Abstract
Salmonella is a leading cause of foodborne diseases, and in recent years, many isolates have exhibited a high level of antibiotic resistance, which has led to huge pressures on public health. Phages are a promising strategy to control food-borne pathogens. In this study, one of our environmental phage isolates, LPSEYT, was to be able to restrict the growth of zoonotic Salmonella enterica in vitro over a range of multiplicity of infections. Phage LPSEYT exhibited wide-ranging pH and thermal stability and rapid reproductive activity with a short latent period and a large burst size. Phage LPSEYT demonstrated potential efficiency as a biological control agent against Salmonella in a variety of food matrices, including milk and lettuce. Morphological observation, comparative genomic, and phylogenetic analysis revealed that LPSEYT does not belong to any of the currently identified genera within the Myoviridae family, and we suggest that LPSEYT represents a new genus, the LPSEYTvirus. This study contributes a phage database, develops beneficial phage resources, and sheds light on the potential application value of phages LPSEYT on food safety.
Keywords: Salmonella, phage, characterization, genome
1. Introduction
Salmonella is one of the most commonly reported microorganisms associated with foodborne disease [1]. Salmonellosis in humans is closely linked with the ingestion of pathogenic Salmonella contaminated vegetables, fruits, and animal products. Salmonella contamination frequently happens during food production (e.g., harvest, packinghouse) and during food handling processes in the kitchen [2]. Although various intervention strategies have been globally developed and implemented, salmonellosis remains one of the most commonly reported anthropozoonoses. In China, the proportion of the foodborne diseases caused by Salmonella was estimated at 22.2% [3]. Similarly, between 2009 and 2015, the Foodborne Disease Outbreak Surveillance System (FDOSS) reported 896 outbreaks caused by Salmonella, accounting for 30% of total foodborne disease outbreaks in the United States [4]. Salmonella enterica serovar Enteritidis (S. Enteritidis) and Salmonella enterica serovar Typhimurium (S. Typhimurium) were the most commonly isolated serovars from outbreaks associated with foodborne salmonellosis [5].
Along food production chains, certain processes can potentially compromise future food safety and human health. For example, the observation that bacterial cross-contamination during transportation and storage results in high levels of multi-drug resistance, where some isolates were even resistant against triclosan [6,7]. The application of antibiotics to reduce the burden of salmonellosis in farm production has caused severe problems in respect to the rapid emergence of antibiotic-resistant strains. A study in Changchun, China, reported that 93.75% of the 48 Salmonella strains isolated from chicken and pork showed resistance to antibacterial agents, and 25% of the isolates were multidrug-resistant strains [8]. Under these circumstances, the development of new strategies to sustainably control food-borne pathogens to improve safety in food production is a current global need.
As an obligate, viral parasites of bacteria, bacteriophage, bind to specific receptors present on bacterial surfaces [9]. As a result, bacteriophages have extreme host-specificity so that they only infect targeted bacteria without affecting other non-targeted bacteria and to preserve the microbiota [10,11,12]. Meanwhile, bacteriophage possesses other advantages, such as self-replicating potential, rapid killing, widespread distribution, which make phage-based biocontrol an attractive alternative against bacterial pathogens in the food production chain. Phage application has been approved by Food and Drug Administration (FDA) and Food Safety and Inspection Service of the U.S. Department of Agriculture (USDA), and a variety of commercial products are available, targeting Salmonella, including SalmoFreshTM [13] and PhageGuard S [14]. These can be used as antimicrobial agents in food raw materials and processed products.
Bacteriophage classification and the genetic backgrounds are critical for the application of phage therapy [15,16,17]. As of 2019, there were, in total, 69 genera of the Myoviridae family certified by the International Committee on Taxonomy of Viruses (ICTV) [18]. In terms of Salmonella phages, there were more than 180 complete genome sequences available in GenBank. For these Salmonella phages which belonged to Myoviridae family, they fell into five genera (recorded in ICTV), including Felixo1virus, P2virus, S16virus, Se1virus, and Spn3virus [18]. Other characterized Salmonella phages of the Myoviridae family remained unclassified. In the early days, both double-stranded or single-stranded RNA or DNA phages were classified according to the taxonomic classification, which was based on morphological similarity and the composition of nucleic acids [19,20]. With the development of sequencing technology, taxonomical classifications have become more precise on the basis of the genomes, transcriptional regime, and gene content, rather than purely based on morphological features [21,22]. Many new genera have been discovered through different methods since then. For example, Lavigne et al. reported that myoviruses could be classified by a cut-off value of 40% homologous proteins into genera [23]. The result obtained by this kind of classification method was consistent with the classification recorded in ICTV and the results from other information-based classifications [23]. In addition, the genes of terminase large subunit and major capsid protein were also used previously in an attempt to classify phages [24,25].
Detailed characterizations of novel Salmonella phage will be significant for exploring beneficial biological reagents against Salmonella in food safety. Genome analysis is also of use to understand the interaction mechanisms between Salmonella and phages and to improve the bacteriophage database and apply bacteriophages as a therapeutic approach.
In this study, LPSEYT, a novel Salmonella lytic phage, was isolated and the lytic potential against Salmonella, morphology, pH stability, and thermal stability of LPSEYT were evaluated. As a biological control agent, the potential efficiency of LPSEYT against Salmonella in food matrices was also tested. Based on genome annotation, comparative genomic and phylogenetic analyses, LPSEYT is safe for application to the food chain, and it is proposed that phage LPSEYT, together with BP63 and UPF_BP2 within the database, could form the basis of a new genus LPSEYTvirus within the Myoviridae family.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
Bacteria strains used in this study are listed in Table S1. Bacterial strains were grown in tryptone soya broth (Becton Dickinson, Sparks, MD, USA) at 37 °C overnight. These strains were applied for phage isolation, propagation, purification, as well as for phage lytic range and lytic activity determination in liquid cultures and in food matrix.
2.2. Bacteriophage Isolation
Phages were isolated from water samples obtained in Wuhan, China, based on modification of the method described by Huang et al. [26]. Water samples were centrifuged at 10,000× g for 10 min, followed by filtration through a 0.22-μm filter (Millipore, Ireland). The filtrate was then mixed with TSB that had been pre-inoculated with overnight-cultured Salmonella as host bacteria. After overnight incubation at 37 °C, the mixture was centrifuged and filtered as described above. The filtrate was detected for the presence of phages by spotting 10 μL onto double-layer agar plates where the top layer consisted of the host bacterial lawn. Any recovered phage samples were plaque purified by picking up single plaques, followed by propagation for at least three times to ensure the purity of phage isolate. The isolated phages were stored at 4 °C for further usage.
2.3. Salmonella Challenge Tests
The efficiency of selected phages virulence were evaluated respectively by lysing S. Enteritidis SGSC 4901, as published previously [27]. In brief, the experimental group was set up by infecting 100 μL freshly cultured S. Enteritidis SGSC 4901 (7 log10 CFU/mL) with 100 μL of diluted phage suspension (5 log10 PFU/mL) in a clear 96-well microplate (Corning Incorporated, USA). As control, 100 μL culture of Salmonella was mixed with 100 μL sterile TSB. The plate was maintained at 37 °C on an orbital shaker at 160 rpm and the absorbance of each sample at 600 nm (A600) was blanked with sterile TSB, and measured every hour for a total duration of 10 h using a Tecan microplate reader (Infinite M200 Pro, Switzerland). Candidate phages were selected based on antimicrobial activity against multiple hosts. The activities of the phages were examined against S. Enteritidis ATCC 13076, S. Enteritidis SGSC 4901, S. Typhimurium ATCC 13311, and S. Typhimurium ST-8 were further evaluated at various range of multiplicity of infections (MOIs) 0.01, 0.1, 1, 10, and 100, as described above.
2.4. Morphology of LPSEYT
Ten microliter of purified phage (9 log10 PFU/mL) was dropped onto the carbon-coated copper mesh grid and incubated at room conditions for 10 min to allow samples to bind. Any excess phage suspension was removed with filter paper after incubation. The phage was negative stained with 2% (w/v) phosphotungstic acid. The grid was then examined with a transmission electron microscope (TEM; Hitachi H-7000FA, Japan) at an acceleration voltage of 75 kV.
2.5. Phage Spot Test
Spot test was carried out according to reported methodology [28]. Overnight cultures of bacteria were mixed with TSB medium supplemented with 0.7% agar and then poured onto a pre-set TSA bottom layer to form bacterial lawn. The phage lysates (5 μL) were spotted onto the lawns, followed by incubation at 37 °C overnight.
2.6. Determination of Phage Adsorption Rate and One-Step Growth
Phage adsorption and one-step growth experiments were carried out according to the respective methods described by Kwiatek et al. [29] and Park et al. [30] with the following modifications. For the one-step growth experiment, briefly, 500 μL of phage suspension (5 log10 PFU/mL) was mixed with 500 μL of bacterial culture (8 log10 CFU/mL) to achieve a multiplicity of infection (MOI) of 0.001. The mixture was incubated at 37 °C for 10 min to allow phage adsorption to take place, and the sample was then centrifuged at 7000× g for 2 min to pellet bacteria. The supernatant was discarded to remove any free phage and the bacterial pellet with binding phage attached was gently washed twice with TSB. After washing, the pellet was re-suspended into 10 mL of TSB and incubated at 37 °C with constant shaking at 160 rpm for 180 min. Two sets of samples were collected at various time intervals. Before the titration, the second set of samples was treated with 1% chloroform to artificially lyse the cell so the appearance of first infective phage particles can be accurately measure to determine the eclipse time [31]. The titer of samples were tested using the double-layer agar plate method [32].
2.7. pH and Temperature Tolerance
In order to determine the influence of pH on the stability of phage, 100 μL of phage suspension (9 log10 PFU/mL) was individually added to a set of 900 μL TSB that have been pH pre-adjusted from 2 to 13. This mix was then incubated 37 °C for 60 min followed by phage titration. To determine the thermal stability, 100 μL of phage suspension (9 log10 PFU/mL) was added into 900 μL of TSB pre-warmed to various temperatures, ranging from 30 to 80 °C. Samples were incubated for 60 min and aliquots were removed for enumeration after 30 and 60 min incubation. The titer of the phage stock was measured through the double-layer agar method.
2.8. Phage Sequencing, Genome Annotation and Comparison
The genomic DNA of phage LPSEYT was extracted and purified according to a previous report [26]. The sequencing of the LPSEYT was performed using the HiSeq platform (Illumina, United States), and the reads obtained assembled by MicrobeTrakr plus (v0.9.1) software. Open reading frames (ORFs) of LPSEYT were predicted using MicrobeTrakr plus (v0.9.1) soft-ware, followed by annotation via BLAST and CD searching against the NCBI non-redundant database, the Conserved Domain Database (CDD), Pfam, SMART, COG, and InterPro databases. Transmembrane domains were identified using TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and signal sequences were identified using SignalP v3.0 (http://www.cbs.dtu.dk/services/SignalP-3.0/). The antimicrobial resistant genes (ARGs) in phage were tested using the database ARG–ANNOT (http://backup.mediterranee-infection.com/article.php?laref=282&titre=arg-annot). The database VFDB (http://www.mgc.ac.cn/VFs/main.htm) was used to detect any potential virulence factors in the phage genome. Whole viral nucleotide sequence similarities between phages were determined by BLASTN analysis at NCBI. Sequences of the terminase large subunit and major capsid protein of LPSEYT and of related phages were aligned employing MAFFT (v7.220), and tree reconstruction was carried out with RAxML (v8.2.4) with 500 bootstrap replicates.
2.9. Infection Effect of Phage LPSEYT against Salmonella in Food
Pasteurized milk was purchased from a local supermarket. For studying the groups, 100 μL of S. Typhimurium ATCC 13311 was pre-cultured in logarithmic growth phase and added into 9.8 mL milk to reach a final viable count of 3 log10 CFU/mL. This was infected with 100 μL of diluted phage suspension to reach a final titer of 6 log10 PFU/mL (MOI = 1000) and 7 log10 PFU/mL (MOI = 10,000), respectively. Phosphate-buffered saline (PBS) was added to milk samples as negative control. Samples were either incubated at 4 °C (refrigerated conditions) or at 25 °C for 6 h. Aliquots of milk sample were removed for bacterial enumeration after 0, 2, 4, 6 h of incubation according to the method of Tomat et al. [33]. Assays were conducted in triplicate.
Lettuce was purchased at local supermarket and stored at 4 °C. Samples were prepared as described previously [26]. The inner leaves of lettuce were washed with 75 % ethanol and allowed to air dry. The leaves were cut into disks of 1.5 cm in diameter using sterile knife, followed by UV treatment on both sides for 1 h to disinfect. The prepared samples were artificially contaminated with 10 μL of Salmonella inoculum in logarithmic growth phase to reach a final viable count of 3 log10 CFU/sample. After drying in a biosafety cabinet, 10 μL of serially diluted LPSEYT was added to a MOI of 1000 (6 log10 PFU/sample) or 10,000 (7 log10 PFU/sample), respectively. The same volumes of PBS were added to the sample as negative control. Then, samples were incubated at either 4 or 25 °C for 6 h. The lettuce samples were suspended in 1 mL PBS, followed by homogenization with sterile bars and vortexed for 30 s [34], and the bacteria was then enumerated with a 2 h interval. Experiments were conducted in triplicate.
2.10. Statistical Analysis
Data sets were analyzed using GraphPad Prism 6 (San Diego, CA, USA). Except as otherwise specified, data were exhibited as the mean ± SD and comparisons between data sets were carried out using the t-test. Bacterial counts recovered from food matrices were analyzed by the repeated measures test with a Tukey’s post-test. Different data were considered to have statistical significance when the p-value was <0.05.
3. Results and Discussion
3.1. Isolation and Lytic Activity of the LPSEYT
A total of 55 phages were isolated from water samples in this study. Among all phage isolates, five phages (LPSEH, LPSEO, LPSER, LPSEX, and LPSEYT) were capable of forming clear visible plaques on all the S. Enteritidis and S. Typhimurium strains tested (Table 1). For two phages, this extended to E. coli strains.
Table 1.
Lytic range of phages LPSEH, LPSEO, LPSER, LPSEX, and LPSEYT.
| Bacterial | Infectivity of Phage | ||||
|---|---|---|---|---|---|
| LPSEH | LPSEO | LPSER | LPSEX | LPSEYT | |
| S. Enteritidis ATCC 13076 | + | + | + | + | + |
| S. Enteritidis SJTUF 10978 | + | + | + | + | + |
| S. Enteritidis SJTUF 10984 | + | + | + | + | + |
| S. Enteritidis LSE4 (LK5) | + | + | + | + | + |
| S. Enteritidis SGSC 4901 | + | + | + | + | + |
| S. Typhimurium ATCC 14028 | + | + | + | + | + |
| S. Typhimurium ATCC 13311 | + | + | + | + | + |
| S. Typhimurium LST2 (ST-8) | + | + | + | + | + |
| S. Typhimurium LST4 (UK-1) | + | + | + | + | + |
| S. Typhimurium LST8 (SL1344) | + | + | + | + | + |
| S. Typhimurium SGSC 4903 | + | + | + | + | + |
| S. Typhi LSX1 (CT18) | + | + | – | + | + |
| S. Typhi LSX2 (Ty2) | + | + | – | + | + |
| S. pullorum CVCC 519 | + | + | + | + | + |
| S. pullorum CVCC 534 | + | – | + | – | + |
| Escherichia coli LEC4 (DH5α) | – | – | + | – | + |
| Escherichia coli LEC6 (BL21) | – | – | – | – | – |
| Escherichia coli LEC10 (D41) | – | – | – | – | + |
| Escherichia coli ATCC 10798 | – | – | – | – | + |
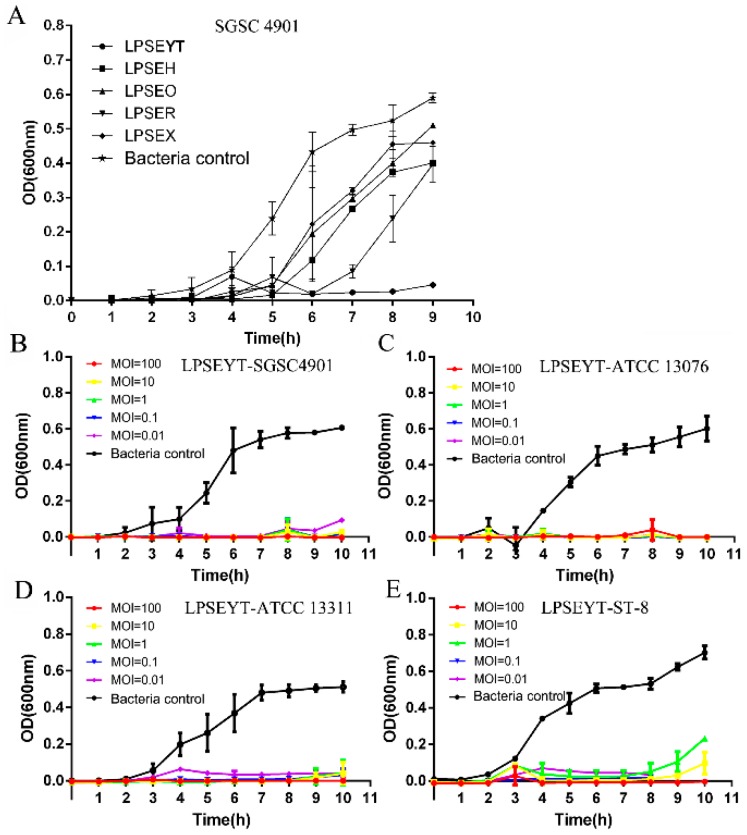
The lytic activity of these five phages were further investigated by infecting S. Enteritidis SGSC 4901 in liquid cultures (Figure 1A). As shown in Figure 1A, these phages suppressed the proliferation of Salmonella relative to the bacteria control with a significant difference after 5 h incubation (p < 0.05). Greater sustained inhibitory effects were observed for LPSEYT to 9 h, while other phages permitted the proliferation of bacteria after 6 h. Hence, LPSEYT was selected for further study. To further establish the lytic capacity of LPSEYT, S. Enteritidis ATCC 13076, S. Enteritidis SGSC 4901, S. Typhimurium ATCC 13311, and S. Typhimurium ST-8 were infected by LPSEYT at multiplicity of infections (MOIs) of 0.01, 0.1, 1, 10, and 100. Restricted growth of Salmonella was observed for all the applied MOIs (p < 0.05; Figure 1B–E). Moreover, no obvious growth for S. Enteritidis ATCC 13076, S. Typhimurium ST-8, S. Enteritidis SGSC 4901, and S. Typhimurium ATCC 13311 were detected within 10 h when phage was administered at an MOI of 1000. By comparison, Salmonella phages FGCSSa1 and PA13076 were reported to be unable to completely lyse their hosts [35,36]. The LPSEYT exhibited vigorous lytic ability against Salmonella for all the MOIs tested; even when applied at an MOI of 0.01, LPSEYT was observed to restrict the growth of Salmonella relative to the initial inoculum over 8 h (p < 0.05), whereas phages fmb-p1 and PA13076 were reported to inhibit Salmonella propagation at MOI = 10,000 and MOI = 100, respectively [36,37]. Considering the commercial feasibility of large-scale application, a lower MOI ratio would be preferred since it would cut down the cost of production and application of the phage product.
Figure 1.
Lytic ability of isolated phages. (A) The antimicrobial efficacy of isolated phages against S. Enteritidis SGSC 4901 was compared at an MOI of 0.01. Bactericidal effects of LPSEYT against (B) S. Enteritidis SGSC 4901, (C) S. Enteritidis ATCC 13076, (D) S. Typhimurium ATCC 13311, and (E) S. Typhimurium ST-8 were evaluated at various range of multiplicity of infections (MOIs) 0.01, 0.1, 1, 10, and 100, respectively. The culture of Salmonella without phage was served as control.
3.2. Morphology and Biological Characteristics of LPSEYT
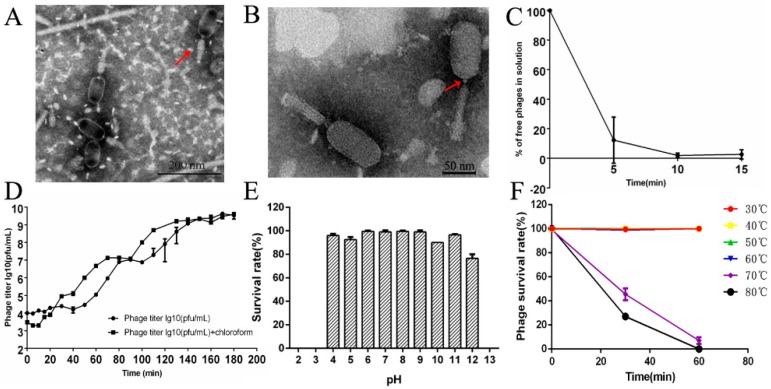
The morphological feature that distinguishes Myoviridae phages from other phage families is the contractile tail [19]. TEMs of phage LPSEYT showed a typical neck and a contractile tail, which suggests it is a member of the Myoviridae family (Figure 2A,B). The capsid of LPSEYT was measured as 90.1 ± 2.9 nm in length and 48.8 ± 3.1 nm in width (n = 10). According to the phage nomenclature proposed by Kropinski et al. [38], the phage name is vB_SalM-LPSEYT. Phage LPSEYT did not produce plaques on Aeromonas hydrophila ZYAH72, Vibrio parahaemolyticus ATCC 33846, Staphylococcus aureus ATCC 29213, and Listeria monocytogenes ATCC 19114. Notably, we found that phage LPSEYT was able to lyse antibiotic-resistant Salmonella strains listed in Table S1.
Figure 2.
Morphology and biochemical characterization of LPSEYT. (A,B) TEM analysis of LPSEYT. (C) Adsorption assay of LPSEYT. (D) One-step growth curve of LPSEYT. (E) pH tolerance of LPSEYT. (F) Temperature tolerance of LPSEYT.
As shown in Figure 2C, the adsorption rate was more than 95% at 10 min. The one-step growth curve showed that phage LPSEYT’s lytic cycle lasted about 80 min (Figure 2D). The eclipse time, latent period, and the burst size were about 10 min, approximately 50 min and 133 ± 23 PFU/cell, respectively (Figure 2D). Compared to literature values, the latent period for LPSEYT is shorter than other myoviruses, such as Salmonella phage vB_SnwM_CGG4-2 (about 58 min), Vibrio harveyi phage VhKM4 (about 60 min), and Stenotrophomonas maltophilia phage φSMA5 (about 80 min) [39,40,41]. At the same time, its burst size was greater than VhKM4 (about 52 PFU/cell), phSE-1 (about 28 PFU/mL), and phSE-2 (about 53 PFU/mL) [28,40], which is likely to be a reflection of high lytic activity. The effects of pH and temperature on the stability of LPSEYT were evaluated in this study. No significant reduction in phage titer was observed after exposing the phage sample to pH 4–11 for 60 min (Figure 2E). The titer of phage LPSEYT decreased below the detection threshold (2 log PFU/mL) when exposed to strong acid (pH < 4) or alkali (pH > 11). For temperature tolerance, the survival rate of LPSEYT was almost 100% over the range 30–60 °C, while phage LPSEYT was undetectable after exposure to 80 °C for 60 min (Figure 2D). We found phage LPSEYT was more tolerant of heat and extreme pH conditions as compared with reports of other Myoviridae phages. Kęsik-Szeloch et al. reported that the titer of Klebsiella pneumonia phage KP15 decreased outside the range of pH 5 to 8 or upon exposure to 60 °C for 15 min [42]. Bao et al. reported that Salmonella phage PC2184 was stable over the pH range of 5 to 8 [36]. LPSEYT is more likely to retain antibacterial activity under challenging conditions, such as those experienced during food processing.
Mitomycin C induction of the Salmonella hosts of LPSEYT resulted in plaque formation likely due to the prophage excision of indigenous lysogens [43]. The presence of the lysogens did not prevent superinfection from LPSEYT, which could have compromised the phages’ application against environmental Salmonella [44]. However, to assess whether LPSEYT could enter the lysogenic cycle, we took advantage of the phage’s ability to replicate in E. coli DH5α, a host that does not excise lysogens upon induction with mitomycin C. We isolated a phage resistant E. coli DH5α after exposure to LPSEYT, and then subjected these to mitomycin C induction. None of the resistant types produced plaques, implying they were not lysogens (Figure S1).
3.3. Investigation of the Infection Effect of Phage LPSEYT against Salmonella in Food
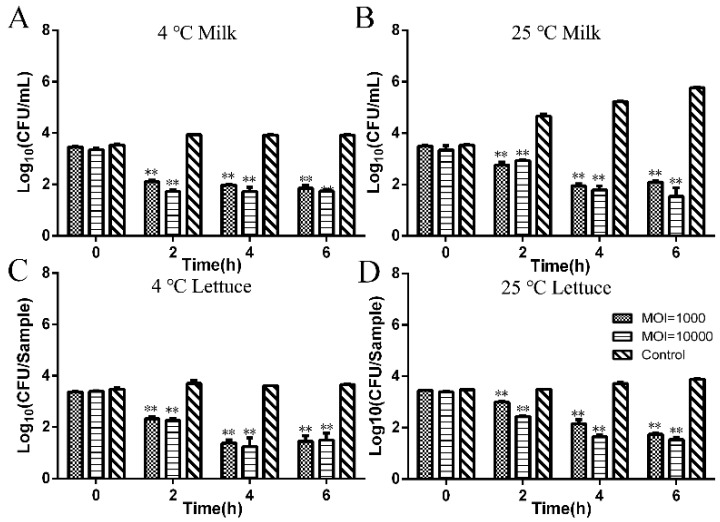
Figure 3 shows the ability of phage LPSEYT to reduce Salmonella in different food matrices under different temperature conditions. We found that phage LPSEYT produced significant reductions in viable Salmonella at both 4 °C (MOI = 1000, 10,000) and 25 °C (MOI = 1000, 10,000) regardless of the food model (p < 0.05). For Salmonella-spiked milk samples administered with phage LPSEYT at a MOI of 1000, the viable Salmonella count reduced by 2.07 log10 CFU/mL at 4 °C (Figure 3A) and 3.67 log10 CFU/mL at 25 °C (Figure 3B), relative to the phage-excluded control. When administered at an MOI of 10,000, phage LPSEYT reduced the Salmonella viable count by 2.19 log10 CFU/mL at 4 °C (Figure 3A) and 4.22 log10 CFU/mL at 25 °C (Figure 3B) after 6 hours of incubation. The antibacterial effect of LPSEYT was more durable than PA13076, as characterized by Bao et al. [36], which inhibited bacteria in milk for up to 5 h. Using a lettuce contamination model, phage LPSEYT applied at MOI = 10,000 produced a reduction in the viable count of Salmonella by 2.2 log10 CFU/sample at 4 °C (Figure 3C) and 2.34 log10 CFU/sample at 25 °C (Figure 3D) after 6 hours of incubation. It is clear that the antimicrobial effect of bacteriophage LPSEYT was greater at 25 than 4 °C. This is not surprising considering phage infection is dependent on the replication of their bacterial host. It would be difficult for phages to replicate effectively given the reduced metabolism of their bacterial host in refrigerated conditions.
Figure 3.
Application of LPSEYT in milk and on lettuce. Phage suspension was added into milk and onto the lettuce at an MOI of 1000 and 10,000. (A) LPSEYT applied in milk at 4 °C. (B) LPSEYT applied in milk at 25 °C. (C) LPSEYT applied on lettuce at 4 °C. (D) LPSEYT applied on lettuce at 25 °C. Data are shown as mean ± SD.
Previous studies have shown that the application of both phage and endolysin derived from phage exhibited greater antimicrobial activity and/or wider spectrum of lysis in comparison to the phage applied alone [45]. Therefore, future studies may focus on the expression of the LPSEYT’s endolysin as a bacterial inhibitor, and a combination of LPSEYT and other natural resources to further enhance the antibacterial effect on different food matrices.
3.4. Sequencing and Bioinformatics Analysis
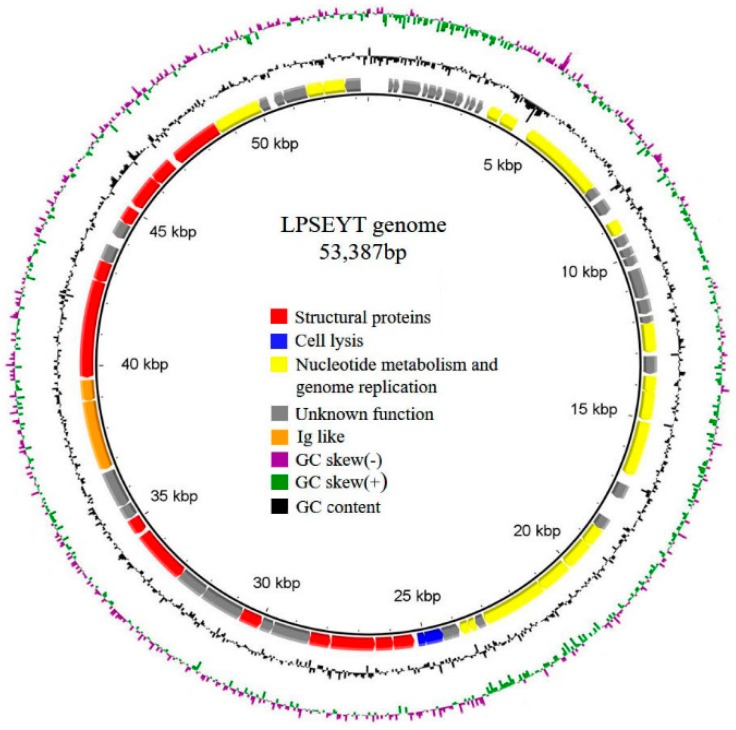
The complete genome of phage LPSEYT is a double-stranded DNA of 53,387 bp with an overall G+C content of 46.01%. Seventy-three ORFs were identified with ATG and GTG as initiation codons. Thirty-four predicted proteins showed significant homology to proteins with known functions in NCBI database (Table S2). Homologues of the remaining thirty-nine ORFs were uncharacterized proteins. Of the coding functions identified, these could classified as structural proteins and proteins involved in cell lysis, nucleotide metabolism, and genome replication (Figure 4). The absence of integrases, repressors, transposases, excisionases, antimicrobial-resistant genes, and virulence factor homologues within the genome supported that phage LPSEYT is a virulent phage and is suitable for safe application. Interestingly, within phage LPSETY’s genome, we identified two proteins containing Ig-like domains, which have been reported in association with phage that binds to human intestinal mucosa and helps the body to fight off bacterial infections [46]. However, the exact function of Ig-like proteins between phage and human immune system remains unclear. The lysis cassette of LPSEYT also drew our attention. The holin is nominally a class III holin but the topology of the transmembrane domain (TMD) of this holin is predicted to be different to the prototype class III holin of bacteriophage T4 [47,48]. Although previously not evident in Salmonella phages, the topology of LPSEYT’s holin is similar to that reported for Escherichia coli phage KBNP1315 and mycobacteriophage Ms6 [49,50]. The endolysin of LPSEYT’s lysis cassette was predicted to have a transmembrane helix in the N-terminal region as the terminal signal–anchor–release (SAR) domain. The characteristics of the N-terminal region of endolysins are important to the mechanism of lysis. It was reported that the N-terminal SAR domain is associated with the permeabilization of endolysin for Gram-negative species so that such endolysins could exhibit exogenous antibacterial activity without outer membrane permeabilization [51]. This could mean that the endolysin of LPSEYT might possess exogenous antibacterial activity.
Figure 4.
Genomic organization of LPSEYT. Patterns are divided into four circles: the full length of the genome is indicated in the first circle; the open reading frame is indicated in the second circle, and ORFs are transcribed in the clockwise or the counterclockwise direction; GC content is indicated in the third circle; while on the fourth circle, GC skew of G-C/G+C is indicated as green and purple, and green means that the value of GC skew is greater than 0 and purple means that the value is less than 0. The open reading frames marked with red, blue, yellow and orange indicate genes encoding structural proteins, cell lysis proteins, nucleotide metabolism and genome replication proteins, and Ig-like proteins, respectively; ORFs with homology to unannotated proteins or hypothetical proteins in the database are indicated in grey.
After search of the NCBI database, 11 phages with similar nucleotide sequences of LPSEYT were found with BLASTN scores of more than 200, including 4 Salmonella phages (Table 2). Comparative genomics analysis indicated that phages LPSEYT, UPF_BP2, and BP63 are closely related. Although Salmonella phages UPF_BP2 and BP63 showed more than 90% query coverage with LPSEYT, characterization of these phages has not been reported (Table 2). The query coverages between the rest of the phages and LPSEYT were less than 2%. Therefore, it was proposed that LPSEYT is a novel phage and might belong to an unknown genus.
Table 2.
Comparison of phages with BLASTN scores of more than 200 against the LPSEYT genome.
| Phage | Country | Accession | Genome Length (bp) | E Value | BLASTN Score | Query Cover | Identify | Ref. |
|---|---|---|---|---|---|---|---|---|
| Salmonella phage UPF_BP2 | Brazil | KX826077.1 | 54,894 | 0 | 9.0 × 104 | 96% | 97% | unpublished |
| Salmonella phage BP63 | Canada | KM366099.1 | 52,437 | 0 | 8.6 × 104 | 98% | 98% | unpublished |
| Erwinia phage vB_EamM-M7 | Switzerland | HQ728263.1 | 84,694 | 2 × 10−14 | 322 | 2% | 65% | [52] |
| Erwinia phage vB_EamM_Asesino | USA | KX397364.1 | 246,291 | 1 × 10−16 | 283 | 1% | 66% | unpublished |
| Cronobacter phage vB_CsaP_GAP52 | Canada | JN882286.1 | 76,631 | 3 × 10−69 | 280 | 1% | 69% | unpublished |
| Acinetobacter phage SH-Ab 15497 | China | MG674163.1 | 43,420 | 1 × 10−30 | 276 | 1% | 66% | unpublished |
| Cronobacter phage vB_CsaP_Ss1 | Ireland | KM058087.1 | 42,205 | 1 × 10−17 | 267 | 1% | 67% | [53] |
| Salmonella phage SE131 | South Korea | MG873442.1 | 89,910 | 4 × 10−55 | 233 | 1% | 67% | unpublished |
| Salmonella phage 7-11 | Canada | HM997019.1 | 89,916 | 4 × 10−54 | 230 | 1% | 67% | [54] |
| Escherichia phage vB_EcoM_PHB05 | China | MF805809.1 | 147,659 | 5 × 10−15 | 211 | <1% | 72% | [55] |
| Enterobacteria phage ECGD1 | China | KU522583.1 | 146,647 | 3 × 10−12 | 204 | <1% | 71% | unpublished |
3.5. Classification of Lytic Phage LPSEYT and a New Genus LPSEYTvirus Can Be Proposed
The established method for classifying myoviruses into a genus is 40% of proteins matching within a 75-bit score by BLASTP [23]. To further investigate the genus of phage LPSEYT, protein homology analysis was performed. In this study, 16 phages that showed more than four homologous proteins with phage LPSEYT were collected and are recorded in Table 3. The result showed that the LPSEYT shared 70 (95%) homologous proteins with phage BP63, 47 (64%) homologous proteins with phage UPF_BP2, and less than 7 homologous proteins with any other phage. The homology analysis of phages BP63 and UPF_BP2 was also performed and, similarly, they could not be classified into any known genus. These results strongly indicate that LPSEYT, BP63, and UPF_BP2 have close ties and can be distinguished from other phage genus. Therefore, LPSEYT, BP63, and UPF_BP2 belong to a new phage genus and can be proposed as LPSEYTvirus.
Table 3.
Phages with homology genes with Salmonella phage LPSEYT by BLASTP.
| Phage name | Phage Family | Genus | Genome Length (kb) | Accession | Homologous Genes with LPSEYT a | Homologous Proteins Rate | Reference |
|---|---|---|---|---|---|---|---|
| Salmonella phage BP63 | Caudovirales | unclassified | 52.437 | NC_031250 | 70 | 95.89% | unpublished |
| Salmonella phage UPF_BP2 | Caudovirales | unclassified | 54.894 | KX826077 | 47 | 64.38% | unpublished |
| Paracoccus phage vB_PmaS_IMEP1 | Siphoviridae | unclassified | 42.093 | NC_026608 | 7 | 9.59% | unpublished |
| Achromobacter phage JWX | Siphoviridae | Steinhofvirus | 49.714 | NC_028768 | 4 | 5.48% | [56] |
| Acinetobacter phage SH-Ab 15497 | unclassified | unclassified | 43.42 | MG674163 | 4 | 5.48% | unpublished |
| Burkholderia phage BcepGomr | Siphoviridae | unclassified | 52.414 | NC_009447 | 4 | 5.48% | unpublished |
| Burkholderia phage KL1 | Siphoviridae | Septimatrevirus | 42.832 | NC_018278 | 4 | 5.48% | [57] |
| Pantoea phage vB_PagS_Vid5 | Siphoviridae | Vidquintavirus | 61.437 | MG948468 | 4 | 5.48% | unpublished |
| Pseudomonas phage 73 | Siphoviridae | Septimatrevirus | 42.999 | NC_007806 | 4 | 5.48% | [58] |
| Pseudomonas phage JG054 | Siphoviridae | Nipunavirus | 57.839 | KX898400 | 4 | 5.48% | unpublished |
| Pseudomonas phage NP1 | Siphoviridae | Nipunavirus | 58.566 | NC_031058 | 4 | 5.48% | unpublished |
| Pseudomonas phage PaMx42 | Siphoviridae | Septimatrevirus | 43.225 | NC_028879 | 4 | 5.48% | [59] |
| Pseudomonas phage vB_PaeS_SCH_Ab26 | Siphoviridae | Septimatrevirus | 43.056 | HG962376 | 4 | 5.48% | [60] |
| Stenotrophomonas phage vB_SmaS-DLP_2 | Siphoviridae | Septimatrevirus | 42.593 | NC_029019 | 4 | 5.48% | [61] |
| Vibrio phage VpKK5 | Siphoviridae | unclassified | 56.637 | NC_026610 | 4 | 5.48% | [62] |
| Xanthomonas phage XAJ2 | unclassified | unclassified | 49.241 | KU197014 | 4 | 5.48% | unpublished |
a The number of homologous proteins between LPSEYT and other phages.
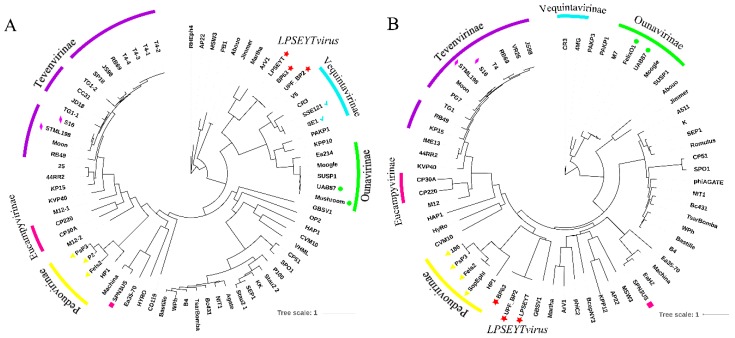
To further validate that the proposed new genus LPSEYTvirus was distinguishable and not overlapping with any established genera of Myoviridae recognized by ICTV, phylogenetic analysis was carried out based on the amino acid sequences of the terminase large subunit and major capsid proteins, which are frequently used to classify phages [24,25]. Since organization and basic features of the terminase large subunit are generally well-conserved in tailed phages, phages with the same genus would expect to have a clustering of their terminase large subunit [63]. The major capsid protein is also suitable for phylogenetic analysis because it is relatively conserved, with little or no evidence of horizontal swapping within the phage head gene regions [64]. Protein sequences of the terminase large subunit and major capsid protein were extracted from phages of the proposed new genus LPSEYTvirus, representative phages of different genera of Myoviridae family (Table S3), and all Salmonella phages of Myoviridae family recorded by ICTV (Table 4). Different subfamilies or genera were shown in the phylogenetic tree (Figure 5A,B). The phylogenetic analysis confirmed the taxonomy listed in Table 4 and Table S3, in which partial myoviruses have been grouped into six subfamilies and Salmonella phages have been classified to five phage genera. As expected, the LPSEYT, BP63, and UPF_BP2 were closely clustered, distinguishing them from other Salmonella phages and other genera of the Myoviridae (Figure 5A,B). Thus, this finding reinforces the proposal that LPSEYT, BP63, and UPF_BP2 belong to a new genus, LPSEYTvirus.
Table 4.
Overview of International Committee on Taxonomy of Viruses (ICTV) recorded genus of Myoviridae that infect Salmonella Spp.
| Family | Subfamily | Genus | Phage | Accession | Country | Genome Length (kb) |
|---|---|---|---|---|---|---|
| Myoviridae | unknown | Spn3virus | Salmonella virus SPN3US | NC_027402 | South Korea | 240.413 |
| Myoviridae | Ounavirinae | Felixo1virus | Salmonella virus FelixO1 | AF320576 | USA | 86.155 |
| Myoviridae | Ounavirinae | Felixo1virus | Salmonella virus HB2014 | unknown | unknown | unknown |
| Myoviridae | Ounavirinae | Felixo1virus | Salmonella virus Mushroom | KP143762 | USA | 87.709 |
| Myoviridae | Ounavirinae | Felixo1virus | Salmonella virus UAB87 | JN225449 | Espanya | 87.603 |
| Myoviridae | Peduovirinae | P2virus | Salmonella virus Fels2 | NC_010463 | USA | 33.693 |
| Myoviridae | Peduovirinae | P2virus | Salmonella virus PsP3 | NC_005340 | USA | 30.636 |
| Myoviridae | Peduovirinae | P2virus | Salmonella virus SopEphi | unknown | unknown | unknown |
| Myoviridae | Tevenvirinae | S16virus | Salmonella virus S16 | NC_020416 | Switzerland | 160.221 |
| Myoviridae | Tevenvirinae | S16virus | Salmonella virus STML198 | NC_027344 | USA | 158.099 |
| Myoviridae | Vequintavirinae | Se1virus | Salmonella virus SE1 | GU070616 | Portugal | 145.964 |
| Myoviridae | Vequintavirinae | Se1virus | Salmonella virus SSE121 | NC_027351 | USA | 147.745 |
Figure 5.
Phylogenetic analysis of selected phages and phages of the new proposed genus LPSEYTvirus based on (A) terminase large subunit and (B) major capsid protein. The relative distances of each main branch are shown in the figure. The arc with the same color is used to exhibit phages that are classified as the same subfamily. The triangle, rhombus, square, checkmark, and roundness represent the genera P2virus, S16virus, Spn3virus, Se1virus, and Felixo1virus, respectively. The star represents the phages of the new proposed genus LPSEYTvirus in this study.
We believe that with more isolates and genomic sequences becoming available within this newly characterized genus, we will gain a better understanding of the biological functions and properties of the proposed new genus.
4. Conclusions
In this study, we isolate and characterize a new lytic phage, LPSEYT, of the Myoviridae family. Phage LPSEYT exhibits high stability against a wide range of pH and temperatures and exhibits a potent anti-Salmonella effect that makes it a promising agent for application in food production. Genome analysis indicates that LPSEYT contains no virulence genes, making it suitable for development for safe use in the food industry. Comparative analysis of the nucleotide sequence and protein-coding content suggests that LPSEYT is a representative of a new genus of the Myoviridae, the LPSEYTvirus.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2076-2607/8/3/400/s1.
Author Contributions
For research articles with several authors, a short paragraph specifying their individual contributions must be provided. The following statements should be used “conceptualization, T.Y. and J.L.; methodology, T.Y, P.Y. and Y.Z.; software, T.Y and J.L; validation, T.Y. and J.L.; formal analysis, T.Y., L.L. and J.L.; investigation, T.Y., Y.Z and J.L.; resources, T.Y. and J.L.; data curation, T.Y., P.Y. and Y.Z.; writing—original draft preparation, Y.T.; writing—review and editing, T.Y., L.L., P.Y., Y.Z., A.M.S., Q.L., X.D., K.L., I.F.C. and J.L.; visualization, T.Y., P.Y. and J.L.; supervision, J.L.; project administration, T.Y. and J.L.; funding acquisition, J.L.”, please turn to the CRediT taxonomy for the term explanation. Authorship must be limited to those who have contributed substantially to the work reported. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31772083), Special Fund for Technology Innovation of Hubei Province (2019AHB074), National Key R&D Program of China (2017YFC1600100), the National Key Research and Development Program of China, the Fundamental Research Funds for the Central Universities (2662017JC040, 2662016QD010), China Scholarship Council (201806765004) and the National Innovation and Entrepreneurship Training Program for Undergraduates (2020284, S201910504071).
Conflicts of Interest
All authors have no conflicts of interest to declare.
References
- 1.WHO . WHO Estimates of the Global Burden of Diseases-Foodborne Disease Burden Epidemiology Reference Group 2007–2015. World Health Organization; Geneva, Switzerland: 2015. [Google Scholar]
- 2.Lynch M.F., Tauxe R.V., Hedberg C.W. The growing burden of foodborne outbreaks due to contaminated fresh produce: Risks and opportunities. Epidemiol. Infect. 2009;137:307–315. doi: 10.1017/S0950268808001969. [DOI] [PubMed] [Google Scholar]
- 3.Wang S., Duan H., Zhang W., Li J.W. Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. Fems Immunol. Med. Microbiol. 2007;51:8–13. doi: 10.1111/j.1574-695X.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- 4.Dewey-Mattia D., Manikonda K., Hall A.J., Wise M.E., Crowe S.J. Surveillance for foodborne disease outbreaks—United States, 2009–2015. MMWR Surveill. Summ. 2018;67:1–11. doi: 10.15585/mmwr.ss6710a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finstad S., O′Bryan C.A., Marcy J.A., Crandall P.G., Ricke S.C. Salmonella and broiler processing in the United States: Relationship to foodborne salmonellosis. Food Res. Int. 2012;45:789–794. doi: 10.1016/j.foodres.2011.03.057. [DOI] [Google Scholar]
- 6.Braoudaki M., Hilton A.C. Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-Resistance to antimicrobial agents. J. Clin. Microbiol. 2004;42:73–78. doi: 10.1128/JCM.42.1.73-78.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braoudaki M., Hilton A.C. Low level of cross-Resistance between triclosan and antibiotics in Escherichia coli K-12 and E. coli O55 compared to E. coli O157. FEMS Microbiol. Lett. 2004;235:305–309. doi: 10.1111/j.1574-6968.2004.tb09603.x. [DOI] [PubMed] [Google Scholar]
- 8.Ren D., Chen P., Wang Y., Wang J., Liu H., Liu H. Phenotypes and antimicrobial resistance genes in Salmonella isolated from retail chicken and pork in Changchun, China. J. Food Saf. 2017;37:1–9. doi: 10.1111/jfs.12314. [DOI] [Google Scholar]
- 9.Rakhuba D.V., Kolomiets E.I., Dey E.S., Novik G.I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010;59:145–155. doi: 10.33073/pjm-2010-023. [DOI] [PubMed] [Google Scholar]
- 10.Bruttin A., Brussow H. Human volunteers receiving Escherichia coli phage T4 orally: A safety test of phage therapy. Antimicrob. Agents Chemother. 2005;49:2874–2878. doi: 10.1128/AAC.49.7.2874-2878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meaden S., Koskella B. Exploring the risks of phage application in the environment. Front. Microbiol. 2013;4:1–8. doi: 10.3389/fmicb.2013.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarker S.A., McCallin S., Barretto C., Berger B., Pittet A.-C., Sultana S., Krause L., Huq S., Bibiloni R., Bruttin A., et al. Oral T4-Like phage cocktail application to healthy adult volunteers from Bangladesh. Virology. 2012;434:222–232. doi: 10.1016/j.virol.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Intralytix: Food Safety. [(accessed on 28 January 2020)]; Available online: http://www.intralytix.com/index.php?page=food.
- 14.Micreos: Food Safety. [(accessed on 28 January 2020)]; Available online: https://www.micreos.com/content/food-safety.aspx.
- 15.Yuan Y., Peng Q., Yang S., Zhang S., Fu Y., Wu Y., Gao M. Isolation of a novel Bacillus thuringiensis phage representing a new phage lineage and characterization of its endolysin. Viruses. 2018;10:611. doi: 10.3390/v10110611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merabishvili M., Pirnay J.-P., Vos D.D. Bacteriophage Therapy. Humana Press; New York, NY, USA: 2018. Guidelines to compose an ideal bacteriophage cocktail; pp. 99–110. [DOI] [PubMed] [Google Scholar]
- 17.Breitbart M., Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 18.ICTV Master Species List 2018a v1. [(accessed on 4 March 2019)]; Available online: https://talk.ictvonline.org/files/master-species-lists/m/msl/7992.
- 19.Ackermann H.W. Phage classification and characterization. Methods Mol. Biol. 2009;501:127–140. doi: 10.1007/978-1-60327-164-6_13. [DOI] [PubMed] [Google Scholar]
- 20.Calendar R. The Bacteriophages. Oxford University Press; New York, NY, USA: 2006. [Google Scholar]
- 21.Eriksson H., Maciejewska B., Latka A., Majkowska-Skrobek G., Hellstrand M., Melefors Ö., Wang J.-T., Kropinski A.M., Drulis-Kawa Z., Nilsson A.S. A suggested new bacteriophage genus, “Kp34likevirus”, within the Autographivirinae subfamily of Podoviridae. Viruses. 2015;7:1804–1822. doi: 10.3390/v7041804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu S., Le S., Tan Y., Zhu J., Li M., Rao X., Zou L., Li S., Wang J., Jin X., et al. Genomic and proteomic analyses of the terminally redundant genome of the Pseudomonas aeruginosa phage PaP1: Establishment of genus PaP1-Like phages. PLoS ONE. 2013;8:e62933. doi: 10.1371/journal.pone.0062933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavigne R., Darius P., Summer E.J., Seto D., Mahadevan P., Nilsson A.S., Ackermann H.W., Kropinski A.M. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 2009;9:224. doi: 10.1186/1471-2180-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopes A., Tavares P., Petit M.-A., Guérois R., Zinn-Justin S. Automated classification of tailed bacteriophages according to their neck organization. BMC Genom. 2014;15:1027. doi: 10.1186/1471-2164-15-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buttimer C., Lucid A., Neve H., Franz C.M., O’Mahony J., Turner D., Lavigne R., Coffey A. Pectobacterium atrosepticum phage vB_PatP_CB5: A member of the proposed genus ‘Phimunavirus’. Viruses. 2018;10:394. doi: 10.3390/v10080394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C., Virk S.M., Shi J., Zhou Y., Willias W.S., Morsy M.K., Abdelnabby H.E., Liu J., Wang X., Li J. Isolation, characterization, and application of bacteriophage LPSE1 against Salmonella enterica in Ready to Eat (RTE) foods. Front. Microbiol. 2018;9:1–11. doi: 10.3389/fmicb.2018.01046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Islam M., Zhou Y., Liang L., Nime I., Liu K., Yan T., Wang X., Li J. Application of a phage cocktail for control of Salmonella in foods and reducing biofilms. Viruses. 2019;11:841. doi: 10.3390/v11090841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira C., Moreirinha C., Lewicka M., Almeida P., Clemente C., Cunha Â., Delgadillo I., Romalde J.L., Nunes M.L., Almeida A. Bacteriophages with potential to inactivate Salmonella Typhimurium: Use of single phage suspensions and phage cocktails. Virus Res. 2016;220:179–192. doi: 10.1016/j.virusres.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 29.Kwiatek M., Parasion S., Mizak L., Gryko R., Bartoszcze M., Kocik J. Characterization of a bacteriophage, isolated from a cow with mastitis, that is lytic against Staphylococcus aureus strains. Arch. Virol. 2012;157:225–234. doi: 10.1007/s00705-011-1160-3. [DOI] [PubMed] [Google Scholar]
- 30.Park M., Lee J.H., Shin H., Kim M., Choi J., Kang D.H., Heu S., Ryu S. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157: H7. Appl. Enviromental Microbiol. 2012;78:58–69. doi: 10.1128/AEM.06231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams M.H. Bacteriophages. Interscience Publishers Inc.; New York, NY, USA: 1959. Assay of phage by agar layer method; pp. 450–454. [Google Scholar]
- 32.López-Cuevas O., Campo N.C., León-Félix J., González-Robles A., Chaidez C. Characterization of bacteriophages with a lytic effect on various Salmonella serotypes and Escherichia coli O157:H7. Can. J. Microbiol. 2011;57:1042–1051. doi: 10.1139/w11-099. [DOI] [PubMed] [Google Scholar]
- 33.Tomat D., Casabonne C., Aquili V., Balagué C., Quiberoni A. Evaluation of a novel cocktail of six lytic bacteriophages against Shiga toxin-producing Escherichia coli in broth, milk and meat. Food Microbiol. 2018;76:434–442. doi: 10.1016/j.fm.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Spricigo D.A., Bardina C., Cortés P., Llagostera M. Use of a bacteriophage cocktail to control Salmonella in food and the food industry. Int. J. Food Microbiol. 2013;165:169–174. doi: 10.1016/j.ijfoodmicro.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Carey-Smith G.V., Billington C., Cornelius J.A., Hudson J.A., Heinemann J.A. Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol. Lett. 2006;258:182–186. doi: 10.1111/j.1574-6968.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- 36.Bao H., Zhang P., Zhang H., Zhou Y., Zhang L., Wang R. Bio-Control of Salmonella Enteritidis in foods using bacteriophages. Viruses. 2015;7:4836–4853. doi: 10.3390/v7082847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C., Chen Q., Zhang C., Yang J., Lu Z., Lu F., Bie X. Characterization of a broad host-spectrum virulent Salmonella bacteriophage fmb-p1 and its application on duck meat. Virus Res. 2017;236:14–23. doi: 10.1016/j.virusres.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Kropinski A.M., Prangishvili D., Lavigne R. Position paper: The creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 2009;11:2775–2777. doi: 10.1111/j.1462-2920.2009.01970.x. [DOI] [PubMed] [Google Scholar]
- 39.El-Dougdoug N.K., Cucic S., Abdelhamid A.G., Brovko L., Kropinski A.M., Griffiths M.W., Anany H. Control of Salmonella Newport on cherry tomato using a cocktail of lytic bacteriophages. Int. J. Food Microbiol. 2019;293:60–71. doi: 10.1016/j.ijfoodmicro.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Lal T.M., Sano M., Ransangan J. Isolation and characterization of large marine bacteriophage (Myoviridae), VhKM4 infecting Vibrio harveyi. J. Aquat. Anim. Health. 2017;29:26–30. doi: 10.1080/08997659.2016.1249578. [DOI] [PubMed] [Google Scholar]
- 41.Chang H.C., Chen C.R., Lin J.W., Shen G.H., Chang K.M., Tseng Y.H., Weng S.F. Isolation and characterization of novel giant Stenotrophomonas maltophilia phage φSMA5. Public Health Microbiol. 2005;71:1387–1393. doi: 10.1128/AEM.71.3.1387-1393.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kęsik-Szeloch A., Drulis-Kawa Z., Weber-Dąbrowska B., Kassner J., Majkowska-Skrobek G., Augustyniak D., Łusiak-Szelachowska M., Żaczek M., Górski A., Kropinski A.M. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol. J. 2013;10:1–12. doi: 10.1186/1743-422X-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Russell N., Elrod B., Dominguez K. Stress-Induced prophage DNA replication in Salmonella enterica serovar Typhimurium. Infect. Genet. Evol. 2009;9:889–895. doi: 10.1016/j.meegid.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Berngruber T.W., Weissing F.J., Gandon S. Inhibition of superinfection and the evolution of viral latency. J. Virol. 2010;84:10200–10208. doi: 10.1128/JVI.00865-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M., Li M., Lin H., Wang J., Jin Y., Han F. Characterization of the novel T4-like Salmonella enterica bacteriophage STP4-A and its endolysin. Arch. Virol. 2016;161:377–384. doi: 10.1007/s00705-015-2647-0. [DOI] [PubMed] [Google Scholar]
- 46.Barr J.J., Auro R., Furlan M., Whiteson K.L., Erb M.L., Pogliano J., Stotland A., Wolkowicz R., Cutting A.S., Doran K.S., et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. USA. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Settle L.L. Ph.D. Thesis. the Virginia Polytechnic Institute and State University; Blacksburg, VA, USA: 2012. Characterization of the Bacteriophage Felix O1 Endolysin and Potential Application for Salmonella Bioremediation. [Google Scholar]
- 48.Ramanculov E., Young R. Genetic analysis of the T4 holin: Timing and topology. Gene. 2001;265:25–36. doi: 10.1016/S0378-1119(01)00365-1. [DOI] [PubMed] [Google Scholar]
- 49.Lee J.S., Jang H.B., Kim K.S., Kim T.H., Im I.S., Kim S.W., Lazarte J.M.S., Kim J.S., Jung T.S. Complete genomic and lysis-Cassette characterization of the novel phage, KBNP1315, which infects avian pathogenic Escherichia coli (APEC) PLoS ONE. 2015;10:e0142504. doi: 10.1371/journal.pone.0142504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Catalão M.J., Gil F., Moniz-Pereira J., Pimentel M. Functional analysis of the holin-Like proteins of mycobacteriophage Ms6. J. Bacteriol. 2011;193:2793–2803. doi: 10.1128/JB.01519-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim J.A., Shin H., Heu S., Ryu S. Exogenous lytic activity of SPN9CC endolysin against gram-negative bacteria. J. Microbiol. Biotechnol. 2014;24:803–811. doi: 10.4014/jmb.1403.03035. [DOI] [PubMed] [Google Scholar]
- 52.Born Y., Fieseler L., Marazzi J., Lurz R., Duffy B., Loessner M.J. Novel virulent and broad-host-range Erwinia amylovora bacteriophages reveal a high degree of mosaicism and a relationship to Enterobacteriaceae phages. Appl. Enviromental Microbiol. 2011;77:5945–5954. doi: 10.1128/AEM.03022-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Endersen L., Guinane C.M., Johnston C., Neve H., Coffey A., Ross R.P., McAuliffe O., O′Mahony J. Genome analysis of Cronobacter phage vB_CsaP_Ss1 reveals an endolysin with potential for biocontrol of gram-negative bacterial pathogens. J. Gen. Virol. 2014;96:463–477. doi: 10.1099/vir.0.068494-0. [DOI] [PubMed] [Google Scholar]
- 54.Kropinski A.M., Lingohr E.J., Ackermann H.W. The genome sequence of enterobacterial phage 7-11, which possesses an unusually elongated head. Arch. Virol. 2011;156:149–151. doi: 10.1007/s00705-010-0835-5. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y.B., Sun E.C., Yang L., Tong Y.G., Song J.Y., Wu B. Characterization and complete genome sequence analysis of Shiga toxin-producing Escherichia bacteriophage vB_EcoM_PHB05. Chin. Vet. Sci. 2018;48:428–437. [Google Scholar]
- 56.Dreiseikelmann B., Bunk B., Spröer C., Rohde M., Nimtz M., Wittmann J. Characterization and genome comparisons of three Achromobacter phages of the family Siphoviridae. Arch. Virol. 2017;162:2191–2201. doi: 10.1007/s00705-017-3347-8. [DOI] [PubMed] [Google Scholar]
- 57.Lynch K.H., Stothard P., Dennis J.J. Comparative analysis of two phenotypically-similar but genomically-distinct Burkholderia cenocepacia-specific bacteriophages. BMC Genom. 2012;13:223. doi: 10.1186/1471-2164-13-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwan T., Liu J., DuBow M., Gros P., Pelletier J. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J. Bacteriol. 2006;188:1184–1187. doi: 10.1128/JB.188.3.1184-1187.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sepúlveda-Robles O., Kameyama L., Guarneros G. High diversity and novel species of Pseudomonas aeruginosa bacteriophages. Appl. Environ. Microbiol. 2012;78:4510–4515. doi: 10.1128/AEM.00065-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Christiane E., Libera L., Cédric M., Yann B., Guillaume L., Nguetta S.P.A., Serge L., Arsher C., Kouassi A.K., Gilles V., et al. Investigation of a large collection of Pseudomonas aeruginosa bacteriophages collected from a single environmental source in Abidjan, Côte d’Ivoire. PLoS ONE. 2015;10:e0130548. doi: 10.1371/journal.pone.0130548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peters D.L., Lynch K.H., Stothard P., Dennis J.J. The isolation and characterization of two Stenotrophomonas maltophilia bacteriophages capable of cross-taxonomic order infectivity. BMC Genom. 2015;16:664. doi: 10.1186/s12864-015-1848-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lal T.M., Ransangan J. Complete genome sequence of VpKK5, a novel Vibrio parahaemolyticus lytic siphophage. Genome Announc. 2015;3:1–2. doi: 10.1128/genomeA.01381-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanamaru S., Kondabagil K., Rossmann M.G., Rao V.B. The functional domains of bacteriophage T4 terminase. J. Biol. Chem. 2004;279:40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- 64.Hatfull G.F., Hendrix R.W. Bacteriophages and their genomes. Curr. Opin. Virol. 2011;1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.