Abstract
In this study, we report the presence of the plasmid-mediated colistin resistance (PMCR)-encoding gene mcr-1 in an Escherichia coli isolate, INSali25, recovered from lettuce produced and marketed in Portugal. Colistin MIC from the vegetable E. coli isolate—determined by microdilution broth method according to EUCAST guidelines—revealed a non-wild-type phenotype of colistin (MIC 16 mg/L). To understand the genetic background of E. coli INSali25, we performed whole genome sequencing. Plasmid sequencing was also performed after plasmid DNA extraction from the transconjugant TcINSali25 (mcr-1). Directed bioinformatics analysis identified the mcr-1 gene in a 39,998 bp length contig, with an upstream region including the antibiotic resistance gene blaTEM-1 in a partial transposon Tn2, truncated by the insertion sequence IS26 and showing >99% identity with previously described mcr-1-harboring IncHI2 plasmids. Further in silico analysis showed the presence of additional genes conferring resistance to β-lactams (blaTEM-1), aminoglycosides (aadA1, aph(4)-Ia, aph(6)-Id, aac(3)-Iv), macrolides (mdf(A)-type), phenicol (floR-type), tetracycline (tetA), and sulphonamides (sul2). INSali25 isolate belonged to the ST1716 lineage and showed the fimH54 and fumC27 alleles. Lettuce is a vegetable that is commonly consumed fresh and not subjected to any cooking process, which may amplify human food safety risks. Moreover, the occurrence of plasmid-mediated colistin resistance in a sample that was not imported and was acquired in a large retail store reinforces the widespread distribution of mcr-1.
Keywords: mcr-1, colistin resistance, food, vegetables
1. Introduction
Colistin constitutes one of the few therapeutic options available for the treatment of infectious diseases caused by multidrug resistant Gram-negative bacteria [1]. However, plasmid-mediated colistin resistance (PMCR) determinants have been detected in humans and animals, and intra-species transmission of resistant isolates has already been reported [2,3]. Fresh produce has been increasingly implicated in bacterial outbreaks. Indeed, fruits and vegetables can become contaminated with antibiotic resistance bacteria before wholesale distribution [4]. Until now, three studies reported MCR-1-producing Enterobacteriaceae isolated from fresh produce, all originated from Asia. In 2016, four E. coli and two Raoultella ornithinolytica recovered from lettuce and tomato retail vegetables was reported in China [5]; other study described two E. coli isolated in Switzerland from ready-to-eat vegetables imported in 2014 from Thailand and Vietnam [6]. Recently, twenty-three MCR-1-producing E. coli and one MCR-1-producing Enterobacter cloacae were isolated from fresh vegetable samples collected between May 2017 and April 2018, in nine provinces in China [7].
Therefore, the aim of this study was to characterize a PMCR-encoding gene mcr-1 detected in an E. coli isolated from a lettuce sample in Portugal and evaluate its genetic relation with the other reported MCR-1-producing E. coli isolated from fresh produce.
2. Materials and Methods
2.1. Bacterial Isolate
In the scope of the analysis of the antibiotic nonsusceptibility of a collection of Gram-negative isolates recovered from fresh fruits and vegetables, we identified the presence of the PMCR-encoding gene mcr-1 in E. coli INSali25 isolate, which was recovered from lettuce acquired at a retail store [8]. This lettuce, produced in conventional agriculture and marketed in Portugal, was purchased at a retail store in the region of Lisbon and Tagus Valley [8].
2.2. Antimicrobial Susceptibility Testing
Minimum inhibitory concentrations (MICs) were determined by microdilution methods as previously described [9] and interpreted according to European Committee of Antimicrobial Susceptibility Testing (EUCAST, http://www.eucast.org/).
2.3. Transfer Experiments
Conjugation experiments were performed using sodium azide-resistant E. coli J53 as a recipient strain [9]. Transconjugants were selected on McConkey agar supplemented with sodium azide (150 mg/L) and colistin (2 mg/L).
2.4. Genetic Background of E. coli INSali25
To understand the genetic background of E. coli INSali25, we performed whole genome sequencing (WGS) (MiSeq, IlluminaInc, San Diego, CA, USA) and bioinformatics analyses, as previously described [8]. This Whole Genome Shotgun (WGS) project was deposited at DDBJ/ENA/GenBank under the accession LSRK00000000. The version described in this paper is version LSRK02000000.
Plasmid sequencing was also performed on a MiSeq Illumina platform using 150 bp paired-end reads, after plasmid DNA extraction from TcINSali25 (mcr-1) using a NucleoBond Xtra Plus kit (Macherey-Nagel, Dueren, Germany). Plasmid sequence reads were trimmed and filtered according to quality criteria and were mapped back to the contigs generated by WGS—which were used as references by means of CLC Genomics Workbench 10.0 (Qiagen, Aarhus, Denmark). Briefly, the raw FASTQ reads were first processed by quality score trimming (quality score limit = 0.05), removing all reads containing more than 2 ambiguous nucleotides or shorter than 50 bp. Trimmed reads were then de novo assembled with automatic bubble, word size and paired distance detection, using mapping mode, “map reads back to contigs” (including scaffolding and minimum contig length of 400 nucleotides).
Twenty contigs (coverage ≥ 10× and length ≥ 500 bp) were obtained, which were BLAST searched against the Microbial Nucleotide BLAST database for complete plasmids. By mapping against plasmid hits, we were able to obtain a larger contig containing the mcr-1 gene (with 39,988 bp, including contigs 106, 119 and 136). Contigs were analyzed, molecular typed, and studied for the presence of antibiotic resistance and plasmid replicon types, using publicly available Web tools hosted by the Center for Genomic Epidemiology (CGE, https://cge.cbs.dtu.dk/services/).
2.5. Genomic Epidemiological Analysis
The BacWGSTdb database was used for genotyping, source tracking bacterial pathogens, and prediction of closely related plasmids [10].
3. Results and Discussion
Colistin MIC of the vegetable E. coli isolate revealed a non-wild-type phenotype to colistin (MIC 16 mg/L). This isolate was also resistant to other antibiotic classes, such as penicillins, quinolones, aminoglycosides, and phenicols, consistent with a multidrug resistant phenotype. The transferability of the mcr-1 gene was achieved, with the transconjugant TcINSali25 (mcr-1) exhibiting the respective resistance to colistin (4 mg/L).
In silico analysis revealed the presence of additional genes conferring resistance to β-lactams (blaTEM-1), aminoglycosides (aadA1, aph(4)-Ia, aph(6)-Id, aac(3)-Iv), macrolides (mdf(A)-type), phenicol (floR-type), tetracycline (tetA), and sulphonamides (sul2). Furthermore, we found a chromosomal point mutation in the gyrA gene (S83L) which is responsible for resistance to fluoroquinolones. One virulence factor (gap-type) was identified. Moreover, this E. coli isolate displayed a prediction of 93.4% for being a human pathogen, based on the probability scores assigned by PathogenFinder. The MCR-1-producing INSali25 E. coli isolate belonged to the ST1716 lineage and showed the fimH54 and fumC27 alleles using CHTyper. This ST was encountered worldwide mainly in isolates collected from livestock samples (http://enterobase.warwick.ac.uk/species/ecoli/).
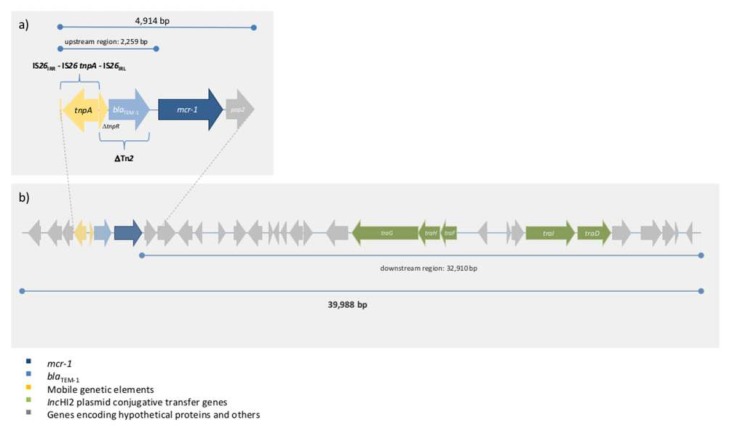
Further bioinformatics analysis directed to the flanking regions of the mcr-1 gene identified, upstream of the mcr-pap2 element, a 2259 bp fragment including the antibiotic resistance gene blaTEM-1 in a partial transposon Tn2 truncated by the insertion sequence IS26 (Figure 1a). The downstream region (32,910 bp) included multiple orfs, among which we highlight the presence of genes encoding plasmid conjugative transfer proteins (Figure 1b). Indeed, in silico plasmid detection and typing analyses revealed the presence of the incompatibility group HI2 (IncHI2) ST4, which has been associated with a wide dissemination of mcr-1 gene [11].
Figure 1.
Schematic representation of mcr-1 gene flanking regions (a) mcr-pap2 element and upstream region including the antibiotic resistance gene blaTEM-1. (b) contig region enclosing mcr-1 gene.
Alignment of pINSali25-MCR contig to complete plasmids from Microbial Nucleotide BLAST database showed >99.6% identity with previously described mcr-1-harboring IncHI2 plasmids (Table 1). Indeed, the top six hits included IncHI2-type mcr-1-harboring plasmids found in isolates collected from animals, humans, and sewage in various countries. Remarkably, p1rc4-mcr1 (NZ_CM008266) from an E. coli isolated from the stool of a Hajj pilgrim returning from a pilgrimage from Saudi Arabia to France [12], showed >99.99% similarity to pINSali25-MCR, highlighting the importance of travelers in the spread of multidrug-resistant bacteria. However, no genetic relation was found between E. coli INSali25 and other reported MCR-1-producing E. coli isolated from fresh produce since all belonged to different MLST and/or plasmid types (Table 2). Furthermore, no E. coli INSali25 closely related isolates were detected among those currently deposited in the public database BacWGSTdb [10].
Table 1.
Comparison of mcr-1.1-harboring INSali25 contig with the top six IncHI2-type mcr-1-harboring plasmids showing the highest identities (>99.6%, E-value 0.0, query coverage >94.0%).
| IncHI2-Type Plasmid (bp) | Strain (MLST 1) |
Isolation Source/ Country/Year |
Identity(%) | Alignment Length (bp) | Antimicrobial Resistance Gene 2 | Plasmid GenBank Acc. No. |
|---|---|---|---|---|---|---|
|
pSE08-00436-1 (264914) |
S. enterica 08-00436 (ST28) |
Chicken skin/Germany/2008 | 99.997 | 32115 | aadA1, aadA2-type, aph(3’)-Ia-type, aph(3’’)-Ib-type, aph(6)-Id, aac(3)-IIa, blaTEM-1B, mcr-1.1, cml-type, catA1-type, sul1, sul2, sul3, tet(A), dfrA1-type | NZ_CP020493.1 |
|
p1rc4-mcr1 (239098) |
E. coli 1RC4 (ST155) |
Hajj pilgrim Stool/France/2014 | 99.994 | 32115 | aadA1, aadA12-type, aph(3’)-Ia, aph(3’’)-Ib, aph(6)-Id, blaTEM-1B, mcr-1.1, mph(A), cml-type, floR-type, sul3-type, tet(A), dfrA4 | NZ_CM008266.1 |
|
pSA186_MCR1 (241600) |
E. coli SA186 (ST131) |
Human patient urine/ Saudi Arabia (Riyadh)/2012 | 99.993 | 30522 | aadA1, aadA2-type, aph(3’)-Ia, aph(3’’)-Ib, aph(6)-Id, blaTEM-1B, mcr-1.1, mph(A), cml-type, floR-type, sul3-type, tet(A), dfrA14 | NZ_CP022735.1 |
|
pRS571-MCR-1.1 (257270) |
E. coli RS571 (ST648) |
rectal swab/Bangladesh: Dhaka/2018 | 99.993 | 29648 | aadA1, aadA2-type, aac(3)-IId-type, aph(3’)-Ia, aph(3’’)-Ib, aph(6)-Id, blaTEM-1B, mcr-1.1, mph(A), cml-type, floR-type, sul3-type, tet(A), dfrA14 | NZ_CP034390.1 |
|
pG3X16-2-2 (265575) |
E. coli G3X16-2 (ST1196) |
Human feces/China: Guangxi/2017 | 99.977 | 30043 | aadA22, aph(3’)-Ia-type, aph(3’’)-Ib, aph(6)-Id, aph(4)-Ia-type, aac(3)-IV, blaTEM-1B, blaCTX-M-65, mcr-1.1, oqxAB-type, Inu(F)-type, mph(A), floR-type, sul1-type, sul2, tet(A)-type, tet(M)-type | NZ_CP038139.1 |
|
pMCR1_025943 (265538) |
E. coli WCHEC025943 (ST410) |
Sewage/China: Sichuan, Chengdu/2017 | 99.977 | 30043 | aadA22, aph(3’)-Ia-type, aph(3’’)-Ib, aph(6)-Id, aph(4)-Ia-type, aac(3)-IV, blaTEM-1B, blaCTX-M-65, mcr-1.1, oqxAB-type, Inu(F)-type, mph(A), floR-type, sul1, sul2, tet(A), tet(M)-type | NZ_CP027202.2 |
1 MLST accordingly with Warwick scheme (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli); 2 ResFinder-3.1 (Selected % ID threshold: 90%; Selected minimum length: 60%).
Table 2.
Comparison of MCR-1-producing E. coli strains isolated from fresh produce.
|
E. coli Strain (MLST a) |
Isolation Source/Country/Year | Antimicrobial Resistance Genes b | Plasmid (bp) | Plasmid Type | Accession Number | Reference |
|---|---|---|---|---|---|---|
|
INSali25 (ST1716) |
Lettuce/Portugal/2015 | aadA1, aac(3)-Iv, aph(4)-Ia,aph(6)-Ia, aph(6)-Id, blaTEM-1B, mcr-1.1, sul2, tet(A), floR-type | pINSali25-MCR (≈250000) |
IncHI2/ST4 | LSRK00000000 | [8]; This study |
|
CTX148 (untypable) |
tomato | mcr-1.1 | pT-CTX148 (57764) |
IncI2 | MK754161 | [7] |
|
TO89 (ST713) |
Tomato/China: Huimin/2017 | mcr-1.1 | pT-89 (∼33) |
IncX4 | SRMK00000000 | [7] |
|
SQB-1-1 (ST2705) |
Romaine lettuce/China: Qingdao/2017 | aph(3’)-Ia, aac(3)-IV-type, aph(4)-Ia, aadA1, aadA2-type, blaCTX-M-14, mcr-1.1, fosA3, mdf(A)-type, mph(A), cmlA1-type, floR-type, tet(M)-type, sul2, sul3 | pSQB-1-1 (≈250000) |
IncHI2/ST3 | SRML00000000 | [7] |
|
H226B (ST167) |
Cha-om imported from Thailand/2014 | mcr-1.1 | pH226B (209401) |
IncHI1 | KX129784 | [6,14] |
|
2SK1 (ST4683) |
Basil leaves imported from Vietnam/2014 | mcr-1, blaCTX-M-65 | - | - | - | [14] |
|
HS20eCTX (ST795) |
Lettuce/China: Guangzhou/2016 | aph(3’)-Ia, aac(3)-IV-type, aph(4)-Ia, aadA1, aadA2-type, blaCTX-M-14, mcr-1.1, fosA3, cmlA1-type, floR-type, sul2, sul3 | pHNHS20EC (250827) |
IncHI2/ST3 | MF135536 | [5] |
|
BS21Ectx (ST2505) |
Lettuce/China: Guangzhou/2016 | mcr-1 | - (∼60) |
IncI2 | - | [5] |
|
6BF21eCTX (ST69) |
Tomato/China: Guangzhou/2016 | mcr-1, bla CTX-M-14 , floR, fosA3, oqxAB | - (∼244) |
IncHI2/ST3 | - | [5] |
|
TS62CTX (ST156) |
Lettuce/China: Guangzhou/2015 | mcr-1 | - (∼60) |
IncI2 | - | [5] |
|
6HS20E (ST48) |
Lettuce/China: Guangzhou/2016 | mcr-1 | - (∼33) |
IncX4 | - | [5] |
a MLST accordingly with Warwick scheme (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli); b Results from ResFinder-3.1 (Selected %ID threshold: 90%; Selected minimum length: 60%) using available plasmid accession numbers; otherwise, results are those from references. - Data not available.
Despite previous reports of food products from animal origin, lettuce is a vegetable that is commonly consumed fresh and not subjected to any cooking process, which severely amplifies the human food safety risks involved [13]. Considering that colistin is a last-resource antibiotic used for the treatment of infections caused by multidrug resistant bacteria, the detection of a mobile colistin resistance gene in a raw vegetable constitutes a serious and unprecedented public health concern [1]. Indeed, the occurrence of plasmid-mediated colistin resistance in a sample that was produced in Portugal and that was acquired in a large retail store reinforces the widespread distribution of mcr-1, and the need to promote a global and concerted strategy to contain the spread of this resistance mechanism.
Author Contributions
V.M. performed bioinformatics analysis, analyzed the data, edited and reviewed the manuscript; D.J.-D. performed experiments, analyzed the data and wrote the original draft; E.F. performed microbiological experiments; M.C. analyzed the data, reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by UIDB/00211/2020 with funding from FCT/MCTES through national funds.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Paterson D.L., Harris P.N. Colistin resistance: A major breach in our last line of defence. Lancet Infect. Dis. 2016;16:132–133. doi: 10.1016/S1473-3099(15)00463-6. [DOI] [PubMed] [Google Scholar]
- 2.Nordmann P., Poirel L. Plasmid-mediated colistin resistance: An additional antibiotic resistance menace. Clin. Microbiol. Infect. 2016;22:398–400. doi: 10.1016/j.cmi.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Sun J., Zhang H., Liu Y.-H., Feng Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018;26:794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Hölzel C.S., Tetens J.L., Schwaiger K. Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: A need for quantitative risk assessment. Foodborne Pathog. Dis. 2018;15:671–688. doi: 10.1089/fpd.2018.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo J., Yao X., Lv L., Doi Y., Huang X., Huang S., Liu J.H. Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli isolates from retail vegetables in China. Antimicrob. Agents Chemother. 2017;61:e01139-17. doi: 10.1128/AAC.01139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zurfuh K., Poirel L., Nordmann P., Nüesch-Inderbinen M., Hächler H., Stephan R. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-β-lactamase-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob. Agents Chemother. 2016;60:2594–2595. doi: 10.1128/AAC.00066-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B.T., Li X., Zhang Q., Shan H., Zou M., Song F.J. Colistin-resistant mcr-positive Enterobacteriaceae in fresh vegetables, an increasing infectious threat in China. Int. J. Antimicrob. Agents. 2019;54:89–94. doi: 10.1016/j.ijantimicag.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Jones-Dias D., Manageiro V., Ferreira E., Barreiro P., Vieira L., Moura I.B., Caniça M. Architecture of class 1, 2, and 3 integrons from Gram negative bacteria recovered among fruits and vegetables. Front. Microbiol. 2016;7:1400. doi: 10.3389/fmicb.2016.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manageiro V., Clemente L., Graça R., Correia I., Albuquerque T., Ferreira E., Caniça M. New insights into resistance to colistin and third-generation cephalosporins of Escherichia coli in poultry, Portugal: Novel blaCTX-M-166 and blaESAC genes. Int. J. Food Microbiol. 2017;263:67–73. doi: 10.1016/j.ijfoodmicro.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Ruan Z., Feng Y. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 2016;44:D682–D687. doi: 10.1093/nar/gkv1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veldman K., van Essen-Zandbergen A., Rapallini M., Wit B., Heymans R., van Pelt W., Mevius D. Location of colistin resistance gene mcr-1 in Enterobacteriaceae from livestock and meat. J. Antimicrob. Chemother. 2016;71:2340–2342. doi: 10.1093/jac/dkw181. [DOI] [PubMed] [Google Scholar]
- 12.Hadjadj L., Riziki T., Zhu Y., Li J., Diene S.M., Rolain J.M. Study of mcr-1 gene-mediated colistin resistance in Enterobacteriaceae isolated from humans and animals in different countries. Genes (Basel) 2017;8:394. doi: 10.3390/genes8120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger C.N., Sodha S.V., Shaw R.K., Griffin P.M., Pink D., Hand P., Frankel G. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 2010;12:2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x. [DOI] [PubMed] [Google Scholar]
- 14.Zurfluh K., Klumpp J., Nüesch-Inderbinen M., Stephan R. Full-length nucleotide sequences of mcr-1-harboring plasmids isolated from extended-spectrum-β-lactamase-producing Escherichia coli isolates of different origins. Antimicrob. Agents Chemother. 2016;60:5589–5591. doi: 10.1128/AAC.00935-16. [DOI] [PMC free article] [PubMed] [Google Scholar]