ABSTRACT
Extracellular vesicles (EV) are increasingly being recognized as important vehicles of intercellular communication and promising diagnostic and prognostic biomarkers in cancer. Despite this enormous clinical potential, the plethora of methods to separate EV from biofluids, providing material of highly variable purity, and lacking knowledge regarding methodological repeatability pose a barrier to clinical translation. Urine is considered an ideal proximal fluid for the study of EV in urological cancers due to its direct contact with the urogenital system. We demonstrate that density-based fractionation of urine by bottom-up Optiprep density gradient centrifugation separates EV and soluble proteins with high specificity and repeatability. Mass spectrometry-based proteomic analysis of urinary EV (uEV) in men with benign and malignant prostate disease allowed us to significantly expand the known human uEV proteome with high specificity and identifies a unique biological profile in prostate cancer not uncovered by the analysis of soluble proteins. In addition, profiling the proteome of EV separated from prostate tumour conditioned medium and matched uEV confirms the specificity of the identified uEV proteome for prostate cancer. Finally, a comparative proteomic analysis with uEV from patients with bladder and renal cancer provided additional evidence of the selective enrichment of protein signatures in uEV reflecting their respective cancer tissues of origin. In conclusion, this study identifies hundreds of previously undetected proteins in uEV of prostate cancer patients and provides a powerful toolbox to map uEV content and contaminants ultimately allowing biomarker discovery in urological cancers.
KEYWORDS: Extracellular vesicles, exosomes, separation, isolation, urine, cancer, biomarkers, proteomics
Introduction
Prostate cancer is the most commonly diagnosed male malignancy in developed countries and the third leading cause of cancer-related mortality [1,2]. An important clinical problem in prostate cancer is the inability of current diagnostic tests to discriminate between indolent and aggressive cancers [3], leading to overdiagnosis and overtreatment [4]. As a result, there is an intense interest in finding biomarkers that improve clinical decision-making in prostate cancer. Despite the high clinical need, only few have been accepted for diagnosis or follow-up in guidelines or routine clinical practice and uncertainty regarding reliability, clinical utility and cost-effectiveness hinder their widespread use [5].
Although still in its infancy compared to the study of circulating tumour cells (CTC) and DNA (ctDNA), the biological role and clinical potential of extracellular vesicles (EV) in cancer have been the focus of increasing scientific interest. Cancer initiation and progression are dependent on the ability of cells to communicate with their local and distant environment through secretory products such as EV, a heterogeneous group of bi-layered, nanometre-sized membrane vesicles, released into the extracellular space by multiple cell types [6]. They are composed of distinct repertoires of proteins, nucleic acids, lipids and metabolites, and their cargo is controlled by specific molecular sorting machineries [7]. In addition to their availability in multiple body fluids, several aspects make them appealing from a biomarker perspective. Their cargo is a spatiotemporal fingerprint of the cell of origin and a reflection of the pathophysiological processes occurring within the source tissue. Moreover, due to their multidimensional cargo, multiple options for biomarker detection exist within the same entity [5]. However, translating EV content into cancer biomarkers remains challenging due to the difficulty of obtaining high-purity EV from complex biological matrices [8]. Urine directly reflects changes in the pathophysiological status of the urogenital system due to its exposure to renal and urothelial cells and prostatic secretions, making it an attractive biofluid for cancer biomarker discovery. However, protein abundance in this matrix spans over five orders of magnitude, with the 20 most abundant proteins, such as high molecular-weight Tamm-Horsfall Protein (THP/UMOD) polymers and albumin, accounting for 50% of the total protein mass [9]. These proteins are co-isolated as a contaminant in urinary EV (uEV) preparations and dominate liquid chromatography–mass spectrometry (LC-MS/MS) analysis, limiting detection of low-abundance uEV proteins. In addition, THP entraps non-EV associated extracellular RNA molecules [10]. Therefore, appropriate steps to control the presence of highly abundant soluble proteins in purified uEV preparations is mandatory to minimize confounding influence of non-EV signals [11]. An efficient and repeatable procedure to separate EV from contaminants is a prerequisite to obtain reliable omics data for clinical and research applications. Our recent review of the literature identified 34 distinct EV separation protocols in 131 studies performed on urine, indicating the absence of standardized protocols [5,12]. Although rigour and standardization are the subject of increasing attention in EV research [8,13–16], repeatability of EV separation from biofluids, as well as the content of the biofluid devoid of EV, have been poorly investigated before researching disease-associated EV-biomarkers [13].
This study presents a floatation-based density gradient protocol to separate EV from urine with minimal THP and soluble protein contamination. This approach is highly repeatable and enables the analysis of EV-enriched and protein-enriched fractions in urine from cancer patients. We applied this protocol to clinically relevant urine samples from patients with prostate cancer (prior to and after treatment) and benign prostatic hyperplasia (BPH). Differential quantitative proteomic analysis of EV-enriched and protein-enriched urine fractions, identified the enrichment of unique and biologically and clinically relevant proteomes in uEV. In addition, by profiling matched prostate cancer tissue-derived EV and uEV, we demonstrated that the uEV proteome is a reflection of that of the tissue EV of origin. This work presents a thoroughly substantiated case favouring the study of uEV as a source of unique biological and disease profiles not uncovered by the conventional analysis of crude urine samples.
Materials & methods
Antibodies
The following antibodies were used for immunostaining: anti-Alix (1:1000 (0.10 µg/mL), 2171 S, Cell Signaling Technology, Beverly, MA, USA), anti-TSG101 (1:1000 (0.20 µg/mL), sc-7964, Santa Cruz Biotechnology, Dallas, Texas, USA), anti-CD9 (1:1000 (0.23 µg/mL), D3H4P, Cell Signaling Technology), anti-Flotillin-1 (1:1000 (0.25 µg/mL), 610820, BD Biosciences, Franklin Lakes, NJ, USA), anti-Syntenin-1 (1:1000 (0.11 µg/mL), ab133267, Abcam, Cambridge, UK), anti-THP (1:800 (0.25 µg/mL), sc-20631, Santa Cruz Biotechnology), anti-GM130 (1:500 (0.50 µg/mL), 610822, BD Biosciences), anti-PMP70 (1:2000 (0.50 µg/mL), P0497, Sigma-Aldrich, St. Louis, MO, USA), anti-green fluorescent protein (GFP) (1:1000 (0.10 µg/mL), MAB3580, Merck Millipore, Billerica, MA, USA), anti-PSA/KLK3 (1:1000 (0.11 µg/mL), D11E1, Cell Signaling Technology), anti-Rab27a (1:1000 (0.40 μg/mL), HPA001333, Sigma-Aldrich), anti-FASN (1:1000 (0.40 μg/mL), HPA056108, Sigma-Aldrich), anti-IDH1 (1:1000 (1.0 μg/mL), AMAB90578, Sigma-Aldrich), anti-AQP2 (1:1000 (1.0 μg/mL), PA5-38004, ThermoFisher Scientific, Waltham, MA, USA), anti-PODXL (1:1000 (0.43 µg/mL), ab150358, Abcam), anti-SLC12A1 (1:500 (2.0 μg/mL), SAB1402357, Sigma-Aldrich, anti-UPK1B (1:400 (0.25 μg/mL), HPA031800, Sigma-Aldrich), sheep anti-mouse horseradish peroxidase-linked antibody (1:3000 (0.14 μg/mL), NA931 V, GE Healthcare life sciences, Uppsala, Sweden), donkey anti-rabbit horseradish peroxidase-linked antibody (1:4000 (0.05 μg/mL), NA934V, GE Healthcare life sciences).
Patients and biological sample collection
Matched urine samples were collected from prostate cancer patients prior to (n = 12) and 3 months after local treatment by radical prostatectomy or intensity-modulated radiation therapy (n = 10). From selected prostate cancer patients (PC; n = 3), urine samples prior to radical prostatectomy and resected prostate cancer tissue samples were collected. In addition, urine samples from healthy controls with BPH (n = 12) and patients diagnosed with urothelial cancer of the bladder (UCC; n = 5) or renal cell carcinoma (RCC; n = 6) were collected prior to treatment. For technical experiments, healthy donor urine samples were used. All samples were second morning whole void urine samples from fasting donors. Pre-treatment prostate cancer samples and BPH control samples were collected immediately following digital rectal examination (DRE), which was performed as 3 finger strokes per prostate lobe. Collection of biological samples was according to the Ethical Committee of Ghent University Hospital approval EC/2015/0260 and in accordance with the guidelines and regulations of the Helsinki Declaration. Participants provided written informed consent.
Values for urinary pH, specific gravity (SG), glucose (GLU), bilirubin (BIL), ketones (KET), blood (BLO), protein (PRO), urobilinogen (URO), nitrite (NIT) and leukocytes (LEU) were analysed with a Multistix 10SG strip-test (Siemens Healthcare, Erlangen, Germany), and these were within normal ranges (pH: 6–7.5; SG: 1.010–1.030; GLU: negative; BIL: negative; KET: negative; BLO: 0–10 erythrocytes/µL; PRO: 0–20 mg/dL; URO: 3.2–16 µmol/L; NIT: negative; LEU: negative). Creatinine was measured using the UC-3500 urine chemistry analyser (Sysmex, Kobe, Japan). Urine samples (50 mL) were centrifuged for 10 min at 1000 g and 4°C (with braking) in accordance with the Eurokup/HKUPP Guidelines [17] using an Eppendorf 5810R (Eppendorf, Hamburg, Germany) benchtop centrifuge with A-4-62 swinging bucket rotor. Cell-free urine supernatants were collected (leaving approximately 0.5 cm urine above the cell pellet) and stored for up to 12 months at −80°C until further use. Detailed patient characteristics and clinical data are summarized in Suppl. table 1.
Urine fractionation by bottom-up Optiprep density gradient centrifugation
Cell-free urine samples (50 mL) were thawed at room temperature, vortexed and concentrated to 500–800 µL using a 10 kDa centrifugal filter device (Centricon Plus-70, Merck Millipore). The concentrated sample was resuspended in Tris buffer containing 10 mM Tris-HCl (pH 7.4), 1 mM EDTA and 0.25 M sucrose to obtain a sample volume of 800 µL. Solutions of 5%, 10% and 20% iodixanol were made by mixing appropriate amounts of homogenization buffer (0.25 M sucrose, 1 mM EDTA, 10 mM Tris-HCL (pH 7.4)) and iodixanol working solution. This working solution was prepared by combining a working solution buffer (0.25 M sucrose, 6 mM EDTA, 60 mM Tris-HCl, pH 7.4) and a stock solution of OptiPrep (60% (w/v) aqueous iodixanol solution, Axis-Shield, Oslo, Norway). The 800 µL concentrated urine sample was resuspended in 3.2 mL working solution, obtaining a 40% iodixanol suspension, and layered on the bottom of a 17 mL Thinwall Polypropylene Tube (Beckman Coulter, Fullerton, CA, USA). A discontinuous bottom-up OptiPrep density gradient (ODG) was prepared by overlaying the urine suspension with 4 mL 20%, 4 mL 10% and 3.5 mL 5% iodixanol solutions, and 1 mL PBS, respectively. Gradient layering was performed by a Biomek 4000 pipetting robot using a custom-made script. The ODG was centrifuged for 18 h at 100,000 g (acceleration: max; deceleration: 9) and 4°C (SW 32.1 Ti rotor with ravg = 11.36 cm and adjusted k-factor = 298.0, Beckman Coulter). Afterwards, ODG fractions of 1 mL were collected from the top of the gradient and pooled (1–5, 6–8, 9–10, 11–12, 13–14, 15–16), diluted to 16 mL in PBS in a 17 mL Thinwall Polypropylene Tube (Beckman Coulter) and centrifuged for 3 h at 100,000 g (acceleration: max; deceleration: max) and 4°C (SW 32.1 Ti rotor with ravg = 11.36 cm and adjusted k-factor = 298.0, Beckman Coulter). The resulting pellets were re-suspended in 100 µL PBS and stored at −80°C. For mass spectrometry-based proteomic experiments, pooled EV-rich fractions 9–10, corresponding to a density of 1.087–1.109 g/mL were used for subsequent size exclusion chromatography (SEC)-based separation of EV from iodixanol as an alternative to pelleting by ultracentrifugation, increasing EV recovery [18,19]. To prepare the SEC column, Sepharose CL-2B (GE Healthcare life sciences) was washed three times by sedimentation in PBS buffer. A nylon net with 20 μm pore size (NY2002500, Merck Millipore) was placed on the bottom of a 10 mL syringe (BD Biosciences, San Jose, CA, USA), followed by stacking of 10 mL washed Sepharose. After adding ODG fractions 9–10, individual fractions of 1 mL eluate were collected. EV-containing fractions 4–7 were pooled and concentrated to 100 µL using Amicon Ultra-2 10K filters (Merck Millipore) and stored at −80°C until further use.
Tumour conditioned medium (TCM) generation and TCM EV separation
Resected prostate cancer tissue specimens were transported in PBS buffer and processed within 2 h of surgery. The tumour tissue was manually sliced to pieces of 2–3 mm3 with sterile surgical scissors. These pieces were washed by placing them on a 70 μm cell strainer and gently flushing three times with 5 mL of PBS buffer at 37°C. The pieces were weighed and then added to a well of a 6-well plate, containing 2 mL of PBS buffer with 40% DMEM containing 0.5% EV-depleted serum (EDS). EDS was obtained by centrifuging (acceleration: max; deceleration: 9) foetal bovine serum at 100,000 g (SW 32.1 Ti rotor with ravg = 11.36 cm and adjusted k-factor = 298.0, Beckman Coulter) for 16 h at 4°C followed by 0.2 μm filtration. The plate was incubated for 3 h on a rotating device at 37°C and 5% CO2. After incubation, tissue pieces were recovered and kept for histology to assess tissue viability. Tissue was fixed in 4% buffered formol for 12 h, followed by a wash with PBS and transfer to 70% ethanol, and then embedded in paraffin, sectioned and stained with haematoxylin and eosin. Tissue viability after incubation was defined as the presence of normal and/or malignant glandular and stromal structures in the absence of ischaemic necrosis.
The supernatant, i.e. TCM, was centrifuged 10 min at 200 g and 20 min at 2000 g (with braking) using an Eppendorf 5810R benchtop centrifuge with A-4-62 swinging bucket rotor to remove cells and cell debris. After the final centrifugation step, supernatant was collected and kept at −80°C until EV separation. Only TCM from viable tissue samples was used for subsequent EV separation and analyses.
A combination of SEC and top-down ODG was used to separate EV from TCM [20,21]. After thawing, TCM (2 mL) was placed on top of a SEC column, followed by elution by PBS and collection of individual fractions of 1 mL eluate. EV-containing fractions 4–7 were pooled and concentrated to 1 mL using Amicon Ultra-2 10K filters (Merck Millipore). Subsequently, a discontinuous top-down ODG was used as described previously [22]. Solutions of 5%, 10%, 20% and 40% iodixanol were made by mixing appropriate amounts of homogenization buffer and iodixanol working solution, as described above. The gradient was formed by layering 4 mL of 40%, 4 mL of 20%, 4 mL of 10% and 3.5 mL of 5% solutions on top of each other in a 17 mL Thinwall Polypropylene Tube (Beckman Coulter). Gradient layering was performed by a Biomek 4000 pipetting robot using a custom-made script. The 1 mL concentrated SEC sample was overlaid on top of the gradient, which was then centrifuged for 18 h at 100,000 g (acceleration: max; deceleration: 9) and 4°C (SW 32.1 Ti rotor with ravg = 11.36 cm and adjusted k-factor = 298.0, Beckman Coulter). Gradient fractions of 1 mL were collected starting from the top of the gradient. Fractions 9 and 10, corresponding to a density of 1.087–1.109 g/mL, were pooled and used for subsequent SEC-based separation of EV from the iodixanol polymer, as described above. EV samples were stored at −80°C until further use.
Density measurement
Density of the ODG fractions was assessed using a standard curve of the absorbance values at 340 nm (SpectraMax Paradigm, Molecular Devices, San Jose, CA, USA) of aqueous dilutions of 5%, 10%, 20% and 40% iodixanol solutions (solutions were diluted 1:1 twice). This standard curve was used to determine the density of fractions collected from a control gradient.
Urinary EV separation by differential ultracentrifugation
Differential ultracentrifugation was performed as described previously by Théry et al. [23]. Cell-free urine samples (50 mL) were thawed at room temperature and vortexed before processing. Next, urine was centrifuged for 30 min at 2000 g (with braking) and 4°C in an Eppendorf 5810R benchtop centrifuge with A-4-62 swinging bucket rotor. The supernatant was transferred to a 38.5 mL Thinwall Polypropylene Tube (Beckman Coulter) and subjected to serial ultracentrifugation (acceleration: max; deceleration: max) at 4°C at 12,000 g (SW 32.1 Ti rotor with ravg = 11.36 cm and adjusted k-factor = 2483.31, Beckman Coulter) and 110,000 g (SW 32.1 Ti rotor with ravg = 11.36 cm and adjusted k-factor = 270.91, Beckman Coulter), for 45 min and 2 h, respectively. The crude pellet was resuspended in 16 mL PBS and passed through a 0.22 µm Whatman syringe filter (GE Healthcare life sciences) and EV were pelleted in a 17 mL Thinwall Polypropylene Tube (Beckman Coulter) by centrifugation at 110,000 g (acceleration: max; deceleration: max) and 4°C for 70 min. An additional washing step was performed by repeating the previous resuspension and pelleting step. The final pellet was re-suspended in 100 µL PBS and stored at −80°C until further use.
Urinary EV separation by Exoquick precipitation
Exoquick-TC (System Biosciences, Palo Alto, CA, USA) was used according to the manufacturer’s instructions. Cell-free urine samples (50 mL) were thawed at room temperature, vortexed before being concentrated to 500 µL using a 10 kDa centrifugal filter device (Centricon Plus-70, Merck Millipore) and mixed with 100 µL of Exoquick-TC solution. The sample was incubated overnight at 4°C after which it was spun down twice at 1500 g (with braking) in an Eppendorf 5810R benchtop centrifuge with A-4-62 swinging bucket rotor for 30 and 5 min, respectively. The pellet was resuspended in 100 µL of PBS and stored at −80°C until further use.
Urinary EV separation by size-exclusion chromatography
SEC was performed as described previously [24]. Cell-free urine samples (50 mL) were thawed at room temperature and vortexed followed by concentration to 1 mL using a 10 kDa centrifugal filter device (Centricon Plus-70, Merck Millipore). To prepare the SEC column, Sepharose CL-2B (GE Healthcare life sciences) was washed three times by sedimentation in PBS buffer. A nylon net with 20 μm pore size (NY2002500, Merck Millipore) was placed on the bottom of a 10 mL syringe (BD Biosciences), followed by stacking of 10 mL washed Sepharose. After adding the 1 mL urine sample, followed by elution by PBS, individual fractions of 1 mL eluate were collected until the urine sample was completely eluted. Individual fractions were stored at −80°C until further use.
Urinary EV separation by top-down Optiprep density gradient centrifugation
Cell-free urine samples (50 mL) were thawed at room temperature and vortexed before being concentrated to 1 mL using a 10 kDa centrifugal filter device (Centricon Plus-70, Merck Millipore). The sample was loaded on top of a discontinuous top-down ODG and processed as described above.
Urinary THP depolymerization by DTT treatment
DTT treatment was performed as a modification of a previously described protocol [25]. Cell-free urine samples (50 mL) were thawed at room temperature and vortexed before processing. Urine was transferred to a 38.5 mL Thinwall Polypropylene Tube (Beckman Coulter) and centrifuged (acceleration: max; deceleration: max) at 4°C and 17,000 g (SW 32.1 Ti rotor with ravg = 11.36 cm and adjusted k-factor = 1752.92, Beckman Coulter) for 10 min to pellet THP polymers. The supernatant was kept at 4°C, while the resulting 1.5 mL pellet was incubated with 1 mL of a freshly prepared dithiothreitol solution (50% DTT in aqua distilled) for 10 min at 37°C and vortexed to depolymerize THP (final DTT concentration: 200 mg/mL). The treated pellet and previous supernatant were mixed in a 38.5 mL Thinwall Polypropylene Tube (Beckman Coulter) and centrifuged again for 10 min at 17,000 g (acceleration: max; deceleration: max) and 4°C.
The resulting supernatant was used for uEV separation by BU ODG, as described above.
Nanoparticle tracking analysis
ODG fractions were analysed by Nanoparticle tracking analysis (NTA) using a NanoSight LM10-HS microscope (Malvern Instruments Ltd, Amesbury, UK) equipped with a 405 nm laser. For each sample, three 60 s videos were recorded at camera level 13. For fluorescent NTA measurements (fNTA) a 488 nm laser and an automatic syringe pump system (infusion speed: 20) were used. An additional 500 nm longpass filter was applied and the camera level was increased to 16. Temperature was monitored during recording. Recorded videos were analysed at detection threshold 3 with NTA Software version 3.3 to determine the concentration and size distribution of measured particles with corresponding standard error. A medium viscosity of 0.929 cP was assumed. For optimal measurements, samples were diluted with PBS until particle concentration was within optimal concentration range of the NTA Software (3 × 108−1 × 109). All size distributions determined with NTA correspond to the hydrodynamic diameters of the particles in suspension.
Generation and spiking of recombinant extracellular vesicles (rEV)
GFP-positive rEV were generated as previously described [19] and spiked in urine to validate BU ODG fractionation and calculate recovery efficiencies of UF, SEC, EQ, DUC, TD ODG and BU ODG.
Protein analysis
Protein concentrations were measured using the fluorometric Qubit Protein Assay (ThermoFisher Scientific, Waltham, MA, USA). Sample preparation was done by 1:1 dilution with SDS 0,4%. Protein measurements were performed using the Qubit Fluorometer 3.0 (ThermoFisher Scientific) according to the manufacturer’s instructions.
Protein samples were dissolved in reducing sample buffer (0.5 M Tris-HCl (pH 6.8), 40% glycerol, 9.2% SDS, 3% 2-mercaptoethanol, 0.005% bromophenol blue) and boiled at 95°C for 5 min in a thermomixer at 900 rpm. Gel loading of proteins was based on the same percentage of the final volume of the pellet/fraction. Proteins were separated by SDS-PAGE (SDS-polyacrylamide gel electrophoresis). SDS-PAGE gels were produced by making a 10% acrylamide separating gel solution (33% acrylamide 30% (Bio-rad, Hercules, CA, USA), 25% separating buffer (0.4% SDS, 18.7% Tris, 6 M HCl in aqua distilled (AD)), 41% AD, 1% ammonium persulphate (APS) and 0.1% tetramethylethylenediamine (TEMED)). The separating gel solution was casted between two glass electrophoresis slides and allowed to polymerize for 30 min. To keep the gel from drying out, 70% ethanol was added on top of the gel. After polymerization, ethanol was removed and a stacking gel solution (15% acrylamide 30% (Bio-rad), 25% stacking buffer (0.4% SDS, 6% Tris, 6 M HCl in AD) 1% APS and 0.1% TEMED) was added on top of the separating gel. A comb was placed in between the two glass slides to form the loading slots and the stacking gel was allowed to harden for 30 min. Gels were placed in a Mini-PROTEAN Tetra Vertical Electrophoresis Cell (Bio-Rad). The electrophoresis cell was filled with running buffer (0.3% glycine, 1.44% Tris, 0.1% SDS in AD) and loaded protein samples and Pageruler prestained protein ladder (Thermo Fisher Scientific) were separated by application of an electric field of 150 V for 60 min. SDS-PAGE gels containing separated proteins were placed in a gel holder cassette and submerged (wet blotting system) in a Criterion Blotter Buffer Tank (Bio-rad) filled with blotting buffer (0.3% glycine, 1.44% Tris, 0.00005% SDS, 20% methanol in AD). Proteins were transferred to a nitrocellulose membrane (Bio-rad) by application of an electric field of 100 V for 45 min. Next, using a molecular weight (MW) ladder as a reference, nitrocellulose membranes were cut into horizontal strips according to the MW of the protein of interest (1 protein per strip) and submerged in blocking buffer (5% non-fat milk powder and 0.5% Tween-20 in PBSD−) for 30 min. Blocked membranes were immunostained overnight at 4°C with primary antibodies. Secondary antibodies were added for 60 min at room temperature after extensive washing with blocking buffer. After final washing, chemiluminescence substrate (WesternBright Sirius, Advansta, Menlo Park, CA, USA) was added and imaging was performed using the Proxima 2850 Imager (IsoGen Life Sciences, De Meern, The Netherlands). Quantification of signal intensity was performed using ImageJ software [26].
Alternatively, separated proteins were stained with Coomassie Brilliant Blue R-250 Staining Solution (Bio-Rad) for 60 min and destained overnight at 4°C in a solution composed of 40% methanol and 10% acetic acid.
Transmission electron microscopy
Samples were qualitatively analysed with transmission electron microscopy (TEM). Samples were deposited on a formvar coated grids stabilized with evaporated carbon film and glow discharged before sample application. Neutral uranyl acetate (2% in AD) was used for staining after which grids were coated with 2% methyl cellulose/uranyl acetate (0,4%) solution. These grids were examined using a Tecnai G2 Spirit transmission electron microscope (FEI, Eindhoven, The Netherlands) operated at 100 kV and images were captured with a Quemesa charge-coupled device camera (Olympus Soft Imaging Solutions GMBH, Munster, Germany).
LC-MS/MS
Samples were processed for LC-MS/MS by filter-aided sample preparation (FASP) [27]. Lysates were prepared by mixing samples with SDT-lysis buffer (2% SDS, 500 mM Tris-HCL (pH 7.6), 0.5 M DTT) at a 4:1 sample to buffer ratio and incubated at 95°C for 5 min. After clarification of lysates by centrifugation (16,000 g for 5 min), samples were mixed with 300 μL UA (8 M urea, 0.1 M Tris-HCl (pH 8.5)) in a Microcon YM-10 centrifugal filter device (Merck KGaA, Darmstadt, Germany). Filters were centrifuged twice (14,000 g for 40 min at 20°C) with the addition of 200 µL UA in between. Proteins were alkylated by addition of 100 μL IAA solution (0.05 M iodoacetamide in UA buffer) and incubated for 30 min at room temperature, followed by centrifugation. This was followed twice by addition of 100 µL UA and twice by addition of 100 µL DB buffer (1 M urea, 0.1 M Tris-HCl (pH 8.5), with centrifugation in between. Filter units were transferred to new collection tubes and proteins were resuspended in 40 µL DB with Trypsin/Lys-C mix (Promega, Madison, WI, USA) for overnight proteolytic digestion at 37°C. Digests were collected by addition of 100 µL DB and centrifugation for 15 min at 14,000 g. This step was repeated once. Collected peptides were acidified with 1% trifluoroacetic acid to a pH of 2–3, followed by desalting with C18-StageTips (C18 Empore Disks, 3 M, St. Paul, MN, USA). Desalted peptides were vacuum dried, dissolved in 0.1% formic acid and analysed by LC-MS/MS. Equal amounts of peptides of each sample (500 ng) were loaded on a nanoflow HPLC system (Easy-nLC1000, Thermo Fisher Scientific) coupled to a Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific) equipped with a nano‐electrospray ionization source. The mobile phase consisted of 0.1% formic acid (solvent A) and acetonitrile/water (95:5 (v/v)) with 0.1% formic acid (solvent B). The peptides were separated with a 50-min gradient from 8% to 35% of solvent B. Before the end of the run, the percentage of solvent B was raised to 100% in 5 min and kept there for 5 min. Full MS scan over the mass-to-charge (m/z) range of 300–1750 with a resolution of 120,000, followed by data dependent acquisition with an isolation window of 2.0 m/z and a dynamic exclusion time of 30 s was performed. The top 12 ions were fragmented by higher energy collisional dissociation (HCD) with a normalized collision energy of 27% and scanned over the m/z range of 200–2000 with a resolution of 15,000. After the MS2 scan for each of the top 12 ions had been obtained, a new full mass spectrum scan was acquired and the process repeated until the end of the 60 min run. Three repeated runs per sample were performed. Tandem mass spectra were searched using the MaxQuant software (version 1.5.2.8) against a database containing both reviewed (SwissProt) and unreviewed (TrEMBL) sequences of homo sapiens, including different isoforms, of UniProtKB release 2018_07. Peptide-spectrum-match- and protein-level false discovery rates were set at 0.01. Carbamidomethyl (C), as a fixed modification, and oxidation (M) and acetylation of the protein N-terminus as dynamic modifications were included. A maximum of two missed cleavages was allowed. The LC-MS profiles were aligned, and the identifications were transferred to non-sequenced or non-identified MS features in other LC-MS runs (matching between runs). The protein was determined as detected in the sample if its identification had been derived from at least two unique peptide identifications. Filtering for contaminating proteins, reverse identification and identification by site was used. Label-free quantification (LFQ) was performed using the MaxLFQ algorithm integrated into the MaxQuant software. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium [28] via the PRIDE [29] partner repository with the dataset identifier PXD015289.
Proteomic data analysis
Identified proteins were analysed and visualized using Perseus software version 1.6.2.2 [30]. For comparison between prostate cancer and benign prostate hyperplasia samples, intensities of proteins were normalized to the prostate-specific marker KLK3/PSA [31]. Proteins showing valid values in at least 70% of at least one group were selected. Missing values were imputed from the observed normal distribution of intensities. For selected analyses, intensities were transformed to z-scores. Unsupervised hierarchical clustering heat maps, using 1-Pearson correlation, were generated using the Morpheus tool. Anosim and principle component analysis were performed using Past3 software [32]. Differences of the mean were evaluated by Student’s t-test with permutation-based false discovery rate (FDR) estimation and q-values smaller than 0.05 were considered statistically significant. Volcano plots were generated with S0 set at 0.1. Pathway enrichment analysis was performed using g:Profiler [33] and data were visualized using Cytoscape and the EnrichmentMap pipeline [34], as previously described [35]. The Vesiclepedia database was explored and Venn diagrams were generated using Funrich software version 3.1.3 [36].
Statistical analysis
Data analysis and visualization was performed using GraphPad Prism version 6 (GraphPad Software, San Diego, CA, USA). Data are expressed as median with interquartile range (IQR). Correlations were calculated using the Pearson product-moment (r) or Spearman rank-order (ρ) correlation method, as indicated in the text. P-values smaller than 0.05 were considered statistically significant.
EV-TRACK
We have submitted all relevant data of our experiments to the EV-TRACK knowledgebase (EV-TRACK ID: EV190064) [12].
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015289. We have submitted all relevant data of our experiments to the EV-TRACK knowledgebase (EV-TRACK ID: EV190064). Full-length western blots for Figures 1 and 8 and Suppl. figs. 2, 3, 4 and 7 are provided in Suppl. Appendix 1.
Figure 1.
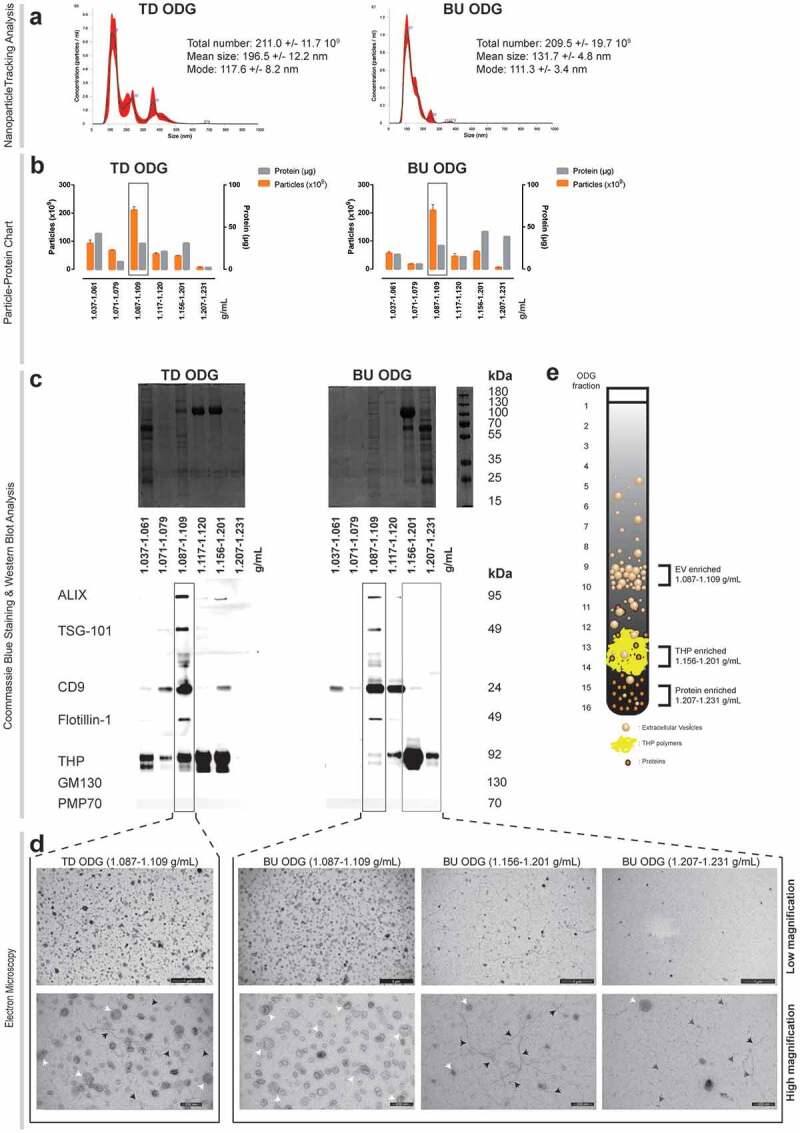
Characterization of urine fractionated by top down (TD) versus bottom up (BU) Optiprep density gradient (ODG) centrifugation. (a) Nanoparticle tracking analysis of EV-enriched fractions. NTA calculated size distributions are depicted as mean (black line) with standard error (red area) and total particle number, mean particle size and mode are shown for each EV-enriched fraction (Fractions 9–10: 1.087–1.109 g/mL). (b) Particle-protein recovery measurements of BU and TD ODG separated urine fractions. Fractions 1–5, 6–8, 9–10, 11–12, 13–14 and 15–16 were pooled and are indicated by their respective density range in the x-axis. EV-enriched fractions are indicated by a black rectangle. (c) Coomassie blue staining and western blot analysis for EV- (ALIX, TSG-101, CD9, Flotillin-1) and non-EV (THP, GM130, PMP70) associated proteins. EV-enriched fractions are indicated by a black rectangle. THP-enriched (Fractions 13–14: 1.156–1.201 g/mL) and protein-enriched (Fractions 15–16: 1.2017–1.231 g/mL) fractions are indicated by a grey rectangle. (d) Transmission electron microscopy of EV, THP and protein-enriched fractions. Low (scale bar: 1000 nm) and high magnification images (scale bar: 200 nm) are provided. White arrows indicate EV, black arrows indicate THP polymers and grey arrows indicate protein complexes. (e) Graphical overview of BU ODG fractionation strategy for urine, resulting in an EV-enriched, THP-enriched and protein-enriched fraction.
Figure 8.
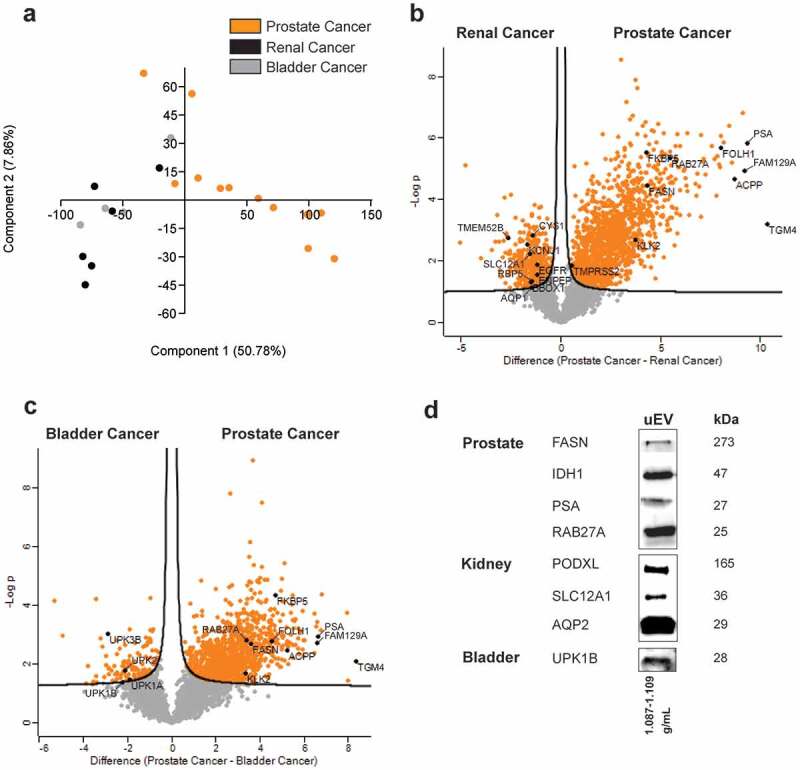
Differences between the urinary EV proteome from men with prostate cancer, renal cancer and bladder cancer. LC-MS/MS data from EV-enriched fractions from prostate, renal and bladder cancer patients are compared by (a) principle component analysis and (b–c) volcano plots. Proteins related to prostate, renal or bladder tissue are highlighted. (d) Western blot validation of selected prostate (cancer)-, kidney- and bladder-enriched proteins in urinary EV.
Results
Differential ultracentrifugation, Exoquick precipitation and size-exclusion chromatography separate EV from urine with low specificity
Mining of the EV-TRACK knowledgebase [12] revealed that urine is a frequently explored biofluid in the search for biomarkers (Suppl. fig. 1a) and that differential ultracentrifugation (DUC), size-exclusion chromatography (SEC) and commercial precipitation-based methods such as Exoquick (EQ) are the most commonly implemented EV separation methods to achieve this (Suppl. fig. 1b). Considering the unique physicochemical properties of urine, we compared the performance of these methods to separate uEV using complementary particle- and protein-based measurement methods. A single pool of cell-free urine from healthy donors was subjected to DUC, EQ and SEC (Suppl. fig. 2). Although all methods recovered particles within the size range of EV, as measured by nanoparticle tracking analysis (NTA) (Suppl fig. 2a), the enrichment of particles relative to co-isolated soluble proteins differed (Suppl. fig. 2b). Western blot analysis revealed that uEV samples obtained by DUC and EQ contain EV-associated proteins ALIX and CD9, while Flotillin-1 was barely detectable. All uEV preparations were clear from contaminating cell organelles as indicated by the absence of PMP70 and GM130. Coomassie blue staining demonstrated co-separation of urinary high-abundance proteins in EV separated by EQ, as indicated by the presence of distinct protein bands for Tamm-Horsfall Protein (THP; 105 kDa), albumin (67 kDa) and IgG (heavy chain: 50 kDa and light chain: 25 kDa), while EV separated from urine by DUC mainly contained contaminating THP. This was confirmed by western blot analysis (Suppl. fig. 2c). An additional washing step in the DUC protocol decreased the amount of THP but also resulted in significant loss in particles and EV-associated proteins ALIX and CD9 (Suppl. fig. 2b,c), as observed previously [37]. Although SEC separated uEV, characterised by ALIX, flotillin-1 and CD9, from the bulk of protein, THP polymers remained present as shown by western blot and electron microscopy analysis (Suppl. fig. 2c,d). In summary, the most commonly implemented separation methods are suboptimal to separate uEV with high specificity.
Bottom-up density gradient centrifugation separates EV from urine with high specificity
We explored density as an alternative dimension of physical particle separation. Urinary ultrafiltrates (UF) were submitted to a discontinuous iodixanol (Optiprep) density gradient (ODG), either aiming for top-down (TD) sedimentation or bottom-up (BU) floatation of uEV to equilibrium density. ODG fractions with distinct physical and biochemical characteristics were identified (Figure 1(a–e)). Particles within the size range of EV (Figure 1(a)) and a density of 1.087–1.109 g/mL (Figure 1(b)) were separated from soluble proteins by both TD and BU ODG (Figure 1(c–d)). Coomassie blue staining identified a distinct protein pattern in these fractions compared to other ODG fractions. Independently of a TD or BU approach, ODG fractions of 1.087–1.109 g/mL were positive for ALIX, TSG101, Flotillin-1 and CD9 (Figure 1(c)) and are therefore hereafter referred to as EV-enriched fractions. All urine fractions were free from contaminating cell organelle markers PMP70 and GM130. In addition, Coomassie blue staining demonstrated separation of EV-enriched proteins from major urinary contaminants, as indicated by the absence of distinct protein bands for THP (105 kDa), albumin (67 kDa) and IgG (heavy chain: 50 kDa and light chain: 25 kDa) in EV-enriched fractions (Figure 1(c)). The TD ODG EV-enriched fraction contained 17.58% of the total contaminating THP protein, based on western blot signal intensity, which was reduced to 2.49% using BU ODG. TEM identified extensive numbers of vesicular structures with a size between 30 and 150 nm, characteristic of EV, in EV-enriched fractions. In addition, TEM analysis confirmed that EV-enriched fractions obtained by TD ODG were contaminated with polymeric networks of THP (Figure 1(d)). In contrast, BU ODG resulted in EV-enriched fractions (1.087–1.109 g/mL) that were efficiently separated from contaminating THP complexes (1.156–1.201 g/mL, hereafter referred to as THP-enriched fraction) and other soluble proteins (1.207–1.231 g/mL, hereafter referred to as protein-enriched fraction) (Figure 1(c,e)).
As described previously [25], treatment of urine with a reducing agent (DTT) released residual uEV from the THP meshwork (1.156–1.201 g/mL) in which they were observed to be trapped, but this neither improved yield nor purity of uEV separated by BU ODG (Figure 1(d); Suppl. fig. 3). Finally, spiking spot urine with GFP-positive recombinant EV (rEV) validated the BU ODG workflow, as demonstrated by the enrichment of rEV in 1.087–1.109 g/mL fractions (Suppl fig. 4a-b). EV recovery efficiency of BU ODG was compared to that of other uEV separation (DUC, EQ, SEC and TD ODG) methods and uEV pre-enrichment by ultrafiltration (UF) alone. This was defined as the number of rEV detected by fNTA after separation divided by the number of rEV spiked (4 × 1010). Recovery efficiency of UF was nearly 60%, while efficiencies of UF followed by SEC, EQ and ODG to separate EV from urine were, respectively, nearly 60%, 30% and 30%. Recovery of EV from EV-enriched ODG density fractions 1.087–1.109 g/mL by a 100,000 g pelleting step instead of SEC was halved from 30% to 15%. DUC recovered only 5% of EV from urine. This was further reduced to 1% after addition of a washing step (Suppl fig. 4c).
In summary, BU ODG allowed the separation of urine into three fractions of interest for biomarker discovery: EV-enriched (1.087–1.109 g/mL), THP-enriched (1.156–1.201 g/mL) and protein-enriched (1.207–1.231 g/mL) (Figure 1(e)).
Bottom-up density gradient separation of urine reveals biologically relevant proteomes with high repeatability
Technical replicates (n = 6) derived from a pool of cell-free urine collected from prostate cancer patients post-DRE were subjected to BU ODG separation and EV-enriched, THP-enriched and protein-enriched fractions (Figure 1(e)) were analysed by mass spectrometry-based proteomics (LC-MS/MS). Correlation analysis based on LFQ intensities revealed high similarity within these fractions with median Pearson’s r coefficients for EV-enriched of 0.97; for THP-enriched of 0.93; and for protein-enriched of 0.93. Significant differences were observed between EV-enriched and both THP- and protein-enriched fractions with median Pearson’s r coefficients down to 0.51 and −0.08, respectively (Figure 2(a)). These observations were confirmed by anosim analysis (R = 1, p = 0.0001) (Figure 2(b)).
Figure 2.
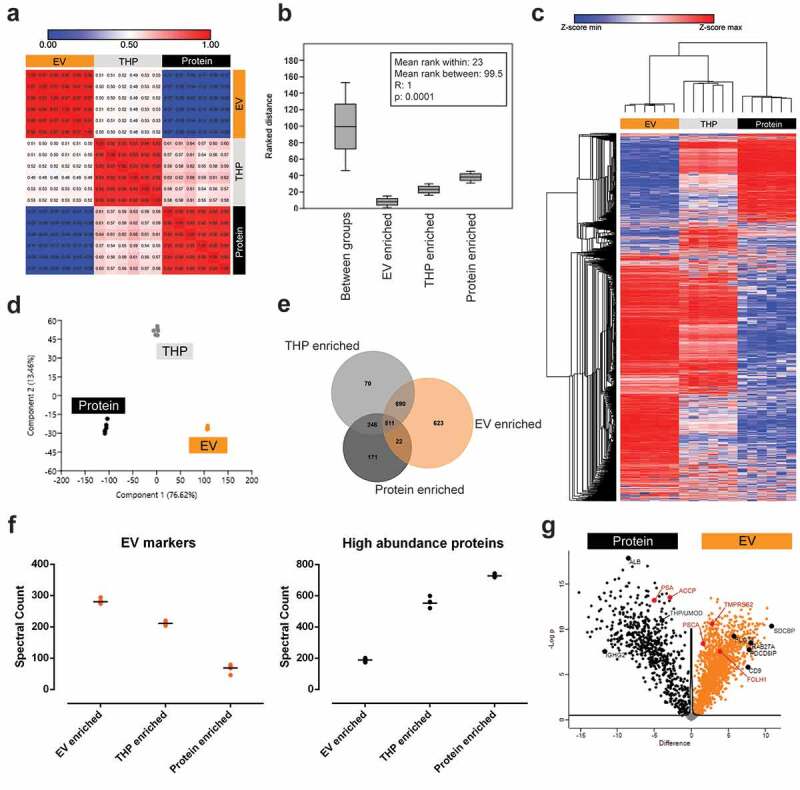
Technical evaluation of BU ODG fractionation of urine by mass spectrometry-based proteomic analysis (LC-MS/MS). LC-MS/MS data from EV-, THP- and protein-enriched fractions are compared by (a) correlation analysis, (b) anosim analysis, (c) hierarchical clustering, (d) principle component analysis, (e) venn diagram, (f) spectral counting of EV-associated proteins (CD9, CD63, CD81, ALIX (PDCD6IP), TSG101, FLOT1, SDCBP, EZR, MSN, ANXA1, ANXA2) and urinary high-abundance proteins (THP, ALB, IGHA1, IGHG1-4, IGHM, TF, HP, A2M, FGA, ORM, complement factors and apolipoproteins) and (g) volcano plot analysis. In (a), correlation is represented as Pearson’s r coefficient. In (g), exemplary proteins of interest are highlighted in black and prostate-specific markers in red.
Technical and methodological repeatability were assessed by unsupervised hierarchical clustering and principal component analysis (PCA). Both analyses showed differential clustering of the three fractions of interest with high similarity of the technical replicates within each cluster (Figure 2(c,d)). With a median coefficient of variation of 0.008 (IQR: 0.005–0.024), 0.019 (IQR: 0.009–0.033) and 0.028 (IQR: 0.013–0.040) for the EV-enriched, THP-enriched and protein-enriched fractions, BU ODG yielded highly repeatable urinary proteomes.
LC-MS/MS repeatedly identified 2333 unique proteins over the three BU ODG fractions of interest (Figure 2(e); Suppl. table 2), of which only 533 (22.86%) were shared between EV and protein-enriched fractions. Of all the identified proteins, 623 (26.7%) were exclusive to the EV-enriched fraction, and this fraction contained 1313 (56.30%) proteins not identified in the protein-enriched fraction. Conversely, 171 (7.33%) unique proteins were identified in the protein-enriched fraction and this fraction contained 417 (17.88%) proteins not identified in the EV-enriched fraction. The THP-enriched fraction contained almost no unique proteins (70; 3.00%) indicating that this is a transition fraction between EV-enriched and protein-enriched fractions, as demonstrated by plotting spectral counts for EV-associated proteins and urinary high abundance proteins for the three fraction types (Figure 2(f)).
Volcano plot analysis and Z-score analysis showed a differential protein enrichment between the different BU ODG fractions and visualized the relative distribution of multiple proteins of interest (Figure 2(g); Suppl fig. 5). Differential protein enrichment between EV-enriched and protein-enriched BU ODG fractions was also reflected in the distribution of known prostate cancer markers (EV-enriched: FOLH1, TMPRSS2, PSCA; Protein-enriched: KLK3/PSA, ACPP), hinting towards biological and clinical relevance of different urine fractions.
Next, Gene Ontology (GO) analysis for biological function of differentially enriched proteins was performed. Functions related to EV biogenesis, protein and nucleic acid metabolism, signalling and cytoskeleton homeostasis were enriched in the EV-enriched fractions. In contrast, cell adhesion, proteolysis, innate immune response, platelet degranulation and extracellular matrix organization pathways were enriched in protein-enriched fractions (Figure 3). These differences were also reflected in the cellular location of these proteins, with EV-enriched proteins mainly being derived from cellular membranes and the cytosol and the protein-enriched fraction consisting of secreted and extracellular matrix proteins (Suppl. fig. 6).
Figure 3.
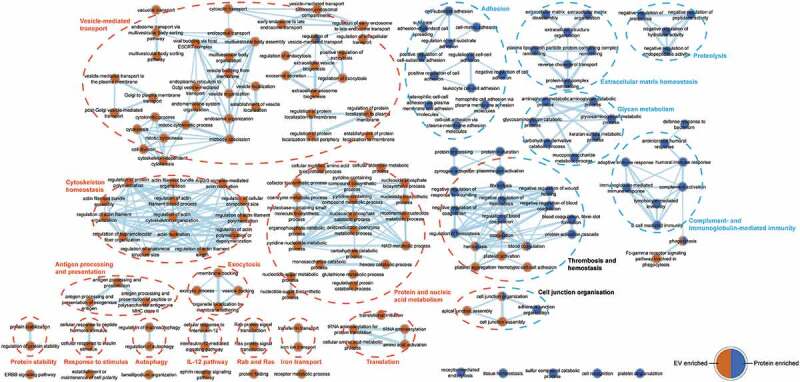
Network map of biological functions enriched among proteins differentially detected in EV-enriched and protein-enriched fractions. Overrepresented gene ontology (GO) biological process (BP) terms among EV- and protein-enriched fractions are represented by orange and blue nodes, respectively. Line thickness represents the number of shared proteins between categories. Identified pathways are clustered by a dotted line; and EV- and protein-enriched fractions are indicated by orange and blue headers, respectively, while shared pathways are indicated by black headers.
In summary, uEV have a unique proteomic composition that is biologically distinct from other biological components present in the biofluid.
Urine from prostate cancer patients is enriched in prostate cancer tissue-specific EV
Matched urine and radical prostatectomy tissue samples from prostate cancer patients were collected, followed by uEV and TCM EV separation. Viability and structure of the prostate cancer tissue after TCM collection was confirmed by histological analysis (Suppl. fig. 7a). Implementation of complementary NTA, TEM and western blot analysis confirmed the separation of TCM EV positive for ALIX, Flotillin-1, CD9 and Syntenin-1 from GM130 and PMP70 contaminants (Suppl. fig. 7b-d). Matched uEV and TCM EV (n = 3) were characterized by mass spectrometry-based proteomics (LC-MS/MS). The majority of proteins identified in TCM EV (Suppl. table 3) was also present in matched uEV (1303/1376; 94.69%), indicating that urine is a suitable biofluid for studying clinical and biological aspects of prostate cancer-derived EV. In addition, enrichment maps of the overlapping proteome between uEV and TCM EV (Suppl. fig. 8–9) independently validated the biological functions and cellular locations identified to be enriched in uEV (Figure 3; Suppl. Fig. 5). While PCA, hierarchical clustering and volcano plot analysis of the whole proteome of both uEV and TCM EV showed differential clustering, individual protein analysis revealed proteins of bladder (UPK1B, UPK2) and kidney (PODXL, AQP2, SLC12A1, etc.) origin in uEV, whereas TCM EV and uEV were equally enriched in prostate-specific markers (KLK2, KLK3/PSA, FOLH1, etc.) (Suppl. fig. 10).
EV separated from urine can be interrogated for prostate cancer markers
Urine from men with prostate cancer prior to (n = 12) and after local treatment (n = 10), and urine from healthy controls with BPH (n = 12), was fractionated in EV- and protein-enriched fractions, which were analysed by LC-MS/MS. Patient and tumour characteristics are summarized in Table 1.
Table 1.
Patient and tumour characteristics. Abbreviations: PSA: prostate-specific antigen, cc: clear cell renal cell carcinoma, pRCC: papillary renal cell carcinoma, chRCC: chromophobe renal cell carcinoma.
| Prostate cancer (n = 12) | Benign prostatic hyperplasia (n = 12) | Bladder cancer (n = 5) | Renal cancer (n = 6) | |
|---|---|---|---|---|
| Age (years) Median (range) |
70.5 (54–78) |
64.5 (51–78) |
69 (65–81) |
61 (50–70) |
| Baseline PSA (ng/mL) Median (range) |
8.9 (4.2–13.9) |
2.8 (1.0–10.4) |
||
| Post-treatment PSA (ng/mL) Median (range) |
0 (0–2.09) |
|||
| Gleason score, n (%) ≤6 7 ≥8 |
3 (25.0%) 5 (41.7%) 4 (33.3%) |
|||
| Tumour stage, n (%) Ta T1 T2 T3a T3b T4 |
– 0 (0%) 7 (58.3%) 3 (25.0%) 2 (16.7%) 0 (0%) |
1 (20%) 1 (20%) 2 (40%) 1 (20%) 0 (0%) 0 (0%) |
– 4 (66.7%) 0 (0%) 1 (16.7%) 0 (0%) 1 (16.7%) |
|
| Nodal stage, n (%) N0 N1 |
11 (91.7%) 1 (8.3%) |
4 (80%) 1 (20%) |
4 (66.6%) 2 (33.3%) |
|
| Local treatment, n (%) Radical Prostatectomy Radiotherapy |
7 (58.3%) 5 (41.7%) |
|||
| Histological subtype ccRCC pRCC chRCC |
3 (50%) 2 (33.3%) 1 (16.7%) |
In total, 3686 and 1996 proteins were, respectively, detected in EV-enriched (Suppl. table 4) and protein-enriched fractions (Suppl. table 5) of all groups. Of these, 3552 (96.36%) and 1913 (95.84%) proteins were, respectively, identified in urine samples from prostate cancer patients prior to treatment.
Using these data, a group of 684 proteins was identified as putative contaminants of uEV preparations based on their selective identification in protein-enriched fractions only (Suppl. table 6). Pathway enrichment analysis of these contaminating proteins revealed their involvement in non-EV associated biological pathways (Suppl. fig. 11), independently validating our previous observations (Figure 3; Suppl. Fig. 5).
Overall, 1134 proteins, including several prostate cancer markers (e.g. KLK3/PSA, FOLH1, TMPRSS2), were detected and quantified in uEV from all 12 prostate cancer patients (Figure 4(a)), while the number of proteins detected in corresponding protein-enriched fractions was lower (489 proteins) (Figure 4(b)). Coverage of membranous prostate cancer markers (e.g. FOLH1, PSCA) was higher in uEV samples, while secreted prostate cancer markers (e.g. KLK3/PSA, MSMB) were more abundant in protein-enriched samples. Proteins were separated into quartiles based on their median abundance (Figure 4(c,d)). As described previously in prostate cancer tissue analyses [38,39], low-abundance proteins were observed in a lower number of patients. Several proteins encoded by prostate cancer driver genes [40–42], including HRAS, AKT1, CUL3, NKX3-1 and PTEN, were detected exclusively in uEV of multiple prostate cancer patients, while others, such as CDH1 and SPINK1, were detected exclusively in protein-enriched fractions. Prostate cancer drivers shared by both fractions, like KRAS, IDH1 and GSTP1, had a generally lower abundance in protein-enriched fractions or were identified in a smaller number of patients (Figure 4(c,d)). As expected, protein products from nuclear transcription factors such as ETS family members, or DNA repair genes (e.g. ATM, BRCA1/2) could not be detected in either urinary fraction.
Figure 4.
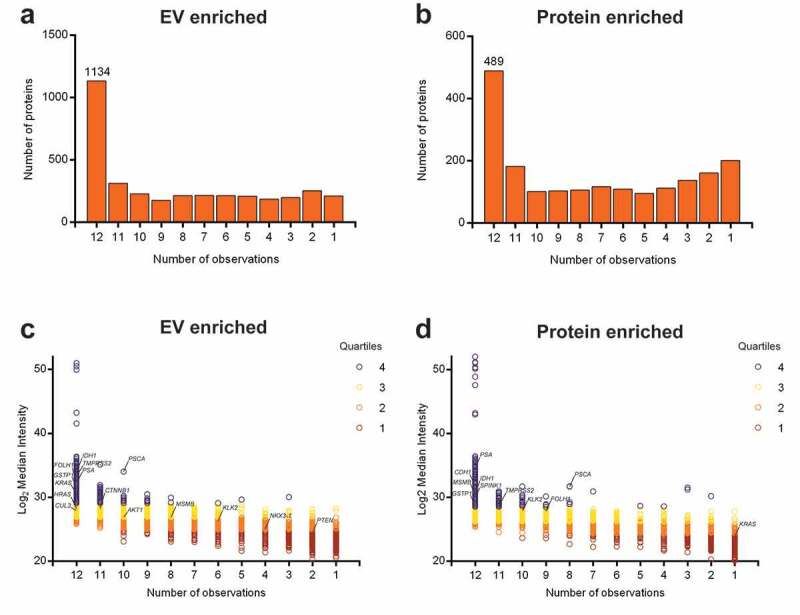
Quantitative protein distribution in EV-enriched and protein-enriched urine fractions from prostate cancer patients. The bar chart shows total counts of proteins detected in a certain number of patient samples for: (a) EV-enriched fractions and (b) protein-enriched fractions. The number on the first bar represents the number of proteins detected in all 12 samples. Protein distribution is measured as Log2 median intensity per protein to the number of patient samples in which it is detected for: (c) EV-enriched fractions and (d) protein-enriched fractions. Colours represent protein abundance in quartiles. Prostate cancer-related proteins are highlighted.
The composition of the EV-enriched (Figure 5(a)) and protein-enriched (Figure 6(a)) urinary proteome changed significantly after local prostate cancer treatment, with a significant reduction in prostate (cancer)-specific proteins (e.g. KLK2, KLK3/PSA, FOLH1, MSMB, ACPP, TGM4, NDRG1, NKX3-1) and androgen-regulated genes (e.g. FKBP5, FAM129A, RAB27A, FASN, NEFH) [43,44] after prostatectomy or radiation (Figures 5(b) and 6(b)). Conversely, post-treatment samples were significantly enriched in EV containing proteins from bladder (e.g. UPK1B, UPK3B) and kidney origin (e.g. PODXL, AQP2, CA4, SLC12A1, SLC12A3, GGT1, KCNJ1, NPHS1) (Figure 5(b)) [45]. In order to identify cancer-specific protein signatures, EV-enriched (Figure 5(c)) and protein-enriched (Figure 6(c)) urine samples, fractionated from men diagnosed with benign prostate hyperplasia (n = 12) were compared with those from prostate cancer patients (n = 12). A comparative LC-MS/MS analysis identified 705 differentially enriched proteins in uEV fractions (Figure 5(d)) and 100 in protein-enriched fractions (Figure 6(d)) which were also more abundant in pre-treatment compared to post-treatment samples. Biological pathway analysis of uEV-enriched proteins in prostate cancer (Figure 7; Suppl. table 7) demonstrated involvement in cancer-associated biological processes such as protein and nucleic acid biosynthesis, autophagy and macromolecule turnover and immune system activation [46]. In contrast, no additional information on prostate cancer biology could be withheld from differentially enriched proteins in protein-enriched fractions (Suppl. fig 12; Suppl. table 8).
Figure 5.
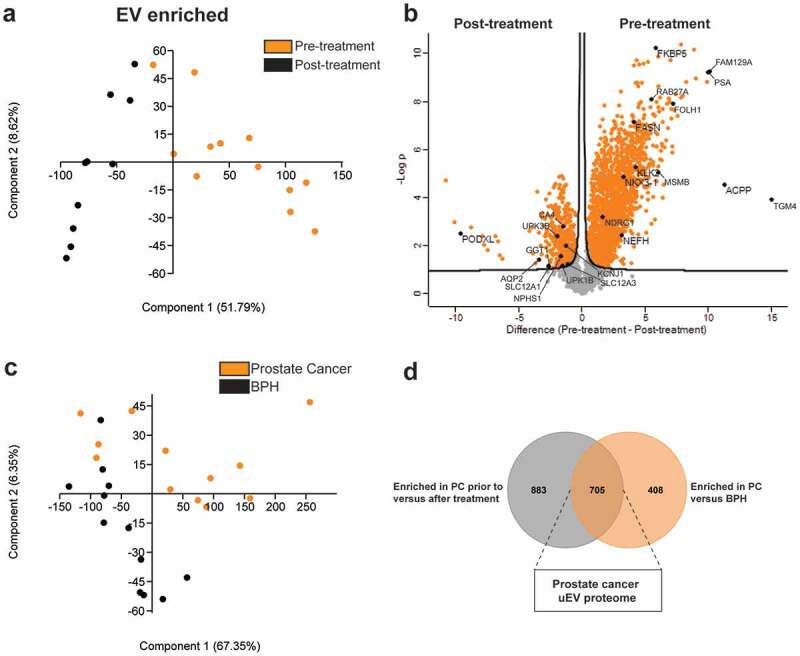
Mapping of the prostate cancer proteome in EV-enriched fractions. LC-MS/MS data from EV-enriched fractions from prostate cancer patients pre- and post-treatment are compared by (a) principle component analysis and (b) volcano plot analysis. Proteins related to prostate (pre-treatment) and bladder and kidney (post-treatment) tissue are highlighted. (c) LC-MS/MS data from EV-enriched fractions from patients with benign prostatic hyperplasia (BPH) and prostate cancer (PC) as compared by principle component analysis. (d) Venn diagram displaying EV-enriched proteins enriched in prostate cancer EV prior to treatment versus after treatment and versus benign prostatic hyperplasia.
Figure 6.
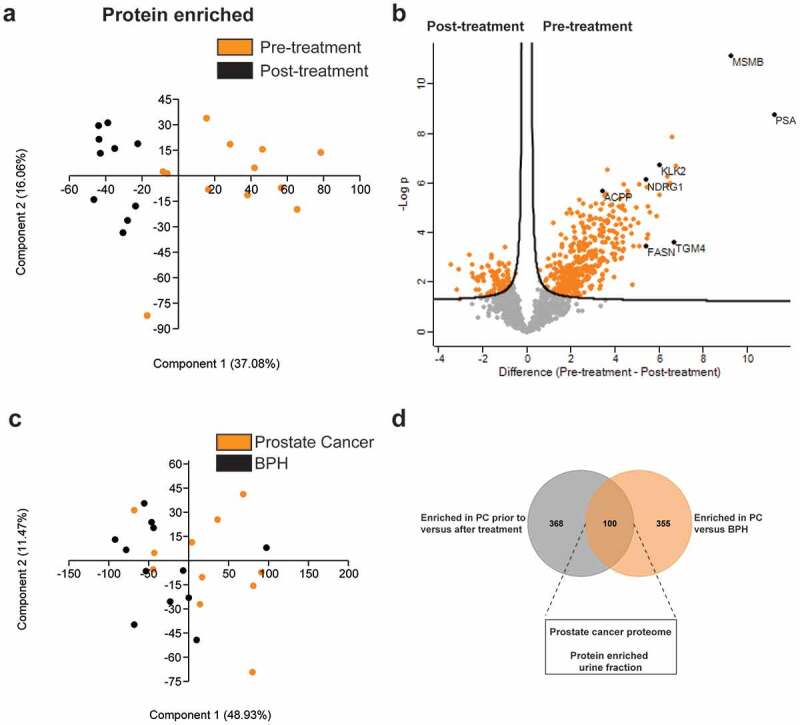
Mapping of the prostate cancer proteome in protein-enriched fractions. LC-MS/MS data from protein-enriched fractions from prostate cancer patients pre- and post-treatment are compared by (a) principle component analysis and (b) volcano plot analysis. Proteins related to prostate (pre-treatment) tissue are highlighted. (c) LC-MS/MS data from protein-enriched fractions from patients with benign prostatic hyperplasia (BPH) and prostate cancer as compared by principle component analysis. (d) Venn diagram displaying proteins enriched in protein-enriched urine fractions in prostate cancer patients prior to treatment versus after treatment and versus patients with benign prostatic hyperplasia (BPH).
Figure 7.
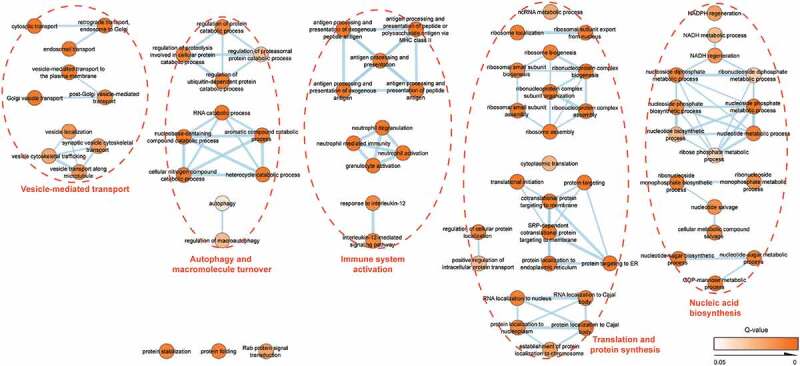
Network map of biological functions enriched in the prostate cancer proteome of EV-enriched fractions. Colour intensity represents increasing statistical significance. Line thickness represents the number of shared proteins between categories. Identified pathways are clustered by a dotted line and indicated by orange headers.
Finally, the uEV proteome of men with prostate cancer was compared to that of patients with other urological cancer types (n = 11), which further confirmed selective enrichment of protein signatures in uEV reflecting their respective cancer tissues of origin (Figure 8(a)) [45]. Specifically, uEV from patients with renal cancer (RCC) were enriched in several proteins overexpressed in RCC (e.g. EGFR, ENPEP, TMEM52B, BBOX1, RBP5, AQP1) compared to other cancers (Figure 8(b)) [45,47–49]. Similarly, bladder cancer uEV were shown to be enriched in UCC-associated proteins (e.g. UPK1A, UPK1B, UPK2, UPK3B) (Figure 8(c)) [45,50]. This could not be observed for protein-enriched fractions (data not shown). The presence of selected prostate- (PSA/KLK3), kidney- (PODXL, SLC12A1, AQP2) and bladder-derived (UPK1B) proteins, as well as biologically relevant proteins involved in prostate cancer pathogenesis (FASN, IDH1, RAB27A) in uEV was independently confirmed by immunoblotting (Figure 8(d)).
Discussion
A repeatable separation method with high specificity is essential to explore EV-specific functions and biomarkers [8,13,22].
We provide a comparative evaluation of the three most commonly implemented methods to separate EV from urine: DUC, SEC and EQ (Suppl. fig. 1). Multiple observations indicate that all three methods separate EV with low specificity due to co-isolation of THP protein complexes. Although well established in the EV research field [12], the tendency of DUC to co-isolate non-EV-associated high-molecular weight proteins and protein complexes [22,51–53], as well as its relatively low recovery efficiency (between 1% and 5% in this study) are widely recognized [19,54–56]. In addition, EV are potentially damaged during the final ultracentrifugation step [57]. SEC-based EV separation has been proposed as a quick and easy method to obtain omics-grade uEV preparations in clinical settings. Although this method separates uEV from the bulk of soluble proteins with a recovery efficiency of nearly 60%, residual contamination with abundant urinary proteins remains (Suppl. fig. 2 and 4c). This is reflected in lower numbers of identified proteins in proteomic studies using SEC-based uEV separation [24,58,59]. Although EQ has a high efficiency to retrieve EV [19], it has been demonstrated that this method co-isolates non-EV associated high-molecular weight proteins and protein complexes that mask the identification of the uEV proteome [22,60].
To overcome the limitations of the most commonly implemented EV separation methods, we introduce density-based fractionation of urine. Our study provides substantial evidence supporting the use of a BU ODG to separate EV, THP protein complexes and soluble proteins with high specificity from urine of healthy donors and patients diagnosed with prostate cancer, bladder cancer and renal cancer for biomarker discovery. Complementary observations confirmed a highly specific fractionation of urine in an EV-enriched fraction, characterized by a density of 1.087 g/mL – 1.109 g/mL containing uEV, and a protein-enriched fraction, characterized by a density of 1.207 g/mL – 1.231 g/mL containing soluble proteins and protein complexes. BU ODG recovered EV from urine with 30% efficiency (Suppl. fig. 4c). EV-enriched fractions obtained by BU ODG show a 16.5-fold, 4.0-fold, 2.1-fold and 1.1-fold enrichment of particle to protein compared to EQ, SEC, DUC and TD ODG (Suppl. fig. 13). Thus, BU ODG separates uEV with the highest specificity compared to the most commonly implemented methods and simultaneously provides the opportunity to explore both EV-enriched fractions and protein-enriched fractions for biomarker discovery (Figure 1(e)).
Indeed, mass spectrometry-based proteomics revealed enrichment of cytosolic and membrane-derived proteins implicated in EV biogenesis, protein and nucleic acid metabolism, signalling and cytoskeleton homeostasis in the EV-enriched fraction, while cell adhesion, proteolysis, innate immune response, platelet degranulation and extracellular matrix organization pathways were enriched in the protein-enriched fraction (Figure 3, Suppl. fig. 5).
Pre-analytical variables in urine processing procedures such as urine collection and storage, and addition of protease inhibitors or chemical agents significantly vary between studies [5]. Although the potential impact of these factors during clinical sample processing on EV studies is increasingly recognized [61], only few have been undertaken using urine [18,62]. For this study, second morning samples from fasting donors were used, omitting urine collection from bladder residues and eliminating dietary interference on urine composition. Bacterial contamination, urinary tract infection, haematuria and proteinuria may affect downstream analysis of uEV [63–65]. Only fresh urine samples with normal values for erythrocytes, protein and nitrite/leukocytes based on strip testing were included in the study. Protease inhibitors were not added, since uEV were demonstrated to be largely resistant to the endogenous proteolytic activity of urine [66]. Clinical urine samples from men with prostate cancer and BPH were collected following DRE, which is generally considered as a critical factor in prostate biomarker studies because it enriches for prostate-derived secretions. This may be especially advantageous for the detection of low-abundance biomarkers. However, omission of DRE in the urine collection process could be beneficial for standardizing collection procedures, removing a certain level of variability introduced by this procedure. Clinical biomarker studies should, therefore, attempt to validate biomarker detection in urine collected without prior DRE, given this does not negatively influence detection sensitivity [67]. The treatment of urine or uEV with the reducing agent dithiothreitol (DTT), zwitterionic detergent CHAPS or urea to denature and remove abundant THP has previously been proposed [68–70]. However, this study demonstrated no significant advantage of DTT treatment on reducing THP contamination or increasing uEV yield when applying a BU ODG. In addition, EV-associated proteins may also be reduced when using DTT, hampering correct folding and functionality, potentially leading to loss of biomarker promise [69]. Finally, normalized analysis of EV in urine is challenging due to variation of uEV concentration because of fluid intake and renal function. It is not clear whether the intrinsic inter- and intrasubject variability of urine is also reflected in uEV [5]. In our study, variations in uEV yield did not correlate well to urinary creatinine levels (Spearman’s ρ = 0.59; p = 0.003) (Suppl. fig. 14a). Instead, we used uEV protein to normalize for uEV yield in proteomics experiments due to its superior correlation to particle count in EV-enriched fractions (Spearman’s ρ = 0.94; p < 0.0001) than urinary creatinine (Suppl. fig. 14b).
Unbiased assessment of the performance of EV separation from urine in terms of technical and methodological repeatability is currently missing in the literature [5,8], impeding the potential utility of EV-derived biomarkers to asses biological processes [16]. The implementation of density-based fractionation requires technical expertise and might be prone to replication bias. Proteomic analysis of EV-enriched and protein-enriched fractions obtained by BU ODG performed in six technical replicates showed a high methodological repeatability (Figure 2). Recent advances in robot-assisted gradient preparation increase the repeatability of density-based fractionation [21]. Furthermore, the use of reference materials such as trackable recombinant EV will assist in mitigating technical variation introduced during sample preparation and analysis [19].
We implemented the BU ODG fractionation strategy to perform the most comprehensive analysis of the uEV proteome in men with benign and malignant prostate disease to date. Overall, we identified a total of 3686 proteins, which represents a more than twofold increase relative to previously published uEV proteomes [24,58,65,71–75], adding, with high specificity, hundreds of proteins to the known human uEV proteome [76] (Table 2 and Suppl. fig. 15). Identified proteins include proteins encoded by prostate cancer driver genes such as NKX3-1 and PTEN, known prostate cancer markers like FOLH1/PSMA and KLK3/PSA and androgen-regulated genes like FKBP5 and FAM129A, among others. A more elaborate comparison of our dataset to previously published ones is complicated due to technical differences in pre-analytical urine collection and processing, EV isolation and LC-MS/MS sample preparation, procedure and bio-informatic data processing.
Table 2.
Overview of previous proteomic studies in urinary EV.
| Study (year) | EV-TRACK ID | Phenotype | Separation method(s) | Protein identifications* | Contaminants (%)*** | EV-METRIC (%) | Reference |
|---|---|---|---|---|---|---|---|
| Smalley et al. (2008) | NA | Healthy control | DUC | 322 | 7 (2.17%) | 22 | [65] |
| Gonzales et al. (2009) | NA | Healthy control; Bartter Syndrome | DUC | 1060 | 40 (3.77%) | 14 | [68,74] |
| Chen et al. (2012) | EV120030 | Healthy control; Bladder Cancer | DUC | 616 | 12 (1.95%) | 33 | [71] |
| Raimondo et al. (2013) | EV130016 | Healthy control; Renal Cancer | DUC | 261 | 10 (3.83%) | 44 | [72] |
| Principe et al. (2013) | EV130224 | Healthy control; Prostate Cancer | DUC | 877 | 139 (15.85%) | 14 | [75] |
| Øverbye et al. (2015) | EV150029 | Benign Prostatic Hyperplasia; Prostate Cancer | DUC | 1664 | NP | 33 | [73] |
| Lozano-Ramos et al. (2015) | EV180054 | Healthy control | UF + SEC | 138 | NP | 14 | [24] |
| Oeyen et al. (2018) | EV170025 | Healthy control | UF + SEC | 423 | 9 (2.13%) | 62 | [58] |
| Vesiclepedia | NA | Multiple | Multiple | 4788** | 274 (5,72%) | NA | [76] |
| Dhondt et al. (2020) | EV190064 | Benign Prostatic Hyperplasia; Prostate Cancer | UF + BU ODG | 3686 | NA | 100 | This study |
* Protein identification based on at least two detected peptides, FDR of 0.01 and availability of UniprotKB identifier
** Known uEV proteome with protein identification based on variable parameters [76]
*** Number of contaminants based on the list of putative contaminants of uEV preparations compiled in this study (Suppl. table 4). This does not include urinary high-abundance proteins co-separated with uEV.
NA: not available/applicable
NP: dataset not public
Next, we implemented multiple approaches to confirm the specificity of the identified uEV proteome for prostate cancer. We separated EV from conditioned medium prepared from prostate cancer tissue (Suppl. fig 7–10) and implemented mass spectrometry-based proteomics to establish the EV proteome of prostate cancer tissue (Suppl. table 3). TCM EV protein signatures were detectable in matched uEV protein signatures hinting towards the identification of prostate cancer-related uEV and demonstrating the suitability of urine as a proximal fluid for studying prostate cancer-derived EV. Differential analysis of the EV proteome from urine samples collected from patients with benign prostate hyperplasia and prostate cancer prior to and after treatment selectively identified cancer-associated biosynthetic and catabolic pathways in uEV obtained from prostate cancer patients prior to treatment (Figure 7). Of note, differential analysis of matched protein-enriched fractions resulted in restricted pathway identification. Several proteins commonly overexpressed in prostate cancer tissue were selectively enriched in uEV from prostate cancer patients prior to treatment versus after treatment and versus men with benign prostate hyperplasia (Suppl. Table 7). These include proteins involved in prostate cancer progression, such as the metalloreductase STEAP2 [77], nuclear export protein XPO1 [78] and citrate metabolic enzyme IDH1 [79]. Enzymes involved in the dysregulated lipid metabolism observed in prostate cancer, like FASN and FABP5 [80,81] were also enriched in uEV. In accordance with our observations, XPO1 and FASN have previously been described to be upregulated in EV separated from prostate cancer cell lines [82]. Other markedly enriched proteins in prostate cancer uEV were Rab GTPases (e.g. Rab7A, Rab27A, Rab27B, etc.), Myosin isoforms (e.g. MYO1E, MYO5A, MYO5C, MYO6, etc.), proteasomal and ribosomal proteins. Many Rab proteins have been shown to be involved in EV biogenesis and are typically identified in EV [7]. Rab27, involved in exocytosis [83], drives invasive growth and metastasis in multiple cancer types [84,85]. Myosins are a family of actin-dependent motor proteins important for cell migration and motility. They are also responsible for cellular EV transport and release [7]. Several myosins, including MYO5A, MYO5C and MYO6, have been found to be overexpressed in prostate cancer and involved in cancer cell migration [86,87]. Finally, using a set of urine samples from patients diagnosed with urological cancers other than prostate cancer, we provided additional evidence of the selective enrichment of protein signatures in uEV reflecting their respective cancer tissues of origin. uEV from patients with renal cell carcinoma were enriched in EGFR, of which overexpression is associated with an unfavourable histologic phenotype [47], ENPEP, involved in renal cell carcinoma angiogenesis [88], and AQP1, a water-channel protein significantly upregulated in clear cell and papillary renal cell carcinoma [49], amongst other proteins, compared to prostate cancer uEV. On the other hand, bladder cancer uEV were enriched in multiple uroplakins (UPK1A, UPK1B, UPK2, UPK3B), transmembrane proteins specifically expressed in urothelium and overexpressed in bladder cancer [50]. Although this small and heterogenous cohort of renal and urothelial cell carcinomas patients was sufficient to demonstrate global differences between uEV of different cancer types, the main research focus of this study was prostate cancer. Therefore, efforts to map the biological role and clinical potential of uEV in other cancer types should be the subject of future research.
While BU ODG fractionation proved to meet necessary standards of reliability and repeatability for EV biomarker discovery, the long turn-around time and low-throughput aspect of a density gradient approach, as well as the need for relatively high sample volumes, limits its use beyond a scientific research setting. We, therefore, propose BU ODG as a standardized method for uEV biomarker discovery, followed by clinical assay development from validated biomarker candidates with clinical value. Microfluidic chip-based technologies, capable of integrated EV separation and analysis, significantly lower the limit of detection, sample volume and analysis time, and might be the key in facilitating the realization of EV-based assays in clinical laboratory and point-of-care settings [5].
Despite the high need, a list of EV contaminants is not yet available to the EV research community [8,13]. BU ODG fractionation applied in this study provides a unique experimental set-up to compile a list of contaminants for uEV based upon differential analysis of EV-enriched fractions versus protein-enriched fractions. We propose a list of 684 putative contaminants of uEV preparations based on their unique identification in the protein-enriched fractions (Suppl. table 6). Of note, abundant proteins such as Tamm-Horsfall Protein (THP/UMOD) and serum albumin (ALB), and several lipoproteins, immunoglobulins and complement factors, are not part of this list of proteins. Despite their abundant enrichment in protein-enriched fractions compared to EV-enriched fractions, their identification is not exclusive to protein-enriched fractions. THP is synthesized by the kidney in the distal convoluted tubules and thick ascending limbs of Henle. Despite its abundance in urine as a high-molecular-weight polymer, monomeric THP is expressed on the luminal surface of kidney epithelial cells as a glycosylphosphatidylinositol (GPI)-linked membrane protein [89]. Therefore, monomeric THP might be a normal constituent of uEV. Alternatively, the co-separation of secretory proteins in EV-enriched fractions might indicate the presence of a protein corona on uEV [8,90], adding an additional layer of complexity towards identifying true uEV content. Although outside the scope of this study, future research on such a potential in vivo EV corona might enhance our understanding of the interaction between EV and soluble factors in biofluids.
In conclusion, we developed a density-based fractionation method for EV separation from urine with high specificity and methodological repeatability. Our approach is applicable to urine samples from multiple clinical conditions. Proteomic analysis of uEV demonstrates that their content mirrors the cytosolic and membrane-bound protein composition of the cells they are derived from. uEV are enriched in multiple prostate cancer markers, oncogenic drivers and androgen-regulated gene products, and provide additional insight into cancer-specific biological processes compared to the soluble urinary proteome. Although disparate uEV protein signatures across several clinical conditions could be identified, this study was not powered for biomarker discovery. Nonetheless, this research represents a thoroughly substantiated case that favours further efforts to map the proteomic content of uEV, separated from prospectively collected clinical samples from well-defined patient cohorts, to generate new candidate biomarkers with potential diagnostic and prognostic value in prostate cancer.
Supplementary Material
Acknowledgments
This work was supported by the Fund for Scientific Spearheads of Ghent University Hospital, Concerted Research Actions from Ghent University, “Stichting tegen Kanker”, the Fund for Scientific Research-Flanders and “Kom op tegen Kanker (Stand up to Cancer), the Flemisch cancer society”. We thank Sofie Degeyter (Laboratory of Experimental Cancer Research, Department of Human Structure and Repair, Ghent University, Ghent, Belgium) for experimental support. We also thank the proteomics and electron microscopy core facilities, supported by biocenter Finland and, respectively, the University of Turku and University of Oulu.
Author Contributions
Author contributions are represented using the Contributor Roles Taxonomy (CRediT) system [91] (Suppl. appendix 2).
Supplementary Material
The supplemental data for this article can be accessed here.
References
- [1].Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–24. [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- [3].Welch HG, Black WC.. Overdiagnosis in cancer. J Natl Cancer Inst. 2010;102:605–613. [DOI] [PubMed] [Google Scholar]
- [4].Carter HB. Prostate cancers in men with low PSA levels–must we find them? N Engl J Med. 2004;350:2292–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dhondt B, Van Deun J, Vermaerke S, et al. Urinary extracellular vesicle biomarkers in urological cancers: from discovery towards clinical implementation. Int J Biochem Cell Biol. 2018;99:236–256. [DOI] [PubMed] [Google Scholar]
- [6].Dhondt B, Rousseau Q, De Wever O, et al. Function of extracellular vesicle-associated miRNAs in metastasis. Cell Tissue Res. 2016;365:621–641. [DOI] [PubMed] [Google Scholar]
- [7].Van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–228. [DOI] [PubMed] [Google Scholar]
- [8].De Wever O, Hendrix A. A supporting ecosystem to mature extracellular vesicles into clinical application. Embo J. 2019;38:e101412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yu Y, Suh MJ, Sikorski P, et al. Urine sample preparation in 96-well filter plates for quantitative clinical proteomics. Anal Chem. 2014;86:5470–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wachalska M, Koppers-Lalic D, van Eijndhoven M, et al. Protein complexes in urine interfere with extracellular vesicle biomarker studies. J Circ Biomarkers. 2016;5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Barreiro K, Holthofer H. Urinary extracellular vesicles. A promising shortcut to novel biomarker discoveries. Cell Tissue Res. 2017;369:217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Van Deun J, Mestdagh P, Agostinis P, et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods. 2017;14:228–232. [DOI] [PubMed] [Google Scholar]
- [13].Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Clayton A, Boilard E, Buzas EI, et al. Considerations towards a roadmap for collection, handling and storage of blood extracellular vesicles. J Extracell Vesicles. 2019;8:1647027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Soekmadji C, Hill AF, Wauben MH, et al. Towards mechanisms and standardization in extracellular vesicle and extracellular RNA studies: results of a worldwide survey. J Extracell Vesicles. 2018;7:1535745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Clayton A, Buschmann D, Byrd JB, et al. Summary of the ISEV workshop on extracellular vesicles as disease biomarkers, held in Birmingham, UK, during December 2017. J Extracell Vesicles. 2018;7:1473707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Human Kidney & Urine Proteome Project Standard protocol for urine collection and storage. Mol Methods Database. 2017;3:8–10. [Google Scholar]
- [18].Vergauwen G, Dhondt B, Van Deun J, et al. Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Sci Rep. 2017;7:2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Geeurickx E, Tulkens J, Dhondt B, et al. The generation and use of recombinant extracellular vesicles as biological reference material. Nat Commun. 2019;10:3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tulkens J, Vergauwen G, Van Deun J, et al. Increased levels of systemic LPS-positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut. 2018. DOI: 10.1136/gutjnl-2018-317726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tulkens J, De Wever O, Hendrix A. Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nat Protoc. 2020 Jan;15(1):40-67. doi: 10.1038/s41596-019-0236-5. [DOI] [PubMed] [Google Scholar]
- [22].Van Deun J, Mestdagh P, Sormunen R, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles. 2014;3 DOI: 10.3402/jev.v3.24858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Théry C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30:3.22.1–3.22.29. [DOI] [PubMed] [Google Scholar]
- [24].Lozano-Ramos I, Bancu I, Oliveira-Tercero A, et al. Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J Extracell Vesicles. 2015;4:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fernández-Llama P, Khositseth S, Gonzales PA, et al. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 2010;77:736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wiśniewski JR, Zougman A, Nagaraj N, et al. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. [DOI] [PubMed] [Google Scholar]
- [28].Vizcaíno JA, Deutsch EW, Wang R, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Perez-Riverol Y, Csordas A, Bai J, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tyanova S, Temu T, Sinitcyn P, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. [DOI] [PubMed] [Google Scholar]
- [31].Wiśniewski JR, Mann M. A proteomics approach to the protein normalization problem: selection of unvarying proteins for MS-based proteomics and western blotting. J Proteome Res. 2016;15:2321–2326. [DOI] [PubMed] [Google Scholar]
- [32].Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4(1): 9pp. [Google Scholar]
- [33].Reimand J, Kull M, Peterson H, et al. g: profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Merico D, Isserlin R, Stueker O, et al. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Reimand J, Isserlin R, Voisin V, et al. Pathway enrichment analysis and visualization of omics data using g: profiler,GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;1 DOI: 10.1038/s41596-018-0103-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pathan M, Keerthikumar S, Chisanga D, et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J Extracell Vesicles. 2017;6:1321455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Webber J, Clayton A. How pure are your vesicles? J Extracell Vesicles. 2013;2:19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sinha A, Huang V, Livingstone J, et al. The proteogenomic landscape of curable prostate cancer. Cancer Cell. 2019;35:414–427.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Iglesias-Gato D, Wikström P, Tyanova S, et al. The proteome of primary prostate cancer. Eur Urol. 2016;69:942–952. [DOI] [PubMed] [Google Scholar]
- [40].Armenia J, Wankowicz SAM, Liu D, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet. 2018;50:645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wedge DC, Gundem G, Mitchell T, et al. Sequencing of prostate cancers identifies new cancer genes, routes of progression and drug targets. Nat Genet. 2018;50:682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Abeshouse A, Ahn J, Akbani R, et al. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu S, Kumari S, Hu Q, et al. A comprehensive analysis of coregulator recruitment, androgen receptor function and gene expression in prostate cancer. Elife. 2017;6 DOI: 10.7554/eLife.28482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shaw GL, Whitaker H, Corcoran M, et al. The early effects of rapid androgen deprivation on human prostate cancer. Eur Urol. 2016;70:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Uhlen M, Fagerberg L, Hallstrom BM, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419–1260419 DOI: 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- [46].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. [DOI] [PubMed] [Google Scholar]
- [47].Minner S, Rump D, Tennstedt P, et al. Epidermal growth factor receptor protein expression and genomic alterations in renal cell carcinoma. Cancer. 2012;118:1268–1275. [DOI] [PubMed] [Google Scholar]
- [48].Nanus DM, Engelstein D, Gastl GA, et al. Molecular cloning of the human kidney differentiation antigen gp160: human aminopeptidase A. Proc Natl Acad Sci U S A. 1993;90:7069–7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Morrissey JJ, Kharasch ED. The specificity of urinary aquaporin 1 and perilipin 2 to screen for renal cell carcinoma. J Urol. 2013;189:1913–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Olsburgh J, Harnden P, Weeks R, et al. Uroplakin gene expression in normal human tissues and locally advanced bladder cancer. J Pathol. 2003;199:41–49. [DOI] [PubMed] [Google Scholar]
- [51].Rood IM, Deegens JKJ, Merchant ML, et al. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010;78:810–816. [DOI] [PubMed] [Google Scholar]
- [52].Tauro BJ, Greening DW, Mathias RA, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. [DOI] [PubMed] [Google Scholar]
- [53].György B, Módos K, Pállinger É, et al. Detection and isolation of cell-derived microparticles are compromised by protein complexes resulting from shared biophysical parameters. Blood. 2011;117:e39–48. [DOI] [PubMed] [Google Scholar]
- [54].Lamparski HG, Metha-Damani A, Yao JY, et al. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270:211–226. [DOI] [PubMed] [Google Scholar]
- [55].Lane RE, Korbie D, Anderson W, et al. Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing. Sci Rep. 2015;5:7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Musante L, Tataruch-Weinert D, Kerjaschki D, et al. Residual urinary extracellular vesicles in ultracentrifugation supernatants after hydrostatic filtration dialysis enrichment. J Extracell Vesicles. 2017;6:1267896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ismail N, Wang Y, Dakhlallah D, et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121:984–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Oeyen E, Van Mol K, Baggerman G, et al. Ultrafiltration and size exclusion chromatography combined with asymmetrical-flow field-flow fractionation for the isolation and characterisation of extracellular vesicles from urine. J Extracell Vesicles. 2018;7 DOI: 10.1080/20013078.2018.1490143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Welton JL, Brennan P, Gurney M, et al. Proteomics analysis of vesicles isolated from plasma and urine of prostate cancer patients using a multiplex, aptamer-based protein array. J Extracell Vesicles. 2016;5:31209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zubiri I, Posada-Ayala M, Sanz-Maroto A, et al. Diabetic nephropathy induces changes in the proteome of human urinary exosomes as revealed by label-free comparative analysis. J Proteomics. 2014;96:92–102. [DOI] [PubMed] [Google Scholar]
- [61].Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2 DOI: 10.3402/jev.v2i0.20360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhou H, Yuen PST, Pisitkun T, et al. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69:1471–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Raimondo F, Chinello C, Stella M, et al. Effects of Hematuria on the proteomic profile of urinary extracellular vesicles: technical challenges. J Proteome Res. 2018;17:2572–2580. [DOI] [PubMed] [Google Scholar]
- [64].Tataruch-Weinert D, Musante L, Kretz O, et al. Urinary extracellular vesicles for RNA extraction: optimization of a protocol devoid of prokaryote contamination. J Extracell Vesicles. 2016;5:30281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Smalley DM, Sheman NE, Nelson K, et al. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J Proteome Res. 2008;7:2088–2096. [DOI] [PubMed] [Google Scholar]
- [66].Mitchell PJ, Welton J, Staffurth J, et al. Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med. 2009;7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Eskra JN, Rabizadeh D, Pavlovich CP, et al. Approaches to urinary detection of prostate cancer. Prostate Cancer Prostatic Dis. 2019;22:362–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Pisitkun T, Shen R-F, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Musante L, Saraswat M, Duriez E, et al. Biochemical and physical characterisation of urinary nanovesicles following CHAPS treatment. PLoS One. 2012;7:e37279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Xu X, Barreiro K, Musante L, et al. Management of Tamm‐Horsfall protein for reliable urinary analytics. PROTEOMICS – Clin Appl. 2019;13:1900018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen CL, Lai YF, Tang P, et al. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J Proteome Res. 2012;11:5611–5629. [DOI] [PubMed] [Google Scholar]
- [72].Raimondo F, Morosi L, Corbetta S, et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol Biosyst. 2013;9:1220. [DOI] [PubMed] [Google Scholar]
- [73].Øverbye A, Skotland T, Koehler CJ, et al. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget. 2015;6:30357–30376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Gonzales PA, Pisitkun T, Hoffert JD, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20:363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Principe S, Jones EE, Kim Y, et al. In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics. 2013;13:1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Pathan M, Fonseka P, Chitti SV, et al. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2018. DOI: 10.1093/nar/gky1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wang L, Jin Y, Arnoldussen YJ, et al. STAMP1 is both a proliferative and an antiapoptotic factor in prostate cancer. Cancer Res. 2010;70:5818–5828. [DOI] [PubMed] [Google Scholar]
- [78].Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer. 2004;4:106–117. [DOI] [PubMed] [Google Scholar]
- [79].Gonthier K, Poluri RTK, Weidmann C, et al. Reprogramming of isocitrate dehydrogenases expression and activity by the androgen receptor in prostate cancer. Mol Cancer Res. 2019;17:1699–1709. [DOI] [PubMed] [Google Scholar]
- [80].Zadra G, Photopoulos C, Loda M. The fat side of prostate cancer. Biochim Biophys Acta - Mol Cell Biol Lipids. 2013;1831:1518–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Senga S, Kobayashi N, Kawaguchi K, et al. Fatty acid-binding protein 5 (FABP5) promotes lipolysis of lipid droplets, de novo fatty acid (FA) synthesis and activation of nuclear factor-kappa B (NF-κB) signaling in cancer cells. Biochim Biophys Acta - Mol Cell Biol Lipids. 2018;1863:1057–1067. [DOI] [PubMed] [Google Scholar]
- [82].Duijvesz D, Burnum-Johnson KE, Gritsenko MA, et al. Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PLoS One. 2013;8:e82589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. [DOI] [PubMed] [Google Scholar]
- [84].Hendrix A, Westbroek W, Bracke M, et al. An ex(o)citing machinery for invasive tumor growth. Cancer Res. 2010;70:9533–9537. [DOI] [PubMed] [Google Scholar]
- [85].Hendrix A, de Wever O. Rab27 GTPases distribute extracellular nanomaps for invasive growth and metastasis: implications for prognosis and treatment. Int J Mol Sci. 2013;14:9883–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Makowska KAA, Hughes REE, White KJJ, et al. Specific myosins control actin organization, cell morphology, and migration in prostate cancer cells. Cell Rep. 2015;13:2118–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Dunn TA, Chen S, Faith DA, et al. A novel role of myosin VI in human prostate cancer. Am J Pathol. 2006;169:1843–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Klatt MG, Kowalewski DJ, Schuster H, et al. Carcinogenesis of renal cell carcinoma reflected in HLA ligands: a novel approach for synergistic peptide vaccination design. Oncoimmunology. 2016;5:e1204504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Rindler MJ, Naik SS, Li N, et al. Uromodulin (Tamm-Horsfall glycoprotein/uromucoid) is a phosphatidylinositol-linked membrane protein. J Biol Chem. 1990;265:20784–20789. [PubMed] [Google Scholar]
- [90].Hadjidemetriou M, Al-Ahmady Z, Mazza M, et al. In vivo biomolecule corona around blood-circulating, clinically used and antibody-targeted lipid bilayer nanoscale vesicles. ACS Nano. 2015;9:8142–8156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015289. We have submitted all relevant data of our experiments to the EV-TRACK knowledgebase (EV-TRACK ID: EV190064). Full-length western blots for Figures 1 and 8 and Suppl. figs. 2, 3, 4 and 7 are provided in Suppl. Appendix 1.
Figure 1.

Characterization of urine fractionated by top down (TD) versus bottom up (BU) Optiprep density gradient (ODG) centrifugation. (a) Nanoparticle tracking analysis of EV-enriched fractions. NTA calculated size distributions are depicted as mean (black line) with standard error (red area) and total particle number, mean particle size and mode are shown for each EV-enriched fraction (Fractions 9–10: 1.087–1.109 g/mL). (b) Particle-protein recovery measurements of BU and TD ODG separated urine fractions. Fractions 1–5, 6–8, 9–10, 11–12, 13–14 and 15–16 were pooled and are indicated by their respective density range in the x-axis. EV-enriched fractions are indicated by a black rectangle. (c) Coomassie blue staining and western blot analysis for EV- (ALIX, TSG-101, CD9, Flotillin-1) and non-EV (THP, GM130, PMP70) associated proteins. EV-enriched fractions are indicated by a black rectangle. THP-enriched (Fractions 13–14: 1.156–1.201 g/mL) and protein-enriched (Fractions 15–16: 1.2017–1.231 g/mL) fractions are indicated by a grey rectangle. (d) Transmission electron microscopy of EV, THP and protein-enriched fractions. Low (scale bar: 1000 nm) and high magnification images (scale bar: 200 nm) are provided. White arrows indicate EV, black arrows indicate THP polymers and grey arrows indicate protein complexes. (e) Graphical overview of BU ODG fractionation strategy for urine, resulting in an EV-enriched, THP-enriched and protein-enriched fraction.
Figure 8.

Differences between the urinary EV proteome from men with prostate cancer, renal cancer and bladder cancer. LC-MS/MS data from EV-enriched fractions from prostate, renal and bladder cancer patients are compared by (a) principle component analysis and (b–c) volcano plots. Proteins related to prostate, renal or bladder tissue are highlighted. (d) Western blot validation of selected prostate (cancer)-, kidney- and bladder-enriched proteins in urinary EV.


