Abstract
Diarrheal illnesses from enteric pathogens are a leading cause of death in children under five in low- and middle-income countries (LMICs). Sanitation is one way to reduce the spread of enteric pathogens in the environment; however, few studies have investigated the effectiveness of sanitation in rural LMICs in reducing pathogens in the environment. In this study, we measured the impact of a sanitation intervention (dual-pit latrines, sani-scoops, child potties delivered as part of a randomized control trial, WASH Benefits) in rural Bangladeshi household compounds by assessing prevalence ratios, differences, and changes in the concentration of pathogen genes and host-specific fecal markers. We found no difference in the prevalence of pathogenic Escherichia coli, norovirus, or Giardia genes in the domestic environment in the sanitation and control arms. The prevalence of the human fecal marker was lower on child hands and the concentration of animal fecal marker was lower on mother hands in the sanitation arm in adjusted models, but these associations were not significant after correcting for multiple comparisons. In the subset of households with ≥10 individuals per compound, the prevalence of enterotoxigenic E. coli genes on child hands was lower in the sanitation arm. Incomplete removal of child and animal feces or the compound (versus community-wide) scale of intervention could explain the limited impacts of improved sanitation.
Introduction
Globally, deaths due to diarrheal illnesses in children under five are decreasing, yet the rate remains high.1 In South Asia, approximately 10% of all deaths in children under five are due to diarrhea.2 These illnesses result from the transmission of enteric pathogens facilitated by inadequate drinking water, sanitation, and hygiene (WASH) conditions. Pathogens can be transmitted from the feces of an infected host to a new host through the fecal-oral route, mediated through environmental reservoirs.3 Specifically, transmission can occur through exposure to contaminated water, soil, hands, fomites, vectors (e.g., flies), and food. Drinking water treatment, sanitation infrastructure, and hygiene interventions use different mechanisms to block these exposure pathways. Of these, sanitation has the potential to block the most upstream fecal-oral transmission pathways by isolating and preventing the spread of feces through soil, water, and flies.
Both humans and animals can serve as hosts for enteric pathogens and contribute to environmental fecal contamination. Recent studies highlight the importance of animal-to-human pathogen transmission pathways.4−6 Globally, livestock produce 4 times the amount of feces as humans and animal feces exposure is of particular concern in low- and middle- income countries, where animal husbandry in the household environment is common.7 Livestock excrete pathogens such as Giardia, Campylobacter, and STEC,8 and a single infected animal may excrete a higher number of pathogens than a single human due to either higher mass concentrations or higher volume of feces.9 Due to the potential hazard posed by animal feces, a broadened scope of sanitation interventions has been suggested that combines animal fecal management (e.g., separating animals from living spaces, removal of animal feces from the household environment with tools, and reducing the movement of animals) with human fecal management (e.g., latrines).4
Determining the levels of fecal contamination in environmental reservoirs, and the impact of sanitation, can provide a mechanistic understanding of how sanitation interventions are effective, or not, at reducing the spread of enteric pathogens. Existing studies have investigated the impact of sanitation on health outcomes, but few have quantified the impact of sanitation on indicators of fecal contamination along exposure routes (soil, hands, water, food, fomites, flies).10−15 Notably, fewer studies have measured the impact of sanitation on the presence of specific pathogens in these reservoirs.15,16
In this study, we quantify the impact of a sanitation intervention (a combined human and animal fecal management intervention that included dual-pit latrines, sani-scoops, and child potties) on enteric pathogen genes and indicators in household environmental reservoirs, within a cluster-randomized controlled trial (WASH Benefits) in rural Bangladesh. We measured pathogen genes (pathogenic Escherichia coli, norovirus, and Giardia lamblia) and microbial source tracking (MST) markers in soil, stored drinking water, and on mother and child hands in a subset of enrolled compounds to assess the impact of the intervention on (1) the prevalence of pathogen genes, (2) the prevalence of human fecal markers, and (3) the prevalence and concentration of animal fecal markers in the domestic environment.
Methods
The study methods (setting, design, sample collection, and pathogen and indicator gene detection) were the same as described in Fuhrmeister et al.17 and are briefly summarized here. The analysis plan was prespecified prior to data analysis and is publicly available on the Open Science Framework (https://osf.io/xrbpz/). Data were analyzed using R and the analysis was independently replicated by two blinded analysts (ERF, YC). This paper complements a previously published paper from the same field trial, which investigated the prevalence and concentration of indicator E. coli in all households enrolled in the control; sanitation; and combined water, sanitation, and handwashing arms four months after the intervention was delivered.13 Another publication documents the relationship between indicator E. coli, microbial source tracking markers, and pathogen genes in the same subset of households sampled as this study.17 Future publications are planned that will report on the prevalence and concentration of indicator E. coli in a subset of sanitation and control households at multiple time points over two years to assess the long-term effect of sanitation.
Study Setting and Design
We conducted our study in a randomly selected subset of households from the sanitation and control arms (300 households per arm) of the WASH Benefits trial in rural Bangladesh.18 WASH Benefits enrolled clusters of households that were geographically pair-matched and then randomly allocated to one of seven intervention or control arms.19 One household was selected per compound (defined as groups of 3–10 households, typically relatives, living in adjacent households surrounding a central courtyard). The sanitation intervention included potties for young children, sani-scoop hoes to remove child and animal feces, and dual-pit latrines in each study compound. Community promoters visited study compounds about 6 times per month to deliver behavior change messages that focused on the use of child potties and latrines for defecation and disposal of human and animal feces. At the time of sample collection, it had been 16–35 months since the latrines were constructed. WASH Benefits enrolled households with a pregnant woman and followed their birth cohort. The interventions were delivered around the time of the cohort’s birth. Therefore, at the time of sample collection, study households contained at least one young child (aged 9–44 months) and any siblings of the birth cohort.
Data Collection
Trained field staff from the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) visited enrolled households, unannounced, between March and October 2015. They collected environmental samples, interviewed the female caregiver of young children regarding household practices related to sanitation and handling of human and animal feces, and conducted environmental spot checks using a structured questionnaire.
Sample Collection
Approximately 720 soil samples, 720 stored drinking water samples, 720 mother hand rinses, and 360 child hand rinses were collected from 600 study households (some households were sampled twice, approximately 4 months apart to capture both the wet and dry seasons; exact sample numbers for each assay are reported in the relevant figures and tables). Soil samples were collected from an area as close to the house entrance as possible by scraping the top layer of soil within a 30 × 30 cm2 stencil into a sterile Whirlpak bag (Nasco, Modesto, CA). For stored water samples, mothers were asked to provide a glass of water as they would give it to their child under five. To collect mother hand rinse samples, mothers placed their left hand into a sterile Whirlpak bag filled with 250 mL of sterile distilled water. The hand was massaged from the outside of the bag for 15 s, followed by 15 s of shaking. The same procedure was repeated with the right hand in the same bag. For child hand rinse samples, respondents placed their child’s hand into a separate Whirlpak bag and followed the same procedure. All samples were placed on ice and transported to the icddr,b field laboratory within 8 h of collection and processed by lab technicians within 12 h of collection.
Detection of Pathogenic E. coli Genes
Previously published methods were used to identify pathogenic E. coli genes.20,21 In brief, IDEXX Colilert-18 was used to determine which samples contained E. coli. Samples that tested positive were then archived for pathogenic E. coli gene analysis. For archiving, broth from up to 20 positive large IDEXX wells was aseptically extracted, pooled, centrifuged, and stored at −80 °C. Samples were transported to UC Berkeley at room temperature (approximately 30 h) and stored at −80 °C upon arrival (2–28 months) prior to sample processing. Multiplex polymerase chain reaction (PCR) reactions were used to detect seven E. coli virulence genes indicative of five pathotypes of E. coli: EAEC (aggR), EPEC/EHEC (eaeA), STEC (stx1/stx2/eaeA), EIEC (ipaH), and ETEC (lt1/st1b) (Table S1). Blanks were included during archiving, DNA extraction, and PCR processing to verify sterile procedures, which resulted in a total of 72 laboratory blanks and 84 extraction blanks (additional information on blanks is available in the Supporting Information).
Filtration and Nucleic Acid Extraction for Quantitative Polymerase Chain Reaction (qPCR) Targets
Field laboratory technicians filtered 50 mL of hand rinse samples and up to 500 mL of stored water (range 100–500 mL) through a 0.45 μm HA filter (Millipore, Burlington, MA) to capture both viruses and bacteria. MgCl2 was added to all samples to increase the retention of viruses.22 Filters were stored at −80 °C before and after transport to UC Berkeley at room temperature. Five grams of soil (wet weight) was weighed and stored at −80 °C until transport to UC Berkeley (under USDA soil import permit, PPQ 525). Filter and soil samples were stored at UC Berkeley at −80 °C between 5 and 24 months prior to processing. DNA and RNA were extracted from soil and filters using modified Mobio PowerWater and PowerViral (both now Qiagen, Germantown, MD) protocols.15,23 A total of 168 blanks were processed during filtration and 119 during nucleic acid extraction. A subset of samples (175 of 2608 total samples, 7%) was spiked with Pseudomonas syringae pv. phaseolicola (pph6) and MS2 (99 of 2608, 4%) to quantify the extraction of nucleic acids. Detailed methods for extraction protocols and quantification of extraction efficiency are available in the Supporting Information of Fuhrmeister et al.17
qPCR Assays
We used qPCR to analyze samples for norovirus GII, G. lamblia, Cryptosporidium spp. specific genes, and microbial source tracking markers (Table S2). To detect human-specific fecal contamination, we used the HumM2 qPCR assay (U.S. Environmental Protection Agency research license #864-14) that targets human-associated Bacteroidales-like microorganisms.24,25 The HumM2 protocol was modified to employ a different master mix designed to reduce inhibition, and the qPCR threshold was selected based on our instrument (StepOnePlus). To detect animal-associated fecal contamination, we used the BacCow assay.26 Both HumM2 and BacCow assays were previously evaluated for sensitivity and specificity in rural Bangladesh.15 The BacCow assay detected Bacteroidales in feces of cows, ducks, goats, and chickens but not humans in this setting.15 HumM2 performed the best out of all human-associated assays tested (HumM2, HF183, and BacHum), although it also amplified, indicating a positive result, in the feces of chickens and goats.15
Initially, a subset of samples (105 child hands, 112 mother hands, 110 stored water, and 109 soil samples) from 50 households (25 control and 25 sanitation) were tested for the presence of norovirus GII, G. lamblia, and Cryptosporidium specific genes. Within this subset, less than 1% of both soil samples and stored water samples tested positive for norovirus and Giardia genes; less than 1% of all sample types were positive for the Cryptosporidium gene. Therefore, we only analyzed remaining hand rinse samples for norovirus and Giardia genes. We did not test any remaining samples for the presence of the Cryptosporidium gene.
Samples were run on a StepOnePlus thermal cycler (Applied Biosystems, Foster City, CA) in triplicate on a 96-well plate (Applied Biosystems, Foster City, CA). Each run contained one standard curve (ranging from 10 to 105 gene copies/2 μL) and three no-template controls. A subset of samples (10 filters and 13 soil samples, ≈1%) were tested for inhibition using the spike and dilute method.27 Filter samples were uninhibited for all DNA and RNA assays. In soil samples, Cryptosporidium spp. and MS2 assays were inhibited in some samples. HumM2, BacCow, G. lamblia, pph6, and Norovirus GII were uninhibited for all samples. A 1:10 dilution for the extraction efficiency of RNA in soil was used for MS2. We decided not to dilute Cryptosporidium due to the resulting increase in the detection limit. A standard curve for quantification for each assay was determined by pooling all standard curves and using a linear mixed-effects model to correct for batch effects.28 Results from inhibition testing and standard curve quantification can be found in the Supporting Information of Fuhrmeister et al.17
Data Analysis for Quantitative Assays
BacCow was the only qPCR assay that produced data in the quantifiable range; nearly all samples amplified below the lowest point on the standard curve (10 gene copies/2 μL) in all other assays. BacCow measurements were quantifiable in 76% of child hands, 67% of mother hands, 10% of stored water, and 49% of soil. For quantitative analysis, hand rinse samples below the limit of detection (BLOD) were assigned half the value of the limit of detection (LOD). Hand rinse samples below the limit of quantification (BLOQ) were assigned the midpoint between the LOD and the limit of quantification (LOQ). To determine which quantities to assign to samples BLOD and BLOQ in stored water and soil samples, which were impacted by differences in processing volume and moisture content, we conducted a sensitivity analysis with four scenarios. The overall results for difference in concentration of BacCow between arms were similar in all scenarios (Table S3). Thus, we chose to assign samples BLOD half the LOD accounting for the volume of water filtered and the moisture content for each sample. Samples BLOQ were assigned the midpoint between the LOD and LOQ. Blanks that amplified for BacCow were BLOQ; samples that amplified in the same region (BLOQ) were treated as BLOD (see the Supporting Information for more details, including Table S4 for the number of samples BLOD and BLOQ).
Data Analysis for Presence Absence Assays
Most samples did not amplify within the quantifiable range for norovirus GII, Giardia, and HumM2. Therefore, these assays were analyzed as presence absence data and samples were considered positive for qPCR targets if at least one of three replicates amplified, even if amplification was BLOQ. For binary analyses of pathogen/source tracking marker presence, all samples with positive lab, extraction, or archiving blanks were removed from the analysis according to the date processed and the lab technician. This resulted in removing 30 child and 33 mother hand samples from norovirus GII and 51 child and 115 mother hands from Giardia gene analyses. In total, 29 child hand, 74 mother hand, 76 stored water, and 92 soil samples were removed from HumM2 analyses. For pathogenic E. coli virulence genes, 6 child hand, 9 mother hand, 11 stored water, and 12 soil samples were removed from eaeA analyses. A total of 3 child hand, 5 mother hand, 5 stored water, and 6 soil samples were removed from stx1/stx2 analyses. A total of 4 child hand, 6 mother hand, 9 stored water, and 8 soil samples were removed from lt1 analyses.
Linear Models
The impact of the sanitation intervention was assessed using generalized linear models with robust standard errors to account for the trial’s clustered study design and did not assume outcome distributions.18 For binary outcomes, we estimated the prevalence, prevalence differences (PD), and prevalence ratios (PR) for pathogen genes and microbial source tracking markers. For BacCow, the only continuous marker, we determined the mean concentration in each arm as well as the difference in concentration between the arms. The analyzed values for BacCow concentrations were log10-transformed.
Randomization in the parent trial resulted in balanced covariates between arms; thus, we used the unadjusted models as our primary analysis.18 We conducted three prespecified subgroup analyses to estimate our parameters of interest in subsets of the data. Data were subset by season (wet vs dry), the number of individuals living in a compound (<10 vs ≥10), and the number of animals in the compound (<20 vs ≥20). Animals included cows, goats, sheep, chickens, non-chicken poultry (ducks, geese), and non-poultry birds (sparrows, crows, pigeons). For the latter two subsets, the cutoffs were determined based on the empirical data distribution. In a deviation from the analysis plan, the wet season was defined as June–September (rather than May–October) to achieve a better balance in sample numbers in the subgroups and to reflect the actual rainfall in 2015. In a secondary analysis, we adjusted our estimates for covariates anticipated to be associated with our outcomes. We included the same covariates as described in the WASH Benefits analysis plan (season, rainfall, and household characteristics) with the addition of sampling condition covariates (sampling location for soil (sun, shade), soil moisture content, cover status of the stored water container, stored water container type, and observation of handwashing during sample collection). We corrected for multiple comparisons, multiplying p-values by four (i.e., the same pathogen evaluated in multiple sample types) using a Bonferroni correction.29
Ethics
Participants provided written informed consent. The study protocol was approved by human subjects committees at the icddr,b (PR-11063), University of California, Berkeley (2011-09-3652), and Stanford University (25863).
Results
Prevalence of Pathogen Genes and Microbial Source Tracking Markers
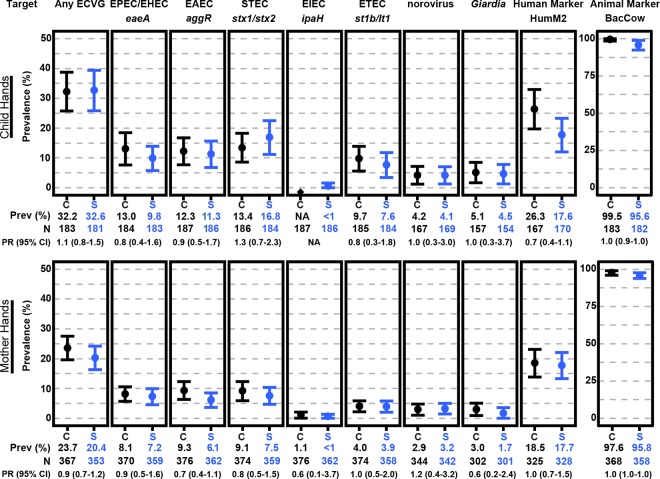
The prevalence of each E. coli virulence gene (ECVG) was consistently higher on child hands than mother hands (Figure 1 and Table S5). On child hands in both study arms, the most prevalent ECVG was stx1/2 (control (C): 13.4 (95% CI: 8.6, 18.3)%; sanitation (S): 16.8 (11.2, 22.5)%). On mother hands, the most prevalent ECVGs were eaeA, aggR, and stx1/2 genes (C and S: 6.1−9.3%). The prevalence of norovirus and G. lamblia genes was 1.7–5.1% in all hand samples in both arms. In other household reservoirs, ECVGs were present in ≈37% (C: 37.5 (33.0, 42.1) %; S: 36.4 (31.3, 41.6)%) of stored water and ≈60% (C: 61.5 (56.4, 66.6) %; S: 58.9 (53.9, 64.0)%) of soil (Figure 2 and Table S5). The most commonly detected ECVG in stored water was stx1/2 (C: 16.6 (12.6, 20.6) %; S: 16.5 (12.6, 20.4)%), and eaeA (C: 38.6 (33.2, 44.1)%; S: 36.7 (31.7, 41.6)%) was the most commonly detected ECVG in soil. The least prevalent ECVG across all sample types was ipaH (≤2%).
Figure 1.
Prevalence (95% CI) and prevalence ratio (95% CI) of pathogenic E. coli virulence genes, norovirus GII gene, G. lamblia gene, and microbial source tracking markers on mother and child hands in the sanitation (S) and control (C) arms.
Figure 2.
Prevalence (95% CI) and prevalence ratio (95% CI) of pathogenic E. coli virulence genes and microbial source tracking markers in stored drinking water and soil in the sanitation (S) and control (C) arms.
Human and animal microbial source tracking markers were detected in all four reservoirs. The prevalence of BacCow was higher than any other target including both pathogen genes and microbial source tracking markers. In total, 99.5 (98.4, 100.0)% and 95.6 (92.4, 98.9)% of child hands in the control and sanitation arms were positive for BacCow and a similar prevalence was also found on mother hands (C: 97.6 (96.0, 99.1)%; S: 95.8 (93.9, 97.7)%). BacCow was detected in 70.0 (65.1, 74.8)% and 67.0 (61.6, 72.4)% of stored water and in 91.2 (88.4, 94.1)% and 90.2 (86.1, 94.2)% of soil in the control and sanitation arms. The prevalence of the human fecal marker (HumM2) was substantially lower than the animal marker in all reservoirs. In the control and sanitation arms, the prevalence of HumM2 on child hands was 26.3 (19.7, 33.0)% and 17.6 (12.0, 23.3)% and a similar level was found on mother hands (C: 18.5 (13.8, 23.1)%; S: 17.7 (13.3, 22.1)%). We detected HumM2 in <5% of stored water samples (C: 3.6 (1.5, 5.6)%; S: 1.6 (0.2, 2.9)%) and in approximately 20% of soil (C: 18.4 (14.3, 22.5)%; S: 22.0 (17.3, 26.6)%). Hand rinses, stored water, and soil samples have different units (per 2 hands, 100 mL of stored water, and g of dry soil); therefore, it is difficult to draw meaningful conclusions regarding the different prevalence results between environmental sample types.
Intervention Impact
Almost all compounds in both the sanitation and control arms contained a latrine (C: 99%; S: 100%) at the time of sample collection (Table 1). Most sanitation arm compounds had a pour-flush latrine (C: 63%; S: 100%) with a functional water seal (C: 40%; S: 97%). Only 3% of intervention arm latrines drained into the environment, whereas more than 20% of control households had latrines draining into the environment or had feces spilling out from the pit. While ownership does not guarantee use, most adults (94%) in the sanitation arm used the latrine during structured observations in the main field trial.30 There were also more compounds with young children defecating in latrines or potties (C, 13%; S, 56%) and using a scoop or hoe to handle animal feces (C, 48%; S, 97%) in the intervention arm.
Table 1. Household Drinking Water, Sanitation, and Animal Hygiene Practicesa.
| household characteristics | control (N = 300) | sanitation (N = 297) |
|---|---|---|
| cover on stored water containerb | 23.2 | 25.4 |
| reported treating waterc | <1 | <1 |
| latrine present in compoundd | 98.7 | 100.0 |
| with slab | 96.3 | 99.7 |
| pour flush | 63.2e | 99.7 |
| functional water seal | 39.7e | 97.0 |
| flow into the environmentf | 21.7e | 3.4 |
| visible feces on slab | 8.4 | 1.7 |
| feces odor | 64.9 | 23.6 |
| urine odor | 19.3 | 2.0 |
| pit emptied since last visit | 16.9 | 3.4 |
| reported using latrine always | ||
| children <3 y | 1.4e | 4.4g |
| men | 84.5h | 90.0i |
| women | 93.2 | 92.6 |
| reported children <5 defecating in potty or latrine | 13.0 | 55.6 |
| human feces visible in the courtyard | <1 | 1.0 |
| reported using scoop or hoe to handle child feces | 36.0 | 36.7 |
| animal feces visible in the courtyard | ||
| chicken/non-chicken poultry | 89.7 | 90.9 |
| cow | 35.7 | 26.9 |
| goat/sheep | 24.3 | 22.2 |
| pig | 0 | 0 |
| dog or cat | <1 | <1 |
| cow patty | 11.0 | 7.4 |
| non-poultry birdsj | 4.7 | 3.7 |
| reported using scoop or hoe to handle animal fecesk | 48.1 | 96.8 |
% of households.
Of households with stored water: 24 control and 29 sanitation households did not have stored drinking water.
Treated with a household water filter.
Characteristics for primary pit latrine used in the compound.
N = 295 households.
Latrine drains directly into the environment or feces spilling out from the pit.
N = 294 households.
N = 282 households.
N = 290 households.
Sparrow, pigeon, and crow.
Of households with animals: 11 control and 15 sanitation households did not have animals.
There was no significant difference in the prevalence of pathogen genes or microbial source tracking markers between the control and sanitation arms across all sample types using the unadjusted models and correcting for multiple comparisons (Figures 1 and 2 and Table S6). On child hands, the prevalence of BacCow was 4% lower in the sanitation arm (prevalence ratio [PR]: 0.96 (95% CI: 0.93, 1.00); corrected p = 0.21). This association was not significant after correcting for multiple comparisons in the unadjusted model but was significant in the adjusted model (PR: 0.95 (0.92, 0.97); corrected p < 0.001). There was a borderline significant difference in the prevalence of the human fecal marker on child hands between arms in the adjusted model (PR: 0.66 (0.44, 0.99); corrected p = 0.19). There was also a borderline significant difference in the prevalence of st1b/lt1 between arms in stored water (PR, 0.64 (0.39, 1.03); corrected p = 0.27) and soil (0.74 (0.54, 1.03); corrected p = 0.29) in the adjusted model. The average log10 concentration of BacCow on mother hands was lower in the sanitation arm in the unadjusted (Δ, −0.16 (95% CI: −0.30, −0.03); corrected p = 0.07) and adjusted models (−0.37 (−0.66, −0.08); corrected p = 0.05), but these associations were not significant after correcting for multiple comparisons (Figure 3 and Table S7).
Figure 3.
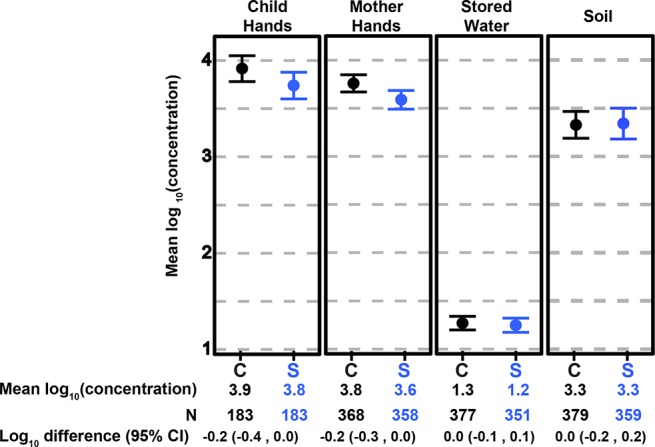
Mean log10 concentration of BacCow on hands, in stored drinking water and soil samples from the sanitation (S) versus control (C) arms. Concentrations are in units of gene copies per 2 hands, 100 mL of water, and gram of dry soil.
In the subgroup analyses, there were no significant associations, correcting for multiple comparisons and separating the data by season (Table S8). In households with ≥20 animals in their compound, there was a borderline significant difference in the prevalence of st1b/lt1 between arms on child hands (0.22 (0.04, 1.06); corrected p = 0.16) and in soil (0.46 (0.23, 0.92); corrected p = 0.21) (Table S9). In households with ≥10 individuals in their compound, the prevalence of st1b/lt1 on child hands was 89% lower in the sanitation arm versus the control arm (0.11 (0.02, 0.77); corrected p = 0.04) (Table S10). We did not find any significant difference in log10 concentration of BacCow in any subgroup analyses, correcting for multiple comparisons, but there were some borderline significant effects on mother hands (Table S11).
Discussion
Prevalence of Pathogen Genes and Microbial Source Tracking Markers
Considering that stored drinking water is a direct exposure route for the ingestion of enteric pathogens, the high prevalence of BacCow (C: 70%; S: 67%) and E. coli virulence genes (C: 38%; S:67%) in stored water in both study arms is a concern. As we did not measure pathogen genes or BacCow in tubewells (source water), we were unable to differentiate contamination in the tubewells from contamination introduced during storage. It is possible that some of the fecal contamination in stored drinking water originated from the source water, as other studies have found indicators of fecal contamination and pathogen genes in these sources.31−34 Another possible mechanism of stored water contamination is through contact with hands,35−37 which we have shown to be contaminated with multiple types of pathogenic E. coli virulence genes (Figure 1). In this same study area, Ercumen et al.13 found that the prevalence of indicator E. coli was significantly higher in stored water (C: 77%; S: 76%) than was found in tubewells (C: 25%; S: 23%);13 thus, contamination during storage is likely responsible for some of the observed high prevalence of BacCow and E. coli virulence genes. Comparing with previously published results on indicator E. coli in all sample types, it is interesting to note that the prevalence of BacCow on hands was higher than the prevalence of indicator E. coli (≈75%), whereas indicator E. coli were more prevalent than BacCow in stored water (88%) and soil (95%).17
Previous studies have investigated the presence of microbial source tracking markers in the household environment in rural India. We found a higher prevalence of the same animal fecal marker (BacCow) in hand rinses and stored water in rural Bangladesh than was observed in Odisha, India.16 We also found a similar level of the human fecal markers on hands as was found in India but a higher prevalence in stored drinking water; however, the results are difficult to compare given that different fecal markers were used based on sensitivity and specificity for each study location.15,38
A comparable study on rural Bangladeshi households found lower prevalence of human fecal contamination using the same human marker (HumM2).15 This study was also conducted in a subset of WASH Benefits households in the sanitation and control arms, but samples were collected at an earlier date (November 2013–March 2014). In the control arm, 9.0% of soil samples, 0% of stored drinking water, and 2.0% of child hands were positive for HumM2 compared to 18.4% soil, 3.6% stored water, and 26.3% of child hands in our study. Taken together, the results indicate a high degree of temporal variability in human fecal contamination. These studies sampled in different years as well as months; the previous study was conducted during the dry season of Bangladesh, while our sampling spanned both seasons. Therefore, differences in the occurrence of HumM2 could be due to both seasonal and yearly variations. Previous studies have found higher levels of fecal contamination in various environmental reservoirs during the wet season in Bangladesh.6 In our study and Boehm et al.’s, the prevalence of viruses on child hands was low (<10%), although the studies investigated different viruses. In the control and sanitation arms, rotavirus was detected in 5.7 and 6.5% of child hands in the earlier Bangladesh study and norovirus was detected in ≈4% of child hands in our study.
Intervention Impact
Overall, the use of the sanitation intervention in the WASH Benefits trial was not sufficient to reduce fecal contamination in the household environment, as evidenced by the similar prevalence of pathogen genes and microbial source tracking markers in both the sanitation and control study arms. This finding is consistent with the results of a previously published indicator bacteria study that found no difference between indicator E. coli concentrations in tubewells, ponds, stored water, food, soil, and on child hands and flies in the sanitation arm compared to those in the control arm.13 One possible explanation provided was that the observed concentrations of indicator E. coli were occluded by “naturalized” E. coli (E. coli naturally found in soil and water that is not derived from fecal sources), which masked the effect of the sanitation intervention. This is still a possibility, although we also did not find a difference in the prevalence of microbial source tracking markers and select pathogen genes between the sanitation and control arms. Another suggested hypothesis from the indicator E. coli study was that spread of human feces, but not animal feces, was reduced by the intervention. The authors reasoned that the indicator E. coli detected could have come predominantly from animal feces, which was still present in intervention households despite the provision of sani-scoops. This is consistent with our finding that the animal fecal marker was substantially more prevalent than the human fecal marker among all sample types in our study. However, given that we saw no significant differences in the human fecal marker (HumM2) between the study arms, apart from a borderline reduction on child hands, we conclude that human fecal contamination was not substantially impacted by the sanitation intervention. Similarly, there was no difference between the prevalence of the animal fecal marker (BacCow) between the two arms except for a small difference in the prevalence on child hands, suggesting no intervention impact on animal fecal contamination.
The WASH Benefits study reported a lower prevalence of childhood diarrhea in households in the sanitation arm,18 while our study found few significant differences in the prevalence of pathogens or fecal indicators in environmental samples collected from a subset of these same households. We expected that because the prevalence of diarrhea was lower in the sanitation arm, containment of feces was better and therefore fewer pathogens would be present in the household environment. One possible explanation for why we did not measure a difference is that pathogen detection limits were too high, resulting in many false negatives and insufficient sensitivity to detect a difference between the two arms. In other words, our study may not have detected whether some of these samples (false negatives) contained pathogen concentrations sufficient to exceed the minimum infective dose. For example, WASH Benefits measured protozoan pathogens in the stool of study participants and found a 25% reduction in the prevalence of Giardia infections in the sanitation arm.39 However, we did not observe a similar reduction in the prevalence of Giardia in environmental reservoirs. The concentration of Giardia in environmental samples is much lower than in the stool of infected individuals, which could have resulted in lower sensitivity to detect a difference in the environmental samples.
Another possibility to explain the discrepancy between the prevalence of diarrhea and pathogens in environmental samples is that we did not measure all pathogens that could be etiological agents of diarrhea. Our study focused on pathogenic E. coli, Giardia, and norovirus, but rotavirus, Shigella, Aeromonas, and Campylobacter jejuni are other possible etiological agents of disease in Bangladesh.40 Also, we did not measure pathogen genes and microbial source tracking markers on fomites or food which could be dominant reservoirs for enteric pathogen transmission.41 The duration between latrine construction and sample collection (16–35 months) should have been sufficiently long for fecal associated organisms and nucleic acids present before construction to degrade in environmental reservoirs, including soil.42−45 Therefore, it is unlikely the genes detected in this study persisted from before the intervention.
A benefit to the pathogenic E. coli detection method used in this study is the enrichment step, which allowed us to detect pathogenic E. coli genes likely from viable organisms. However, there are some limitations to this and the other detection methods used and these could influence the measured impact of the sanitation intervention. The method we used for pathogenic E. coli was not quantitative, and a quantitative method such as qPCR could have determined whether the concentrations of pathogenic E. coli, not just prevalence, were impacted in the trial. However, using qPCR does not guarantee that quantitative results will be produced, as occurred in this study. Despite using qPCR to detect norovirus, Giardia, and HumM2, most of the results were below the limit of quantification. The quantitative analysis of BacCow results in stored water and soil was also impacted by a high proportion of nonquantitative samples in stored water and soil (Table S4), which may have reduced the ability to detect a difference between the study arms. Detection can be improved by processing larger sample volumes; however, sample volumes are constrained in the household setting. For example, stored water samples are restricted to the volume that can be spared at the time of collection and the logistical constraints of transporting samples from the field to the lab. We were unable to filter the targeted 500 mL from all households. This volume was selected to reduce the lower detection limit (from the standard 100 mL sample volume), but in some households, less than 500 mL was available at the time of collection or in other cases filters clogged before the full sample was filtered. The variable filtration volumes resulted in variable detection limits; however, the impact on detection limits was similar in sanitation and control arms. Therefore, we do not believe that this was a limitation in our ability to detect a difference between arms. More effort is needed to develop methods with lower detection limits compatible with complex environmental samples. While there have been recent developments in aptamer-based detection (nucleotides that bind to a target), with potential for lower detection limits, very few have been tested in environmental matrices.46
Our results are generally in agreement with a previous study in rural India that found no reduction in pathogens and microbial source tracking markers in household reservoirs despite a 27% increase in latrine coverage.16 One notable finding in our study was a slightly lower (5%) prevalence of BacCow on child hands in the sanitation arm, although the prevalence was still high (100 versus 96% in control and sanitation arms, respectively). However, the lower prevalence of BacCow was not reflected in the difference in log10 concentration of BacCow on child hands. The lower prevalence of BacCow could be due to the use of sani-scoops for animal feces removal in 97% of the sanitation arm households. We observed a lower prevalence of cow feces in sanitation versus control compound courtyards (C, 36%; S, 27%), although poultry feces were still visible in 90% of courtyards overall (Table 1). We also observed 89% lower prevalence of st1b/lt1 genes (indicative of ETEC) in sanitation versus control arm households with ≥10 individuals. More research is needed to understand differences in sanitation practices in households with more individuals, which could be impacted by different social dynamics in larger families. It should be noted that while our results are in agreement with previous rural studies, they may not be generalizable to more urban settings.
Poor child and animal fecal management could be a main reason we did not see lower fecal contamination in household reservoirs in the sanitation arm. While there were more children using a potty or latrine in the sanitation arm, it was still just over half of the households (C: 13%; S: 56%). Interestingly, the percent of households that reported hygienic disposal of child feces in the sanitation arm is higher in this study than the 21% of sanitation households that reported disposing of child feces in a latrine approximately one year prior.15 Although this is a promising result and could be due to more exposure to community promoters and children reaching an appropriate age to use the latrine, there is still significant room for improvement. Similar practices are also evident in the management of animal feces. The animal fecal marker (BacCow) was prevalent in all environmental reservoirs in both study arms, and a concurrent study showed that increased concentrations of BacCow were associated with the increased prevalence of some pathogen genes on mother hands.17 Another reason we did not see the impact of the sanitation intervention could be due to the compound scale of the intervention. Intercompound transmission can occur through a variety of pathways,47 and pathogens can be transferred from schools, neighboring compounds, agriculture fields, and other places of work.
It is also possible that the quality of the latrine may not have been sufficient to isolate human fecal contamination from the household environment. The latrine improvements in this study provided a hygienic barrier between feces and the environment through the metrics evaluated such as functional water seal and reduced flow from overfull pits or latrines draining directly into the environment. However, Bangladesh has a high water table and leakage from latrines into groundwater is possible but was not measured in this study.48 It should be noted that the sanitation intervention did not include specific procedures for pit emptying or treatment offsite, although pit emptying is unlikely to be a main source of exposure as very few pit latrines in the sanitation arm were emptied during the study period (3%).
In this study, access to latrines in both arms was high (C: 98.7%; S: 100%), but the management of child and animal feces was still incomplete. Modeling could be a useful tool to estimate the impact of further improvements in the management of child and animal feces on the concentration of pathogens in the environment and the prevalence of diarrhea in children under five. Quantitative microbial risk assessment, including exposure models,41,49 can be combined with pathogen measurements to predict the number of pathogens children ingest in their household environment and estimate the resulting prevalence of diarrhea. Our study provides an opportunity to calibrate a model using actual data on environmental pathogen concentrations and diarrheal prevalence. Other recent studies in Bangladesh can provide more detailed information on child behavior and interactions with the environment,50,51 providing a foundation for exposure models to investigate fecal contamination pathways that result in pathogen ingestion by children in these households. Previous work has taken a similar approach via calibrating a QMRA model to field observations of diarrheal prevalence to model the reduction in infection risk due to different levels of use of household water treatment.52−54 Modeling the impacts of varying strategies of child and animal fecal management interventions could be used to explore which interventions and degree of compliance are needed to significantly reduce fecal contamination levels in the household environment. Finally, this study was not designed to measure fecal pathogen levels in potential reservoirs outside of the household environment to which children may be exposed such as open drains, fecal sludge dumping sites, open defecation sites, and animal feces for fertilizer. Additional research is needed to better understand exposure in public versus private realms.41,55,56
Acknowledgments
The research was supported by Grant R01HD078912 from the NIH and in part by Grant 0PPGD759 from the Bill & Melinda Gates Foundation to the University of California, Berkeley. ERF was also supported by the National Science Foundation Graduate Research Fellowship under Grant DGE 1106400 and DGE 1752814.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.9b04835.
Details on PCR and qPCR methods (Tables S1−S2), sensitivity analysis (Table S3), limits of detection (Table S4), and generalized linear model results (Tables S5−S11). (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- GBD 2015 Mortality and Causes of Death Collaborators. Global, Regional, and National Life Expectancy, All-Cause Mortality, and Cause-Specific Mortality for 249 Causes of Death, 1980-2015: A Systematic Analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. 10.1016/S0140-6736(16)31012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childhood Mortality-UNICEF Data; UNICEF, 2019. https://data.unicef.org/topic/child-survival/under-five-mortality/.
- Wagner E. G.; Lanoix J. N.. Excreta Disposal for Rural Areas and Small Communities; World Health Organization, 1958; Vol. 39. [PubMed] [Google Scholar]
- Penakalapati G.; Swarthout J.; Delahoy M. J.; McAliley L.; Wodnik B.; Levy K.; Freeman M. C. Exposure to Animal Feces and Human Health: A Systematic Review and Proposed Research Priorities. Environ. Sci. Technol. 2017, 51, 11537–11552. 10.1021/acs.est.7b02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A. R.; Pickering A. J.; Harris M.; Doza S.; Islam M. S.; Unicomb L.; Luby S.; Davis J.; Boehm A. B. Ruminants Contribute Fecal Contamination to the Urban Household Environment in Dhaka, Bangladesh. Environ. Sci. Technol. 2016, 50, 4642–4649. 10.1021/acs.est.5b06282. [DOI] [PubMed] [Google Scholar]
- Ercumen A.; Pickering A. J.; Kwong L. H.; Arnold B. F.; Parvez S. M.; Alam M.; Sen D.; Islam S.; Kullmann C.; Chase C.; Ahmed R.; Unicomb L.; Luby S. P.; Colford J. M. Animal Feces Contribute to Domestic Fecal Contamination: Evidence from E. coli Measured in Water, Hands, Food, Flies, and Soil in Bangladesh. Environ. Sci. Technol. 2017, 51, 8725–8734. 10.1021/acs.est.7b01710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendes D. M.; Yang P. J.; Lai A.; Hu D.; Brown J. Estimation of Global Recoverable Human and Animal Faecal Biomass. Nat. Sustain. 2018, 1, 679–685. 10.1038/s41893-018-0167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahoy M. J.; Wodnik B.; McAliley L.; Penakalapati G.; Swarthout J.; Freeman M. C.; Levy K. Pathogens Transmitted in Animal Feces in Low- and Middle-Income Countries. Int. J. Hyg. Environ. Health 2018, 221, 661–676. 10.1016/j.ijheh.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels M. E.; Shrivastava A.; Smith W. A.; Sahu P.; Odagiri M.; Misra P. R.; Panigrahi P.; Suar M.; Clasen T.; Jenkins M. W. Cryptosporidium and Giardia in Humans, Domestic Animals, and Village Water Sources in Rural India. Am. J. Trop. Med. Hyg. 2015, 93, 596–600. 10.4269/ajtmh.15-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S. R.; Arnold B. F.; Salvatore A. L.; Briceno B.; Ganguly S.; Colford J. M.; Gertler P. J. The Effect of India’s Total Sanitation Campaign on Defecation Behaviors and Child Health in Rural Madhya Pradesh: A Cluster Randomized Controlled Trial. PLoS Med. 2014, 11, e1001709 10.1371/journal.pmed.1001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasen T.; Boisson S.; Routray P.; Torondel B.; Bell M.; Cumming O.; Ensink J.; Freeman M.; Jenkins M.; Odagiri M.; Ray S.; Sinha A.; Suar M.; Schmidt W. P. Effectiveness of a Rural Sanitation Programme on Diarrhoea, Soil-Transmitted Helminth Infection, and Child Malnutrition in Odisha, India: A Cluster-Randomised Trial. Lancet Global Health 2014, 2, e645–e653. 10.1016/S2214-109X(14)70307-9. [DOI] [PubMed] [Google Scholar]
- Pickering A. J.; Djebbari H.; Lopez C.; Coulibaly M.; Alzua M. L. Effect of a Community-Led Sanitation Intervention on Child Diarrhoea and Child Growth in Rural Mali: A Cluster-Randomised Controlled Trial. Lancet Global Health 2015, 3, e701–e711. 10.1016/S2214-109X(15)00144-8. [DOI] [PubMed] [Google Scholar]
- Ercumen A.; Pickering A. J.; Kwong L. H.; Mertens A.; Arnold B. F.; Benjamin-Chung J.; Hubbard A. E.; Alam M.; Sen D.; Islam S.; Rahman M. Z.; Kullmann C.; Chase C.; Ahmed R.; Sarker P. M.; Unicomb L.; Rahman M.; Ram P. K.; Clasen T.; Luby S. P.; Colford J. M. Jr. Do Sanitation Improvements Reduce Fecal Contamination of Water, Hands, Food, Soil, and Flies? Evidence from a Cluster-Randomized Controlled Trial in Rural Bangladesh. Environ. Sci. Technol. 2018, 52, 12089–12097. 10.1021/acs.est.8b02988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclar G. D.; Penakalapati G.; Amato H. K.; Garn J. V.; Alexander K.; Freeman M. C.; Boisson S.; Medlicott K. O.; Clasen T. Assessing the Impact of Sanitation on Indicators of Fecal Exposure along Principal Transmission Pathways: A Systematic Review. Int. J. Hyg. Environ. Health 2016, 219, 709–723. 10.1016/j.ijheh.2016.09.021. [DOI] [PubMed] [Google Scholar]
- Boehm A. B.; Wang D.; Ercumen A.; Shea M.; Harris A. R.; Shanks O. C.; Kelty C.; Ahmed A.; Mahmud Z. H.; Arnold B. F.; Chase C.; Kullmann C.; Colford J. M.; Luby S. P.; Pickering A. J. Occurrence of Host-Associated Fecal Markers on Child Hands, Household Soil, and Drinking Water in Rural Bangladeshi Households. Environ. Sci. Technol. Lett. 2016, 3, 393–398. 10.1021/acs.estlett.6b00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odagiri M.; Schriewer A.; Daniels M. E.; Wuertz S.; Smith W. A.; Clasen T.; Schmidt W. P.; Jin Y.; Torondel B.; Misra P. R.; Panigrahi P.; Jenkins M. W. Human Fecal and Pathogen Exposure Pathways in Rural Indian Villages and the Effect of Increased Latrine Coverage. Water Res. 2016, 100, 232–244. 10.1016/j.watres.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmeister E. R.; Ercumen A.; Pickering A. J.; Jeanis K. M.; Ahmed M.; Brown S.; Arnold B. F.; Hubbard A. E.; Alam M.; Sen D.; Islam S.; Kabir M. H.; Kwong L. H.; Islam M.; Unicomb L.; Rahman M.; Boehm A. B.; Luby S. P.; Colford J. M.; Nelson K. L. Predictors of Enteric Pathogens in the Domestic Environment from Human and Animal Sources in Rural Bangladesh. Environ. Sci. Technol. 2019, 53, 10023–10033. 10.1021/acs.est.8b07192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby S. P.; Rahman M.; Arnold B. F.; Unicomb L.; Ashraf S.; Winch P. J.; Stewart C. P.; Begum F.; Hussain F.; Benjamin-Chung J.; Leontsini E.; Naser A. M.; Parvez S. M.; Hubbard A. E.; Lin A.; Nizame F. A.; Jannat K.; Ercumen A.; Ram P. K.; Das K. K.; Abedin J.; Clasen T. F.; Dewey K. G.; Fernald L. C.; Null C.; Ahmed T.; Colford J. M. Effects of Water Quality, Sanitation, Handwashing, and Nutritional Interventions on Diarrhoea and Child Growth in Rural Bangladesh: A Cluster Randomised Controlled Trial. Lancet Global Health 2018, 6, e302–e315. 10.1016/S2214-109X(17)30490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold B. F.; Null C.; Luby S. P.; Unicomb L.; Stewart C. P.; Dewey K. G.; Ahmed T.; Ashraf S.; Christensen G.; Clasen T.; Dentz H. N.; Fernald L. C. H.; Haque R.; Hubbard A. E.; Kariger P.; Leontsini E.; Lin A.; Njenga S. M.; Pickering A. J.; Ram P. K.; Tofail F.; Winch P. J.; Colford J. M. Cluster-Randomised Controlled Trials of Individual and Combined Water, Sanitation, Hygiene and Nutritional Interventions in Rural Bangladesh and Kenya: The WASH Benefits Study Design and Rationale. BMJ Open 2013, 3, e003476 10.1136/bmjopen-2013-003476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar K. B.; Potgieter N.; Barnard T. G. Development of a Rapid Screening Method for the Detection of Pathogenic Escherichia coli Using a Combination of Colilert Quanti-Trays/2000 and PCR. Water Sci. Technol. 2010, 10, 7–13. 10.2166/ws.2010.862. [DOI] [Google Scholar]
- Pickering A. J.; Julian T. R.; Marks S. J.; Mattioli M. C.; Boehm A. B.; Schwab K. J.; Davis J. Fecal Contamination and Diarrheal Pathogens on Surfaces and in Soils among Tanzanian Households with and without Improved Sanitation. Environ. Sci. Technol. 2012, 46, 5736–5743. 10.1021/es300022c. [DOI] [PubMed] [Google Scholar]
- Victoria M.; Guimaraes F.; Fumian T.; Ferreira F.; Vieira C.; Leite J. P.; Miagostovich M. Evaluation of an Adsorption-Elution Method for Detection of Astrovirus and Norovirus in Environmental Waters. J. Virol. Methods 2009, 156, 73–76. 10.1016/j.jviromet.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Mattioli M. C.; Pickering A. J.; Gilsdorf R. J.; Davis J.; Boehm A. B. Hands and Water as Vectors of Diarrheal Pathogens in Bagamoyo, Tanzania. Environ. Sci. Technol. 2013, 47, 355–363. 10.1021/es303878d. [DOI] [PubMed] [Google Scholar]
- Shanks O. C.; Kelty C. A.; Sivaganesan M.; Varma M.; Haugland R. A. Quantitative PCR for Genetic Markers of Human Fecal Pollution. Appl. Environ. Microbiol. 2009, 75, 5507–5513. 10.1128/AEM.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks O. C.; Domingo J. W. S.; Lu J.; Kelty C. A.; Graham J. E. Identification of Bacterial DNA Markers for the Detection of Human Fecal Pollution in Water. Appl. Environ. Microbiol. 2007, 73, 2416–2422. 10.1128/AEM.02474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kildare B. J.; Leutenegger C. M.; McSwain B. S.; Bambic D. G.; Rajal V. B.; Wuertz S. 16S RRNA-Based Assays for Quantitative Detection of Universal, Human-, Cow-, and Dog-Specific Fecal Bacteroidales: A Bayesian Approach. Water Res. 2007, 41, 3701–3715. 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- Cao Y.; Griffith J. F.; Dorevitch S.; Weisberg S. B. Effectiveness of QPCR Permutations, Internal Controls and Dilution as Means for Minimizing the Impact of Inhibition While Measuring Enterococcus in Environmental Waters. J. Appl. Microbiol. 2012, 113, 66–75. 10.1111/j.1365-2672.2012.05305.x. [DOI] [PubMed] [Google Scholar]
- Sivaganesan M.; Haugland R. A.; Chern E. C.; Shanks O. C. Improved Strategies and Optimization of Calibration Models for Real-Time PCR Absolute Quantification. Water Res. 2010, 44, 4726–4735. 10.1016/j.watres.2010.07.066. [DOI] [PubMed] [Google Scholar]
- Abdi H.The Bonferonni and Sidak Corrections for Multiple Comparisons. In Encyclopedia of Measurment and Statistics; Salkind N. J., Ed.; Sage Publications: Thousand Oaks, CA, 2007. [Google Scholar]
- Parvez S. M.; Azad R.; Rahman M.; Unicomb L.; Ram P. K.; Naser A. M.; Stewart C. P.; Jannat K.; Rahman M. J.; Leontsini E.; Winch P. J.; Luby S. P. Achieving Optimal Technology and Behavioral Uptake of Single and Combined Interventions of Water, Sanitation Hygiene and Nutrition, in an Efficacy Trial (WASH Benefits) in Rural Bangladesh. Trials 2018, 19, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby S. P.; Gupta S. K.; Sheikh Ma.; Johnston R. B.; Ram P. K.; Islam M. S. Tubewell Water Quality and Predictors of Contamination in Three Flood-Prone Areas in Bangladesh. J. Appl. Microbiol. 2008, 105, 1002–1008. 10.1111/j.1365-2672.2008.03826.x. [DOI] [PubMed] [Google Scholar]
- Wu J.; Yunus M.; Islam M. S.; Emch M. Influence of Climate Extremes and Land Use on Fecal Contamination of Shallow Tubewells in Bangladesh. Environ. Sci. Technol. 2016, 50, 2669–2676. 10.1021/acs.est.5b05193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. S.; Siddika A.; Khan M. N.; Goldar M. M.; Sadique M. A.; Kabir A. N.; Huq A.; Colwell R. R. Microbiological Analysis of Tube-Well Water in a Rural Area of Bangladesh. Appl. Environ. Microbiol. 2001, 67, 3328–3330. 10.1128/AEM.67.7.3328-3330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson A. S.; Layton A. C.; Mailloux B. J.; Culligan P. J.; Williams D. E.; Smartt A. E.; Sayler G. S.; Feighery J.; McKay L. D.; Knappett P. S. K.; Alexandrova E.; Arbit T.; Emch M.; Escamilla V.; Ahmed K. M.; Alam M. J.; Streatfield P. K.; Yunus M.; van Geen A. Comparison of Fecal Indicators with Pathogenic Bacteria and Rotavirus in Groundwater. Sci. Total Environ. 2012, 431, 314–322. 10.1016/j.scitotenv.2012.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriewer A.; Odagiri M.; Wuertz S.; Misra P. R.; Panigrahi P.; Clasen T.; Jenkins M. W. Human and Animal Fecal Contamination of Community Water Sources, Stored Drinking Water and Hands in Rural India Measured with Validated Microbial Source Tracking Assays. Am. J. Trop. Med. Hyg. 2015, 93, 509–516. 10.4269/ajtmh.14-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering A. J.; Davis J.; Walters S. P.; Horak H. M.; Keymer D. P.; Mushi D.; Strickfaden R.; Chynoweth J. S.; Liu J.; Blum A.; Rogers K.; Boehm A. B. Hands, Water, and Health: Fecal Contamination in Tanzanian Communities with Improved, Non-Networked Water Supplies. Environ. Sci. Technol. 2010, 44, 3267–3272. 10.1021/es903524m. [DOI] [PubMed] [Google Scholar]
- Trevett A. F.; Carter R. C.; Tyrrel S. F. Water Quality Deterioration: A Study of Household Drinking Water Quality in Rural Honduras. Int. J. Environ. Health Res. 2004, 14, 273–283. 10.1080/09603120410001725612. [DOI] [PubMed] [Google Scholar]
- Odagiri M.; Schriewer A.; Hanley K.; Wuertz S.; Misra P. R.; Panigrahi P.; Jenkins M. W. Validation of Bacteroidales Quantitative PCR Assays Targeting Human and Animal Fecal Contamination in the Public and Domestic Domains in India. Sci. Total Environ. 2015, 502, 462–470. 10.1016/j.scitotenv.2014.09.040. [DOI] [PubMed] [Google Scholar]
- Lin A.; Ercumen A.; Benjamin-Chung J.; Arnold B. F.; Das S.; Haque R.; Ashraf S.; Parvez S. M.; Unicomb L.; Rahman M.; Hubbard A. E.; Stewart C. P.; Colford J. M.; Luby S. P. Effects of Water, Sanitation, Handwashing, and Nutritional Interventions on Child Enteric Protozoan Infections in Rural Bangladesh: A Cluster-Randomized Controlled Trial. Clin. Infect. Dis. 2018, 67, 1515–1522. 10.1093/cid/ciy320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff K. L.; Nataro J. P.; Blackwelder W. C.; Nasrin D.; Farag T. H.; Panchalingam S.; Wu Y.; Sow S. O.; Sur D.; Breiman R. F.; Faruque A. S.; Zaidi A. K.; Saha D.; Alonso P. L.; Tamboura B.; Sanogo D.; Onwuchekwa U.; Manna B.; Ramamurthy T.; Kanungo S.; Ochieng J. B.; Omore R.; Oundo J. O.; Hossain A.; Das S. K.; Ahmed S.; Qureshi S.; Quadri F.; Adegbola R. A.; Antonio M.; Hossain M. J.; Akinsola A.; Mandomando I.; Nhampossa T.; Acacio S.; Biswas K.; O’Reilly C. E.; Mintz E. D.; Berkeley L. Y.; Muhsen K.; Sommerfelt H.; Robins-Browne R. M.; Levine M. M. Burden and Aetiology of Diarrhoeal Disease in Infants and Young Children in Developing Countries (the Global Enteric Multicenter Study, GEMS): A Prospective, Case-Control Study. Lancet 2013, 382, 209–222. 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Robb K.; Null C.; Teunis P.; Yakubu H.; Armah G.; Moe C. L. Assessment of Fecal Exposure Pathways in Low-Income Urban Neighborhoods in Accra, Ghana: Rationale, Design, Methods, and Key Findings of the SaniPath Study. Am. J. Trop. Med. Hyg. 2017, 97, 1020–1032. 10.4269/ajtmh.16-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Booth D. J.; Campbell R. G.; Gulden R. H.; Hart M. M.; Powell J. R.; Klironomos J. N.; Pauls K. P.; Swanton C. J.; Trevors J. T.; Dunfield K. E. Cycling of Extracellular DNA in the Soil Environment. Soil Biol. Biochem. 2007, 39, 2977–2991. 10.1016/j.soilbio.2007.06.020. [DOI] [Google Scholar]
- England L. S.; Holmes S. B.; Trevors J. T. Review: Persistence of Viruses and DNA in Soil. World J. Microbiol. Biotechnol. 1998, 14, 163–169. 10.1023/A:1008865609698. [DOI] [Google Scholar]
- Olson M. E.; Goh J.; Phillips M.; Guselle N.; McAllister T. A. Giardia Cyst and Cryptosporidium Oocyst Survival in Water, Soil, and Cattle Feces. J. Environ. Qual. 1999, 28, 1991. 10.2134/jeq1999.00472425002800060040x. [DOI] [Google Scholar]
- Ogorzaly L.; Bertrand I.; Paris M.; Maul A.; Gantzer C. Occurrence, Survival, and Persistence of Human Adenoviruses and F-Specific RNA Phages in Raw Groundwater. Appl. Environ. Microbiol. 2010, 76, 8019–8025. 10.1128/AEM.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdinasab M.; Hayat A.; Marty J. L. Aptamer-Based Assays and Aptasensors for Detection of Pathogenic Bacteria in Food Samples. TrAC, Trends Anal. Chem. 2018, 107, 60–77. 10.1016/j.trac.2018.07.016. [DOI] [Google Scholar]
- Eisenberg J. N. S.; Scott J. C.; Porco T. Integrating Disease Control Strategies: Balancing Water Sanitation and Hygiene Interventions to Reduce Diarrheal Disease Burden. Am. J. Public Health 2007, 97, 846–852. 10.2105/AJPH.2006.086207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naser A. M.; Doza S.; Rahman M.; Ahmed K. M.; Gazi M. S.; Alam G. R.; Karim M. R.; Khan G. K.; Uddin M. N.; Mahmud M. I.; Ercumen A.; Rosenbaum J.; Annis J.; Luby S. P.; Unicomb L.; Clasen T. F. Sand Barriers around Latrine Pits Reduce Fecal Bacterial Leaching into Shallow Groundwater: A Randomized Controlled Trial in Coastal Bangladesh. Environ. Sci. Technol. 2019, 53, 2105–2113. 10.1021/acs.est.8b04950. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Moe C. L.; Null C.; Raj S. J.; Baker K. K.; Robb K. A.; Yakubu H.; Ampofo J. A.; Wellington N.; Freeman M. C.; Armah G.; Reese H. E.; Peprah D.; Teunis P. F. M. Multipathway Quantitative Assessment of Exposure to Fecal Contamination for Young Children in Low-Income Urban Environments in Accra, Ghana: The SaniPath Analytical Approach. Am. J. Trop. Med. Hyg. 2017, 97, 1009–1019. 10.4269/ajtmh.16-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong L.; Ercumen A.; Pickering A.; Unicomb L.; Davis J.; Luby S. Hand- and Object-Mouthing of Rural Bangladeshi Children 3–18 Months Old. Int. J. Environ. Res. Public Health 2016, 13, 563. 10.3390/ijerph13060563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong L. H.; Ercumen A.; Pickering A. J.; Unicomb L.; Davis J.; Luby S. P. Age-Related Changes to Environmental Exposure: Variation in the Frequency That Young Children Place Hands and Objects in Their Mouths. J. Exposure Sci. Environ. Epidemiol. 2019, 30, 205–216. 10.1038/s41370-019-0160-3. [DOI] [PubMed] [Google Scholar]
- Enger K. S.; Nelson K. L.; Rose J. B.; Eisenberg J. N. S. The Joint Effects of Efficacy and Compliance: A Study of Household Water Treatment Effectiveness against Childhood Diarrhea. Water Res. 2013, 47, 1181–1190. 10.1016/j.watres.2012.11.034. [DOI] [PubMed] [Google Scholar]
- Enger K. S.; Nelson K. L.; Clasen T.; Rose J. B.; Eisenberg J. N. S. Linking Quantitative Microbial Risk Assessment and Epidemiological Data: Informing Safe Drinking Water Trials in Developing Countries. Environ. Sci. Technol. 2012, 46, 5160–5167. 10.1021/es204381e. [DOI] [PubMed] [Google Scholar]
- Mellor J.; Abebe L.; Ehdaie B.; Dillingham R.; Smith J. Modeling the Sustainability of a Ceramic Water Filter Intervention. Water Res. 2014, 49, 286–299. 10.1016/j.watres.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medgyesi D.; Brogan J.; Sewell D.; Creve-Coeur J.; Kwong L.; Baker K.; Medgyesi D. N.; Brogan J. M.; Sewell D. K.; Creve-Coeur J. P.; Kwong L. H.; Baker K. K. Where Children Play: Young Child Exposure to Environmental Hazards during Play in Public Areas in a Transitioning Internally Displaced Persons Community in Haiti. Int. J. Environ. Res. Public Health 2018, 15, 1646. 10.3390/ijerph15081646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medgyesi D.; Sewell D.; Senesac R.; Cumming O.; Mumma J.; Baker K. K. The Landscape of Enteric Pathogen Exposure of Young Children in Public Domains of Low-Income, Urban Kenya: The Influence of Exposure Pathway and Spatial Range of Play on Multi-Pathogen Exposure Risks. PLoS Neglected Trop. Dis. 2019, 13, e0007292 10.1371/journal.pntd.0007292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.