Abstract
Objective: Oncogenes have been shown to be drivers of non-small cell lung cancer (NSCLC), yet the tumor suppressing genes involved in lung carcinogenesis remain to be systematically investigated. This study aimed to identify tumor suppressing ubiquitin pathway genes (UPGs) that were critical to lung tumorigenesis.
Methods: The 696 UPGs were silenced by an siRNA screening in NSCLC cells; the potential tumor suppressing UPGs were analyzed, and their clinical significance was investigated.
Results: We reported that silencing of 11 UPGs resulted in enhanced proliferation of NSCLC cells, and four UPGs (UBL3, TRIM22, UBE2G2, and MARCH1) were significantly downregulated in tumor samples compared to that in normal lung tissues and their expression levels were positively associated with overall survival (OS) of NSCLC patients. Among these genes, UBL3 was the most significant one. UBL3 expression was decreased in tumor samples compared to that in paired normal lung tissues in 59/86 (68.6%) NSCLCs, was correlated with TNM stage and sex of NSCLC patients, and was significantly higher in non-smoking patients than in smoking patients. Silencing UBL3 accelerated cell proliferation and ectopic expression of UBL3 suppressed NSCLC in vitro and in vivo.
Conclusions: These results showed that UBL3 represented a tumor suppressor in NSCLC and may have potential for use in therapeutics and for the prediction of clinical outcome of patients.
Keywords: Lung cancer, ubiquitin pathway genes, UBL3, tumor suppressor, prognosis
Introduction
Lung cancer is the most common cause of cancer-related mortality, which kills 1.8 million people worldwide each year1. Lung cancer comprises small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC divides into large cell carcinoma, lung adenocarcinoma (LUAD), and lung squamous cell carcinoma (LUSC)2. Comprehensive sequencing efforts over the past decade have confirmed that lung cancer is a disease of the genome3–5. Abnormalities in oncogenes and tumor suppressors act as drivers to initiate the onset and progression of NSCLCs. However, druggable driver molecules are found in only a minority of patients; hence, efforts should be made to uncover the driver genes in the majority of lung cancers2.
The ubiquitin (Ub)-dependent modification system is one of the protein degradation systems within the cell and is involved in a variety of cellular processes. In this process, Ub is activated by a Ub-activating enzyme, E1, and transferred to a Ub-conjugating enzyme, E2, which attaches Ub to a target protein through the ε-amino group of a lysine residue with the help of a Ub-protein ligase, E3. The ubiquitinated target protein is then recognized and degraded by the 26S proteasome6. Ub-like proteins (UBLs) usually have a UBL domain and have been reported to act as post-translational modifiers. The modification of proteins by UBLs proceeds through an enzyme cascade similar to that used by Ub. As most UBLs have far fewer known substrates than are known for Ub, they require a more limited number of E2-type conjugating enzymes and E3-type ligases7. Small Ub-like modifier (SUMO)8 and neuronal precursor cell-expressed developmentally downregulated protein-8 (Nedd8)9 are two well-known UBLs and have been reported to play important roles in cancer. Both SUMO and Nedd8 are two well-studied UBLs: SUMO is involved in several cellular activities, including cell cycle control, nuclear transport and responses to viral infections, and Nedd8 functions in the regulation of E3 ubiquitin-protein ligases. These two UBLs are implicated in tumorigenesis8,9. Some oncogenic Ub-pathway genes (UPGs) have been reported in NSCLCs10–12, but the roles of tumor suppressing UPGs in this deadly disease remain to be elucidated. In this study, we conducted a systematic silencing of the E1, E2, E3, and deubiquitinases in NSCLC cells to identify tumor suppressing UPGs that were crucial for lung cancer cell proliferation.
Materials and methods
Patient samples
The study was approved by the local Research Ethics Committees of all the participating sites; all lung cancer samples were collected with informed consent. The diagnosis of lung cancer was confirmed by at least two pathologists. Tissue samples were obtained at the time of surgery and quickly frozen in liquid nitrogen. Tumor samples contained a tumor cellularity of greater than 60%, and the matched control samples contained no tumor cells.
Cell culture and siRNA library
The NSCLC cell lines A549, H1975, and H460 were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). These cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) or Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 µg/mL streptomycin (GIBCO BRL, Grand Island, NY, USA). Human siGENOME SMARTpool siRNA libraries, containing 696 UPGs (Catalog numbers G-004705-05, G-005615-05, G-005625-05, G-005635-05), were purchased from Thermo Scientific Dharmacon (Lafayette, CO, USA). The small interfering RNAs (siRNAs) were transfected into cells using DharmFECT transfection reagent (#T-2001-01), and cell viability was assessed by CellTiter-Glo Reagent (Promega, Fitchburg, WI, USA) according to the manufacturer’s instructions. Each SMARTpool experiment was performed in duplicate. The Z-score was calculated as follows: z = (x − m)/s, where x is the raw score to be standardized, m is the mean of the plate, and s is the standard deviation (SD) of the plate13. The Z-score of a gene reflects its requirement for cell proliferation, and a Z-score of ≤ −2 indicates that the gene is required for cell proliferation; silencing of the gene significantly inhibits cell proliferation. However, a Z score of ≥ 2 indicates that the gene inhibits lung cancer cell proliferation and that inhibition of the gene significantly promotes cell proliferation.
Cell cycle
To assess the cell cycle distribution, cells were harvested and washed in phosphate-buffered saline (PBS), fixed in 70% ethanol and incubated at 4 °C overnight. Cells were centrifuged and washed with PBS containing 1% FBS, followed by treatment with 1% RNase A for 15 min at 37 °C and staining with 50 µg/mL propidium iodide (Sigma-Aldrich, St. Louis, MO, USA). The fluorescence intensity was measured by flow cytometry (FACSVantage Diva, BD Biosciences, San Jose, CA, USA).
Antibodies
The antibodies used included mouse anti-β-actin (#A5441; Sigma-Aldrich; 1:5000 for Western blot), mouse anti-HA (#AE008; ABclonal, Cambridge, MA, USA; 1:2000 for Western blot), rabbit anti-ubiquitin-like 3 (UBL3) (#A4028; Abclonal; 1:1000 for Western blot and 1:100 for immunohistochemistry (IHC)), rabbit anti-Ki67 (#ab15580; Abcam, Cambridge, MA, USA; 1:400 for IHC), rabbit anti-p27 (#sc-528; Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:1000 for Western blot), rabbit anti-Cyclin D1 (#2922; Cell Signaling Technology, Beverly, MA, USA; 1:1000 for Western blot), and mouse anti-Cyclin E (#4129; Cell Signaling Technology; 1:1000 for Western blot).
The siRNA and plasmid transfection
The siRNA was purchased from GenePharma Co., Ltd. (Shanghai, China), and the sequences were as follows: UUCUCCGAACGUGUCACGUTT (siNC); GAGAGUAAUUGUUGUGUAA (#siUBL3-1); and CGGCGGAUAUGAUAAAUUU (#siUBL3-2). The HA-UBL3 vector was constructed based on the pCS2 plasmid. Cells were transfected with siRNA or plasmids using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA).
Lentivirus-mediated cell transfection and transduction
For lentiviral particle production, pCDH-UBL3 constructs were co-transfected with psPAX2 and pMD2G into HEK293T cells. The culture medium was replaced with fresh medium after 6 h, and supernatant was harvested 48 h and 72 h post-transfection. A549-luciferase cells were infected with viral particles in the presence of 8 µg/mL Polybrene®. One week after infection, cells were sorted by flow cytometry.
Western blot
Cells were lysed in RIPA buffer supplemented with protease inhibitor cocktail (Sigma-Aldrich). Proteins (20 µg) were subjected to 10%–15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), electrophoresed, and transferred to nitrocellulose membranes. After blocking with 5% nonfat milk in Tris-buffered saline, membranes were washed and incubated with the indicated primary and secondary antibodies. Immunoreactions were detected using a Luminescence Image Analyzer LAS 4000 (GE, Fairfield, CO, USA).
Animal studies
Animal studies were approved by the Institutional Review Board of the Chinese Academy of Medical Sciences Cancer Institute and Hospital, with the approval ID of NCC2019A188. The methods were performed in accordance with the approved guidelines. Six-week-old SCID-beige mice were maintained under specific pathogen-free (SPF) conditions. The mice were numbered, injected into the lateral tail veins with A549-luciferase cells (5 × 105) stably expressing pCDH-UBL3 or pCDH-control plasmid, and randomized into groups (n = 13 per group). After 45 days, tumors were monitored with the IVIS Spectrum Imaging System (Caliper Life Sciences; Hopkinton, MA, USA). For IHC, sections were fixed in formalin and embedded in paraffin, incubated with primary antibodies overnight and incubated with the indicated secondary antibodies. Immunoreactions were detected using 3,3′-diaminobenzidine (DAB, Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China) and hematoxylin.
Online data availability
The Oncomine14 cancer microarray database (www.oncomine.org), including the Hou Lung, Selamat Lung, Su Lung, StearmanLung, Landi Lung, Okayama Lung, Lee lung, Kuner Lung, and Zhu Lung datasets, with the accession codes GSE19188, GSE32867, GSE7670, GSE2514, GSE10072, GSE31210, GSE8894, GSE10245, and GSE14814, respectively, were used. The transcriptome data and clinical data of 517 patients with LUAD and 501 patients with LUSC were downloaded from the Cancer Genomics Hub (https://xenabrowser.net/datapages/). TCGA database was accessed using the accession code phs000178. The online survival analysis software containing the microarray data of patients with NSCLC15 (http://kmplot.com/analysis/index.php?p=service&cancer=lung) was used. The correlation of UBL3 expression with OS was evaluated by entering UBL3 into the database and analyzed by setting different clinical parameters, and the Kaplan-Meier survival plots and log-rank P values were obtained from the webpage. The baseline demographic characteristics of the patients are shown by Gyorffy et al.15.
Statistical analysis
All statistical analyses were conducted using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA). All experiments were repeated at least three times, and the data were presented as the mean ± SD unless otherwise noted. Differences between groups of data were evaluated for significance using Student’s t-test for unpaired data or one-way analysis of variance (ANOVA). The survival curve for each group was estimated by the Kaplan-Meier method and log-rank test. P values less than 0.05 indicated statistical significance.
Results
Identification of 11 tumor suppressing UPGs in NSCLCs
To systematically identify UPGs that are crucial for lung cancer cell proliferation, we conducted a systematic silencing of the UPGs in NSCLC cell lines. The 696 siRNAs for UPGs in the Dharmacon human siGENOME SMARTpool library were transfected into the NSCLC lines A549 and H1975. Cell viability was measured 72 h after transfection, and robust Z-scores from duplicate experiments for each SMARTpool were determined13. While genes with a Z score ≤ −2 have been analyzed in other work16, UPGs with a Z-score ≥ 2 (indicating that cell proliferation was increased when the gene was silenced) were assessed here and 11 UPGs were identified (Figure 1A and Supplementary Figure S1). These included UBL3, USP26, UBL4, UBE2G2, FBXO42, RNF10, TRIM6, LOC51136, RNF113A, MARCH1, and TRIM22.
Figure 1.
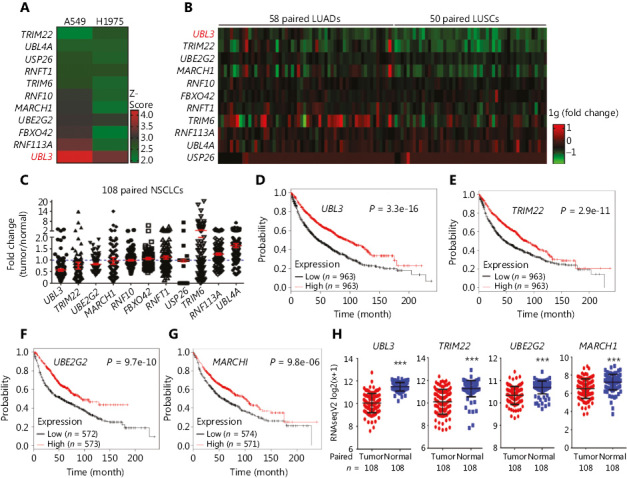
Identification of UBL3 as a potential tumor suppressor in non-small cell lung cancer (NSCLC) cells. (A) Heat map showing 11 candidates with Z-scores ≥ 2 in both A549 and H1975 cells. (B, C) Heat map and scatter plot of the expression of the 11 candidates in 58 paired lung adenocarcinoma (LUAD) tissues and 50 paired lung squamous cell carcinoma (LUSC) tissues. The fold change indicates the tumor/normal ratio. (D–G) The association between the expression levels of the four genes and the prognosis of patients as determined by online survival analysis software. (H) The expression levels of the four genes in TCGA datasets. ***P < 0.0001.
Unveiling UBL3 in NSCLCs
We analyzed the expression levels of the above UPGs in The Cancer Genome Atlas (TCGA) datasets containing 58 paired LUADs and 50 paired LUSCs (Supplementary Table S1; Figure 1B and 1C), and reported that the expressions of five genes (UBL3, TRIM22, UBE2G2, MARCH1, and RNF10) were downregulated in these NSCLCs. The association between the expression levels of these five genes and the OS of the patients was analyzed using online survival analysis software15 (http://kmplot.com/analysis/index.php?p=service&cancer=lung). We found that the expression of four genes, UBL3, TRIM22, UBE2G2, and MARCH1, was positively associated with the OS of the patients, and the median OS of patients with high levels of these genes was significantly longer than that of patients with low levels of these genes (Figure 1D–G). In TCGA datasets, the expression levels of the four genes in tumors were lower than those in normal tissues at the mRNA level (Figure 1H), and UBL3 represented the most significant one (Figure 1H). Therefore, UBL3 was selected for further investigation.
The expression of UBL3 in NSCLC
To evaluate the expression of UBL3 in NSCLC, the Oncomine Cancer Microarray Database14 (www.oncomine.org) was explored. In the Hou Lung17, Selamat Lung18, Su lung19, Stearman Lung20, Landi Lung21, and Okayama Lung22 datasets, the UBL3 mRNA levels were significantly reduced in tumor tissues compared to those in the counterpart normal lung tissues (Figure 2A–F). We investigated UBL3 RNA levels in TCGA dataset containing 1, 018 NSCLC tumor samples and 110 normal lung specimens and found significantly reduced expression of UBL3 in NSCLC tumor samples compared to that in normal lung tissue samples (Figure 2G). In both lung adenocarcinoma (LUAD, n = 517) and lung squamous cell carcinoma (LUSC, n = 501), the expression of UBL3 was significantly higher in normal tissues than in tumor lung samples (Figure 2H and 2I; P < 0.0001). In paired tumor adjacent samples from TCGA datasets, UBL3 expression in tumor tissues was lower than that in the adjacent lung samples (Figure 2J and 2K, P < 0.0001). In addition, the UBL3 copy number loss was found in 632 (62.1%) of 1,017 TCGA patients (Figure 2L).
Figure 2.
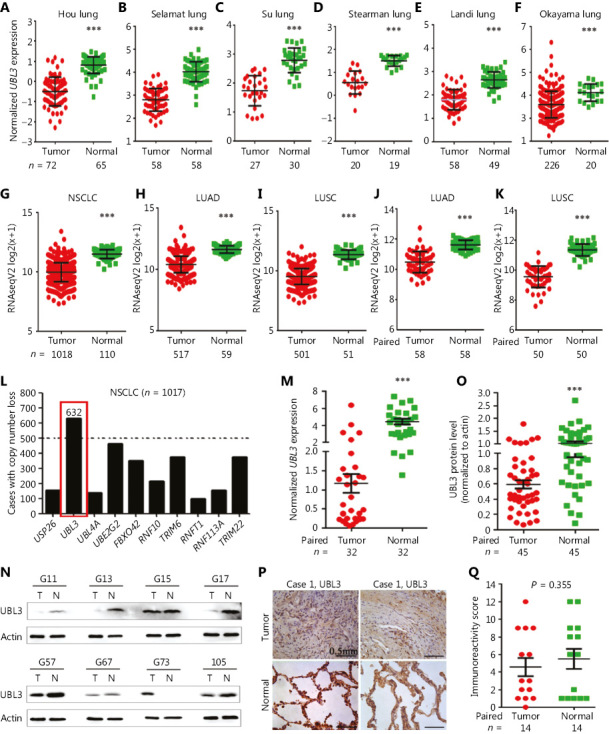
The expression of UBL3 in patients with NSCLC in the Oncomine and TCGA datasets. The expression of UBL3 in the Hou Lung (A), Selamat Lung (B), Su Lung (C), Stearman lung (D), Landi Lung (E), and Okayama lung (F) Oncomine datasets and the NSCLC (G), lung adenocarcinoma (LUAD) (H), and lung squamous cell carcinoma (LUSC) (I) of TCGA datasets is shown. (J, K) UBL3 expression levels in tumor tissues and the counterpart adjacent normal lung tissues from the TCGA LUAD (J) and LUSC (K) datasets. (L) Copy number loss analysis of 10 candidates in the TCGA database. (M–Q) The expression of UBL3 was tested by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (M), Western blot (N, O), and immunohistochemistry (P, Q) assays of lung cancer patients of our cohort. Size bar, 0.5 mm. The densitometry analysis of the Western blot results (O) and the immunoreactivity score of UBL3 were calculated (Q). P values, Student’s t-test. ***P < 0.0001.
We tested the expression of UBL3 in 86 NSCLCs (Table 1) by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Figure 2M), Western blot analysis (Figure 2N and 2O), and immunohistochemistry (IHC) (Figure 2P and 2Q), and observed a decrease in UBL3 expression in tumor tissues of 59/86 (68.6%) patients, demonstrating the suppressed expression of UBL3 in NSCLCs.
Table 1.
Summary of the baseline demographic characteristics of the 86 patients
| Characteristics | Cases, n | UBL3-low, n (%) | P |
|---|---|---|---|
| Total | 86 | 59 (68.6) | |
| Gender | |||
| Male | 48 | 36 (75) | 0.402 |
| Female | 20 | 13 (65) | |
| Not recorded | 18 | 10 (55.6) | |
| Age (years) | |||
| < 65 | 47 | 33 (70.2) | 0.691 |
| ≥ 65 | 20 | 15 (75) | |
| Not recorded | 19 | 11 (57.9) | |
| Smoking status | |||
| Smoker | 39 | 27 (69.2) | 0.547 |
| Non-smoker | 29 | 22 (75.9) | |
| Not recorded | 18 | 10 (55.6) | |
| Histology | |||
| LUAD | 39 | 29 (74.4) | 0.729 |
| LUSC | 23 | 18 (78.3) | |
| LUAD + LUSC | 4 | 0 (0) | |
| SCLC | 2 | 2 (100) | |
| Not recorded | 18 | 10 (55.6) | |
| TNM Stage | |||
| I-II | 36 | 23 (63.9) | 0.150 |
| III-IV | 30 | 24 (80) | |
| Not recorded | 20 | 12 (60) |
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma. P values were calculated using a two-sided Fisher’s exact test. “UBL3-low” indicates that the levels of UBL3 were lower in tumor tissues than in normal tissues.
UBL3 expression and prognosis of smoker patients with NSCLC
The association between the UBL3 expression levels and tobacco smoking status of the patients was explored in TCGA and Oncomine databases. In TCGA datasets, the expression of UBL3 was significantly higher in non-smoking NSCLC patients than in former and current smoking NSCLCs (Figure 3A). In the Oncomine Lee Lung dataset23, UBL3 in non-smoking patients was also higher than in smoking patients with NSCLC (Figure 3B). We analyzed the potential association between UBL3 expression and patient prognosis using the online survival analysis software. The results showed that smoking patients with NSCLC with high levels of UBL3 had a favorable prognosis, but non-smoking patients with high levels of UBL3 had much better prognosis (Figure 3C and 3D). These results suggest that the expression of UBL3 in NSCLC may be modulated by tobacco smoke and related carcinogens and thus involved in lung carcinogenesis. This possibility could be further investigated in the future.
Figure 3.
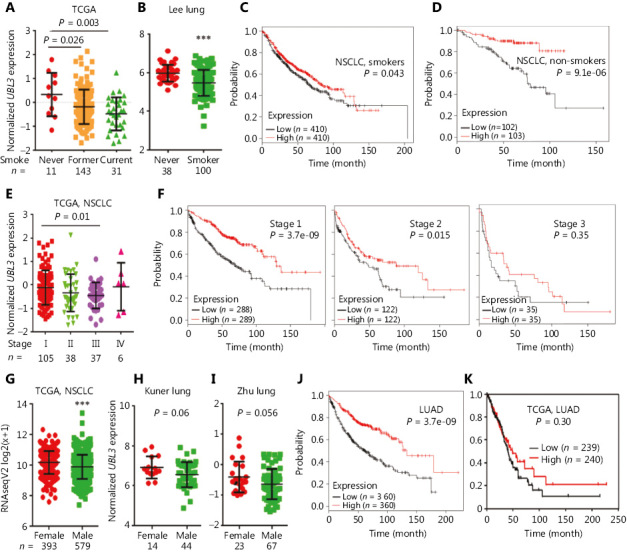
The association between UBL3 expression and clinical characteristics. The expression of UBL3 in smoking and non-smoking patients with NSCLC in TCGA dataset (A) and Oncomine Lee Lung dataset (B). UBL3 expression and prognosis of smoking (C) and non-smoking (D) patients with NSCLC as determined by online survival analysis software. (E) The expression of UBL3 in patients with NSCLC at different stages. (F) Overall survival (OS) of patients with NSCLCs at different stages with high or low expression of UBL3. (G) Expression of UBL3 in female and male patients with NSCLC in TCGA (G) and the Oncomine datasets Kuner Lung (H) and Zhu Lung (I). The Kaplan-Meier survival curve of LUAD patients with high or low levels of UBL3 expression as determined by online survival analysis software (J) and TCGA database analysis (K). ***P < 0.0001.
The association between UBL3 expression and clinical characteristics
We also explored the association between UBL3 expression levels and TNM stage in patients in TCGA database, and the data showed that the expression of UBL3 was significantly higher in stage I NSCLCs than in stage II and III NSCLCs (Figure 3E). The prognostic significance of UBL3 was further analyzed using stage-matched samples. Among stage I and stage II patients, those with higher expression of UBL3 had much longer survival time than those with lower levels of UBL3 (Figure 3F). Among patients with stage III NSCLCs (Figure 3F, right panel), those with higher UBL3 had slightly but not statistically significantly shorter OS than those with lower expression of UBL3. The expression of UBL3 was significantly higher in female NSCLC patients than in male NSCLC patients in TCGA dataset (Figure 3G). In Oncomine datasets24,25, UBL3 expression in female patients was also higher than in male patients (Figure 3H and 3I), although the difference was not statistically significant, and was possibly attributed to less smoking in women than in men. Analysis with online survival analysis software indicated that LUAD patients with lower UBL3 expression had significantly worse prognosis than those with higher UBL3 expression (Figure 3J). Similarly, in TCGA datasets, LUAD patients with lower UBL3 expression had poorer prognosis, although the difference was not statistically significant (Figure 3K). These data demonstrated that UBL3 might play a role in the pathogenesis of NSCLC.
Knockdown of UBL3 promotes lung cancer cell proliferation
We found that silencing UBL3 accelerated cell proliferation, with Z-scores of 4.22 and 3.42 in A549 and H1975 cells, respectively (Figure 1A and Supplementary Figure S1). We verified the effects of siRNA against UBL3 (siUBL3) on lung cancer cells, and found that siUBL3 treatment resulted in a reduction of UBL3 protein expression and increased viability of A549 and H1975 cells (Figure 4A). Knockdown of UBL3 led to a significant increase in cell growth (Figure 4B). In contrast, exogenous expression of UBL3 (by transient transfection) significantly inhibited the proliferation (Figure 4C) and growth (Figure 4D) of cells. We further showed that knockdown of UBL3 promoted the colony-forming activity of A549 cells (Figure 4E). Silencing UBL3 in H1975 cells led to cell cycle arrest at the S phase (Figure 4F), downregulation of p27, Cyclin D1, and upregulation of Cyclin E (Figure 4G), indicating that UBL3 played a role in the control of the replication checkpoint.
Figure 4.
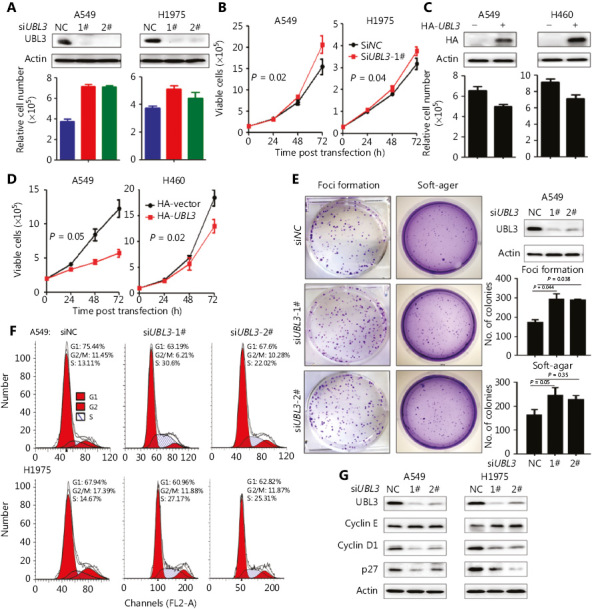
UBL3 inhibits NSCLC cell proliferation. (A) A549 and H1975 cells were transfected with siUBL3 (1# and 2#) and lysed 72-h later for the detection of UBL3 expression by Western blot assays (upper panel) and assessment of cell viability (lower panel). Error bars, SD. (B) A549 and H1975 cells were transfected with siUBL3-1, and cell proliferation was assessed by a Trypan Blue dye exclusion assays. (C, D) A549 and H460 cells were transfected with HA-tagged UBL3, and cell proliferation was assessed by a Trypan Blue exclusion assay. (E) Foci formation and soft agar assays of A549 cells transfected with siUBL3. Error bars, SD; P values, Student’s t-test. (F) A549 and H1975 cells were transfected with siUBL3 and the cell cycle distribution was assessed by propidium iodide staining of DNA followed by flow cytometric analysis. (G) A549 and H1975 cells were transfected with siUBL3, lysed 72 h later, and the lysates were subjected to Western blot assay using indicated antibodies.
UBL3 inhibits tumor growth in vivo
To investigate the role of UBL3 in tumor growth in vivo, the pCDH-UBL3 plasmid was transfected into A549-luciferase cells (Figure 5A), which were then injected into SCID-beige mice via the tail vein (n = 13 for each group). To characterize the growth of the xenograft tumors, the luciferase intensity in the mice was measured by an IVIS Spectrum system 45 days after cell inoculation. We found that UBL3 significantly decreased the tumor burden (Figure 5B) and extended the life span of the mice (Figure 5C). Western blot assays of tumor lysates indicated overexpression of UBL3 in vivo (Figure 5D). Hematoxylin-eosin (HE) and Ki67 staining of lung sections confirmed the tumor-inhibitory effect of UBL3 (Figure 5E). Cell cycle analysis showed that overexpression of UBL3 induced arrest at G1 phase, without accumulation of sub-G1 content (Figure 5F and 5G).
Figure 5.
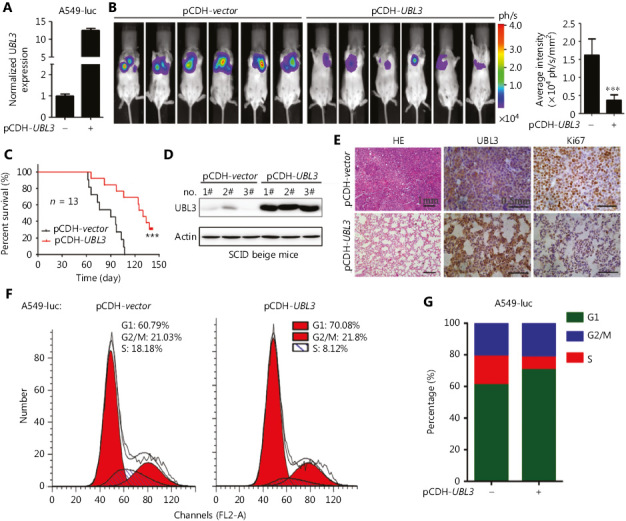
UBL3 suppresses lung cancer in vivo. (A) qRT-PCR analysis of UBL3 mRNA levels in pCDH-UBL3-expressing A549-luciferase cells. (B) A total of 5 × 105 pCDH-UBL3-expressing A549-luciferase cells were injected into SCID-beige mice and the relative luciferase intensity was determined by an IVIS Spectrum system at the indicated time point (B). Error bars, SD; P values, Student’s t-test. The Kaplan-Meier survival curve of the mice is shown (C). P value, log-rank test. Mice were sacrificed, and lung tissues were lysed for Western blot assays using the indicated antibodies (D) or subjected to hematoxylin-eosin (HE) or immunohistochemical staining (E). Scale bar, 1 mm for HE and 0.5 mm for IHC. (F, G) A549-luc cells with pCDH-vector or pCDH-UBL3 were assessed by propidium iodide staining of DNA followed by flow cytometric analysis (F), and cell cycle distribution was assessed (G).
Discussion
In this study, we carried out an siRNA screen for 696 UPGs found in the human genome, and reported 11 candidates that were able to promote cell proliferation when they were silenced in A549 and H1975 cells (Figure 1). In TCGA database, the expressions of four potential tumor suppressor genes (UBL3, TRIM22, UBE2G2, and MARCH1) were significantly downregulated in tumor samples compared to that in normal lung tissues. Using online survival analysis software15, we found that the expression levels of these four genes were positively associated with the OS of patients with NSCLC, with UBL3 as the most significant one. These results provided rationales for us to further investigate the role of UBL3 in lung carcinogenesis.
UBL3 has been identified in Drosophila melanogaster26 and is a membrane protein localized by prenylation27. UBL3 acts as a post-translational modification factor that regulates protein sorting to small extracellular vesicles (sEVs)28. In melanocytic cells, UBL3 may be regulated by MITF29. A genome-wide association study demonstrated that UBL3 harbors a susceptibility locus for biliary atresia30. Additionally, UBL3 was identified to harbor a novel susceptibility locus for BRCA1/2-negative, high risk breast cancers in Asian women31. UBL3 was upregulated in human prostate cancer cells (LNCaP cells) exposed to silvestrol32. UBL3 was identified as part of a 7-gene signature (UBL3, FGF3, BMI1, PDGFRA, PTPRF, RFC4, and NOL7) for predicting relapse and survival in early-stage patients with cervical carcinoma33. Promoter hypermethylation and downregulation were found in UBL3 in a northeast Indian population with esophageal cancer34. An in-frame fusion of MAP3K8-UBL3 was reported as a distinct genetic driver in a unique subgroup of spitzoid neoplasms35. Previous studies showed that UBL3 was abnormally expressed in BRCA1/2-negative, high risk breast cancers (BRCAX)31, cervical carcinoma33, and esophageal cancer34. However, the role of UBL3 in lung cancer remains poorly understood. Here, we found that UBL3 expression was reduced in tumor samples from both TCGA and Oncomine datasets and in 86 NSCLC samples from our laboratory (Figure 2, Table 1). UBL3 expression levels were much lower in patients with stage II or III NSCLC than in patients with stage I NSCLC (Figure 3E). LUAD patients with higher levels of UBL3 had more favorable prognosis than those patients with lower levels of UBL3 (Figure 3). We investigated the function of UBL3 in NSCLC cells and demonstrated that silencing of UBL3 promoted, whereas overexpression of UBL3 suppressed, NSCLC cell proliferation in vitro and in vivo (Figure 4 and Figure 5). These data indicated that UBL3 could be a tumor suppressor in NSCLCs. However, why UBL3 was suppressed in tumor samples and how it suppressed cell proliferation, warrant further investigation.
Tobacco is responsible for about 85% of lung cancer deaths (1.53 million) each year worldwide1. Tobacco smoke causes genomic mutations in cancers36; promotes cell proliferation; inhibits programmed cell death; facilitates angiogenesis, invasion and metastasis; and enhances tumor-promoting inflammation37,38. We found that the expression of UBL3 was significantly higher in non-smoking than in smoking patients and that non-smoking patients with NSCLC with high levels of UBL3 had much more favorable prognosis than those who smoked (Figure 3). The effects of tobacco smoke on UBL3 expression need to be investigated in the future.
Collectively, the above results suggested that UBL3 may be involved in lung carcinogenesis by altering the expression of oncoproteins/tumor suppressors, thus affecting critical pathways to promote cell proliferation and disease progression. These possibilities and whether UBL3 could be a therapeutic target for lung cancer warrant further investigation.
Supporting Information
Acknowledgments
This work was supported by the National Key Research and Development Program of China (Grant No. 2016YFC0905501), the National Natural Science Funds for Distinguished Young Scholar (Grant No. 81425025), the Key Project of the National Natural Science Foundation of China (Grant No. 81830093), the CAMS Innovation Fund for Medical Sciences (Grant No. CIFMS; 2019-I2M-1-003), and the National Natural Science Foundation of China (Grant No. 81672765 and 81802796). The study sponsor had no role in the design of the study; the data collection, analysis, or interpretation; the writing of the article; or the decision to submit for publication.
Conflict of interest statement
No potential conflicts of interest are disclosed.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;(553):446–54. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 3.George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;(524):47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Network TCGAR. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;(511):543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Network TCGAR. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;(489):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connection. An Intern Med. 2006;(145):676–84. doi: 10.7326/0003-4819-145-9-200611070-00010. [DOI] [PubMed] [Google Scholar]
- 7.Veen AGvd, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem. 2012;(81):323–57. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 8.Seeler J-S, Dejean A. Sumo and the robustness of cancer. Nat Rev Cancer. 2017;17:184. doi: 10.1038/nrc.2016.143. [DOI] [PubMed] [Google Scholar]
- 9.Zhou L, Jiang Y, Luo Q, Li L, Jia L. Neddylation: a novel modulator of the tumor microenvironment. Mol Cancer. 2019;18:77. doi: 10.1186/s12943-019-0979-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan J, Deng Q, Jiang C, Wang X, Niu T, Li H, et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene. 2014;(34):3957–67. doi: 10.1038/onc.2014.327. [DOI] [PubMed] [Google Scholar]
- 11.Snoek BC, de Wilt LH, Jansen G, Peters GJ. Role of e3 ubiquitin ligases in lung cancer. World J Clin Oncol. 2013;4:58–69. doi: 10.5306/wjco.v4.i3.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu YQ, Wang XL, Cheng X, Lu YZ, Wang GZ, Li XC, et al. Skp1 in lung cancer: clinical significance and therapeutic efficacy of its small molecule inhibitors. Oncotarget. 2015;(6):34953–67. doi: 10.18632/oncotarget.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;(24):167–75. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 14.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;(9):166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Zhao X, Zhou Y, Wang M, Zhou G. Identification of an E3 ligase-encoding gene, RFWD3, in non-small cell lung cancer. Front Med. 2019 doi: 10.1007/s11684-019-0708-6. [DOI] [PubMed] [Google Scholar]
- 17.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;(22):1197–211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA, Jr, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;(167):1763–75. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;(72):100–11. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 23.Lee ES, Son DS, Kim SH, Lee J, Jo J, Han J, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;(14):7397–404. doi: 10.1158/1078-0432.CCR-07-4937. [DOI] [PubMed] [Google Scholar]
- 24.Kuner R, Muley T, Meister M, Ruschhaupt M, Buness A, Xu EC, et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer. 2009;(63):32–8. doi: 10.1016/j.lungcan.2008.03.033. [DOI] [PubMed] [Google Scholar]
- 25.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;(28):4417–24. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chadwick BP, Kidd T, Sgouros J, Ish-Horowicz D, Frischauf AM. Cloning, mapping and expression of UBL3, a novel ubiquitin-like gene. Gene. 1999;(233):189–95. doi: 10.1016/s0378-1119(99)00138-9. [DOI] [PubMed] [Google Scholar]
- 27.Downes BP, Saracco SA, Lee SS, Crowell DN, Vierstra RD. MUBs, a family of ubiquitin-fold proteins that are plasma membrane-anchored by prenylation. J Biol Chem. 2006;(281):27145–57. doi: 10.1074/jbc.M602283200. [DOI] [PubMed] [Google Scholar]
- 28.Ageta H, Ageta-Ishihara N, Hitachi K, Karayel O, Onouchi T, Yamaguchi H, et al. UBL3 modification influences protein sorting to small extracellular vesicles. Nat Commun. 2018;9:3936. doi: 10.1038/s41467-018-06197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;(21):665–76. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Barcelo MM, Yeung MY, Miao XP, Tang CS, Cheng G, So MT, et al. Genome-wide association study identifies a susceptibility locus for biliary atresia on 10q24.2. Human Mol Genetics. 2010;(19):2917–25. doi: 10.1093/hmg/ddq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JY, Kim J, Kim SW, Park SK, Ahn SH, Lee MH, et al. BRCA1/2-negative, high-risk breast cancers (BRCAX) for Asian women: genetic susceptibility loci and their potential impacts. Sci Rep. 2018;8:15263. doi: 10.1038/s41598-018-31859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mi Q, Kim S, Hwang BY, Su BN, Chai H, Arbieva ZH, et al. Silvestrol regulates g2/m checkpoint genes independent of p53 activity. Anticancer Res. 2006;(26):3349–56. [PubMed] [Google Scholar]
- 33.Huang L, Zheng M, Zhou QM, Zhang MY, Yu YH, Yun JP, et al. Identification of a 7-gene signature that predicts relapse and survival for early stage patients with cervical carcinoma. Med Oncol. 2012;(29):2911–8. doi: 10.1007/s12032-012-0166-3. [DOI] [PubMed] [Google Scholar]
- 34.Singh V, Singh LC, Vasudevan M, Chattopadhyay I, Borthakar BB, Rai AK, et al. Esophageal cancer epigenomics and integrome analysis of genome-wide methylation and expression in high risk northeast Indian population. Omics. 2015;(19):688–99. doi: 10.1089/omi.2015.0121. [DOI] [PubMed] [Google Scholar]
- 35.Quan VL, Zhang B, Mohan LS, Shi K, Isales MC, Panah E, et al. Activating structural alterations in MAPK genes are distinct genetic drivers in a unique subgroup of spitzoid neoplasms. Am J Surg Pathol. 2019;(43):538–48. doi: 10.1097/PAS.0000000000001213. [DOI] [PubMed] [Google Scholar]
- 36.Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;(354):618–22. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobus SL, Warren GW. The biologic effects of cigarette smoke on cancer cells. Cancer. 2014;(120):3617–26. doi: 10.1002/cncr.28904. [DOI] [PubMed] [Google Scholar]
- 38.Hecht SS. Lung carcinogenesis by tobacco smoke. Int J Cancer. 2012;(131):2724–32. doi: 10.1002/ijc.27816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


