Abstract
Recurrent neural networks (RNNs) were designed for dealing with time-series data and have recently been used for creating predictive models from functional magnetic resonance imaging (fMRI) data. However, gathering large fMRI datasets for learning is a difficult task. Furthermore, network interpretability is unclear. To address these issues, we utilize multitask learning and design a novel RNN-based model that learns to discriminate between classes while simultaneously learning to generate the fMRI time-series data. Employing the long short-term memory (LSTM) structure, we develop a discriminative model based on the hidden state and a generative model based on the cell state. The addition of the generative model constrains the network to learn functional communities represented by the LSTM nodes that are both consistent with the data generation as well as useful for the classification task. We apply our approach to the classification of subjects with autism vs. healthy controls using several datasets from the Autism Brain Imaging Data Exchange. Experiments show that our jointly discriminative and generative model improves classification learning while also producing robust and meaningful functional communities for better model understanding.
1. Introduction
Functional magnetic resonance imaging (fMRI) has become an important tool for investigating neurological disorders and diseases. In addition, machine learning has begun to play a large role, in which classification models are learned and interpreted to discover potential fMRI biomarkers for disease. Traditional approaches for building classification models from resting-state fMRI first parcellate the brain into a number of regions of interest (ROIs) and use functional connectivity between the ROIs as inputs to a classification algorithm [1]. Recently with the advent of deep learning, temporal inputs based on the time-series data combined with recurrent neural network (RNN) models have been explored for predicting from fMRI [7,8,14]. Such RNN models are attractive for processing fMRI as they were designed for dealing with sequential data. However, the large sample sizes required for effective deep learning are difficult to gather for fMRI data, particularly for many different patient populations or types of studies.
One way to handle the limited data problem is to apply multitask learning [4]. The idea in multitask learning is that shared information across related tasks is jointly learned in order to improve the learning of each individual task. For a classification task based on fMRI data, e.g., distinguish subjects with a given disease from healthy individuals, the amount of labeled data is often limited. Thus, we propose to apply multitask learning to improve the learning of a target discriminative task by jointly learning an auxiliary generative model for the fMRI data, which does not require any annotation. Moreover, simultaneous learning of the generative model will assist in interpreting the discriminative model.
Specifically, we propose to jointly learn a discriminative task while also learning to generate the input fMRI time-series by using an RNN with long short-term memory (LSTM). Generative RNN models have been extensively used in natural language processing, e.g., for text generation [9], but application to the medical imaging field has been limited. Furthermore, multitask learning with discriminative and generative components have been combined in many different neural network architectures, notably generative adversarial networks, but such a joint learning approach utilizing the RNN framework has only begun to be explored and under the context of adversarial training for a target generative task [2].
In this paper, we design a novel RNN-based model with LSTM to simultaneously learn a discriminative and generative task by utilizing the state information in a shared LSTM layer. Using fMRI ROI time-series as inputs, we interpret the LSTM block as modeling the coordination of functional activity in the brain and the nodes of the LSTM as representing functional communities, i.e., groupings of the input brain ROIs that work together to both generate the fMRI time-series and perform the discriminative task. We apply the proposed network for classification of ASD vs. healthy controls, validating on multiple datasets from the Autism Brain Imaging Data Exchange (ABIDE) I dataset. Compared to several recent methods, we achieve some of the highest accuracy reported on single-site ABIDE data. Finally, we evaluate the generative results by analyzing the robustness of the extracted functional communities and validate influential communities for classification in the context of ASD.
2. Methods
2.1. Network Architecture
LSTM Block for Communities
The LSTM module was designed to learn long-term dependencies in sequential data [10]. An LSTM cell is composed of 4 neural network layers with K nodes that modulate two state vectors, the hidden state and the cell state . The state vectors are updated using input from the current time point and state information from the previous time point ht−1 and ct−1:
| (1) |
| (2) |
| (3) |
where for layer l ∈ {i, f, o, c}, Wl are the weights for the input, Ul are the weights for the hidden state, and bl are the bias parameters.
The proposed network first takes the fMRI ROI time-series as inputs to an LSTM layer (Fig. 1, blue path). The purpose of this layer is to discover meaningful groupings of the ROIs, i.e. functional communities, that are important for both generating and classifying the input data. The LSTM block acts as a model for the interaction between R individual ROIs and K functional communities formed by the brain network to generate community activity. The activity generated by each functional community k is then represented by the hidden state ht (k) and cell state ct (k), which will serve as inputs to the rest of the network.
Fig. 1:
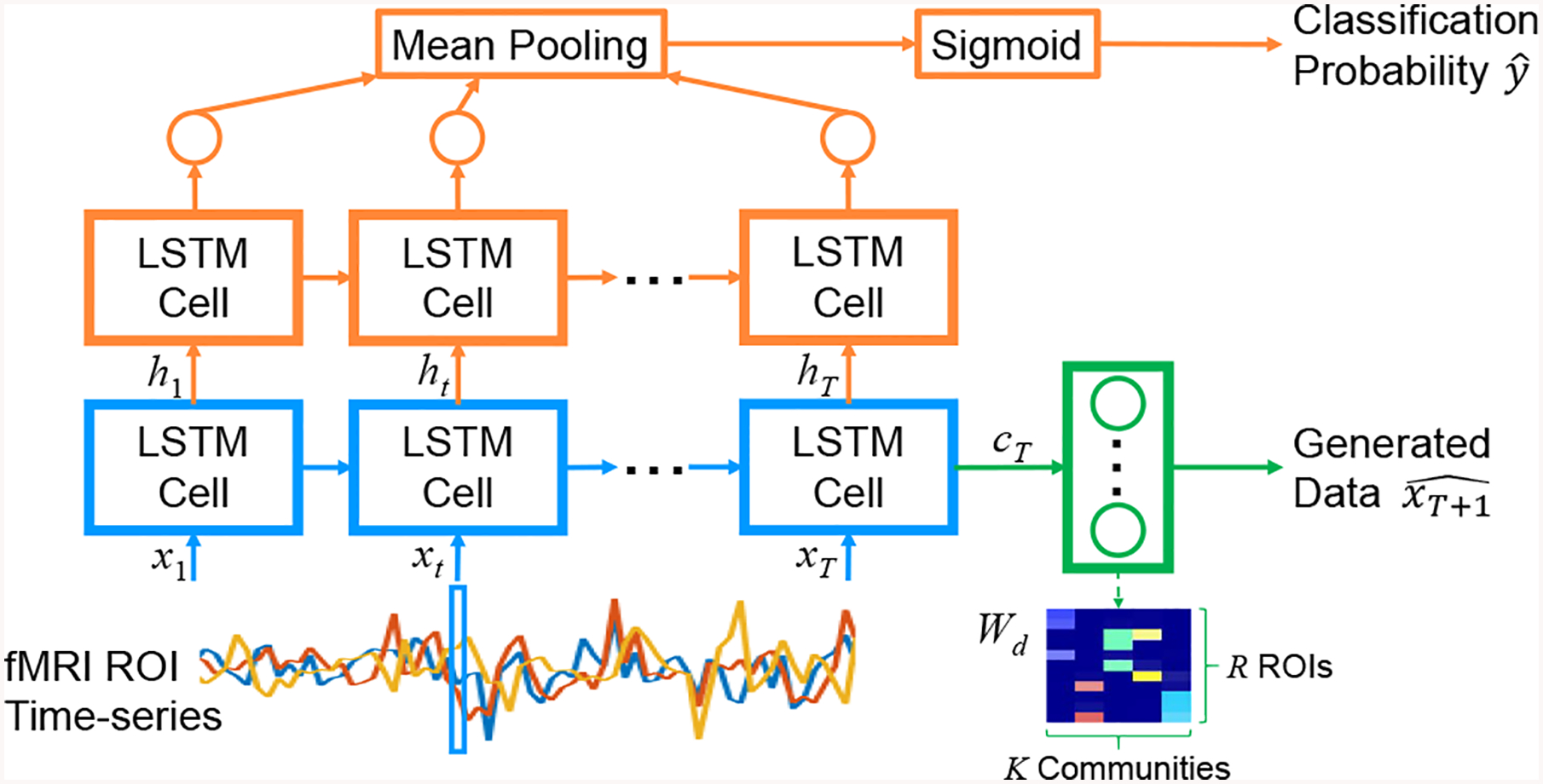
Architecture of our jointly discriminative and generative RNN: LSTM for functional communities (blue), discriminative path (orange), and generative path (green).
Standard community detection methods for fMRI perform clustering based on functional connectivity, where highly positively correlated ROIs are grouped into a community. In our approach, we propose defining a functional community by the interactions modeled in the LSTM and the generated ROI data (see Sec.2.2). To ensure that ROIs within a community have positive ties as in standard approaches, we constrain the input weights Wl to be non-negative.
Discriminative Path
The discriminative portion of the network aims to classify subjects with ASD vs. typical controls (Fig. 1, orange path). The architecture is similar to the network in [7]. The difference is our approach first processes the input time-series through an LSTM layer that learns to represent functional communities of the ROI data. The hidden state of the LSTM cell at each time point is then fed to another LSTM layer, followed by a shared 1-node dense layer, mean pooling layer, and sigmoid activation to give the probability of ASD.
Generative Path
The generative portion of the network looks to generate the data at the next time point xT+1 of an input time-series with length T (Fig. 1, green path). The input is first processed by the same LSTM layer for functional communites as in the discriminative network. The final cell state cT of the LSTM cell is then passed to a dense layer with R nodes to produce the predicted ROI values for the next time point . To enforce that communities exert a positive influence on their members, we constrain Wd to be non-negative.
Model Training
The discriminative and generative paths are tied together during training with the loss function , where LG is the loss for the generative model, LD is the loss for the discriminative model, y ∈ {0, 1} is the true label (1 denoting ASD), is the predicted probability of ASD, and λ is a hyperparameter to balance the two losses. For regularization, we include dropout layers before the shared dense layer and mean pooling layer in the discriminative network and before the dense layer in the generative network.
2.2. Extraction of Functional Communities
As described above, we propose interpreting each node of the first LSTM block as representing a functional community, where community activity is summarized by state vectors ht and ct. Since it is difficult to analyze the interactions between ROIs and communities via all the layers of the LSTM block, we propose defining the communities based on their influence on each individual ROI. Recall that the generative path uses the cell state cT as input to a dense layer to generate the next ROI values . From a graph structure perspective, a community is defined by densely connected nodes, i.e. each member of a community is strongly influenced by that community, but also the community is strongly influenced by its members. Thus, we will use the weights to denote the membership between individual ROIs and their functional communities. Row r of Wd represents the influence of each community on ROI r, while column k of Wd represents the influence of each ROI on community k. To provide hard membership assignments, we perform k-means clustering with 2 clusters on the membership weights in column k of Wd and assign the extracted ROIs in the cluster with larger weights to community k (Fig. 1, lower right).
3. Experiments
3.1. Data
We used resting-state fMRI data from the four ABIDE I [6] sites with the largest sample sizes: New York University (NY), University of Michigan (UM), University of Utah School of Medicine (US), and University of California, Los Angeles (UC). We selected preprocessed data from the Preprocessed Connectomes Project [5] using the Connectome Computation System pipeline, global signal regression and band-pass filtering, and the Automated Anatomical Labeling (AAL) parcellation with 116 ROIs. The extracted mean time-series of each ROI was standardized (subtracted mean, divided by standard deviation) for each subject.
Since the number of subjects per site is small for neural network training, we augmented the datasets by extracting all possible consecutive subsequences with length T = 30 (i.e., 1 min. scantime) from each subject, producing inputs of size 30×116. Thus, we augmented the data by a factor of ~150–250 for a total of ~14000–38000 samples per site. At test time, the predicted probability of ASD for a given subject was set to the proportion of subsequences labeled as ASD.
3.2. Experimental Methods
Models for classification of ASD vs. control were trained for each individual ABIDE site. We implemented the following LSTM-based networks which all take the ROI time-series data as input: the proposed joint discriminative/generative LSTM network (LSTM-DG); the same network but using the hidden state for both data generation and class discrimination (LSTM-H); the same network but with no generative constraint, i.e. only the discriminative loss (LSTM-D); and a single layer discriminative LSTM network as proposed in [7] (LSTM-S). Models were implemented in Keras, with 50 nodes for the first LSTM (for functional communities) and 20 nodes for the second LSTM. Optimization was performed using the Adam optimizer, with binary cross-entropy for LD, mean squared error for LG, a batch size of 32, and early stopping based on validation loss and a patience of 10 epochs. For joint discriminative/generative networks, we set λ = 0.1 so that LG and LD are on similar scales. We also implemented a traditional learning pipeline for resting-state fMRI (FC-SVM) [1]: the functional connectivity based on Pearson correlation was input to a linear support vector machine with L2 regularization, using nested cross-validation to choose the penalty hyperparameter. All implemented models were trained and tested on the augmented datasets. In addition, we compared published results for the same ABIDE datasets and AAL atlas, including another time-series modeling approach using hidden markov models (HMM) [11] and another neural network approach based on stacked autoencoders and deep transfer learning (DTL) [13].
To assess our implemented models, we used 10-fold cross-validation (CV), keeping all data from the same subject within the same partition (training, validation, or test). We measured model classification performance by computing the accuracy (ACC), true positive rate (TPR), true negative rate (TNR), and area under the receiver operating characteristic curve (AUC). Paired one-tailed t-tests were used to compare model performance over all folds and datasets.
For the generative results, with no ground truth for functional communities, we instead evaluated the robustness of extracted communities and compared a tensor decomposition approach for finding overlapping communities. For each sample, we calculated the correlation matrix of the R ROI time-series, then generated a tensor T with dimension R×R×S, where S is the number of samples. We then used non-negative PARAFAC [3] to decompose where K is the number of communities, contains the membership weight of each ROI to community contains the membership weight of each sample to community k, and ○ is the vector outer product. Similar to our approach, we set K = 50 communities and use k-means clustering to assign hard ROI memberships to each community. Then for each approach, we computed the correlation of the membership weights and the Dice similarity coefficient (DSC) of hard membership assigments between community k in fold 1 and all communities in fold f ≠ 1. The robustness of community k in fold 1 compared to fold f was measured as the maximum correlation/DSC computed in fold f. We then assessed overall community robustness between fold 1 and f using the average correlation/DSC over all communities.
We also performed validation of the functional communities in the context of the ASD classification task using Neurosynth [15], which correlates over 14000 fMRI studies with 1300 descriptors. The influence of a community for classification was denoted by the sum of absolute weights across all nodes in the second LSTM block for the discriminative task. A binary mask of the extracted ROIs for an important discriminative community was then input to Neurosynth to assess neurocognitive processes associated with ASD classification.
3.3. Classification Results
Classification results for each ABIDE site are in Tables 1 and 2. Our LSTM-DG model produced the highest accuracy for 3 of the 4 sites and second highest for US, in which the LSTM-H variation of our model (generative path from hidden state) performed best. Furthermore, LSTM-DG produced the highest or nearly highest AUC for each site. Overall, our LSTM-DG consistently outperformed all non-generative implemented models (ACC p < 0.05) and showed potential for improved classification compared to LSTM-H (ACC p = 0.08). Moreover, LSTM-DG was the only method to significantly outperform LSTM-S (ACC p = 0.04, TNR p = 0.04), the original LSTM model for fMRI classification. The results demonstrate the effectiveness of our proposed LSTM-DG method to improve classification by jointly learning the generative fMRI time-series model.
Table 1:
NY and UM Classification Results
| Model | NY (184 subjects, 42.3% ASD) | UM (143 subjects, 46.2% ASD) | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean (Std) ACC (%) | Mean (Std) TPR (%) | Mean (Std) TNR (%) | AUC | Mean (Std) ACC (%) | Mean (Std) TPR (%) | Mean (Std) TNR (%) | AUC | |
| LSTM-S [7] | 69.5 (11.0) | 52.4 (26.5) | 83.1 (12.0) | 0.720 | 69.8 (11.4) | 56.7 (24.2) | 74.0 (25.3) | 0.740 |
| FC-SVM [1] | 70.7 (8.2) | 54.8 (21.5) | 83.2 (11.8) | 0.783 | 69.2 (12.0) | 46.7 (18.9) | 89.8 (12.8) | 0.713 |
| HMM [11] | 70.6 (6.6) | 61.6 | 66.7 | 0.712 | 73.4 (10.5) | 68.5 | 76.9 | 0.738 |
| DTL [13] | - | - | - | - | 67.2 | 68.9 | 67.6 | 0.67 |
| LSTM-D | 70.7 (11.0) | 48.9 (27.1) | 86.7 (16.1) | 0.746 | 67.0 (12.0) | 52.9 (22.2) | 78.6 (25.6) | 0.738 |
| LSTM-H | 68.0 (7.7) | 52.0 (19.8) | 80.1 (10.1) | 0.779 | 69.2 (11.4) | 57.9 (14.5) | 78.7 (18.1) | 0.777 |
| LSTM-DG | 72.2 (14.7) | 57.4 (25.5) | 84.1 (12.2) | 0.772 | 74.8 (10.0) | 60.8 (12.8) | 85.6 (14.5) | 0.774 |
Table 2:
US and UC Classification Results
| Model | US (101 subjects, 57.4% ASD) | UC (99 subjects, 54.6% ASD) | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean (Std) ACC (%) | Mean (Std) TPR (%) | Mean (Std) TNR (%) | AUC | Mean (Std) ACC (%) | Mean (Std) TPR (%) | Mean (Std) TNR (%) | AUC | |
| LSTM-S [7] | 67.5 (15.4) | 79.8 (25.3) | 56.2 (41.8) | 0.659 | 62.7 (14.8) | 74.4 (31.5) | 51.5 (32.5) | 0.691 |
| FC-SVM [1] | 67.3 (13.5) | 86.2 (13.6) | 43.5 (27.6) | 0.721 | 61.7 (18.0) | 73.3 (20.6) | 47.7 (31.7) | 0.624 |
| DTL [13] | 70.4 | 72.5 | 67.0 | 0.73 | 62.3 | 55.9 | 68.0 | 0.60 |
| LSTM-D | 64.7 (17.8) | 75.3 (32.2) | 61.8 (39.6) | 0.682 | 63.6 (8.8) | 71.8 (27.3) | 51.3 (30.5) | 0.662 |
| LSTM-H | 76.4 (13.9) | 85.6 (18.0) | 65.8 (22.2) | 0.757 | 61.6 (11.4) | 66.6 (14.5) | 54.7 (18.1) | 0.705 |
| LSTM-DG | 73.2 (14.7) | 82.8 (25.5) | 61.8 (12.2) | 0.746 | 67.4 (10.0) | 67.5 (12.8) | 62.2 (14.5) | 0.715 |
3.4. Learned Functional Communities
Results for extracted communities by tensor-based community detection (CD, blue) and the proposed LSTM approach (orange) are plotted in Fig. 2. Our LSTM method produced consistently smaller communities with more uniform size compared to CD, with an average of 11 ROIs compared to 16. Furthermore, our LSTM approach consistently generated communities with higher correlation of membership weights and higher DSC of hard community assignments across CV folds for all sites, with a 15% increase in average correlation and 11% increase in average DSC. Thus, our proposed network produced smaller and more robust functional communities than CD, giving our model the potential for more reliable interpretation of further analyses on the functional communities.
Fig. 2:
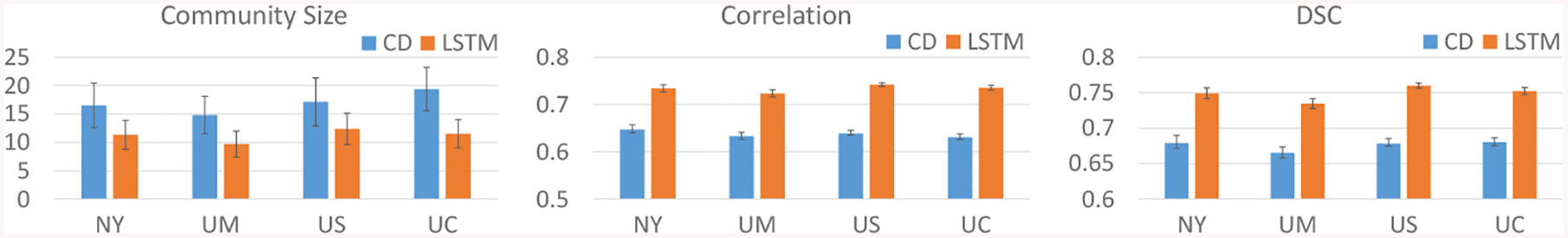
Size (left) and robustness of extracted functional communities across CV folds measured by correlation of membership weights (middle) and DSC of hard assignments (right). CD = tensor-based community detection, LSTM = proposed network.
The top 3 influential communities for the ASD classification of the largest dataset (NY) were extracted from the best CV fold and analyzed in Neurosynth. ASD is characterized by impaired social skills and communiciation; thus, we expect to find communities related to associated neurological functions. The top extracted community (Fig. 3, yellow) includes the temporal lobe and ventromedial prefrontal cortex, which are associated with social and language processes. The second community (Fig. 3, green) includes the ventromedial prefrontal cortex, hippocampus, and amygdala, which are associated with memory. The third community (Fig. 3, pink), containing the ventromedial prefrontal cortex and ventral striatum, is involved in reward processing and decision making. Dysfunction of all these brain regions and processes in ASD have previously been shown [12].
Fig. 3:
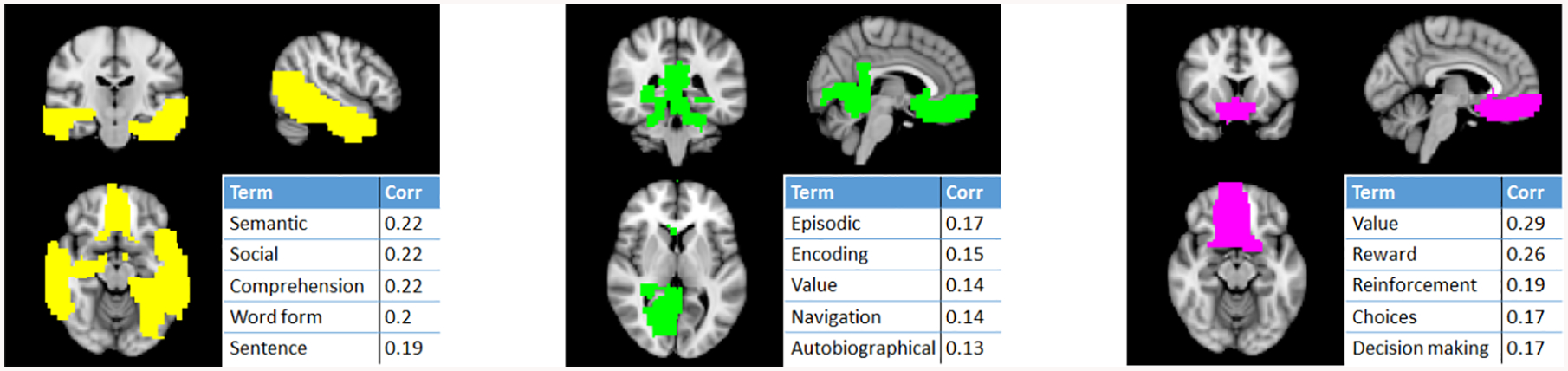
Top 3 influential communities for ASD classification of the NY dataset and the top associated neurocognitive terms from Neurosynth.
4. Conclusions
We have presented a novel RNN-based network for jointly learning a discriminative task and a generative model for fMRI time-series data. We achieved higher ASD classification performance on several datasets, demonstrating the advantage of joint learning. Finally, we showed that functional communities defined by the LSTM nodes provide robust representations of brain activity and facilitate interpretation of the ASD classification model. Understanding functional network organization will offer insights into brain disease as well as healthy cognition.
Acknowledgments
This work was supported by NIH grants R01MH100028 and R01NS035193.
References
- 1.Abraham A, Milham MP, Martino AD, Craddock RC, Samaras D, Thirion B, Varoquaux G: Deriving reproducible biomarkers from multi-site resting-state data: An autism-based example. Neuroimage 147, 736–745 (2017) [DOI] [PubMed] [Google Scholar]
- 2.Adate A, Tripathy B: S-lstm-gan: Shared recurrent neural networks with adversarial training. In: 2nd International Conference on Data Engineering and Communication Technology (2019) [Google Scholar]
- 3.Carroll J, Chang J: Analysis of individual differences in multidimensionalscaling via an n-way generalization of eckart-young decomposition. Psychometrika (1970) [Google Scholar]
- 4.Caruana R: Multitask learning. Machine Learning (1997) [Google Scholar]
- 5.Craddock C, Benhajali Y, Chu C, Chouinard F, Evans A, Jakab A, …, Bellec P: The neuro bureau preprocessing initiative: open sharing of preprocessed neuroimaging data and derivatives. In: Neuroinformatics (2013) [Google Scholar]
- 6.Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, …, Milham, M.P.: The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular Psychiatry (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dvornek NC, Ventola P, Pelphrey KA, Duncan JS: Identifying autism from resting-state fmri using long short-term memory networks. In: MLMI 2017. LNCS 10541 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Güçlü U, van Gerven MAJ: Modeling the dynamics of human brain activity with recurrent neural networks. Front Comput Neurosci (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graves A: Generating sequences with recurrent neural networks. https://arxiv.org/abs/1308.0850 (2014)
- 10.Hochreiter S, Schmidhuber J: Long short-term memory. Neural Computation (1997) [DOI] [PubMed] [Google Scholar]
- 11.Jun E, Kang E, Choi J, Suk HI: Modeling regional dynamics in low-frequency fluctuation and its application to autism spectrum disorder diagnosis. NeuroImage (2019) [DOI] [PubMed] [Google Scholar]
- 12.Kaiser M, Hudac C, Shultz S, Lee S, Cheung C, Berken A, …, Pelphrey K : Neural signatures of autism. Proc Natl Acad Sci U S A (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Parikh NA, He L: A novel transfer learning approach to enhance deep neural network classification of brain functional connectomes. Front. Neurosci (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Fan Y: Brain decoding from functional mri using long short-term memory recurrent neural networks. MICCAI 2018 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD: Large-scale automated synthesis of human functional neuroimaging data. Nature Methods (2011), www.neurosynth.org [DOI] [PMC free article] [PubMed] [Google Scholar]


