Abstract
Drought stress causes the greatest soybean [Glycine max (L.) Merr.] yield losses among the abiotic stresses in rain-fed U.S. growing areas. Because less than 10% of U.S. soybean hectares are irrigated, combating this stress requires soybean plants which possess physiological mechanisms to tolerate drought for a period of time. Phenotyping for these mechanisms is challenging, and the genetic architecture for these traits is poorly understood. A morphological trait, slow or delayed canopy wilting, has been observed in a few exotic plant introductions (PIs), and may lead to yield improvement in drought stressed fields. In this study, we visually scored wilting during stress for a panel of 162 genetically diverse maturity group VI-VIII soybean lines genotyped with the SoySNP50K iSelect BeadChip. Field evaluation of canopy wilting was conducted under rain-fed conditions at two locations (Athens, GA and Salina, KS) in 2015 and 2016. Substantial variation in canopy wilting was observed among the genotypes. Using a genome-wide association mapping approach, 45 unique SNPs that tagged 44 loci were associated with canopy wilting in at least one environment with one region identified in a single environment and data from across all environments. Several new soybean accessions were identified with canopy wilting superior to those of check genotypes. The germplasm and genomic regions identified can be used to better understand the slow canopy wilting trait and be incorporated into elite germplasm to improve drought tolerance in soybean.
Keywords: soybean, Glycine max, drought tolerance, canopy wilting, genome-wide association study (GWAS)
Soybean [Glycine max (L.) Merrill] is the world’s leading oilseed crop used to produce vegetable oil, protein feed for livestock, biodiesel, and many soyfoods. Drought stress is the most significant abiotic threat to the agricultural productivity of soybean around the world, and can reduce yield by more than 40% (Specht et al. 1999; Purcell and Specht 2004).
Loss of turgor and leaf droop, known commonly as canopy wilting, is an often-observed response to drought stress in soybean. Some exotic soybean types exhibit a slow or delayed canopy wilting response to drought, which may reflect favorable underlying plant mechanisms to access soil moisture, conserve soil moisture prior to stress, or use water more efficiently. One mechanism related to water conservation is to restrict transpiration early in the growing season whenever vapor pressure deficit (VPD) is high, so that plants can utilize saved soil water during pod filling when drought stress in soybean is usually more detrimental to yield. Plant introduction (PI) 416937 is a Japanese maturity group (MG) VI introduction identified in the 1980s as exhibiting slower wilting under water deficit conditions than existing cultivars (Sloane et al. 1990). This PI has an extensive lateral root system, high root surface area (Goldman et al. 1989; Sloane et al. 1990; Hudak and Patterson 1996; Pantalone and Rebetzke 1996), and low stomatal conductance (Tanaka et al. 2010). In a study that evaluated PI 416937 in high VPD conditions, it reached a maximum transpiration rate near 2.0 kPa (unit of pressure), whereas other genotypes continued to increase transpiration rates at much greater than 2.0 kPa (Fletcher et al. 2007). This indicated that water conservation during vegetative growth may be the basis for the slow wilting trait found in PI 416937. PI 471938, a MG V introduction from Nepal, also exhibits the slow wilting trait, but the basis for this trait is unknown (Sadok et al. 2012; Bagherzadi et al. 2017). PI 471938 was also previously identified as expressing N2 fixation tolerance to soil drying (Sinclair et al. 2000; Devi and Sinclair 2013; Riar et al. 2018). These two plant introductions are being used as sources of slow wilting in applied breeding programs (Devi et al. 2014; Carter et al. 2016). Two additional MG III PIs were also previously identified that have reduced yield loss under drought stress and delayed leaf wilting (Pathan et al. 2014).
The canopy wilting trait has been mapped using linkage and genome-wide association mapping approaches. Charlson et al. (2009) mapped four QTL on chromosomes (Chr) 8, 13, 14, and 17 that collectively explained 47% of phenotypic variation in a KS4895 × ‘Jackson’ RIL population (Johnson 1958; Hwang et al. 2015a). Eight QTL were identified for canopy wilting under field and greenhouse conditions in Du et al. (2009) using 184 RILs derived from Kefeng No. 1 × Nannong1138-2. Using 150 RILs derived from the hybridization of ‘Benning’ (Boerma et al. 1997) and PI 416937, seven QTL were identified by Abdel-Haleem et al. (2012) that explained 75% of the variation in canopy wilting. Hwang et al. (2015b) identified eight QTL clusters that had QTL from at least two of five different RIL populations (93705 KS4895 × Jackson, 08705 KS4895 × Jackson, KS4895 × PI 424140, ‘A5959’ × PI 416937, and Benning × PI 416937) responsible for canopy wilting. With these same populations, Hwang et al. (2016) performed a meta-analysis on nine QTL to refine map positions and reduce confidence intervals for the eight QTL clusters reported by Hwang et al. (2015b). Kaler et al. (2017b) used a genome-wide association analysis of 373 MG IV genotypes to identify 61 single nucleotide polymorphism (SNP) markers associated with canopy wilting, which tagged 51 different genetic loci. Of the 373 genotypes tested, 185 genotypes had lower canopy wilting scores across environments than PI 416937 (Kaler et al. 2017b).
There are approximately 170,000 soybean accessions maintained in germplasm collections worldwide, and the USDA maintains a collection of around 20,000 accessions. However, only a limited number of these genotypes have been screened for stress tolerance, and few have been identified as tolerant and used in soybean breeding programs to improve drought tolerance (Carter et al. 1999, 2004, 2016; Devi et al. 2014; Sinclair et al. 2016). Identification of new accessions with beneficial alleles for drought tolerance related traits, including slow canopy wilting, could help in the development of drought tolerant soybean cultivars. To aid in the search for beneficial alleles, the SoySNP50K and SoySNP6K iSelect BeadChips are available for high-throughput genotyping that supports QTL mapping efforts. In addition, the entire USDA soybean germplasm collection has been genotyped with the SoySNP50K iSelect BeadChips (Song et al. 2013, 2014, and 2015).
Genome-wide association studies (GWAS) allow for the opportunity to identify the genomic regions for traits of interest by utilizing diverse soybean germplasm and populations. Use of association panels can increase the mapping resolution compared to traditional QTL mapping (Deshmukh et al. 2014). Population structure in these panels can occur if some of the genotypes are more related to each other compared to the rest of the population. Failure to correct for population stratification in GWAS models can lead to false positives, especially if the trait of interest is correlated with the structure of the panel (Wang et al. 2005). Several studies in soybean have been reported using the GWAS approach with SNP markers for many different traits, such as seed composition (Hwang et al. 2014; Vaughn et al. 2014; Bandillo et al. 2015; Cao et al. 2017), salt tolerance (Patil et al. 2016; Zeng et al. 2017), carbon isotope composition (Dhanapal et al. 2015; Kaler et al. 2017a), ureide concentration (Ray et al. 2015), agronomic traits (Zhang et al. 2015; Contreras-Soto et al. 2017; Li et al. 2017), chlorophyll traits (Dhanapal et al. 2016), local adaptation (Bandillo et al. 2017), insect resistance (Chang and Hartman 2017), and canopy wilting (Kaler et al. 2017b). These studies have provided a useful way to identify potential genomic regions with high resolution and candidate genes or QTL for traits of interest.
The objectives of this study were to: i) evaluate a genetically diverse panel of soybean genotypes in repeated field experiments for canopy wilting, ii) identify new germplasm that possesses the slow canopy wilting trait, and iii) elucidate genomic regions responsible for canopy wilting using an association mapping approach.
Materials And Methods
Plant materials and panel selection
When selecting the panel, approximately 600 soybean accessions were chosen initially from the USDA collection based on geographic origin and low annual precipitation. The 600 were truncated to 169 accessions by examining the diversity among the accessions based on SNP genotype profiles. Only PIs with less than 85% similarity to each other based on these SNP genotypes were included in the panel. An additional 40 newly developed breeding lines with enhanced drought-related traits, as well as drought tolerant and susceptible checks, were added to bring the total number of genotypes in the panel to 209. These 209 genotypes were derived from 30 countries and range from MG III-IX. To minimize the maturity effect on the canopy wilting, only canopy wilting scores from 162 MG VI-VIII lines (groups commonly grown in the southeastern USA) were used in analyses in this study.
Genotype data and quality control
All but nine of the genotypes in the panel were previously genotyped with the SoySNP50K iSelect BeadChips (Song et al. 2013). These nine accessions were genotyped using the same procedure at the USDA Soybean Genomics and Improvement Lab in Beltsville, MD (Song et al. 2013). Briefly, 15 seed from each of these nine accessions were grown in a single 32 oz. styrofoam cup in a greenhouse at the University of Georgia, Athens, GA, USA. After approximately two weeks, leaf tissue was harvested and bulked in a 50 mL tube. The tissue was then placed in a lyophilizer for two days, and ground into a fine powder using a Geno/Grinder (SPEX SamplePrep, Metuchen, New Jersey, USA). DNA was extracted and then genotyped with the 50K chip. All SNP marker data were downloaded from SoyBase (Grant et al. 2010) for the remaining accessions. A total of 42,079 SNP markers were available from these genotyping efforts. The final number employed for the analyses was 34,892 for all but one environment (File S1), with the Kansas 2015 environment employing 34,397 markers after removing markers with minor allele frequencies below 0.05. The number of markers used varied slightly in the Kansas 2015 environment due to the different number of accessions evaluated being slightly lower. The physical positions of Glyma.Wm82.a2 reference genome were used to determine the locations of the SNPs used in the analysis.
Evaluation of canopy wilting
Canopy wilting was evaluated in Athens, GA, USA in 2015 (GA-15) and 2016 (GA-16) and Salina, KS, USA in 2015 (KS-15) and 2016 (KS-16) in rain-fed field plots after extended periods of little or no rainfall (File S2). Genotypes were planted in two-row plots in each environment in a randomized complete block design with three replications. Experiments were sown on June 16, 2015 (GA-15), June 8, 2016 (GA-16), June 10, 2015 (KS-15), and June 8, 2016 (KS-16). All plots for the four environments were planted with 0.76 m row spacing at a seeding density of 32 seed m-2. For GA-15 and GA-16, the plots were 2.43 m in length, and for KS-15 and KS-16 the plots were 3.65 m long.
Wilting was rated in increments of 5 on a scale from 0 to 100: 0 = no wilting present; 20 = slight wilting and some rolling in the top of the canopy; 40 = somewhat severe leaf rolling at the top of the canopy, moderate wilting of leaves throughout the rest of the canopy, and some loss of petiole turgidity; 60 = severe wilting of leaves throughout the entire canopy, with advanced loss of petiole turgidity; 80 = plants with petioles severely wilted and dead leaves throughout much of the canopy; and 100 = plant death. Volumetric water content (VWC) was measured with a single Decagon GS1 soil moisture probe placed approximately 30 cm below the soil surface in one corner of the field plots to measure available soil water at the time the scores were recorded.
Wilting scores in Athens, GA were taken by two raters in 2015 and three raters in 2016. In 2015, a single canopy wilting rating was taken on 29 July (most genotypes in vegetative stages, 17% VWC) for the Athens, GA plots and four ratings were taken in 2016 between 25 August and 16 September (most genotypes in pod filling stage, ∼5–8% VWC). One rater recorded canopy wilting scores for Salina, KS in both years. In 2015, four ratings were taken between 12 August and 28 September (most genotypes in vegetative stage for the first rating and at the pod filling stage for the last rating, ∼18–20% VWC), and in 2016 three ratings were taken between 26 July and 4 August (during flowering, VWC data not available). Mean ratings for an individual plot over dates and raters were employed as the phenotypic wilting score given that correlations between raters and rating dates were generally high (data not shown). All 162 entries were evaluated in GA-15, GA-16, and KS-16. However, because of seed availability and quality, only 142 entries were evaluated in KS-15.
Population structure analyses
Population structure was determined using fastSTRUCTURE (Raj et al. 2014), principal coordinate analysis, and by constructing a dendrogram based on the 50K SNP data. The fastSTRUCTURE program was run in default settings with the simple option testing for subpopulations (K) ranging from K = 2 - 10. As part of the fastSTRUCTURE package, the python script ChooseK was used to choose the number of subpopulations that maximize the marginal likelihood. Principal coordinate analysis was performed using the GAPIT R package (Lipka et al. 2012) and visualized with TIBCO Spotfire (TIBCO Software Inc., Palo Alto, CA, USA). The neighbor-joining clustering algorithm in TASSEL version 5.0 (Bradbury et al. 2007) was used to build a dendrogram, which was visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Statistical analyses
Analyses of variance (ANOVA) were conducted using PROC GLM in SAS version 9.4 (SAS Institute 2014). A model for canopy wilting scores was created with genotype treated as a fixed effect, and environment, replication within environment, and genotype by environment interaction treated as random effects. Broad-sense heritability was calculated on an entry-mean basis after Holland et al. (2010) with the variance components being calculated with PROC MIXED of SAS version 9.4 using a model where all variables were treated as random. Correlations of genotype means were calculated using PROC CORR in SAS version 9.4. Best linear unbiased predictions (BLUPs) were calculated for canopy wilting scores across all environments using JMP Pro (JMP, Version 13, SAS Institute Inc., Cary, NC, USA). The model was built by treating genotype, environment, genotype by environment interaction, and replication within environment as random variables using the Standard Least Squares personality and REML method. For individual environments, only genotype and replication were used and treated as random to calculate BLUPs. Use of BLUP values for each genotype across and within environments helped to account for variation caused by environmental factors and missing data. These BLUPs were used as the phenotype values for subsequent GWAS analyses.
Genome-wide association analyses
Genome-wide association analyses were performed using Fixed and random model Circulating Probability Unification (FarmCPU) (Liu et al. 2016). This R package uses a multiple loci linear mixed model (MLMM) which incorporates the most significant SNP markers as covariates in a modified mixed linear model (MLM), and uses fixed and random effect models iteratively to eliminate confounding between kinship and the markers being tested. This method helps to improve statistical power to detect significant markers associated with a particular phenotype and is computationally efficient. A total of 34,892 genome-wide SNP markers were used for the analysis after removing markers with minor allele frequencies (MAF) below 0.05. The number of markers used varied slightly in the KS-15 environment (34,397 markers) due to a smaller number of accessions evaluated. The differences in accession number affected which SNP markers were included for each genotype file when the MAF of the marker was close to 0.05. Manhattan plots were visualized with the ‘qqman’ R package (Turner 2018) using p-values generated from the FarmCPU output.
A Bonferroni threshold (P < 2.83E-07, -log10(P) > 6.55) is overly strict when the linkage disequilibrium among genetic markers is large, which is generally the case with soybean (Hyten et al. 2007). Therefore, a p-value threshold of (P < 0.0001; -log10(P) > 4) was used, which is less stringent than the Bonferroni-corrected threshold, but more stringent than the threshold used in Kaler et al. (2017b) and many other soybean GWAS studies using SoySNP50K genotype data. This threshold was used to identify SNPs that were significantly associated with the canopy wilting trait.
Pairwise estimates of Dʹ and r2 were calculated by chromosome using Haploview version 4.2 software (Barrett et al. 2005). Linkage disequilibrium (LD) blocks were estimated using the Solid Spine of LD option using Dʹ > 0.8 to extend the spine. Significant SNPs associated with canopy wilting were considered part of the same locus (genomic region) controlling the trait if they were in the same LD block. Allelic effects were calculated by taking the difference in mean canopy wilting score between the two alleles at a particular SNP, and the direction, negative or positive, of the allelic effect estimates were relative to the alphabetical order of the nucleotides at each particular marker. For example, if the nucleotides at a particular SNP are “A” and “C”, then a positive (above zero) allelic effect (e.g., 3) indicates that possessing the “C” allele will increase the canopy wilting score by three, which is unfavorable. A negative (below zero) effect value indicates that a line possessing the second nucleotide alphabetically for this SNP would have a lower canopy wilting score, which is favorable. For this study, the allelic effects are based on BLUP values, not actual canopy wilting scores, so while the overall effect is relevant, it does not directly apply to raw canopy wilting scores. The amount of phenotypic variation explained (R2) for all significant SNPs in a given environment was calculated using a simple linear regression in R with lm(BLUP ∼ SNP1 + SNP2 + …).
Breeding values for accessions evaluated were calculated by adding the allelic effects for all SNPs significantly associated (P < 0.0001; -log10(P) > 4) with canopy wilting in each individual environment and with the across all environments BLUPs. Breeding values from across individual environments (GA-15, GA-16, KS-15, and KS-16) were also summed. The allelic effect for a given accession was considered below zero (favorable) if the allele contributed to lower canopy wilting scores. In contrast, if the allele increased canopy wilting score, it was considered an above zero (unfavorable) value. Therefore, a more negative breeding value indicated an accession had a sum of allelic effects across all significant SNPs that was more favorable toward reduced canopy wilting. If the allele at a particular SNP was heterozygous or missing for a genotype, it was omitted in the breeding value calculation.
Candidate gene identification
SNPs from the GWAS that met the threshold of -log10(P) > 4 were used to identify nearby candidate genes. Candidate genes and their functional annotation were identified using the Glyma2.1 gene models in SoyBase for models within plus or minus 10 kb of the SNP physical position.
Data availability
SNP marker genotypes for accessions included in the association panel are in File S1 and canopy wilting scores are in File S2. All other datasets generated and/or analyzed for this study are not publicly available, but are available from the corresponding author based on a reasonable request. Supplemental material available at figshare: https://doi.org/10.25387/g3.11441340.
Results
Canopy wilting
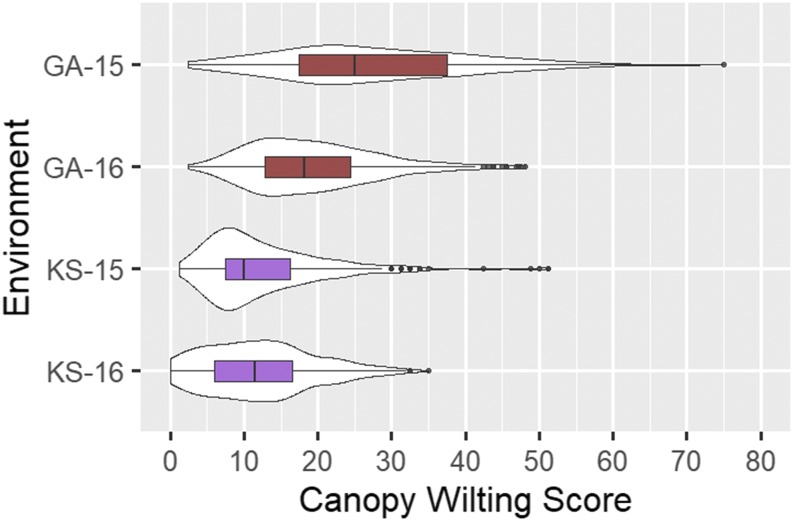
Substantial variation for canopy wilting was observed among the genotypes within the panel across the four environments tested. In general, canopy wilting scores were higher (more severe wilting) in Athens, GA compared to Salina, KS in both 2015 and 2016 (Figure 1). Genotypes, environments, and their interactions were statistically significant (P < 0.05) for canopy wilting scores (Table 1). Correlations (data not shown) of canopy wilting scores based on genotype means among the environments ranged from r = 0.42 (KS-15/KS-16) to r = 0.65 (GA-15/KS-16). Broad sense heritability of canopy wilting on an entry-mean basis for each environment was 60% (GA-15), 73% (GA-16), 30% (KS-15), 72% (KS-16), and 34% across all environments.
Figure 1.
Violin plots with boxplots inside showing the distribution of canopy wilting scores. Environments are named as Location-Year, with Georgia (GA) and Kansas (KS) as locations, and 2015 (15) and 2016 (16) as years.
Table 1. Summary of analyses of variance (ANOVA) for effects of genotype (G), environment (E), and their interaction based on canopy wilting scores. The G × E MS was used as the denominator of the F Value for significance testing.
| Source | DF | F Value | P > F |
|---|---|---|---|
| Genotype (G) | 161 | 10.1 | <0.0001 |
| Environment (E) | 3 | 648.2 | <0.0001 |
| G × E | 483 | 2.1 | <0.0001 |
The 162 genotypes were ranked from lowest to highest canopy wilting score within each environment, and then over the four environments (Tables 2 and S1). Numerically, 78 genotypes exhibited less wilting than the slow canopy wilting check, PI 416937. Thirty-eight and 44 lines exhibited numerically greater wilting than fast wilting cultivars Benning and Hutcheson, respectively (Table S1).
Table 2. Canopy wilting scores for the 10 genotypes with the lowest and highest scores based on mean ranking across environments along with two check genotypes. Each environment was ranked individually, and the mean of those rankings was used to rank all of the 162 genotypes tested. Canopy wilting scores shown are the mean of all replications within each respective environment. A full table of all accessions tested and their canopy wilting scores is provided in the supplementary materials (Table S1).
| Canopy Wilting Score | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession | Name | Countrya | MGb | ALL-PANEL | GA-15-PANEL | GA-16-PANEL | KS-15-PANEL | KS-16-PANEL | Rank | Beneficial Allelesc | Breeding Valued |
| Slow wilting | |||||||||||
| PI603535 | Hei zong huang dou | China | VIII | 6 | 10 | 6 | 6 | 1 | 1 | 18 | −31.80 |
| PI603513A | Xiao niu mao huang | China | VIII | 7 | 9 | 7 | 6 | 5 | 2 | 23 | −19.12 |
| PI603529 | Hei huang dou | China | VIII | 8 | 12 | 10 | — | 2 | 3 | 15 | 15.55 |
| PI603513B | — | China | VIII | 8 | 13 | 10 | — | 3 | 4 | 28 | −17.33 |
| PI603534A | Da niu mao huang | China | VII | 8 | 7 | 12 | 8 | 5 | 5 | 26 | −15.62 |
| PI219698 | Kulat | Pakistan | VI | 10 | 18 | 11 | — | 3 | 6 | 25 | −32.46 |
| PI532458 | Ba yue bao | China | VIII | 10 | 23 | 10 | 3 | 3 | 7 | 22 | −25.01 |
| PI269518B | (Koolat) | Pakistan | VI | 11 | 21 | 9 | — | 3 | 8 | 22 | −25.16 |
| PI567405 | Wei zi dou | China | VI | 9 | 10 | 12 | 7 | 6 | 9 | 27 | 5.93 |
| PI603521 | Huang dou | China | VIII | 10 | 18 | 8 | 9 | 5 | 10 | 16 | −14.86 |
| Checks | |||||||||||
| PI416937 | Houjaku Kuwazu | Japan | VI | 18 | 39 | 16 | 8 | 11 | 79 | 23 | −34.10 |
| PI595645 | Benning | United States | VII | 23 | 38 | 24 | 11 | 20 | 132 | 23 | −1.84 |
| Fast wilting | |||||||||||
| PI424131 | Buffalo | Zimbabwe | VII | 27 | 36 | 26 | 23 | 25 | 153 | 24 | −28.20 |
| PI430737 | Oribi | Zimbabwe | VII | 31 | 49 | 27 | — | 16 | 154 | 21 | −11.27 |
| PI567377B | (Ba yue zha) | China | VI | 34 | 63 | 32 | 13 | 26 | 155 | 22 | −17.65 |
| PI159096 | 41S77 | South Africa | VII | 31 | 52 | 29 | 25 | 17 | 156 | 20 | −33.96 |
| PI381663 | Kakira 1 | Uganda | VI | 35 | 55 | 45 | 20 | 20 | 157 | 25 | −34.39 |
| NCC06-1090 | — | United States | VI | 32 | 39 | 39 | 24 | 28 | 158 | 11 | 13.74 |
| PI639573 | — | Burundi | VIII | 33 | 39 | 37 | 30 | 25 | 159 | 26 | −34.27 |
| PI599333 | Musen | United States | VI | 33 | 53 | 32 | 20 | 27 | 160 | 16 | 7.09 |
| PI417562 | 54.S.30 DL/64/185 | South Africa | VI | 36 | 46 | 36 | 36 | 25 | 161 | 17 | −11.48 |
| PI330635 | — | South Africa | VII | 39 | 53 | 40 | — | 25 | 162 | 23 | −20.70 |
Country of origin of the accession based on GRIN data.
Maturity group.
Number of alleles from all significant SNPs with an effect that reduces canopy wilting score.
Breeding value determined by adding the allelic effects for all significant SNPs individually by environment, and then summing the breeding values across individual environments.
Population structure
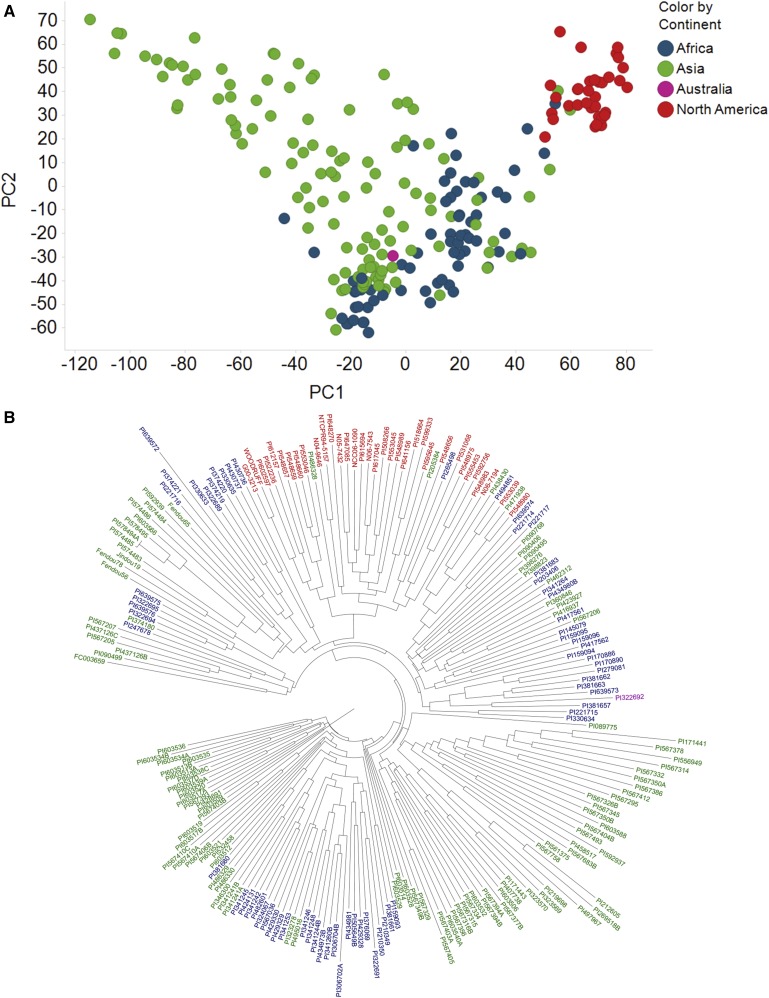
All population structure analyses were conducted using the full panel of 209 genotypes initially selected from MG III to IX for other drought tolerance related studies conducted in Steketee et al. (2019), which include the 162 genotypes evaluated for canopy wilting in this study. The first two principal coordinates were visualized and colored by continent of origin (Figure 2A). All of the North American lines represent genotypes from the USA, and the majority (88/118) of Asian genotypes were from China. The genotypes of U.S. origin were more tightly clustered than were genotypes from China (Figure 2A). The first four principal coordinates explained approximately 19% of the variation in the data set, and were used as covariates in the GWAS model to help correct for potential population stratification (Figure S1). The North American genotypes had shorter genetic distances between accessions compared to Asian and African genotypes based on the neighbor joining dendrogram analysis, indicating they are more closely related, which concurs with the principal coordinates analysis. Genotypes from Asia tended to group close to one another based on this analysis, but also intermixed with lines of African origin (Figure 2B). A continuous increase in marginal likelihood with increasing K was observed for the fastSTRUCTURE analysis, meaning as each sequential K value (increasing from 2 to 10) was tested it was deemed the K value that best characterized the population structure of the panel. This indicated that little structure was apparent for this population, because fastSTRUCTURE was not able to settle on an optimal K value to describe this panel within K = 2 - 10 (Westbrook et al. 2015) (Figure S2).
Figure 2.
(A) Plot of first and second principal coordinates for a diverse panel of soybean accessions evaluated in drought tolerance related studies. Each individual soybean genotype is colored by their continent of origin. (B) Dendrogram using neighbor joining clustering algorithm in TASSEL visualized in FigTree. Genotypes are colored by their continent of origin: red = North America, blue = Africa, green = Asia, and purple = Australia.
GWAS of canopy wilting trait
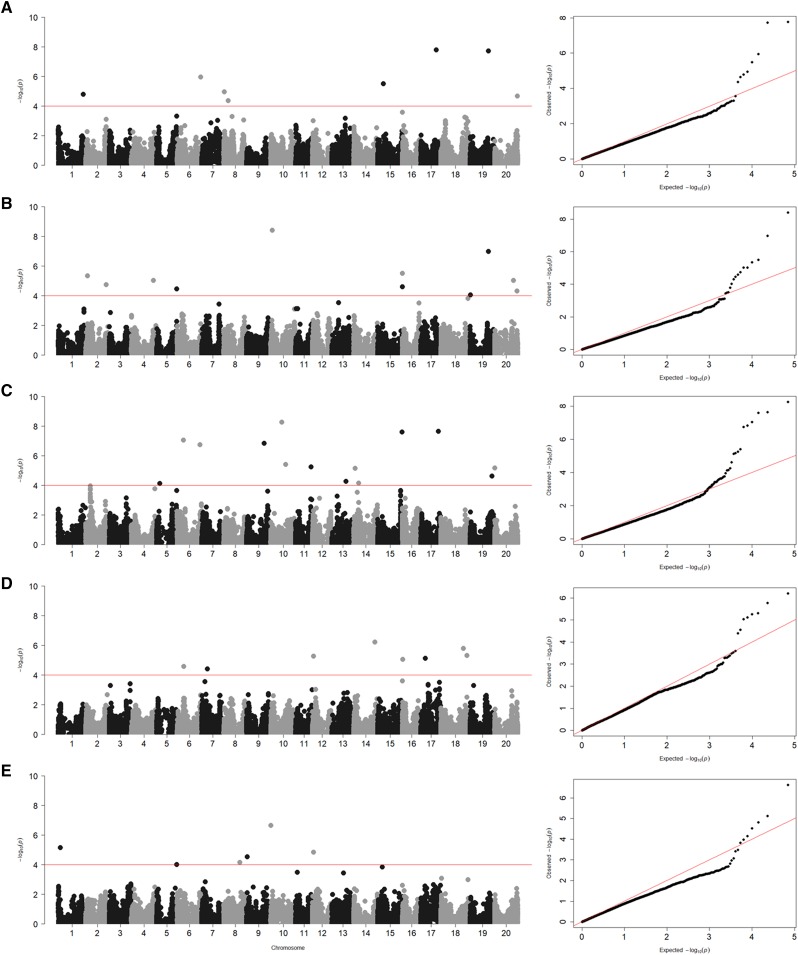
Across and within environments, 45 unique SNPs were identified that tagged 44 loci associated with canopy wilting from the GWAS analysis (Figure 3 and Table 3). One of these SNPs (ss715634688) on Chr 19 was significant (P < 0.0001; -log10(P) > 4) both in individual environments and with the BLUP value calculated using the canopy wilting scores across all environments. One additional region, locus 32 on Chr 16 was identified in two of the four environments.
Figure 3.
Genome-wide Manhattan plots for (A) ALL, (B) GA-15, (C) GA-16, (D) KS-15, and (E) KS-16. The X-axis is the genomic position of SNPs by chromosome across the soybean genome, and the Y-axis is the -log10 of the p-values obtained from the GWAS model. Significance threshold -log10(P) > 4 (red line). The quantile-quantile (QQ) plots to the right of each Manhattan plot show the expected vs. observed p-values of each SNP tested in the GWAS models.
Table 3. SNPs that met significance level of -log10(P) > 4 for the GWAS of canopy wilting.
| Locusa | Chr.b | Pos.c | SNP | -log10(P) | MAFd | Effecte | Envf | SoyBase QTLg |
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 4015639 | ss715579324 | 5.13 | 0.27 | −1.62 | KS-16 | |
| 2 | 1 | 51961463 | ss715580187 | 4.78 | 0.09 | 2.13 | ALL | |
| 3 | 2 | 3165348 | ss715581823 | 5.34 | 0.06 | −3.65 | GA-15 | Canopy wilt 3-4, 3-8, 3-11, 6-1; mqCanopy wilt-001 |
| 4 | 2 | 42073473 | ss715582842 | 4.73 | 0.37 | 1.97 | GA-15 | Canopy wilt 6-3 |
| 5 | 4 | 46096228 | ss715588277 | 5.02 | 0.38 | −1.97 | GA-15 | |
| 6 | 5 | 6207961 | ss715592288 | 4.13 | 0.06 | −3.19 | GA-16 | Canopy wilt 3-5 |
| 7 | 5 | 41387548 | ss715591700 | 4.45 | 0.21 | 1.91 | GA-15 | |
| 8 | 6 | 13090474 | ss715592991 | 7.05 | 0.06 | 2.97 | GA-16 | |
| 9 | 6 | 14258126 | ss715593189 | 4.56 | 0.06 | 1.51 | KS-15 | |
| 10 | 6 | 47633030 | ss715594738 | 6.75 | 0.28 | −1.87 | GA-16 | Canopy wilt 3-12 |
| 11 | 6 | 49189084 | ss715595012 | 5.94 | 0.37 | 1.50 | ALL | |
| 12 | 7 | 11177483 | ss715596171 | 4.40 | 0.32 | 0.71 | KS-15 | |
| 13 | 8 | 1699023 | ss715599875 | 4.94 | 0.24 | 1.40 | ALL | |
| 14 | 8 | 9837263 | ss715602901 | 4.36 | 0.15 | −1.42 | ALL | |
| 15 | 8 | 34471238 | ss715601484 | 4.14 | 0.20 | 1.79 | KS-16 | |
| 16 | 9 | 1769730 | ss715603168 | 4.53 | 0.18 | −1.93 | KS-16 | |
| 17 | 9 | 36942176 | ss715603680 | 6.84 | 0.31 | −2.01 | GA-16 | |
| 18 | 10 | 270252 | ss715606054 | 6.63 | 0.47 | 1.81 | KS-16 | |
| 19 | 10 | 3598580 | ss715606348 | 8.41 | 0.41 | 3.20 | GA-15 | |
| 20 | 10 | 23376136 | ss715605804 | 8.26 | 0.27 | −3.31 | GA-16 | |
| 21 | 10 | 30831897 | ss715606157 | 5.41 | 0.15 | 3.43 | GA-16 | |
| 22 | 11 | 31929823 | ss715610250 | 5.25 | 0.48 | −1.45 | GA-16 | |
| 23 | 12 | 2053039 | ss715611755 | 5.26 | 0.08 | −1.38 | KS-15 | |
| 24 | 12 | 2839426 | ss715612002 | 4.82 | 0.35 | −1.43 | KS-16 | |
| 25 | 13 | 29459954 | ss715614803 | 4.25 | 0.10 | −1.89 | GA-16 | |
| 26 | 14 | 3078346 | ss715618273 | 5.13 | 0.43 | 1.55 | GA-16 | Canopy wilt 1-2 |
| 27 | 14 | 10057919 | ss715617366 | 4.15 | 0.07 | 2.76 | GA-16 | |
| 28 | 14 | 43597753 | ss715618915 | 6.21 | 0.36 | −0.90 | KS-15 | |
| 29 | 15 | 12437556 | ss715620442 | 5.49 | 0.34 | 1.29 | ALL | |
| 30 | 15 | 50499617 | ss715622647 | 7.59 | 0.24 | 2.16 | GA-16 | |
| 31 | 15 | 51622014 | ss715622805 | 4.60 | 0.23 | −1.75 | GA-15 | |
| 32 | 16 | 164715 | ss715623538 | 5.49 | 0.48 | 1.81 | GA-15 | |
| 16 | 517535 | ss715625192 | 5.04 | 0.09 | −1.29 | KS-15 | ||
| 33 | 17 | 9384325 | ss715628378 | 5.12 | 0.43 | 0.75 | KS-15 | Canopy wilt 1-3, 3-10, 3-13; mqCanopy wilt-006 |
| 34 | 17 | 31794022 | ss715626726 | 7.78 | 0.12 | 2.51 | ALL | Canopy wilt 5-2 |
| 35 | 17 | 36227875 | ss715626968 | 7.64 | 0.09 | 3.47 | GA-16 | |
| 36 | 18 | 46860521 | ss715631153 | 5.78 | 0.13 | −1.26 | KS-15 | |
| 37 | 18 | 54371258 | ss715632037 | 5.31 | 0.09 | 1.47 | KS-15 | |
| 38 | 19 | 809326 | ss715636293 | 4.04 | 0.28 | −1.80 | GA-15 | |
| 39 | 19 | 38109922 | ss715634688 | 7.73 | 0.15 | −2.16 | ALL | Canopy wilt 2-7, 4-3 |
| 19 | 38109922 | ss715634688 | 6.98 | 0.15 | −3.05 | GA-15 | Canopy wilt 2-7, 4-3 | |
| 40 | 19 | 45307395 | ss715635460 | 4.61 | 0.34 | 1.47 | GA-16 | Canopy wilt 5-4, 6-2 |
| 41 | 20 | 294010 | ss715637218 | 5.17 | 0.46 | 1.55 | GA-16 | |
| 42 | 20 | 39013106 | ss715637991 | 5.03 | 0.36 | −1.56 | GA-15 | |
| 43 | 20 | 46438247 | ss715638748 | 4.30 | 0.46 | −1.71 | GA-15 | |
| 44 | 20 | 47435005 | ss715638900 | 4.65 | 0.48 | −1.46 | ALL |
If multiple SNPs were identified in the same linkage disequilibrium (LD) block they were deemed part of the same locus (genomic region).
Chromosome.
Glyma.Wm82.a2 physical position.
Minor allele frequency.
Allelic effects were calculated by taking the difference in mean canopy wilting score between the two alleles at a particular SNP, and the direction, negative or positive, of the allelic effect estimates are relative to the alphabetical order of the nucleotides at each particular marker.
Environment written as location-year-population.
Canopy wilting QTL identified on SoyBase in which loci from our study are located within.
The quantile-quantile (QQ) plots follow the expected diagonal, with a sudden uptick for statistically significant (P < 0.0001; -log10(P) > 4) SNP markers in each environment (Figure 3). This linear pattern of expected vs. observed p-values also does not have a slope greater than one, which indicates the first four principal coordinates included in the GWAS model adequately accounted for population stratification in this panel of genotypes. The deviation of the markers from this diagonal occurs at or greater than –log10(P) = 4 in each environment, which was the threshold we used to determine if a SNP marker was significantly associated with the canopy wilting trait.
Allelic effects across all significant (P < 0.0001; -log10(P) > 4) SNPs ranged from -3.65 to 3.47 (Table 3). The R2 for all significant SNPs in a given environment was 54% (GA-15), 45% (GA-16), 53% (KS-15), 30% (KS-16), and 36% (ALL). The number of beneficial alleles each genotype possessed was determined by counting the number of alleles with effects that reduced canopy wilting score from all the significant SNPs. Overall, the number of beneficial alleles ranged from 11 to 31. The 10 slowest wilting genotypes had 15 to 27 beneficial alleles, while the 10 fastest wilting genotypes had 11 to 26 beneficial alleles (Table 2 and Table S1). Summed breeding values across the individual environments ranged from -42.32 to 17.17 overall (Table 2 and Table S1). Negative (below zero) breeding values indicate that the genotype had a sum of allelic effects across the significant SNPs that was more favorable toward reduced canopy wilting scores. Positive (above zero) breeding values indicate that the genotype had a sum of allelic effects across the significant SNPs that was less favorable toward reduced canopy wilting scores.
Identification of candidate genes for the canopy wilting trait
The median distance between SNP markers used in the GWAS was 9 kb, and the mean distance was 26 kb. Although identifying all gene models in LD with significant SNPs would be ideal, we focused our efforts on models in close proximity (within plus or minus 10 kb), which approximately spans this distance between markers. Eighty-seven candidate genes were found within plus or minus 10 kb of the 45 significant (P < 0.0001; -log10(P) > 4) SNPs for the GWAS (Table S2).
Discussion
Canopy wilting
In this study, we evaluated 162 soybean genotypes in four environments for canopy wilting score after extended periods of drought stress, and most of these genotypes were never evaluated previously for drought tolerance related traits. There was substantial genetic variation for canopy wilting within each environment (Figure 1). Canopy wilting is a complex, quantitative trait (Charlson et al. 2009; Hwang et al. 2016), and our phenotypic data further confirm this notion.
Slow wilting PI 603535, a MG VIII accession from China, had a mean score of 6 across all environments. Eight of the 10 genotypes with the slowest wilting ranking originated from China. Among the fast wilting genotypes in the panel, PI 330635, a MG VII accession from South Africa, had a mean canopy wilting score of 39 across environments (Table 2). PI 416937 was included as a slow wilting check in the studies, and many accessions had lower mean wilting scores (less wilting) than this check genotype. Seventy-eight genotypes had lower canopy wilting scores than PI 416937 (Table S1). However, the mean canopy wilting score across all environments evaluated for PI 416937 was 18, which is also a relatively low score (Table S1). These newly identified PIs could be germplasm sources as parents that could be exploited by soybean breeders to improve the canopy wilting trait, especially the accessions with favorable alleles different than PI 416937, and with negative breeding values. In Kaler et al. (2017b), 185 of the 373 genotypes they tested had scores lower than PI 416937. This study and Kaler et al. (2017b) demonstrate there is more variation and potential for improvement of the canopy wilting trait than previously reported, but testing these new slow wilting genotypes in more environments is necessary to further confirm they will consistently exhibit this trait in different locations and drought stress severities.
Physiological mechanisms for canopy wilting and relationship to other traits
Slow canopy wilting could lead to less yield reduction during drought stress in soybeans. A previous study proposed three different combinations of physiological mechanisms that could lead to delayed canopy wilting (Ries et al. 2012). One is a combination of high water use efficiency (WUE), high radiation use efficiency (RUE), and conservation of soil moisture. Genotypes in this group would utilize transpired water for biomass production more efficiently, and higher RUE would be expected in both drought stressed and optimal growing conditions. The second combination is low stomatal conductance, low RUE, low WUE, and conservation of soil moisture. The genotypes in this group would have low transpiration which would reduce potential photosynthetic capacity, and would be better at conserving water during drought stress conditions. However, this second combination of physiological attributes could reduce overall yield potential, especially in well-watered environments. Deeper rooting is a third mechanism that could delay canopy wilting in soybean (Ries et al. 2012). Given these advantages and trade-offs for different physiological traits, identifying soybean germplasm with the optimal combination to reduce canopy wilting during drought stress will be different depending on the target environment.
Much like canopy wilting in soybean, evaluation of other crops for drought tolerance commonly uses secondary traits for indirect selection which can show relationships with yield under stressed conditions. Leaf rolling reduces exposed leaf area, and thereby decreases transpiration and reduces light interception, and can be observed in crops such as maize, rice, and wheat. The earlier leaf rolling occurs in a given day or longer duration of rolling indicates the plant is experiencing more stress (Rauf et al. 2016). Therefore, ratings and selections can be made to identify plants with reduced leaf rolling during drought periods to improve drought tolerance. In maize, another trait that is evaluated to improve drought adaptation is the anthesis-silking interval (ASI). This trait is negatively correlated with grain yield under drought conditions, and has been a breeding target due to the ease of measurement and moderate heritability (Tuberosa 2012). Additional traits such as stay green, root architecture, and canopy temperature depression can impact a plant’s ability to tolerate drought stress and have been evaluated in a number of crop species (Tuberosa 2012). Slow canopy wilting in soybean is potentially related to other secondary physiological mechanisms that can be evaluated to improve our understanding of soybean drought physiology and potentially improve yield under stressed conditions.
Relationship of canopy wilting, days to flowering, and maturity group
Canopy wilting scores were overall higher in Georgia compared to Kansas in both 2015 and 2016 (Figure 1). Given that the genotypes we evaluated consisted of genetically diverse genotypes (most of which were plant introductions) with some variation in phenology (flowering time, height, root mass, etc.) these scores could potentially be affected by varying degrees of competition for water resources from neighboring plots due to these factors.
Days to flowering (DTF) was recorded in the GA-15 and GA-16 environments as the number of days from planting until 50% of the plants in a plot reached the R1 (first bloom) stage of development. Maturity groups (MG) for all accessions were obtained from the USDA GRIN website or were provided by the breeder who developed the line. Correlations between wilting score and MG (r = -0.34 to -0.05), and wilting score and DTF (r = -0.30 – 0.00) in single environments were relatively low. Across all environments, the correlation for wilting score and MG was r = -0.20 and for wilting score and DTF was r = -0.22. There did not appear to be a relationship between mean rank across environments and MG or DTF in this study (Figure S3). In our study, canopy wilting was primarily evaluated during the early reproductive growth stages, with the exception of GA-15 and first rating in KS-15, which were rated during the late vegetative growth stages for most genotypes. Water stress during the early reproductive growth stages has the greatest impact on reducing soybean yield from plants producing fewer pods, and in turn, less seed (Manavalan et al. 2009). Therefore, slow canopy wilting during reproductive growth stages could be a good indicator of drought tolerance and ability to maintain yield potential during water stress.
Genotype by environment interaction and heritability
Although genotype by environment interactions were significant (P < 0.05) (Table 1) and the severity of wilting experienced in the four environments varied (Figure 1), the correlations between wilting scores across environments were relatively high (r = 0.42-0.65), indicating that the genotypes tested from this panel wilted similarly across environments. Heritability across environments was also moderate to high, with heritability comparable to those observed in previously canopy wilting QTL mapping and GWAS studies (Charlson et al. 2009; Abdel-Haleem et al. 2012; Hwang et al. 2015b; Kaler et al. 2017b).
Population structure analyses with panel
Soybean was first domesticated in China (Hyten et al. 2006), and the accessions of Chinese origin from the panel had the least tight cluster in the principal coordinates plot compared to other countries, and exhibited the greatest distance between accessions in the dendrogram, indicating they had the most diversity of the accessions tested (Figure 2). Lines from the USA were tightly clustered in the principal coordinates plot, and had short distances apart from one another in the dendrogram (Figures 2). The genetic base used in North American soybean breeding has been characterized as being narrow, with only a few common ancestors explaining the majority of diversity for these breeding materials (Carter et al. 2004). The tight clustering based on principal coordinates and short distance between accessions in the dendrogram is a reflection of this narrow genetic diversity of elite U.S. soybean breeding lines. Given that the panel of soybean genotypes used for this study was explicitly chosen to be genetically diverse based on genome-wide 50K SNP data, the lack of ability for fastSTRUCTURE to select an optimal number of K groups was expected. Based on the combination of the results of these population structure analyses, we determined that this panel had little or moderate population structure present that would affect the GWAS and cause false positives. As is commonly done with association mapping, we did include the first four principal coordinates in our GWAS model as a way to help reduce the possibility of potential population structure affecting the mapping results (Price et al. 2006).
Comparisons of genetic mapping for canopy wilting to previous studies
We identified 45 unique SNPs that tagged 44 loci that are associated with canopy wilting using a genome-wide association mapping approach. One of these SNPs was found both in an individual environment and when using the BLUP value calculated using the canopy wilting scores across all environments. On Chr 16, two physically close SNPs found in two different environments within the same LD block were significantly associated with canopy wilting. Overall, significant SNPs were identified on 19 of the 20 soybean chromosomes, with only Chr 3 not having any marker-trait associations (Table 3). The R2 for all significant SNPs in a given environment was 54% (GA-15), 45% (GA-16), 53% (KS-15), 30% (KS-16), and 36% across all environments.
Several reports of QTL or genomic regions that control canopy wilting in soybean have been previously reported, and many are numbered with their approximate physical locations on the SoyBase website. For the GWAS results, Loci 3 is within the QTL interval for a meta-QTL identified in Hwang et al. (2016). Loci 4, 6, 10, 34, and 40 were within QTL found in Hwang et al. (2015b). Loci 26 and 33 are within Canopy wilt 1-2 and Canopy wilt 1-3 QTL identified, respectively, in Charlson et al. (2009). On Chr 19, Loci 39 was within the Canopy wilt 2-7 QTL identified in Abdel-Haleem et al. (2012). Of the 45 SNPs identified from GWAS associated with canopy wilting in the this study, a total of nine were found in or near the same location as canopy wilting QTL identified from the linkage mapping studies described above (Table 3). The overlapping regions and consistent QTL across mapping studies help provide validation that these loci are associated with the canopy wilting trait and could be the targets of improvement efforts.
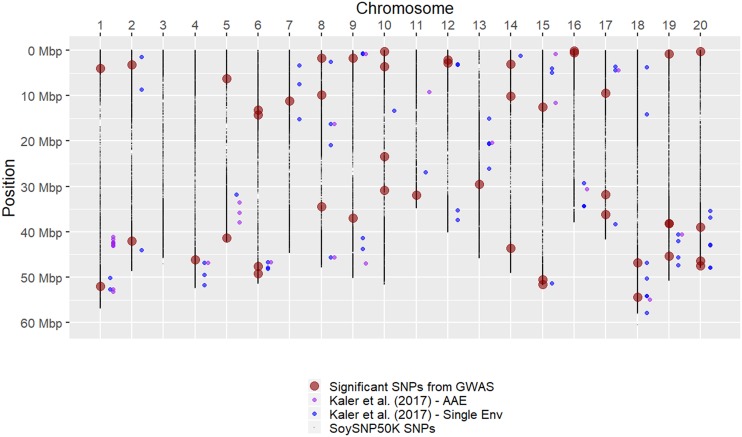
Kaler et al. (2017b) used an association mapping approach to identify 61 SNP markers tagging 51 different loci for canopy wilting. The significant SNPs found in our study and Kaler et al. (2017b) were compared (Figure 4). Twelve SNPs tagging 12 genomic regions were found on Chr 1, 4, 6, 9, 12, 15, 18, 19, and 20 in our study that are near SNPs identified in Kaler et al. (2017b). The main difference in our study compared to Kaler et al. (2017b) is that later maturity group soybeans (VI-VIII vs. IV) were used in our study. The genomic regions that are consistent across maturity groups and many different environments show promise as selection targets for improving canopy wilting under drought stress. Directing research efforts toward the genomic regions found in common between the current and previous GWAS studies could yield favorable alleles for the improvement of the canopy wilting trait in soybean. Because many genomic regions were identified, a simple marker-assisted selection approach for improvement of this trait may not be feasible. An alternative, and perhaps more effective, approach would be to utilize genomic selection with whole genome markers to improve canopy wilting in applied breeding programs.
Figure 4.
Location and comparison of SNPs significantly associated with canopy wilting based on association mapping results. Physical positions are based on the Glyma.Wm82.a2 version of the soybean genome. SNPs identified in GWAS that met -log10(P) > 4 significance threshold are shown as large red colored circles. Average of all environments (AAE) and single environment (Env) significant SNPs from Kaler et al. (2017b) are shown as purple and blue circles, respectively. Position locations were converted from version 1 to 2 of the soybean genome assembly for the Kaler et al. (2017b) SNPs, so that comparisons were made using the same physical positions. ss715637687 found in AAE for Kaler et al. (2017b) is not in version 2 of soybean genome assembly, and therefore not included in this comparison.
Candidate genes at canopy wilting significant genomic regions
The SNP with the greatest absolute allelic effect (3.65) was found on Chr 2 (ss715581823) and has a MAF of 0.06 (Table 3). This SNP is located near Glyma.02g034000, which has an aldehyde dehydrogenase annotation (Table S2). Aldehyde dehydrogenase genes have been previously shown to play a role in response to abiotic stresses of soybeans, and can be highly induced by drought stress in soybean leaves (Wang et al. 2017). Loci 12 (Glyma.07g110100) and 33 (Glyma.17g118400) have gene models which encodes a RING superfamily protein (Table S2), and locus 33 was co-located with previously identified canopy wilting QTL. RING-type E3 ubiquitin ligases including DREB2A-interacting proteins DRIP1 and DRIP2 have been previously shown to play a role in drought response (Qin et al. 2008). GmRFP1 functions as a RING-type E3 ubiquitin ligase and is down-regulated by drought and cold stress, but is induced by ABA and salt stress suggesting it may be involved in abiotic stress response (Du et al. 2010). Given their relationship with drought stress response and improvement, these gene models could be targeted for understanding and improving canopy wilting in soybean.
Conclusions
Using 162 genetically diverse maturity group VI-VIII soybean genotypes, a genome-wide association mapping approach identified 45 unique SNPs tagging 44 loci that are significantly associated with the canopy wilting trait. One of these SNPs was identified in an individual environment, as well as across environments. Of these 45 SNPs, nine were found in or near the locations as previous canopy wilting QTL identified from other linkage mapping studies. In addition, 12 SNPs mapped to 12 genomic regions on 11 chromosomes were found near regions identified in a previous association mapping study for canopy wilting. Candidate genes located at these genomic regions were identified that could help to understand the functions of these genes to improve canopy wilting in soybean. The genomic regions discovered across environments, in addition to the new slow wilting germplasm identified with favorable alleles, can be exploited by breeders to improve soybean drought tolerance.
Acknowledgments
The authors would like to thank Miles W. Ingwers for his help with canopy wilting rating in Georgia in 2016, and the soybean breeding and molecular genetics labs at UGA and KSU for their help with field maintenance at the Georgia and Kansas locations, respectively. Thanks also go to Nicole Bachleda and Tatyana Nienow for their technical support. Clinton J. Steketee was supported by a United Soybean Board Fellowship. We also thank the United Soybean Board Abiotic Stress Project for partial funding of this research.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11441340.
Communicating editor: S. Pearce
Literature Cited
- Abdel-Haleem H., Carter T. E., Purcell L. C., King C. A., Ries L. L. et al. , 2012. Mapping of quantitative trait loci for canopy-wilting trait in soybean (Glycine max L. Merr). Theor. Appl. Genet. 125: 837–846. 10.1007/s00122-012-1876-9 [DOI] [PubMed] [Google Scholar]
- Bagherzadi L., Sinclair T. R., Zwieniecki M., Secchi F., Hoffmann W. et al. , 2017. Assessing water-related plant traits to explain slow-wilting in soybean PI 471938. J. Crop Improv. 31: 400–417. 10.1080/15427528.2017.1309609 [DOI] [Google Scholar]
- Bandillo N. B., Anderson J. E., Kantar M. B., Stupar R. M., Specht J. E. et al. , 2017. Dissecting the genetic basis of local adaptation in soybean. Sci. Rep. 7: 17195 10.1038/s41598-017-17342-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandillo N., Jarquin D., Song Q., Nelson R., Cregan P. et al. , 2015. A population structure and genome-wide association analysis on the USDA soybean germplasm collection. Plant Genome 8: 1–13. 10.3835/plantgenome2015.04.0024 [DOI] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J., and Daly M. J., 2005. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Boerma H. R., Hussey R. S., Phillips D. V., Wood E. D., Rowan G. B. et al. , 1997. Registration of “Benning” soybean. Crop Sci. 37: 1982 10.2135/cropsci1997.0011183X003700060061x [DOI] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y. et al. , 2007. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Cao Y., Li S., Wang Z., Chang F., Kong J. et al. , 2017. Identification of major quantitative trait loci for seed oil content in soybeans by combining linkage and genome-wide association mapping. Front. Plant Sci. 8: 1222 10.3389/fpls.2017.01222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter T. E., Nelson R. L., Sneller C. H., and Cui Z., 2004. Genetic diversity in soybean, pp. 303–416 in Soybeans: Improvement, Production, and Uses, ASA, CSSA, and SSSA, Madison, WI. [Google Scholar]
- Carter T. E., De Souza P. I., and Purcell L. C., 1999. Recent advances in breeding for drought and aluminum resistance in soybean, pp. 106–125 in World Soybean Conference VI, edited by Kauffman H. Champaign, IL. [Google Scholar]
- Carter T. E., Todd S. M., and Gillen A. M., 2016. Registration of ‘USDA-N8002’ soybean cultivar with high yield and abiotic stress resistance traits. J. Plant Regist. 10: 238–245. 10.3198/jpr2015.09.0057crc [DOI] [Google Scholar]
- Chang H.-X., and Hartman G. L., 2017. Characterization of insect resistance loci in the USDA soybean germplasm collection using genome-wide association studies. Front. Plant Sci. 8: 670 10.3389/fpls.2017.00670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson D. V., Bhatnagar S., King C. A., Ray J. D., Sneller C. H. et al. , 2009. Polygenic inheritance of canopy wilting in soybean. [Glycine max (L.) Merr.] Theor. Appl. Genet. 119: 587–594. 10.1007/s00122-009-1068-4 [DOI] [PubMed] [Google Scholar]
- Contreras-Soto R. I., Mora F., de Oliveira M. A. R., Higashi W., Scapim C. A. et al. , 2017. A genome-wide association study for agronomic traits in soybean using SNP markers and SNP-based haplotype analysis. PLoS One 12: e0171105 10.1371/journal.pone.0171105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh R., Sonah H., Patil G., Chen W., Prince S. et al. , 2014. Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 5: 244 10.3389/fpls.2014.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi M. J., and Sinclair T. R., 2013. Nitrogen fixation drought tolerance of the slow-wilting soybean PI 471938. Crop Sci. 53: 2072–2078. 10.2135/cropsci2013.02.0095 [DOI] [Google Scholar]
- Devi J. M., Sinclair T. R., Chen P., and Carter T. E., 2014. Evaluation of elite southern maturity soybean breeding lines for drought-tolerant traits. Agron. J. 106: 1947–1954. 10.2134/agronj14.0242 [DOI] [Google Scholar]
- Dhanapal A. P., Ray J. D., Singh S. K., Hoyos-Villegas V., Smith J. R. et al. , 2016. Genome-wide association mapping of soybean chlorophyll traits based on canopy spectral reflectance and leaf extracts. BMC Plant Biol. 16: 174 10.1186/s12870-016-0861-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanapal A. P., Ray J. D., Singh S. K., Hoyos-Villegas V., Smith J. R. et al. , 2015. Genome-wide association study (GWAS) of carbon isotope ratio (δ13C) in diverse soybean [Glycine max (L.) Merr.] genotypes. Theor. Appl. Genet. 128: 73–91. Erratum: 375–376. 10.1007/s00122-014-2413-9 [DOI] [PubMed] [Google Scholar]
- Du Q. L., Cui W. Z., Zhang C. H., and Yu D. Y., 2010. GmRFP1 encodes a previously unknown RING-type E3 ubiquitin ligase in soybean (Glycine max). Mol. Biol. Rep. 37: 685–693. 10.1007/s11033-009-9535-1 [DOI] [PubMed] [Google Scholar]
- Du W., Yu D., and Fu S., 2009. Detection of quantitative trait loci for yield and drought tolerance traits in soybean using a recombinant inbred line population. J. Integr. Plant Biol. 51: 868–878. 10.1111/j.1744-7909.2009.00855.x [DOI] [PubMed] [Google Scholar]
- Fletcher A. L., Sinclair T. R., and Allen L. H., 2007. Transpiration responses to vapor pressure deficit in well watered ‘slow-wilting’ and commercial soybean. Environ. Exp. Bot. 61: 145–151. 10.1016/j.envexpbot.2007.05.004 [DOI] [Google Scholar]
- Goldman I. L., Carter T. E., and Patterson R. P., 1989. Differential genotypic response to drought stress and subsoil aluminum in soybean. Crop Sci. 29: 330–334. 10.2135/cropsci1989.0011183X002900020020x [DOI] [Google Scholar]
- Grant D., Nelson R. T., Cannon S. B., and Shoemaker R. C., 2010. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 38: D843–D846. 10.1093/nar/gkp798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J. B., Nyquist W. E., and Cervantes-Martinez C. T., 2010. Estimating and interpreting heritability for plant breeding: An update, Plant Breeding Reviews, edited by Janick J. John Wiley and Sons, Inc., Oxford, UK: 10.1002/9780470650202.ch2 [DOI] [Google Scholar]
- Hudak C. M., and Patterson R. P., 1996. Root distribution and soil moisture depletion pattern of a drought-resistant soybean plant introduction. Agron. J. 88: 478–485. 10.2134/agronj1996.00021962008800030020x [DOI] [Google Scholar]
- Hwang S., King C. A., Chen P., Ray J. D., Cregan P. B. et al. , 2016. Meta-analysis to refine map position and reduce confidence intervals for delayed-canopy-wilting QTLs in soybean. Mol. Breed. 36: 91 10.1007/s11032-016-0516-5 [DOI] [Google Scholar]
- Hwang S., King C. A., Davies M. K., Charlson D. V., Ray J. D. et al. , 2015a Registration of the KS4895 × Jackson soybean mapping population, AR93705. J. Plant Regist. 9: 266–271. 10.3198/jpr2014.05.0034crmp [DOI] [Google Scholar]
- Hwang S., King C. A., Ray J. D., Cregan P. B., Chen P. et al. , 2015b Confirmation of delayed canopy wilting QTLs from multiple soybean mapping populations. Theor. Appl. Genet. 128: 2047–2065. 10.1007/s00122-015-2566-1 [DOI] [PubMed] [Google Scholar]
- Hwang E.-Y., Song Q., Jia G., Specht J. E., Hyten D. L. et al. , 2014. A genome-wide association study of seed protein and oil content in soybean. BMC Genomics 15: 1 10.1186/1471-2164-15-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten D. L., Choi I. Y., Song Q., Shoemaker R. C., Nelson R. L. et al. , 2007. Highly variable patterns of linkage disequilibrium in multiple soybean populations. Genetics 175: 1937–1944. 10.1534/genetics.106.069740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten D. L., Song Q., Zhu Y., Choi I.-Y., Nelson R. L. et al. , 2006. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA 103: 16666–16671. 10.1073/pnas.0604379103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H. W., 1958. Registration of soybean varieties, VI. Agron. J. 50: 690–691. 10.2134/agronj1958.00021962005000110016x [DOI] [Google Scholar]
- Kaler A. S., Dhanapal A. P., Ray J. D., King C. A., Fritschi F. B. et al. , 2017a Genome-wide association mapping of carbon isotope and oxygen isotope ratios in diverse soybean genotypes. Crop Sci. 57: 3085–3100. 10.2135/cropsci2017.03.0160 [DOI] [Google Scholar]
- Kaler A. S., Ray J. D., Schapaugh W. T., King C. A., and Purcell L. C., 2017b Genome-wide association mapping of canopy wilting in diverse soybean genotypes. Theor. Appl. Genet. 130: 2203–2217. 10.1007/s00122-017-2951-z [DOI] [PubMed] [Google Scholar]
- Li S., Cao Y., He J., Zhao T., and Gai J., 2017. Detecting the QTL-allele system conferring flowering date in a nested association mapping population of soybean using a novel procedure. Theor. Appl. Genet. 130: 2297–2314. 10.1007/s00122-017-2960-y [DOI] [PubMed] [Google Scholar]
- Lipka A. E., Tian F., Wang Q., Peiffer J., Li M. et al. , 2012. GAPIT: Genome association and prediction integrated tool. Bioinformatics 28: 2397–2399. 10.1093/bioinformatics/bts444 [DOI] [PubMed] [Google Scholar]
- Liu X., Huang M., Fan B., Buckler E. S., and Zhang Z., 2016. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 12: e1005767 10.1371/journal.pgen.1005767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavalan L. P., Guttikonda S. K., Tran L.-S., and Nguyen H. T., 2009. Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 50: 1260–1276. 10.1093/pcp/pcp082 [DOI] [PubMed] [Google Scholar]
- Pantalone V., and Rebetzke G., 1996. Phenotypic evaluation of root traits in soybean and applicability to plant breeding. Crop Sci. 36: 456–459. 10.2135/cropsci1996.0011183X003600020039x [DOI] [Google Scholar]
- Pathan S. M., Lee J.-D., Sleper D. A., Fritschi F. B., Sharp R. E. et al. , 2014. Two soybean plant introductions display slow leaf wilting and reduced yield loss under drought. J. Agron. Crop Sci. 200: 231–236. 10.1111/jac.12053 [DOI] [Google Scholar]
- Patil G., Do T., Vuong T. D., Valliyodan B., Lee J. D. et al. , 2016. Genomic-assisted haplotype analysis and the development of high-throughput SNP markers for salinity tolerance in soybean. Sci. Rep. 6: 19199 10.1038/srep19199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A. et al. , 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Purcell L. C., and Specht J. E., 2004. Physiological traits for ameliorating drought stress, pp. 569–620 in Soybeans: Improvement, Production, and Uses, ASA, CSSA, and SSSA, Madison, WI. [Google Scholar]
- Qin F., Sakuma Y., Tran L.-S. P., Maruyama K., Kidokoro S. et al. , 2008. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707. 10.1105/tpc.107.057380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Stephens M., and Pritchard J. K., 2014. FastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 197: 573–589. 10.1534/genetics.114.164350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauf S., Al-khayri J. M., Zaharieva M., Monneveux P., and Khalil F., 2016. Breeding strategies to enhance drought tolerance in crops, pp. 397–445 in Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits, Springer, Cham. [Google Scholar]
- Ray J. D., Dhanapal A. P., Singh S. K., Hoyos-Villegas V., Smith J. R. et al. , 2015. Genome-wide association study of ureide concentration in diverse maturity group IV soybean [Glycine max (L.) Merr.] accessions. G3 (Bethesda) 5: 2391–2403. 10.1534/g3.115.021774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riar M. K., Cerezini P., Manandhar A., Sinclair T. R., Li Z. et al. , 2018. Expression of drought-tolerant N fixation in heterogeneous inbred families derived from PI471938 and Hutcheson soybean. Crop Sci. 58: 364–369. 10.2135/cropsci2017.02.0089 [DOI] [Google Scholar]
- Ries L. L., Purcell L. C., Carter T. E., Edwards J. T., and King C. A., 2012. Physiological traits contributing to differential canopy wilting in soybean under drought. Crop Sci. 52: 272–281. 10.2135/cropsci2011.05.0278 [DOI] [Google Scholar]
- Sadok W., Gilbert M. E., Raza M. A. S., and Sinclair T. R., 2012. Basis of slow-wilting phenotype in soybean PI 471938. Crop Sci. 52: 1261–1269. 10.2135/cropsci2011.11.0622 [DOI] [Google Scholar]
- SAS Institute, 2014 The SAS system for Windows. Release 9.4. Cary, NC.
- Sinclair T. R., Devi J. M., and Carter T. E., 2016. Limited-transpiration trait for increased yield for water-limited soybean: From model to phenotype to genotype to cultivars, pp. 129–146 in Crop Systems Biology: Narrowing the gaps between crop modelling and genetics, edited by Yin X., and Struik P. C.. Springer International Publishing, Cham: 10.1007/978-3-319-20562-5_6 [DOI] [Google Scholar]
- Sinclair T. R., Purcell L. C., Vadez V., Serraj R., King C. A. et al. , 2000. Identification of soybean genotypes with N fixation tolerance to water deficits. Crop Sci. 40: 1803–1809. 10.2135/cropsci2000.4061803x [DOI] [Google Scholar]
- Sloane R. J., Patterson R. P., and Carter T. E., 1990. Field drought tolerance of a soybean plant introduction. Crop Sci. 30: 118–123. 10.2135/cropsci1990.0011183X003000010027x [DOI] [Google Scholar]
- Song Q., Hyten D. L., Jia G., Quigley C. V., Fickus E. W. et al. , 2013. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS One 8: e54985 10.1371/journal.pone.0054985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q., Hyten D. L., Jia G., Quigley C. V., Fickus E. W. et al. , 2015. Fingerprinting soybean germplasm and its utility in genomic research. G3 (Bethesda) 5: 1999–2006. 10.1534/g3.115.019000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Q., G. Jia, C. Quigley, E. Fickus, D. Hyten et al., 2014 Soybean BARC- SoySNP6K Beadchip—a tool for soybean genetics research., pp. Abstract no.: P306 in Poster presented at: Plant and Animal Genome XXII Conference [Google Scholar]
- Specht J., Hume D., and Kumudini S., 1999. Soybean yield potential—A genetic and physiological perspective. Crop Sci. 39: 1560–1570. 10.2135/cropsci1999.3961560x [DOI] [Google Scholar]
- Steketee C. J., Sinclair T.R., Riar M.K., Schapaugh W.T., and Li Z., 2019. Unraveling the genetic architecture for carbon and nitrogen related traits and leaf hydraulic conductance in soybean using genome-wide association analyses. BMC Genomics 20: 811 10.1186/s12864-019-6170-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Fujii K., and Shiraiwa T., 2010. Variability of leaf morphology and stomatal conductance in soybean [Glycine max (L.) Merr.] cultivars. Crop Sci. 50: 2525–2532. 10.2135/cropsci2010.02.0058 [DOI] [Google Scholar]
- Tuberosa R., 2012. Phenotyping for drought tolerance of crops in the genomics era. Front. Physiol. 3: 347 10.3389/fphys.2012.00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S. D., 2018 qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Software 3:731. 10.21105/joss.00731 [DOI]
- Vaughn J. N., Nelson R. L., Song Q., Cregan P. B., and Li Z., 2014. The genetic architecture of seed composition in soybean is refined by genome-wide association scans across multiple populations. G3 (Bethesda) 4: 2283–2294. 10.1534/g3.114.013433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Y. S., Barratt B. J., Clayton D. G., and Todd J. A., 2005. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 6: 109–118. 10.1038/nrg1522 [DOI] [PubMed] [Google Scholar]
- Wang W., Jiang W., Liu J., Li Y., Gai J. et al. , 2017. Genome-wide characterization of the aldehyde dehydrogenase gene superfamily in soybean and its potential role in drought stress response. BMC Genomics 18: 518 10.1186/s12864-017-3908-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook J., Chhatre V., Wu L., Chamala S., Neves L. et al. , 2015. A consensus genetic map for Pinus taeda and Pinus elliottii and extent of linkage disequilibrium in two genotype-phenotype discovery populations of Pinus taeda. G3 (Bethesda) 5: 1685–1694. 10.1534/g3.115.019588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng A., Chen P., Korth K., Hancock F., Pereira A. et al. , 2017. Genome-wide association study (GWAS) of salt tolerance in worldwide soybean germplasm lines. Mol. Breed. 37: 30 10.1007/s11032-017-0634-8 [DOI] [Google Scholar]
- Zhang J., Song Q., Cregan P. B., and Jiang G.-L., 2015. Genome-wide association study, genomic prediction and marker-assisted selection for seed weight in soybean (Glycine max). Theor. Appl. Genet. 129: 117–130. 10.1007/s00122-015-2614-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
SNP marker genotypes for accessions included in the association panel are in File S1 and canopy wilting scores are in File S2. All other datasets generated and/or analyzed for this study are not publicly available, but are available from the corresponding author based on a reasonable request. Supplemental material available at figshare: https://doi.org/10.25387/g3.11441340.