Abstract
Listeria monocytogenes, a human foodborne pathogen that causes listeriosis with high-rate mortality, has been reported to be resistant to commonly used antibiotics. New antibiotics or cocktails of existing antibiotics with synergistic compounds are in high demand for treating this multi-drug-resistant pathogen. Fosfomycin is one of the novel and promising therapeutic antibiotics for the treatment of listeriosis. However, some L. monocytogenes strains with the FosX gene were recently reported to survive from the fosfomycin treatment. This work aims to identify FosX inhibitors that can revive fosfomycin in treating resistant L. monocytogenes. Since structures and activities of the FosX protein in L. monocytogenes have been well studied, we used an integrated computational and experimental approach to identify FosX inhibitors that show synergistic effect with fosfomycin in treating resistant L. monocytogenes. Specifically, automated ligand docking was implemented to perform virtual screening of the Indofine natural-product database and FDA-approved drugs to identify potential inhibitors. An in vitro bacterial growth inhibition test was then utilized to verify the effectiveness of identified compounds combined with fosfomycin in inhibiting the resistant L. monocytogenes strains. Two phenolic acids, i.e., caffeic acid and chlorogenic acid, were predicted as high-affinity FosX inhibitors from the ligand-docking platform. Experiments with these compounds indicated that the cocktail of either caffeic acid (1.5 mg/mL) or chlorogenic acid (3 mg/mL) with fosfomycin (50 mg/L) was able to significantly inhibit the growth of the pathogen. The finding of this work implies that the combination of fosfomycin with either caffeic acid or chlorogenic acid is of potential to be used in the clinical treatment of Listeria infections.
Introduction
Listeria monocytogenes is a Gram-positive, foodborne pathogen, which could cause listeriosis disease, especially in immune-compromised people.1,2 In particular, L. monocytogenes was found to infect and adversely affect patients’ liver and spleen.3−5 In addition, L. monocytogenes can penetrate the blood–brain barrier and blood–placenta barrier to harm the central neural system of pregnant women and infants.1,4 Unfortunately, L. monocytogenes can also grow in stress conditions, such as under 4 °C, high salt, and low pH conditions.6,7 This poses a big challenge to the food preservation in food processing industries.1,8,9 The ready-to-eat (RTE) food is more likely to cause the outbreak of listeriosis.8,10 Based on the data posted by the Center for Disease Control and Prevention (CDC), there was at least one multistate listeriosis outbreak in the United States in the last 9 years.11 Since L. monocytogenes is an intracellular pathogen, the antibiotics used to treat listeriosis should be able to be transported into host cells.9,12,13 Penicillin, ampicillin, and amoxicillin were the commonly used antibiotics in the treatment of listeriosis.13 However, antibiotic resistance genes have been continuously found in Listeria strains. For example, the strain that has resistance to penicillin G was isolated from vegetables in 2016.1,14 To solve this problem, either new antibiotics or the cocktails of existing antibiotics with inhibitors of antibiotic resistance proteins should be explored.
Fosfomycin was found to be effective against clinical isolates of L. monocytogenes and used as a novel therapeutic antibiotic for clinical treatment of listeriosis.15 As a natural product, fosfomycin is a broad-spectrum phosphonic acid antibiotic produced by several Streptomyces and Pseudomonas species. Fosfomycin acts as a phosphoenolpyruvate (PEP) analogue to form a covalent bond with the active cysteine-115 side chain of the MurA enzyme. This leads to the inhibition of the biosynthesis of peptidoglycan, which further interferes with cell wall formation and leads to cell death.16 In addition, fosfomycin is able to penetrate the blood–brain barrier and reach clinically relevant concentrations. Thus, it has the potential to eliminate L. monocytogenes, which causes neuron damage.17
L. monocytogenes was previously reported with a limited uptake of fosfomycin with certain in vitro standard susceptibility assays.18 In contrast, further investigations indicated that the pathogen highly expressed the virulence gene prfA and thereby induced the uptake of fosfomycin through an Hpt transporter in infection-mimic or in vivo conditions.19,20 However, stronger fosfomycin resistance was found in the L. monocytogenes isolates with fosfomycin resistance proteins detected.21 In particular, the resistant gene FosX (LMO1702,402bp) was identified and expressed in L. monocytogenes EGDe (strain ATCC BAA-679), a typical well-studied strain.22 The FosX enzyme catalyzed the hydrolysis of fosfomycin and resulted in fosfomycin resistance in L. monocytogenes EGDe.2,21 Unfortunately, the FosX gene in the EGDe strain would be transferred to other Listeria strains via gene recombination (e.g., conjugation) in a short time period.23 Few effective inhibitors have been found for fosfomycin resistance proteins. Phosphonoformate (Foscarnet) is one of the identified inhibitors for FosA, which belongs to the same metalloenzyme superfamily as FosX.24 However, the safety and efficacy of the compound was not approved for the treatment of Listeria infection. Since there are many FDA-approved natural compounds in the existing database,25 it is possible to find inhibitors against FosX. However, thorough experimental screening for all of these compounds would be time-consuming and costly. An efficient and affordable approach is needed to address this issue.
Automated molecular docking is the most commonly used computational approach that evaluates the binding of small-molecule ligands like compounds to a target receptor with a known three-dimensional (3D) crystal structure.26 Molecular docking provides an avenue for a high-throughput virtual screening of ligands, and it has been widely implemented in drug discovery research for hit identification.27−31 Docking programs have been improved recently to provide more accurate prediction on ligand–target binding by optimizing docking algorithms and scoring functions.32 Among these existing docking programs, Molsoft ICM was evaluated with 93% accuracy in flexible docking and 90% successful rate in covalent docking. This was significantly better than the performance of Autodock, DOCK, FlexX, Gold, FITTED, and MOE.27,32,33 Furthermore, ICM was ranked the first place for docking pose and energy prediction in the drug design data resource (D3R) challenge for both 2017 and 2018.34−36 The ICM software has succeeded in identifying inhibitors of pancreatic endoplasmic reticulum kinase, P300 HAT, STAT3 dimer, tumor marker AKR1B10, and type-II kinase.37−41 Molsoft ICM was thus used in this study to identify the compounds that inhibit the FosX protein.
Generation of a docking receptor is a crucial step for virtual screening; fortunately, crystal structures and the mechanism of FosX protein in L. monocytogenes EGDe were well studied, in which various ligands, e.g., sulfate ions, citric acid, or glycerol, were co-crystallized with the protein.22 The enzyme is a homodimer in a domain-swapped conformation and has two identical catalytic sites formed by paired βαβββ motifs. The H7 residue from one subunit joins H69 and E188 from the opposite subunit to form a cup-shaped manganese ion-binding site.22 The enzyme adds water to the carbon-1 position of fosfomycin, initiating the breaking of the epoxide ring, thereby eliminating the activity of the antibiotic. The catalytic mechanism is as follows: the Mn (II) ions provide electrophilic assistance, and T9 supplies a proton to the oxirane oxygen and E44 residues acting as a general base, resulting in addition of water and breaking of the epoxide ring.21,22 Superimposition of the structures of the FosX protein with various ligands (PDB codes 2P7P, 2P7K, and 2P7L) indicates that ligand binding might not initiate a global conformational change, particularly in loop K94-G104 and the C terminal (Figure S1A). Furthermore, residues within the catalytic sites, e.g., E44, T9, E118, Y108, Y67, H69, H7, and C55, of the three co-crystal complexes are conserved (Figure S1B). Therefore, the ligand-bound receptor is rational for ligand–protein docking. In all, the chain EF of the FosX·Mn(II)·SO42– complex (PDB code 2p7p), indicating no missing loops and conserved residues (Figure S2), was modified as the receptor for virtual screening. The top compounds were then validated by in vitro wet-lab experiments.
Results
Caffeic Acid and Chlorogenic Acid Were Identified as FosX Inhibitors by Virtual Screening
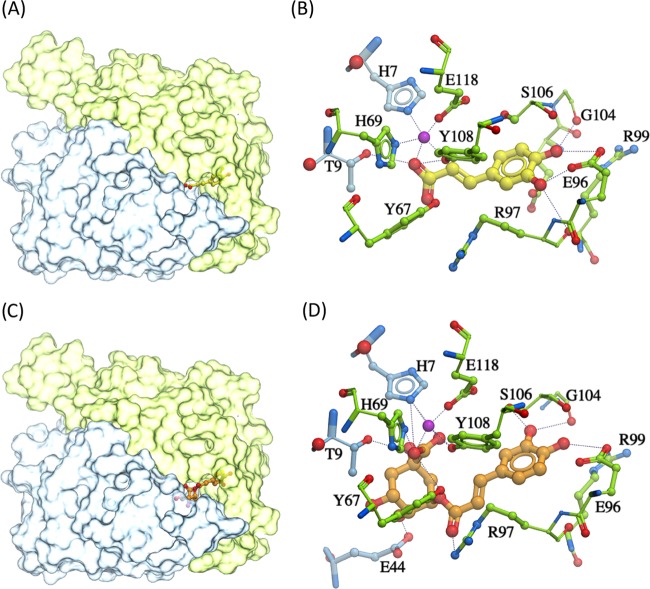
Among the 4821 compound candidates from the Indofine and FDA-approved database, the top compounds that bound to the catalytic site of the FosX protein and had a stronger binding affinity than fosfomycin are listed in Table 1. It was hypothesized that treating L. monocytogenes with fosfomycin and one of these compounds should be able to eliminate the resistant Listeria strain. Among these compounds, caffeic acid and chlorogenic acid had the lowest scores (−21.1 and −17.9, respectively), which indicated a relatively higher binding affinity than any other compounds, including fosfomycin. Furthermore, both caffeic acid and chlorogenic acid were expected to bind at the catalytic site of the FosX protein. Figure 1 displays docking conformations of caffeic acid and chlorogenic acid, in which the two compounds occupy the fosfomycin-binding site and overlap with the sulfate ion in the crystal structure 2p7p. Caffeic acid was predicted to be interacting with Mn2+ and forming H-bonds with the T9 side chain and G104 backbone on the FosX protein, thereby preventing the substrate fosfomycin access. Chlorogenic acid might have further H-bond binding with side chains of E96 and C55. Therefore, these two phenolic acid compounds could be potential competitive inhibitors of the FosX enzyme. Considering the solubility impact on the drug efficacy in the in vitro susceptibility assay and the toxicity of the identified compounds, only chlorogenic acid and caffeic acid were further validated experimentally.
Table 1. Top Compounds with Stronger Binding Affinities with the FosX Protein than Fosfomycina.
| name | score | Hbond | Hphob | VwInt | Eintl | Dsolv | SolEl | mfScore | dTSsc | x log P3-AA | solubility |
|---|---|---|---|---|---|---|---|---|---|---|---|
| chicoric acid | –22.0132 | –5.89096 | –3.48015 | –19.7111 | 0.516713 | 16.0599 | –0.6382 | –42.8735 | 1.776343 | 2 | NA |
| marein | –21.2412 | –19.7007 | –4.36088 | –34.2541 | 19.88202 | 44.29087 | 25.80296 | –95.8018 | 1.96109 | 0.7 | NA |
| caffieic acid | –21.1092 | –7.84353 | –2.36317 | –19.0568 | 1.295344 | 12.20363 | 13.40166 | –82.837 | 1.221385 | 1.2 | 1 mg/mL |
| N-carbamimidoyl-2-(2,6-dichlorophenyl)acetamide | –19.6856 | –5.75583 | –2.79432 | –16.2979 | 0 | 12.17383 | 3.206563 | –18.1895 | 0.507306 | 2.2 | NA |
| chlorogenic acid | –17.9415 | –13.4456 | –3.72354 | –23.0387 | 17.36799 | 21.76005 | 17.98263 | –148.516 | 1.791441 | –0.4 | 40 mg/mL |
| 2′,5′-dihydroxy-4-methoxychalcone | –15.8542 | –5.88801 | –4.98774 | –29.9978 | 1.992762 | 17.59366 | 27.25938 | –24.5201 | 1.427804 | 3.5 | NA |
| 4,2′,5′-trihydroxychalcone | –14.5402 | –5.80241 | –4.17045 | –27.9698 | 2.67159 | 18.61023 | 23.24728 | –12.7586 | 1.333933 | 3.2 | NA |
| plumbagin | –13.577 | –8.81215 | –3.17567 | –9.75784 | 2.856866 | 14.22039 | 12.46037 | –102.235 | 1.45787 | 2.3 | NA |
| l-mimosine | –12.9844 | –10.075 | –2.20455 | –16.8497 | 0.878521 | 19.13007 | 20.54147 | –102.074 | 1.560315 | –3.5 | slightly soluble in water |
| fosfomycin | 1.3945 | –3.67585 | –1.54743 | –6.8822 | 0.186757 | 14.94213 | 8.325324 | –41.929 | 1.229387 |
Note: the score of each ligand indicates the ligand–receptor binding affinity (the lower the better). Other scores evaluate various energies between the ligand and protein, e.g., hydrogen bond energy (Hbond), hydrophobic energy in exposing a surface to water (Hphob), van der Waals interaction energy (VwInt), internal conformation energy of the ligand (Eintl), desolvation of exposed H-bond donors and acceptors (Dsolv), solvation electrostatics energy change upon binding (SolEl), loss of entropy (dTSsc), and potential of mean force score (mfScore).42x log P3-AA values and solubility were obtained from Pubchem.43
Figure 1.
Docked conformation of caffeic acid/chlorogenic acid. (A) Caffeic acid (yellow sticks) docked within the catalytic site of the dimeric FosX protein; (B) predicted ligand–residue interactions of caffeic acid (yellow) and the FosX protein; (C) chlorogenic acid (orange sticks) docked in the catalytic site of the FosX protein; and (D) molecular interactions of chlorogenic acid (orange) with the FosX protein. Two subunits of the FosX protein are colored distinctly (green and blue); the manganese ion is shown in purple. Hydrogen bonds are represented as black dashes.
Minimum Inhibitory Concentrations (MICs) of Fosfomycin, Chlorogenic Acid, and Caffeic Acid
The MIC of fosfomycin for L. monocytogenes EGDe was determined to be greater than 100 mg/L, which is consistent with a previous study.2 The tested strain was resistant to fosfomycin. Since FosX is the only fosfomycin resistance gene that is proven to be present in L. monocytogenes EGDe, the FosX gene should be expressed.21 The tested strain was used for further tests. The MIC of chlorogenic acid was around 6 mg/mL. The MIC of caffeic acid was around 2.25 mg/mL. Therefore, all of the concentrations of chlorogenic acid and caffeic acid used in the cell growth study were lower than the tested MICs of fosfomycin.
Chlorogenic Acid had Synergistic Inhibition Effect with Fosfomycin
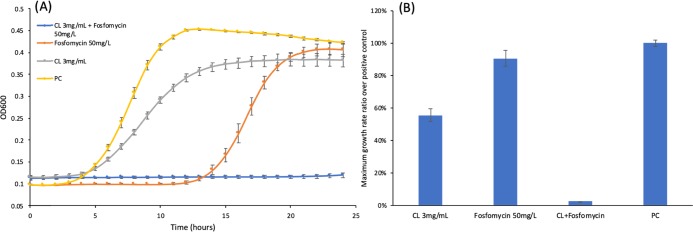
Based on the calculation from eq 1, the growth reduction rate of 50 mg/L fosfomycin was around 5% when compared to the positive control group. The growth reduction rate of the 3 mg/mL chlorogenic acid treatment was around 17.7%. However, the chlorogenic acid (3 mg/mL) and fosfomycin (50 mg/L) co-treated group had a growth reduction of 98%. This is 19.6-fold of the reduction from the fosfomycin treatment group (50 mg/L fosfomycin). As shown in Figure 2A, the growth curve of the chlorogenic acid and fosfomycin co-treated group does not show an obvious log phase, which indicates that the growth of L. monocytogenes is inhibited. The maximum growth rate of L. monocytogenes shown in Figure 2B also indicates a synergistic effect of chlorogenic acid and fosfomycin. Compared to the group treated with 50 mg/L fosfomycin, the group treated with the same concentration of fosfomycin together with 3 mg/mL chlorogenic acid had a maximum growth rate that was only 2.46% of that of the group that had been treated with only fosfomycin. Additionally, the co-treatment of fosfomycin (50 mg/L) with a lower concentration of chlorogenic acid (Figure S4A: 2.7 mg/mL and Figure S4B: 2.4 mg/mL) could also inhibit the growth of L. monocytogenes in the first 18–19 h, but L. monocytogenes had obvious growth after 20 h. These results indicated that if co-treated with 3 mg/mL chlorogenic acid, the MIC of fosfomycin would be decreased to around 50 mg/L. A previous study had shown that the Listeria strain is susceptible to fosfomycin if the MIC of fosfomycin is no more than 64 mg/L.15 The chlorogenic acid co-treatment thus had reverted the fosfomycin-resistant strain to a fosfomycin-susceptible strain.
Figure 2.
Cell growth test with chlorogenic acid and fosfomycin. (A) Comparison of the growth curves of L. monocytogenes under different chemical treatment conditions. (B) Maximum growth rate normalized with the positive control of L. monocytogenes. CL: chlorogenic acid; triplicate samples; error bar: standard deviation; and PC: positive control (L. monocytogenes overnight culture was diluted with 5% dimethyl sulfoxide (DMSO)).
Inhibition and Synergistic Inhibition Effect of Caffeic Acid
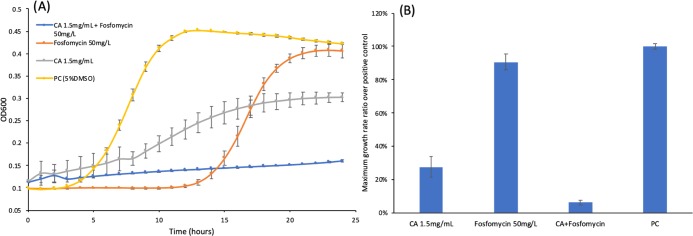
Similar to chlorogenic acid, caffeic acid also showed synergistic effect with fosfomycin on growth inhibition of L. monocytogenes. The growth curves of L. monocytogenes treated with fosfomycin, caffeic acid, or both are shown in Figure 3A. As mentioned in Section 3.3, the 50 mg/L fosfomycin-treated group exhibited a 5% growth reduction compared to the positive control. On the other hand, the co-treatment of 50 mg/L fosfomycin and 1.5 mg/mL caffeic acid was able to impose 85.2% reduction in the growth compared to the positive control. This was around 17-fold of the growth reduction from the fosfomycin-treated group (50 mg/L fosfomycin). In addition, the co-treatment of fosfomycin (50 mg/L) with a lower concentration of caffeic acid (Figure S5A: 1.25 mg/mL and Figure S5B: 1 mg/mL) also indicated the synergistic effect to inhibit growth of L. monocytogenes, but the lower concentration of caffeic acid could not completely inhibit the growth of L. monocytogenes, even at the early stage of treatment. Furthermore, the growth reduction for the treatment with 1.5 mg/mL caffeic acid was around 42.16%, which showed that caffeic acid has the potential to inhibit the growth of L. monocytogenes. The maximum growth rates shown in Figure 3B also indicated that caffeic acid could reduce the rounded maximum growth rate to 0.28, but the co-treated group still had the lowest rounded maximum growth rate among all experimental groups, which was around 0.06.
Figure 3.
Cell growth test with caffeic acid and fosfomycin. (A) Comparison of the growth curves of L. monocytogenes under different chemical treatment conditions. (B) Maximum growth rate ratio over the positive control of L. monocytogenes. CA: caffeic acid; triplicate samples; error bar: standard deviation; and PC: positive control (L. monocytogenes overnight culture was diluted with 5% DMSO).
Discussion
Chlorogenic acid and caffeic acid are phenolic acids, which are secondary metabolites of plants like blueberry44−46 among others. Studies have shown that chlorogenic acid and caffeic acid have antimicrobial and antioxidant effects.47,48 Chlorogenic acid gained substantial attention due to its various biological and pharmacological effects, e.g., anti-diabetics, anti-obesity, anti-oxidation, anti-inflammation, anti-hypertension, antimicrobial activities, and metabolism regulation. It was suggested that chlorogenic acid had bactericidal effects against some pathogens through various mechanisms.49 It has been shown that the treatment of chlorogenic acid at the concentrations of 2MIC or 4MIC could induce cell constituents’ release, reduce intracellular ATP concentration, and reduce the intracellular pH.45 Furthermore, chlorogenic acid could limit oxidation of polyunsaturated lipids and delayed the growth of L. monocytogenes.(44) Besides L. monocytogenes, chlorogenic acid also had growth inhibitory effects on other bacteria, such as Salmonella enteritidis and Staphylococcus aureus.(46,48) Similarly, caffeic acid was also shown to have inhibitory effects on L. monocytogenes, S. aureus, Escherichia coli, and Bacillus cereus.(45,50−52) The compound can increase membrane permeability, resulting in the release of cell contents and access of hydrophobic antibiotics.45,53
This study showed a general consistency in the growth inhibitory effect of chlorogenic acid and caffeic acid with the aforementioned findings. Furthermore, this study explored a potential mechanism for these compounds to inhibit the FosX protein and then eliminate the resistance of L. monocytogenes to fosfomycin. Different from the traditional high-throughput drug screening methods, computational docking was implemented to identify the potential inhibitors of the target protein FosX. Among the 4821 bioactive natural compounds or FDA-approved drugs, chlorogenic acid and caffeic acid were predicted by the ligand–protein docking to have the highest affinity with the catalytic sites of the FosX protein. In particular, they can outperform fosfomycin in binding to the FosX protein. This enables fosfomycin to function properly as an antibiotic to eliminate L. monocytogenes. Our experimental results demonstrated that each of these two phenolic acids showed a strong synergistic activity with fosfomycin to inhibit the growth of L. monocytogenes EGDe resistant to fosfomycin. The co-treatment of fosfomycin (50 mg/L) and chlorogenic acid (3 mg/mL) was able to reduce the growth of L. monocytogenes by 98% from the positive control conditions, which was much higher than the sum of the growth reduction rates of 22.7% (fosfomycin (5%) + chlorogenic acid (17.7%)) of monotreatment groups. Similarly, the cocktail of fosfomycin (50 mg/L) and caffeic acid (1.5 mg/mL) reduced the growth of L. monocytogenes by 85.2% versus 5% by fosfomycin and 42.16% by caffeic acid. Thus, both chlorogenic acid and caffeic acid exhibited excellent synergistic effects with fosfomycin on inhibiting the growth of fosfomycin-resistant L. monocytogenes. Although there are a lot of studies on chlorogenic acid and caffeic acid, no previous research indicated that these two phenolic compounds had synergistic effects in combination with fosfomycin against L. monocytogenes. The novel drug combinations provide a potential medical treatment of Listeria infection, particularly caused by fosfomycin-resistant strains.
The toxicities of the two compounds have been evaluated. Based on the information provided by NIH, chlorogenic acid and caffeic acid could be considered as relatively nontoxic.54 Additionally, probiotic bacteria are not sensitive to chlorogenic acid and thus it may be practically used as a natural safeguard food additive to replace the synthetic antibiotics, thereby reducing the medicinal cost.49
While all of these experimental results imply the effectiveness of chlorogenic acid and caffeic acid in eliminating the resistance of L. monocytogenes to fosfomycin, mechanisms other than their inhibitory effect on the FosX protein may exist. The mechanism of the synergistic effect of chlorogenic acid or caffeic acid with fosfomycin should be studied systematically using a transcriptomic study or phenotypic microarray study. Furthermore, the kinetic study of FosX under the treatment of identified FosX inhibitors could be considered for future research to verify the protein–ligand binding of caffeic acid and chlorogenic acid with the fosfomycin resistance protein FosX.49
Conclusions
Chlorogenic acid and caffeic acid were predicted as potential high-affinity inhibitors of the FosX protein through molecular docking. In the in vitro bacterial growth test, chlorogenic acid/caffeic acid and fosfomycin combinations had synergistic effects against the growth of a fosfomycin-resistant L. monocytogenes strain, L. monocytogenes EGDe. The mechanism might be that the two compounds prevent degradation of fosfomycin and thus contribute to maintaining its antibacterial activity. Our study identified two compounds that might be used together with fosfomycin against fosfomycin-resistant L. monocytogenes in the future clinical use. The synergistic effect of the combination in this study suggests more laboratory and clinical studies to quantify the doses of drugs in the regimen against L. monocytogenes.
Methods and Materials
Identification of FosX Protein Inhibitors using Ligand–Protein Docking
The fosfomycin resistance gene, FosX, in the L. monocytogenes EGDe strain was obtained from UniPlot with ID Q8Y6I2. The X-ray structure of the FosX protein was found in the Protein Data Bank with PDB code 2p7p and was chosen as the model for virtual screening described in this study. Furthermore, the bound sulfate ion in the structure predicts the position of fosfomycin or its hydrolysate product, which is a good marker to evaluate the accuracy of molecular docking.22 Molecular docking was carried out using ICM-Pro 3.7b (Molsoft, San Diego, CA). The FosX protein structure (2p7p) was input into ICM through a PDB search tool. The E and F monomers of FosX were modified as the receptor for virtual screening by deleting A, B, C, and D monomers, sulfate ions, and water molecules. The ligand-binding pocket was identified using pocket prediction tools with tolerance 4.6 and selected to cover the enzymatic active site. The default docking box and maps were generated around the pocket. The initial docking probe was manually removed from the docking box. The FDA-approved drug and Indofine database containing 4821 compounds were docked into the pocket with highest docking effort 10. Ligands with relatively lower scores and having overlap with the sulfate ion were selected for the hit list. Additionally, the five Lipinski rules have been used to filter library according to their physicochemical properties in virtual screening, and chemical solubility was considered as a very important factor in the experiment.55
Bacterial Strain and Culture Conditions
The L. monocytogenes EGDe strain used in this study was purchased from American Type Culture Collection (ATCC, Manassas, VA). Before each experiment, a single colony of L. monocytogenes EGDe was cultured overnight in 5 mL of brain heart infusion (BHI, Sigma-Aldrich Inc., St. Louis, MO) broth at 37 °C with 200 rpm agitation.
Chemical Stock Solution Preparation and Storage
The 50 mg/mL fosfomycin sodium (analytical standard, Sigma-Aldrich Inc., St. Louis, MO) stock solution was prepared by dissolving fosfomycin sodium in distilled water. The chemicals, chlorogenic acid, and caffeic acid, were identified by the ligand–protein docking (as shown in Section 3.1). The chlorogenic acid (≥95%, Sigma-Aldrich Inc., St. Louis, MO) stock solution was prepared by dissolving the chemical in DMSO (Sigma-Aldrich Inc., St. Louis, MO) with a final concentration of 60 mg/mL. The caffeic acid stock solution was prepared by dissolving caffeic acid (≥98%, Sigma-Aldrich Inc., St. Louis, MO) in DMSO with the final concentration of 50 mg/mL. All stock solutions were sterilized using a 0.22 μm syringe filter and stored under −20 °C until further use.
Bacterial Minimum Inhibitory Concentration (MIC) Assay
The chemical stock solutions were diluted using a 2-fold serial dilution method.56 Then, 10 μL of the diluted/undiluted stock solution was placed in a 96-well plate with 190 μL of fresh BHI-diluted L. monocytogenes EGDe overnight culture (1:1000 dilution). The plate was incubated in a microplate reader (BioTek Instruments, Inc., Winooski, VT) at 37 °C with 0.05 linear agitation. The absorbance at 600 nm (OD600) was monitored by the microplate reader every hour for 24 h. The lowest chemical concentration among the groups that showed no increase of OD600 over 24 h was determined as MIC.
Bacterial Cell Growth Study and Maximum Growth Rate Calculation
The overnight culture of L. monocytogenes was diluted 1000-fold in fresh BHI for the cell growth study. The stock solution of fosfomycin, chlorogenic acid, and caffeic acid was diluted to the concentrations that were 20-fold to the final concentration of each well. In the co-treated group, 10 μL of the diluted fosfomycin sodium solution and 10 μL of chlorogenic acid/caffeic acid diluted solution were added into the corresponding well in a 96-well plate. In the cocktail of fosfomycin and chlorogenic acid/caffeic acid, 10 μL of the diluted chlorogenic acid/caffeic acid solution and 10 μL of BHI was added into each well. Then, 180 μL of diluted L. monocytogenes overnight culture (1:1000 dilute in BHI) was added into each experimental group. For the negative control group, 10 μL of DMSO and 190 μL of fresh BHI were added into each well. For the positive control group, 10 μL of DMSO, 10 μL of fresh BHI, and 180 μL of 0.1% L. monocytogenes overnight culture were added into each well. The 96-well plate holding negative control groups, positive control groups, and experimental groups was placed into a microplate reader and incubated at 37 °C with 0.05 linear agitation. The OD600 was measured by the microplate reader and recorded by Gen 5 software (version 3.00.19, BioTek Instruments, Inc., Winooski, VT) every hour for 24 h. The growth reduction rate (i.e., r) was calculated as follows
| 1 |
where PC stands for the positive control condition. The maximum growth rate of each group was calculated and recorded by Gen 5 software. The maximum growth rate of the positive control group was normalized to 1, and all growth rates of other experimental groups were normalized based on the growth rate of the positive control group.
Acknowledgments
The authors would like to thank the support provided by Villanova University to acquire the ICM license.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c00352.
Analysis of X-ray crystal structures of FosX protein; cell growth test of lower concentrations of chlorogenic acid/caffeic acid and 50 mg/L fosfomycin (PDF)
Author Contributions
∥ F.Z. and T.Z. contributed equally.
The authors declare no competing financial interest.
Notes
All data generated or analyzed during this study are included in this article (and its Supporting Information files).
Supplementary Material
References
- Radoshevich L.; Cossart P. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 2018, 16, 32–46. 10.1038/nrmicro.2017.126. [DOI] [PubMed] [Google Scholar]
- Scortti M.; Han L.; Alvarez S.; Leclercq A.; Moura A.; Lecuit M.; Vazquez-Boland J. Epistatic control of intrinsic resistance by virulence genes in Listeria. PLoS Genet. 2018, 14, e1007525 10.1371/journal.pgen.1007525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen B. N.; Portnoy D. A. An Inducible Cre-lox System to Analyze the Role of LLO in Listeria monocytogenes Pathogenesis. Toxins 2020, 12, 38 10.3390/toxins12010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaguezeski A. M.; da Silva A. S.; Gomes T. M. A.; Bottari N. B.; Lopes T. F.; Cechin R. A.; Morsch V. M.; Schetinger M. R. C.; Giongo J. L.; de A. Vaucher R. Experimental listeriosis: A study of purinergic and cholinergic inflammatory pathway. Vet. Microbiol. 2020, 241, 108528 10.1016/j.vetmic.2019.108528. [DOI] [PubMed] [Google Scholar]
- Wang G.; Zhao H.; Zheng B.; Li D.; Yuan Y.; Han Q.; Tian Z.; Zhang J. TLR2 Promotes Monocyte/Macrophage Recruitment Into the Liver and Microabscess Formation to Limit the Spread of Listeria monocytogenes. Front. Immunol. 2019, 10, 1388 10.3389/fimmu.2019.01388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Ceruso M.; Jiang Y.; Datta A. R.; Carter L.; Strain E.; Pepe T.; Anastasi A.; Fratamico P. Construction of Listeria monocytogenes mutants with in-frame deletions in the phosphotransferase transport system (PTS) and analysis of their growth under stress conditions. J. Food Sci. 2013, 78, M1392–M1398. 10.1111/1750-3841.12181. [DOI] [PubMed] [Google Scholar]
- Liu Y. Construction of Listeria monocytogenes mutants with in-frame deletions in putative ATP-binding cassette (ABC) transporters and analysis of their growth under stress conditions. J. Microb. Biochem. Technol. 2012, 04, 141–146. 10.4172/1948-5948.1000085. [DOI] [Google Scholar]
- Jordan K.; Leong D.; Ordóñez A. Á.. Listeria monocytogenes in the Food Processing Environment; Springer International Publishing: Switzerland, 2015. [Google Scholar]
- Tilocca B.; Costanzo N.; Morittu V. M.; Spina A. A.; Soggiu A.; Britti D.; Roncada P.; Piras C. Milk microbiota: Characterization methods and role in cheese production. J. Proteomics 2020, 210, 103534 10.1016/j.jprot.2019.103534. [DOI] [PubMed] [Google Scholar]
- Garg N.; Abdel-Aziz S. M.; Aeron A.. Microbes in Food and Health; Springer International Publishing: Switzerland, 2016. [Google Scholar]
- CDC Listeria Outbreaks. https://www.cdc.gov/listeria/outbreaks/index.html (accessed July 2019).
- Aparna V.; Shiva M.; Biswas R.; Jayakumar R. Biological macromolecules based targeted nanodrug delivery systems for the treatment of intracellular infections. Int. J. Biol. Macromol. 2018, 110, 2–6. 10.1016/j.ijbiomac.2018.01.030. [DOI] [PubMed] [Google Scholar]
- Temple M. E.; Nahata M. C. Treatment of Listeriosis. Ann. Pharmacother. 2000, 34, 656–661. 10.1345/aph.19315. [DOI] [PubMed] [Google Scholar]
- de Vasconcelos Byrne V.; Hofer E.; Vallim D. C.; de Castro Almeida R. C. Occurrence and antimicrobial resistance patterns of Listeria monocytogenes isolated from vegetables. Braz. J. Microbiol. 2016, 47, 438–443. 10.1016/j.bjm.2015.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepe J. A.; Torres M. J.; Smani Y.; Parra-Millán R.; Pachón J.; Vazquez-Barba I.; Aznar J. In vitro and intracellular activities of fosfomycin against clinical strains of Listeria monocytogenes. Int. J. Antimicrob. Agents 2014, 43, 135–139. 10.1016/j.ijantimicag.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Falagas M. E.; Athanasaki F.; Voulgaris G. L.; Triarides N. A.; Vardakas K. Z. Resistance to fosfomycin: Mechanisms, Frequency and Clinical Consequences. Int. J. Antimicrob. Agents 2019, 53, 22–28. 10.1016/j.ijantimicag.2018.09.013. [DOI] [PubMed] [Google Scholar]
- Falagas M. E.; Vouloumanou E. K.; Samonis G.; Vardakas K. Z. Fosfomycin. Clin. Microbiol. Rev. 2016, 29, 321–347. 10.1128/CMR.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxler R.; von Graevenitz A.; Funke G.; Wiedemann B.; Stock I. Natural antibiotic susceptibility of Listeria species: L. grayi, L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri and L. welshimeri strains. Clin. Microbiol. Infect. 2000, 6, 525–535. 10.1046/j.1469-0691.2000.00168.x. [DOI] [PubMed] [Google Scholar]
- Chico-Calero I.; Suarez M.; Gonzalez-Zorn B.; Scortti M.; Slaghuis J.; Goebel W.; Vazquez-Boland J. A.; Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 431–436. 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortti M.; Lacharme-Lora L.; Wagner M.; Chico-Calero I.; Losito P.; Vazquez-Boland J. A. Coexpression of virulence and fosfomycin susceptibility in Listeria: molecular basis of an antimicrobial in vitro-in vivo paradox. Nat. Med. 2006, 12, 515–517. 10.1038/nm1396. [DOI] [PubMed] [Google Scholar]
- Fillgrove K. L.; Pakhomova S.; Newcomer M. E.; Armstrong R. N. Mechanistic diversity of fosfomycin resistance in pathogenic microorganisms. J. Am. Chem. Soc. 2003, 125, 15730–15731. 10.1021/ja039307z. [DOI] [PubMed] [Google Scholar]
- Fillgrove K. L.; Pakhomova S.; Schaab M. R.; Newcomer M. E.; Armstrong R. N. Structure and mechanism of the genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes. Biochemistry 2007, 46, 8110–8120. 10.1021/bi700625p. [DOI] [PubMed] [Google Scholar]
- Lomonaco S.; Nucera D.; Filipello V. The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect., Genet. Evol. 2015, 35, 172–183. 10.1016/j.meegid.2015.08.008. [DOI] [PubMed] [Google Scholar]
- Ito R.; Tomich A. D.; McElheny C. L.; Mettus R. T.; Sluis-Cremer N.; Doi Y. Inhibition of Fosfomycin Resistance Protein FosA by Phosphonoformate (Foscarnet) in Multidrug-Resistant Gram-Negative Pathogens. Antimicrob. Agents Chemother. 2017, 61, e01424-17 10.1128/AAC.01424-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patridge E.; Gareiss P.; Kinch M. S.; Hoyer D. An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discovery Today 2016, 21, 204–207. 10.1016/j.drudis.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Meng X. Y.; Zhang H. X.; Mezei M.; Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comput.-Aided Drug Des. 2011, 7, 146–157. 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen D. B.; Decornez H.; Furr J. R.; Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discovery 2004, 3, 935–949. 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- Kaushik A. C.; Kumar S.; Wei D. Q.; Sahi S. Structure Based Virtual Screening Studies to Identify Novel Potential Compounds for GPR142 and Their Relative Dynamic Analysis for Study of Type 2 Diabetes. Front. Chem. 2018, 6, 23 10.3389/fchem.2018.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik A. C.; Gautam D.; Nangraj A. S.; Wei D.-Q.; Sahi S. Protection of Primary Dopaminergic Midbrain Neurons Through Impact of Small Molecules Using Virtual Screening of GPR139 Supported by Molecular Dynamic Simulation and Systems Biology. Interdiscip. Sci.: Comput. Life Sci. 2019, 11, 247–257. 10.1007/s12539-019-00334-x. [DOI] [PubMed] [Google Scholar]
- Varghese G. K.; Abraham R.; Chandran N. N.; Habtemariam S. Identification of Lead Molecules in Garcinia mangostana L. Against Pancreatic Cholesterol Esterase Activity: An In Silico Approach. Interdiscip. Sci.: Comput. Life Sci. 2019, 11, 170–179. 10.1007/s12539-017-0252-5. [DOI] [PubMed] [Google Scholar]
- Chinnasamy S.; Selvaraj G.; Selvaraj C.; Kaushik A. C.; Kaliamurthi S.; Khan A.; Singh S. K.; Wei D. Q. Combining in silico and in vitro approaches to identification of potent inhibitor against phospholipase A2 (PLA2). Int. J. Biol. Macromol. 2020, 144, 53–66. 10.1016/j.ijbiomac.2019.12.091. [DOI] [PubMed] [Google Scholar]
- Lionta E.; Spyrou G.; Vassilatis D.; Cournia Z. Structure-Based Virtual Screening for Drug Discovery: Principles, Applications and Recent Advances. Curr. Top. Med. Chem. 2014, 14, 1923–1938. 10.2174/1568026614666140929124445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpino A.; Ferenczy G. G.; Keserű G. M. Comparative Evaluation of Covalent Docking Tools. J. Chem. Inf. Model. 2018, 58, 1441–1458. 10.1021/acs.jcim.8b00228. [DOI] [PubMed] [Google Scholar]
- Bursulaya B. D.; Totrov M.; Abagyan R.; Brooks C. L. 3rd Comparative study of several algorithms for flexible ligand docking. J. Comput.-Aided Mol. Des. 2003, 17, 755–763. 10.1023/B:JCAM.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- Pagadala N. S.; Syed K.; Tuszynski J. Software for molecular docking: a review. Biophys. Rev. 2017, 9, 91–102. 10.1007/s12551-016-0247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P. C.-H.; Abagyan R.; Totrov M. Ligand-biased ensemble receptor docking (LigBEnD): a hybrid ligand/receptor structure-based approach. J. Comput.-Aided Mol. Des. 2018, 32, 187–198. 10.1007/s10822-017-0058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Blais J.; Ron D.; Cardozo T. Structural Determinants of PERK Inhibitor Potency and Selectivity. Chem. Biol. Drug Des. 2010, 76, 480–495. 10.1111/j.1747-0285.2010.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers E. M.; Yan G.; Mukherjee C.; Orry A.; Wang L.; Holbert M. A.; Crump N. T.; Hazzalin C. A.; Liszczak G.; Yuan H.; Larocca C.; Saldanha S. A.; Abagyan R.; Sun Y.; Meyers D. J.; Marmorstein R.; Mahadevan L. C.; Alani R. M.; Cole P. A. Virtual Ligand Screening of the p300/CBP Histone Acetyltransferase: Identification of a Selective Small Molecule Inhibitor. Chem. Biol. 2010, 17, 471–482. 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. J.; Leung K. H.; Chan D. S. H.; Wang Y. T.; Ma D. L.; Leung C. H. Identification of a natural product-like STAT3 dimerization inhibitor by structure-based virtual screening. Cell Death Dis. 2014, 5, e1293 10.1038/cddis.2014.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S.; Matsunaga T.; Kuwata K.; Zhao H.-T.; El-Kabbani O.; Kitade Y.; Hara A. Chromene-3-carboxamide derivatives discovered from virtual screening as potent inhibitors of the tumour maker, AKR1B10. Bioorg. Med. Chem. 2010, 18, 2485–2490. 10.1016/j.bmc.2010.02.050. [DOI] [PubMed] [Google Scholar]
- Kufareva I.; Abagyan R. Type-II Kinase Inhibitor Docking, Screening, and Profiling Using Modified Structures of Active Kinase States. J. Med. Chem. 2008, 51, 7921–7932. 10.1021/jm8010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves M. A.; Totrov M.; Abagyan R. Docking and scoring with ICM: the benchmarking results and strategies for improvement. J. Comput.-Aided Mol. Des. 2012, 26, 675–686. 10.1007/s10822-012-9547-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Chen J.; Cheng T.; Gindulyte A.; He J.; He S.; Li Q.; Shoemaker B. A.; Thiessen P. A.; Yu B.; Zaslavsky L.; Zhang J.; Bolton E. E. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernin A.; Dubois-Brissonnet F.; Roux S.; Masson M.; Bosc V.; Maillard M.-N. Phenolic compounds can delay the oxidation of polyunsaturated fatty acids and the growth of Listeria monocytogenes: structure-activity relationships. J. Sci. Food Agric. 2018, 98, 5401–5408. 10.1002/jsfa.9082. [DOI] [PubMed] [Google Scholar]
- Li G.; Wang X.; Xu Y.; Zhang B.; Xia X. Antimicrobial effect and mode of action of chlorogenic acid on Staphylococcus aureus. Eur. Food Res. Technol. 2014, 238, 589–596. 10.1007/s00217-013-2140-5. [DOI] [Google Scholar]
- Shen X.; Sun X.; Xie Q.; Liu H.; Zhao Y.; Pan Y.; Hwang C.-A.; Wu V. C. H. Antimicrobial effect of blueberry (Vaccinium corymbosum L.) extracts against the growth of Listeria monocytogenes and Salmonella enteritidis. Food Control 2014, 35, 159–165. 10.1016/j.foodcont.2013.06.040. [DOI] [Google Scholar]
- Lou Z.; Wang H.; Zhu S.; Ma C.; Wang Z. Antibacterial Activity and Mechanism of Action of Chlorogenic Acid. J. Food Sci. 2011, 76, M398–M403. 10.1111/j.1750-3841.2011.02213.x. [DOI] [PubMed] [Google Scholar]
- Luís Â.; Silva F.; Sousa S.; Duarte A. P.; Domingues F. Antistaphylococcal and biofilm inhibitory activities of gallic, caffeic, and chlorogenic acids. Biofouling 2014, 30, 69–79. 10.1080/08927014.2013.845878. [DOI] [PubMed] [Google Scholar]
- Naveed M.; Hejazi V.; Abbas M.; Kamboh A. A.; Khan G. J.; Shumzaid M.; Ahmad F.; Babazadeh D.; FangFang X.; Modarresi-Ghazani F.; WenHua L.; XiaoHui Z. Chlorogenic acid (CGA): A pharmacological review and call for further research. Biomed. Pharmacother. 2018, 97, 67–74. 10.1016/j.biopha.2017.10.064. [DOI] [PubMed] [Google Scholar]
- Wen A.; Delaquis P.; Stanich K.; Toivonen P. Antilisterial activity of selected phenolic acids. Food Microbiol. 2003, 20, 305–311. 10.1016/S0740-0020(02)00135-1. [DOI] [Google Scholar]
- Vaquero M. J. R.; Alberto M. R.; de Nadra M. C. M. Influence of phenolic compounds from wines on the growth of Listeria monocytogenes. Food Control 2007, 18, 587–593. 10.1016/j.foodcont.2006.02.005. [DOI] [Google Scholar]
- Stojković D.; Petrović J.; Soković M.; Glamočlija J.; Kukić-Marković J.; Petrović S. In situ antioxidant and antimicrobial activities of naturally occurring caffeic acid, p-coumaric acid and rutin, using food systems. J. Sci. Food Agric. 2013, 93, 3205–3208. 10.1002/jsfa.6156. [DOI] [PubMed] [Google Scholar]
- Perumal S.; Mahmud R.; Ismail S. Mechanism of Action of Isolated Caffeic Acid and Epicatechin 3-gallate from Euphorbia hirta against Pseudomonas aeruginosa. Pharmacogn. Mag. 2017, 13, 311–315. 10.4103/pm.pm_309_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice R.Chlorogenic Acid and Caffeic Acid Review of Toxicological Literature; 1998.
- Zhu T.; Cao S.; Su P. C.; Patel R.; Shah D.; Chokshi H. B.; Szukala R.; Johnson M. E.; Hevener K. E. Hit identification and optimization in virtual screening: practical recommendations based on a critical literature analysis. J. Med. Chem. 2013, 56, 6560–6572. 10.1021/jm301916b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balouiri M.; Sadiki M.; Ibnsouda S. K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.