Abstract
Innovative tools are essential for advancing malaria control and depend on an understanding of molecular mechanisms governing transmission of malaria parasites by Anopheles mosquitoes. CRISPR/Cas9-based gene disruption is a powerful method to uncover underlying biology of vector-pathogen interactions and can itself form the basis of mosquito control strategies. However, embryo injection methods used to genetically manipulate mosquitoes (especially Anopheles) are difficult and inefficient, particularly for non-specialist laboratories. Here, we adapted the ReMOT Control (Receptor-mediated Ovary Transduction of Cargo) technique to deliver Cas9 ribonucleoprotein complex to adult mosquito ovaries, generating targeted and heritable mutations in the malaria vector Anopheles stephensi without injecting embryos. In Anopheles, ReMOT Control gene editing was as efficient as standard embryo injections. The application of ReMOT Control to Anopheles opens the power of CRISPR/Cas9 methods to malaria laboratories that lack the equipment or expertise to perform embryo injections and establishes the flexibility of ReMOT Control for diverse mosquito species.
Keywords: CRISPR/Cas9, ovary translocation, reverse genetics
To solve the growing problem of mosquito-borne disease, we need improved and more efficient gene editing methods. Targeted disruption of genes in mosquito vectors of human pathogens is a powerful method to uncover the underlying biology of mosquito pathogen transmission that can inform the development and application of new approaches to mosquito and disease control (Dong et al. 2018; Ling and Raikhel 2018; Duvall et al. 2019; Raji et al. 2019; Matthews et al. 2019). Genome manipulation technologies can themselves form the basis of mosquito and pathogen control strategies (Kyrou et al. 2018; Binh Pham et al. 2019). While many techniques based on CRISPR/Cas9 have improved studies in mosquito genetics, these techniques rely on embryo injection of the Cas9 ribonucleo-protein complex (RNP), which requires specialized and expensive equipment and training. To get around this bottleneck, a strategy termed ReMOT Control (Receptor-Mediated Ovary Transduction of Cargo) was developed for delivery of gene-editing moieties to the arthropod germline from the hemocoel, allowing targeted and heritable mutations to be made by adult injection instead of embryo microinjection (Chaverra-Rodriguez et al. 2018). Proof-of-principle experiments validating ReMOT Control were conducted in Aedes aegypti, but extension of the method to species more recalcitrant to editing (such as Anopheles mosquitoes) was not established.
In this study, we have adapted the ReMOT Control technique for efficient gene editing in Anopheles stephensi a major vector of Plasmodium parasites in India and Southeast Asia and so a major object for malaria research. A transgenic line of An. stephensi containing two fluorescent markers was already being used to establish protocols Cas9-mediated gene editing and so this line provided a straight-forward first target for ReMOT Control in Anopheles. We found that once optimized, gene editing by ReMOT Control in Anopheles mosquitoes was comparable to the technique in Ae. aegypti and as efficient in editing as standard embryo injections. As Anophelines are substantially more difficult to manipulate at the embryo stage than Aedes mosquitoes, ReMOT Control represents a much-needed improvement in gene editing for malaria vectors. We expect that with ReMOT Control gene editing will become as commonly used for reverse genetics in Anopheles mosquitoes as it has become in Aedine vectors of human arboviruses.
Materials and Methods
Mosquitoes
Anopheles stephensi wild type (Liston strain) and the Anopheles stephensi transgenic line VgCp26.10 (AP26) were reared at 27°, 75 ± 10% relative humidity, 12h light: 12h dark in a walk-in environmental chamber. Larvae were fed with a slurry of 1:2 by volume Tetramin:baker’s yeast. Adults were provided ad libitum with 10% sterilized sugar on a cotton ball. For injection experiments female mosquitoes were fed on anonymous human blood (Biospecialty Corp.) using a water-jacketed membrane feeding system.
Embryo microinjections
Anopheles stephensi females from a homozygous transgenic line VgCp26.10 (Binh Pham et al. 2019) were used to generate heterozygous embryos for injection. Blood-fed An. stephensi mosquitoes were induced to lay eggs 3-5 days after a blood meal by combining 6-10 females in a narrow Drosophila vial with cotton and Whatman filter paper wet with isotonic buffer (150mM NaCl, 5mM KCl; 10mM HEPES; 2.5mM CaCl2; pH 7.2). At approximately 1 hr and 15 min after laying, a paintbrush was used to transfer embryos that were sufficiently melanized to a small piece of filter paper wet with isotonic buffer. Under a dissecting scope, the outer membrane was removed with jeweler’s forceps from the embryos and embryos were aligned with posterior poles to the left while maintaining moisture on the paper. After alignment of 30-70 embryos, the paper was dried briefly, embryos were transferred to toupee tape on a plastic cover slip and an oil mixture (1:1 Halocarbon 700 oil: Halocarbon 27 oil) was used to cover the embryos to prevent further desiccation. Quartz needles were pulled using a Sutter P2000 needle puller and were used with a Femtojet injector (Eppendorf) and InjectMan micromanipulator. Following injection, oil was removed from embryos with a Kim Wipe and embryos were submerged on the tape into a petri dish with isotonic buffer and transferred back to an insectary to hatch. Hatching was monitored daily for 14 days and hatched larvae were immediately transferred to a pan with food. The injection mix from successful experiments comprised 200ng/uL Streptococcus pyogenes Cas9 (PacBio) and 100ng/µL each of three single guide RNAs initially described for use to target green fluorescent protein (EGFP) in human cell (12, Table S4). Injected embryos (generation 0, G0) that survived to adulthood were outcrossed: families comprised five G0 males allowed to mate individually to 10 wild-type females each for 2 days, then combined and pools comprised age-matched G0 females batch mated to wild-type males. G1 progeny resulting from families and pools were screened for both red and cyan fluorescence in the eyes.
Adult injections
Adult females with ages ranging from 5 to 22 days old were blood-fed using a glass water jacketed membrane feeder. The next day, females were immobilized by incubation at 4° until motionless, then placed on ice and sorted. Females with visible blood-meals were injected intrathoracically using a glass needle drawn from a glass capillary (World Precision Instruments) using a needle puller (Sutter P2000) and aspirator assembly (A5177, Sigma) until we could observe visible distention of the abdomen, diuresis or liquid emerging from the injection site (approximately 200 nl).
Adult injection mix preparation
P2C-Cas9 and P2C-EGFP proteins were expressed from pET28a-P2C-Cas9 and pRSET-P2C-EGFP respectively by recombinant BL21 E.coli (NEB) as described previously in detail (Chaverra-Rodriguez et al. 2018). Single guide RNAs (sgRNAs) were prepared using a PCR template amplified by CRISPR_F primers designed for each ECFP target and CRISPR_R primers (Table S4). In vitro transcription was used to produce sgRNAs from PCR template using either MegaScript T7 or MegaScript RNAi kits with at least 1000ng of PCR template. Two to four volumes of the in vitro transcription reaction were required to achieve the high quantities of guide RNA required for adult injections. In addition to P2C-Cas9 and sgRNAs, saponin was included in some injection mixes. Saponin dilutions were prepared fresh prior to each injection from dry crude extract of Quillaja bark containing >20% saponin (Sigma-Aldrich). All concentrations of saponin reported here represent concentration of crude extract.
The final step in P2C-Cas9 expression and purification from transformed Escherichia coli is dialysis of the protein in a pH 8.0 buffer consisting 50 mM Tris-HCl pH 8.0, 200 mM KCl, 0.1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and 1mM dithiothrietol (DTT). Injections using this buffer produced a high mortality in An. stephensi females, but we found that concentrated stocks of P2C-Cas9 protein often precipitated when water, 1X PBS or guide RNA solutions were added to the protein solution. However, if the P2C-Cas9 protein was diluted in a large volume (300-500 µL) of 50 mM Tris-HCl pH 8.0, 200 mM KCl and small volumes (∼<20 µL) of highly concentrated (∼3-10 µg/µL) sgRNA were added and allowed to complex, the buffer of the RNP could then be exchanged with 1X PBS using an Amicon 10K filter column, which retains the large protein and both bound and unbound guide RNA. Following buffer exchange, saponin, water and additional sgRNA were added to produce solutions of 0.5-0.75 µg/µL P2C-Cas9, 0.75-1.2 µg/µL of total guide RNA (from a premixed solution of the three sgECFPs), 50 mg/L saponin extract from Quillaja bark and less than 5 µM residual KCl.
Larval screening
G0 and G1 larvae were screened at larval instar stage 3 or 4. Larvae were immobilized by applying larvae to a wet filter paper in a BÜchner funnel attached to vacuum filtering flask. Larvae were kept slightly wet during screening under UV fluorescence microscopy with a Leica dissecting scope or Zeiss Axiozoom.
Imaging
To determine the optimal time for P2C-mediated delivery, adult females were injected with P2C-EGFP (Chaverra-Rodriguez et al. 2018) 24 hr before and 4, 24, and 48 hr after a bloodmeal. Adult female ovaries were dissected 72 hr post-blood meal, mounted on a slide in SlowFade Gold Antifade Mountant (Invitrogen) between two double layers of scotch tape to prevent the ovaries from being squashed by the plastic coverslip. The coverslip was sealed in place using nail polish on the edges of the slip and ovaries were visualized on Olympus BX41. All images were captured using 311ms exposure for 200X and 1030ms exposure for 40X images.
Molecular analysis of mutant individuals
Mutant G0 and G1 mosquitoes identified by visual screening were collected individually and G1 progeny from mutant G0 were collected in groups of five individuals, frozen and stored at -20°. Genomic DNA was extracted using Wizard Genomic DNA Purification Kit or Qiagen DNeasy Blood & Tissue kits following the protocol for extraction of DNA from animal tissues. Genomic DNA was used as a template for PCR using the NEB Phusion enzyme and using primers against the transgenes and surrounding DNA (Figures 3A, S4). Diagnostic gel bands were extracted using Zymoclean Gel DNA Recovery kit (Zymo, Irvine) cloned into pJet1.2/blunt using the CloneJET PCR Cloning Kit (ThermoFisher) and sequenced. Sequence analyses were performed using the DNAstar Lasergene suite.
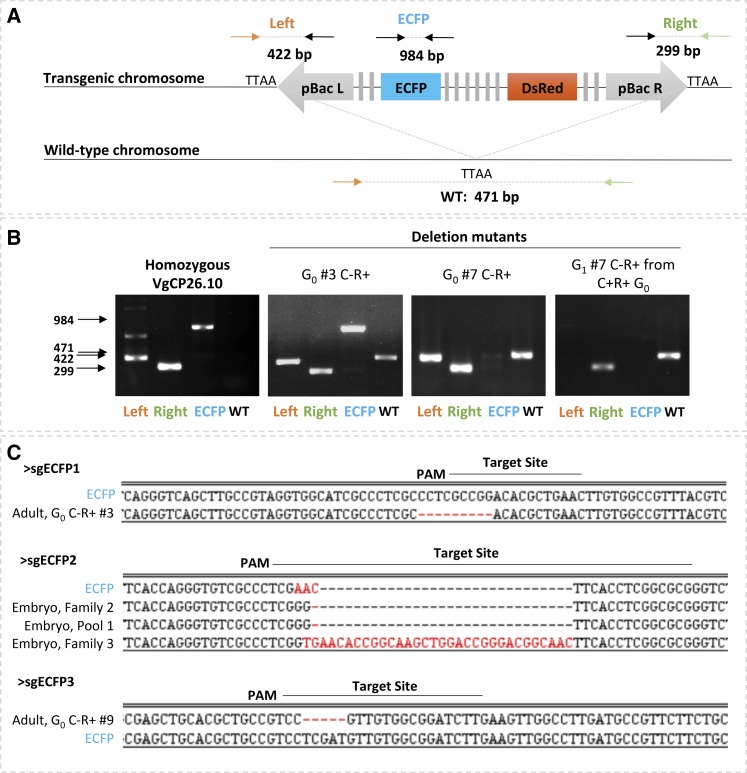
Figure 3.
Molecular characterization of ECFP negative G0 and G1 reveals both small and large deletions. A) Schematic of PCR strategy for mutant genotype characterization. Left and Right Primer sets produce amplicons of 422 and 299 base pairs, respectively, that confirm the presence of the transgene at the 26.10 genomic location. ECFP amplicon, 984 bp, spans all three target sites. Primers against genomic DNA (orange and green arrows) produce a 471bp diagnostic band from a non-transgenic chromosome. B) Representative gel images from individuals with intact transgene on both chromosomes (first panel), diagnostic amplicons for both transgenic and wild-type chromosome (second panel), amplicons diagnostic for a deletion that interrupts the primer binding site for ECFP amplification (third panel) and amplicons diagnostic for a deletion that interrupts primer sites for the ECFP amplicon and the Left amplicon (fourth panel). C) Alignments of small deletion mutants from both embryo and adult injections to ECFP. Abbreviations are the same as Figure 1 and WT: wild-type, C: cyan, R: Red, PAM: protospacer motif.
Data availability
All data are described in the text and/or included in tables and figures within the manuscript and in the supplement uploaded into figshare as follows:
Figure S1: Alignment of amplicon sequences from PCR B validate the presence of the transgene in 17 ECFP knockout individuals. Table S1: Results from screening single G0 male families (1,2,3 etc.) and pools (Px) of G0 females injected as embryos with 200 ng/µL Cas9 and three sgRNAs targeting ECFP (300ng/µL total). Table S2: Results from screening single G0 male families (1,2,3 etc.) and Pools (PX) of G0 females injected as embryos with 200 ng/µL Cas9 and three sgRNAs targeting ECFP (total 75ng/µL). Table S3: Summary of injection components used for G0 embryo injections and resulting gene-editing detected in G1. Table S4: Primers used for current study. Supplemental material available at figshare: https://doi.org/10.25387/g3.11827914.
Results and Discussion
ECFP targeting, mutation detection scheme, and validation by embryo microinjection
To establish a method for site-specific editing in An. stephensi, we used ReMOT Control to target one of the two fluorescent marker genes in the transgenic An. stephensi line VgCp26.10 (Binh Pham et al. 2019), which offered several advantages for establishment of mutagenesis methods over the endogenous eye color genes previously used. The gene encoding kynurenine 5-monooxygenase (kmo or kynurenine hydroxylase, kh) was used as a visible marker to develop both embryo injection and adult injection methods in Ae. aegypti (Basu et al. 2015; Chaverra-Rodriguez et al. 2018). However, the kmo knockout is associated with substantially decreased fitness in An. stephensi, while the VgCp26.10 transgenic line is robust and is not likely to suffer from the phenotypic loss of a fluorescent marker (Gantz et al. 2015; Binh Pham et al. 2019). A mutation in the one dominant visible fluorescent marker gene (enhanced cyan fluorescence protein, ECFP) is easy to detect in hemizygous live mosquitoes by fluorescence imaging while simultaneous expression of the intact linked marker gene (Discosoma species Red, DsRed) confirms presence of the transgene rather than the wild-type empty locus (Li and Handler 2019).
Initial validation of targeted knockdown of ECFP in the VgCp26.10 transgenic line by embryo microinjection provided a benchmark to assess ReMOT Control knockout efficiency. Homozygous transgenic embryos (generation 0, G0) were injected with Cas9 plasmid expression cassette, mRNA, or protein, and three single guide RNAs originally designed to target EGFP in human cells (Zhang et al. 2014) but which also cut ECFP due to a high degree of sequence conservation between the two genes. Surviving G0 embryos injected with 200 ng/µL Cas9 protein and 300 ng/µL sgRNAs produced G1 offspring lacking cyan fluorescence in the eyes but expressing DsRed in four out of 32 single male G1 families and one female pool, resulting in frequencies of 12.5% families producing edited offspring and 1.5–24% of G1 offspring edited within those families (Table 1 and Table S1). ECFP knockout individuals comprised 1.5% of total G1 screened across all families. Two male families produced individuals with loss of both cyan and red fluorescence, indicating that a modification occurred that not only interrupted ECFP expression, but also interrupted expression of the DsRed marker that was over 7 kilobases away (Figure 1B). Such large deletions are not unprecedented, but are likely under-reported in studies where PCR amplicons of limited size are used to detect deletions (Kosicki et al. 2018). Injection of embryos with a reduced concentration of total gRNA (75ng/µL) produced fewer visually detectible mutations; G1 individuals with ECFP disruption were only identified from one pool of females out of 11 total female pools and single male families and constituted 0.13% of G1 individuals screened (Tables S1 and S2). No knock-outs were detectable in G1 larvae when G0 embryos were injected with 500 ng/µL of Cas9 transcript or 500 ng/µL of plasmid with Cas9 expressed from the Hsp70 promoter (pDCC6, Gokcezade et al. 2014) (Table S3).
Table 1. Injection data and comparison of ECFP mutation by adult and embryo injections.
| Injection Mix | G-1 | G0 Fluorescence Phenotype | G1 Fluorescence Phenotype | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein | sgECFP | Saponin | G-1 Cross | No. Injected | Injection time PBM (hours) | No. on EC day | No. | Cyan+ | Cyan- | Cyan- | Cyan Mosaic | G0 C+R+ Outcross | No. | Cyan+ | Cyan- | Cyan- |
| DsRed+ | DsRed- | DsRed+ | DsRed+ | DsRed+ | DsRed- | DsRed+ | ||||||||||
| Adult injections: P2C-Cas9/gRNA mix injections into VgCp26.10 colony females (G-1) crossed with wild type males | ||||||||||||||||
| 0.5 µg/µL | 0.75 µg/µL | + | VgCp26.10 Females X WT Males | 300 | 24 | 212 | 425 | 106 | 317 | 2 (0.47%) | 0 | Female | 295 | 103 | 192 | 0 |
| Male | 622 | 324 | 298 | 0 | ||||||||||||
| 0.75 µg/µL | 1.2 µg/µL | + | VgCp26.10 Females X WT Males | 83 | 48 | 60 | 194 | 160 | 24 | 10 (5.2%) | 1* (1.2%) | Female | 52 | 26 | 26 | 0 |
| Male | 102 | 38 | 53 | 11 (10.8%) | ||||||||||||
| 0.75 µg/µL | 1.2 µg/µL | + | VgCp26.10 Females X WT Males | 63 | 47 | 33 | 161 | 52 | 107 | 2 (1.2%) | 1** (0.62%) | Female | 61 | 29 | 32 | 0 |
| Male | 84 | 35 | 49 | 0 | ||||||||||||
| 0.75 µg/µL | 1.2 µg/µL | + | VgCp26.10 Females X WT Males | 78 | 46 | 46 | 71 | 41 | 30 | 0 | 2** (2.8%) | Female | 113 | 30 | 83 | 0 |
| Male | ND | |||||||||||||||
| Sap+ Totals: | 524 | 351 | 851 | 359 | 478 | 14 (1.7%) | 4 (0.47%) | 1329 | 585 | 733 | 11 (0.82%) | |||||
| 0.75 µg/µL | 1.1 µg/µL | — | VgCp26.10 Females X WT Males | 97 | 28 | 78 | 184 | 114 | 70 | 0 | 0 | Female | 217 | 96 | 118 | 3 (1.4%) |
| Male | 97 | 49 | 48 | 0 | ||||||||||||
| 0.75 µg/µL | 1.1 µg/µL | — | VgCp26.10 Females X WT Males | 71 | 25 | 43 | 173 | 61 | 112 | 0 | 0 | Female | 41 | 15 | 26 | 0 |
| Male | 207 | 109 | 98 | 0 | ||||||||||||
| Sap - Totals: | 168 | 121 | 357 | 175 | 182 | 0 | 0 | 562 | 269 | 290 | 3 (0.53%) | |||||
| Totals: | 692 | 472 | 1208 | 534 | 660 | 0 | 0 | 1891 | 854 | 1023 | 14 (0.74%) | |||||
| Embryo injections: Cas9/gRNA mix injections into homozygous VgCp26.10 transgenic embryos (G0) | ||||||||||||||||
| 0.2 µg/µL | 0.075 µg/µL | — | VgCp26.10 Intercross | 309 | Not Screened | 2320 | 2317 | 3 (0.13%) | 0 | |||||||
| 0.2 µg/µL | 0.3 µg/µL | — | VgCp26.10 Intercross | 382 | Not Screened | 5646 | 5506 | 52 (0.92%) | 88 (1.5%) | |||||||
| Totals: | 691 | 7966 | 7823 | 55 (0.69%) | 88 (1.1%) | |||||||||||
individual had one cyan+ eye and one cyan- eye.
these individuals had markedly reduced cyan fluorescence.
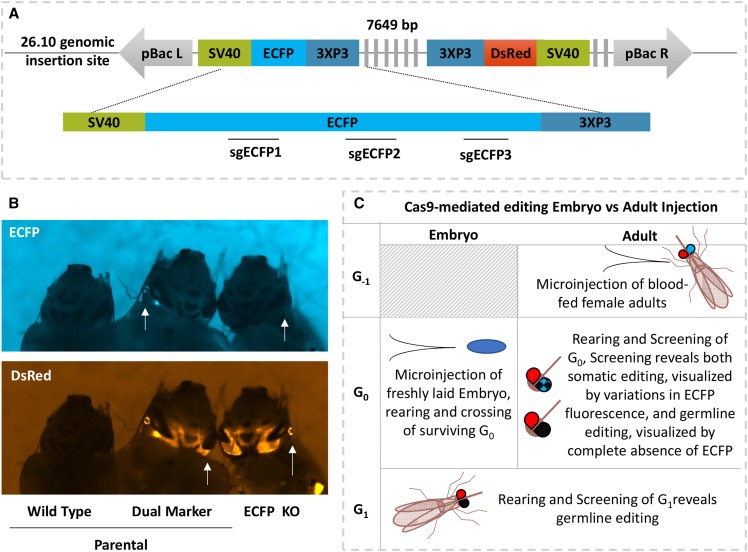
Figure 1.
Schematic of knock-down approach. A) Schematic representation of transgene present in transgenic line VgCp26.10. The transgene has an ECFP and DsRed marker genes. Three guide RNAs (sgECFP) target Cas9 to the ECFP gene. B) Representative fluorescence image of larvae showing parental wild-type, parental transgenic and exceptional Cyan-, DsRed+ phenotypes. C) Schematic comparison of embryo and adult injection approaches for ECFP knock-out and detection of mutants. The shaded box represents not done. Abbreviations are bp: base pair(s), pBac L/R: piggyBac left/right arm, SV40: simian virus 40 3′ untranslated region, DsRed: Discosoma species Red, ECFP: enhanced cyan fluorescent protein, sgECFP: single guide RNAs against ECFP, KO: knock-out, Gx: generation X.
Optimization of injection components for adult injection
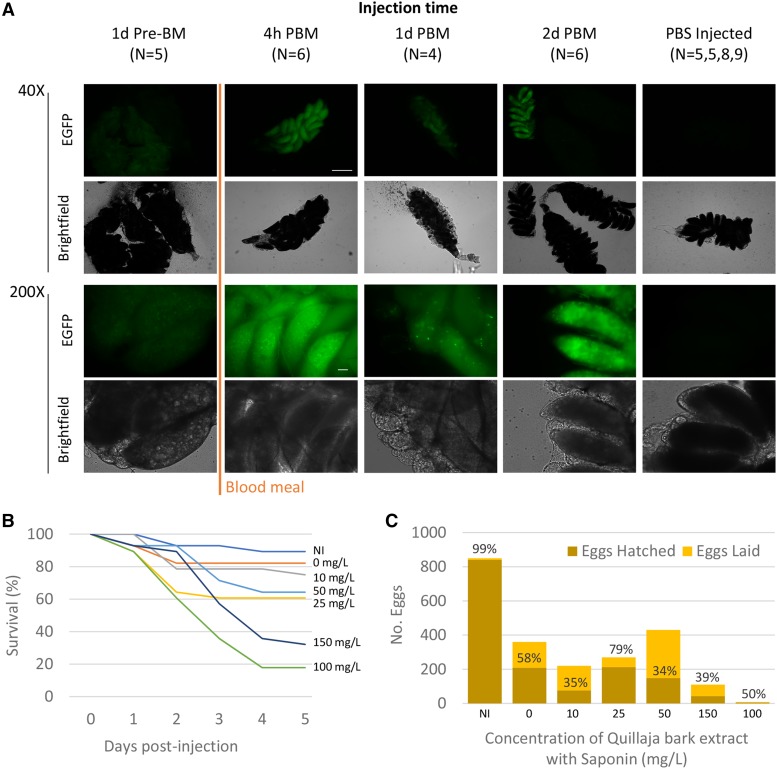
In order to accomplish heritable mutagenesis in An. stephensi by ReMOT Control, we utilized the ovary targeting P2C peptide derived from Drosophila melanogaster yolk protein 1 (DmYP1), which we recently reported in Ae. aegypti to mediate delivery of Cas9 RNP into vitellogenic oocytes (Chaverra-Rodriguez et al. 2018). However, we found that the Aedes injection parameters were not directly translatable to An. stephensi. Timing of injection of the P2C-Cas9 RNP relative to a blood-meal was reported to impact the developmental stage at which genome modifications are made in Ae. aegypti. Additionally, An. stephensi have lower tolerance in general to injected components. We identified the injection conditions and timing that would maximize RNP uptake into the ovaries in a way that balanced survival and egg laying with concentrations of RNP effective for editing. We first injected females with P2C-EGFP fusion protein and visualized uptake into the ovary by fluorescence microscopy (Figure 2A). Green fluorescence was more intense and present in all oocytes dissected from injected females when P2C-EGFP was injected within two days of blood feeding compared to decreased fluorescence when the protein was injected prior to a blood meal. Only 50% of ovary pairs had visible fluorescence in females injected 48 hr after a blood meal, perhaps because injected females were past the peak of vitellogenesis (Nirmala et al. 2006; Isaacs et al. 2011) but those with visible fluorescence were comparable in intensity to earlier injections. Thus, experimental injections were performed within the first two days of a blood meal.
Figure 2.
Optimization of injection components. A) Fluorescent and bright-field imaging of 72 hr (h) post-blood meal (PBM) ovaries following injection of P2C-EGFP at 1 day (d) before and 4 hr, 1 day and 2 days post-blood meal. Scale Bar represents 1mm in 40X images and 100uM in 200X images. B) Survival measurement following adult injections with different concentrations of Quillaja bark extract. C) Eggs and hatching counts from eggs laid by females injected with different concentrations of Saponin from Quillaja bark for endosome disruption. Percentages above bars represent the percentage hatching from eggs laid. Abbreviations are the same as Figure 1 and h: hour, N: number ovary pairs, pre-BM: before blood meal, PBS: phosphate buffered saline.
In previous work, we used chloroquine to facilitate exit of the Cas9 RNP from the endosome after uptake, but previous work in the field of drug delivery has demonstrated that saponins (Quallaja bark extract) act as potent endosomal escape reagents and ongoing work in our lab optimizing ReMOT Control for Aedes suggested that it might be as or more effective (Fuchs et al. 2009; Gilabert-Oriol 2014). For Anopheles ReMOT editing, we therefore investigated the use of saponin for endosome escape. In order to identify the highest concentration of saponin that would result in survival and viable offspring of injected adults, we performed a set of preliminary injections using different concentrations of the extract in 1X phosphate-buffered saline (PBS, Figure 2B and C). Injection itself caused substantial decrease in survival and egg laying, but a concentration of 50 mg/L saponin could be injected and still have a high enough oviposition and hatch rate to have enough G0 progeny to screen. Importantly, we also determined that An. stephensi adults could not tolerate the dialysis buffer, which is used in the last step of purification of P2C-Cas9 from E. coli (and which was the buffer used for injections in Ae. aegypti). P2C-Cas9 precipitated out of solution when diluted in water or 1X PBS. We found that the buffer of P2C-Cas9 in complex with sgRNA did not precipitate and so could be column exchanged with water resulting in a final neutral injection solution with less than 5uM KCl remaining from the dialysis buffer, and that was tolerated by injected adults.
ECFP marker knockout by ReMOT Control
P2C-Cas9 RNP with ECFP guide RNAs sgECFP 1, 2 and 3 were injected along with saponin (50 mg/L) into VgCp26.10 adult females (G-1) crossed to wild-type males, such that G0 and G1 progeny with parental phenotypes are either positive for both ECFP and DsRed, or wild-type. Expected mutant phenotypes from successful editing would be ECFP-, DsRed+. We recovered ECFP-, DsRed+ G0 individuals from three out of the four injection groups in which saponin was included, with a total of twelve ECFP-, DsRed + individuals out of 851 total G0 screened (1.4%). No mutant individuals were recovered from the G0 progeny in injections without saponin. The VgCp26.10 colony was not completely homozygous at the generation of adult injection, resulting in wild-type G0, so the efficiency of editing is more accurately understood in terms of number of alleles edited rather than the number of individuals. In this case a total of 377 transgenic alleles were available for editing in the G0 progeny of saponin/P2C-Cas9 RNP injected G-1 females as visually detected by DsRed fluorescence in heterozygous G1 individuals and 14 (3.7%) were edited, whereas 0% of the 534 alleles were edited in the G0 progeny of G-1 females injected without saponin. This adjustment is relevant to the extension of this technique to the mutation of endogenous mosquito genes that will be present in two copies. In these applications, especially for genes that lack a visible phenotype, editing is sufficiently efficient to be able to screen for these mutants by PCR-based methods.
ECFP-, DsRed+ individuals that survived to adulthood were individually outcrossed to wild-type mosquitoes resulting in G1 progeny, of which approximately half were ECFP-,DsRed+ and half were wild-type, consistent with Mendelian segregation of a transgenic chromosome and a non-transgenic chromosome (Χ2 = 0.0397, P = 0.84) along with one transgenic mutant and one wild-type. We conclude from this that ECFP knockouts detected at G0 are complete knockouts and likely occurred as Cas9 cleavage of the DNA in the oocyte prior to embryogenesis (Table 2). Four G0 progeny were designated mosaics. One of these was ECFP positive in only one eye and the three others had markedly decreased ECFP fluorescence, which is unexpected in individuals with a transgene at a fixed location (Table 1). Individuals mosaic for mutation in a targeted gene is expected in G0, as the P2C-Cas9 editing complex can be active at different stages of development. In studies targeting the KMO gene for mutation, this manifests in patchy or pink color in the eyes instead of black eyes (Gantz et al. 2015). In our case, from the presence of fluorescence in only one eye, we infer that editing at an early enough stage of development that a distinct pattern of fluorescence loss is seen. Conversely, in individuals with markedly decreased fluorescence, we speculate that ECFP was knocked out in some cells of the eye and not in others. We were not initially screening for mosaicism in ECFP expression, so it is likely that mosaic presence of ECFP mutations in G0 is more prevalent. The presence of mosaicism in G0 progeny from adults injected with RNP demonstrates that, as in analogous experiments in Ae. aegypti, the complex is delivered to the G-1 ovaries, but in some cases is not immediately active, but becomes active following initiation of embryo development. G0 Individuals identified as mosaics were outcrossed but produced no progeny.
Table 2. G1 progeny from Cyan negative G0 demonstrate knockouts are heritable.
| G0 Cross | G1 Fluorescence Phenotype | |||
|---|---|---|---|---|
| No. | Cyan+ | Cyan- | Cyan- | |
| DsRed+ | DsRed- | DsRed+ | ||
| C-R+ G0 Male #1 X WT Females | 246 | 0 | 123 | 123 |
| C-R+ G0 Male #2 X WT Females | 163 | 0 | 86 | 77 |
| C-R+ G0 Male #3 X WT Females | 275 | 0 | 141 | 134 |
| C-R+ G0 Male #5 X WT Females | 119 | 0 | 58 | 61 |
| C-R+ G0 Male #6 X WT Females | 339 | 0 | 170 | 169 |
| C-R+ G0 Male #9 X WT Females | 116 | 0 | 56 | 60 |
ECFP+, DsRed+ individuals, which are parental in phenotype, were retained from each injection group, separated by sex and then out-crossed en masse to wild-type mosquitoes to test for the presence of germline mutations in the germline ECFP coding sequence. Wild-type G0 were excluded from further crosses. ECFP-, DsRed+ G1 offspring were recovered from one male outcross from a saponin injection group and one female outcross from a saponin negative injection group, 0.82% and 0.53% of total G1 from each group respectively. The presence of ECFP-, DsRed+ progeny from ECFP+, DsRed+ G0 mosquitoes likely represent editing of the G0 germline without visible editing of the somatic tissues.
A panel of four primer pairs was used to characterize ECFP-, DsRed+ modifications (Figure 3A). Two primer sets amplify across the genomic DNA and the left and right piggyBac arms of the transgene and so validate the presence of the transgene at the 26.10 locus. Similarly, a PCR amplicon using a primer set across the transgene insertion site indicates the presence of a wild-type chromosome at that locus. A primer set designed to amplify 984 bp across the ECFP open reading frame and all three sgRNA target sites diagnosed the mutation character at this site: out of the 19 ECFP- knockout individuals that were molecularly characterized, only two produced diagnostic bands with ECFP primers and sequencing of the amplicons revealed small indels (Figure 3B second panel, Figure 3C and Figure S1). In 17 cases, PCR and sequencing further confirmed the presence of the transgene at the original locus, but no diagnostic band was seen from the ECFP open reading frame (Figure 3B third panel, Figure S1). In one of these, no diagnostic band was seen for amplification of either the ECFP open reading frame or the left arm, but the right arm was validated as intact, indicating a disruption of at least 1600 nucleotides of sequence (Figure 3B fourth panel). These results suggest that large deletions commonly occurred using ReMOT Control, similar to results observed from embryo injection of the same components.
Altogether, we demonstrate with these experiments that heritable targeted knockouts can be achieved by ReMOT Control in An. stephensi, and that targeted editing occurs both before and after embryogenesis as evidenced both by the presence of mosaics in the G0 progeny and the recovery of ECFP-,DsRed+ G1 individuals from ECFP+,DsRed+ G0 outcrosses. Compared to embryo injection, ReMOT Control will allow non-specialist laboratories to conduct gene editing experiments in Anopheles mosquitoes and increase the number of reverse genetic studies in this malaria vector. The translation of ReMOT Control from Ae. aegypti to An. stephensi mosquitoes required modifications of the injection parameters, including timing, concentrations, and endosome escape strategies. The use of saponin dramatically increased the efficiency of knockout, consistent with results of analogous injections of the endosomal escape reagent chloroquine in the application of ReMOT Control to Ae. aegypti. It is possible that many reagents that mediate endosomal escape are usable for ReMOT control, which will further the adaptability of this technique. The ligand P2C is broadly effective in mosquitoes (Chaverra-Rodriguez et al. 2018), and may be useful in other insect species as well. In organisms where P2C is not effective in delivery, other ligands may be identified from proteins intrinsic to that species that are specifically targeted to the germline. We anticipate that such developments along with the straight-forward set of modifications for the adaptation to An. stephensi demonstrated here, will lead to the application of P2C-Cas9 to genetic studies not only to other important malaria vectors, but also to other species currently recalcitrant to gene editing techniques.
Acknowledgments
This work was funded by NSF/BIO grant 1645331, NIH/NIAID grants R21AI111175 and R01AI128201, USDA/NIFA grant 2014-10320, USDA Hatch funds (Accession #1010032; Project #PEN04608), and a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds to JLR, USDA/NIFA grant 2017-67012-26101 to VMM, Wolfson Foundation and Royal Society fellowship RSWF\R1\180013, and NIH/NIAID grants R21AI138074 and R21AI129507 and the John S. Dunn Foundation Collaborative Research Award to GLH, and NIH/NIAID grant R01AI29746 to AAJ. D.C.R. was partially supported by a Fulbright Fellowship and by Colciencias.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11827914.
Communicating editor: A. Bashirullah
Literature Cited
- Basu S., Aryan A., Overcash J. M., Samuel G. H., Anderson M. A. E. et al. , 2015. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc. Natl. Acad. Sci. USA 112: 4038–4043. 10.1073/pnas.1502370112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T. B., Phong C. H., Bennett J. B., Hwang K., Jasinskiene N. et al. , 2019. Experimental population modification of the malaria vector mosquito, Anopheles stephensi. PLOS Genet. 15: e1008440 10.1371/journal.pgen.1008440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaverra-Rodriguez D., Macias V. M., Hughes G. L., Pujhari S., Suzuki Y. et al. , 2018. Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat. Commun. 9: 3008 10.1038/s41467-018-05425-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Simões M. L., Marois E., and Dimopoulos G., 2018. CRISPR/Cas9 -mediated gene knockout of Anopheles gambiae FREP1 suppresses malaria parasite infection. PLOS Pathog. 14: e1006898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall L. B., Ramos-Espiritu L., Barsoum K. E., Glickman J. F., and Vosshall L. B., 2019. Small-Molecule Agonists of Ae. aegypti Neuropeptide Y Receptor Block Mosquito Biting. Cell 176: 687–701.e5. 10.1016/j.cell.2018.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs H., Bachran D., Panjideh H., Schellmann N., Weng A. et al. , 2009. Saponins as Tool for Improved Targeted Tumor Therapies. Curr. Drug Targets 10: 140–151. 10.2174/138945009787354584 [DOI] [PubMed] [Google Scholar]
- Gantz V. M., Jasinskiene N., Tatarenkova O., Fazekas A., Macias V. M. et al. , 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 112: E6736–E6743. 10.1073/pnas.1521077112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilabert-Oriol R., 2014. Development of a platform technology for enhanced endo/lysosomal escape of targeted toxins by structurally specific oleanane saponins. Department of Biology, Chemistry, Pharmacy Freie Universität, Berlin. [Google Scholar]
- Gokcezade J., Sienski G., and Duchek P., 2014. Efficient CRISPR/Cas9 Plasmids for Rapid and Versatile Genome Editing in Drosophila. G3 (Bethesda) 4: 2279–2282. 10.1534/g3.114.014126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs A. T., Li F., Jasinskiene N., Chen X., Nirmala X. et al. , 2011. Engineered resistance to Plasmodium falciparum development in transgenic Anopheles stephensi. PLoS Pathog. 7: e1002017 10.1371/journal.ppat.1002017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M., Tomberg K., and Bradley A., 2018. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36: 765–771. 10.1038/nbt.4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrou K., Hammond A. M., Galizi R., Kranjc N., Burt A. et al. , 2018. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36: 1062–1066. 10.1038/nbt.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., and Handler A. M., 2019. CRISPR/Cas9-mediated gene editing in an exogenous transgene and an endogenous sex determination gene in the Caribbean fruit fly, Anastrepha suspensa. Gene 691: 160–166. 10.1016/j.gene.2018.12.055 [DOI] [PubMed] [Google Scholar]
- Ling L., and Raikhel A. S., 2018. Serotonin signaling regulates insulin-like peptides for growth, reproduction, and metabolism in the disease vector Aedes aegypti. Proc. Natl. Acad. Sci. USA 115: E9822–E9831. 10.1073/pnas.1808243115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. J., Younger M. A., and Vosshall L. B., 2019. The ion channel ppk301 controls freshwater egg-laying in the mosquito Aedes aegypti. eLife 8 10.7554/eLife.43963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirmala X., Marinotti O., Sandoval J. M., Phin S., Gakhar S. et al. , 2006. Functional characterization of the promoter of the vitellogenin gene, AsVg1, of the malaria vector, Anopheles stephensi. Insect Biochem. Mol. Biol. 36: 694–700. 10.1016/j.ibmb.2006.05.011 [DOI] [PubMed] [Google Scholar]
- Raji J. I., Melo N., Castillo J. S., Gonzalez S., Saldana V. et al. , 2019. Aedes aegypti Mosquitoes Detect Acidic Volatiles Found in Human Odor Using the IR8a Pathway. Curr. Biol. 29: 1253–1262.e7. 10.1016/j.cub.2019.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ge X., Yang F., Zhang L., Zheng J. et al. , 2014. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci. Rep. 4: 5405 10.1038/srep05405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are described in the text and/or included in tables and figures within the manuscript and in the supplement uploaded into figshare as follows:
Figure S1: Alignment of amplicon sequences from PCR B validate the presence of the transgene in 17 ECFP knockout individuals. Table S1: Results from screening single G0 male families (1,2,3 etc.) and pools (Px) of G0 females injected as embryos with 200 ng/µL Cas9 and three sgRNAs targeting ECFP (300ng/µL total). Table S2: Results from screening single G0 male families (1,2,3 etc.) and Pools (PX) of G0 females injected as embryos with 200 ng/µL Cas9 and three sgRNAs targeting ECFP (total 75ng/µL). Table S3: Summary of injection components used for G0 embryo injections and resulting gene-editing detected in G1. Table S4: Primers used for current study. Supplemental material available at figshare: https://doi.org/10.25387/g3.11827914.