures for subjects generating protective responses following HBV vaccination.
Baseline Might Predict Outcome for Malaria Vaccination
Malaria remains a major global health problem (caused by Plasmodium sp. infection), causing an estimated 228 million cases and 405 000 deaths in 2018 alone [34]. Decades of research have thus far have resulted in only a single licensed subunit vaccine candidate, the RTS,S/AS01E compound [35]. Despite being recommended by the WHO for pilot implementation studies ongoing in Africa since April 2019, the efficacy of the vaccine against clinical malaria is, at best, moderate and of limited duration [35]. At present, it is unknown why the RTS,S vaccine protects only a proportion of immunized subjects; specifically, 30–55% over 1 year and 26–36% with a booster dose over 3–4 years [35]. Vaccines based on live-attenuated Plasmodium falciparum parasites through irradiation [36] or chemoprophylaxis with antimalarial agents [37,38] are promising alternatives that have shown up to 100% efficacy in small clinical trials in naïve adults [39]. Major knowledge gaps in the development of more effective malaria vaccines are the absence of immune CoP and an understanding of the specific mechanisms of protective immunity. The best correlates for either of these malaria vaccines is the titer of IgG antibodies against the main vaccine antigen [the circumsporozite protein (CSP)], thought to prevent liver-stage infection and thus the subsequent blood-stage infection causing clinical disease [34]. However, unlike the anti-HBs antibody titers following HBV vaccination, the positive predictive value of vaccine-induced IgG titers is too low to be considered a CoP [40]. Of relevance to the concept of determining an immune baseline at the time of vaccination to predict vaccine outcomes, peripheral blood monocyte-to-lymphocyte ratios at baseline have been negatively correlated with RTS,S/AS01 efficacy in a completed large Phase 2B randomized prevention clinical trial in 894 African children (primary outcome: frequency of first case of malaria meeting the primary case definition) (NCT00380393)i [14]; this suggested that baseline proportions of white blood cells might be considered a predictive parameter to assess the response to RTS,S/AS01 vaccination a priori.
Baseline Predictors and Potential Determinants Shared across Vaccines
A recent study derived a ten-gene blood transcriptional surrogate (TGSig) of the temporally stable cell frequency-based baseline signature predictive of antibody response to influenza virus vaccination in the NIH cohort highlighted earlier [41]. The authors showed that peripheral blood mRNA expression of TGSig was predictive of influenza virus vaccination responses in three of four independent US locations over four different vaccination seasons. Gene expression of this TGSig was also predictive of antibody responses to the yellow fever (YF) vaccine (a live-attenuated virus) given to YF-naïve individuals, further supporting the notion that this particular baseline signature might capture predictive information beyond antigen-specific prior exposure [41]. In the same study, the authors used simultaneous protein and transcriptome analysis [cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq)] of single peripheral blood mononuclear cells from high and low influenza virus vaccination responders (differentiated based on antibody titers produced in response to influenza vaccination). The results revealed a complex network where TGSig expression correlated with cell cycle activation and type I IFN response status in a cellular circuit comprising plasmacytoid dendritic cells (pDCs), class-switched B cells, and T lymphocytes. This interactive circuit of cells was known to be fully activated only after an immune challenge such as an infection, yet healthy high vaccine responders displayed an elevated activation status at baseline [41]. The mechanisms that maintain this activated state and restrain the full-blown systemic response before vaccination and inflammatory stimulation remain to be determined. However, these results suggest that baseline predictors might be shared across different types of vaccines in both naïve and pre-exposed populations.
Based on the aforementioned, it is reasonable to speculate that baseline immune status might impact responses to a wider range of vaccines. However, larger-scale studies are needed to define and assess such predictors, by integrating simultaneous assessment of genetics, microbiome, intrinsic factors (age, sex etc.), and nongenetic immune status across different vaccines. While meta-analysis of immune profiling data from heterogeneous studies involving different vaccines and populations can be powerful [9], currently the data modality shared across most studies is largely limited to blood transcriptional profiling and/or assessing the frequency of major circulating immune cell populations. Thus, further and more robust targeted studies, involving randomized vaccination trials of individuals with or without certain baseline immune signatures, are needed to delineate the extent to which the baseline can predict and influence outcomes for different types of vaccines.
Modulating Baseline Might Modulate Vaccine Outcome
The concept of ‘modulating baseline to modulate vaccine outcome’ (Figure 1 , Key Figure) might be empirically realized through targeted modulation of the immune status baseline. For example, the administration of antibiotics can change an individual’s microbiome frequency and composition as well as, potentially, their inflammatory status at baseline (e.g., perhaps due to an underlying pathology); this in turn might impact antibody responses to influenza virus vaccination [6]. Moreover, vaccines themselves can alter immune baseline status and thus also potentially the outcomes of subsequent vaccinations (Box 3 ) [42., 43., 44., 45.]. For example, the bacille Calmette–Guérin (BCG) vaccine is one of the most widely used vaccines worldwide. In addition to providing moderate protection against TB, it can have nonspecific (heterologous) immunomodulatory effects beyond the target pathogen (TB), including responses to other vaccines [42] and infections such as malaria [46] and possibly even SARS-CoV-2 [47,48]. For example, BCG vaccination is associated with, and can increase the antibody responses to, HBV, polio virus type 1, Pneumococcus sp., and influenza virus vaccination [43., 44., 45.]. The underlying mechanisms for BCG’s impact on subsequent vaccinations have not been fully elucidated but could involve shifting the responsiveness of immune cells to cytokines at baseline [49,50]. Further, cytomegalovirus (CMV) infection is known to affect multiple components of the innate and adaptive immune system [51] and has been associated with enhanced immune responses (higher antibody titers) of young, healthy humans to influenza virus vaccination relative to non-CMV infected subjects; this, however, has not been observed in older adults, suggesting that the impact of CMV might be age dependent, an aspect that certainly merits further investigation [52]. By contrast, Epstein–Barr virus (EBV) infection, also known to affect multiple components of innate and adaptive immunity, is associated with reduced responses (lower antibody titers) to the routine infant vaccines relative to non-infected subjects [53].
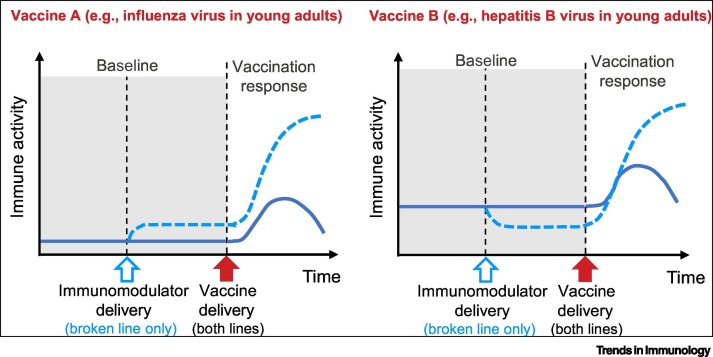
Figure 1.
Key Figure. Model for Modulating Immune Baseline Status to Modulate Vaccine Responses.
For Vaccine A, which could be influenza virus vaccination in young adults [52], an immunomodulator that increases the baseline activity of an immune pathway (blue broken line) could increase a vaccine-induced immune response. For Vaccine B, which could be hepatitis B virus vaccination [11., 12., 13.], an immunomodulator that decreases the baseline activity of a distinct immune pathway (blue broken line) could increase a vaccine-induced antibody response. Note that the targets of the baseline modulation and the immune activity associated with the vaccine response could be completely distinct; this is a simplified illustration and is not meant to indicate that changes induced by baseline modulation would have to occur in the same particular cell or pathway as the immune activity (e.g., antibody or T cell responses).
Box 3. Modulating Baseline before Birth.
Vaccination as a means of modulating the immune baseline has been suggested to occur before birth. For example, MF59-adjuvanted influenza virus vaccination during pregnancy was found to alter the immune cytokine production profile in the nasal mucosal fluid of 4-week-old infants; specifically, significant upregulation of TGF-β1 but downregulation of IL-12p70, IFN-γ, IL-5, eotaxin-1, TARC, MDC, and IL-8 was documented compared with those vaccinated after pregnancy [62]. While the implications of this finding for infant vaccine responses have not been investigated, these data highlight that the concept of immunomodulation at baseline prior to vaccination might be relevant for maternal immunizations as well [63].
Alt-text: Box 3
It is important to note that increased immune activation at baseline does not always lead to better vaccine responses, likely because the type, quantity, and cellular specificity of the activation and other host factors such as age matter. For example, an activated immune state in natural killer (NK) cells, proinflammatory monocytes, and differentiated T/B cell subsets was associated with reduced responses (lower antibody titers) to the YF vaccine prior to vaccination in an African population relative to controls [54]. Moreover, this was consistent with the aforementioned studies on HBV vaccination in the elderly, where a more pronounced inflammatory gene expression profile at baseline predicted a poorer response to vaccination relative to those having a less pronounced inflammatory gene expression profile [11., 12., 13.]. Furthermore, oral administration of the immunosuppressant rapamycin has been shown to increase the immune response to influenza virus vaccination (higher antibody response) in older adults relative to those not having received rapamycin [55]. In addition to rapamycin, other small molecules and biologics with immunomodulatory function, such as imiquimod, are currently approved for clinical use [56., 57., 58.]. Thus, the tools to modulate immune baseline status to potentially improve the outcome of vaccination are already available. We posit that, based on these observations, modulation of the baseline to tune vaccine outcome in a vaccine- and population-dependent manner should be possible [59] (Figure 1). This concept is indirectly supported by data from several infectious diseases where differences in host baseline status at the time of infection may be linked to different clinical outcomes.
A Path towards Targeted Modulation of Immune Baseline to Improve Vaccine Outcomes
Many of the baseline predictive signatures identified so far are associated with innate immune functions and thus may reflect recent activation of functionally interacting cells prior to the time of vaccination [19,41,60]. This hints at the possibility that acute modulation of the baseline immune status not long before vaccination could alter the outcome. The concept of deliberately modulating the baseline to improve the outcome of vaccination has, however, never been directly assessed. Specifically, while vaccine adjuvants administered simultaneously with vaccines can enhance the response to some vaccines, to our knowledge the optimal timing of administration of an adjuvant in relation to the vaccine antigen has not been determined. The original choice to combine adjuvant and antigen in one administration has obvious practical benefits in that the number of injections can be reduced. However, given that many vaccines require multiple doses to provide protection, the presumed advantage of co-administration of adjuvant and antigen may be small, or even become disadvantageous, if modulating the baseline prior to vaccination can ultimately reduce the number of vaccine doses administered. To begin assessing this approach experimentally, one could administer the specific approved agents discussed earlier, such as rapamycin or other vaccines such as BCG, or even alter specific components of the microbiome prior to administration of the vaccine, to then assess vaccination outcomes. It might also be possible to determine whether better vaccine responses are elicited when adjuvants are administered separately from the antigen in terms of time (before, after vaccination) and/or space (separate routes or sites of administration). It is obviously also important to assess the safety and reactogenicity of the adjuvant given alone. Of note, this approach may not even require a change in existing vaccine formulations, but rather the addition of immunomodulators prior to the administration of existing vaccine/adjuvant formulations.
Concluding Remarks
Extensive work is needed to further test and validate the concept of baseline predictors and the feasibility and utility of targeted modulation of the immune baseline before vaccinations. Many questions remain (see Outstanding Questions). Future studies comparing the effects of the administration of immunomodulators, the timing of the administration of modulators [i.e., defining the gap between the administration of the modulator(s) (including adjuvants) and the vaccine], and the temporal stability of the modulated baseline states should ideally lead to effective strategies for the development of better vaccines. We posit that this strategy merits further attention particularly when considering the most vulnerable populations, including infants, the elderly, and immunosuppressed individuals, as well as those living in low- and middle-income countries.
Clinician’s Corner.
Although largely unexplored, baseline immune status might also predict the clinical outcome for infectious diseases. Some examples are as follows.
-
•
The risk of severe infections early in life can be predicted from baseline innate immune phenotypes measured at birth [64].
-
•
Certain immune signatures at baseline can predict not only the response to malaria vaccination but the clinical outcomes of acute infection [65].
-
•
Ebola virus infections have a case fatality rate of approximately 50%, with some individuals succumbing to disease but others recovering, or even controlling the infection in an asymptomatic manner [66]. Is the differential pathogenesis of Ebola virus infection also due to variations in baseline immune status? If so, could immunomodulators enhance protection and thus transform individuals who would have otherwise died from Ebola virus infection into those with asymptomatic infections [67]?
-
•
HIV-1 infection has a bell-shaped pathogenesis curve ranging from infected individuals who are rapid progressors, to moderate progressors, to elite controllers [68]. Could baseline predictive signatures for HIV-1 immune responses be identified, leading to novel prevention and control measures?
-
•
This concept is not limited to infectious diseases. While there is an ongoing revolution in cancer immunotherapy using checkpoint inhibitors, responses are highly variable across patient populations. For example, less than 15% of subjects across the spectrum of cancers who have received checkpoint immunotherapy benefit clinically [69]. Predictive signatures could ideally identify responders and nonresponders before therapy is commenced [70].
Alt-text: Clinician’s Corner
Outstanding Questions.
Do baseline signatures reflect causal mechanisms or do they merely represent a correlate of yet-to-be-determined factors and biological networks?
Are predictive baseline signatures similar or distinct across different populations (e.g., the young, elderly, pregnant, different ethnicities)?
How long do various baseline states last? How robust are they to perturbations such as infection and shifts in the microbiome?
Could separate administration of adjuvant and antigen (vs the current standard co-administration) potentiate vaccine responses? If so, could this approach lead to protective one-dose vaccine regimens?
Alt-text: Outstanding Questions
Acknowledgments
We thank Ellysia Hollams and Tara McLaren for help with figure design. J.S.T. is at the Multiscale Systems Biology Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases (NIAID), NIH, Center for Human Immunology (CHI), NIH and supported by the Intramural Research Program of the NIAID and Intramural Programs of the NIH Institutes supporting the CHI. The work on malaria vaccines by C.D. and G.M. was supported in part by the EU FP7-funded SysMalVac project (grant number: 305869-2) and is part of the ISGlobal’s Program on the Molecular Mechanisms of Malaria, which is partially supported by the Fundación Ramón Areces, and we acknowledge support from the Spanish Ministry of Science and Innovation through the ‘Centro de Excelencia Severo Ochoa 2019–2023’ Program (CEX2018-000806-S) and support from the Generalitat de Catalunya through the CERCA Program. G.M. is supported by the Department of Health, Catalan Government grant (SLT006/17/00109). P.VD. is full professor at and supported by the University of Antwerp, Belgium. A.M. is Research Director at and supported by the Fonds de la Recherche Scientifique, FRS-FNRS, Belgium. M.S. is Associate Professor at and supported by the BC Children’s Hospital Foundation, the Canadian Child Health Clinician Scientist Program and the Michael Smith Foundation for Health Research. W.K. is supported by the Human Vaccines Project (https://www.humanvaccinesproject.org) philanthropic donors. Both T.R.K. and R.B.O. are supported by Telethon Kids and Perth Children’s Hospital Foundation and NIH/NIAID U19AI118608-02. Several of the authors have been investigators on projects funded by vaccine manufacturers. All such funds have been paid to their institute; none were received as personal payments.
Glossary
- Cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq)
analyzes, at the single-cell level, gene expression profiles of that cell via mRNA and protein amounts.
- Checkpoint immunotherapy
targets immune checkpoint molecules with the aim of enabling immune cells to attack malignant cells (in the case of cancer).
- Class-switched B cells
B cells that have rearranged their genetic information, ceasing to produce surface IgM antibodies to secrete other classes of antibodies such as IgG, IgA, etc.
- Imiquimod
used as a therapy to treat, for example, warts, superficial basal cell carcinoma, and actinic keratosis. It is believed to act by modulating the local immune environment.
- Intrinsic versus extrinsic variables
age, sex, and genetics are ‘intrinsic’ variables and factors such as the microbiome and past exposures including previous vaccination or infection are ‘extrinsic’ variables.
- MF59
squalene (a natural oil compound made in, e.g., the skin) adjuvant added to vaccines to increase immune responses (e.g., antibody titers).
- Systems vaccinology
the approach of systems biology (i.e., highly multiplexed immune profiling and data-driven computational modeling) applied to the study of vaccine responses.
Resource
iNCT00380393: this trial is listed in ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT00380393
References
- 1.Plotkin S. History of vaccination. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12283–12287. doi: 10.1073/pnas.1400472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koff W.C. Accelerating next-generation vaccine development for global disease prevention. Science. 2013;340:1232910. doi: 10.1126/science.1232910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koff W.C. Toward a human vaccines project. Nat. Immunol. 2014;15:589–592. doi: 10.1038/ni.2871. [DOI] [PubMed] [Google Scholar]
- 4.Lynn D.J., Pulendran B. The potential of the microbiota to influence vaccine responses. J. Leukoc. Biol. 2018;103:225–231. doi: 10.1189/jlb.5MR0617-216R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh J.Z. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagan T. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178:1313–1328.e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raeven R.H.M. Systems vaccinology and big data in the vaccine development chain. Immunology. 2019;156:33–46. doi: 10.1111/imm.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsang J.S. Utilizing population variation, vaccination, and systems biology to study human immunology. Trends Immunol. 2015;36:479–493. doi: 10.1016/j.it.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.HIPC-CHI Signatures Project Team and HIPC-I Consortium Multicohort analysis reveals baseline transcriptional predictors of influenza vaccination responses. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aal4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsang J.S. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fourati S. Pre-vaccination inflammation and B-cell signalling predict age-related hyporesponse to hepatitis B vaccination. Nat. Commun. 2016;7:10369. doi: 10.1038/ncomms10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartholomeus E. Transcriptome profiling in blood before and after hepatitis B vaccination shows significant differences in gene expression between responders and non-responders. Vaccine. 2018;36:6282–6289. doi: 10.1016/j.vaccine.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Qiu S. Significant transcriptome and cytokine changes in hepatitis B vaccine non-responders revealed by genome-wide comparative analysis. Hum. Vaccin. Immunother. 2018;14:1763–1772. doi: 10.1080/21645515.2018.1450122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warimwe G.M. Peripheral blood monocyte-to-lymphocyte ratio at study enrollment predicts efficacy of the RTS,S malaria vaccine: analysis of pooled Phase II clinical trial data. BMC Med. 2013;11:184. doi: 10.1186/1741-7015-11-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hannoun C. The evolving history of influenza viruses and influenza vaccines. Expert Rev. Vaccines. 2013;12:1085–1094. doi: 10.1586/14760584.2013.824709. [DOI] [PubMed] [Google Scholar]
- 16.Krammer F., Palese P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 2015;14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 17.Furman D. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc. Natl. Acad. Sci. U. S. A. 2014;111:869–874. doi: 10.1073/pnas.1321060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belongia E.A. Repeated annual influenza vaccination and vaccine effectiveness: review of evidence. Expert Rev. Vaccines. 2017;16:1–14. doi: 10.1080/14760584.2017.1334554. [DOI] [PubMed] [Google Scholar]
- 19.Brodin P. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newport M.J. Genetic regulation of immune responses to vaccines in early life. Genes Immun. 2004;5:122–129. doi: 10.1038/sj.gene.6364051. [DOI] [PubMed] [Google Scholar]
- 21.Nakaya H.I. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bucasas K.L. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J. Infect. Dis. 2011;203:921–929. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frasca D. Unique biomarkers for B-cell function predict the serum response to pandemic H1N1 influenza vaccine. Int. Immunol. 2012;24:175–182. doi: 10.1093/intimm/dxr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy R.B. The composition of immune cells serves as a predictor of adaptive immunity in a cohort of 50- to 74-year-old adults. Immunology. 2016;148:266–275. doi: 10.1111/imm.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jurchott K. Highly predictive model for a protective immune response to the A(H1N1)pdm2009 influenza strain after seasonal vaccination. PLoS One. 2016;11 doi: 10.1371/journal.pone.0150812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews S.F. High preexisting serological antibody levels correlate with diversification of the influenza vaccine response. J. Virol. 2015;89:3308–3317. doi: 10.1128/JVI.02871-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He X.S. Baseline levels of influenza-specific CD4 memory T-cells affect T-cell responses to influenza vaccines. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schillie S.F., Murphy T.V. Seroprotection after recombinant hepatitis B vaccination among newborn infants: a review. Vaccine. 2013;31:2506–2516. doi: 10.1016/j.vaccine.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Van Damme P. Hepatitis B vaccines. In: Vesikari T., Van Damme P., editors. Pediatric Vaccines and Vaccinations: A European Textbook. Springer; 2017. pp. 109–116. [Google Scholar]
- 30.WHO Hepatitis B vaccines. Wkly Epidemiol. Rec. 2009;84:405–419. [PubMed] [Google Scholar]
- 31.Shepard C.W. Hepatitis B virus infection: epidemiology and vaccination. Epidemiol. Rev. 2006;28:112–125. doi: 10.1093/epirev/mxj009. [DOI] [PubMed] [Google Scholar]
- 32.Mast E. 4th edn. Saunders; 2004. Hepatitis B Vaccine. [Google Scholar]
- 33.Leuridan E., Van Damme P. Hepatitis B and the need for a booster dose. Clin. Infect. Dis. 2011;53:68–75. doi: 10.1093/cid/cir270. [DOI] [PubMed] [Google Scholar]
- 34.WHO World Malaria Report 2019. 2019. https://www.who.int/publications-detail/world-malaria-report-2019 Published online December 4, 2019.
- 35.RTS,S Clinical Trials Partnership Efficacy and safety of the RTS,S/AS01 malaria vaccine during 18 months after vaccination: a Phase 3 randomized, controlled trial in children and young infants at 11 African sites. PLoS Med. 2014;11 doi: 10.1371/journal.pmed.1001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishizuka A.S. Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nat. Med. 2016;22:614–623. doi: 10.1038/nm.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roestenberg M. Protection against a malaria challenge by sporozoite inoculation. N. Engl. J. Med. 2009;361:468–477. doi: 10.1056/NEJMoa0805832. [DOI] [PubMed] [Google Scholar]
- 38.Mordmuller B. Sterile protection against human malaria by chemoattenuated PfSPZ vaccine. Nature. 2017;542:445–449. doi: 10.1038/nature21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jongo S.A. Safety, immunogenicity, and protective efficacy against controlled human malaria infection of Plasmodium falciparum sporozoite vaccine in Tanzanian adults. Am. J. Trop. Med. Hyg. 2018;99:338–349. doi: 10.4269/ajtmh.17-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ubillos I. Baseline exposure, antibody subclass, and hepatitis B response differentially affect malaria protective immunity following RTS,S/AS01E vaccination in African children. BMC Med. 2018;16:197. doi: 10.1186/s12916-018-1186-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotliarov Y. Broad immune activation underlies shared set point signatures for vaccine responsiveness in healthy individuals and disease activity in patients with lupus. Nat. Med. 2020 doi: 10.1038/s41591-020-0769-8. Published online February 24, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmermann P. The influence of neonatal bacille Calmette–Guerin (BCG) immunisation on heterologous vaccine responses in infants. Vaccine. 2019;37:3735–3744. doi: 10.1016/j.vaccine.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 43.Ota M.O. Influence of Mycobacterium bovis bacillus Calmette–Guerin on antibody and cytokine responses to human neonatal vaccination. J. Immunol. 2002;168:919–925. doi: 10.4049/jimmunol.168.2.919. [DOI] [PubMed] [Google Scholar]
- 44.Scheid A. Adjuvant effect of bacille Calmette–Guerin on hepatitis B vaccine immunogenicity in the preterm and term newborn. Front. Immunol. 2018;9:29. doi: 10.3389/fimmu.2018.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmermann P., Curtis N. The influence of BCG on vaccine responses – a systematic review. Expert Rev. Vaccines. 2018;17:547–554. doi: 10.1080/14760584.2018.1483727. [DOI] [PubMed] [Google Scholar]
- 46.Bassat Q. Making sense of emerging evidence on the non-specific effects of the BCG vaccine on malaria risk and neonatal mortality. BMJ Glob. Health. 2020;5 doi: 10.1136/bmjgh-2020-002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Vrieze J. Can a century-old TB vaccine steel the immune system against the new coronavirus? Science. 2020 Published online March 23, 2020. https://www.sciencemag.org/news/2020/03/can-century-old-tb-vaccine-steel-immune-system-against-new-coronavirus. [Google Scholar]
- 48.Miller A. Correlation between universal BCG vaccination policy and reduced morbidity and mortality for COVID-19: an epidemiological study. medRxiv. 2020 Published online March 28, 2020. https://www.medrxiv.org/content/10.1101/2020.03.24.20042937v1. [Google Scholar]
- 49.Freyne B. Neonatal BCG vaccination influences cytokine responses to Toll-like receptor ligands and heterologous antigens. J. Infect. Dis. 2018;217:1798–1808. doi: 10.1093/infdis/jiy069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Namakula R. Monocytes from neonates and adults have a similar capacity to adapt their cytokine production after previous exposure to BCG and β-glucan. PLoS One. 2020;15 doi: 10.1371/journal.pone.0229287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bayard C. Coordinated expansion of both memory T cells and NK cells in response to CMV infection in humans. Eur. J. Immunol. 2016;46:1168–1179. doi: 10.1002/eji.201546179. [DOI] [PubMed] [Google Scholar]
- 52.Furman D. Cytomegalovirus infection enhances the immune response to influenza. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aaa2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holder B. Epstein–Barr virus but not cytomegalovirus is associated with reduced vaccine antibody responses in Gambian infants. PLoS One. 2010;5 doi: 10.1371/journal.pone.0014013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muyanja E. Immune activation alters cellular and humoral responses to yellow fever 17D vaccine. J. Clin. Invest. 2014;124:3147–3158. doi: 10.1172/JCI75429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mannick J.B. mTOR inhibition improves immune function in the elderly. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3009892. [DOI] [PubMed] [Google Scholar]
- 56.Roukens A.H. Intradermal hepatitis B vaccination in non-responders after topical application of imiquimod (Aldara) Vaccine. 2010;28:4288–4293. doi: 10.1016/j.vaccine.2010.04.029. [DOI] [PubMed] [Google Scholar]
- 57.Hung I.F. Topical imiquimod before intradermal trivalent influenza vaccine for protection against heterologous non-vaccine and antigenically drifted viruses: a single-centre, double-blind, randomised, controlled Phase 2b/3 trial. Lancet Infect. Dis. 2016;16:209–218. doi: 10.1016/S1473-3099(15)00354-0. [DOI] [PubMed] [Google Scholar]
- 58.Caimi A.T. Novel imiquimod nanovesicles for topical vaccination. Colloids Surf. B Biointerfaces. 2019;174:536–543. doi: 10.1016/j.colsurfb.2018.11.031. [DOI] [PubMed] [Google Scholar]
- 59.Alter G., Sekaly R.P. Beyond adjuvants: antagonizing inflammation to enhance vaccine immunity. Vaccine. 2015;33:B55–B59. doi: 10.1016/j.vaccine.2015.03.058. [DOI] [PubMed] [Google Scholar]
- 60.Orru V. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grohskopf L.A. Prevention and control of seasonal influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices–United States, 2018–19 influenza season. MMWR Recomm. Rep. 2018;67:1–20. doi: 10.15585/mmwr.rr6703a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bischoff A.L. Airway mucosal immune-suppression in neonates of mothers receiving A(H1N1)pnd09 vaccination during pregnancy. Pediatr. Infect. Dis. J. 2015;34:84–90. doi: 10.1097/INF.0000000000000529. [DOI] [PubMed] [Google Scholar]
- 63.Marchant A. Maternal immunisation: collaborating with mother nature. Lancet Infect. Dis. 2017;17:e197–e208. doi: 10.1016/S1473-3099(17)30229-3. [DOI] [PubMed] [Google Scholar]
- 64.Goetghebuer T. Initiation of anti-retroviral therapy before pregnancy reduces the risk of infection-related hospitalization in HIV-exposed uninfected infants born in a high-income country. Clin. Infect. Dis. 2018;68:1193–1203. doi: 10.1093/cid/ciy673. [DOI] [PubMed] [Google Scholar]
- 65.Tran T.M. A molecular signature in blood reveals a role for p53 in regulating malaria-induced inflammation. Immunity. 2019;51:750–765. doi: 10.1016/j.immuni.2019.08.009. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malvy D. Ebola virus disease. Lancet. 2019;393:936–948. doi: 10.1016/S0140-6736(18)33132-5. [DOI] [PubMed] [Google Scholar]
- 67.Liu X. Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol. 2017;18:4. doi: 10.1186/s13059-016-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goulder P., Deeks S.G. HIV control: is getting there the same as staying there? PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haslam A., Prasad V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw. Open. 2019;2 doi: 10.1001/jamanetworkopen.2019.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krieg C. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 2018;24:144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]