Abstract
Camelina sativa (L.) Crantz an oilseed crop of the Brassicaceae family is gaining attention due to its potential as a source of high value oil for food, feed or fuel. The hexaploid domesticated C. sativa has limited genetic diversity, encouraging the exploration of related species for novel allelic variation for traits of interest. The current study utilized genotyping by sequencing to characterize 193 Camelina accessions belonging to seven different species collected primarily from the Ukrainian-Russian region and Eastern Europe. Population analyses among Camelina accessions with a 2n = 40 karyotype identified three subpopulations, two composed of domesticated C. sativa and one of C. microcarpa species. Winter type Camelina lines were identified as admixtures of C. sativa and C. microcarpa. Eighteen genotypes of related C. microcarpa unexpectedly shared only two subgenomes with C. sativa, suggesting a novel or cryptic sub-species of C. microcarpa with 19 haploid chromosomes. One C. microcarpa accession (2n = 26) was found to comprise the first two subgenomes of C. sativa suggesting a tetraploid structure. The defined chromosome series among C. microcarpa germplasm, including the newly designated C. neglecta diploid née C. microcarpa, suggested an evolutionary trajectory for the formation of the C. sativa hexaploid genome and re-defined the underlying subgenome structure of the reference genome.
Keywords: Camelina, Domestication, cryptic species, Reference genome, Subgenome, related species
Camelina sativa (L.) Crantz is an ancient oilseed of the Brassicaceae family that contributed to the human diet from the Bronze to the Middle Ages (Hjelmqvist 1979; Hovsepyan and Willcox 2008; Larsson 2013) before losing favor to higher yielding relatives. More recently it has shown potential to become a low-input high value oil crop for the food and feed industry (Faure and Tepfer 2016). Several advantages of this species have been reported (Brown et al. 2016; Ye et al. 2016) including the ability to yield well on dry and marginal lands and its unique seed quality traits (Gugel and Falk 2006), particularly its balanced omega fatty acids (Simopoulos 2002). However, improvements can be made to the crop such as increasing seed size for improved harvestability and reducing the glucosinolate content, which is an anti-nutritional in animal feed (Schuster and Friedt 1998; Amyot et al. 2018). Biologically, Camelina species have two crop habits, annual spring and biennial winter types (Berti et al. 2016). Most of the domesticated C. sativa are spring type, whereas the majority of its wild relatives are winter type. Genetic diversity is vital for developing a robust breeding strategy to identify and incorporate the necessary variation for further crop improvement. Thus far, different molecular approaches have been explored to study a range of Camelina germplasm including, RAPD (Vollmann et al. 2005), AFLP (Ghamkhar et al. 2010), SSR (Manca et al. 2013), and SNP marker analyses (Singh et al. 2015); all the studies concluded that there were low levels of genetic diversity available within spring type C. sativa compared to other oilseed crop species.
The genus Camelina has been reported in the literature to contain anywhere from 6 to 11 species, suggesting some taxonomic confusion (Warwick and Al-Shehbaz 2006; Brock et al. 2019). Latterly there appear to be between six and seven commonly accepted species belonging to the genus which range in chromosome number and ploidy level; namely C. sativa (2n= 6x = 40), Camelina microcarpa Andrz. ex DC. (2n = 12, 2n= 4x = 26, 2n = 6x = 40) (Martin et al. 2017), Camelina hispida (Boiss.) Hedge (2n = 2x = 14), Camelina rumelica Velen. (2n = 4x = 26), Camelina neglecta (2n = 2x = 12) (Brock et al. 2019) and Camelina laxa C.A. Mey. (2n = 2x= 12) (Galasso et al. 2015). The seventh species Camelina alyssum is more contentious since current accessions available within genebanks appear indistinguishable from and are inter-fertile with C. sativa; therefore, it was suggested that C. alyssum is a synonym of C. sativa, although this has yet to be adopted by genebanks (Martin et al. 2017; Al-Shehbaz 1987). Although there was a well-documented chromosome series for C. microcarpa until recently there were no reported sub-species; however, Brock et al. (2019) suggested that the smallest C. microcarpa karyotype (2n = 12) should be re-classified as a new species, Camelina neglecta. Currently cultivated C. sativa is considered to be hexaploid with 20 chromosomes in a haploid set, while at least one of the related species (e.g., C. microcarpa) has the same chromosome number (Francis and Warwick 2009) most have lower numbers. The genome sequence of C. sativa suggested a neopolyploid that had evolved from three lower chromosome number species, specifically one n = 6 and two n = 7 species (Kagale et al. 2014). Camelina species such as C. neglecta, C. laxa and C. hispida possess the same haploid chromosome numbers as subgenomes of the hexaploid and recent work has proposed that C. neglecta and C. hispida could indeed be extant progenitors of C. sativa (Mandáková et al. 2019). The study of these lower ploidy species could be instrumental in defining the relationship among the species as well as uncovering the polyploidization history of Camelina (Brock et al. 2019). Defining the relationships between these species at the subgenome level may also help to identify those species that are potential novel sources of allelic variation for introgression into C. sativa.
Camelina microcarpa has been of interest in studies of Camelina diversity as it is believed to be the closest extant relative to domesticated C. sativa and could help in understanding the domestication process in Camelina species, as well as providing novel variation (Brock et al. 2018). The collections of C. microcarpa species in different genebanks suggest that it has a diverse range of origin including the Mediterranean region, Armenia (Brock et al. 2018), Germany, Poland, Czechia, Slovakia and Georgia (Martin et al. 2017; Smejkal 1971). Diversity studies, analyses of genome size and chromosome number along with the success of hybridization efforts between C. microcarpa and C. sativa (Séguin-Swartz et al. 2013; Martin et al. 2019) suggested the close relationship between these two species (Brock et al. 2018; Martin et al. 2017). However, not all the results were so encouraging with varying levels of hybridization success depending on the genotype (Séguin-Swartz et al. 2013). These results were likely due to confusion with the classification of C. microcarpa accessions, either due to disparities in chromosome number and/or crosses being attempted with completely different species such as C. neglecta (Brock et al. 2019; Martin et al. 2017). Such anomalies could have led to an assumption of higher diversity within C. microcarpa species, with the discovery of C. neglecta in particular there is a need to better understand the relationship between the different accessions of C. microcarpa and C. sativa for potential utilization of such germplasm in Camelina breeding programs.
Estimation of genome size using flow cytometry and chromosome counts are common tools to infer ploidy in a species (Johnston et al. 2005; Martin et al. 2017; Brock et al. 2018; Séguin-Swartz et al. 2013). Complementary genomic tools can assist in clearly defining evolutionary relationships between species and in the case of Camelina, the available reference genome for C. sativa can facilitate such analyses (Kagale et al. 2014). Here, we explored genetic diversity using predominantly genotyping by sequencing (GBS) in different Camelina species, with a focus on C. microcarpa. The analyses of these related species suggested a group of C. microcarpa lines could represent a novel cryptic species. In addition, the subgenome structure of the C. sativa reference genome was re-defined and will provide a basis for utilization of the related species in C. sativa breeding. For example, this study identified a range of potentially valuable minor alleles from C. microcarpa, including those in three flowering related genes which may have impacted the Camelina domestication process.
Materials and methods
Plant materials
This study included a collection of 160 C. sativa, 27 C. microcarpa, two C. alyssum, one C. neglecta, one C. laxa, one C. hispida and two C. rumelica to establish the genetic relationship among the accessions (Table S1). The accessions were mainly obtained from Plant Genetic Resources of Canada in Saskatoon (http://pgrc3.agr.gc.ca/). One accession, “Midas”, was a commercial Canadian variety and 12 accessions were commercial varieties from the United States and Europe. Five accessions are breeding lines from the Agriculture and Agri-Food Canada Saskatoon Research and Development Centre (provided by Dr. Christina Eynck) and the remainder of the lines were thought to originate from eastern Europe and the Russian-Ukraine region and were donated from the National Centre for Plant Genetic Resources of Ukraine in Kharkiv.
Flow cytometry analysis
The relative genome sizes of six different Camelina species were measured using flow cytometry according to the method described in Garcia et al. (2004). Approximately 1 cm2 of leaf tissue of both sample and an internal standard was placed in a plastic petri dish with 2 ml of Galbraith buffer (Galbraith et al. 1983), the mixture was chopped up with a razor blade and the solution was supplemented with 200 µg of ribonuclease A, before being filtered through a filter with a pore size of 30 µm. Propidium iodide was then added at a concentration of 60 µg/ml. The stained solution was kept at 4° for 2 hr and allowed to incubate at room temperature for an hour before taking measurements. DNA content of the nuclei from each species was estimated using fluorescence measurements with a green laser (532 nm) in a CyFlow Space Flow Cytometer (Partec). Camelina sativa (TMP23992) having known ploidy level and genome size (Kagale et al. 2014; Martin et al. 2017) was used as an internal standard to estimate the genome size of lower ploidy species. For all accessions three biological replicates were used.
Chromosome counts
For this study, seeds from six accessions (C. sativa TMP23992, C. neglecta PI650135, C. hispida PI650133, C. microcarpa CN119243, C. microcarpa TMP24026 and C. microcarpa TMP23999) were germinated on moist filter paper in Petri dishes at room temperature. Chromosome counts were carried out based on the protocols detailed in Harrison and Heslop-Harrison (1995) and Snowdon et al. (1997) with minor modifications. Growing root tips (1-2 cm) were collected into tubes containing 0.04% 8-hydroxyquinoline solution (290 mg 8-hydroxyquinoline powder dissolved in 1 L H2O via treatment at 60° for 2 hr, then stored at -4° until use). The root-tip-containing solution was incubated in the dark for 2 hr at room temperature followed by incubation at 4° for 2 hr. Cells were fixed with Carnoy’s I solution (3 parts ethanol to 1 part glacial acetic acid) for 2 days at room temperature. After fixation the root tips were stored in 70% ethanol at -20°. The fixed root tips were rinsed twice for 10 min with distilled water to remove the fixative and incubated in 0.1 M pH 4.5 citrate solution (1.47 g trisodium citrate-dihydrate (Na3C6H5O70.2H2O) and 1.05 g citric acid monohydrate (C6H8O7.H2O) in 500 mL water) for 15 min at room temperature followed by incubation in enzyme solution (0.25 g (5%) Onozuka R-10 cellulase and 0.05 g (1%) pectinase in 5 mL citrate solution) for another 30-40 min at 37°. Root tips were washed with distilled water for 30 min and placed onto a slide with a few drops of Carnoy’s I solution. On the slide, the root tissue was scrambled with a pin and left until the solution dried. Finally, a drop of DAPI staining solution VECTASHIELD Antifade Mounting Medium with DAPI (4,6-diamidino-2-phenylindole; product number H-1200 from Vector Laboratories) was added and covered with a coverslip before observing under UV fluorescence using a Leica DRME microscope at 1000 × magnification.
DNA extraction
Immature leaf samples were collected for DNA extraction. Leaf tissue was stored at -80° prior to DNA extraction. All the samples were freeze-dried for at least 48 hr before lysis. DNA extractions were performed using a CTAB method (2% CTAB, 100mM Tris-HCl, 20mM EDTA, 1.4M NaCl) (Murray and Thompson 1980). After DNA extraction, samples were treated with RNase at 37° to remove RNA contamination. Quantification of DNA was performed with Quant-iT PicoGreen dsDNA Assay Kit (ThermoFisher Scientific) through fluorescence measured (485nm/535nm, 0.1s) using the Victor XPlate Reader (PerkinElmer).
Library preparation and DNA sequencing
Genotyping was performed by an established GBS method (Poland et al. 2012). After DNA normalization (20 ng/ul), 200 ng of DNA were digested with PstI and MspI at 37° for 2 hr. Next, adapters were ligated to the restriction digested DNA fragments using T4 DNA ligase at 22° for 2 hr. The products were inactivated before multiplexing and 96 samples were pooled into a single library. After pooling, the library was amplified with a short extension time (30 sec) and purified using a QIAquick PCR Purification Kit (Qiagen). The final libraries were quantified using a Bioanalyzer (Agilent Technologies) to confirm the fragment size and quality of the library. Sequencing of 35 C. sativa, 9 C. microcarpa, 1 C. rumelica and one C. alyssum were completed on an Illumina HiScan SQ module (paired-end 100 bp reads) and the remainder were sequenced on an Illumina HiSeq2500 platform (paired-end 125 bp reads).
DNA sequence analysis
An existing pipeline was used to demultiplex the reads and trim the reads for adapters, short reads and poor quality data using Trimmomatic (Bolger et al. 2014). Leading and trailing bases with quality below 15 and reads shorter than 55 bp were removed prior to mapping to the reference genome. The trimmed sequence reads were aligned with the reference genome of hexaploid C. sativa (Kagale et al. 2014) using Bowtie2 (Langmead and Salzberg 2012). In bowtie2 mapping, –local with -sensitive parameters were used with –score-min of L,0,0.8. In addition, a custom perl script was used to extract the single best unique hits. Obtained binary files (BAM) were used for variant calling as well as mapping sequence distribution. BEDTools (Quinlan and Hall 2010) was used to extract mapped reads and calculate the frequency of mapped reads along 100 Kb bins in the genome. Circos (Krzywinski et al. 2009) was used to plot the distribution of mapped reads along the C. sativa reference genome for the diploid, tetraploid and hexaploid Camelina genotypes. UnifiedGenotyper with standard parameters from the Genome Analysis Toolkit (McKenna et al. 2010) was used to call SNPs.
Population differentiation
Obtained SNPs were analyzed for average dissimilarity between genotypes and Principle Coordinate Analysis (PCoA) was performed utilizing AveDissR Package (Yang and Fu 2017) in the R program (R Core Team 2017). Population structure was determined using Bayesian technique in STRUCTURE (Pritchard et al. 2000) with a burn-in period of 150,000 steps and 150,000 MCMC replicates where parallelization was performed with StrAuto tool (Chhatre and Emerson 2017). To determine optimal K, three replications were run with each value of K from 1 to 10. The value of K was converted into LnP(K) to obtain the plateau of ΔK. The optimal K was determined using the online version of “Structure harvester” (Earl 2012). PowerMarker (Liu and Muse 2005) was used to calculate gene diversity, Polymorphic Information Content (PIC) and Nei’s (1983) based genetic distance between the genotypes. MEGA 7 (Kumar et al. 2016) was used to construct the Neighbor Joining (NJ) tree among the genotypes. The phylogenetic tree was confirmed through the use of the maximum likelihood method (Tamura and Nei 1993) in MEGA 7 using bootstrap consensus tree (Felsenstein 1985) inferred from 1000 replicates, no significant differences were noted between the alternate tree structures (Figure S5). Analysis of Molecular Variance (AMOVA) and pairwise FST were calculated using GeneAlEx 6.5 (Peakall and Smouse 2006, 2012).
Subgenome dominance
Data previously published by Kagale et al. (2016) was re-analyzed. The expression data from 12 tissues of C. sativa were arranged according to the re-defined subgenome structure and filtered for expression less than 0.01 TPM for all replicates. The 12 tissues were Germinating Seed (GS), Cotyledon (C), Young leaf (YL), Root (R), Stem (S), Senescing leaf (SL), Bud (BUD), Flower (F), Early seed development (ESD), Early mid seed development (EMSD), Late mid seed development (LMSD) and Late seed development (LSD). Filtering provided data for a range of expressed triplicated genes, from 9149 in LSD to 12634 triplets in Root (Table S10), which were analyzed for subgenome dominance in C. sativa. The analysis was performed using analysis of variance techniques where effects due to replication were kept as random. Genes that were expressed significantly (P-value <0.05) higher in any subgenome compared to the other two were considered dominant.
Data availability
Supplemental data (Tables S1-S10; Figures S1-S6), as well as variant data, are provided through figshare: https://doi.org/10.25387/g3.11299280.
Results
Identification of ploidy series among Camelina species
GBS was performed for 193 Camelina accessions, high-quality sequence reads were aligned to the reference genome of C. sativa, DH55 (Kagale et al. 2014). The number of reads per line and alignment rate is summarized in Table S2. As expected, consistent read coverage was found across all 20 linkage groups of the reference genome for all accessions of C. sativa and C. alyssum. However, for particular Camelina accessions the results showed biased read mapping across the reference linkage groups (Figure 1, Table S2, Figure S6). In particular the C. neglecta accession (PI650135) aligned significantly to six chromosomes; whereas, C. microcarpa accessions aligned to either thirteen or 20 chromosomes. For a proportion of the C. microcarpa lines showing read alignment to thirteen chromosomes it was observed that the read depth was somewhat higher for six of those chromosomes, which represented the first of the three sub-genomes of the C. sativa hexaploid (Table S2). In light of the observed bias in read mapping, flow cytometry and chromosome counts were performed to measure the relative size of the nuclear genome content as well as to infer the ploidy level for a subset of the different Camelina accessions (Table 2, Figure 2, Figure S1). Camelina sativa (TMP23992) a well-characterized hexaploid with a genome size estimated to be 1.50 pg/2C (Martin et al. 2017) was used as an internal standard to measure the absolute genome size of lower ploidy Camelina species.
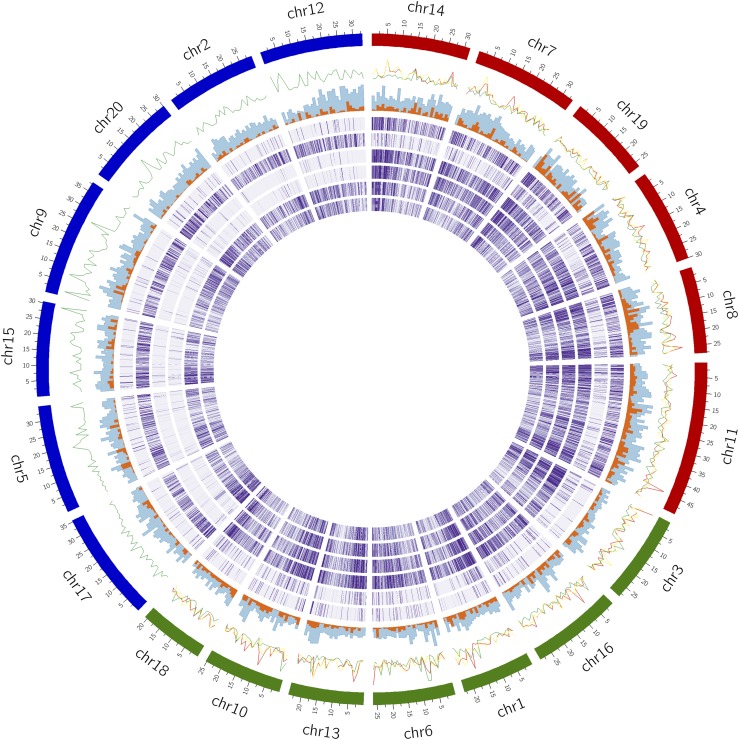
Figure 1.
Identification of ploidy in Camelina species using genotyping by sequencing (GBS) data. From outer to inner track: 1) Clockwise three subgenomes of C. sativa reference genome in red, green and blue; 2) FST distribution across the genome: C. sativa vs. C. microcarpa “Type 1” in green, C. sativa vs. C. microcarpa “Type 2” in red and C. microcarpa “Type 1” vs. C. microcarpa “Type 2” in yellow; 3) SNP distribution of Camelina species in 1 Mb bins in blue and filtered SNPs in orange; 4-9) Heat maps showing read alignment of diploid genotype C. neglecta (PI650135), C. hispida (PI650133), tetraploid C. microcarpa (CN119243), C. microcarpa “Type 2” (TMP23999), C. microcarpa “Type 1” (TMP26172) and C. sativa (TMP23992) to the reference genome.
Table 2. Genome size estimation of different Camelina species using flow cytometry.
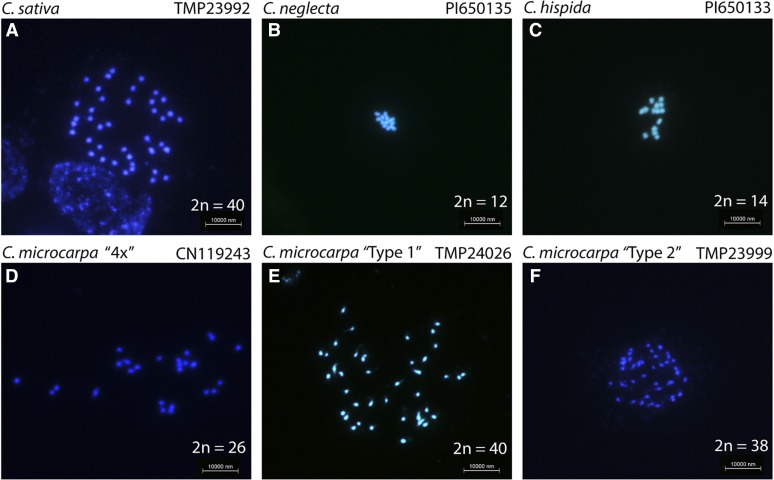
Figure 2.
Chromosome counts for different Camelina species. a) C. sativa TMP23992 (2n = 40); b) C. neglecta PI650135 (2n = 12); c) C. hispida PI650133 (2n = 14); d) C. microcarpa “4x” CN119243 (2n = 26); e) Camelina microcarpa “Type 1” TMP24026 (2n = 40); and f) C. microcarpa “Type 2” TMP23999 (2n = 38).
For the known diploid C. neglecta (2n = 12) genotype (PI650135) (previously C. microcarpa) the GBS data mapped to only six chromosomes thus correlated well with the expected results. This line also had the lowest genome size (0.43 pg/2C) in comparison to C. sativa (1.50 pg/2C). Also as expected the diploid species, C. hispida was found to have 2n = 14 chromosomes with a relatively similar genome size of 0.59 pg/2C as of diploid C. neglecta. For the C. hispida GBS reads, there was a significant bias in mapping with just over 57% of the reads mapped to the third subgenome of the reference C. sativa genome (Figure 1, Figure S6). This might indicate an affinity of C. hispida with the third subgenome of reference C. sativa (Mandáková et al. 2019).
More interestingly, of the C. microcarpa lines where the GBS data aligned with 13 linkage groups from the reference genome, only one genotype (CN119243) possessed a lower genome size (0.95 pg/2C) in comparison to the hexaploids, and based on the read alignments as well as chromosome counts was inferred to be tetraploid (2n = 26) (Figure 1 and 2). Seven genotypes from C. microcarpa (hereafter referred to as “Type 1”) showed consistent read coverage across all chromosomes from the reference genome of C. sativa, while GBS data from 18 C. microcarpa genotypes (hereafter referred to as “Type 2”) aligned with only 13 linkage groups but with a somewhat higher read coverage in the first subgenome (Table S2). Camelina microcarpa (TMP24026), representing the “Type 1” group, had 2n = 40 chromosomes, as expected. However, C. microcarpa (TMP23999), representing the “Type 2” group, had an estimated DNA content (1.49 pg/2C) similar to that of C. sativa yet was found to have 38-40 chromosomes, most likely 2n = 38 (Figure 2). Estimates for this latter line were slightly confounded by the large variation in size between chromosomes and are hence presented with reasonable but not 100% certainty. Sub-genome 1 of C. sativa, with only six chromosomes possesses a larger “fusion” chromosome (Csa-11), it would seem likely that the unidentified six chromosome sub-genome of Type 2 C. microcarpa has a similar “fusion” chromosome which would interfere with accurate chromosome counts; see Figure 3a.
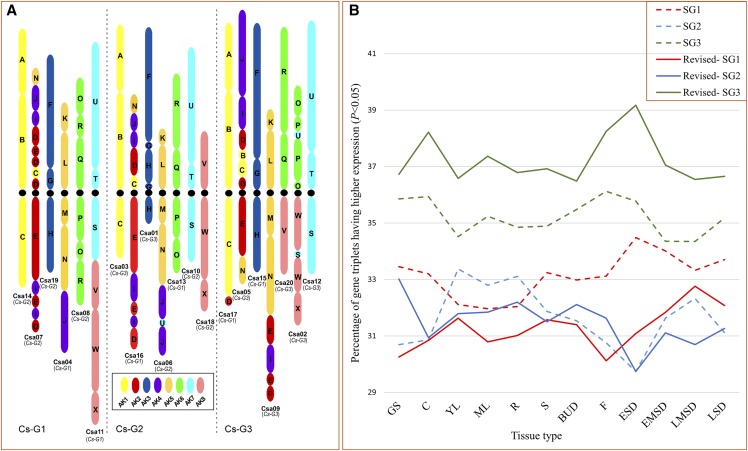
Figure 3.
Re-defining the Camelina sativa subgenome composition. a) Newly defined subgenome architecture of C. sativa; b) Evidence of genome dominance based on refined subgenome structure and gene expression data (GS: Germinating Seed, C: Cotyledon, YL: Young Leaf, ML: Senescing Leaf, R: Root, S: Stem, BUD: Bud, F: Flower, ESD: Early Seed Development; EMSD: Early Mid Seed Development, LMSD: Late Mid Seed Development and LSD: Late Seed Development).
Of the 13 chromosomes showing read alignment for the C. microcarpa “Type 2” group, six chromosomes were shared with the diploid species C. neglecta and seven with subgenome 2 of C. sativa, while the apparently missing chromosomes comprise subgenome 3, to which reads from the diploid C. hispida also align. These results suggested two different types of higher chromosome number C. microcarpa accessions (Type 1: 2n = 40 and Type 2: 2n = 38) with similar genome sizes; one which shares the genome organization as that of the reference C. sativa genome and the second which shares only two subgenomes with that of the reference. Thus, representatives of diploid, tetraploid and two different hexaploid Camelina “species” could be differentiated. The tetraploid C. rumelica (TMP24027) (Martin et al. 2017), previously suggested as a progenitor of C. sativa (Mandáková et al. 2019), had a higher nuclear genome content (1.26 pg/2C) than the tetraploid C. microcarpa (CN119243; 2n = 26). The read alignment data of C. rumelica mapped to all chromosomes with no observable pattern; this ambiguity with regards to its relationship to the subgenomes of C. sativa would not be expected if C. rumelica was indeed a progenitor genome (Table S2, Figure S6). Further accessions of this line would need to be tested.
A refined subgenome structure for C. sativa
The increase in ploidy level in Camelina species from 2n = 12 in C. neglecta to 2n = 26 and 2n = 40 in C. microcarpa might be expected to correspond to the three subgenomes of C. sativa as defined in the reference genome (Kagale et al. 2014); however, this was not the case. The original assignment of reference pseudo-molecules to each of the subgenomes used synteny analyses to identify the most parsimonious route, minimizing genome-restructuring events, from the ancestral karyotype of the Brassicaceae to the modern day C. sativa genome (Kagale et al. 2014). However, it was recognized at the time that some linkage groups, for example Csa14 and Csa03, shared the same basic chromosome structure and their subgenome assignment was more difficult. Thus based on the GBS read alignments and the assumption that the simplest path to the hexaploid genome is through the hybridization of identified lower chromosome number species the subgenome structure has been refined. More explicitly it was assumed that C. neglecta is an extant relative of subgenome 1, the tetraploid C. microcarpa CN119243 represents the second stage in the evolutionary path and is composed of subgenome 1 and 2, and finally hexaploid C. microcarpa (2n = 40) is a direct ascendant of C. sativa, comprised of all three subgenomes; where the origin of the third subgenome is still unclear, although likely a relative of C. hispida. Thus the new genome organization is as follows Subgenome 1 (SG1) contains Csa14, Csa07, Csa19, Csa04, Csa08 and Csa11, which are shared with the diploid C. neglecta (formerly C. microcarpa); SG2 is composed of Csa03, Csa16, Csa01, Csa06, Csa13, Csa10 and Csa18 that along with SG1 are in common with the tetraploid C. microcarpa CN119243; and finally SG3 that is found in all C. sativa lines consists of Csa17, Csa05, Csa15, Csa09, Csa20, Csa02 and Csa12, which are also shared with C. hispida (Figure 1, Figure 3a). As shown in Figure 3a the majority of the re-assignments were between SG1 and SG2, with four chromosomes changing in each instance, only two chromosomes from SG3 were re-assigned. There was no suggestion of chromosomal rearrangements, although this will have to be confirmed through either genetic mapping and/or genome sequencing of the lower ploidy species. It was noted that one scaffold assigned to SG3 was found to have a high read depth when reads were aligned from C. microcarpa “Type 2”, which was an anomaly in the mapping pattern and could indicate a miss-assembly, which again will need to be confirmed through sequencing. The refined subgenome organization was used for all subsequent analyses.
Population differentiation in Camelina species
Depending upon the distribution of the read alignments against the reference genome and corroborated by the chromosome counts and nuclear DNA content, only one genotype each belonged to C. neglecta, tetraploid C. microcarpa, C. hispida and C. laxa; two genotypes were classified as C. rumelica, and two as C. alyssum; seven genotypes were hexaploid C. microcarpa with 20 chromosomes, while, 18 genotypes belonged to C. microcarpa “Type 2” with putatively 19 chromosomes and a novel hexaploid structure compared to the C. sativa reference genome (e.g., TMP23999); the remaining 160 genotypes were classified as C. sativa with 20 chromosomes (Table S1).
Prior to filtering, variant calling in all 193 genotypes yielded 102,744 SNPs across the C. sativa reference genome where a significant proportion of SNPs were from the related species (Table S3). Due to the presence of these distant relatives and the presumption of novel alleles being captured, raw SNPs were filtered for a minor allele frequency of greater than 1% among all samples and after allowing varying levels of missing data points (Figure S2), SNPs with 20% of the genotypes with missing data were selected, providing 4803 variants including indels for all the Camelina species studied (Figure 1). These SNPs were further filtered for indels yielding 4268 SNPs which were used to study population structure and genetic diversity in Camelina species.
The SNP distribution across the subgenomes reflected the genome composition of the total collection of accessions; with the first subgenome having a greater number of SNPs in comparison to the second and third; and the third subgenome having the lowest number of SNPs (Table 1). Gene diversity was found to be low for all chromosomes, similarly the PIC values were low; however, the range for these parameters was high across all chromosomes (Table 1). These results were somewhat skewed due to the genotypes from C. microcarpa “Type 2” and other related species which led to lower coverage in the third subgenome therefore an independent analysis was performed with the 169 genotypes with the same 20 chromosomes as that of the reference genome (Table S4). Removing the related Camelina species reduced the overall number of SNPs but also filtered out less polymorphic loci leading to higher average gene diversity and average PIC values for each of the chromosomes. Likewise, the analysis among the genotypes of domesticated C. sativa species (162 genotypes) including C. alyssum and C. sativa ssp. pilosa suggested an overall gene diversity of 0.181 and PIC value of 0.15 (Table S5).
Table 1. Genetic diversity parameters for 193 Camelina genotypes belonging to 8 species. The numbers in parenthesis indicate range.
| Subgenome | Chromosome | Total SNP | Filtered SNP | Gene Diversity | PIC |
|---|---|---|---|---|---|
| SGI | Chr14 | 5754 | 263 | 0.117 (0.021-0.499) | 0.103 (0.020-0.375) |
| Chr7 | 6280 | 235 | 0.130 (0.021-0.499) | 0.114 (0.021-0.374) | |
| Chr19 | 5209 | 298 | 0.111 (0.021-0.500) | 0.098 (0.020-0.375) | |
| Chr4 | 5462 | 271 | 0.127 (0.021-0.500) | 0.111 (0.021-0.375) | |
| Chr8 | 5535 | 309 | 0.101 (0.021-0.500) | 0.091 (0.020-0.375) | |
| Chr11 | 9593 | 550 | 0.120 (0.021-0.500) | 0.105 (0.021-0.410) | |
| Subtotal | 37833 | 1926 | 0.118 (0.021-0.500) | 0.104 (0.020-0.410) | |
| SGII | Chr3 | 3642 | 166 | 0.117 (0.021-0.498) | 0.102 (0.021-0.374) |
| Chr16 | 4333 | 207 | 0.135 (0.021-0.500) | 0.118 (0.021-0.375) | |
| Chr1 | 3406 | 195 | 0.112 (0.021-0.495) | 0.101 (0.020-0.372) | |
| Chr6 | 3477 | 153 | 0.146 (0.021-0.500) | 0.126 (0.021-0.375) | |
| Chr13 | 3337 | 146 | 0.110 (0.021-0.499) | 0.097 (0.021-0.375) | |
| Chr10 | 3614 | 208 | 0.119 (0.021-0.500) | 0.104 (0.021-0.375) | |
| Chr18 | 2740 | 167 | 0.111 (0.021-0.495) | 0.099 (0.021-0.373) | |
| Subtotal | 24549 | 1242 | 0.122 (0.021-0.498) | 0.107 (0.021-0.374) | |
| SGIII | Chr17 | 5200 | 139 | 0.102 (0.021-0.397) | 0.094 (0.021-0.318) |
| Chr5 | 4993 | 156 | 0.137 (0.021-0.500) | 0.120 (0.021-0.375) | |
| Chr15 | 4726 | 152 | 0.082 (0.021-0.406) | 0.075 (0.021-0.324) | |
| Chr9 | 6603 | 186 | 0.084 (0.022-0.499) | 0.076 (0.022-0.374) | |
| Chr20 | 5031 | 105 | 0.089 (0.021-0.494) | 0.079 (0.021-0.372) | |
| Chr2 | 4451 | 122 | 0.099 (0.021-0.498) | 0.089 (0.021-0.374) | |
| Chr12 | 6450 | 188 | 0.106 (0.021-0.494) | 0.093 (0.021-0.372) | |
| Subtotal | 37454 | 1048 | 0.100 (0.021-0.470) | 0.089 (0.021-0.359) | |
| Scaffolds | 2908 | 52 | |||
| Total SNPs | 102744 | 4268 | 0.114 (0.020-0.500) | 0.101 (0.000-0.410) |
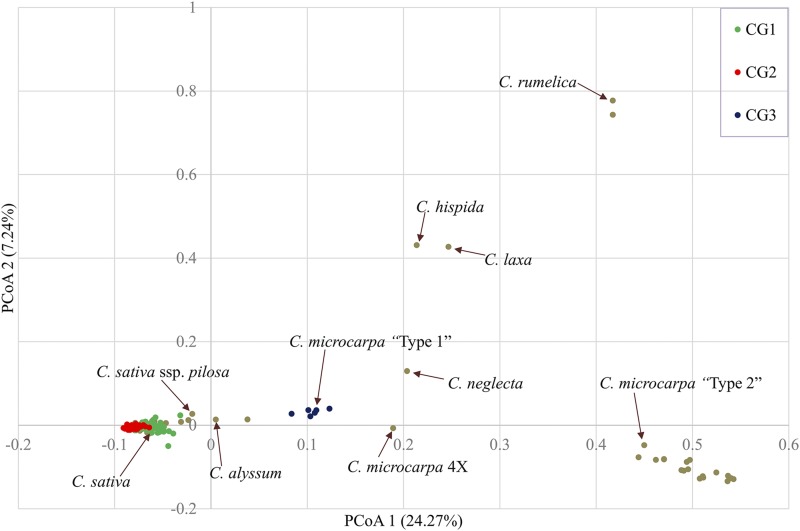
Principle coordinate analysis (PCoA) differentiated the related species from the C. sativa population including C. alyssum and C. sativa ssp. pilosa (Figure 4). The first coordinate explains 24.27% of the variation, which differentiated C. sativa from other Camelina relatives; the second coordinate explains 7.24% of variation, which differentiated more distant relatives such as C. rumelica, C. laxa and C. hispida from C. sativa and C. microcarpa. The PCoA result suggested that C. alyssum followed by C. microcarpa “Type 1” genotypes were quite similar to domesticated C. sativa, while C. microcarpa “Type 2”, C. hispida, C. laxa and C. rumelica species were clearly divergent. This analysis mainly differentiated between species; however, separate analysis of Camelina species with 20 chromosomes was used to differentiate among C. sativa genotypes, and to suggest some sub-population structure (Figure S3).
Figure 4.
Principle coordinate analysis of 193 Camelina genotypes based on 4268 SNPs. The different colors represent three subpopulations defined by the STRUCTURE analysis.
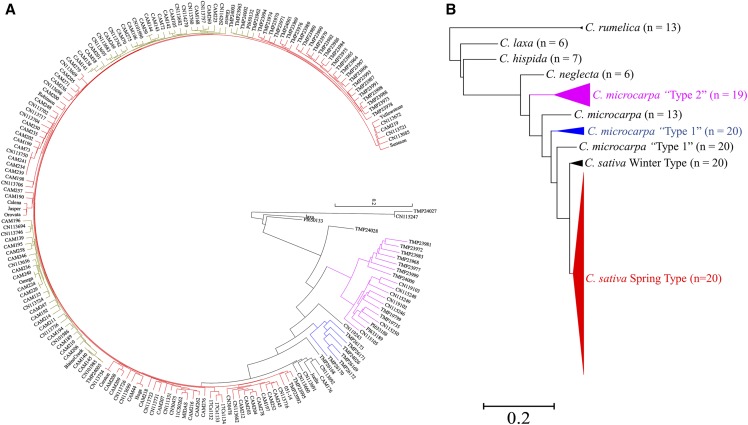
The results from the PCoA were mirrored in the generation of a Neighbor Joining (NJ) tree showing the phylogenetic relationships among the 193 Camelina genotypes (Figure 5). All the domesticated Camelina genotypes were closely related to each other, forming a separate large cluster. The NJ tree showed that the related species, which all share a vernalisation requirement, were clustered next to a number of Camelina lines which were winter types, including C. alyssum (CAM176), C. sativa ssp. pilosa (CN113692) and the line Joelle (North Dakota State University) (Figure 5). Tetraploid C. microcarpa CN119243 formed a separate cluster and was basal to the C. sativa sub-populations, the diploid C. neglecta (PI650135) was basal to all higher chromosome number accessions. One C. microcarpa genotype (TMP26168) had a very similar genomic organization as the reference genome; however, was categorized as C. microcarpa “Type 1” and formed a separate single cluster. Camelina microcarpa “Type 2” species formed their own separate cluster, but showed further sub-population structure, separating into two groups with 11 and 7 genotypes, respectively. Two genotypes belonging to C. rumelica formed a separate cluster along with C. laxa and C. hispida and suggesting these had diverged sometime earlier from the progenitors of domesticated Camelina species.
Figure 5.
Genetic relationship among Camelina accessions as determined by NJ tree construction based on 4268 SNPs. a) Relationship among 193 Camelina accessions; b) Summary of the relationship among different species of Camelina (number in parenthesis indicate number of chromosomes in a haploid set).
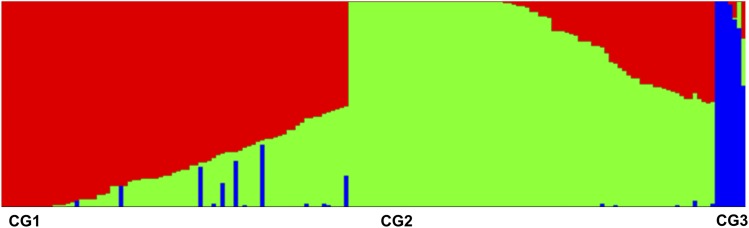
The PCoA and NJ suggested some sub-structure among the domesticated C. sativa accessions, which was further assessed using the Bayesian clustering approach of STRUCTURE (Pritchard et al. 2000). This analysis was performed with the hexaploid Camelina accessions with 20 chromosomes only (n = 169) and suggested two populations confirming the separation of C. microcarpa “Type 1” accessions from C. sativa. The peak of delta K also suggested further population differentiation at K = 3, which identified two sub-populations among the C. sativa accessions. Assuming this three population structure and, based on a Q value cut-off of 70%, 124 genotypes were clustered into three subpopulations with 45 genotypes found to be an admixture of these subpopulations (Table S6, Figure S4). As shown in Figure 6, 162 Camelina genotypes were found in two sub-populations CG1 (red), CG2 (green) and C. microcarpa “Type 1” formed subpopulation CG3 (blue). The genotypes belonging to CG1 and CG2 were spring type whereas the genotypes belonging to CG3 were winter type. One genotype (TMP26168) belonging to C. microcarpa “Type 1” was found to be an admixture of CG3, CG2 and CG1, which confirmed its unique status, noted in the NJ tree analyses. The winter type C. alyssum (CAM176) was also an admixture of CG1, CG2 and CG3, with a higher contribution from subpopulation CG1. Other winter types such as C. sativa ssp. pilosa (CN113692) and C. sativa (Joelle) were grouped with CG1. All the winter type Camelina lines were found to have a contribution of alleles from subpopulation CG3, representing C. microcarpa “Type 1” (Table S6).
Figure 6.
Population structure of Camelina species. CG1 (Red) and CG2 (Green) represent C. sativa genotypes, and CG3 (Blue) represents C. microcarpa “Type 1”.
Pairwise FST values were calculated among the three subpopulations (124 genotypes), excluding the lines showing admixture. The results suggested that spring type Camelina species of subpopulations CG1 and CG2 were closely related with an FST of 0.065. FST values between the two spring Camelina sub-populations and C. microcarpa “Type 1” indicated greater differentiation between the species, with values of 0.302 and 0.349, respectively (Table 3). However, a separate analysis of pairwise FST with all the genotypes irrespective of admixture suggested a lower FST value (0.263) (Table S7d). For all the subpopulation the third subgenome showed higher differentiation among subpopulations in comparison to the other subgenomes (Table S7). The FST analysis between C. sativa and C. microcarpa “Type 1” also suggested strong selection for alleles in C. sativa on chromosome Csa06 in a relatively small region (6Mb to 9 Mb region) (Figure 1).
Table 3. Pairwise FST among three subpopulations of Camelina species. CG1 (58 genotypes) and CG2 (60 genotypes) represent C. sativa genotypes and CG3 (6 genotypes) represents C. microcarpa “Type 1” accessions.
| CG1 | CG2 | CG3 | |
|---|---|---|---|
| CG1 | 0.000 | ||
| CG2 | 0.065 | 0.000 | |
| CG3 | 0.302 | 0.349 | 0.000 |
Related Camelina species as a reservoir of minor alleles
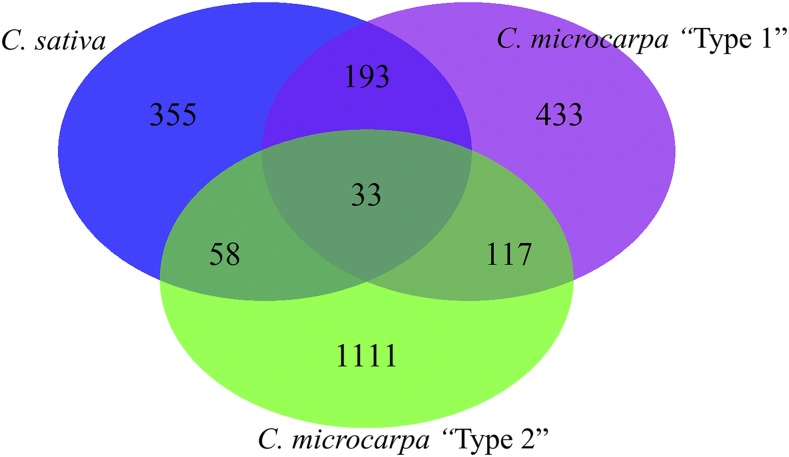
Although, this study included a number of species, approximately 96% of the total samples were either classified as C. sativa, C. microcarpa “Type 1” or C. microcarpa “Type 2”. Among the 4268 filtered SNPs, the number of minor alleles (less than 5% homozygous) were identified for each of the three species, to assess their potential as a source of novel alleles. Such minor alleles were found for 2300 SNPs; only 33 were shared by all three species (Figure 7). Of the minor alleles, 1111 were unique to C. microcarpa “Type 2”, 433 were unique to C. microcarpa “Type 1” and 355 were unique to C. sativa species. The distribution of minor alleles along the subgenomes suggested the first subgenome of both C. sativa and C. microcarpa “Type 2” contained the highest number of minor alleles, while the third subgenome for C. microcarpa “Type 1” contained more minor alleles (Table S8).
Figure 7.
Venn diagram showing distribution of minor alleles in different species of Camelina.
Minor alleles not present in the domesticated C. sativa were explored to identify mutations that may have helped to shape the existing C. sativa accessions through selection for changes to particular genes. Of all the SNPs with minor alleles 536 were within the genic region of 355 genes. Of these, 275 genes had orthologs in Arabidopsis thaliana (Table S8a), although there was no apparent bias for particular functional category, three genes were found to have an influence on flowering time and photoperiod response and could be interesting candidates for manipulating phenology (Table S8b).
Discussion
The current study exploited GBS data and the reference genome of C. sativa to characterize variation among Camelina species, which not only identified a potentially novel Camelina species but also suggested refinements to the underlying subgenome structure of C. sativa. The hexaploid structure of C. sativa was clear from the genome assembly of Kagale et al. (2014); however, the differentiation of the three subgenomes was complicated by the high degree of synteny between particular chromosomes. Phylogenetic analyses of a set of unanchored genome scaffolds of C. neglecta (PI650135) (Toro 2017) also suggested changes to the first subgenome of C. sativa genome, which concurred with the GBS data presented in this study. By alignment of GBS data from the diploid C. neglecta (2n = 12), a presumed tetraploid (C. microcarpa; 2n = 26) and multiple hexaploids (2n = 40) a step-wise hybridization path to the current C. sativa genome was suggested, implicating the diploid and tetraploid line as potential progenitor species of C. sativa. The third subgenome shares significant homology to C. hispida, implying this may represent an extant progenitor of the final subgenome, which is in agreement with the recent work of Mandáková et al. (2019).
After redefining the subgenome composition of C. sativa, there was a slight change in distribution of gene coverage, with a higher number of genes now present on the third subgenome (33.7% compared to 32.7% of total annotated genes) and a slight decrease in the number of genes for the second subgenome (30.2% compared to 31.1% of total genes) (Table S9). Although there was no change in number of genes retained in triplicate, in light of the re-definition of the karyotype, subgenome dominance was re-analyzed based on the previously published gene expression data from Kagale et al. (2016). Depending on the tissue type between 9,188 (late seed development) and 12,688 (root) triplicated orthologous gene sets were analyzed for evidence of genome dominance in C. sativa (Table S10). As found in Kagale et al. (2016) the results suggest dominance of the third subgenome over the other two; however, the impact was far more pronounced (Figure 3b). For all tissue types, the third subgenome had a greater number of genes with higher expression in comparison to both the first and second subgenome, deviating from a hypothetical 1:1:1 ratio of number of genes significantly expressing higher in any one subgenome (χ2 test, P-value < 0.05). There were some tissue specific patterns observed with regards to SG1 and SG2: the second subgenome was found to dominate the first subgenome until flowering, after which the first subgenome dominated the second. However, the ratio of the total number of expressed genes for the third subgenome with either first or second subgenome was not particularly high (∼1.11-1.27), suggesting limited gene silencing, and might reflect the young neopolyploid status of Camelina as suggested by Kagale et al. (Kagale et al. 2014). The marked dominance of the third subgenome, or by inference the genome added last in the stepwise evolution of C. sativa, is in concordance with evidence from other polyploid species with similar evolutionary trajectories (Ramírez-González et al. 2018; Edger et al. 2019; Mandáková et al. 2019).
The chromosome numbers for C. neglecta, C. hispida, C. sativa and C. microcarpa “Type 1” were consistent with previous reports (Martin et al. 2017; Brock et al. 2018). However, C. microcarpa “Type 2” was suggested to have n = 19 chromosomes, noticeably the sequences from this genome mapped to only two of the C. sativa subgenomes, suggesting a hexaploid derived from progenitors with 6, 7 and 6 chromosomes. The available tetraploid (n = 13) which could be a progenitor of both “Type 1” and “Type 2” C. microcarpa suggests two different routes to the formation of the higher ploidy hexaploid genomes in the Camelina genus. The mapping of C. hispida (n = 7) to the third subgenome of C. sativa (Figure 1), also indicated by the results of Mandáková et al. (2019) could suggest hybridization of the tetraploid with C. hispida in the formation of modern hexaploid C. sativa. As yet, the origin of the third subgenome for C. microcarpa “Type 2” remains elusive, although it shares some homology with subgenome 1, suggesting it could be a relative of C. neglecta. The current study did not find clear association of the tetraploid C. rumelica with specific subgenomes of the reference C. sativa, suggesting that greater genetic distance and possibly chromosomal rearrangement separate the two species (Čalasan et al. 2019).
The genetic characterization of the accessions confirmed the low level of differentiation among C. sativa lines (Vollmann et al. 2005; Singh et al. 2015; Luo et al. 2019; Gehringer et al. 2006), yet there was some indication of sub-structure within the C. sativa population. A significant number of recently collected accessions, which originated from the Russian/Ukraine border populated CG1 and could provide a source of some limited variation in C. sativa breeding, but the related hexaploid species offer the potential of much more diversity. It appears that some of this variation may have begun to be captured, in particular with the generation of C. sativa types with a vernalisation requirement. Similarly, it was noted that one apparent C. microcarpa “Type 1” line showed evidence of shared alleles across the three defined sub-populations, including those seemingly specific to C. sativa. The evolutionary history of Camelina hexaploids may have played a role in limiting variation with a smaller number of SNPs found in the second subgenome, which may reflect a small number of hybridization events from which this subgenome was derived. Although C. sativa and C. microcarpa both evolved through polyploidy, C. microcarpa “Type 1” has maintained a greater collection of minor alleles, implicating the influence of selection on a crop which has been subjected to less intensive breeding than most, or again could result from a polyploidization bottleneck. The frequency of minor alleles was higher in the first subgenome of domesticated C. sativa in comparison to C. microcarpa “Type 1” (Table S8) and might indicate further differentiation of C. sativa subpopulations or relate to age of divergence of the subgenomes. The study of minor allele frequencies has been used to understand domestication and potential bottlenecks created during the process, enabling the identification of genes under selection that may underlie QTL controlling traits of interest (Ross-Ibarra et al. 2007). The current study identified a number of genes carrying minor alleles in the wild relative that may represent genes under selection in the crop, further comprehensive sequence analyses and trait association will determine the value of such variation.
Acknowledgments
This work was supported through funding from the Global Institute of Food Security, Saskatoon for the project “Developing Camelina sativa as a modern crop platform”. ASM and LV are supported by Emmy Noether DFG grant MA6473/1-1. The authors would like to thank Dr. V. Ryabchoun and Dr. R. Boguslavsky from the National Center for Plant Genetic Resources of Ukraine at Kharkiv for providing seeds of wild species of Camelina for this project. Similarly, we also would like to thank Tina Bundrock for technical assistance in the flow cytometry analysis.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11299280.
Communicating editor: A. Eckert
Literature Cited
- Al-Shehbaz I. A., 1987. Camelina. J. Arnold Arbor. 68: 234–240. [Google Scholar]
- Amyot L., McDowell T., Martin S. L., Renaud J., Gruber M. Y. et al. , 2018. Assessment of Antinutritional Compounds and Chemotaxonomic Relationships between Camelina sativa and Its Wild Relatives. J. Agric. Food Chem. 67: 796–806. 10.1021/acs.jafc.8b04724 [DOI] [PubMed] [Google Scholar]
- Berti M., Gesch R., Eynck C., Anderson J., and Cermak S., 2016. Camelina uses, genetics, genomics, production, and management. Ind. Crops Prod. 94: 690–710. 10.1016/j.indcrop.2016.09.034 [DOI] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock J. R., Donmez A. A., Beilstein M. A., and Olsen K. M., 2018. Phylogenetics of Camelina Crantz. (Brassicaceae) and insights on the origin of gold-of-pleasure (Camelina sativa). Mol. Phylogenet. Evol. 127: 834–842. 10.1016/j.ympev.2018.06.031 [DOI] [PubMed] [Google Scholar]
- Brock J. R., Mandakova T., Lysak M. A., and Al-Shehbaz I. A., 2019. Camelina neglecta (Brassicaceae, Camelineae), a new diploid species from Europe. PhytoKeys 115: 51–57. 10.3897/phytokeys.115.31704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. D., Hori T. S., Xue X., Ye C. L., Anderson D. M. et al. , 2016. Functional Genomic Analysis of the Impact of Camelina (Camelina sativa) Meal on Atlantic Salmon (Salmo salar) Distal Intestine Gene Expression and Physiology. Mar. Biotechnol. (NY) 18: 418–435. 10.1007/s10126-016-9704-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čalasan A. Ž., Seregin A. P., Hurka H., Hofford N. P., and Neuffer B., 2019. The Eurasian steppe belt in time and space: Phylogeny and historical biogeography of the false flax (Camelina Crantz, Camelineae, Brassicaceae). Flora 260: 151477 10.1016/j.flora.2019.151477 [DOI] [Google Scholar]
- Chhatre V. E., and Emerson K. J., 2017. StrAuto: automation and parallelization of STRUCTURE analysis. BMC Bioinformatics 18: 192 10.1186/s12859-017-1593-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D. A., 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4: 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Edger P. P., Poorten T. J., VanBuren R., Hardigan M. A., Colle M. et al. , 2019. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 51: 541–547. 10.1038/s41588-019-0356-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure J.-D., and Tepfer M., 2016. Camelina, a Swiss knife for plant lipid biotechnology. OCL Oilseeds Fats Crops Lipids. Nat. Genet. 51: 765. [Google Scholar]
- Felsenstein J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Francis, A., and S. I. Warwick, 2009 The Biology of Canadian Weeds. 142. Camelina alyssum (Mill.) Thell.; C. microcarpa Andrz. ex DC.; C. sativa (L.) Crantz. Canadian Journal of Plant Science 89: 791–810.
- Galasso I., Manca A., Braglia L., Ponzoni E., and Breviario D., 2015. Genomic fingerprinting of Camelina species using cTBP as molecular marker. Am. J. Plant Sci. 6: 1184–1200. 10.4236/ajps.2015.68122 [DOI] [Google Scholar]
- Galbraith D. W., Harkins K. R., Maddox J. M., Ayres N. M., Sharma D. P. et al. , 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051. 10.1126/science.220.4601.1049 [DOI] [PubMed] [Google Scholar]
- Garcia S., Sanz M., Garnatje T., Kreitschitz A., McArthur E. D. et al. , 2004. Variation of DNA amount in 47 populations of the subtribe Artemisiinae and related taxa (Asteraceae, Anthemideae): karyological, ecological, and systematic implications. Genome 47: 1004–1014. 10.1139/g04-061 [DOI] [PubMed] [Google Scholar]
- Gehringer A., Friedt W., Lühs W., and Snowdon R. J., 2006. Genetic mapping of agronomic traits in false flax (Camelina sativa subsp. sativa). Genome 49: 1555–1563. 10.1139/g06-117 [DOI] [PubMed] [Google Scholar]
- Ghamkhar K., Croser J., Aryamanesh N., Campbell M., Kon’kova N. et al. , 2010. Camelina (Camelina sativa (L.) Crantz) as an alternative oilseed: molecular and ecogeographic analyses. Genome 53: 558–567. 10.1139/G10-034 [DOI] [PubMed] [Google Scholar]
- Gugel R. K., and Falk K. C., 2006. Agronomic and seed quality evaluation of Camelina sativa in western Canada. Can. J. Plant Sci. 86: 1047–1058. 10.4141/P04-081 [DOI] [Google Scholar]
- Harrison G., and Heslop-Harrison J., 1995. Centromeric repetitive DNA sequences in the genus Brassica. Theor. Appl. Genet. 90: 157–165. 10.1007/BF00222197 [DOI] [PubMed] [Google Scholar]
- Hjelmqvist, H., 1979 Beiträge zur Kenntnis der prähistorischen Nutzpflanzen in Schweden: Verlag nicht ermittelbar.
- Hovsepyan R., and Willcox G., 2008. The earliest finds of cultivated plants in Armenia: evidence from charred remains and crop processing residues in pisé from the Neolithic settlements of Aratashen and Aknashen. Veg. Hist. Archaeobot. 17: 63–71. 10.1007/s00334-008-0158-6 [DOI] [Google Scholar]
- Johnston J. S., Pepper A. E., Hall A. E., Chen Z. J., Hodnett G. et al. , 2005. Evolution of genome size in Brassicaceae. Ann. Bot. (Lond.) 95: 229–235. 10.1093/aob/mci016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Koh C., Nixon J., Bollina V., Clarke W. E. et al. , 2014. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 5: 3706 10.1038/ncomms4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Nixon J., Khedikar Y., Pasha A., Provart N. J. et al. , 2016. The developmental transcriptome atlas of the biofuel crop Camelina sativa. Plant J. 88: 879–894. 10.1111/tpj.13302 [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R. et al. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., and Tamura K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson M., 2013. Cultivation and processing of Linum usitatissimum and Camelina sativa in southern Scandinavia during the Roman Iron Age. Veg. Hist. Archaeobot. 22: 509–520. 10.1007/s00334-013-0413-3 [DOI] [Google Scholar]
- Liu K., and Muse S. V., 2005. PowerMaker: An integrated analysis environment for genetic maker analysis. Bioinformatics 21: 2128–2129. 10.1093/bioinformatics/bti282 [DOI] [PubMed] [Google Scholar]
- Luo Z., Brock J., Dyer J. M., Kutchan T. M., Augustin M. et al. , 2019. Genetic diversity and population structure of a Camelina sativa spring panel. Front. Plant Sci. 10: 184 10.3389/fpls.2019.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca A., Pecchia P., Mapelli S., Masella P., and Galasso I., 2013. Evaluation of genetic diversity in a Camelina sativa (L.) Crantz collection using microsatellite markers and biochemical traits. Genet. Resour. Crop Evol. 60: 1223–1236. 10.1007/s10722-012-9913-8 [DOI] [Google Scholar]
- Mandáková T., Pouch M., Brock J. R., Al-Shehbaz I. A., and Lysak M. A., 2019. Origin and Evolution of Diploid and Allopolyploid Camelina Genomes was Accompanied by Chromosome Shattering. Plant Cell 31: 2596–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. L., Lujan-Toro B. E., Sauder C. A., James T., Ohadi S. et al. , 2019. Hybridization rate and hybrid fitness for Camelina microcarpa Andrz. ex DC (♀) and Camelina sativa (L.) Crantz (Brassicaceae). Evol. Appl. 12: 443–455. 10.1111/eva.12724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. L., Smith T. W., James T., Shalabi F., Kron P. et al. , 2017. An update to the Canadian range, abundance, and ploidy of Camelina spp. (Brassicaceae) east of the Rocky Mountains. Botany 95: 405–417. 10.1139/cjb-2016-0070 [DOI] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K. et al. , 2010. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M. G., and Thompson W. F., 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., and Takezaki N., 1983. Estimation of genetic distances and phylogenetic trees from DNA analysis. Proc. 5th World Cong. Genet. Appl. Livstock Prod. 21: 405–412. [Google Scholar]
- Peakall R., and Smouse P. E., 2006. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6: 288–295. 10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R., and Smouse P. E., 2012. GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539. 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J. A., Brown P. J., Sorrells M. E., and Jannink J.-L., 2012. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7: e32253 10.1371/journal.pone.0032253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., and Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., and Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-González, R., P. Borrill, D. Lang, S. Harrington, J. Brinton et al., 2018 The transcriptional landscape of polyploid wheat. Science 361: eaar6089. 10.1126/science.aar6089 [DOI] [PubMed]
- R Core Team, 2017 R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org
- Ross-Ibarra J., Morrell P. L., and Gaut B. S., 2007. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl. Acad. Sci. USA 104: 8641–8648. 10.1073/pnas.0700643104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster A., and Friedt W., 1998. Glucosinolate content and composition as parameters of quality of Camelina seed. Ind. Crops Prod. 7: 297–302. 10.1016/S0926-6690(97)00061-7 [DOI] [Google Scholar]
- Séguin-Swartz G., Nettleton J. A., Sauder C., Warwick S. I., and Gugel R. K., 2013. Hybridization between Camelina sativa (L.) Crantz (false flax) and North American Camelina species. Plant Breed. 132: 390–396. 10.1111/pbr.12067 [DOI] [Google Scholar]
- Simopoulos A. P., 2002. The importance of the ratio of omega-6/omega-3 essential fatty acids. Biomed. Pharmacother. 56: 365–379. 10.1016/S0753-3322(02)00253-6 [DOI] [PubMed] [Google Scholar]
- Singh R., Bollina V., Higgins E. E., Clarke W. E., Eynck C. et al. , 2015. Single-nucleotide polymorphism identification and genotyping in Camelina sativa. Mol. Breed. 35: 35 10.1007/s11032-015-0224-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smejkal M., 1971. Revision der tschechoslowakischen Arten der Gattung Camelina Crantz (Cruciferae). Preslia 43: 318–337. [Google Scholar]
- Snowdon R., Köhler W., Friedt W., and Köhler A., 1997. Genomic in situ hybridization in Brassica amphidiploids and interspecific hybrids. Theor. Appl. Genet. 95: 1320–1324. 10.1007/s001220050699 [DOI] [Google Scholar]
- Tamura K., and Nei M., 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10: 512–526. [DOI] [PubMed] [Google Scholar]
- Toro B. E. L., 2017. Genome Assembly of Camelina microcarpa Andrz. Ex DC, A step towards understanding genome evolution in Camelina, Carleton University, Ottawa, Ontario. [Google Scholar]
- Vollmann J., Grausgruber H., Stift G., Dryzhyruk V., and Lelley T., 2005. Genetic diversity in camelina germplasm as revealed by seed quality characteristics and RAPD polymorphism. Plant Breed. 124: 446–453. 10.1111/j.1439-0523.2005.01134.x [DOI] [Google Scholar]
- Warwick S. I., and Al-Shehbaz I. A., 2006. Brassicaceae: Chromosome number index and database on CD-Rom. Plant Syst. Evol. 259: 237–248. 10.1007/s00606-006-0421-1 [DOI] [Google Scholar]
- Yang M. H., and Fu Y. B., 2017. AveDissR: An R function for assessing genetic distinctness and genetic redundancy. Appl. Plant Sci. 5: 1700018 10.3732/apps.1700018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C. L., Anderson D. M., and Lall S. P., 2016. The effects of camelina oil and solvent extracted camelina meal on the growth, carcass composition and hindgut histology of Atlantic salmon (Salmo salar) parr in freshwater. Aquaculture 450: 397–404. 10.1016/j.aquaculture.2015.08.019 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplemental data (Tables S1-S10; Figures S1-S6), as well as variant data, are provided through figshare: https://doi.org/10.25387/g3.11299280.