Abstract
Plant growth-promoting rhizobacterial strain S58 was isolated from the tobacco rhizosphere. It showed strong antagonism against a battery of plant pathogenic fungi and bacteria, and controlled wheat sharp eyespot and tobacco wildfire diseases efficiently. Further tests showed that strain S58 solubilized organic phosphate and produced siderophore, protease, ammonia, and indole-3-acetic acid. In Arabidopsis thaliana, it promoted plant growth and changed root system architecture by restricting the growth of primary roots and increasing lateral root numbers. We relied on morphological, biochemical, physiological characteristics, and molecular phylogenic analysis to identify strain S58 as Pseudomonas mediterranea. The complete genome of strain S58 has a single circular chromosome of 6,150,838 bp with a 61.06% G+C content. The bacterial genome contained 5,312 predicted genes with an average length of 992.90 bp. A genome analysis suggested that P. mediterranea S58 was a rich cyclic lipopeptide (CLP)-producing strain that possessed seven non-ribosomal peptide gene clusters for CLP synthesis. Leaf inoculation of the bacterial culture and supernatants triggered cell death-like immunity in tobacco. Quantitative real-time PCR assays showed that the strain S58 induced the expression of pattern-triggered immunity and cell death marker genes, but not jasmonic acid marker genes. The results suggested that P. mediterranea S58 is a novel, versatile plant growth-promoting agent with multiple beneficial traits for plants.
Keywords: Pseudomonas mediterranea, biological control, cyclic lipopeptide, cell death
1. Introduction
Beneficial microbes have received a great deal of attention as a sustainable alternative for improving plant health. Pseudomonas is a well-known plant growth-promoting rhizobacterial (PGPR) genus for its multifarious plant beneficial functions. The mechanisms of a class of Pseudomonas species, such as P. fluorescens, P. protegens, P. chlororaphis, and P. putida, have been studied intensively and used widely in agricultural applications [1,2]. The PGPR Pseudomonas strains exert positive effects on plants in many ways, such as direct antagonism against pathogens, induction of plant resistance and immunity, alteration of plant growth morphology, toleration of environmental stress, and utilization of minerals [3,4]. Cyclic lipopeptides (CLPs) and polyketides that are synthesized by non-ribosomal peptide synthetases (NRPS) and by polyketide synthases (PKS), respectively, are major antimicrobial secondary metabolites produced by PGPR Pseudomonas [5,6,7]. Pseudomonas isolates, such as P. kilonensis F113, P. protegens Pf-5, and P. fluorescens SBW25 and 2P24 produced 2,4-diacetylphloroglucinol (DAPG), phenazines, pyoluteorin, pyrrolnitrin, viscosin, orfamide A, and amphisin to inhibit a broad range of plant pathogens directly [8,9,10,11]. In addition to secondary metabolite-mediated, antagonistic effects on pathogens, the biological control activity of Pseudomonas strains is also linked to the induction of induced systemic resistance (ISR) and siderophore-mediated competition for iron [12]. P. simiae WCS417 and other strains colonized plant root systems and induced ISR to prime host immunity, which provided enhanced protection against a broad spectrum of plant pathogens [12]. Some other Pseudomonas bacteria promoted host plant growth by solubilizing phosphorus, fixing atmospheric nitrogen, and synthesizing phytohormones [13,14,15,16]. Therefore, the identification of PGPR and biological control agents (BCAs) from different matrices, mainly from the plant rhizosphere, is a significant research direction for improving plant health and stress resistance.
P. mediterranea, along with its closely related species P. corrugata, are recognized as two causal agents isolated from tomato pith necrosis [17,18]. Actually, P. mediterranea and P. corrugata are rather ubiquitous and richly present in the plant rhizosphere, and they are characterized as having a high metabolic versatility to promote plant health [17]. Many strains of the two species, especially P. corrugata, have been used widely as biological control agents in the management of fungal and bacterial diseases of plants. P. corrugata 2140, which is isolated from the wheat rhizosphere, reduced wheat take-all disease, and the Pythium root rot in sugarbeet [19,20]. The P. corrugata CFBP 5454 inhibited the growth of plant pathogenic fungi and bacteria by CLP production, which is controlled by a LuxR transcriptional regulator and the quorum sensing system [21,22]. In comparison, P. mediterranea has been used rarely as PGPR, but some strains such as CFBP 5447T have been well-studied to efficiently synthesize medium-chain-length polyhydroxyalkanoate elastomers (mcl-PHA) and extracellular products [23].
In this study, we reported a new isolate of P. mediterranea from the tobacco rhizosphere. The strain S58 exhibited versatile plant growth-promoting traits, such as direct antagonism against a class of plant pathogens, solubilization of organic phosphate, production of siderophores, protease, ammonia, and indole-3-acetic acid, and the induction of plant immunity. Genome sequencing and comparative genomic analysis revealed that it possessed an arsenal of secondary metabolites. The data we obtained will increase the knowledge of the plant growth-promoting potential in P. mediterranea, and they will allow for the further study of adaptation mechanisms of P. mediterranea S58.
2. Materials and Methods
2.1. Bacterial Isolation and Identification
Soil samples were collected from the tobacco rhizosphere in Kunming, China. Two grams of soil sample was suspended in 18 mL sterile distilled water and spread on Lysogeny broth (LB) media for bacterial isolation. The individual colonies with different colors or morphology were selected for further purification and confirmation. The bacterial cells were stained with 1% (w/v) phosphotungstic acid first and then visualized with a transmission electron microscope (Hitachi-H600, Japan). The physiological and biochemical characteristics were determined by GEN III MicroPlates™ (Biolog, Hayward, CA, USA), according to the manufacturer’s protocol. The utilization pattern was monitored on an OmniLog® Incubator/Reader (Biolog, Hayward, CA, USA). This assay was repeated twice. Oligonucleotide primers 27F (5′-GAGAGTTTGATCCTGGCTCAG-3′) and 1494R (5′-CTACGGCTACCTTGTTACGA-3′) were used for 16S rDNA amplification, as described previously [24]. The PCR product was sequenced at GENEWIZ Ltd., China. 16S rDNA, gyrB, rpoB, and rpoD gene sequences were extracted from the PseudoMLSA Database (http://microbiologia.uib.es/bioinformatica/) and used for a phylogenetic analysis [25]. The accession numbers of gyrB, rpoB, and rpoD in strain S58 are HG325842, HG325844, HG325843, respectively. MEGA X was used for multiple alignments, and the neighbor-joining and p-distance methods were used to construct phylogenetic trees [26].
2.2. Antagonistic Test
Eight plant pathogens that included fungi and bacteria (Figure 1) were chosen to test the antagonistic capacity of strain S58. First, fresh fungal disks were inoculated onto the Potato Dextrose Agar (PDA) plate center, and 5 µL of saturated strain S58 culture (ca. OD600 =1.8) was dotted along the two sides of the fungal disks at a distance of 30 mm. The inhibitory zones were measured after incubating at 25 °C for 3–4 d. For antibacterial tests, 5 mL of saturated pathogenic bacteria were mixed with 45 mL LB media that were pre-melted and incubated at 50 °C and poured onto plates. Five µL of saturated strain S58 culture was dropped onto the plate center. The inhibitory zones were measured after incubating at 28 °C for 2–3 d. All of the tests were performed in triplicate.
Figure 1.
In vitro antimicrobial activity of strain S58 against different phytopathogens. (a) Four plant fungal pathogens (upper) and four plant bacterial pathogens were chosen to test the antimicrobial activity of strain S58; (b) Antimicrobial activity was estimated by measuring the diameter (mm) of the clear zone of growth inhibition. Results are expressed as the mean and standard deviation.
2.3. Disease Control Assay
For fungal disease, wheat (cv. Mingxian 169) seeds were disinfected with 5% sodium hypochlorite for 5 min and soaked in LB culture of strain S58 at 5 × 108 CFU/mL with 0.02% Silwet L-77 for 30 min. Fresh LB medium was used as mock. Ten seeds each were planted in 14 mL Falcon™ tubes with sterilized soil. A 15 mm-diameter fungal disk of Rhizoctonia solani on PDA was placed on the top of the soil. The mock covered with fungal disks was referred to as disease control, and the mock that was covered with fresh PDA disks was referred to as the healthy control. It was maintained at room temperature for 7 d, and then they were dug out for disease measurements.
For bacterial disease, four- week-old Nicotiana benthamiana was used for tobacco wildfire assays. A fresh lawn of strain S58 was scraped from overnight plates with a sterile loop and suspended in 10 mM MgCl2. The suspension was sprayed on the leaves of N. benthamiana at 5 × 108 CFU/mL with 0.02% Silwet L-77 and kept in the chamber for 24 h. A buffer of 10 mM MgCl2 was used as mock. A fresh pathogen suspension of Pseudomonas syringae pv. tabaci was then sprayed at 107 CFU/mL with 0.02% Silwet L-77 on the leaves that were pre-inoculated with strain S58. The mock sprayed with the pathogen suspension was referred to as disease control, and the mock that was sprayed with 10 mM MgCl2 again was referred to as the healthy control. Inoculated plants were maintained at 70% humidity at 22°C with 16 h of light and 8 h of darkness. The symptoms were recorded at seven days post-inoculation. The assays were repeated twice.
2.4. Identification of Plant Growth-Promoting Traits
Strain S58 was characterized by typical plant growth-promoting traits qualitatively. Phosphate solubilization tests were performed [27], and the synthesis of siderophores was analyzed [28]. Enzymatic activity of protease, amylase, and cellulose was measured by the clearing zone technique [29]. Production of ammonia and indole-3-acetic acid (IAA) was detected as usual [30]. For the IAA assay, 100 μg/mL IAA was used as positive control.
Arabidopsis thaliana Col-0 was used to test the plant growth-promoting capacity. First, the seeds were surface-sterilized and then sown on Murashige and Skoog (MS) agar plates for stratification for 2 d at 4 °C [31]. The seed plates were moved into a plant growth chamber for vertical incubation at 21 °C with 16/8 h photoperiod. The bacterial suspension (250 µL, 108 cfu/mL) was applied to the agar 5 cm below the seedlings. The same volume of MgSO4 (10 mM) was applied as a mock treatment. Primer root length and lateral root numbers were determined after 10 d. All of the tests were repeated three times.
2.5. Genome Sequencing and Comparative Genomic Analysis
Purified genomic DNA was used for library construction and sequencing by GENEWIZ Ltd., China. The complete genome was sequenced using an Illumina Hiseq and PacBio RSII platform, and then it was assembled using HGAP software. The coding genes were annotated using the National Center for Biotechnology Information (NCBI) nr database by Diamond. The functional annotation of genes was based mainly on protein sequence alignment using the databases NR, swiss-prot, Pfam, EggNOG, GO, and KEGG. The comparative analysis of the chromosomes between strain S58 and related Pseudomonas species was performed using GenomeMatcher software (http://www.ige.tohoku.ac.jp/joho/gmProject/gmhomeJP.html) and the bl2seq program [32]. The ANI calculator (www.ezbiocloud.net/tools/ani) was used to calculate the average nucleotide identity (ANI) [33]. A Ring Image Generator (BRIG) was used for genome comparison [34]. Secondary metabolite gene clusters were predicted using antiSMASH 5.0, a web-based analysis platform (http://antismash.secondarymetabolites.org/) [35]. The Bacterial Pan-Genome Analysis (BPGA) pipeline was used for the pan-genome analyses [36]. The cut-off value was set up to 50% to obtain the pan and core genomes. Venn diagrams were drawn using OrthoVenn2, which is a web server for the comparison and analysis of whole-genome orthologous clusters [37].
2.6. Assay of Cell Death
To test cell death elicitation, the bacterial suspension of strain S58 was infiltrated into the leaves of four-wk-old N. benthamiana, Nicotiana tabacum, and tomato (Lycopersicon esculentum cv. Moneymaker) at 107 CFU/mL, 5 × 107 CFU/mL, 108 CFU/mL, 5 × 108 CFU/mL, respectively. Inoculated plants were maintained at 70% humidity at 22°C with 16/8 h photoperiod for visualized cell death. To test the ability of the supernatant of strain S58 to trigger cell death, 100 mL saturated LB culture of strain S58 was centrifuged at 8000 rpm for 20 min at 4 °C. The supernatant was collected after being filtered through a 0.2 µm membrane. The filtrates were lyophilized and then extracted with methanol two times. Finally, it was dissolved in 2 mL methanol at 50 times concentration. Leaves of N. benthamiana were inoculated with this extract. To determine the thresh-hold concentration of cell death, the 50 times extract was diluted with methanol to 10 times and five times concentration. Leaves were photographed at 2 d post-inoculation. Each experiment was repeated twice.
2.7. Quantitative Reverse Transcription PCR
Fresh bacterial cells of strain S58 were suspended in 10 mM MgCl2 and infiltrated into the leaves of N. benthamiana at 108 CFU/mL. After 12 h, total RNA of the inoculation area was isolated using the Tri-Reagent (Omega Bio-tek, USA) and treated with RNase-free DNase (Tiangen, China). The RNA integrity was determined by 1.2% agarose gel electrophoresis. Purified RNA (2 μg) was used to prepare cDNA using M-MLV reverse transcriptase (Tiangen, China) with random primers, according to the manufacturer’s instructions. qRT-PCR was performed in 96-well plates on an ABI QuantStudio6 Flex real-time PCR system (Applied Biosystems, USA). The reaction mix was performed using 5 μL of FastStart Universal SYBR Green Master (New England Biolabs, USA), 2 μL of 2 μM primer mix (final concentration at 0.4 μM), 2 μL of a diluted 1:10 cDNA, and water to complete a final volume of 10 μL. Cycling conditions were 95 °C for 10 min, and 40 cycles at 95 °C for 15 s, and 60 °C for 1 min. Primer sequences are shown in Table S1. The housekeeping gene EF1a was used as a reference. Fold changes in expression were calculated using the delta-delta Ct (ddCt) method as usual. Three biological and three technical replicates of the experiments were performed.
2.8. Statistical analysis
All of the experiments were performed in triplicate, and the mean and SD are shown. Tukey’s HSD test (p < 0.05) was used for statistical analysis of the results.
3. Results
3.1. Antagonism Against Phytopathogens
We screened a bench of bacterial isolates, in which strain S58 showed the broadest range in the antimicrobial spectrum. It inhibited the growth of fungi and bacteria, which included most of the pathogens that cause soil-borne diseases, such as Rhizoctonia solani that causes wheat sharp eyespot, Phytophthora nicotianae that causes tobacco black shank, Fusarium graminearum that causes corn stalk rot, and Rastonia solanacearum that causes bacterial wilt. S58 also inhibited pathogens that cause seed-borne diseases, such as Xanthomonas oryzae that causes rice leaves blight, Clavibacter michiganensis that causes tomato bacterial canker, and Acidovorax citrulli that causes cucurbit bacterial fruit blotch. Pathogens of aerial-borne diseases, such as Magnaporthe oryzae that causes rice blight, was also inhibited (Figure 1a). Overall, strain S58 had stronger antagonistic capacity against phytobacteria than against pathogenic fungi based on quantitation of the inhibition zone (Figure 1a). It inhibited the growth of X. oryzae and C. michiganensis, especially, that had inhibition zones that extended up to 30 mm (Figure 1a). These results indicated that the strong, broad antagonism of strain S58 might result in potent biological control. The inhibition by this phenotype suggested that strain S58 produced strong diffusible antimicrobial metabolites.
3.2. Biological Control of Plant Diseases
Given the notably inhibitory effect on plant pathogens, the biocontrol potential of strain S58 was determined in a pot experiment. At 7 dpi of planting, wheat seedings were dug to quantify black shank symptom. The group that served as the healthy control survived and were symptomless (Figure 2a). The disease control group treated with R. solani developed tan cortical rot and lesions on the roots and basal leaf sheaths (Figure 2a). The lesions were elliptic or “eye” shaped with a tan center surrounded by a dark brown margin and had an average length of 12.0±2.4 mm. Surprisingly, wheat seedings pre-soaked with strain S58 and then treated with R. solina had smaller lesions with an average length of 3.2±1.1 mm (Figure 2a). This demonstrated that strain S58 protected plants from R. solani, an important soil-borne disease.
Figure 2.
Disease management by strain S58. (a) Wheat sharp eyespot disease controlled by strain S58; (b) tobacco wildfire disease controlled by strain S58. The healthy control groups were treated with medium or buffer. S58+R. solani and S58+P. syringae represent that the plants were pretreated with strain S58 prior to being treated with the pathogens. Disease control groups were only treated with pathogens R. solani and P. syringae. Wheat represents wheat cv. Mingxian 169 and Nb represents N. benthamiana. The symptoms have been described in the text.
Tobacco wildfire is a foliar disease that is caused by the plant pathogenic bacterium Pseudomonas syringae pv. tabaci. The healthy control group exhibited no symptoms, but the disease control group exhibited chlorotic halos and necrotic spots that coalesced and expanded irregularly (Figure 2b). Infected leaves became brown and distorted. Significantly, the leaves pre-treated with strain S58 only had small circular pale-green areas and did not develop chlorosis and necrosis (Figure 2b, Figure S1). Strain S58 inhibited the growth of a number of phytobacteria, but not P. syringae pv. tabaci (Figure S2). The results suggested that direct antagonism against pathogens was not the only way that the strain S58 controlled plant disease.
3.3. Plant-Growth Promoting Activities
We used selective media to test a series of plant growth-promoting traits qualitatively. The formation of clear hydrolytic halos around the colonies demonstrated that strain S58 produced siderophores, amylase, protease, and solubilized organic phosphorus (Figure 3abcd). The development of a brownish color indicated the production of ammonia (Figure 3e). The Salkowski reagent was used for the detection of IAA. The reagent treated with the bacterial culture of the strain S58 became dark pink, which suggested the formation of an IAA complex and a reduction of Fe3+ (Figure 3f).
Figure 3.
In vitro test of plant growth-promoting traits of strain S58. The transparent zone around the colonies are visualized due to (a) siderophore production, (b) amylase production, (c) protease production, and (d) phosphate solubilization. (e) Ammonia production is shown from the visualized brown, yellow color treated with strain S150 (lower two replicates) compared with the upper left water control. (f) Indole-3-acetic acid (IAA) production is shown from the visualized pink color treated with strain S58 (lower two replicates) compared with the upper left positive control (100 mg/mL IAA) and upper right negative control (LB medium).
The plant growth-promoting effect of strain S58 studied in Arabidopsis thaliana seedlings that grew vertically on agar-solidified medium, in which the seeds were challenged with strain S58. As time went by, the primary root length (PRL) of seedlings exposed to the strain S58 was reduced, and the lateral root number (LRN) increased (Figure 4a). After 10 d of cocultivation, the PRL in the mock control grew up to 5 cm, which was the distance of the inocula from the seeds (Figure 4b). PRL in the treatment with strain S58 was reduced by approximately 50% (Figure 4b). At the same time, strain S58-treated roots formed approximately 5.5-fold more LRN compared with the mock control, and the fresh weight increased three-fold also (Figure 4b). These results showed that S58 promoted plant growth and stimulated plant biomass production by affecting the architecture of the root system.
Figure 4.
Effects of strain S58 on plant growth and root system architecture of Arabidopsis thaliana Col-0 seedlings. (a) The root architecture changed on the S58 plates at 5 dpi and 10 dpi; (b) Root length, fresh weight, and lateral roots were measured at 10 dpi. Stars indicate statistically significant differences (Tukey’s HSD test; p < 0.01).
3.4. Identification of Strain S58
Strain S58 was characterized as a multi-functional beneficial bacterium, as shown above. We then made a comprehensive classification for it. The colonies of strain S58 on LB medium were round, wrinkled with a mucous texture, and often produced a diffuse yellowish non-fluorescent pigment. The growth on KB and under UV did not reveal a fluorescent pigment. The bacterial cells of strain S58 were rod-shaped, 0.4–1.2 μm in diameter, and 1.3–4.3 μm long. Only one flagellum at one polar was observed under a transmission electron microscope (Figure S3). From the results of the GEN III MicroPlates™ assay (Table S2), strain S58 had a very similar biochemical profile to Pseudomonas mediterranea [18], such as utilizing L-alanine, aminobutyrate, D-fructose, D-galactose, D-gluconate, D-glucose, L-glutamate, lactate, L-malate, D-mannitol, D-mannose, mucate, L-proline, quinate, D-saccharate, sucrose, D-trehalose, etc. and not utilizing D-arabitol, D-cellobiose, L-fucose, gentobiose, L-histidine, lactose, maltose, D-melibiose, D-raffinose, L-rhamnose, D-sorbitol, L-tartrate, tricarballylate, D-turanose, etc.
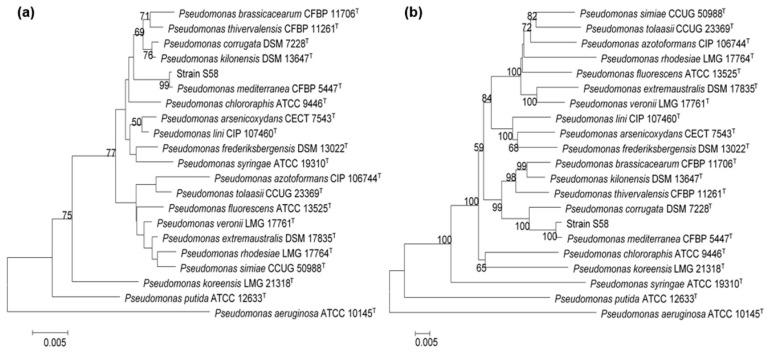
To carry out a more accurate identification of the bacterium, we performed a phylogenic analysis of the 16S rRNA gene. The 16S rDNA sequence of strain S58 had 99.04% identity with that of P. mediterranea type strain CFBP 5447T. The phylogenic tree of 16S rRNA genes showed that strain S58 clustered with the type strain of P. mediterranea with a high bootstrap score (>99%) (Figure 5a). For an accurate determination, a multi-locus sequence analysis (MLSA) using the 16S rRNA gene along with the sequences of the housekeeping genes rpoB, rpoD, and gyrB was conducted. The results indicated that strain S58 had the same phylogenetic distribution as analyzed with the 16S rRNA gene (Figure 5b).
Figure 5.
Phylogenetic tree showing the relationship of strain S58 with other Pseudomonas spp. type strains. (a) Phylogenetic analysis based on the 16S rRNA gene sequences; (b) Phylogenetic analysis based on four core housekeeping genes (16S rRNA, gyrB, rpoB, rpoD). P. aeruginosa ATCC 10145T was used as an outgroup. Bootstrap values from 1000 replicates are indicated at the nodes.
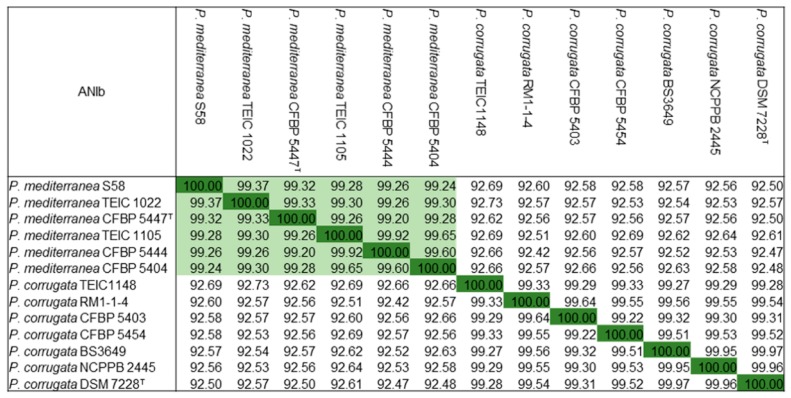
P. mediterranea and P. corrugata are the most closely related species. To establish further the phylogenetic relationship of strain S58, we compared the available genomes of representative strains in the two species by calculating the average nucleotide identity (ANI) values. Strain S58 had higher ANI values >99% with P. mediterranea strains that included the type strain P. mediterranea CFBP 5447T. However, all P. corrugata strains showed ANI values <94%, which have been proposed for defining the boundary between species (Figure 6). In conclusion, morphological, physiological, and biochemical characteristics along with phylogenetic and ANI analyses revealed that strain S58 belonged to P. mediterranea. A culture of S58 was deposited in the China General Microorganism Culture Collection (CGMCC17043).
Figure 6.
Average Nucleotide Identity based on BLAST (ANIb) for most of the genome-released strains of P. mediterranea and P. corrugata. Green shading indicates the same species of P. mediterranea.
3.5. Comparative Genomic Analysis
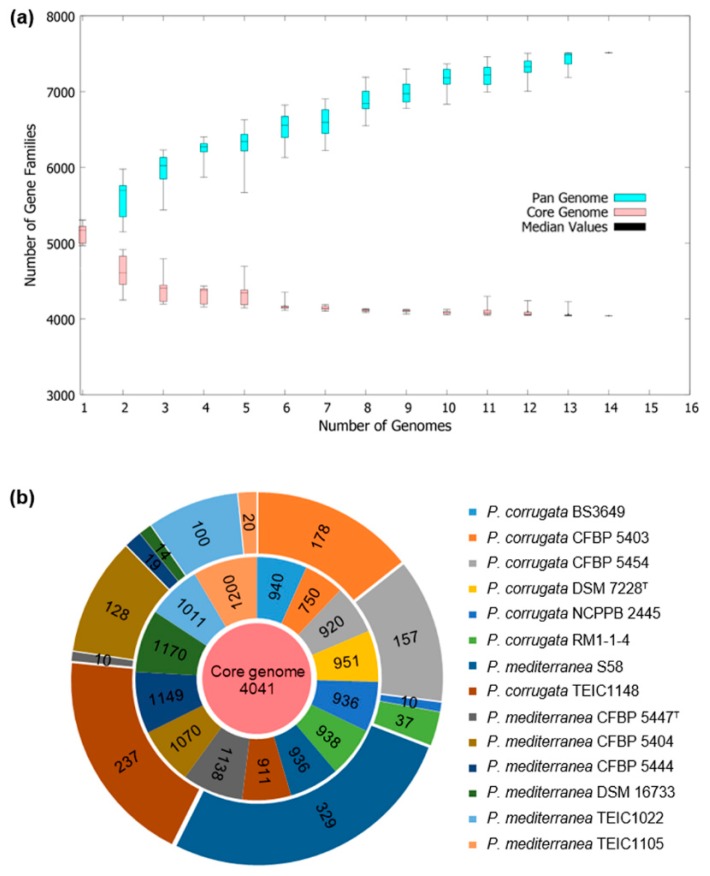
A summary of the general sequence characteristics is given in Table 1. The size of the genome of P. mediterranea S58 was 6,150,838 bp with a 61.06% G+C content, and it was predicted to contain 5594 coding sequences (CDSs) that included 5312 genes with an average length of 992.90 bp (Table 1). The total sequence of the CDSs covered 87.62% of the entire genome. In addition, 282 genes were assigned pseudogenes due to a missing N- and C-terminus or a frameshift mutation. A total of 67 tRNA, 15 rRNAs, and four non-coding RNA genes were predicted to occur on the chromosome. A genome circle map of P. mediterranea S58 and the closely related three strains (i.e., P. mediterranea CFBP 5447T [38], P. corrugata DSM 7228T [39], and P. corrugata RM1-1-4 [40]) was generated using BRIG (Figure S4). Conservation and variation in gene content between genomes were visualized from the image. It should be noted that the CFBP 5447T and DSM 7228T genomes are draft genomes, and only the genome of RM1-1-4 was closed completely. Therefore, regions that were not included in the BRIG alignment most likely represented regions not sequenced in one or more genomes. To identify phylogenomics and orthologous genes common to these two related species, we carried out a pan-genome and core genome analysis of all 13 strains included in P. mediterranea and P. corrugata (Figure 7). The pan-genome of all the strains had 18,446 genes, which was the sum of the core genes and dispensable genes, which included unique genes and accessory genes (Figure 7a). Of the identified genes, 4041 were core genes and 329 were unique to strain S58 (Figure 7b).
Table 1.
General features of the P. mediterranea S58 genome.
| Features | Chromosome |
|---|---|
| Size (bp) | 6,486,667 |
| G+C content (%) | 61.06 |
| Number of total CDSs | 5594 |
| Number of genes | 5312 |
| Pseudogenes | 282 |
| tRNAs | 67 |
| rRNA genes | 15 |
| ncRNAs | 4 |
| Contigs | 1 |
| Total CDSs size (bp) | 5,683,341 |
| Coding % | 87.62 |
| Average CDS length (nt) | 992.90 |
Figure 7.
Pan and core genome analysis of the strains of P. mediterranea and P. corrugata. (a) Pan-genome and core genome plot show the progression of the pan (blue line) and core (pink line) genomes as more genomes are added for analysis; (b) The flower plot shows the numbers of core genes (inner circle), accessory genes (middle circle), and unique genes (outer circle).
Given the versatile plant growth-promoting traits of P. mediterranea S58, the potential to produce secondary metabolites was analyzed using the online pipeline antiSMASH 5.0. Ten putative gene clusters involved in the biosynthesis of different natural products were found, which included seven nonribosomal peptide-synthetase (NRPS) clusters that encoded antimicrobial cyclic lipopeptides (CLPs) (Table 2). Moreover, one bacteriocin biosynthetic gene cluster was found. No polyketide-encoding gene clusters were predicted. Interestingly, two clusters that encoded crochelin A and Orfamide B were present only in P. mediterranea S58, but not in P. corrugata RM1-1-4, which was the only complete sequenced strain in the P. mediterranea and P. corrugata group (Table 2). A pqqFABDE operon (E3Z27_25820- E3Z27_25845) was present, which might be involved in gluconic acid (GA) biosynthesis used for solubilization of phosphates [41]. It was tested that P. mediterranea S58 could produce plant growth hormone IAA. A high identity (89.94%) homolog of aldehyde dehydrogenase (AldA) was found in the genome, which was reported as a part of a novel IAA synthesis pathway in P. syringae strain DC3000 [42]. Further genome analysis revealed that P. mediterranea S58 contained genes related to other plant growth-promoting traits, such as the production of siderophores, glutamine synthetase, protease, chitinase, and 1-aminocyclopropane-1-carboxylic acid deaminase.
Table 2.
List of predicted secondary metabolite biosynthetic gene clusters of P. mediterranea S58.
| Cluster Type/Metabolites | S58 | RM1-1-4 1 | |
|---|---|---|---|
| Location | Gene ID | ||
| NRPS-like/Fragin | 177591-219516 | E3Z27_00825-E3Z27_00960 | + |
| Arylpolyene/APE Vf | 476416-520027 | E3Z27_02080-E3Z27_02290 | + |
| Bacteriocin | 1422090-1432977 | E3Z27_06235-E3Z27_06295 | + |
| NAGGN | 1834296-1849084 | E3Z27_08105-E3Z27_08160 | + |
| NRPS/Fengycin | 2221816-2245051 | E3Z27_09835-E3Z27_-09940 | + |
| NRPS/Crochelin A | 2691862-2766373 | E3Z27_12005-E3Z27_12180 | - |
| NRPS-like/Entolysin | 2967333-3015201 | E3Z27_12950-E3Z27_13125 | + |
| NRPS/Orfamide B | 3271309-3337076 | E3Z27_14180-E3Z27_14400 | - |
| NRPS/Siderophore | 3444389-3463297 | E3Z27_14930-E3Z27_15000 | + |
| NRPS/Syringomycin | 3672567-3850583 | E3Z27_15910-E3Z27_16205 | + |
1 represents P. corrugata RM1-1-4.
3.6. Strain S58 Triggered Cell Death and Plant Immunity
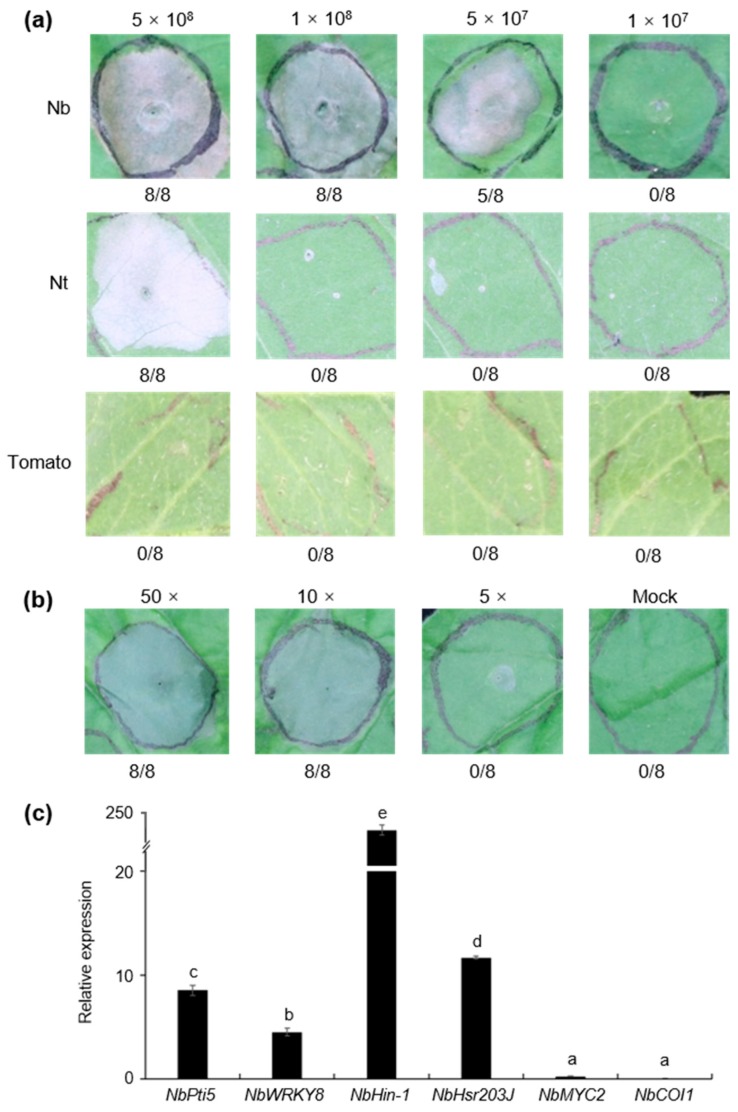
A test of hypersensitive response (HR)-like cell death was performed by infiltrating 5 × 108 CFU/mL of bacteria into the leaves of Nicotiana benthamiana. Significant necrosis was present at 24 h post-inoculation (Figure 8a). The titer was determined for the lowest concentration of P. mediterranea S58 to trigger cell death. The results showed that the threshold concentration for cell death was 5 × 107 CFU/mL (Figure 8a). We determined further whether P. mediterranea S58 triggered cell death in N. tabacum and tomato (Lycopersicon esculentum cv. Moneymaker). Interestingly, P. mediterranea S58 only induced cell death in N. tabacum at a high concentration of infiltrate of 5 × 108 CFU/mL but failed to elicit any cell death-like symptoms in tomato at any of the tested gradients (Figure 8a). We then asked whether the cell death was induced by extracellular components of P. mediterranea S58. The concentrated supernatant of the bacterial culture was tested for cell death, in which tenfold and fivefold concentrations elicited visible cell death (Figure 8b).
Figure 8.
Cell death-like immunity triggered by P. mediterranea S58. (a) Cell death assays with strain S58 at a concentration gradient on different plants. The fraction under each image indicates the number of times that the cell death occurred relative to the number of test inoculations. Nb, Nt, and tomato represent N. benthamiana, Nicotiana tabacum,and L. esculentum cv. Moneymaker, respectively; (b) Cell death assays with the supernatant of strain S58 at different concentrations in N. benthamiana; (c) Expression of immunity marker genes was tested by qRT-PCR. Relative expression levels were normalized to the reference gene NbEF1α. Means with the same letters are not different statistically based on a Tukey test (α = 0.05).
Cell death is a process by which the plants produce immune responses to restrict microbial infection. A series of gene markers was selected to determine immunity using qRT-PCR. The expressions of NbPti5 and NbWRK7, which are two pattern-triggered immunity (PTI) marker genes that are generally induced by the flagellin protein and its flg22 peptide epitope, were up-regulated by P. mediterranea S58 at 12 dpi. This suggested that the flagellin protein of the P. mediterranea S58 might be a Microbe-Associated Molecular Pattern (MAMP) to trigger PTI. Unsurprisingly, NbHin-1 and NbHsr203J, which are two cell death marker genes, were up-regulated sharply by P. mediterranea S58. The expression of cell death markers was significantly higher than PTI markers. However, the jasmonic acid (JA) signaling genes (NbMYC2 and NbCOI1) that were involved in rhizobacteria-mediated Induced Systemic Resistance (ISR) were not affected at all by P. mediterranea S58. All these results suggested that P. mediterranea S58 triggered potent, cell death-like, local innate immunity in tobacco.
4. Discussion
Pseudomonas spp. are major plant-associated bacteria that occupy different niches and exhibit varied effects on plants [2]. They can act as pathogens, such as P. syringae that cause diseases in most higher plants, or they can act as plant growth-promoting agents, such as P. fluorescens that controls plant fungal and bacterial diseases [4]. Some P. mediterranea strains, a weak and opportunistic pathogen, were isolated in association with tomato pith necrosis [18]. In addition, a number of P. mediterranea strains have been isolated from non-diseased plants, mainly from rhizospheres and roots. These strains showed multiple beneficial traits in plants, such as the production of antimicrobial products and plant hormones [39]. S58 is one strain of P. mediterranea that was isolated from the tobacco rhizosphere. In a comprehensive in vitro test, it inhibited the growth of a wide range of plant pathogens and produced siderophores, protease, ammonia, and indole-3-acetic acid. It also controlled wheat and tobacco diseases efficiently, promoted the development of Arabidopsis thaliana, and induced cell death-like immunity. All of those results suggested that P. mediterranea S58 is a potent PGPR with versatile beneficial characters.
It is noteworthy that Pseudomonas species or strains are closely related, but their identification and taxonomy cannot be defined simply by relying on a single approach [39]. P. mediterranea is a member of the P. corrugata subgroup of the P. fluorescens group based on MLSA studies that used the 16S rRNA gene and rpoB, rpoD, and gyrB gene sequences [43,44]. In this study, we performed morphological, biochemical, physiological characterization, and MLSA phylogenic analysis. The results distinctly showed that strain S58 belonged in the species P. mediterranea. Further analysis of the ANI values of the phylogenetically related species and strains also revealed that strain S58 belonged to P. mediterranea. Although it has been almost two decades since P. mediterranea was identified as a new species, only six strains had been sequenced prior to this study. However, all of the released sequences are draft genomes, which indicates that we have only a superficial understanding of the molecular mechanisms; this restricts the application of P. mediterranea as bioderemediators or PGPR.
The complete genome of P. mediterranea S58 provides insights into the genes involved in rhizo competence. The core genome of 13 genome sequences from P. mediterranea and P. corrugata was composed of 4041 orthologous genes, which is larger in comparison with a previous analysis that used 10 representative genome sequences of the P. fluorescens group (2789 orthologous genes) [44]. In addition, 1265 dispensable genes (including 329 unique and 936 accessory genes) that we found in the S58 genome are not present in any other genomes of strains within this comparison. The most dispensable genes in P. mediterranea S58 suggests that it has more specific and versatile metabolisms. We need to note that strain S58 triggered cell death in tobacco (Figure 8a), which was confirmed by up-regulated expressions of NbHin-1 and NbHsr203J, two typical cell death marker genes (Figure 8c). The pathogen P. syringae induced type III secretion system (T3SS)-dependent cell death in non-host plants [45]. Some T3SS effector and Harpin proteins contributed to cell death [45]. An in-depth investigation of the proteomes of P. mediterranea S58 using all T3SS components revealed that it did not contain any T3SS homologs that included effector and Harpin proteins, which are identical to the findings in other P. corrugata and P. mediterranea isolates [39]. This suggested that other secreted proteins, phytotoxins, or secondary metabolites, but not T3SS-related proteins, conferred the phenotype with cell death capability. Syringotoxin and syringomycin E isloated from P. syringae triggered cell death in tobacco [7]. Recently, cormycin A, a cyclic lipodepsipeptide (CLP) isolated from the culture of P. corrugata NCPPB 2445, also induced cell death in tobacco [46]. It is a CLP-rich bacterium; hence, P. mediterranea S58 might rely on some CLPs to induce hypersensitive cell death in plants. Our study opens up opportunities for the exploitation of the molecular mechanisms of the versatile PGPR traits in P. mediterranea S58 and related species.
5. Patents
The work reported in the manuscript has been granted a Chinese invention patent (ZL201910911436).
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/3/334/s1, Figure S1: Symptoms of tobacco wildfire disease, Figure S2: No antagonism of strain S58 against Pseudomonas syringae pv. tabaci, Figure S3: Electron micrograph of the cells of strain S58, Figure S4: Genome comparison of strain S58, Pseudomonas corrugata, and Pseudomonas mediterranea. The circles from inner to external: GC Content, GC Skew, P. mediterranea S58 (pink), P. mediterranea CFBP 5447T (green), P. corrugata DSM 7228T (blue), and P. corrugata RM1-1-4 (light blue), Table S1: Sequences of the gene-specific primer pairs used in the study, Table S2: Physiological and biochemical indexes of strain S58.
Author Contributions
Conceptualization, H.-L.W.; investigation, Y.G., J.W. and Z.X.; writing—original draft preparation, H.-L.W.; writing—review and editing, Y.G., J.W., Z.X. and H.-L.W.; funding acquisition, Z.X. and H.-L.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Science and Technology Program of the Yunnan Tobacco (2017YN08), the Program of Science and Technology of Beijing, China (Z191100004019025), the National Key R&D Program of China (2019YFD1002001), and the Central Public-interest Scientific Institution Basal Research Fund (Y2019XK07, 1610132020039).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Raaijmakers J.M., Weller D.M., Thomashow L.S. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl. Environ. Microbiol. 1997;63:881–887. doi: 10.1128/AEM.63.3.881-887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramamoorthy V., Raguchander T., Samiyappan R. Enhancing resistance of tomato and hot pepper to Pythium diseases by seed treatment with fluorescent pseudomonads. Eur. J. Plant Pathol. 2002;108:429–441. doi: 10.1023/A:1016062702102. [DOI] [Google Scholar]
- 3.Rosier A., Bishnoi U., Lakshmanan V., Sherrier D.J., Bais H.P. A perspective on inter-kingdom signaling in plant-beneficial microbe interactions. Plant Mol. Biol. 2016;90:537–548. doi: 10.1007/s11103-016-0433-3. [DOI] [PubMed] [Google Scholar]
- 4.Vacheron J., Desbrosses G., Bouffaud M.L., Touraine B., Moenne-Loccoz Y., Muller D., Legendre L., Wisniewski-Dye F., Prigent-Combaret C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013;4:356. doi: 10.3389/fpls.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stringlis I.A., Zhang H., Pieterse C.M.J., Bolton M.D., de Jonge R. Microbial small molecules—weapons of plant subversion. Nat. Prod. Rep. 2018;35:410–433. doi: 10.1039/C7NP00062F. [DOI] [PubMed] [Google Scholar]
- 6.Ghequire M.G., De Mot R. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol. Rev. 2014;38:523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 7.Geudens N., Martins J.C. Cyclic lipodepsipeptides from Pseudomonas spp. - biological swiss-army knives. Front. Microbiol. 2018;9:1867. doi: 10.3389/fmicb.2018.01867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almario J., Bruto M., Vacheron J., Prigent-Combaret C., Moenne-Loccoz Y., Muller D. Distribution of 2,4-diacetylphloroglucinol biosynthetic genes among the Pseudomonas spp. reveals unexpected polyphyletism. Front. Microbiol. 2017;8:1218. doi: 10.3389/fmicb.2017.01218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramette A., Frapolli M., Fischer-Le Saux M., Gruffaz C., Meyer J.M., Défago G., Sutra L., Moënne-Loccoz Y. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 2011;34:180–188. doi: 10.1016/j.syapm.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Nowak-Thompson B., Gould S.J., Loper J.E. Identification and sequence analysis of the genes encoding a polyketide synthase required for pyoluteorin biosynthesis in Pseudomonas fluorescens Pf-5. Gene. 1997;204:17–24. doi: 10.1016/S0378-1119(97)00501-5. [DOI] [PubMed] [Google Scholar]
- 11.Wu X.G., Duan H.M., Tian T., Yao N., Zhou H.Y., Zhang L.Q. Effect of the hfq gene on 2,4-diacetylphloroglucinol production and the PcoI/PcoR quorum-sensing system in Pseudomonas fluorescens 2P24. FEMS Microbiol. Lett. 2010;309:16–24. doi: 10.1111/j.1574-6968.2010.02009.x. [DOI] [PubMed] [Google Scholar]
- 12.Berendsen R.L., van Verk M.C., Stringlis I.A., Zamioudis C., Tommassen J., Pieterse C.M.J., Bakker P.A.H.M. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genomics. 2015;16:539. doi: 10.1186/s12864-015-1632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrade L.F., de Souza G.L., Nietsche S., Xavier A.A., Costa M.R., Cardoso A.M., Pereira M.C., Pereira D.F. Analysis of the abilities of endophytic bacteria associated with banana tree roots to promote plant growth. J. Microbiol. 2014;52:27–34. doi: 10.1007/s12275-014-3019-2. [DOI] [PubMed] [Google Scholar]
- 14.Luo S., Xu T., Chen L., Chen J., Rao C., Xiao X., Wan Y., Zeng G., Long F., Liu C., et al. Endophyte-assisted promotion of biomass production and metal-uptake of energy crop sweet sorghum by plant-growth promoting endophyte Bacillus sp. SLS18. Appl. Microbiol. Biotechnol. 2012;93:1745–1753. doi: 10.1007/s00253-011-3483-0. [DOI] [PubMed] [Google Scholar]
- 15.Taurian T., Anzuay M.S., Angelini J.G., Tonelli M.L., Ludueña L., Pena D., Ibáñez F., Fabra A. Phosphate-solubilizing peanut associated bacteria: Screening for plant growth-promoting activities. Plant Soil. 2010;329:421–431. doi: 10.1007/s11104-009-0168-x. [DOI] [Google Scholar]
- 16.Cavalcante J.J., Vargas C., Nogueira E.M., Vinagre F., Schwarcz K., Baldani J.I., Ferreira P.C., Hemerly A.S. Members of the ethylene signalling pathway are regulated in sugarcane during the association with nitrogen-fixing endophytic bacteria. J. Exp. Bot. 2007;58:673–686. doi: 10.1093/jxb/erl242. [DOI] [PubMed] [Google Scholar]
- 17.Catara V. Pseudomonas corrugata: Plant pathogen and/or biological resource? Mol. Plant Pathol. 2007;8:233–244. doi: 10.1111/j.1364-3703.2007.00391.x. [DOI] [PubMed] [Google Scholar]
- 18.Catara V., Sutra L., Morineau A., Achouak W., Christen R., Gardan L. Phenotypic and genomic evidence for the revision of Pseudomonas corrugata and proposal of Pseudomonas mediterranea sp. nov. Int. J. Syst. Evol. Microbiol. 2002;52:1749–1758. doi: 10.1099/00207713-52-5-1749. [DOI] [PubMed] [Google Scholar]
- 19.Ryder M.H., Rovira A.D. Biological control of take-all of glass house-grown wheat using strains of Pseudomonas corrugata isolated from wheat field soil. Soil Biol. Biochem. 1993;25:311–320. doi: 10.1016/0038-0717(93)90129-Y. [DOI] [Google Scholar]
- 20.Schmidt C.S., Agostini F., Leifert C., Killham K., Mullins C.E. Influence of inoculum density of the antagonistic bacteria Pseudomonas fluorescens and Pseudomonas corrugata on sugar beet seedling colonisation and suppression of Pythium damping off. Plant Soil. 2004;265:111–122. doi: 10.1007/s11104-005-8943-9. [DOI] [PubMed] [Google Scholar]
- 21.Licciardello G., Caruso A., Bella P., Gheleri R., Strano C.P., Anzalone A., Trantas E.A., Sarris P.F., Almeida N.F., Catara V. The LuxR regulators PcoR and RfiA co-regulate antimicrobial peptide and alginate production in Pseudomonas corrugata. Front. Microbiol. 2018;9:521. doi: 10.3389/fmicb.2018.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Licciardello G., Strano C.P., Bertani I., Bella P., Fiore A., Fogliano V., Venturi V., Catara V. N-acyl-homoserine-lactone quorum sensing in tomato phytopathogenic Pseudomonas spp. is involved in the regulation of lipodepsipeptide production. J. Biotechnol. 2012;159:274–282. doi: 10.1016/j.jbiotec.2011.07.036. [DOI] [PubMed] [Google Scholar]
- 23.Licciardello G., Catara A.F., Catara V. Production of polyhydroxyalkanoates and extracellular products using Pseudomonas corrugata and P. mediterranea: A review. Bioengineering. 2019;6:105. doi: 10.3390/bioengineering6040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/JB.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennasar A., Mulet M., Lalucat J., García-Valdés E. PseudoMLSA: A database for multigenic sequence analysis of Pseudomonas species. BMC Microbiol. 2010;10:118. doi: 10.1186/1471-2180-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nautiyal C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999;170:265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- 28.Schwyn B., Neilands J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 29.Cattelan A.J., Hartel P.G., Fuhrmann J.J. Screening of plant growth—Promoting rhizobacteria to promote early soybean growth. Soil Sci. Soc. Amer. 1999;63:1670–1680. doi: 10.2136/sssaj1999.6361670x. [DOI] [Google Scholar]
- 30.Cappuccino J.C., Sherman N. Negative staining. In: Cappuccino J.C., Sherman N., editors. Microbiology: A Laboratory Manual. 3rd ed. Cumming Publ.; New York, NY, USA: 1992. pp. 125–179. [Google Scholar]
- 31.Zamioudis C., Mastranesti P., Dhonukshe P., Blilou I., Pieterse C.M.J. Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiol. 2013;162:304–318. doi: 10.1104/pp.112.212597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohtsubo Y., Ikeda-Ohtsubo W., Nagata Y., Tsuda M. GenomeMatcher: A graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008;9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon S.H., Ha S.M., Lim J.M., Kwon S.J., Chun J.A. large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 34.Alikhan N.F., Petty N.K., Ben Zakour N.L., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S.Y., Medema M.H., Weber T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaudhari N.M., Gupta V.K., Dutta C. BPGA—An ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016;6:24373. doi: 10.1038/srep24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu L., Dong Z., Fang L., Luo Y., Wei Z., Guo H., Zhang G., Gu Y.Q., Coleman-Derr D., Xia Q., et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47:W52–W58. doi: 10.1093/nar/gkz333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Licciardello G., Bella P., Devescovi G., Strano C.P., Sarris P.F., Catara A.F., Venturi V., Catara V. Draft genome sequence of Pseudomonas mediterranea strain CFBP 5447T, a producer of filmable medium-chain-length polyhydroxyalkanoates. Genome Announc. 2014;2:e01260-14. doi: 10.1128/genomeA.01260-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trantas E.A., Licciardello G., Almeida N.F., Witek K., Strano C.P., Duxbury Z., Ververidis F., Goumas D.E., Jones J.D., Guttman D.S., et al. Comparative genomic analysis of multiple strains of two unusual plantpathogens: Pseudomonas corrugata and Pseudomonas mediterranea. Front. Microbiol. 2015;6:811. doi: 10.3389/fmicb.2015.00811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zachow C., Müller H., Laireiter C.M., Tilcher R., Berg G. Complete genome sequence of Pseudomonas corrugata strain RM1-1-4, a stress protecting agent from the rhizosphere of an oilseed rape bait plant. Stand. Genomic Sci. 2017;12:66. doi: 10.1186/s40793-017-0278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oteino N., Lally R.D., Kiwanuka S., Lloyd A., Ryan D., Germaine K.J., Dowling D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015;6:745. doi: 10.3389/fmicb.2015.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClerklin S.A., Lee S.G., Harper C.P., Nwumeh R., Jez J.M., Kunkel B.N. Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog. 2018;14:e1006811. doi: 10.1371/journal.ppat.1006811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulet M., Lalucat J., García-Valdés E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010;12:1513–1530. doi: 10.1111/j.1462-2920.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- 44.Loper J.E., Hassan K.A., Mavrodi D.V., Davis E.W., II, Lim C.K., Shaffer B.T., Elbourne L.D., Stockwell V.O., Hartney S.L., Breakwell K., et al. Comparative genomics of plant-associated Pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8:e1002784. doi: 10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei H.L., Collmer A. Defining essential processes in plant pathogenesis with Pseudomonas syringae pv. tomato DC3000 disarmed polymutants and a subset of key type III effectors. Mol. Plant Pathol. 2018;19:1779–1794. doi: 10.1111/mpp.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scaloni A., Dalla Serra M., Amodeo P., Mannina L., Vitale R.M., Segre A.L., Cruciani O., Lodovichetti F., Greco M.L., Fiore A., et al. Structure, conformation and biological activity of a novel lipodepsipeptide from Pseudomonas corrugata: Cormycin A. Biochem. J. 2004;384:25–36. doi: 10.1042/BJ20040422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.