Short abstract
The current study was conducted to investigate the role of long non-coding RNA PVT1 in hyperglycemia-triggered human osteoarthritis (OA) chondrocytes. Cartilage from knee OA patients with and without diabetes, as well as normal cartilage, was obtained. Isolated human chondrocytes were treated with 30 nM of Glc with or without pioglitazone. The expression levels of PVT1, miR-26b, and type II collagen were determined by RT-PCR. Type II collagen was detected by immunocytochemistry and chondrocytes were stained with Alcian blue. Moreover, the interaction among PVT1, miR-26b, and CTGF was examined using bioinformatics, FISH, RIP, RNA-pull down, and luciferase reporter assays. Over-expression of PVT1 and miR-26b were performed and expressions of CTGF, TGF-β1, SMAD3, MMP-13, and type II collagen proteins were examined. Significantly higher expression of PVT1 was observed in diabetic OA. High Glc induced the elevated expression of PVT1, CTGF, TGF-β1, IL-6, and MMP-13, as well as decreased expression of type II collagen and miR-26b. These alterations could be reversed by pioglitazone. PVT1 acted as a sponge for miR-26b to facilitate CTGF expression. Over-expression of PVT1 increased the expressions of CTGF, TGF-β1, SMAD3, and MMP-13 and decreased expression of type II collagen. Our findings confirmed that PVT1 is involved in the hyperglycemia-induced collagen degradation, via the miR-26b-CTGF-TGF-β1-axis.
Keywords: Diabetic osteoarthritis, long non-coding RNA PVT1, connective tissue growth factor, competing endogenous RNA
Introduction
Osteoarthritis (OA) is a common chronic joint disease characterized by gradual irreversible loss of articular cartilage, breakdown of subchondral bone, and synovium inflammation.1 OA mainly affects mass-bearing joints, especially the knee joints, and results in pain and motion restriction.2 It is estimated that 9.6% of men and 18.0% of women worldwide aged above 60 yr suffer from symptomatic OA.3 It is expected to be the fourth leading cause of disability by 2020, and so it represents a major public health challenge in the near future.4
In recent years, OA has been identified to have a strong positive association with Glc metabolism disruption and diabetes mellitus (DM).5,6 Several studies have found that patients with type 2 DM have accelerated cartilage matrix degeneration and meniscus lesions in comparison diabetes-free individuals.7,8 In addition, our previous study demonstrated that expressions of matrix metalloproteinases (MMPs) were significantly elevated in type 2 DM than those in diabetes-free OA patients and healthy controls.9 Also, the phenomenon that high concentration of Glc affects chondrocytes and cartilage metabolism has also been observed in experimental studies. One study found that 25 nM Glc attenuated proteoglycan synthesis induced by IGF-1 in rabbit chondrocytes.10 Rosa et al. also indicated that high Glc induced initiation of the catalytic alternations through inducing higher expressions MMP-1 and MMP-13 in human chondrocytes.11 In a recent study, hyperglycemia was found to result in cartilage damage in diabetic mice that could be reversed by pioglitazone.12
Long noncoding RNA (lncRNA) is a type of RNA greater than 200 nucleotides in length acting as non-protein coding RNA, whereas microRNAs (miRNAs) comprise a large family of single-stranded, small, non-coding RNAs with a sequence length of 19–23 nucleotides.13 LncRNA and miRNA dys-regulation are associated with aberrant biological processes promoting a variety of diseases.13 With the development of bioinformatics and functional genomics tools, abnormal lncRNA and miRNA expressions were discovered in OA.14,15 However, the possible roles of lncRNA and miRNA in diabetic OA still remain unknown.
The lncRNA plasmacytoma variant translocation 1 (PVT1) is located at 8q24.21.16 PVT1 was originally identified as a common retroviral integration site in murine leukemia.17 PVT1 has been identified to play important roles in many diseases including various cancers,18 liver fibrosis,19 and sepsis,20 among others. Moreover, studies have also found that PVT1 expression was associated with clinicopathological characteristics in many diseases.
miR-26b is located in the human chromosome 2q35 and in the fourth intron of the C-terminal domain small phosphatase 1 (CTDSP1) gene.21 Different from PVT1, many studies have shown that miR-26b plays an inhibiting role in the development and progression of many cancers and liver fibrosis.22,23 However, studies on PVT1 and miR-26b on diabetic OA are rare.
Connective tissue growth factor (CTGF), also known as cellular communication network factor 2 (CCN2), is a cysteine-rich matricellular protein involved in the control of biological processes, such as cell proliferation, differentiation, adhesion, and angiogenesis, as well as multiple pathologies including OA.24 CTGF is found to be up-regulated in adjacent areas of cartilage surface damage in OA as well as in synovial fluid of OA that stimulates the production of inflammatory cytokines.25 CTGF also activates NF-κB and increases the production of chemokines and cytokines that leading to the reduction of proteoglycan contents in joint cartilage.26 Furthermore, CTGF could subsequently control TGF-β signaling,27 which plays pivotal roles in the development of OA.28 Altered TGF-β signaling is frequently observed in patients with OA,29 and inhibition of TGF-β signaling is considered to be a promising therapeutic target for this disease.30
In the present study, we hypothesized that LncRNA PVT1 might act as a competing endogenous RNA (ceRNA) by sponging miR-26b to abolish the inhibitory effect of miR-26b on CTGF, thereby activating the TGF-β1 signaling pathway and then promoting diabetic OA. To address this, we detected PVT1 expression in human OA cartilage with or without diabetes as well as in human chondrocytes. Furthermore, we investigated the interaction between PVT1, miR-26b, and CGTF. Moreover, we also explored the role of the PVT1 in regulating CGTF/TGF-β axis in hyperglycemia treated human chondrocytes.
Materials and methods
Cartilage specimen collection and ethics statement
Articular cartilage was obtained from primary OA patients undergoing total knee replacement surgery (n = 40, 20 males and 20 females, aged 50–70 yr), including 20 patients with diabetes and 20 patients without diabetes. Normal cartilage samples were obtained from 15 donors (8 males and 7 females, aged 52–67 yr) following trauma or death. All samples of cartilage were collected from knees and stored at −80°C after promptly frozen by liquid nitrogen. There were no significant differences regarding age and sex distribution among the control and OA groups. Informed consents were obtained from all patients or their relatives prior to specimen collection. All protocols were carried out based on the Declaration of Helsinki, and the experiments were approved by the Ethics Committee of our hospital.
Isolation and culture of primary chondrocytes
To prepare the primary chondrocytes, the collected cartilage was minced and pre-treated with trypsin for 10 min. Then, the tissue slices were digested overnight (>15 h) with collagenase II in DMEM medium containing 10% FCS. The isolated cells were then passed through a filter to remove the residual cartilage matrix fragments, followed by centrifugation at 2000 g for 5 min. Afterward, cells were re-suspended in DMEM medium supplemented with 10% FCS and antibiotics consisting of 100 U/ml penicillin and 100 μg/ml streptomycin. During experiments, the cells were subsequently cultured in Ham’s F-12 (regular Glc medium, RGM, 10 mM Glc) or in the same medium supplemented with D-Glc to yield a final Glc concentration of 30 mM (high Glc medium, HGM).
Quantitative RT-PCR assay
Total RNA was extracted from the cartilage by using TRIzol reagent (Invitrogen, US). The cDNA Synthesis Kit (Takara, China) was used for the synthesis of cDNA according to the manufacturer’s instructions. RNA was then reverse transcribed into cDNAs using the Reverse Transcription System Kit (Takara, China). The cDNA templates were amplified by qRT-PCR using SYBR Green PCR Mix (TaKaRa) with the following primers: PVT1-F, 5′-TGAGAACTGTCCTTA CGTGACC -3′; PVT1-R, 5′-AGAGCACCAAGACT GGCTCT-3′; miR-26b-F, 5′-CCGGGACCCAGTTC AAGTAA-3′, R,5′-CCCCGAGCCAAGTAATGGA G-3′; COL2A1-F, 5′-ATGACAATCTGGCTCCCAA CACTGC-3′, R-5′-GACCGGCCCTATGTCCACAC CGAAT-3′; U6-F, 5′-CTCGCTTCGGCAGCACA-3′, R,5′- AACGCTTCACGAATTTGCGT-3′; GAPDH-F, 5′-ACCACAGTCCATGCCATCAC-3′, R,5′-TCCACC ACCCTGTTGCTGTA-3′. Comparative quantification was determined using the 2−ΔΔCt method. Expressions of PVT1 and miR-26b were normalized to U6 small nuclear RNA, whereas COL2A1 was normalized to GAPDH. The qRT-PCR results were analyzed and expressed as relative mRNA levels of the CT value, which was then converted into fold-change.
Measurement of CGTF, TGF-β1, SMAD3, and MMP-13
Human chondrocytes (1 × 106/ml) were cultured in six-well plates in Ham’s F-12 medium with 10% FBS. Chondrocytes (> 85% confluent) were cultured in regular Glc medium or high Glc medium in the presence or absence of pioglitazone (1, 10, or 50 mM). The levels of CGTF, TGF-β1, IL-6, and MMP-13 in culture media were quantified by using the commercially available CGTF, TGF-β1, IL-6, and MMP-13 specific ELISA kits according to the manufacturer’s instructions (R&D Systems, USA) at 24 h.
Alcian blue staining
Cells were washed three times with PBS and fixed with 4% PFA for 30 min. Subsequently, cells were incubated with 0.1 mol/l hydrochloric acid solution until its pH decreases to 1.0, followed by 1% Alcian blue staining overnight. Finally, cells were washed with 0.1 mol/l hydrochloric acid solution. Depositions were observed and captured under an inverted microscope.
Immunocytochemistry of type II collagen
The cells were fixed with 4% paraformaldehyde, dehydrated and paraffin-embedded as described above. The sections were then incubated with blocking solution [PBS/10% goat serum (Beijing Biosynthesis Biotechnology Co., Ltd., Beijing, China)/0.1% Triton X-100] for 30 min, and were then incubated with monoclonal rabbit anti-human Abs against collagen type II (1:200; Beijing Biosynthesis Biotechnology Co., Ltd.) for 2 h at room temperature (25°C) and washed with PBS three times. Subsequently, the sections were incubated with HRP-conjugated secondary Ab for 1 h at room temperature (1:1,000; ProteinTech Group, Inc.) and washed with PBS three times. Subsequently, the sections were incubated with DAB solution for 30 min at room temperature and washed with deionized water for 5 min. Finally, the sections were stained with hematoxylin to identify the nuclei. Images were captured.
Luciferase reporter assay
The CTGF mutant 3′‑UTR was generated by replacing the seed regions of the miR‑26b binding sites with 5′‑TTGGTT‑3′ and PVT1 mutant was generated using site-directed mutagenesis. Subsequently, the mutant sequence was cloned into the firefly luciferase‑expressing vector pGL3 (Shanghai GenePharma Co., Ltd.). As for luciferase assay, the chondrocytes were seeded in 24‑well plates at 4 × 104 cells/well the day before transfection and transfected with the CTGF wild type or mutant 3′‑UTR reporter vector (Shanghai GenePharma Co., Ltd.), PVT1 or PVT1 mutant using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were harvested and lysed 48 h after transfection and the luciferase activity was assayed using the Dual‑Luciferase Reporter system (Promega Corporation, Madison, WI, USA). The β‑lactamase gene of the pGL3 luciferase vector was used for the normalization of the luminescence levels. Three independent experiments were performed.
Fluorescence in situ hybridization
DNA oligo probes (GenePharma) labeled with Cy5 for LncPVT1 (50-GGTGCCATC GGAAACCCTGGAT ATTGCAGACA-30-Cy5) and FAM for miR-26b (50-AC ACAGCTCTTCCATATCTCCAG-30-FAM) were utilized in the fluorescence in situ hybridization (FISH) assays, whereas the nuclei were counterstained with 4,6-diamidino- 2-phenylindole (DAPI). All procedures were carried out on the basis of the manufacturer’s instructions (GenePharma), and all images were acquired using a Leica SP5 confocal microscope (Leica Microsystems, Mannheim, Germany).
RNA immunoprecipitation
RNA immunoprecipitation (RIP) was performed to investigate whether PVT1 and miR-26b were in the same RNA-induced silencing complex (RISC) complex. Briefly, cell lysates of human chondrocytes were incubated with human anti-CTGF (Ago2) or anti-IgG (Millipore) overnight at 4°C. Normal IgG was used as a negative control. RNA-protein complexes were immunoprecipitated with protein A agarose beads and RNA was extracted by using TRIzol (Invitrogen). The resulting purified RNA was subjected to qRT-PCR analysis.
Biotin-coupled probe RNA pull down assay
To pull down the miRNA by LncRNA, human chondrocytes transfected with miR-26b mimics were lysed and incubated with biotin-coupled probe of LncRNA PVT1 which was pre-bound on magnetic beads. For 2 h, target RNA was pulled by the RNeasy Mini Kit (QIAGEN, Germany). Biotin-coupled probe of miR-26b were processed through the same protocol.
Synthesis and transfection of interfering RNA
We last explored the potential mechanisms of PVT1 and miR-26b in diabetic OA through over-expressing PVT1 and miR-26b over-expressions. The full-length PVT1 cDNA fragments were cloned into the pcDNA 3.1 plasmid (Invitrogen, USA), generating pcDNA3.1-PVT1.An empty pcDNA3.1 vector was used as the control. The human chondrocytes were transfected with the respective constructs using Lipofectamine™ 2000 (Invitrogen, USA), following the manufacturer’s instructions. A miR-26b mimic (Ribobio, Guangzhou, China) was also constructed to be transfected into chondrocytes. After transfection for 48 h, the human chondrocytes were harvested for qRT-PCR and western blot analysis to examine knockdown or over-expression efficiency.
Western blot analysis
Total protein was extracted from chondrocytes using a RIPA lysis buffer kit (Santa Cruz, TX, USA). Protein lysates were separated on 10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore Corp. Billerica, MA, USA). After being blocked in 5% fat-free milk overnight at 4°C, the membranes were then incubated with the following primary Abs: MMP-13 (1:1000, Abcam, UK), TGF-β1 (1:1000, Abcam), SMAD3 (1:1000, Abcam), CTGF (1:1000, Abcam) and COL2A1 (1:1000, Abcam) at 4°C overnight. Then the membranes were incubated with the secondary Ab IgG at room temperature for 1 h. The band intensity was quantified with software Image J 6.0. GAPDH was used as the loading control.
Statistical methods
GraphPad 6.0 statistical software was used for statistical analysis. The significance was determined with the Student’s t test or one-way ANOVA. The least significant difference (LSD) test was used for post-hoc analysis. The measurement data were represented as mean ± SD. P < 0.05 indicated statistically significant differences.
Results
PVT1 was up-regulated in diabetic OA cartilage and negatively correlated with miR-26b and type II collagen
The expressions of PVT1 in cartilage tissues and cultured chondrocytes were analyzed by qRT-PCR. qRT-PCR analysis demonstrated that PVT1 was expressed at higher levels in diabetic OA cartilage than non-diabetic knee OA cartilage (P < 0.01 vs. non-diabetic OA) and normal cartilage (P < 0.01 vs. normal control) (Figure 1a). On the other hand, miR-26b was expressed at lower levels in diabetic OA cartilage than non-diabetic knee OA cartilage (P < 0.05 vs. non-diabetic OA) and normal cartilage (P < 0.01 vs. normal control) (Figure 1b). PVT1 expressions in diabetic OA cartilage were significantly and negatively correlated with miR-26b (r = −0.524, P = 0.018) (Figure 1c) and COL2AI expression (Figure 1e). However, PVT1 expressions in diabetic OA cartilage were not significantly associated with miR-26b (r = −0.104, P = 0.662) (Figure 1d) and COL2AI expressions (r = −0.283, P = 0.226) (Figure 1f).
Figure 1.
(a) Comparison of PVT1 expression among diabetic OA, non-diabetic OA, and normal control cartilage. (b) Comparison of miR-26b expression among diabetic OA, non-diabetic OA, and normal control cartilage. (c) Correlation of PVT1 expression with miR-26b expression in diabetic OA cartilage. (d) Correlation of PVT1 expression with miR-26b expression in non-diabetic OA cartilage. (e) Correlation of PVT1 expression with type II collagen expression in diabetic OA cartilage. (f) Correlation of PVT1 expression with type II collagen expression in non-diabetic OA cartilage.
Effects of high Glc and pioglitazone on PVT1, miR-26b expressions, chondrocyte properties, and type II collagen in HGM-induced human chondrocytes
We first explored the effects of pioglitazone on PVT1, miR-26b expressions, and type II collagen. The expression of PVT1 was time-dependently increased in human chondrocytes (Figure 2a). On the other hand, the expressions of miR-26b and type II collagen mRNA were time-dependently decreased (Figure 2b and c). As also shown in Figure 2, pioglitazone could dose dependently inhibit high Glc-induced PVT1 expression (Figure 2d). Furthermore, pioglitazone could dose dependently reverse high Glc-inhibited miR-26b and collagen II expression (Figure 2e and f). High Glc significantly decreased the formation of Alcian blue deposit (Figure 3a and b) and type II collagen expressions (Figure 3a and b), which could be reversed by pioglitazone (Figure 3(a) and (b)).
Figure 2.
Chondrocytes (1 × 106/ml) were cultured in FGM RGM and HGM for 24 h with or without pioglitazone (1, 10, and 50 mM). Cell lysates were collected. The expressions of PVT1 (a, d), miR-26b (b, e), and type II collagen (c, f) in the culture supernatants were measured by RT-PCR. Data were expressed as mean ± SD for three independent experiments. **P < 0.01 versus control; *P < 0.05 versus control; #P < 0.05 versus HGM group.
Figure 3.
(a) Effect of high Glc and pioglitazone (50 µM) on the Alcian blue staining and type II collagen. (b) Quantitative analysis of Alcian blue staining. (c) Quantitative analysis of type II collagen (**P < 0.001).
Effects of high Glc and pioglitazone on the expression of CGTF, TGF-β1, IL-6, and MMP-13 in HGM-induced human chondrocytes
We next evaluated the effect of pioglitazone on HGM-induced catalytic condition and inflammatory responses as well as the expressions of CGTF and TGF-β1 in human chondrocytes. As depicted in Figure 4, the protein expression of CGTF (Figure 4a), TGF-β1 (Figure 4b), IL-6 (Figure 4c), and MMP-13 (Figure 4d) were significantly increased in HGM-induced human chondrocytes compared with FGM and HGM. Furthermore, pioglitazone could dose dependently inhibit high Glc-induced production of CGTF, TGF-β1, IL-6, and MMP-13 (Figure 4).
Figure 4.
Chondrocytes (1 × 106/ml) were cultured in FGM RGM and HGM for 24 h with or without pioglitazone (1, 10, and 50 mM). Cell lysates were collected. The expressions of CGTF (a), TGF-β1 (b), IL-6 (c), and MMP-13 (d) in the culture supernatants were measured by ELISA. Data were expressed as mean ± SD for three independent experiments. **P < 0.01 versus control; *P < 0.05 versus control; #P < 0.05 versus HGM group.
LncRNA PVT1 abolishes the interaction of miR-26b with the targets of CTGF
We performed bioinformatics analysis (DIANA TOOLS-LncBase v.2) to predict the potential target miRNAs, and we focused on miR-26b. The predicted complementary binding sites at the 3’-UTR are shown in Figure 5a. Bioinformatics analysis also showed that CTGF were potential target genes of miR-26b (Figure 5c). As shown next, the luciferase assay confirmed that CTGF was a target of miR-26b (StarBase v 2.0) (Figure 5b). Luciferase activity assay showed that miR-26b mimic led to a notable decrease in luciferase activity in PVT1-WT reporter compared with the mimic NC group, whereas had no obvious effect on luciferase activity in PVT1-MUT reporter (Figure 5d). In order to further identify the interaction among PVT1, miR-26b, and CTGF in human chondrocytes, we next carried out RNA pull-down, RIP, and FISH assays.
Figure 5.
(a) Bioinformatics analysis of matching sequence of miR‑26b within 3′‑UTR of CTGF. MuT CTGF 3′‑UTR is the mutation of the match sequence of 3′‑UTR of CTGF with miR‑26b (StarBase v.2.0). (b) Luciferase reporter assay revealed that miR‑26b binds to the 3′‑UTR of WT CTGF, not MUT CTGF. Relative luciferase activity was quantified, and the data were presented as mean ± SD. *P < 0.05 vs. respective NC groups. 3′‑UTR, 3′‑untranslated region; CTGF, connective tissue growth factor; miR, microRNA; NC, negative control; WT, wild type; MUT, mutant. (c) The predicted binding sites between PVT1 and miR-26b (DIANA TOOLS-LncBase v.2). (d) Luciferase activity was measured in human chondrocytes co-transfected with mimic NC or miR-26b mimic and PVT1-wt or PVT1-mut reporter at 48 h after transfection. PVT1 interacted directly with miR-26b (*P < 0.05, **P < 0.01).
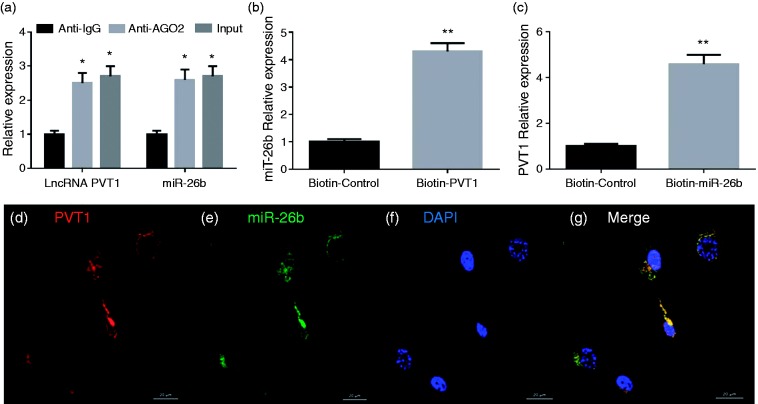
RIP assays disclosed that PVT1 and miR-26b expressions were substantially enriched by Ago2 Ab compared with control IgG Ab (Figure 6a). The biotin-coupled probe pull-down assay was then performed and the results showed miR-26b was detected in the PVT1 pulled-down pellet compared with the control group (Figure 6b). Also, PVT1 was detected in the miR-26b pulled-down pellet compared with the control group (Figure 6c). FISH technology demonstrated that PVT1 (red fluorescence) and miR-26b (green fluorescence) could be visualized in the cells, and co-localization was observed in human chondrocytes (Figure 6d to g).
Figure 6.
(a) Relative PVT1 and miR-26b expression presented as fold enrichment in Ago2 relative to normal IgG immune-precipitates. RIP assays disclosed that PVT1 and miR-26b expressions were substantially enriched by Ago2 Ab compared with control IgG Ab. (b) The biotin-coupled probe pull-down assay was performed and the results showed miR-26b was detected in the LncRNA PVT1 pulled-down pellet compared with the control group. (c) LncRNA PVT1 was detected in the biotin-miR-26b vector compared with the control group (d–g). PVT1 and miR-26b were co-localized in human chondrocytes by FISH using confocal microscope. PVT1 was stained red (d), miR-26b was stained green (e), nuclei were stained blue (DAPI) (f), and overlapped expression was mixed (g) (scale bar, 20 µm).
LncRNA PVT1 on collagen production, MMP-13, and CTGF/TGF-β1 signaling activation
Last we transfected the LncRNA PVT1 and miR-26b mimic into the human chondrocytes in high Glc medium. After 48 h, we found PVT1 over-expression further up-regulated the protein expression of MMP-3, CTGF, TGF-β1, Smad3, and decreased type collagen II expression compared with control. On the other hand, miR-26b mimic had the opposite effect. Co-transfection of pcDNA3.1-PVT1 and miR-26b mimic had no significant differences regarding these protein expressions compared with control, indicating the function of pcDNA3.1-PVT1 was attenuated by miR-26b and vice versa (Figure 7). This data demonstrated that the hyperglycemia-induced type II collagen reduction, and CTGF/TGF-β signaling activation, was facilitated by PVT1 over-expression but attenuated by miR-26b over-expression (original data are shown in Supplemental Table 1).
Figure 7.
(a) Western Blot analysis of type II collagen (b), CTGF (c), TGF-β1 (d), SMAD3 (e), and MMP-13 (f) followed by pcDNA3.1-PVT1 and miR-26b mimic after 48 h in high Glc medium.
Discussion
Our current study showed that PVT1 promoted collagen degradation through binding miR‑26b and subsequently leading to the elevated expression of CTGF and TGF-β signaling. Bioinformatics and biochemical analyses demonstrated that PVT1 was able to directly bind to miR‑26b. In addition, bioinformatics prediction and dual reporter luciferase assay revealed that miR‑26b directly targeted the CTGF 3′‑UTR. Over-expression of PVT1 increased the expressions of CTGF, TGF-β1, SMAD3, and MMP-13 and decreased expression of type II collagen. Collectively, PVT1 plays an important role in the hyperglycemia-induced collagen degradation in human chondrocytes by attenuating the miR‑26b expression level to activate CTGF/TGF-β signaling.
OA is an aging-related degenerative disease that severely influences the elders’ life quality. However, there have been few clinical approaches available so far. In recent years, authors looking to embrace a consensus of opinion related to the pathogenesis of OA in their works have been devoting increasingly more attention to the importance of a global metabolism component.31 Advances in the basic and clinical sciences over these years have substantiated the belief that OA is not merely a local problem associated with cartilage damage, but that a wider approach is needed—including a systemic angle—in view of a significant contribution of the diabetes.32 Generally, risk factors for OA included age, mass, genetics and stimuli such as growth factors and chemokines.33 However, research has pointed out that diabetes, especially type II DM, can be the only predictor for OA.34,35
LncRNAs function through chromatin regulation, histone modification, chromatin remodeling, genomic imprinting, and as ceRNAs in post-transcriptional regulation.36 The ceRNA theory hypothesized that all types of RNA transcripts could communicate with each other through a new “language” mediated by microRNA (miR/miRNA) response elements.37
The lncRNA PVT1 has been extensively studied in several types of cancer as mentioned at the beginning. Recent studies have also demonstrated that PVT1 played important roles both in OA and diabetes. Li found silencing PVT1 inhibited the apoptosis of OA chondrocytes, and over-expression of PVT1 promoted the apoptosis of normal chondrocytes.38 In addition, high expression of PVT1 was observed in OA cartilage and IL-1β-stimulated chondrocytes, and down-regulation of PVT1 expression markedly inhibited IL-1β-induced production of MMP-3, MMP-9, and MMP-13.39 On the other hand, PVT1 has also been involved in the progression of diabetic nephropathy through ECM accumulation.40 Furthermore, PVT1 is associated with end-stage renal disease in both type 1 and type 2 diabetes.41,42
miR-26b has been implicated to be a protector and found to be down-regulated during OA progression.43 miR-26b over-expression significantly decreased the histologic scores of OA and intra-articular injection of miR-26b attenuated OA progression in vivo.43 On the other hand, miR-26b was found to be down-regulated in the disease of diabetes both in human and animal studies.44
In the present study, PVT1 was revealed to be significantly up-regulated in diabetic OA in clinical cartilage tissue as well as high Glc medium. Subsequently, a loss-of-function experiment demonstrated that the over-expression of PVT1 promoted collagen degradation. Our bioinformatics analysis revealed that PVT1 harbors predictive binding sites for miR-26b. Furthermore, miR-26b was capable of targeting the CTGF that binds to TGF-β1 and can activate TGF-β1 expression.
In conclusion, our study demonstrates that in hyperglycemia condition, PVT1 promotes collagen degradation of human chondrocytes by acting as an endogenous sponge of miR-26b, and further activating CTGF/TGF-β1 signaling. There still remains a need to further seek the intensified mechanisms responsible for LncRNA PVT1 involved in diabetes-induced OA, thus developing new and improved therapeutic strategies for this chronic condition.
Supplemental Material
Supplemental material, INI881778 Supplemental Material for Long non-coding RNA PVT1, a molecular sponge of miR-26b, is involved in the progression of hyperglycemia-induced collagen degradation in human chondrocytes by targeting CTGF/TGF-β signal ways by Luo-Bin Ding, Yao Li, Guang-Yuan Liu, Tai-Hang Li, Feng Li, Jian Guan and Hua-Jun Wang in Innate Immunity
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Natural Science Foundation of Guangdong Province (Grant Number 2016A030313100), the National Natural Science Foundation of China (Grant Number 81601219), and the China Postdoctoral Science Foundation (Grant Numbers 2015M582480 and 2017T100660).
References
- 1.Hermann W, Lambova S, Muller-Ladner U. Current treatment options for osteoarthritis. Curr Rheumatol Rev 2018; 14: 108–116. [DOI] [PubMed] [Google Scholar]
- 2.Bay-Jensen AC, Hoegh-Madsen S, Dam E, et al. Which elements are involved in reversible and irreversible cartilage degradation in osteoarthritis? Rheumatol Int 2010; 30: 435–442, [DOI] [PubMed] [Google Scholar]
- 3.Hunter DJ, Felson DT. Osteoarthritis. BMJ Clin Res 2006; 332: 639–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martel-Pelletier J, Barr AJ, Cicuttini FM, et al. Osteoarthritis. Nat Rev Dis Primers 2016; 2: 16072. [DOI] [PubMed] [Google Scholar]
- 5.Courties A, Sellam J. Osteoarthritis and type 2 diabetes mellitus: what are the links? Diabetes Res Clin Pract 2016; 122: 198–206. [DOI] [PubMed] [Google Scholar]
- 6.Williams MF, London DA, Husni EM, et al. Type 2 diabetes and osteoarthritis: a systematic review and meta-analysis. J Diabetes Complications 2016; 30: 944–950. [DOI] [PubMed] [Google Scholar]
- 7.Neumann J, Hofmann FC, Heilmeier U, et al. Type 2 diabetes patients have accelerated cartilage matrix degeneration compared to diabetes free controls: data from the Osteoarthritis Initiative. Osteoarthritis Cartilage 2018; 26: 751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumann J, Guimaraes JB, Heilmeier U, et al. Diabetics show accelerated progression of knee cartilage and meniscal lesions: data from the Osteoarthritis Initiative. Skeletal Radiol 2019; 48: 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo S, Shi Q, Chen J, et al. Expression and significance of MMPs in synovial fluid, serum and PBMC culture supernatant stimulated by LPS in osteoarthritis patients with or without diabetes. Exp Clin Endocrinol Diabetes 2019; 127: 195–202. [DOI] [PubMed] [Google Scholar]
- 10.Kelley KM, Johnson TR, Ilan J, et al. Glc regulation of the IGF response system in chondrocytes: induction of an IGF-I-resistant state. Am J Physiol 1999; 276: R1164–R1171. [DOI] [PubMed] [Google Scholar]
- 11.Rosa SC, Rufino AT, Judas FM, et al. Role of Glc as a modulator of anabolic and catabolic gene expression in normal and osteoarthritic human chondrocytes. J Cell Biochem 2011; 112: 2813–2824. [DOI] [PubMed] [Google Scholar]
- 12.Chen YJ, Chan DC, Lan KC, et al. PPARγ is involved in the hyperglycemia-induced inflammatory responses and collagen degradation in human chondrocytes and diabetic mouse cartilages. J Orthop Res 2015; 33: 373–381. [DOI] [PubMed] [Google Scholar]
- 13.Marques-Rocha JL, Samblas M, Milagro FI, et al. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J 2015; 29: 3595–3611. [DOI] [PubMed] [Google Scholar]
- 14.Jiang SD, Lu J, Deng ZH, et al. Long noncoding RNAs in osteoarthritis. Joint Bone Spine 2017; 84: 553–556. [DOI] [PubMed] [Google Scholar]
- 15.Kopańska M, Szala D, Czech J, et al. MiRNA expression in the cartilage of patients with osteoarthritis. J Orthop Surg Res 2017; 12: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao M, Feng Y, Liu C, et al. Prognostic values of long noncoding RNA PVT1 in various carcinomas: an updated systematic review and meta-analysis. Cell Prolif 2018; 51: e12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeidler R, Joos S, Delecluse HJ, et al. Breakpoints of Burkitt's lymphoma t(8;22) translocations map within a distance of 300 kb downstream of MYC. Genes Chromosomes Cancer 1994; 9: 282–287. [DOI] [PubMed] [Google Scholar]
- 18.Lu D, Luo P, Wang Q, et al. lncRNA PVT1 in cancer: a review and meta-analysis. Clin Chim Acta 2017; 474: 1–7. [DOI] [PubMed] [Google Scholar]
- 19.Zheng J, Yu F, Dong P, et al. Long non-coding RNA PVT1 activates hepatic stellate cells through competitively binding microRNA-152. Oncotarget 2016; 7: 62886–62897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng F, Qi Y, Dong C, et al. PVT1 regulates inflammation and cardiac function via the MAPK/NF-κB pathway in a sepsis model. Exp Ther Med 2018; 16: 4471–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verghese ET, Drury R, Green CA, et al. MiR-26b is down-regulated in carcinoma-associated fibroblasts from ER-positive breast cancers leading to enhanced cell migration and invasion. J Pathol 2013; 231: 388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato M, Goto Y, Matsushita R, et al. MicroRNA-26a/b directly regulate La-related protein 1 and inhibit cancer cell invasion in prostate cancer. Int J Oncol 2015; 47: 710–718. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, Dong C, Yang J, et al. MicroRNA-26b-5p inhibits mouse liver fibrogenesis and angiogenesis by targeting PDGF receptor-beta. Mol Ther Nucleic Acids 2019; 16: 206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramazani Y, Knops N, Elmonem MA, et al. Connective tissue growth factor (CTGF) from basics to clinics. Matrix Biol 2018; 68–69: 44–66. [DOI] [PubMed] [Google Scholar]
- 25.Omoto S, Nishida K, Yamaii Y, et al. Expression and localization of connective tissue growth factor (CTGF/Hsc24/CCN2) in osteoarthritic cartilage. Osteoarthritis Cartilage 2004; 12: 771e8. [DOI] [PubMed] [Google Scholar]
- 26.Tu M, Yao Y, Qiao FHH, et al. The pathogenic role of connective tissue growth factor in osteoarthritis. Biosci Rep. 2019; 39: BSR20191374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang X, Muhammad H, McLean C, et al. Connective tissue growth factor contributes to joint homeostasis and osteoarthritis severity by controlling the matrix sequestration and activation of latent TGFβ. Ann Rheum Dis 2018; 77: 1372–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waly NE, Refaiy A, Aborehab NM. IL-10 and TGF-β: roles in chondroprotective effects of glucosamine in experimental osteoarthritis. Pathophysiology 2017; 24: 45–49. [DOI] [PubMed] [Google Scholar]
- 29.Van der Kraan PM. The changing role of TGFβ in healthy, ageing and osteoarthritic joints. Nat Rev Rheumatol 2017; 13: 155–163. [DOI] [PubMed] [Google Scholar]
- 30.Zhen G, Wen C, Jia X, et al. Inhibition of TGF-β signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nat Med 2013; 19: 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Jordan JM. Epidemiology of osteoarthritis. Clin Geriatr Med 2010; 26: 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan W, Li X. Impact of diabetes and its treatments on skeletal diseases. Front Med 2013; 7: 81–90. [DOI] [PubMed] [Google Scholar]
- 33.Sandell LJ. Etiology of osteoarthritis: genetics and synovial joint development. Nat Rev Rheumatol 2012; 8: 77–89 [DOI] [PubMed] [Google Scholar]
- 34.Wen CY, Chen Y, Tang HL, et al. Bone loss at subchondral plate in knee osteoarthritis patients with hypertension and type 2 diabetes mellitus. Osteoarthritis Cartilage 2013; 21: 1716–1723. [DOI] [PubMed] [Google Scholar]
- 35.Schett G, Kleyer A, Perricone C, et al. Diabetes is an independent predictor for severe osteoarthritis: results from a longitudinal cohort study. Diabetes Care 2013; 36: 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol 2017; 1008: 1–46. [DOI] [PubMed] [Google Scholar]
- 37.Grüll MP, Massé E. Mimicry, deception and competition: the life of competing endogenous RNAs. Wiley Interdiscip Rev RNA 2019; 10: e1525. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Li S, Luo Y, et al. LncRNA PVT1 regulates chondrocyte apoptosis in osteoarthritis by acting as a sponge for miR-488-3p. DNA Cell Biol 2017; 36: 571–580. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Y, Zhao J, Guo X, et al. Long non-coding RNA PVT1, a molecular sponge for miR-149, contributes aberrant metabolic dysfunction and inflammation in IL-1β-simulated osteoarthritic chondrocytes. Biosci Rep. 2018; 38: BSR20180576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One 2011; 6: e18671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Millis MP, Bowen D, Kingsley C, et al. Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes 2007; 56: 3027–3032 [DOI] [PubMed] [Google Scholar]
- 42.Hanson RL, Craig DW, Millis MP, et al. Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes 2007; 56: 975–983. [DOI] [PubMed] [Google Scholar]
- 43.Hu J, Wang Z, Pan Y, et al. MiR-26a and miR-26b mediate osteoarthritis progression by targeting FUT4 via NF-κB signaling pathway. Int J Biochem Cell Biol 2018; 94: 79–88. [DOI] [PubMed] [Google Scholar]
- 44.Liang YZ, Li JJ, Xiao HB, et al. Identification of stress-related microRNA biomarkers in type 2 diabetes mellitus: a systematic review and meta-analysis. J Diabetes Epub ahead of print 17 January 2018. DOI: 10.1111/1753-0407.12643. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, INI881778 Supplemental Material for Long non-coding RNA PVT1, a molecular sponge of miR-26b, is involved in the progression of hyperglycemia-induced collagen degradation in human chondrocytes by targeting CTGF/TGF-β signal ways by Luo-Bin Ding, Yao Li, Guang-Yuan Liu, Tai-Hang Li, Feng Li, Jian Guan and Hua-Jun Wang in Innate Immunity