Abstract
Objectives
Decreased gut microbial gene richness (MGR) and compositional changes, is associated with adverse metabolism in overweight or moderate obesity, but lacks characterization in severe obesity. Bariatric surgery (BS) improves metabolism and inflammation in severe obesity and is associated with gut microbiota modifications. Here, we characterized severe obesity associated dysbiosis (i.e. MGR, microbiota composition and functional characteristics) and assessed whether BS would rescue these changes.
Design
Sixty-one severely obese subjects, candidates for adjustable gastric banding (AGB, n=20) or Roux-en-Y-gastric bypass (RYGB, n=41), were enrolled. Twenty-four subjects were followed at 1, 3 and 12 months post-BS. Gut microbiota and serum metabolome were analyzed using shotgun metagenomics and LC-MS. Confirmation groups were included.
Results
Low gene richness (LGC) was present in 75% of patients and correlated with increased trunk-fat mass and comorbidities (type-2 diabetes, hypertension and severity). Seventy-eight metagenomic species (MGS) were altered with LGC, among which 50% were associated with adverse body composition and metabolic phenotypes. Nine serum metabolites (including Glutarate, 3-Methoxyphenylacetic Acid, and L-Histidine) and functional modules containing protein families involved in their metabolism were strongly associated with low MGR. BS increased MGR one-year post-surgery, but most RYGB patients remained with low MGR 1-year post-BS, despite greater metabolic improvement than AGB patients.
Conclusions
We identified major gut microbiota alterations in severe obesity, which includes decreased MGR and related-functional pathways linked with metabolic deteriorations. The lack of full rescue post-BS calls for additional strategies to improve the gut microbiota ecosystem and microbiome-host interactions in severe obesity; ClincialTrials.gov number, NCT01454232.
Keywords: intestinal tract, gut microbiota, gastric surgery, severe obesity, metabolomics
Introduction
Among the complex obesity causes [1] and its related-diseases [2] (type-2 diabetes (T2D) and cardiometabolic diseases), the gut microbiota appears to be a relevant contributor and is likely a pivotal factor between changes in lifestyle and host biology ([3] for review). Gut dysbiosis was identified in overweight and moderate obesity [4,5] as evidenced by substantial modifications in gut microbiota composition (with enrichment or decrease in specific bacterial groups) and low microbial gene richness (MGR) [4,5], which are associated with metabolic alterations (insulin-resistance, low-grade inflammation, and adipocyte hypertrophy [4,5]). However, gut microbiota characteristics have been scarcely explored in extreme forms of obesity although severe (BMI>35kg/m2) and morbid obesity (BMI>40kg/m2) have progressed worldwide, reaching 2.3% and 5% in men and women, respectively. While some severely obese patients remain metabolically healthy [6], in general, reaching a BMI>35kg/m2 induces a significant rise in chronic disorders [1]. Furthermore, healthy obese individuals often develop metabolic alterations and comorbidities with time [6].
Furthermore, severe/morbid obese individuals represent the only eligible candidates for bariatric surgery (BS), a treatment which has dramatically increased worldwide [7] as it reduces cardiovascular risks and improves metabolic conditions [8]. BS represents a good model to understand the intestinal contribution to health improvements by comparing adjustable gastric banding (AGB), a procedure solely inducing caloric restriction due to gastric volume reduction (equivalent to a successful diet intervention); and Roux-en-Y-Gastric bypass (RYGB), which by contrast, drastically rearranges the digestive tract architecture and adds intestinal malabsorption to food intake reduction [9]. MGR is modulated by dietary interventions and increased by 30% after a short-term dietary restriction (with fiber enrichment) in overweight/moderately obese individuals [5]. Few studies addressed microbiota evolution using whole shotgun metagenomics (WGS) [10–12] in paired subjects followed at several time points post-BS. Particularly, MGR evolution post-BS as well as its relation with other characteristics (clinical improvements, or systemic metabolomics) have been scarcely assessed. Importantly, 50% of these beneficial associations, depend on post-BS dietary modifications [13], therefore confirming the need to compare microbiota modifications after different BS techniques, namely AGB and RYGB, where food reduction in terms of total calorie do not differ [14].
Herein, we used WGS and aimed to examine i) whether MGR worsens in severe obesity and how it relates to aggravation of comorbidities, and ii) whether different BS types could differentially correct severe obesity-related gut microbial characteristics, including changes in MGR, composition, and function.
Materials and methods
Clinical cohorts
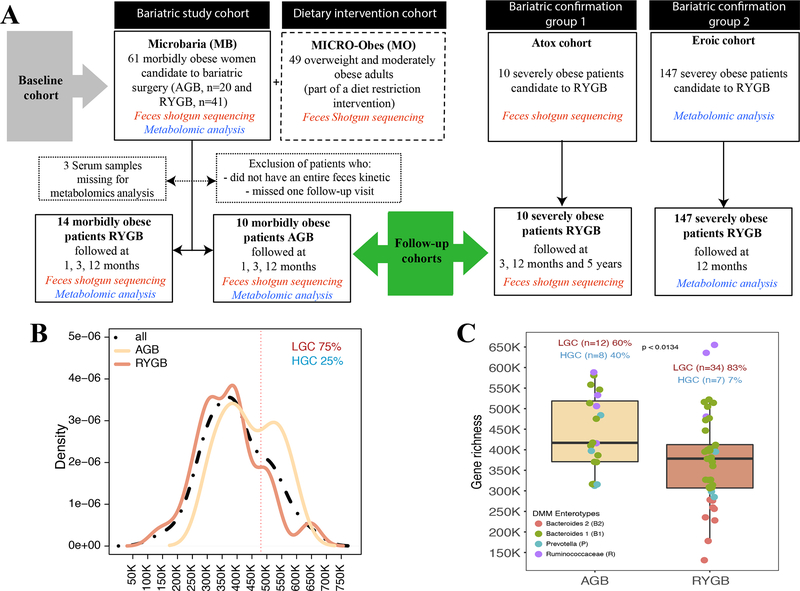
We prospectively included 61 severely obese women (Microbaria (MB) at Pitié-Salpêtrière Hospital Obesity Unit, Paris (Figure 1A), as described [15]. Patients were assigned for AGB or RYGB following international BS guidelines (i.e. BMI ≥40kg/m2, or ≥35kg/m2 with at least one severe obesity-related comorbidity) and patients’ preferences, a decision subsequently validated by a multidisciplinary panel. RYGB was frequently chosen for more severely diseased individuals.
Figure 1: Microbial gene richness (MGR) in severe obesity:
A: Study flow-chart: baseline (MB or MB+MO) and MB follow-up cohorts. Two independent confirmation cohorts (EROIC and ATOX) were used for data confirmation, B: Microbial gene Richness (MGR) bimodal distribution in MB baseline cohort. C: Baseline MGR in AGB and RYGB patients, including four enterotype characteristics in each surgery groups.
Clinical, anthropometric, and biological evaluations were obtained at baseline (T0) and during follow-up at one (T1), three (T3), and twelve months (T12) post-surgery [16]. T2D, glucose intolerant status and dyslipidemia status (definitions in supplemental material) were acknowledged. Dual-x-ray-absorptiometry (DXA) estimated body composition (Hologic Discovery W, software v12.6, 2; Bedford, MA) [16], which included total fat-free mass (FFM), total-fat mass (FM), trunk-fat mass, (all in kg or %), and gynoid-fat partitioning. Patients filled questionnaires to record general health, medications, birth mode, and the Bristol Stool Scale (BSS). Blood samples were collected after a-12 hour overnight fast at all described time points to measure biochemical parameters using routine techniques for glucose homeostasis and lipid profiles. An oral glucose tolerance test (OGTT) was performed in a subset of patients (n=21, 34%) at T0 to assess glucose and insulin AUC, and the Stumvoll index was used to characterize glucose tolerance [17]. All patients undergo the same preparation pathway that last on average 6 to 12 months, where they initially are advised to have an equilibrate diet. On average their T0 visit was performed 3 months prior to their surgery. We do not advise them to modify their diet or undergo weight loss, but rather stabilize their weight.
Feces were collected at each visit using a standardized method [18]. No patients had received antibiotic treatment for three months pre-BS, nor had any past history of acute or chronic gastrointestinal diseases. Ethical approval was obtained from the Pitié-Salpêtrière Hospital Research Ethics Committee (CPP-Ile-de-France). All subjects signed the informed written consent and the protocol was registered at clinicaltrial.gov (NCT01454232).
To examine gut microbiota across a broader range of BMI and metabolic complications, we used the previously described [5] MICRO-Obes (MO) cohort (Figure 1A) similar in age and composed mainly of women (84%), who are overweight/moderately obese patients without T2D or any medical treatment. Two confirmation and independent cohorts were added including patients with severe obesity undergoing RYGB. A group of 10 severely obese individuals (i.e. ATOX cohort) who underwent RYGB and were further followed at T0, 3, 12 months and 5 years after their surgery, for whom we sampled feces and performed WGS to analyze MGR and MGS associated signatures. This cohort was initially designed for characterization of longer-term microbiota evolution. Therefore, we had access to only 4 patients at 1 years and the whole group at 5 years. Another independent confirmation cohort of 147 severely obese individuals (i.e. EROIC cohort: 64 T2D and 83 obese non T2D patients) were followed at T0 and 12 months post-surgery and for whom we sampled blood and performed metabolomics analysis (Figure 1A).
MB, ATOX and MO gut microbiota were sequenced using the same WGS methodology and bioinformatics processing. Similar clinical phenotyping were also acquired in both cohorts.
Gut microbiota analysis by quantitative metagenomics
Participants collected fecal samples in two 20ml-tubes within 24 hours before each visit. Samples were either stored immediately at −80°C or briefly conserved in home freezers, in anaerobic conditions, before transport to the laboratory where they were immediately frozen at −80°C following guidelines [18].
Total fecal DNA from 182 MB samples was extracted, sequenced, and analyzed. DNA extraction used quenching solutions to protect DNA from degradation by DNases and a bead-beating step that ensures the lysis of particularly robust cells [19,20]. A barcoded fragment library was prepared for each sample and DNA sequencing data were generated using SOLiD 5500×l sequencers. An average of 68.72 million 35-base-long single reads (sd. 26M) was obtained for the samples. The same methodology applies also for ATOX.
Primary analysis, from reads quality cleaning to read mapping over reference 3.9 million gene reference catalog [21], was performed using METEOR Studio [5]. Secondary analyses, from gene abundance normalization to MGS projection [21] and statistical analysis, were performed using the momr R package. Supplemental material and methods describe in details the bioinformatics processing.
Serum metabolomics
Serum metabolomics were performed for 58 MB patients at baseline (Figure 1A). Serum samples were extracted using cold acetonitrile (ACN) containing labeled mix of 16 amino acids at 12.5μg/ml and processed as described in supplemental materials and methods.
LC-MS analysis was carried out on a UPLC® Waters Acquity (Waters Corp, Saint-Quentin-en-Yvelines, France) coupled to a Q-Exactive™ (Thermo Fisher Scientific, Illkirch, France). Chromatographic conditions were adapted to screen microbiota-derived metabolites as described [22]. Data were curated, normalized and annotated yielding 242 different metabolites. Details about preprocessing and processing steps are reported in supplemental materials and methods.
Statistical analyses
Statistical analyses were performed in R using public and in-house packages. Non-parametric statistics were performed when variables displayed non-normality. All tests were corrected for multiple testing using Benjamini-Hochberg. Results were considered significant at an FDR<5% (unless specified otherwise). Paired testing was performed for comparing samples across time. Graphics were built using R core and ggplot plots.
Results
Gut microbiota richness and clinical phenotypes in severe obesity
In sixty-one obese women (MB) with BMI>35, MGR exhibited a bimodal distribution [4,5]. Using the same methodology and gene cut-off (480,000) as previously [4,5] to create low gene count (LGC)/high gene count (HGC) classes, the vast majority of patients (75%) belonged to the LGC group (Figure 1B), a dramatic increase compared to overweight/moderate obesity [4,5]. At baseline, RYGB and AGB patients had similar overall clinical characteristics and BMI, except for increased DEXA-quantified trunk-fat mass and prevalence of obstructive sleep apnea in RYGB patients (Supplemental Table 1). MGR was significantly higher in AGB patients compared to RYGB patients (p=0.013) (Figure 1C).
LGC inversely correlated with metabolic alterations: triglycerides (p=0.049), uricemia (an indirect insulin-resistance marker, p=0.038) and systemic inflammation markers fibrinogen (p=0.048) and neutrophil count (p=0.042) (Supplemental Table 2) [4,5]. Beyond previous findings, MGR was identified to be inversely correlated with detrimental body composition (i.e. trunk-fat mass (r=−0.27 p=0.04)) and was significantly decreased in T2D patients (p=0.014), as compared to normoglycemic patients. MB cohort included 9 T2D patients (among the 61) and 8 of them were upon metformin treatment, the first treatment given to control glucose homeostasis, as recommended by the international recommendations for T2D therapy [23]. The only one without metformin did not need any anti-diabetic drugs to remain below the target of 6.5%. MGR decreased in patients with elevated blood pressure (p=0.05), and we observed a trend (p=0.058) toward more features of metabolic syndrome (IDF definition), in LGC patients (Supplemental Table 2). The BSS revealed that softer stools were associated with decreased MGR (p=0.005, rho=−0.42) (Supplemental Table 2). We did not observe any effect of birth mode (vaginal or C-section), PPI use, or smoking on MGR.
To gain further insight into microbiome composition in addition to MGR, we used the DMM approach of Holmes et-al [24], to characterize enterotype composition of Microbaria cohort. We herein showed the presence of the same four enterotypes described recently [25] explaining 40% of the variation in microbiome composition (R2=0.4; PERMANOVA tests) (Supplemental Figure 1), yet there was no significant association of enterotype composition and BSS at baseline (Fisher’s test p=0.97 (AGB) and 0.57 (RYGB)). Interestingly, we observed that patients with the B2 enterotype were those with the lower MGR, whereas those with the Ruminococcaceae enterotype (although few in numbers) were indeed those with the higher MGR. B2 enterotype was mostly observed in T2D patients at baseline.
Gene richness worsens with aggravated obesity
We further aimed to gain insight of MGR in a broader BMI range spanning overweight to morbid obesity by examining its distribution and bioclinical relationships. For this, we pooled clinical information from the MB and MO (overweight/moderate obesity) cohorts examined in our center [5], (Figure 1A). Compared to MO, MB patients, were younger, all women that displayed worse body composition (i.e. increased total-fat and trunk-fat mass), and more frequent cardiometabolic complications: T2D, hypertension (HTA), increased insulin-resistance (at fasting and during the OGTT), and increased CRP and IL6 (Table 1).
Table 1:
MB and MO baseline patient’s clinical characteristics.
| MB (n=61) | MO (n=49) | p | q | |||
|---|---|---|---|---|---|---|
| N | Baseline | N | Baseline | |||
| Anthropometry | ||||||
| Age, (years) | 56 | 36.9 ± 9.86 | 49 | 42.2 ± 12 | 0.028 | 0.035 |
| Sex (% female) (N=61) | 61 | 100 | 49 | 84 | 0.004 | 0.005 |
| Weight, (kg) (N=61) | 61 | 123 ± 18 | 49 | 91.5 ± 13.8 | 1,47E-17 | 1,33E-16 |
| BMI, (kg/m2) (N=61) | 61 | 45.6 ± 5.23 | 49 | 33.2 ± 3.8 | 1,44E-26 | 3,89E-25 |
| Fat free mass, (%) (N=59) | 59 | 47.7 ± 3.48 | 49 | 57.7 ± 6.3 | 8,93E-16 | 4,80E-15 |
| Fat mass, (%) (N=59) | 59 | 49.9 ± 3.73 | 49 | 39.4 ± 6.6 | 4,92E-15 | 2,22E-14 |
| Trunc fat mass (kg) (N=59) | 59 | 29 ± 4.39 | 47 | 16.9 ± 4.4 | 6,73E-24 | 9,10E-23 |
| Leptin (mg/ml) (N=58) | 58 | 78.9 ± 28.2 | 49 | 50.7 ± 22 | 6,89E-08 | 1,69E-07 |
| Basal metabolic rate (kg/24h) | 58 | 1970 ± 315 | 21 | 1540 ± 257 | 2,21E-07 | 4,90E-07 |
| Obesity related diseases | ||||||
| Hypertension (%) | 61 | 69 | 49 | 0 | 5,40E-05 | 9,60E-05 |
| Dyslipidemia (%) | 61 | 83.6 | 49 | 53 | 0.001 | 0.002 |
| Diabetes (%) | 61 | 14.7 | 49 | 0 | 0.014 | ns |
| Glucose intolerance (%) | 61 | 37.7 | 49 | 24.5 | 0.022 | ns |
| Biology | ||||||
| Total Cholesterol (mmol/l) | 61 | 4.78 ± 1.01 | 49 | 5.3 ± 0.8 | 0.003 | 0.004 |
| HDL-cholesterol (mmol/l) | 61 | 1.15 ± 0.35 | 49 | 1.4 ± 0.36 | 7,40E-05 | 0.00013 |
| LDL-cholesterol (mmol/l) | 61 | 3.08 ± 0.89 | 49 | 3.3 ± 0.76 | 0.07 | 0.07 |
| Triglycerides (mmol/l) | 61 | 1.22 ± 0.49 | 49 | 1.3 ± 0.83 | ns | ns |
| Glycemia (mmol/l) | 61 | 5.42 ± 1.31 | 49 | 5.22 ± 0.39 | ns | ns |
| Insulin (mUI/l) | 60 | 22.3 ± 15.7 | 49 | 8.9 ± 4.3 | 4,23E-12 | 1,43E-11 |
| HOMA-IR | 56 | 5.4 ± 4.57 | 56 | 1.17 ± 0.56 | 7,40E-17 | 4,99E-16 |
| Glucose AUC (mmol/l 120min) | 19 | 898 ± 199 | 40 | 799 ± 135 | 0.06 | 0.07 |
| Insulin AUC (mUI/l 120min) | 12 | 10800 ± 5230 | 39 | 4560 ± 2860 | 1,80E-05 | 3,70E-05 |
| Adiponectine (μg/ml) | 59 | 4.45 ± 1.74 | 49 | 14.1 ± 6.28 | 1,90E-14 | 7,58E-14 |
| Adipocyte volume (pl) | 59 | 874 ± 194 | 49 | 684 ± 140 | 4,30E-08 | 1,17E-07 |
| ALAT (IU/L) | 61 | 26.5 ± 16.1 | 48 | 29.5 ± 15.1 | 0.046 | 0.056 |
| ASAT (IU/L) | 61 | 25.4 ± 7.63 | 48 | 29.1 ± 17.6 | 0.45 | 0.47 |
| IL6 (pg/ml) | 58 | 4.6 ± 2.25 | 49 | 2.08 ± 2 | 1,72E-10 | 5,16E-10 |
| CRP (mg/L) | 57 | 8.73 ± 6.17 | 49 | 5.01 ± 5.67 | 2,00E-05 | 4,00E-05 |
Statistical tests performed (T, Mann-Whitney, and Chi2 tests), adapted to the variable distribution; Benjamini-Hochberg method, used for multiple testing adjustments (q).
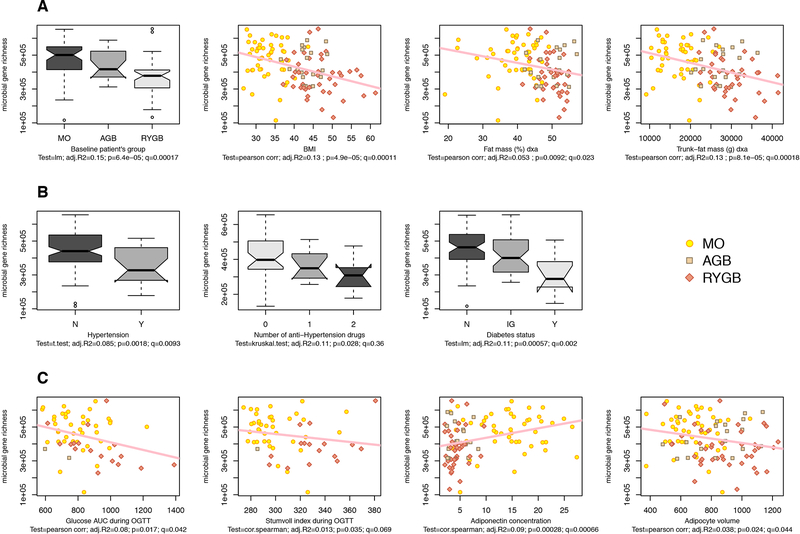
In the entire population (MB+MO, N=110 subjects, BMI [26–61 kg/m2], fat-mass [16.5–81 kg]), MGR was inversely correlated with fat-mass (p=0.0002), leptin (p=0.0072), fasting insulin (p=0.00019), HOMA-IR (p=0.00005), triglyceride levels (p=0.0024), and systemic inflammation (IL6 and CRP (p=0.019, p=0.038)). MGR decreased from MO to AGB to RYGB patients, respectively, and was significantly and inversely correlated with BMI, total-fat mass, and trunk-fat mass (Figure 2A), and positively associated with gynoid-fat distribution (p=0.037). MGR decreased with HTA and its severity (evidenced by drugs number to achieve normal blood pressure), glucose intolerance and T2D (Figure 2B). MGR negatively correlated with glucose intolerance related-parameters (OGTT glucose-AUC and OGTT-Stumvoll index) and subcutaneous adipocyte volume and positively associated with adiponectin (Figure 2C). There was no gender effect on MGR, confirming previous observations [4,5].
Figure 2: Links between MGR and bio-clinical characteristics in MO+MB subjects:
(A) MGR relationships with anthropometric parameters (B) MGR relation with metabolic comorbidities (hypertension and hypertension treatments and diabetes), N=non-diabetes, IG =glucose intolerance, D=diabetes, (C) MGR relation with OGTT-derived glucose-tolerance parameters (AUC area under the curve of glucose after OGTT with 75g glucose, and Stumvoll index), Adiponectin and adipocyte volume. Pearson correlations are performed (p=p-value, q=FDR and r2; Statistics include linear models (lm), Pearson and Spearman correlations, t-test and Kruskal-Wallis when appropriate.
Among these subjects, we considered all 786 metagenomic species (MGS), which represent co-abundant groups of genes with at least 500 genes as described [21]. The association between corpulence and the gut microbiome was also observed at the level of MGS abundance as observed with a PCoA analysis. The first two principal components described 23% of the total variance and the second component mostly associated with MGR (Supplemental Figure 2A,B), which demonstrates important ecosystem differences according to the degree of obesity and richness. From overweight to morbid obesity, the loss of MGR is linked with adverse body composition, adipocyte hypertrophy and overt metabolic complications.
Richness-linked metagenomics species associate with metabolic deteriorations in severe obesity
We examined whether some of the 786 MGS [21] were specifically associated with parameters linked to clinical phenotypes in severe obesity. Out of this list, about 29% (n=226) significantly associated with MGR (FDR<0.05). We focused on the most MGR-correlated MGS (n=78 with FDR<0.001, Figure 3A and Supplemental Table 3), of which only 18 were previously found associated with LGC [4] in less obese individuals. Whereas the vast majority of these 78 MGS associated positively with MGR (rho>0.47), three correlated negatively (rho<−0.51); 19 of them were annotated at the species level. Enrichment analysis (Fisher test), compared to the overall MGS catalogue (n=786), indicates Firmicutes (FDR<6.7e-05) as the most prevalent phylum associated with MGR and Clostridiales most prevalent at the order level (FDR<5.5e-06). Importantly, we confirmed this MGS signature of low MGR in the ATOX independent cohort composed of severely obese individuals who underwent RYGB (Figure 1A). Most of these MGS (50/78) associated with low MGR (Supplemental Figure 3). We also confirmed the significant association between low MGR and increasing BMI, and trunk-fat mass. Likewise, MGR was significantly lower in patients with comorbidities (T2D, hypertension, use of anti-hypertensive drugs (data not shown).
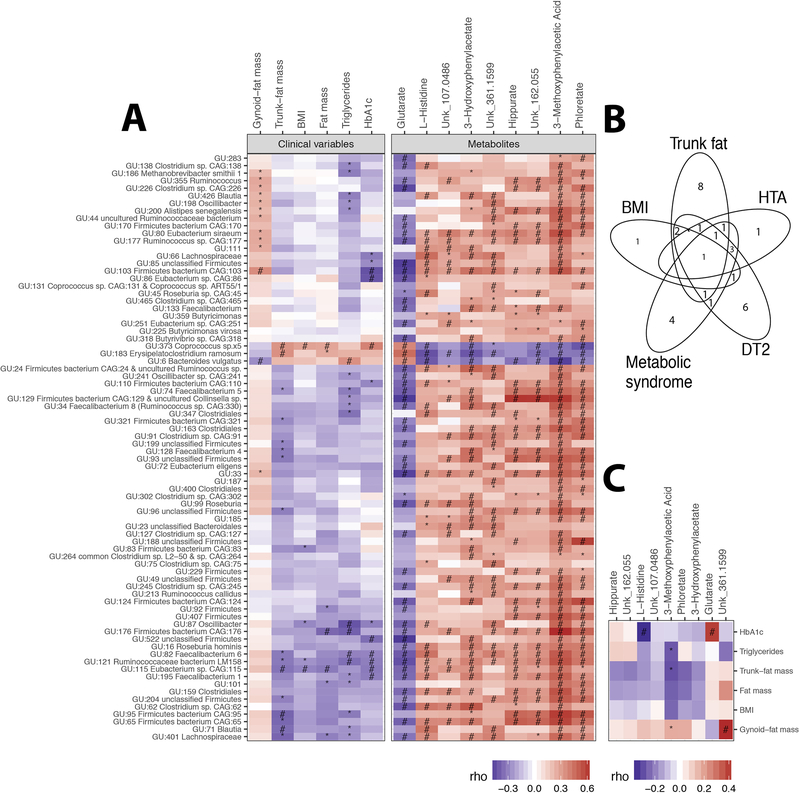
Figure 3: MGR-associated MGS at baseline.
A: Heatmap of Spearman pairwise correlation coefficients between MGR-associated-MGS abundance and metabolic variables (body composition and corpulence and metabolic traits), and MGR-associated serum metabolites B: Venn diagram of metabolic parameters associated with MGR-related MGS. C: Heatmap of Spearman pairwise correlation coefficients between metabolic phenotypes and targeted serum metabolites. P-value significance denoted by (*) and FDR significance by (#).
We found relevant associations between these 78 MGR-linked MGS and clinical variables (Supplemental Table 3): five and seven MGS were significantly associated with BMI and fat-mass, respectively (two of them resisted multiple-testing: GU:373 Coprococcus_sp5 and GU:115 Eubacterium_sp_CAG_115). 16 MGS were associated with trunk-fat distribution, including the two BMI-associated MGS, GU:373 Coprococcus_sp5 and GU:115 Eubacterium_sp_CAG_115 (Figure 3A), and a few were associated with either T2D, HTA or metabolic syndrome as shown in Figure 3B.
Looking at the overall patterns of MGS-phenotype associations, we observed relevant MGS subset positively associated with metabolic parameters and corpulence traits, such as total and trunk-fat mass, triglyceride and Hba1c (which included GU:6 Bacteroides vulgatus, GU:183 Erysipelatoclostridium ramosum and GU:373 Coprococcus_sp5, the latter was also associated with a cluster of clinical comorbidities (metabolic syndrome, HTA, T2D)) and one cluster negatively associated with BMI, total-fat and trunk-fat, triglycerides and Hba1c (GU:115 Eubacterium_sp_CAG_115, GU:121 Ruminococcaceae bacterium and GU:82 Faecalibacterium_6) (Figure 3A). Five MGS from the Firmicutes phylum were associated with three distinct metabolic alterations (T2D, metabolic syndrome and HTA), namely GU:373 Coprococcus_sp5; GU:195 Faecalibacterium 1; GU: 66 Lachnospiraceae; GU: 82 Faecalibacterium 6; GU: 86 Eubacterium sp.CAG:86; GU:163 Clostridiales) (Supplemental Table 3). Despite its smaller sample size, we observed similar trends of associations, in the confirmation cohort (ATOX group), between MGR-linked MGS and metabolic clinical variables: GU:6 Bacteroides vulgatus was positively associated with Hba1c (Supplemental Figure 4).
In severe obesity, among the most MGR-linked MGS, 50% are associated with metabolic variables, among which, 20.5% are associated with both adverse body composition and metabolic alterations.
Metabolites associated with MGR and related bacterial functions
At baseline, we identified 9 serum metabolites (4% of the measured metabolites) significantly associated with MGR (Spearman correlations, FDR<0.05) (Supplemental Table 4 and Supplemental Figure 5). One metabolite (Glutarate) correlated negatively (rho=−0.4; p<0.0017) while eight metabolites (3-Methoxyphenylacetic Acid, Phloretate, Hippurate, 3-Hydroxyphenylacetate, L-Histidine and three unknown) correlated positively with MGR (0.4<rho<0.66). These metabolites were strongly associated with MGR-linked MGS (Figure 3A). 3-Methoxyphenylacetic Acid displayed the strongest correlation, particularly with the MGS GU:6 B.vulgatus, GU:183 Erysipelatoclostridium ramosum and GU:373 Coprococcus_sp5.
We defined functional modules containing protein families from the KEGG repository involved in the above-cited metabolites metabolism. These modules were projected on targeted MGS based on functional annotations of the 3.9M gene catalog [21]. These projections showed functional linkages of target MGSs with Glutarate, Hippurate and L-Histidine. The enzyme succinate-semialdehyde dehydrogenase/glutarate-semialdehyde dehydrogenase (K00135) that oxidizes glutarate-semialdehyde to glutarate, was encoded in only 3 out of 75 positively MGR-linked MGS. GU:347 encodes two of the enzymes involved in D-lysine degradation to ketoadipate (K00128, K00832), which can be channeled to acetyl-CoA through decarboxylation to glutarate according to C-13 labelling experiments in Pseudomonas putida [26,27]. Nine MGS among the 78 MGR-linked MGS encode for the hippurate hydrolase enzyme, which is responsible for hippurate degradation to glycine and benzoate. Among these, two of the three MGS were negatively associated with MGR (GU:183 and GU:373). Under the assumption of a microbial origin of hippurate, this result, together with the positive association of hippurate serum levels with MGR, would be in line with a role of these MGS in lowering hippurate in LGC patients. The most complete functional profile was observed for L-Histidine biosynthesis, (8 target MGSs having the complete pathway) and degradation (8 target MGS having 3/4 KOs of the pathway) (Supplemental Figure 6).
Looking at associations with clinical variables, 3-Methoxyphenylacetic Acid positively correlated with gynoid-fat distribution and negatively with trunk-fat mass and triglycerides. L-Histidine was decreased in patients with HTA and/or metabolic syndrome (Supplemental Table 4). Glutarate was increased with higher HbA1c, as well as in diabetics and glucose impaired patients (Figure 3C). Overall, we show novel relationships between low MGR, related-MGS, systemic metabolite concentrations, and related-MGS metabolite pathways in severe obesity.
Partial microbial recovery post-bariatric interventions
We investigated whether MGR would change post-BS in a patient subset from the MB cohort (10 AGB and 14 RYGB) with follow-up at 1, 3 and 12 months post-BS (Figure 1A). This subset had similar clinical characteristics compared to the 61 patients at baseline (Supplemental Table 1 right) and improved MGR post-BS (Table 2). RYGB patients improved their body composition, metabolic, and inflammatory profile more so than AGB patients (Table 2) one year post-BS [28].
Table 2:
Follow-up clinical characteristics evolution after RYBG and AGB
| Gastric bypass (n=41) |
Ajustable gastric banding AGB (n=10) |
|||||||
|---|---|---|---|---|---|---|---|---|
| T 0 | T 1M | T 3M | T 12M | T 0 | T 1M | T 3M | T 12M | |
| Anthropometry | ||||||||
| Weight, (kg) | 123 ± 16.4 | 113 ± 15 (*#) | 103 ± 14.9 (*#) | 85.3 ± 17 (*#) | 113 ± 7.12 | 108 ± 9 (*) | 101 ± 8.93 (*#) | 93.7 ± 5.97 (*#) |
| BMI, (kg/m2) | 46.3 ± 6.22 | 42.4 ± 6.09 (*#) | 39.3 ± 6.21 (*#) | 32.2 ± 6.95 (*#) | 43 ± 2.22 | 40.8 ± 3.26 (*) | 38.2 ± 3.45 (*#) | 35.7 ± 2.34 (*#) |
| Fat free mass, (kg) | 58.2 ± 6.88 | 53.2 ± 6.6 (*#) | 51.4 ± 5.1 (*#) | 48.6 ± 5.3 (*#) | 54.9 ± 4.7 | 52.8 ± 4.5 | 51.2 ± 3.1 (*#) | 49.5 ± 3.4 (*#) |
| Fat mass, (kg) | 60.9 ± 11 | 56.5 ± 9.6 (*#) | 49.7 ± 10 (*#) | 34.7 ± 11.5 (*#+) | 55.7 ± 6.4 | 51.8 ± 6.9 (*) | 47.4 ± 7. (*#) | 42.4 ± 5.1 (*#+) |
| Trunc fat mass, (kg) | 30.4 ± 5.59 | 27.6 ± 5.3 (*#) | 23.9 ± 4.4 (*#) | 15.6 ± 5.9 (*#) | 26.7 ± 3.4 | 24.3 ± 3.6 (*) | 22 ± 4.2 (*#) | 18.8 ± 3.4 (*#) |
| Leptin, (mg/ml) | 81.4 ± 28.2 | 51.9 ± 28.2 (*#) | 39.8 ± 25.5 (*#) | 27.6 ± 27.2 (*#) | 74.1 ± 26.5 | 48 ± 20 (*) | 31.7 ± 9.43 (*#) | 40.3 ± 15.7 (*#) |
| Obesity related diseases | ||||||||
| Hypertension, (%) | 57,1 | 50 | 57,1 | 21,4 | 20 | 40 | 40 | 30 |
| Dyslipedmia, (%) | 100 | 92.9 (*#) | 85.7 (*#) | 57,1 | 80 | 80 (*) | 80 | 30 |
| Diabetes, (%) | 35.7 (+) | 7,14 | 7,14 | 7,14 | 0 | 0 | 0 | 0 |
| Glucose intolerance, (%) | 50 | 42,9 | 35,7 | 21,4 | 40 | 10 | 20 | 0 (*) |
| Biology | ||||||||
| Total cholesterol, (mmol/l) | 5.24 ± 1.33 | 4.45 ± 0.97 (*#) | 4.48 ± 0.77 (*#) | 4.38 ± 0.65 (*#) | 4.57 ± 0.68 | 4.6 ± 0.98 | 4.48 ± 0.85 | 4.73 ± 0.69 |
| HDL-cholesterol, (mmol/l) | 1.15 ± 0.46 | 1.01 ± 0.35 (*) | 1.08 ± 0.32 | 1.45 ± 0.35 (*#) | 1.12 ± 0.29 | 1.1 ± 0.24 | 1.1 ± 0.19 | 1.38 ± 0.3 (*#) |
| LDL-cholesterol, (mmol/l) | 3.38 ± 1.23 | 2.79 ± 0.95 | 2.85 ± 0.76 | 2.5 ± 0.63 (*#) | 2.98 ± 0.52 | 3.07 ± 0.82 | 2.97 ± 0.7 | 3 ± 0.64 |
| Triglycerides, (mmol/l) | 1.58 ± 0.58 (+) | 1.43 ± 0.4 (+§) | 1.23 ± 0.32 (*#+) | 0.946 ± 0.34 (*#) | 1.01 ± 0.43 (+) | 0.939 ± 0.42 (+§) | 0.887 ± 0.33 (+) | 0.767 ± 0.28 (*#) |
| Fasting glucose, (mmol/l) | 5.98 ± 1.83 | 5.33 ± 1.18 (*#) | 4.89 ± 0.79 (*#) | 4.64 ± 0.94 (*#) | 5.34 ± 0.6 | 4.9 ± 0.2 (*) | 4.93 ± 0.52 | 4.69 ± 0.37 (*#) |
| Insulin, (mmol/l) | 22 ± 9.34 | 12.2 ± 5.75 (*#) | 12.5 ± 5.53 (*#) | 6.95 ± 3.4 (*#+) | 27.8 ± 27 | 26.5 ± 32.6 | 13.2 ± 6.35 (*#) | 11.8 ± 3.02 (*#+) |
| HBA1c, (%) | 6.21 ± 0.88 (+) | 5.89 ± 0.71 (*#) | 5.68 ± 0.55 (*#) | 5.72 ± 0.61 (*#+) | 5.61 ± 0.41 (+) | 5.4 ± 0.4 | 5.31 ± 0.44 (*#) | 5.24 ± 0.37 (*#+) |
| HOMA-IR | 3.28 ± 1.27 | 1.82 ± 0.79 (*#) | 1.79 ± 0.77 (*#) | 0.707 ± 0.21 (*#) | 3.91 ± 3.54 | 3.53 ± 3.85 | 1.91 ± 0.85 (*#) | 0.696 ± 0.06 (*#) |
| Adiponectine, (μg/ml) | 3.89 ± 1.79 | 4.89 ± 1.72 (*#) | 5.19 ± 3.05 (*#) | 6.91 ± 2.4 (*#) | 4.14 ± 1.34 | 4.01 ± 1.54 | 4.44 ± 1.38 | 5.17 ± 1.73 (*#) |
| ALAT, (IU/L) | 31.7 ± 23.4 | 56.1 ± 26.7 (*#+§) | 47.7 ± 33.9 (*#+§) | 24.5 ± 8.45 (+) | 27.7 ± 22.7 | 19.2 ± 9.79 (+§) | 17.1 ± 6.82 (+§) | 15.9 ± 4.86 (*#+) |
| ASAT, (IU/L) | 26.4 ± 7.98 | 37.9 ± 9.76 (*#+§) | 41.4 ± 27.7 (*#+§) | 25.9 ± 4.37 | 26.3 ± 7.67 | 22 ± 4.81 (+§) | 20.5 ± 2.92 (*#+§) | 23.1 ± 4.63 |
| GGT, (IU/L) | 46.6 ± 28.3 | 52.3 ± 32.8 (+§) | 31.7 ± 23.6 (*#) | 25.1 ± 14.9 (*#) | 30.1 ± 12.8 | 21.6 ± 7.39 (+§) | 19.8 ± 7.33 (*#) | 21.1 ± 8.02 (*#) |
| IL6, (pg/ml) | 4.44 ± 1.85 | 3.75 ± 1.08 | 4.12 ± 1.52 | 2.61 ± 1.43 (*#) | 3.78 ± 2.86 | 6.36 ± 4 | 3.25 ± 1.54 | 29.6 ± 75 |
| CRP, (mg/L) | 7.51 ± 3.27 | 3.44 ± 2.57 (*#) | 4.79 ± 3.83 | 1.33 ± 1.78 (*#+) | 7.35 ± 4.57 | 4.05 ± 2.65 | 3.2 ± 2.23 (*#) | 3.33 ± 2.34 (*#+) |
Significant differences between baseline and follow-up time points for each surgery group
and
when FDR significance.
significant differences between RYGB and AGB at the same time points and
when FDR significance.
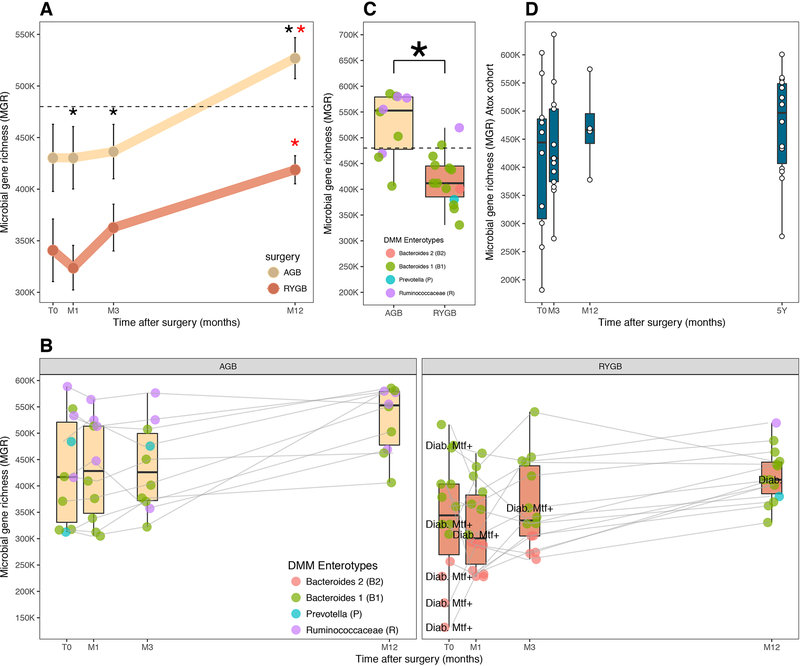
MGR increased from baseline as soon as M3, and reached statistical significance at M12 in both surgeries (Figure 4A,B). Despite greater metabolic improvement post-RYGB than post-AGB, MGR remained significantly lower in RYGB during the one-year follow-up even at M12 (Figure 4B,C). At baseline, RYGB patients had a more dysbiotic microbiome compared to AGB patients (average AGB-MGR=430322 genes, s.e.=32556; average RYGB-MGR=340743, s.e.=30347), and these differences were maintained at M12, (average AGB-MGR=526922 genes, s.e.=19899; average RYGB-MGR=418822, s.e.=13650) (Figure 4B). This points to similar patterns of richness recovery after both surgeries (at M12, average MGR display a 1.39-fold and 1.27-fold increase post-RYGB and post-AGB respectively), which was not significantly different between surgery groups (Wilcoxon p=0.86). We also observed inter-individual variability in MGR evolution with some patients remaining quite stable during the follow-up while others in either surgery group demonstrated a major increase (Figure 4B). Baseline HCG patients remained stable or increased further post-BS. Four patients switched from LGC to HGC at M12 (three of whom were from the AGB group; 30%). All T2D patients (receiving metformin at baseline) also increased their MGR post-surgery along the first year follow-up, with one patient remaining on metformin treatment until M3 thus suggesting that richness recovery post-BS is independent of metformin treatment (Figure 4B). Importantly, we observed that most patients changed Enterotype composition after RYGB, in particular most B2 patients at baseline switched to B1 enterotype (Figure 4B).
Figure 4: Microbial composition post-bariatric surgery.
A: Mean changes in MGR in RYGB and AGB from baseline to Month 1 (M1) Month 3 (M3) and Month 12 (M12) B: Evolution of richness and enterotype composition of 24 patients with kinetics follow-up at M1, M 3 (M3) and M12 C: MGR with enterotype distribution at month 12 in AGB (N=10) and RYGB (N=14) patients with kinetics follow-up, D: richness evolution confirmed in another independent RYGB (ATOX) cohort followed at 5 years (*) significance between AGB and RYGB and (*) significance between baseline and T12M
The increase in MGR post-BS was confirmed in the independent bariatric group (ATOX). MGR increased from baseline to 1 year following a similar pattern as the Microbaria patients. Most interestingly, MGR stabilized from 1 to 5 years post-RYGB suggesting that the major gut microbiota modifications seen during the first year, do not occur afterwards. Like in the Microbaria patients, despite an increasing MGR, few patients switched to high MGR at one or 5 years (Figure 4D).
At the ecosystem level, RYGB patients were more similar to each other than to AGB patients regardless of the time points (Supplemental Figure 7A). For each patient, we computed the intra-subject similarity between all-time points and unrelated samples (Supplemental Figure 7A,B). BS modified the gut microbial ecosystem and these alterations occurred along the entire follow-up as seen by the trajectories from M1 to M12, which were significantly different between AGB and RYGB (p<0.013). RYGB altered the microbiome to a higher extent than AGB (lower intra-sample similarity, p<0.001).
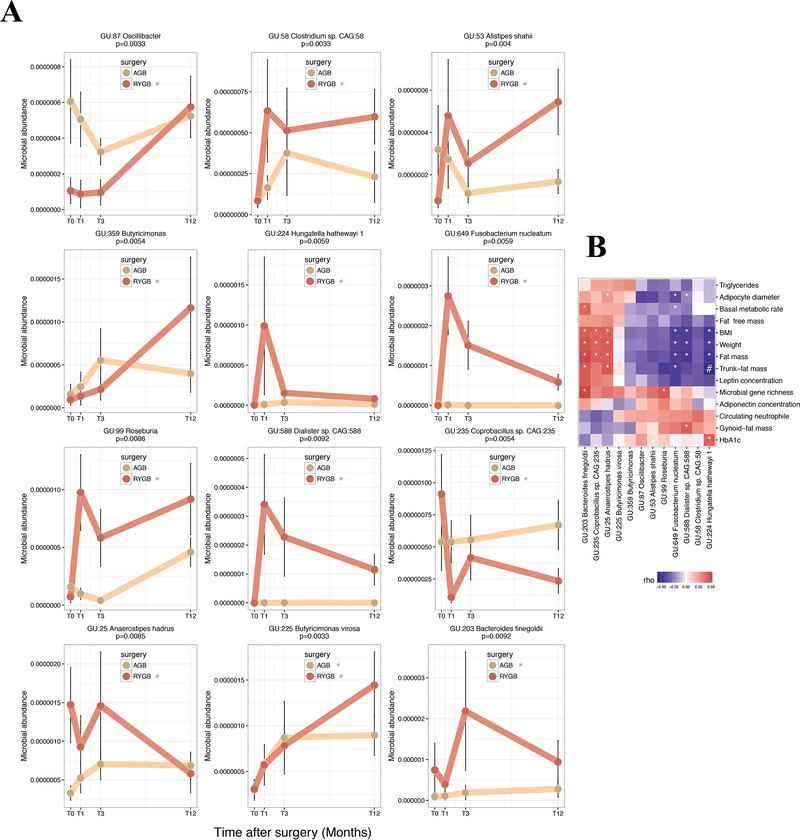
Next, we examined the specific MGS changes. Among the 786 MGS described above, 11 and 2 MGS were significantly modified 12 months after RYGB and AGB surgeries (p<0.01), respectively; one of them (GU:225 Butyricimonas virosa) was altered by both interventions (Figure 5A and Supplemental Table 5). Among the 78 MGR-linked MGS, only three increased significantly (FDR<0.05) at M12 in the whole group (GU:99 Roseburia, GU:225 Butyricimonas virosa, GU:359 Butyricimonas) (Supplemental Table 3), but none were significant when tested within each surgery sub-group. Importantly, 10 of the 11 MGS significantly modified post-RYGB in MB, followed the same pattern in the ATOX confirmation cohort (Supplemental Figure 8).
Figure 5: Significant MGS change post bariatric surgery:
A: 12 MGS significantly differ between baseline and T12M post both surgeries. Brown: RYBG (11 MGS); beige: AGB (2 MGS; 1 is common to RYGB-MGS); (*) significant p-value (in brown for RYGB and beige for AGB between T0 and T12) B: Heatmap of Spearman pairwise correlation coefficients between MGS delta and improvement in clinical outcomes. (*) significant p-value and (#) significant FDR.
Among these 12 MGS altered post-BS, we found significant relationships of their changes post-BS with the changes of clinical variables between M12 and baseline (Figure 5B and Supplemental Table 5). Half of these MGS were associated with at least two corpulence and body composition variables. Out of these species, GU:224 Hungatella hathewayi, GU:649 Fusobacterium nucleatum and GU:588 Dialister sp, displayed the strongest negative associations with the change of BMI, gynoid or trunk-fat, while GU:203 Bacteroides finegoldii, GU:25 Anaerostipes hadrus and GU:235 Coprobacillus sp. were positively related to the change of these phenotypes. The change of GU:224 Hungatella hathewayi also correlated with HbA1c levels and GU:235 Coprobacillus sp diminished with HTA resolution. Importantly, five of the baseline MGR-associated MGS changed post-BS.
Among the circulating metabolites, 30 were significantly modified post-BS at M12 with 50% decreased (mostly related to amino acid, branched-chain amino-acid, phenylalanine, and tryptophan pathway metabolites). Among these, only one (Unk_107.0486, still unknown) was significantly associated with MGR at baseline (Supplemental Table 6, Supplemental Figure 9). In our confirmation group (EROIC) (Figure 1A), we could map 18 of the 30 annotated metabolites, significantly modified in the MB cohort. All followed a similar pattern post BS in patients with or without T2D (supplemental Figure 10).
The changes of these metabolites one-year post-BS (Microbaria group) significantly associated with changes in bio-clinical parameters (mostly related to corpulence and body composition Supplemental Figure 11). We observed a cluster of metabolites (such as glycine, acetylglycine, methylmalonate) increasing post-RYGB that negatively and significantly correlated with the decrease in body corpulence and adipocyte diameter. The changes in methylmalonate and glycine were also significantly associated with the change of two BS-modified MGS (i.e. GU:203 Bacteroides finegoldii and GU:235 Coprobacillus_sp), which were also associated with body composition improvement. In the EROIC confirmation cohort, we confirmed the same trends of associations between metabolites changes and improvement of clinical variables for the 18 mapped metabolites one year post-RYGB. Acetyl glycine and glycine that increased post-BS were negatively associated with improved weight and body composition in both groups (Supplemental Figure 12).
Discussion
Here, we report in severe obesity a major decrease in microbial gene richness (MGR) associated with specific MGS-related signatures confirmed in two independent cohorts. We described related microbiome functional alterations and low MGR-related metabolomics signatures. We showed in severe obesity the presence of four enterotypes recently described in 40 individuals ranging from underweight to moderate obesity [25]. Herein, very few severely obese patients harbor a Ruminococcaceae enterotype (i.e. those who displayed the higher MGR). MGS associated with metabolic comorbidities at baseline were different from those significantly changing one-year post-BS. Strikingly, despite major weight loss and metabolic improvements post-BS, low MGR was not fully rescued. MGR significantly increased after both surgery types, but most patients remained with low MGR at one year, the time where the improvements induced post-BS reach their peak. This phenotype remained similar even after 5 years. A switch of enterotypes was observed for many patients post-BS with a reduction of B2 enterotype associated with low bacterial load [25]
Low MGR is much more prevalent (75% of patients displayed LGC) in severe obesity compared to lean or overweight/moderate obesity where LGC concerned 23% [4] to 40% [5] of subjects. Decreased MGR appears thus as a marker of disease severity as in inflammatory intestinal diseases. While confirming associations between LGC, altered metabolism, and low-grade inflammation [4,5], we show new links between MGR and trunk-fat, a detrimental fat-depot more strongly associated with cardiometabolic risks (in particular T2D status), than BMI per se [29]. The negative association between MGR and adipocyte size further illustrates the interaction with adipose tissue phenotype similar to our reported link between decreased A.muciniphila and adipocyte hypertrophy [30]. Since most T2D patients included herein (8/9) received metformin therapy at baseline, we cannot disentangle the effect of the drug from that of T2D status on low MGR. Yet, T2D patients in our study received association of anti-diabetic drugs suggesting a more severe disease compared to subjects previously explored in Forslund and Backed’s studies where confounding effect of metformin on microbiota composition was investigated [31,32]. However, in Forslund study [32], patients with or without metformin did not have any significant differences in MGR, therefore we do not believe that the observed low MGR is drug related but maybe a marker of disease severity.
Vandeputte et al reported that B2 enterotype was associated to decreased cell count, increased cell moisture and decreased gene richness [25]. Here, we did not observe any relation between Enterotypes and Bristol Stool Score linked to cell moisture, yet we observed an association between BSS and MGR as previously reported [33]. We did not assess cell count but interestingly B2 patients, being in the RYGB group, were those with the lowest MGR. Enterotypes could therefore be used as a proxy to cell count measures [25]. Noteworthy, previous data showed that patients with severe obesity display slower (rather than increased) transit time [34], which is not modified after bariatric surgery [35]. BSS may be an indirect marker of intestinal inflammation as described previously by the association between increased BSS and calprotectine an intestinal inflammatory marker [36]. It is concordant with a previous work from our lab, where we demonstrated that severe obesity was characterized by increased intestinal inflammation as seen by infiltration of immune cells in the jejunum as compared to lean individuals [37]. Finally, this is also concordant with previous results where decreased MGR was observed in patients with BSS’s scores above 3, which herein concerns most of the MB cohort [38]. We did not observe links with smoking, birth mode nor ppi as reported elsewhere, and we suggest that this may be due to the already very altered states of the microbiome in severely obese subjects.
Importantly, among 786 reference gene catalogued-MGS, one third linked with MGR (among which (n=78; 10%) were also highly associated with comorbidities, most of which seem to be a specific signature of severe obesity. Half were MGS associated with both trunk-fat mass and metabolic alterations, such as increased triglycerides and impaired glucose-tolerance. Among the MGS grouped together using hierarchical clustering, one cluster, which included B.vulgatus, was strongly and positively associated with adverse metabolism, confirming findings linking B.vulgatus and insulin-resistance [39]. In obese women treated with prebiotics, B.vulgatus decreases along with fat-mass reduction and improved glucose homeostasis [40]. Increased B.vulgatus induces adverse health outcomes such as colitis in rodents [41]. B.vulgatus is enriched in patients with active Crohn’s disease [42], and induces the production of a glycoprotein involved in mucosal defense which is triggered by inflammation [43]. Deeper analysis of B. vulgatus’s contribution in metabolism is now warranted. In the same MGS cluster, we also found GU:373 Coprococcus_sp.x5. Rodent data indicates that Coprococcus increases in fructose-fed animals that develop metabolic syndrome. Coprococcus decreases in animals after either antibiotic treatment or fecal transfer that improved their metabolic phenotype [44]. In contrast, another MGS cluster was composed of species strongly reduced with increasing trunk-fat. This cluster includes Clostridiales (Lachnospiraceae and Ruminicoccaceae), the genera Blautia, Faecalibacterium, which has anti-inflammatory properties [45], and other Firmicutes. This observation paves the way to understanding the potential interaction or imbalance between these MGS and adverse body composition and metabolism. More insight is needed into other MGS clusters with unknown annotation, to further explore these bacterial taxa associated with metabolic deterioration.
We also highlighted metabolomic interactions with MGR and its MGS-signatures. 3-Methoxyphenylacetic Acid, positively associated with MGR and negatively with trunk-fat mass and with B.vulgatus, appeared influenced by polyphenols. 3-Methoxyphenylacetic Acid production results from polyphenol and flavonoid fermentation [46]. Whether this metabolite mediate beneficial health effects via gut microbiota processing of food components needs further evaluation. Histidine is an essential amino acid, found in protein-enriched diets and serum Histidine is decreased in obese individuals [47], concordant with our result where Histidine decreased with obesity severity. It was associated with 16 MGR-related MGS contained genes encoding Histidine production/degradation pathway. We found key enzymes changes in the Histidine degradation pathway (KO1745, KO1712, KO1468, KO1479) that may lead to L-glutamate production, a precursor of GABA [12], which is linked to down-regulation of pro-inflammatory cytokines and inflammatory diseases [48]. Glutarate was also negatively associated with MGR-related MGS and positively with HbA1C [49] and could be an interesting target to further investigate for its potential link with low-grade inflammation maintenance and altered metabolism.
If previous studies demonstrated changes in gut microbiota composition post-BS, most were performed using less resolutive techniques (qPCR, 16S rDNA sequencing) in [50–53] small subject groups or at only one time point post-BS. Very few studies used whole metagenome sequencing [10–12,54] combined with large scale multi-omic approaches in paired patients followed at several time points for one year, when the nadir of weight loss and metabolic improvement occurs, nor compared the effects of the different techniques.
Using WGS, we showed that low MGR and related-functional alterations in severe obesity are only partially rescued one-year post-BS, and these changes marginally involve metagenomic and functional signature alterations seen at baseline. MGR increased only progressively and became significant one year after both surgeries, concordant with previous reports in (i) 8 RYGB patients, showing a non-significantly increased MGR at one year, [10]; (ii) increased MGR 3 months post-sleeve in a cohort similar in size to ours, using shotgun-sequencing [12], and (iii) increased richness 6 months post-RYGB using 16S-pyrosequencing [50]. Noteworthy, metformin, which represents the first line of diabetes treatment [23] and not a marker of T2D severity, was given at baseline to most of our T2D patients (8/9). Despite metformin, those patients increased MGR thus suggesting that low MGR cannot be solely attributed to metformin.
A major finding of our work is that despite the significant MGR increase, most patients remained with low MGR one-year post-BS and even more post-RYGB although the bioclinical improvements are more important to those observed post-AGB. These results question the overall contribution of gut microbiota changes in explaining weight loss and metabolism improvement. We nevertheless found bacterial species associated with improved metabolic traits at one year. In addition, evidences of gut microbiota contribution originate from feces transfer experiments. Transferring feces from either RYGB operated mice or humans into germ-free mice reproduced some clinical improvements [11,52]. Nevertheless, the effects in terms of metabolic improvement or magnitude of weight loss were always much smaller after the feces transfer than that observed by the surgery itself. Altogether, these elements probably suggest that whereas gut microbiota components may contribute to some aspects of metabolic improvements, many other mechanisms are involved. Notably, causal compositional modifications could relate to changes in hormonal secretion, bile acids availability [53], or other mechanisms [9] associated with metabolic improvements post-BS. Our study also shows that more than increased MGR, the switch of enterotypes post-RYGB could be an important feature in improved metabolic outcomes.
When looking at MGS profiles, AGB induced a few significant modifications, some of which are in the opposite direction to those seen post-RYGB. Compared with RYGB, AGB solely restricts food intake and induces weight loss with limited effects on digestive tract ecology or hormonal changes [8]. Sleeve gastrectomy [12] has been shown to significantly change gut microbiota. Post-RYGB, 9 MGS significantly increased whereas 2 MGS decreased at M12, but they were not similar from those associated with clinical variables and MGR at baseline. This suggests a specific switch of the gut microbiota induced by the surgical change of digestive physiology [9]. Among these MGS changing post-RYGB, Hungatella hathewayi (a Clostridiales member of the phylum Firmicutes) was associated with improved trunk-fat mass and HbA1c. This species has not previously been linked to metabolic diseases, but is associated with sepsis, where it is increased in the blood [55]. Alistipes shahii increased post-BS and was associated with metabolic improvements confirming previous data [50]. Two species were significantly decreased post-RYGB, including Anaerostipes hadrus, a butyrate producing species. When adding a strain of this species into a colitis-induced rodent model, it exacerbated the disease severity with associated severe dysbiosis [56]. Conversely, adding this strain in healthy mice demonstrated favorable outcomes. These observations illustrate that specific strains could be associated with adverse outcomes when present in a dysbiotic microbiota.
BS significantly modulated 30 metabolites, some of which were associated with MGS changes and clinical improvement. Glycine increased post-surgery concordant with previously observed increased glycine levels in the urine metabolome of obese patients post-BS [57]. Compared to lean individuals, glycine decreases in obesity [58]. We found that acetyl-glycine was associated with corpulence improvements and Hungatella hathewayi, which was linked to Hba1c. Acetyl-glycine has been associated with a reduced risk of T2D development [59], therefore we could postulate that increased acetyl-glycine levels post-RYGB could be linked to improved health status and the reduction of T2D incidence/T2D remission post-BS [8]. Finally, in these post-BS patients improving their insulin-resistance, we observed a reduction in branched-chain amino-acid which is similar to the known relation between increased BCAA and insulin-resistance [39].
We acknowledge that our study presents some limitations. We display numerous associations between changes in MGS, metabolites and clinical metabolic and corpulence phenotype that should be further tested in vitro or in germ free mice models. However, the strengths of our study lays in the high throughput analysis using multi-omic data in well phenotyped subjects followed at different kinetic time points post-BS. Furthermore, we had access to independent cohorts where we could replicate our findings. We also display that MGR increases at one year and then further stabilize in the longer-term at 5 years. Another limitation was the absence of available food intake data. However, food reduction in terms of total calorie, lipid or carbohydrate intake is neither different between AGB and RYGB at one and 3 months post-BS [14], nor between sleeve and RYGB at 3 and 12 months [16]. Therefore, we do not believe that the observed differences in MGR or microbiota characteristics before, but also one-year post-BS, originate from diet differences between the two surgical groups.
To conclude, severe obesity is characterized by very low MGR and specific low MGR related-MGS signature are linked with visceral adiposity, adipocyte hypertrophy, and metabolic and inflammatory consequences. We identified new metagenomic signatures, functional modifications, and serum metabolites associated with decreased MGR and demonstrate that the 4 enterotypes are also found in severe obesity. It remains unknown whether this low MGR is a cause or consequence of obesity and of its duration but it represents a good biomarker of gut microbiota alteration, eventually useful for patient stratification. Lifestyle factors such as diet and corpulence explain a large part of microbiome composition variability as compared to host genetic [60], but this needs to be deciphered in severe obesity. After bariatric surgery, enterotype modification was observed but most patients remain with very low MGR, despite digestive tract modifications and clinical major improvements. Interventions, such as fecal transfer experiment, showed increased MGR and metabolic improvements in metabolic syndrome individuals with low MGR at baseline [61]. Thus, a perspective of this work, would be to use strategies aiming at restoring gut microbiota ecosystem before or during the BS intervention and examine whether these interventions could further improve further MGR and/or clinical outcomes post-BS.
Supplementary Material
1. What is already known about this subject?
Moderate obesity is characterized by decreased microbial gene richness (MGR) (20–40% of the patients) associated with altered metabolic risk and a shift in MGS signature. This has not been explored in severe obesity. Some studies in limited number of subjects showed changes in gut microbiota but none explored precisely MGR and combined related metagenomics and metabolomics signatures after 6 months follow-up.
2. What are the new findings?
This is the first comprehensive study performed in severe obesity demonstrating a very high prevalence of patients (75%) with decreased MGR, which associates with overt metabolic complications. We describe novel metabolomic and MGS signatures that are specific to decreased MGR found in severe obesity. Bariatric surgery (both AGB and RYGB) improves MGR, but it is partially restored in most patients, and most remain with low MGR despite major metabolic improvement and weight loss in all patients. Clusters of metabolites (such as glycine, acetylglycine, methylmalonate) increasing post-RYGB, were linked with improved body composition. Importantly, even longer periods post-RYGB (i.e. 5-years) do not further increase MGR.
3. How might it impact on clinical practice in the foreseeable future?
Our results question whether specific interventions (specialized diets, pre/probiotics, or gut microbiota transfers) may be useful to consider prior or post-bariatric surgery, in severely obese individuals, in order to further improve MGR and metabolic health post-bariatric interventions.
Acknowledgements
The authors wish to thank Ms. Valentine Lemoine for patient recruitment and Dr. Florence Marchelli, who contributed to clinical and biological data collections in patients and data base constitution. The authors thank Profs Jean-Michel Oppert and Christine Poitou for contribution to patient recruitment and the paramedic staff from the Nutrition department, Pitié-Salpêtrière hospital. The investigation was performed at Clinical Center of Human Nutrition (Paris-Ile de France). The authors thank Timothy Swartz for English language review of the manuscript.
Financial support
This project is supported by the « Programme Hospitalier de Recherche Clinique » (PHRC Microbaria AOM10285/ P100111 to KC). JAW received a grant from Institut Appert, from Nestlé research and from Aviesan alliance nationale pour les sciences de la vie et de la santé ITMO santé publique. Partners have received funding from the European Union’s Seventh Framework Program (FP7) for research, technological development and demonstration under grant agreement HEALTH-F4-2012-305312 (Metacardis) and from the French “Investissement d’Avenir” FORCE and the Metagenopolis grant ANR-11-DPBS-0001. Clinical investigation is performed at the Human Nutrition Research Center (CRNH Ile de France), Pitié-Salpêtrière Hospital.
Abbreviations
- AGB
adjustable gastric banding
- RYGB
Roux-en-Y gastric bypass
- MGS
metagenomic species
- MGR
microbial gene richness
- WGS
whole metagenome sequencing
- LGC
low gene count
- HGC
high gene count
- BSS
Bristol stool scale
Footnotes
Conflict of interest: none to declare
Shotgun Metagenomics Profiling: list accession number of repository for EBI. PRJEB23292
Writing Assistance: none
References
- 1.Kopelman PG. Obesity as a medical problem. Nature 2000;404:635–43. doi: 10.1038/35007508 [DOI] [PubMed] [Google Scholar]
- 2.Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008;57:1470–81. doi: 10.2337/db07-1403 [DOI] [PubMed] [Google Scholar]
- 3.Dao MC, Clément K. Gut microbiota and obesity: Concepts relevant to clinical care. Eur J Intern Med Published Online First: 27 October 2017. doi: 10.1016/j.ejim.2017.10.005 [DOI] [PubMed] [Google Scholar]
- 4.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013;500:541–6. doi: 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 5.Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature 2013;500:585–8. doi: 10.1038/nature12480 [DOI] [PubMed] [Google Scholar]
- 6.Blüher M Are metabolically healthy obese individuals really healthy? Eur J Endocrinol Eur Fed Endocr Soc 2014;171:R209–219. doi: 10.1530/EJE-14-0540 [DOI] [PubMed] [Google Scholar]
- 7.Angrisani L, Santonicola A, Iovino P, et al. Bariatric Surgery and Endoluminal Procedures: IFSO Worldwide Survey 2014. Obes Surg 2017;27:2279–89. doi: 10.1007/s11695-017-2666-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sjöström L, Peltonen M, Jacobson P, et al. Bariatric surgery and long-term cardiovascular events. JAMA J Am Med Assoc 2012;307:56–65. doi: 10.1001/jama.2011.1914 [DOI] [PubMed] [Google Scholar]
- 9.Aron-Wisnewsky J, Doré J, Clement K. The importance of the gut microbiota after bariatric surgery. Nat Rev Gastroenterol Hepatol 2012;9:590–8. doi: 10.1038/nrgastro.2012.161 [DOI] [PubMed] [Google Scholar]
- 10.Palleja A, Kashani A, Allin KH, et al. Roux-en-Y gastric bypass surgery of morbidly obese patients induces swift and persistent changes of the individual gut microbiota. Genome Med 2016;8:67. doi: 10.1186/s13073-016-0312-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tremaroli V, Karlsson F, Werling M, et al. Roux-en-Y Gastric Bypass and Vertical Banded Gastroplasty Induce Long-Term Changes on the Human Gut Microbiome Contributing to Fat Mass Regulation. Cell Metab 2015;22:228–38. doi: 10.1016/j.cmet.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu R, Hong J, Xu X, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med 2017;23:859–68. doi: 10.1038/nm.4358 [DOI] [PubMed] [Google Scholar]
- 13.Furet J-P, Kong L-C, Tap J, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes 2010;59:3049–57. doi: 10.2337/db10-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aron-Wisnewsky J, Verger EO, Bounaix C, et al. Nutritional and Protein Deficiencies in the Short Term following Both Gastric Bypass and Gastric Banding. PloS One 2016;11:e0149588. doi: 10.1371/journal.pone.0149588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kayser BD, Lhomme M, Dao MC, et al. Serum lipidomics reveals early differential effects of gastric bypass compared to banding on phospholipids and sphingolipids independent of differences in weight loss. Int J Obes 2005. Published Online First: 10 March 2017. doi: 10.1038/ijo.2017.63 [DOI] [PubMed] [Google Scholar]
- 16.Verger EO, Aron-Wisnewsky J, Dao MC, et al. Micronutrient and Protein Deficiencies After Gastric Bypass and Sleeve Gastrectomy: a 1-year Follow-up. Obes Surg Published Online First: 24 July 2015. doi: 10.1007/s11695-015-1803-7 [DOI] [PubMed] [Google Scholar]
- 17.Stumvoll M, Mitrakou A, Pimenta W, et al. Use of the oral glucose tolerance test to assess insulin release and insulin sensitivity. Diabetes Care 2000;23:295–301. [DOI] [PubMed] [Google Scholar]
- 18.Thomas V, Clark J, Doré J. Fecal microbiota analysis: an overview of sample collection methods and sequencing strategies. Future Microbiol Published Online First: 8 September 2015. doi: 10.2217/fmb.15.87 [DOI] [PubMed] [Google Scholar]
- 19.Suau A, Bonnet R, Sutren M, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 1999;65:4799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godon JJ, Zumstein E, Dabert P, et al. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol 1997;63:2802–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen HB, Almeida M, Juncker AS, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 2014;32:822–8. doi: 10.1038/nbt.2939 [DOI] [PubMed] [Google Scholar]
- 22.Boudah S, Olivier M-F, Aros-Calt S, et al. Annotation of the human serum metabolome by coupling three liquid chromatography methods to high-resolution mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2014;966:34–47. doi: 10.1016/j.jchromb.2014.04.025 [DOI] [PubMed] [Google Scholar]
- 23.Inzucchi SE, Bergenstal RM, Buse JB, et al. Management of hyperglycaemia in type 2 diabetes, 2015: a patient-centred approach. Update to a position statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diabetologia 2015;58:429–42. doi: 10.1007/s00125-014-3460-0 [DOI] [PubMed] [Google Scholar]
- 24.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PloS One 2012;7:e30126. doi: 10.1371/journal.pone.0030126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandeputte D, Kathagen G, D’hoe K, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 2017;551:507–11. doi: 10.1038/nature24460 [DOI] [PubMed] [Google Scholar]
- 26.Revelles O, Espinosa-Urgel M, Fuhrer T, et al. Multiple and interconnected pathways for L-lysine catabolism in Pseudomonas putida KT2440. J Bacteriol 2005;187:7500–10. doi: 10.1128/JB.187.21.7500-7510.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perfetti R, Campbell RJ, Titus J, et al. Catabolism of pipecolate to glutamate in Pseudomonas putida. J Biol Chem 1972;247:4089–95. [PubMed] [Google Scholar]
- 28.Puzziferri N, Roshek TB, Mayo HG, et al. Long-term follow-up after bariatric surgery: a systematic review. JAMA 2014;312:934–42. doi: 10.1001/jama.2014.10706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahima RS, Lazar MA. Physiology. The health risk of obesity--better metrics imperative. Science 2013;341:856–8. doi: 10.1126/science.1241244 [DOI] [PubMed] [Google Scholar]
- 30.Dao MC, Everard A, Aron-Wisnewsky J, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut Published Online First: 22 June 2015. doi: 10.1136/gutjnl-2014-308778 [DOI] [PubMed] [Google Scholar]
- 31.Wu H, Esteve E, Tremaroli V, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med 2017;23:850–8. doi: 10.1038/nm.4345 [DOI] [PubMed] [Google Scholar]
- 32.Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015;528:262–6. doi: 10.1038/nature15766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falony G, Joossens M, Vieira-Silva S, et al. Population-level analysis of gut microbiome variation. Science 2016;352:560–4. doi: 10.1126/science.aad3503 [DOI] [PubMed] [Google Scholar]
- 34.Nguyen NQ, Debreceni TL, Burgess JE, et al. Impact of gastric emptying and small intestinal transit on blood glucose, intestinal hormones, glucose absorption in the morbidly obese. Int J Obes 2005. Published Online First: 30 January 2018. doi: 10.1038/s41366-018-0012-6 [DOI] [PubMed] [Google Scholar]
- 35.Carswell KA, Vincent RP, Belgaumkar AP, et al. The effect of bariatric surgery on intestinal absorption and transit time. Obes Surg 2014;24:796–805. doi: 10.1007/s11695-013-1166-x [DOI] [PubMed] [Google Scholar]
- 36.Adam B, Koldehoff M, Ditschkowski M, et al. Endoscopic and Histological Findings Are Predicted by Fecal Calprotectin in Acute Intestinal Graft-Versus-Host-Disease. Dig Dis Sci 2016;61:2019–26. doi: 10.1007/s10620-016-4112-7 [DOI] [PubMed] [Google Scholar]
- 37.Monteiro-Sepulveda M, Touch S, Mendes-Sá C, et al. Jejunal T Cell Inflammation in Human Obesity Correlates with Decreased Enterocyte Insulin Signaling. Cell Metab 2015;22:113–24. doi: 10.1016/j.cmet.2015.05.020 [DOI] [PubMed] [Google Scholar]
- 38.Vandeputte D, Falony G, Vieira-Silva S, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2016;65:57–62. doi: 10.1136/gutjnl-2015-309618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedersen HK, Gudmundsdottir V, Nielsen HB, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016;535:376–81. doi: 10.1038/nature18646 [DOI] [PubMed] [Google Scholar]
- 40.Dewulf EM, Cani PD, Claus SP, et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013;62:1112–21. doi: 10.1136/gutjnl-2012-303304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rath HC, Wilson KH, Sartor RB. Differential induction of colitis and gastritis in HLA-B27 transgenic rats selectively colonized with Bacteroides vulgatus or Escherichia coli. Infect Immun 1999;67:2969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dicksved J, Halfvarson J, Rosenquist M, et al. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J 2008;2:716–27. doi: 10.1038/ismej.2008.37 [DOI] [PubMed] [Google Scholar]
- 43.Gersemann M, Becker S, Nuding S, et al. Olfactomedin-4 is a glycoprotein secreted into mucus in active IBD. J Crohns Colitis 2012;6:425–34. doi: 10.1016/j.crohns.2011.09.013 [DOI] [PubMed] [Google Scholar]
- 44.Di Luccia B, Crescenzo R, Mazzoli A, et al. Rescue of Fructose-Induced Metabolic Syndrome by Antibiotics or Faecal Transplantation in a Rat Model of Obesity. PloS One 2015;10:e0134893. doi: 10.1371/journal.pone.0134893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731–6. doi: 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao K, Xu A, Krul C, et al. Of the major phenolic acids formed during human microbial fermentation of tea, citrus, and soy flavonoid supplements, only 3,4-dihydroxyphenylacetic acid has antiproliferative activity. J Nutr 2006;136:52–7. [DOI] [PubMed] [Google Scholar]
- 47.Gralka E, Luchinat C, Tenori L, et al. Metabolomic fingerprint of severe obesity is dynamically affected by bariatric surgery in a procedure-dependent manner. Am J Clin Nutr 2015;102:1313–22. doi: 10.3945/ajcn.115.110536 [DOI] [PubMed] [Google Scholar]
- 48.Aggarwal S, Ahuja V, Paul J. Attenuated GABAergic Signaling in Intestinal Epithelium Contributes to Pathogenesis of Ulcerative Colitis. Dig Dis Sci Published Online First: 30 June 2017. doi: 10.1007/s10620-017-4662-3 [DOI] [PubMed] [Google Scholar]
- 49.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 2014;5:e00889. doi: 10.1128/mBio.00889-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kong L-C, Tap J, Aron-Wisnewsky J, et al. Gut microbiota after gastric bypass in human obesity: increased richness and associations of bacterial genera with adipose tissue genes. Am J Clin Nutr 2013;98:16–24. doi: 10.3945/ajcn.113.058743 [DOI] [PubMed] [Google Scholar]
- 51.Zhang H, DiBaise JK, Zuccolo A, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A 2009;106:2365–70. doi: 10.1073/pnas.0812600106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liou AP, Paziuk M, Luevano J-M, et al. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med 2013;5:178ra41. doi: 10.1126/scitranslmed.3005687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryan KK, Tremaroli V, Clemmensen C, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 2014;509:183–8. doi: 10.1038/nature13135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graessler J, Qin Y, Zhong H, et al. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J 2013;13:514–22. doi: 10.1038/tpj.2012.43 [DOI] [PubMed] [Google Scholar]
- 55.Randazzo A, Kornreich A, Lissoir B. A Clostridium hathewayi isolate in blood culture of a patient with an acute appendicitis. Anaerobe 2015;35:44–7. doi: 10.1016/j.anaerobe.2015.07.003 [DOI] [PubMed] [Google Scholar]
- 56.Zhang Q, Wu Y, Wang J, et al. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci Rep 2016;6:27572. doi: 10.1038/srep27572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Narath SH, Mautner SI, Svehlikova E, et al. An Untargeted Metabolomics Approach to Characterize Short-Term and Long-Term Metabolic Changes after Bariatric Surgery. PloS One 2016;11:e0161425. doi: 10.1371/journal.pone.0161425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 2009;9:311–26. doi: 10.1016/j.cmet.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Floegel A, Stefan N, Yu Z, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 2013;62:639–48. doi: 10.2337/db12-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018;555:210–5. doi: 10.1038/nature25973 [DOI] [PubMed] [Google Scholar]
- 61.Kootte RS, Levin E, Salojärvi J, et al. Improvement of Insulin Sensitivity after Lean Donor Feces in Metabolic Syndrome Is Driven by Baseline Intestinal Microbiota Composition. Cell Metab 2017;26:611–619.e6. doi: 10.1016/j.cmet.2017.09.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.