Abstract
During Drosophila melanogaster metamorphosis, arrested immature neurons born during larval development differentiate into their functional adult form. This differentiation coincides with the downregulation of two zinc-finger transcription factors, Chronologically Inappropriate Morphogenesis (Chinmo) and the Z3 isoform of Broad (Br-Z3). Here, we show that br-Z3 is regulated by two microRNAs, let-7 and miR-125, that are activated at the larval-to-pupal transition and are known to also regulate chinmo. The br-Z3 3′UTR contains functional binding sites for both let-7 and miR-125 that confers sensitivity to both of these microRNAs, as determined by deletion analysis in reporter assays. Forced expression of let-7 and miR-125 miRNAs leads to early silencing of Br-Z3 and Chinmo and is associated with inappropriate neuronal sprouting and outgrowth. Similar phenotypes were observed by the combined but not separate depletion of br-Z3 and chinmo. Because persistent Br-Z3 was not detected in let-7-C mutants, this work suggests a model in which let-7 and miR-125 activation at the onset of metamorphosis may act as a failsafe mechanism that ensures the coordinated silencing of both br-Z3 and chinmo needed for the timely outgrowth of neurons arrested during larval development. The let-7 and miR-125 binding site sequences are conserved across Drosophila species and possibly other insects as well, suggesting that this functional relationship is evolutionarily conserved.
Keywords: heterochronic, temporal identity, let-7, miR-125, Chinmo, Br-Z3
The heterochronic microRNAs (miRNAs) lin-4 and let-7 were originally identified because they ensured that cells adopted proper temporal cell fates during the development of the nematode Caenorhabditis elegans (C. elegans) (reviewed in Luhur et al. 2013; Faunes and Larrain 2016). As in C. elegans, these miRNAs display distinctive temporal expression profiles during the development of most animal species (Pasquinelli et al. 2000). In Drosophila melanogaster (D. melanogaster), for example, expression of let-7 and lin-4 ortholog miR-125 is activated during the larval-to-pupal transition and then persists through metamorphosis and adulthood (Sempere et al. 2003; Chawla et al. 2016). This temporal control is regulated by the late larval pulse of the insect molting steroid hormone Ecdysone, which along with its nuclear receptor, Ecdysone Receptor (EcR), directly activates the let-7-Complex (let-7-C), a polycistronic locus that encodes Drosophila let-7, miR-125, and a third miRNA, miR-100, (Sokol et al. 2008; Chawla and Sokol 2012). Once activated, let-7-C is transcribed in cells throughout the central nervous system (CNS), where a key joint target of let-7 and miR-125 is Chronologically Inappropriate Morphogenesis (Chinmo) (Wu et al. 2012), whose post-transcriptionally controlled downregulation ensures that cells in the Mushroom Body (MB) adopt proper cell fate (Zhu et al. 2006; Liu et al. 2015). In addition to chinmo, let-7 also represses abrupt (ab) allowing the timely remodeling of neuromuscular junctions (NMJs) during metamorphosis (Caygill and Johnston 2008). Chinmo and Abrupt are closely related members of the Broad-Complex/Tramtrack/Bric-a-brac (BTB) family of zinc finger (ZF) transcription factors (Spokony and Restifo 2007). Whether let-7-C miRNAs function in cells other than the MB and NMJ to regulate additional targets during nervous system remodeling remains unknown.
The bulk of the late larval CNS is composed of arrested immature neurons born from neural stem cells known as neuroblasts (NBs) (reviewed in Venkatasubramanian and Mann 2019). There are between 25 and 30 distinct NBs in each thoracic neuromere of the ventral nerve cord (VNC) and these NBs divide during the course of larval development to produce clusters of lineally related neurons. Because neurons in each cluster extend a primary neurite to a common destination, these clusters, or “adult lineages”, can be labeled and distinguished from one another based on cell number and neurite morphology (Truman et al. 2004). Terminal differentiation of these adult neurons is activated at the onset of metamorphosis, when individual neurons become distinguishable from one another by taking on unique morphologies and connections needed to generate the functional adult nervous system. While mechanisms by which sibling neurons acquire distinct identities have been uncovered in some lineages like the MB and olfactory lobe for example, less is known about this process in VNC lineages. Early and late born neurons in most VNC lineages, however, can be distinguished by the respective expression of Chinmo and the Z3 isoform of another member of the BTB-ZF family, Broad (Br), both of which persist until the onset of metamorphosis (Maurange et al. 2008; Zhou et al. 2009). While the Chinmo-to-Br-Z3 switch is associated with decreased cell size of VNC neurons, the functional roles of Chinmo and Br-Z3 as well as the mechanisms controlling their expression are less clear.
Here, we report that the 3′UTR of br-Z3 contains functional let-7 and miR-125 binding sites and that, consistently, transcriptional activation of let-7-C coincides with downregulation of both Br-Z3 and Chinmo in the VNC. By using the mosaic analysis with a repressible cell marker (MARCM) technique (Lee et al. 2000b), we also find a birth-order dependent onset of let-7-C in many VNC adult lineages. Forced premature expression of let-7-C miRNAs leads to early silencing of Br-Z3 and Chinmo that is associated with premature neuronal sprouting and outgrowth. Similar phenotypes were observed by simultaneous depletion of br-Z3 and chinmo, suggesting that timely onset of let-7-C ensures stage-specific neuronal remodeling.
Materials and Methods
Molecular biology and cell culture methods
broad cDNAs containing the miR-125 binding site were recovered by RNA ligase-mediated rapid amplification of 5′ cDNAs (RLM-RACE) using the GeneRacer Kit (Life Technologies). A ∼4.5kb cDNA was cloned from 5 μg of total larval RNA using an oligo located 42 bp away from the miR-125 binding site. This cDNA was sequenced completely (Figure S1), and the sequence has been deposited in GenBank (accession number MN990459). Cell culture experiments and luciferase assays were performed as described in Wu et al., 2012, using psiCHECK-2 plasmids containing br-Z3 3′UTR (described here) or w 3′UTR (described in Wu et al. 2012).
Transgenes and plasmids
br-Z3 3′UTR reporters: A 2,030 bp fragment containing the br-Z3 3′UTR was PCR amplified, subcloned, sequenced to confirm absence of errors, and inserted into pP(wORF::MCS) (Wu et al. 2012) or psiCHECK2 (Promega) for analysis in vivo or in cell culture, respectively. MiRNA binding sites were deleted using Quickchange Site-Directed Mutagenesis Kit (Agilent) with oligos that removed basepairs 354-413 (let-7 binding sites) and/or basepairs 1332-1341 (miR-125 binding site) of the br-Z3 3′UTR.
UAS-br-Z3 shRNAi transgenes: br-Z3 siRNAs were identified by applying rules described in Fellmann et al. 2011 to sequence unique to the br-Z3 open reading frame. Oligos encoding one of the identified br-Z3 siRNAs (TTGTTGCTGTTGTTGTTCGCG) was annealed and subcloned into the EcoRI and NheI sites of pWalium20.
Drosophila strains
Strains used included: (1) P{wORF::w3′UTR} (Wu et al. 2012), (2) P{wORF::br-Z33′UTR} (Luhur et al., 2014), (3) P{wORF::br-Z33′UTRΔlet-7}, (4) P{wORF::br-Z33′UTRΔmir-125}, (5) P{wORF::br-Z33′UTRΔlet-7+miR-125}, (6) let-7-CKO2 (Wu et al. 2012), (7) npr6 (Kiss et al. 1988), (8) hs-Z3 (Bayer et al. 1997), (9) {let-7-Cp12.5kb::lacZ}attP2 (Chawla and Sokol 2012), (10) {let-7-Cp12.5kb::lacZ}attP40 (11) let-7-CΔ3miR::optGal4 (Wu et al. 2012), (12) {UAS-BrZ3shRNAi233}attPVK00033 (this study), (13) FRT40A, UAS-mCD8::GFP, chinmo1 (Zhu et al. 2006), (14) UAS-let-7-C (Wu et al. 2012), (15) UAS-Δlet-7-C (Wu et al. 2012), (16) UAS-chinmo-SV40, (17) UAS-Br-Z3-SV40, (18) w, GAL4C155, hsFLP, UAS-mCD8::GFP; FRT40A, tubP-GAL80, (19) w, GAL4C155, hsFLP, UAS-mRFP.LG-28a; FRT40A, tubP-GAL80, and (20) FRT40A. New transgenics were generated by Rainbow Transgenic Services (California USA).
MARCM strains and clone generation
Stocks containing either w, GAL4C155, hsFLP, UAS-mCD8::GFP; FRT40A, tubP-GAL80 or w, GAL4C155, hsFLP, UAS-mCD8::mRFP.LG28a; FRT40A, tubP-GAL80 were used to generate GFP and RFP labeled clones, respectively. Some MARCM strains also contained 2nd or 3rd chromosome insertions of {let-7-Cp12.5kb::lacZ}attP2. Flies of the appropriate strain were allowed to lay eggs for 6 hr, and then transferred to new vials. Resulting embryos were usually aged for 18 hr and then heat shocked at 38° for 30 min to generate neuroblast clones in adult lineages of 18-24 hr old animals. After heat shock, animals were reared at 25° degrees until dissection. Nervous systems were generally dissected during larval and pupal stages. Pre-wandering larvae were chosen as 3rd instar larvae that remained in the food, while wandering larvae were those that had left the food. Pupae were staged by picking white prepupae every hour and then aging them on humid Petri dishes for 12, 18, and 24 hr.
Antibody generation
To generate Br-Z3 antibodies, the 271 amino acid Z3-specific portion of the Br-Z3 protein was analyzed to find regions that maximize hydrophilicity, antigenicity, and surface probability, that contain turns, and that do not contain glycosylation sites. Based on this analysis, two peptides (residues 601-614, SLKRHFQDKHEQSD, and residues 622-636, CHRRYRTKNSLTTHK, of the BR-PA polypeptide sequence) were synthesized, coupled to KLH, and injected into rabbits (ProSci Inc, Poway, CA). Resulting polyclonal antisera were affinity-purified and found to be specific for Broad-Z3 by immunofluorescence (see Figure S3).
Immunostaining and microscopy
Antibodies used for immunostaining included: mouse anti-nc82 (1:50, Developmental Studies Hybridoma Bank), rat-anti-Chinmo (1:500; Wu et al., 2012), rabbit anti-Br-Z3 (1:250; this study), rabbit anti-GFP (1:1000; Invitrogen), chicken anti-GFP (1:500; Rockland Immunochemicals), rabbit anti-dsRed (1:500; Clontech), mouse anti-mCherry (1:500; Clontech), rabbit anti-β-gal (1:500; MP Biomedicals), chicken anti-β-gal (1:500; Abcam). Most primary antibodies were preabsorbed against fixed embryos. Secondary goat antibodies were conjugated to Alexa Fluor 488, 568 or 633 (Molecular Probes). Slides were mounted in Vectashield (Vector Labs) or dehydrated through an ethanol series and mounted in DPX. Images were collected on a Leica SP5 (Light Microscopy Imaging Center, Indiana University). Confocal stacks were merged using Leica LSM software. Samples that were directly compared were prepared under identical conditions and imaged in parallel.
Statistical analysis
All statistical analyzes were performed using Prism (GraphPad, Version 7.0). First datasets were tested for normality using D’Agostino-Pearson test. For comparisons of two datasets, an Unpaired t-test was performed. Significance is indicated as follows: n, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Data availability
Strains and plasmids are available upon request. The sequence of the broad-Z3 cDNA found in Supplemental Figure 1 has been also deposited in GenBank (accession number MN990459). The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25387/g3.11858517.
Results
broad-Z3 3′UTR contains functional let-7 and miR-125 binding sites
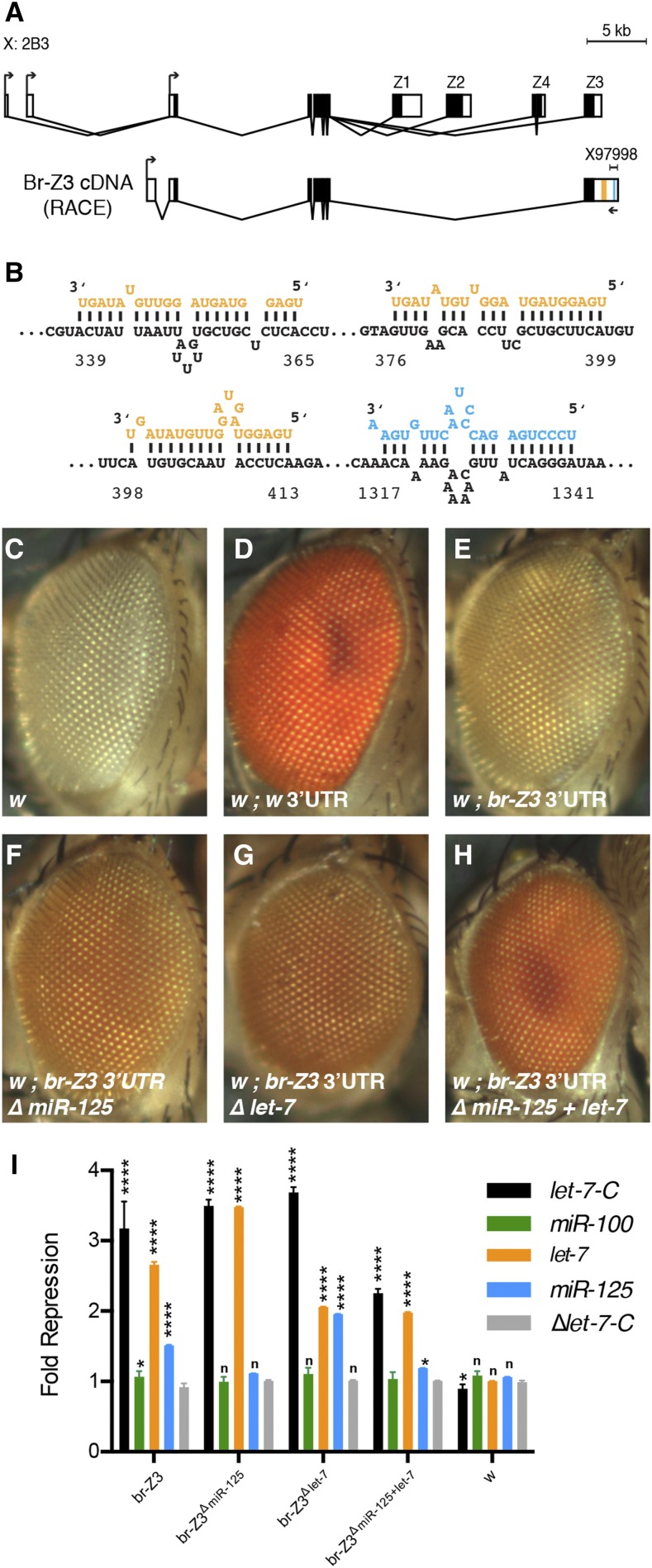
We searched available miRNA target predictions to identify additional let-7-C targets and found that PicTar predicted a putative miR-125 binding site in a transcript annotated as broad-RF (Grün et al. 2005). broad (br) is a complicated locus that produces four protein isoforms (Z1-Z4) that share a common BTB domain plus one of four different zinc finger domains (Dibello et al. 1991; Bayer et al. 1996). These alternate zinc finger domains are encoded by a series of 3′ exons that also contain isoform-specific 3′UTR sequences (Figure 1A). Since the identified binding site was located in sequence from two incomplete cDNAs (X97998 and AY060896), we performed 5′ RACE to identify full-length br transcripts containing this predicted site using oligos located 40 nucleotides (nts) downstream of it. A 4.5 kb Broad-Z3-encoding cDNA was isolated that contained a previously unannotated first exon as well as unannotated 3′UTR sequence in the last exon (Figure 1A, Figure S1). This cDNA was consistent with RNA-seq data indicating that the last br exon contains an extended 3′UTR (Graveley et al. 2011). To determine whether the 3′UTR sequence in our novel cDNA might contain additional binding sites for let-7-C miRNAs, we searched it for sequences complementary to miR-100, let-7 and miR-125 using RNAhybrid (Rehmsmeier et al. 2004) and found three putative let-7 binding sites along with the PicTar-predicted miR-125 site (Figure 1B), all of which were conserved among eleven Drosophila species and possibly other insects as well (Figure S2). This data therefore raised the possibility that Broad-Z3 is regulated by both miR-125 and let-7.
Figure 1.
broad-Z3 3′UTR contains functional let-7 and miR-125 binding sites. (A) Organization of the broad locus (top) and RACE-identified br-Z3 transcript (bottom). Locations of Z1-Z4 encoding exons and predicted let-7 and miR-125 binding sites (orange and blue lines, respectively) are shown. X97998 indicates the location of a partial broad cDNA predicted to contain a miR-125 binding site originally reported in Makunin et al. 1996. (B) Sequences of predicted let-7-C binding sites in br-Z3. let-7, miR-125, and br-Z3 3′UTR sequences are shown in orange, blue, and black, respectively, and the br-Z3 3′UTR sequence is numbered relative to the stop codon. (C-H) Eyes of white mutant flies harboring reporter transgenes containing a white (D), br-Z3 (E), or mutated br-Z3 3′UTR (F-H). (I) Fold repression of luciferase reporters containing full-length wildtype br-Z3 3'UTR, mutated br-Z3 3′UTR, or white 3′UTR in cell cultures in which let-7-C miRNAs were ectopically expressed either together or individually. ****, P < 0.0001; *P < 0.05; n.s. = not significant. Error bars = ± SD.
To determine whether these predicted miRNA binding sites were functional, we analyzed the br-Z3 3′UTR in an in vivo reporter system that relies on the white (w) eye pigmentation gene (Luhur et al., 2014). Variation in the expression of this gene is easily detected, since it leads to gradations in eye pigment levels that vary from white to orange to bright red. For example, eye pigment levels of w mutant flies are fully restored with a transgene containing the w open reading frame (ORF) fused to its usual w 3′UTR (Figure 1 C, D). In contrast, eye pigment levels are only slightly increased in flies harboring the w open reading frame (ORF) fused to a 2 kb portion of the br-Z3 3′UTR identified in our cDNA (Figure 1E), suggesting that this 3′UTR mediates post-transcriptional repression of w. To test whether let-7-C miRNAs are responsible for this repression, we generated mutant versions of the white reporter in which either the miR-125 site or all three let-7 sites were deleted. Pigment levels were darker in flies harboring mutant transgenes relative to those carrying the wildtype br 3′UTR (Figure 1E-G), indicating that both miR-125 and let-7 sites were functional. Deletion of all four sites led to a further increase in pigment (Figure 1H), indicating that miR-125 and let-7 sites are nonredundant. Taken together, these data indicated that endogenous let-7 and miR-125 repressed reporter levels via binding sites in the br-Z3 3′UTR.
To obtain further support that the br-Z3 3′UTR was responsive to let-7-C miRNAs, we used a cell culture assay to test whether ectopic let-7-C miRNAs repressed expression of a luciferase reporter containing the br-Z3 3′UTR. Expression of pri-let-7-C, which encodes all three miRNAs, as well as just let-7 or miR-125 alone, resulted in repression of luciferase (Figure 1I). The identified let-7 and miR-125 binding sites in the br-Z3 3′UTR mediated this repression, since their deletion led to a significant reduction in the amount of repression. This analysis also indicated that the br-Z3 3′UTR might contain additional let-7 sites, since the mutated reporter was still responsive to ectopic let-7. Nevertheless, we concluded that the identified let-7 and miR-125 binding sites in the br-Z3 3′UTR were functional.
BTB-ZFs and let-7-C display inverse temporal expression profiles in the larval CNS
In order to investigate the in vivo relationship between br-Z3 and let-7-C miRNAs, we generated rabbit antibodies against Br-Z3. This antisera labeled adult lineage neurons in the CNS (Figure S3). This expression profile was consistent with the reported staining patterns of both the Broad-core antisera that recognizes all four isoforms as well as previously generated mouse Broad-Z3 antisera (Mugat et al. 2000; Spokony and Restifo 2009; Zhou et al. 2009), supporting the veracity of our results. To further confirm the specificity of this staining pattern, we also stained the CNS tissue dissected from br mutant null allele npr6 (Kiss et al. 1988), which affects all Broad isoforms, as well as CNS tissue from larvae in which Br-Z3 was overexpressed using a verified heatshock-inducible Z3 (hs-Z3) transgene (Crossgrove et al. 1996; Bayer et al. 1997). Br-Z3 staining was sharply reduced in the adult lineages of npr6 mutants and elevated in hs-Z3 CNS tissue (Figure S3). We also found that adult lineage clones expressing a br-Z3-specific shRNA transgene described later in the paper displayed a loss of anti-Br-Z3 staining (See Figure 5A insets). Altogether, these results indicated that our new antisera recognized Br-Z3.
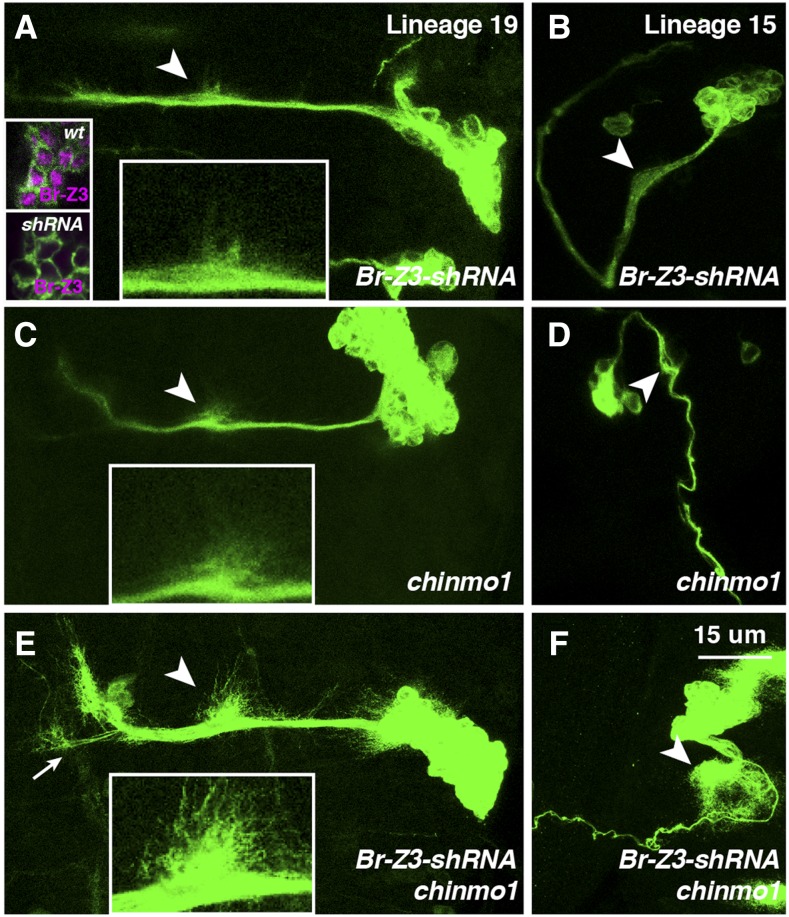
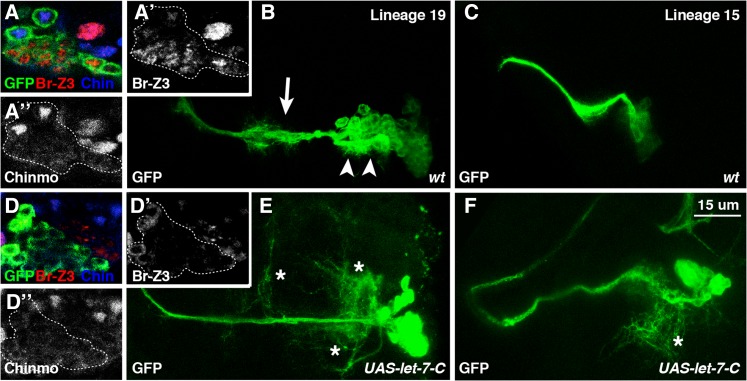
Figure 5.
Co-depletion of Br-Z3 and Chinmo leads to premature adult lineage elaboration. (A-L) Elav-Gal4, UAS-mCD8::GFP labeled UAS-Br-Z3-shRNA (A-B), chinmo1 (C-D), and UAS-Br-Z3-shRNA; chinmo1 (E-F), lineage 19 (A, C, E) and lineage 15 (B, D, F) clones stained for GFP (green). Insets in A show Br-Z3 expression in wildtype (top) and UAS-Br-Z3-shRNA (bottom) clones, indicating both the efficacy of the shRNA as well as the specificity of the antibody. Arrowheads indicate secondary outgrowths and are enlarged in insets at bottom. Clones were generated in newly hatched larvae and analyzed at WL. Scale bar: 15 μm.
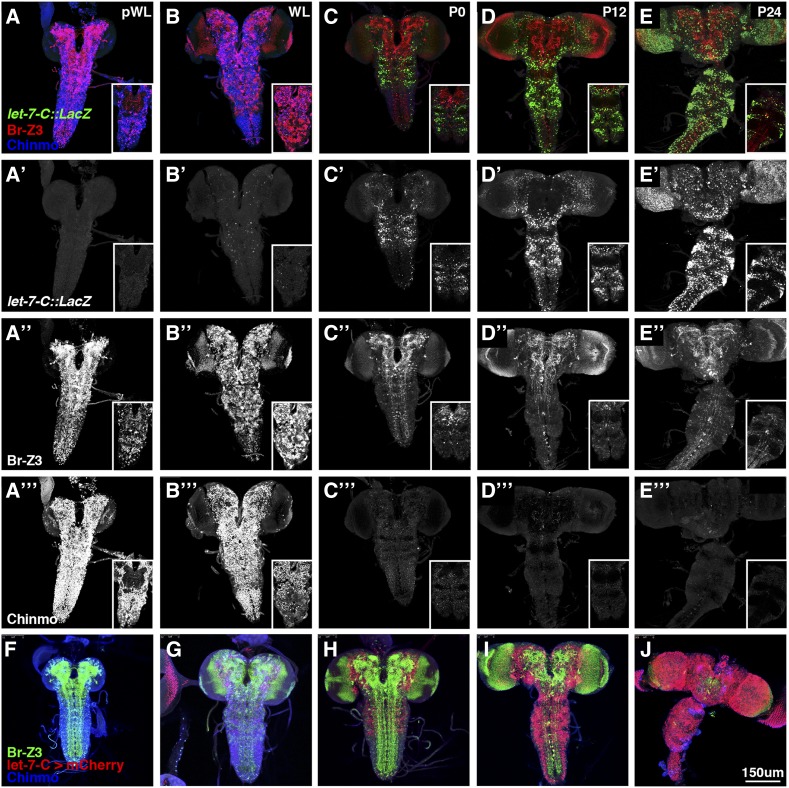
We then used these antibodies to compare the temporal dynamics of let-7-C expression with Br-Z3 as well as Chinmo, a previously identified target of let-7 and miR-125 (Wu et al. 2012) throughout the entire CNS during the larval-to-pupal transition. let-7-C expression was monitored using a previously verified let-7-C transcriptional reporter, let-7-Cp12.5kb::lacZ, which contains a 12.5kb fragment of the first let-7-C intron upstream of the LacZ reporter gene (Chawla and Sokol 2012). Consistent with previous Northern blots of let-7-C miRNAs in the CNS (Chawla and Sokol 2012; Wu et al. 2012), the let-7-C transcriptional reporter was first detected at the wandering 3rd larval instar stage in cells scattered throughout the central brain (CB) and VNC (Figure 2A-E). This pattern intensified and expanded by pupariation (P0), when let-7-C expression was most robustly detected in the thoracic segments (Figure 2C’ inset). Expression continued to expand at twelve (P12) and twenty-four hours (P24) after pupariation in the CB and VNC. Expression of let-7-C in the optic lobe (OL) was only first weakly detected at P12 and then more broadly at P24. As expected, the onset of let-7-C transcription in the CB and VNC coincided with a decrease in Br-Z3 and Chinmo. Both Br-Z3 and Chinmo were detected throughout the CB and VNC in pre-wandering and wandering third instar larvae. However, by P0, both proteins were significantly reduced and Chinmo, in particular, appeared to be almost entirely switched off. While Br-Z3 displayed a similar downregulation to Chinmo in the VNC, it persisted in the CB in the P0-P24 samples and expanded in the OL, consistent with previous reports (Riddiford et al. 2018), although in a pattern that was largely non-overlapping with let-7-C. To confirm this analysis, we compared a second let-7-C transcriptional reporter, let-7-CΔ3miR::optGal4 (Wu et al. 2012) with Br-Z3 and Chinmo, and found a similar expression pattern (Figure 2F-J). Thus, the inverse expression of let-7-C and the two BTB-ZF proteins in the VNC suggested that let-7-C miRNAs might dampen the expression of both Chinmo and Br-Z3 during the larval-to-pupal transition.
Figure 2.
let-7-C expression inversely correlates with Br-Z3 and Chinmo expression in the CNS during the larval-to-pupal transition. (A-E). Dissected CNS from (A) pre-wandering larva (pWL), (B) wandering larva (WL), (C) white prepupa (P0), (D)12 hr-old pupa (12hr), and (E) 24 hr-old pupa (P24) stained for let-7-Cp12.5kb::lacZ transcriptional reporter (green in A-E, white in A’-E’), Br-Z3 (red in A-E, white in A’’-E’’) and Chinmo (blue in A-E, white in A’’’-E’’’). Insets: single confocal sections of the VNC at each timepoint. (F-J) Dissected CNS from (F) pre-wandering larva (pWL), (G) wandering larva (WL), (H) white prepupa (P0), (I)12 hr-old pupa (12hr), and (J) 24 hr-old pupa (P24) stained for let-7-CΔ3miR::optGal4 > mcherry (red), Br-Z3 (green) and Chinmo (blue). Scale bars: A-J, 150µm.
let-7-C is activated in a birth-order dependent pattern in adult lineages
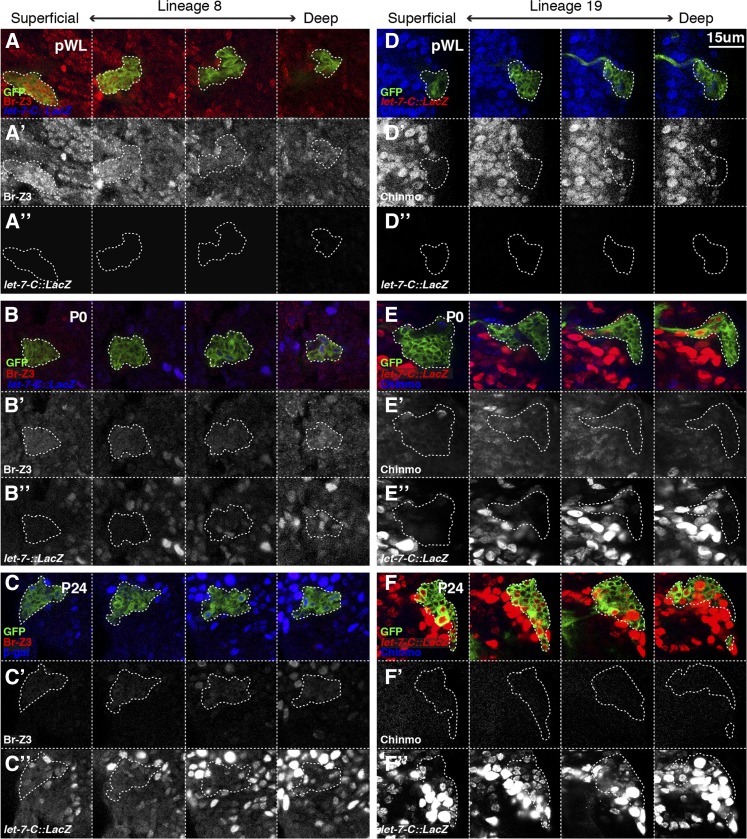
To more finely characterize the temporal dynamics of let-7-C, Br-Z3, and Chinmo at the cellular level, we compared their expression in individually labeled adult lineage in the VNC before, at, and after pupariation. Adult lineages can be distinguished based on stereotypical location and neurite morphology (Truman et al. 2004) and, for our initial analysis, we focused in comparing let-7-C with Br-Z3 in lineage 8 and with Chinmo in lineage 19. Lineage 8 is a ventrolateral cluster located in the anterior half of hemimeres found in segments T1-T3, while Lineage 19 is situated dorsolaterally at the posterior border of hemimeres in segments T1-A1. In the VNC, mitotic neuroblasts and their most recently born daughters are located near the surface of the tissue whereas older born daughters are more deeply located. Consistent with the previously reported birth-order-dependent expression of Broad and Chinmo (Maurange et al. 2008), we found that Br-Z3+ cells were superficially located in lineage 8 whereas Chinmo+ cells were more deeply located in lineage 19 (Figure 3A’, D’). While let-7-C::lacZ was absent from labeled clones in pre-wandering larvae (Figure 3A, D), it was detected in a subset of cells in labeled lineages analyzed at pupariation (Figure 3B”, E”). These cells were located in the deep layer, indicating that let-7-C is first activated in older-born neurons in both lineage 8 and 19. Chinmo as well as Br-Z3 were almost entirely absent by this stage, indicating that Br-Z3 downregulation precedes let-7-C onset in newly born neurons and suggesting a let-7-C independent Br-Z3 silencing mechanism, at least in lineage 8. By P24, let-7-C was detected in cells ranging from the deep to superficial layers (Figure 3C”), indicating that the initial activation of let-7-C in older neurons had expanded to include more recently born neurons as well.
Figure 3.
let-7-C expression inversely correlates with Br-Z3 and Chinmo expression in adult lineage clones during the larval-to-pupal transition. (A-C) Elav-Gal4, UAS-mCD8::GFP-labeled lineage 8 clone stained for GFP (green in A-C), Br-Z3 (red A-C, white in A’-C’), and the let-7-Cp12.5kb::lacZ transcriptional reporter (blue in A-C, white in A”-C”). Single confocal sections from the apical to basal layers show late-to-early born neurons at pWL (A), P0 (B), and P24 (C). (D-F) Elav-Gal4, UAS-mCD8::GFP-labeled lineage 19 clone stained for GFP (green in D-F), Chinmo (blue in D-F, white in D’-F’), and the let-7-Cp12.5kb::lacZ transcriptional reporter (red in D-F, white in D”-F”). Single confocal sections from the apical to basal layers show late-to-early born neurons at pWL (D), P0 (E), and P24 (F). Scale bar: A-F, 15µm.
To determine whether let-7-C displayed a similar pattern of onset in other adult lineages, we quantified the number of let-7-C+ neurons within adult lineages located throughout the VNC at P0 and analyzed, on average, 80 clones per lineage. The majority of adult lineages (0, 1, 2, 3, 5, 6, 10, 11, 12, 13, 14, 17, 21, and 20/22) displayed a pattern similar to the one described above for lineages 8 and 19. In these cases, we found that fewer than 50% of the labeled cells were let-7-C+ at P0 and that these let-7-C+ cells were located in the deep layer, indicating that they were older-born neurons. In addition, we identified lineages that displayed one of two additional patterns. In lineages 4, 15, and 24, we found that all cells were let-7-C+ at P0. Lineage 15 and 24 are distinctive because they are composed entirely of motoneurons that innervate the adult leg (Baek and Mann 2009) and contain relatively few cells (10-30) compared to interneuron lineages (50-100) due to early termination of neuroblast division (Truman et al., 2004). Finally, in lineages 7, 9, 16, 18, and 23, we found that greater than 50% but not all cells were let-7-C+ and that these cells were located in the deeper layers. Thus, with the possible exception of motoneuron lineages, we concluded that let-7-C is activated in a birth order dependent manner in adult lineages throughout the VNC.
Precocious let-7-C represses Br-Z3 and Chinmo and alters neuronal morphology
Given the ability of let-7-C miRNAs in the MB to regulate Chinmo expression and temporal identity (Wu et al. 2012), we wanted to determine whether precocious let-7-C onset in adult lineages could lead to premature termination of Br-Z3 and Chinmo expression. We therefore compared wildtype and UAS-let-7-C-expressing MARCM-labeled clones induced in newly hatched larvae for Br-Z3 and Chinmo expression, focusing again on lineage 19. Within wildtype clones analyzed at the wandering third instar stage, Chinmo and Br-Z3 displayed a complementary expression pattern (Figure 4A, A’ and A’’). While UAS-let-7-C clones analyzed at the same timepoint contained the same number of cells as wildtype clones, they in contrast displayed a complete loss of both Chinmo and Br-Z3 proteins (compare Figure 4A and 4D). These results indicated that let-7-C miRNAs could repress Chinmo and Br-Z3 expression in adult lineages.
Figure 4.
Precocious let-7-C expression drives premature adult lineage elaboration. (A-F) Elav-Gal4, UAS-mCD8::GFP labeled wild-type (A-C) and UAS-let-7-C (D-F) lineage 19 (A, B, D, E) and lineage 15 (C, F) clones stained for GFP (green in A-F), Br-Z3 (red in A, D, white in A’, D’), and Chinmo (blue in A, D, white in A”, D”). Clones were generated in newly hatched larvae and analyzed at WL. Arrow and arrowheads in B indicate contralateral and ipsilateral neurite bundles respectively. Asterisks in E and F indicate outgrowth. Scale bar: 15 μm.
To determine the consequences of let-7-C expression in adult lineages, we analyzed the effect of this premature let-7-C expression on the neuronal morphology of specific adult lineages. During the growth of adult specific neurons, primary projections are sent out to initial targets and subsequent projections follow this same path (Truman et al., 2004). The neurite is continuously maintained throughout larval development until metamorphosis, when secondary outgrowth and elaboration establishes their adult connections. By comparing neurite morphology in wildtype and UAS-let-7-C-expressing MARCM-labeled clones in multiple lineages, we found that premature let-7-C expression resulted in inappropriate outgrowth. For example, lineage 19 in the second thoracic hemisegment contains two primary axon bundles, one contralateral across the midline (arrow in Figure 4B) and another with ipsilateral arbor appearance (arrowheads in Figure 4B). In comparison to wildtype clones, UAS-let-7-C clones displayed a clear outgrowth of neurite bundles in the interstitial region of the contralateral axons as well as more complex processes on the ipsilateral arbors (asterisks in Figure 4E). These phenotypes displayed by UAS-let-7-C-expressing lineage 19 clones in larvae were almost identical to the morphologies displayed by wild type clones at 18 hr APF or even later timepoints (data not shown), raising the posibility that premature expression of let-7-C miRNAs leads to precocious neuronal remodeling.
In addition to lineage 19 phenotypes, we also characterized neurite morphology in a motoneuron-specific lineage that prematurely expressed the UAS-let-7-C transgene (Figure 4C). Lineage 15 exclusively contains motoneurons that project out of the CNS to the leg imaginal disc (Truman et al., 2004; Baek and Mann 2009). However, in comparison to wildtype lineage 15 clones, UAS-let-7-C clones displayed bundles surrounded by a dense fibrous neuropil (Figure 4C, F, arrowhead). The feature of elaborated branches in motoneuron lineages with higher order is a known characteristic associated with the differentiation of adult neurons (Brierley et al. 2009). Thus, as with intereuron lineages, premature expression of let-7-C miRNAs also leads to inappropriate outgrowth of motoneuron lineages.
Since premature expression of let-7-C miRNAs led to inappropriate neurite outgrowth in larvae, we wondered whether loss of let-7-C miRNAs mutants pupae displated a delay in neuronal remodeling. To address this possibility, we generated MARCM-labeled clones in newly hatched larvae that were homozygous mutant for a let-7-CKO2, a verified let-7-C null allele (Wu et al. 2012), and examined secondary sprouting at multiple timepoints during the beginning of metamorphosis (6, 12, 18, 24 hr APF) focusing on lineage 19. The general morphology in let-7-C mutant clones, however, did not display an obvious phenotype; secondary outgrowth started a few hours APF and proceeded on the same pace as wild type, with elaboration and sprouting appearing as usual (data not shown). Consistent with this observation, we failed to detect persistent Br-Z3 or Chinmo expression in let-7-CKO2 mutant adult lineages. Together, these observations indicate the presence of a complementary, let-7-C independent mechanism that promotes the timely outgrowth of adult lineages.
Premature downregulation of Br-Z3 and Chinmo alters neuronal morphology
Given that let-7-C miRNAs can co-target both br-Z3 and chinmo, the UAS-let-7-C phenotypes may be due to the repression of either one or both of these genes. To distinguish these possibilities, we compared the phenotypic consequences of eliminating chinmo and br-Z3, either alone or in combination, in adult lineages. The generation of double mutant homozygous clones that would be needed for this analysis is technically challenging, since br and chinmo genes are located on different chromosomes, the X and 3rd respectively. To circumvent this challenge, we decided to generate chinmo null clones that also expressed a UAS transgene that specifically targeted the br-Z3 transcript. We therefore designed a UAS-br-Z3-shRNAi transgene and tested it first by generating MARCM-labeled lineage 19 clones. Br-Z3 protein was not detectable in UAS-br-Z3-shRNAi clones, confirming that this transgene effectively targeted Br-Z3 as well as further indicating the specificity of the Z3 antibody (Figure 5A inset). In addition, lineage 19 and lineage 15 clones in which Br-Z3 was knocked down displayed relatively subtle outgrowth phenotypes in the neurites of adult lineages (Figure 5A, B). Similarly, homozygous chinmo1 null lineage 19 and lineage 15 clones had wild-type-like larval neurite morphology (Figure 5C, D). Consistent with published reports (Maurange et al., 2008), Br-Z3 and Chinmo were normally expressed in chinmo1 or UAS-br-Z3-shRNAi clones respectively indicating that these two BTB-ZFs did not affect each other’s expression (data not shown). Since the ectopic let-7-C phenotypes described above were not due to depletion of either one of these targets alone, we wondered whether simultaneous elimination of both Chinmo and Br-Z3 would phenocopy the effects of let-7-C overexpression. We therefore generated chinmo1/UAS-br-Z3-shRNAi adult lineage clones at the beginning of L1. When analyzed in wandering larvae, double mutant lineage 19 clones displayed more elaborate secondary neurite outgrowth than clones of either single mutant (Figure 5E). Similarly, dual depletion in lineage 15 also caused an increased complexity of sprouting in this motoneuron lineage (Figure 5F). We also analyzed the phenotypes of dual chinmo and br-Z3 depletion in additional adult lineages, and found many lineages that displayed premature outgrowth during the larval phase, including lineages 7, 11, and 24 (data not shown). The data suggested that the presence of both Chinmo and Br-Z3 prevents neurite elaboration before the pupal period. Furthermore, overexpression of let-7-C represses Chinmo and Br-Z3 together and leads to phenotypes similar to the combined loss of both proteins.
Discussion
Here we provide evidence that co-transcribed miRNAs let-7 and miR-125 regulate Br-Z3. This conclusion is based on identification of a br-Z3 transcript isoform that contains predicted let-7 and miR-125 binding sites. These sites confer let-7/miR-125 sensitivity to the br-Z3 3′UTR as determined by the consequences of their deletion. Consistently, premature expression of let-7 and miR-125 eliminates the normal expression of Br-Z3 as well as a second, previously verified let-7/miR-125 target, Chinmo, in the developing larval CNS. This forced expression of let-7 and miR-125 leads to inappropriate neuronal outgrowth, a phenotypic effect that is very similar to the consequences of the combined but not separate depletion of br-Z3 and chinmo. Taken together, this work suggests a model in which let-7-C activation in the CNS during the larval-to-pupal transition ensures the coordinated silencing of both br-Z3 and chinmo that is needed for the timely outgrowth during metamorphosis of neurons arrested during larval development.
The observation that Br-Z3 and Chinmo downregulation precedes the first detection of let-7-C transcription (see Figure 2 and 3) indicates a complementary, let-7-C-independent mechanism that is also responsible for Br-Z3 and Chinmo offset. Consistent with this, we failed to detect the persistence of Br-Z3 or Chinmo expression in let-7-C mutant adult lineages. Further supporting the absence of persistent Br-Z3 in let-7-C mutants, we failed to detect morphological defects in either the dendrite sensory neurons in the periphery or the lateral clock neurons in the central brain of let-7-C mutants, even though these two sets of neurons display axonal defects when Br-Z3 is ectopically expressed (Zhou et al. 2009; Scott et al. 2011). In addition, given the expression of let-7-C throughout lineages 15 and 24, we also carefully assessed the morphologies of the adult leg motoneurons in let-7-C mutant adults but found no obvious defects. Taken together, these observations suggest that let-7-C regulation of Br-Z3 is a failsafe mechanism to ensure the proper timing of neuronal remodeling when circumstances compromise let-7-C-independent control of Br-Z3.
The relationship between let-7-C miRNAs and br-Z3 identified here extends the known connections between these miRNAs and the Ecdysone pathway and may have significance in other insects. In Drosophila melanogaster, the direct stimulation of br and let-7-C by Ecdysone at the onset of metamorphosis play complementary roles to activate pupal programs and repress larval ones (Bayer et al. 1996; Sempere et al. 2002; Sokol 2012; Truman and Riddiford 2019). The expression profile of Br-Z3 is therefore unusual relative to other Broad isoforms in that it transiently appears during embryogenesis in most central neurons and then persists through most of larval development in a subset of these cells while Br-Z1 and Br-Z4, in contrast, are detected only at the onset of metamorphosis (Zhou et al. 2009). While the function of neural Br-Z3 is not fully understood, available evidence suggests that the absence of Br-Z3 is needed for the remodeling of larval neurons to generate adult arbors. This raises the possibility that Br-Z3 might participate in maintaining neurons in an arrested, immature state, perhaps by competitively inhibiting any stochastic expression of other Br isoforms. Late larval Ecdysone expression would then lead to both the induction of Br-Z1 and -Z4 isoforms and the simultaneous elimination of Br-Z3 via let-7-C-dependent and -independent mechanisms. The conservation of the let-7 and miR-125 binding sites in broad-Z3 across Drosophila species and possibly also in other insects suggests that the functional relationship between br-Z3 and let-7-C is conserved.
Recent work has also highlighted the intimate connection between Ecdysone signaling and temporal cell fate determination pathways acting in the Drosophila nervous system. The ordered production of neurons in the mushroom body lineage, for example, was recently shown to involve a let-7-C dependent negative feedback loop containing Chinmo and the Ecdysone Receptor: Chinmo activates EcR, which in turn represses Chinmo by activating let-7-C (Marchetti and Tavosanis 2017). Similarly, EcR was also recently shown to be important for promoting the transition between early-born Chinmo-expressing neurons and late-born Broad-expressing neurons in type II neuroblasts lineages in the CNS (Syed et al. 2017). Our work here establishing a connection between let-7-C and Broad, a well-known component of the Ecdysone signaling, further extends the relationship between these two pathways.
Along with Abrupt and Chinmo, Br-Z3 is the third identified BTB-ZF target of let-7-C miRNAs in Drosophila (Caygill and Johnston 2008; Wu et al. 2012). These three BTB-ZFs are not only closely related to one another by sequence (Spokony and Restifo 2007), but also appear to play similar roles. All three are dosage sensitive factors that are expressed in subsets of neurons during larval development. As most clearly illustrated by genetic analysis of abrupt in larval dendritic arborization neurons (Li et al. 2004; Sugimura et al. 2004), these BTB-ZFs may collectively function to maintain neurons in an arrested juvenile state and are downregulated during the larval-to-pupal transition in order for neurons to achieve their mature adult form. Since BTB-ZFs are known to heterodimerize (Bonchuk et al. 2011), persistence of larval BTB-ZFs may inappropriately interact and interfere with the function of pupally expressed BTB-ZF cohorts. Interestingly, the temporal expression profile of let-7-C miRNAs is shared by isoforms of a fourth BTB-ZF, Fruitless, which are activated during metamorphosis and are closely related in sequence to Abrupt, Broad, and Chinmo (Lee et al. 2000a; Spokony and Restifo 2007). These observations raise the possibility that let-7-C miRNAs prevent the persistent expression of Abrupt, Broad, and Chinmo that could interfere with Fruitless activity, a hypothesis that future experiments can address.
Acknowledgments
We thank the Bloomington Drosophila Stock Center (supported by grant NIHP4OOD018537), the Drosophila Genome Resource Center (supported by grant NIH2P40OD010949), and the Developmental Studies Hybridoma Bank (created by the NICHD of the NIH) for reagents; the Light Microscopy Imaging Center (supported by grant NIH1S10OD024988-01) for access to confocal microscopes; and the National Institute of Mental Health (Awards R01MH087511 and R03MH115429) for financial support.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11858517.
Communicating editor: M. Arbeitman
Literature Cited
- Baek M., and Mann R. S., 2009. Lineage and birth date specify motor neuron targeting and dendritic architecture in adult Drosophila. J. Neurosci. 29: 6904–6916. 10.1523/JNEUROSCI.1585-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer C. A., Holley B., and Fristrom J. W., 1996. A switch in broad-complex zinc-finger isoform expression is regulated posttranscriptionally during the metamorphosis of Drosophila imaginal discs. Dev. Biol. 177: 1–14. 10.1006/dbio.1996.0140 [DOI] [PubMed] [Google Scholar]
- Bayer C. A., von Kalm L., and Fristrom J. W., 1997. Relationships between protein isoforms and genetic functions demonstrate functional redundancy at the Broad-Complex during Drosophila metamorphosis. Dev. Biol. 187: 267–282. 10.1006/dbio.1997.8620 [DOI] [PubMed] [Google Scholar]
- Bonchuk A., Denisov S., Georgiev P., and Maksimenko O., 2011. Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. J. Mol. Biol. 412: 423–436. 10.1016/j.jmb.2011.07.052 [DOI] [PubMed] [Google Scholar]
- Brierley D. J., Blanc E., Reddy O. V., Vijayraghavan K., and Williams D. W., 2009. Dendritic targeting in the leg neuropil of Drosophila: the role of midline signalling molecules in generating a myotopic map. PLoS Biol. 7: e1000199 10.1371/journal.pbio.1000199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill E. E., and Johnston L. A., 2008. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr. Biol. 18: 943–950. 10.1016/j.cub.2008.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla G., Deosthale P., Childress S., Wu Y. C., and Sokol N. S., 2016. A let-7-to-miR-125 MicroRNA Switch Regulates Neuronal Integrity and Lifespan in Drosophila. PLoS Genet. 12: e1006247 10.1371/journal.pgen.1006247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla G., and Sokol N. S., 2012. Hormonal activation of let-7-C microRNAs via EcR is required for adult Drosophila melanogaster morphology and function. Development 139: 1788–1797. 10.1242/dev.077743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossgrove K., Bayer C. A., Fristrom J. W., and Guild G. M., 1996. The Drosophila Broad-Complex early gene directly regulates late gene transcription during the ecdysone-induced puffing cascade. Dev. Biol. 180: 745–758. 10.1006/dbio.1996.0343 [DOI] [PubMed] [Google Scholar]
- DiBello P. R., Withers D. A., Bayer C. A., Fristrom J. W., and Guild G. M., 1991. The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics 129: 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faunes F., and Larrain J., 2016. Conservation in the involvement of heterochronic genes and hormones during developmental transitions. Dev. Biol. 416: 3–17. 10.1016/j.ydbio.2016.06.013 [DOI] [PubMed] [Google Scholar]
- Fellmann C., Zuber J., McJunkin K., Chang K., Malone C. D. et al. , 2011. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol. Cell 41: 733–746. 10.1016/j.molcel.2011.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M. et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D., Wang Y. L., Langenberger D., Gunsalus K. C., and Rajewsky N., 2005. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLOS Comput. Biol. 1: e13 10.1371/journal.pcbi.0010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss I., Beaton A. H., Tardiff J., Fristrom D., and Fristrom J. W., 1988. Interactions and developmental effects of mutations in the Broad-Complex of Drosophila melanogaster. Genetics 118: 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Foss M., Goodwin S. F., Carlo T., Taylor B. J. et al. , 2000a Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J. Neurobiol. 43: 404–426. [DOI] [PubMed] [Google Scholar]
- Lee T., Marticke S., Sung C., Robinow S., and Luo L., 2000b Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron 28: 807–818. 10.1016/S0896-6273(00)00155-0 [DOI] [PubMed] [Google Scholar]
- Li W., Wang F., Menut L., and Gao F. B., 2004. BTB/POZ-zinc finger protein abrupt suppresses dendritic branching in a neuronal subtype-specific and dosage-dependent manner. Neuron 43: 823–834. 10.1016/j.neuron.2004.08.040 [DOI] [PubMed] [Google Scholar]
- Liu Z., Yang C. P., Sugino K., Fu C. C., Liu L. Y. et al. , 2015. Opposing intrinsic temporal gradients guide neural stem cell production of varied neuronal fates. Science 350: 317–320. 10.1126/science.aad1886 [DOI] [PubMed] [Google Scholar]
- Luhur A., Chawla G., and Sokol N. S., 2013. MicroRNAs as components of systemic signaling pathways in Drosophila melanogaster. Curr. Top. Dev. Biol. 105: 97–123. 10.1016/B978-0-12-396968-2.00004-X [DOI] [PubMed] [Google Scholar]
- Luhur A., Chawla G., Y.-C Wu, Li J., Sokol and N. S., 2014. Drosha-independent DGCR8/Pasha Pathway Regulates Neuronal Morphogenesis Proc Natl Acad Sci U S A 111: 1421–1426. 10.1073/pnas.1318445111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makunin I. V., Kholodilov N. G., and Shestoapal S. A., 1996. Nucleotide sequences from proximal part of broad-complex locus of D. melanogaster. Drosoph. Inf. Serv. 77: 100–101. [Google Scholar]
- Marchetti, G., and G. Tavosanis, 2017 Steroid Hormone Ecdysone Signaling Specifies Mushroom Body Neuron Sequential Fate via Chinmo. Curr Biol 27: 3017–3024 e3014. 10.1016/j.cub.2017.08.037 [DOI] [PubMed]
- Maurange C., Cheng L., and Gould A. P., 2008. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133: 891–902. 10.1016/j.cell.2008.03.034 [DOI] [PubMed] [Google Scholar]
- Mugat B., Brodu V., Kejzlarova-Lepesant J., Antoniewski C., Bayer C. A. et al. , 2000. Dynamic expression of broad-complex isoforms mediates temporal control of an ecdysteroid target gene at the onset of Drosophila metamorphosis. Dev. Biol. 227: 104–117. 10.1006/dbio.2000.9879 [DOI] [PubMed] [Google Scholar]
- Pasquinelli A. E., Reinhart B. J., Slack F., Martindale M. Q., Kuroda M. I. et al. , 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89. 10.1038/35040556 [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M., and Giegerich R., 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517. 10.1261/rna.5248604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford L. M., Truman J. W., and Nern A., 2018. Juvenile hormone reveals mosaic developmental programs in the metamorphosing optic lobe of Drosophila melanogaster. Biol. Open 7: 1–13. 10.1242/bio.034025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. A., Williams D. W., and Truman J. W., 2011. The BTB/POZ zinc finger protein Broad-Z3 promotes dendritic outgrowth during metamorphic remodeling of the peripheral stretch receptor dbd. Neural Dev. 6: 39 10.1186/1749-8104-6-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere L. F., Dubrovsky E. B., Dubrovskaya V. A., Berger E. M., and Ambros V., 2002. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev. Biol. 244: 170–179. 10.1006/dbio.2002.0594 [DOI] [PubMed] [Google Scholar]
- Sempere L. F., Sokol N. S., Dubrovsky E. B., Berger E. M., and Ambros V., 2003. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev. Biol. 259: 9–18. 10.1016/S0012-1606(03)00208-2 [DOI] [PubMed] [Google Scholar]
- Sokol N. S., 2012. Small temporal RNAs in animal development. Curr. Opin. Genet. Dev. 22: 368–373. 10.1016/j.gde.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol N. S., Xu P., Jan Y. N., and Ambros V., 2008. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 22: 1591–1596. 10.1101/gad.1671708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spokony R. F., and Restifo L. L., 2007. Anciently duplicated Broad Complex exons have distinct temporal functions during tissue morphogenesis. Dev. Genes Evol. 217: 499–513. 10.1007/s00427-007-0159-y [DOI] [PubMed] [Google Scholar]
- Spokony R. F., and Restifo L. L., 2009. Broad Complex isoforms have unique distributions during central nervous system metamorphosis in Drosophila melanogaster. J. Comp. Neurol. 517: 15–36. 10.1002/cne.22119 [DOI] [PubMed] [Google Scholar]
- Sugimura K., Satoh D., Estes P., Crews S., and Uemura T., 2004. Development of morphological diversity of dendrites in Drosophila by the BTB-zinc finger protein abrupt. Neuron 43: 809–822. 10.1016/j.neuron.2004.08.016 [DOI] [PubMed] [Google Scholar]
- Syed M. H., Mark B., and Doe C. Q., 2017. Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. eLife 6: 1–24. 10.7554/eLife.26287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman J. W., and Riddiford L. M., 2019. The evolution of insect metamorphosis: a developmental and endocrine view. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374: 20190070 10.1098/rstb.2019.0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman J. W., Schuppe H., Shepherd D., and Williams D. W., 2004. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development 131: 5167–5184. 10.1242/dev.01371 [DOI] [PubMed] [Google Scholar]
- Venkatasubramanian L., and Mann R. S., 2019. The development and assembly of the Drosophila adult ventral nerve cord. Curr. Opin. Neurobiol. 56: 135–143. 10.1016/j.conb.2019.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. C., Chen C. H., Mercer A., and Sokol N. S., 2012. let-7-Complex MicroRNAs Regulate the Temporal Identity of Drosophila Mushroom Body Neurons via chinmo. Dev. Cell 23: 202–209. 10.1016/j.devcel.2012.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Williams D. W., Altman J., Riddiford L. M., and Truman J. W., 2009. Temporal patterns of broad isoform expression during the development of neuronal lineages in Drosophila. Neural Dev. 4: 39 10.1186/1749-8104-4-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Lin S., Kao C. F., Awasaki T., Chiang A. S. et al. , 2006. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell 127: 409–422. 10.1016/j.cell.2006.08.045 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The sequence of the broad-Z3 cDNA found in Supplemental Figure 1 has been also deposited in GenBank (accession number MN990459). The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25387/g3.11858517.