ABSTRACT
Streptococcus pneumoniae (S. pneumoniae) is one of the main causative agents of pneumococcal diseases. To date, more than 90 distinct serotypes have been identified. Implementation of vaccines has caused a drastic reduction in vaccine-serotype pneumococcal diseases but increase in cases due to non-vaccine serotype has been observed in Malaysia. However, further investigation on different serotype incidence in Malaysia is needed and the rate of pneumococcal vaccination for new-born babies in Malaysia remains low. The recent emergence of drug-resistant S. pneumoniae (DRSP) has also been a global concern, especially penicillin resistance. This study determined the serotypes of S. pneumoniae strains (n = 95) isolated from nasopharyngeal specimens from children admitted to UMMC from 2013 to 2015. In accordance with previous studies, PCR result showed 40% of NT isolates were successfully typed as 3 less common serotypes, namely 9N/L, 17A, and 23B. The repetitive-element PCR (REP-PCR) result revealed genetic variations among the strains whereby five major clusters were observed at the similarity of 80% by clustering analysis based on fingerprint data. Penicillin-binding proteins (pbps) of selected isolates were studied by PCR and sequencing. Three strains with ≤19-mm diameter zone for Oxacillin Disc Diffusion (ODD) test previously were recorded to have mutation on all pbp1a, pbp2b, and pbp2x with MIC of 4 µg/ml, which were penicillin-intermediate resistance according to the CLSI breakpoints.
KEYWORDS: Molecular detection, genotyping, characterization, Streptococcus pneumoniae, REP-PCR
Introduction
Streptococcus pneumoniae (S. pneumoniae) is one of the leading causes of bacterial infections, ranging from self-limiting respiratory tract infections to severe invasive infections such as meningitis. It causes more than half a million of morbidity and mortality in children worldwide, particularly in developing countries and the majority of death incidences were reported from Africa and Asia [1]. Pneumococcal diseases are common among children <5 years of age and the elderly, who have weakened or impaired immune system.
Among the many infections caused by S. pneumoniae, the pneumococcal pneumonia remains one of the major problems that plagued immunocompromised individuals. Pneumonia is an acute lower respiratory tract infection (LRTI) that could lead to fatal disease outcome if not treated in time [2]. Pneumonia occurs when the lungs are infected by bacteria, viruses, or fungi, causing the alveoli to inflame and filled up with fluid or pus, decreasing the oxygen supply to the bloodstream. S. pneumoniae is commonly existing as an ‘asymptomatic’ colonizer in human hosts, i.e. the carrier state [3]. This organism is usually transmitted through respiratory droplets or direct contact, colonizing the nasopharynx. Although mostly remained as colonizers, S. pneumoniae is capable of causing disease, or even severe invasive infection, through the expression of virulence factors [3]. Moreover, the ability of the pneumococcus to acquire and disseminate antimicrobial resistance determinants not only complicates the treatment of pneumococcal infection but also aids the persistence of this bacterium in the environment and human hosts.
Currently, more than 90 distinct serotypes of S. pneumoniae have been identified based on its capsular polysaccharide (CPS) with distinct chemical structures and patterns [4]. CPS is also one of the virulence factors which shield pneumococci from host immune system and increase their pathogenicity. Due to its immunogenicity, CPS is being used as the target to develop pneumococcal vaccines.
To date, there are four different types of S. pneumoniae vaccines available in the market. The polysaccharide vaccine, Pneumovax™ 23 (PPSV23) was introduced since the mid-1980s for patients above 65 years of age, whereas 3 types of pneumococcal conjugate vaccines (PCV) were also developed, i.e. 7-valent (PCV7), 10-valent (PCV10), and 13-valent (PCV13). The most commonly used vaccines are PPSV23 and PCV13. PCV13 was introduced back in 2010 and it covers the most common pneumococcal serotypes that cause invasive diseases. These vaccines are remarkably effective and they contribute to the reduction in PCV-serotype pneumococcal diseases as demonstrated in previous studies [5–7]. Moreover, the widespread vaccination also results in significant herd immunity, hence indirectly diminishing the incidence in non-vaccinated individuals [8]. In Malaysia, pneumonia was reported to be the third leading cause of death in children under five in 2016 according to Department of Statistics [9]. However, less than 20% of the new-born babies receive the pneumococcal vaccine, which is not an ideal situation [10].
Penicillin resistant S. pneumoniae (PRSP) was first detected in the 1960s and the emergence of multi-drug resistant strains has become a global concern. This is probably due to the altered antibiotic affinities within penicillin-binding proteins (pbps) [11–13]. Resistance to other classes of antibiotics such as macrolides and sulfonamides has also been recorded [14–16]. The recent vaccine-induced serotype replacement phenomenon has also caused obvious changes in the antibiotic resistance patterns of pneumococci in the community [17].
The emergence of PRSP and increase of pneumococcal incidence are an alarming issue in Malaysia [18]. However, the lack of information and surveillance data about serotypes of disease-causing S. pneumoniae causes difficulty in appropriate antibiotic prescription and vaccine use. Hence, it is important to determine the local serotypes and antibiotic resistance patterns of S. pneumoniae isolates in children. This study determined the serotype distribution, genetic relatedness, and antibiotic susceptibility profiles in S. pneumoniae isolated from nasopharyngeal swabs/secretion of patients <12 years of age, who were admitted to University Malaya Medical Center (UMMC) from 2013 to 2015, showing symptoms of infection and signs of LRTI.
Materials and methods
Bacterial isolates
S. pneumoniae (n = 95) were obtained from the collection of cultures from University of Malaya Medical Center (UMMC). These isolates were previously isolated from respiratory specimens (nasopharyngeal swab/secretion) from pediatric patients <12 years of age with LRTI, between the years of 2013 to 2015. Patient’s data showed that children were admitted to UMMC with symptoms of infection (fever, cough and/or runny nose) and signs of LRTI (shortness of breath, tachypnea, recessions, crepitations, and/or rhonchi). Nonetheless, not all patients had the strict definition of pneumonia which is signs of a LRTI with focal or diffuse infiltrates, silhouette sign, pleural effusion, or air bronchogram. All the isolates were recovered on Columbia agar supplemented with 5% horse blood and incubated for 24 h at 37°C, with 5% CO2. This study was approved by the University of Malaya Research Ethics Committee (UMREC) (MECID: 20146–336).
DNA extraction and identification
Heat lysis extraction method was employed as previously described with modification [19]. Briefly, colony sweeps from overnight cultures from blood agar that suspended in 1 ml of saline water were centrifuged. The supernatant was discarded and 100 µl of ultra-pure water was added into the tube thereafter heated at 95°C for 10 min and snapped cooled on ice for 5 min. It was then centrifuged and stored at −20°C. All the isolates were then confirmed as S. pneumoniae by amplifying the internal fragments of a house-keeping gene, recP via specific primer (recP-S, 5ʹ – GCC AAC TCA GGT CAT CCA GG – 3ʹ and recP-AS, 5ʹ – TGC AAC CGT AGC ATT GTA AC – 3ʹ). The PCR was performed as previously described [20].
Serotyping of S. pneumoniae isolates
The serotypes of part of the isolates have been determined by multiplex-PCR assays in a previous study [21]. In the present study, the serotyping of strains was conducted using multiple-PCR assays as described previously [21,22]. Additional primer sequences that are specific to the less common serotypes, namely 9N/L, 13, 16A, 17A, 21, 23A, 23B, 24A/F, 28A/F, 29F, and 41A/F were added in this study to type the non-typeable (NT) strains and the primer sequences are listed in Table 1 [22–25]. The NT isolates were first tested by using multiplex-PCR scheme with modifications. The additional multiplex reaction set was performed with the incorporation of four additional primer pairs that target serotypes 9N/L, 21, 24A/F, and 29. The primer concentrations for all four additional primers were 1.5 mM, respectively, and in all reactions, a concentration of 2-mM MgCl2 was used. The remaining NT isolates after multiplex-PCR were further tested by conventional singleplex PCR with serotype-specific primers. A primer pair that targets the highly conserved cpsA locus was included in each assay as internal positive control (Table 1) and synthetic positive control has been used for each serotype. The amplified DNA products were resolved by gel electrophoresis in 0.5X TAE buffer using 1.5% agarose gels (Sigma-Aldrich, Germany) at 100 V for 80 min. The sizes of the PCR products were compared with 100bp DNA ladder (Promega, USA) as molecular size standard. The strain was classified as non-typeable (NT) if no amplification from the multiplex-PCRs was observed.
Table 1.
Oligonucleotide primers and amplified fragment length in molecular serotyping of S. pneumoniae isolates.
| Serotype | Primer sequence (5ʹ–3ʹ) | Amplicon size (bp) |
|---|---|---|
| 9N/L | F – GAA CTG AAT AAG TCA GAT TTA ATC AGC R – ACC AAG ATC TGA CGG GCT AAT CAA T |
516 |
| 13 | F – GAT GGG AAA ATA CGA TAT GCT C R – AAC TCC ATG ACA AAA CTC CAG C |
309 |
| 16A | F – GAT CCG CTC ACG GTA TGG ACT A R – AAT TTT GCT GTC AGC CAA TAA G |
320 |
| 17A | F – TAG ACT TCT TAG AGC CTA TTG TGG R – ATA ACA GTT TGC GCT ATT GGT C |
318 |
| 21 | F – TTC TTA AAA ATT ACG CCT ATA ATC TCT CTT R – GGT ACA TTT TCT TCA CAG ACT TAT AAT CAC |
831 |
| 23A | F – TAT TCT AGC AAG TGA CGA AGA TGC G R – CCA ACA TGC TTA AAA ACG CTG CTT TAC |
722 |
| 23B | F – TTG TTA GTG GTA TTA AAT TGG GGA CTA CTA GG R – ATA CCT ATC TGA AGT GTT ATT AAC CCA CCA AC |
216 |
| 24A/F | F – TCT CAA CCA AGA TAC AGA TTT TGA TTT TAC TC R – TAT AAA CCT TTA GTA AAC ACT CTG CTT GAT CG |
686 |
| 28A/F | F- CAG AGT TTG GTC GAG GTT CCT A R – GTG ATT TCC GTC GTT GAT TGA G |
327 |
| 29 | F – CTA GCG CAA AGT TGG GAG TT R – AAG CGA GAA TCA GTT TGT CCA |
217 |
| 41A/F | F- GTA GTT ACT GGC CCT TTC TTA TTC C R – TAG CGA GAA ACT ATC TGC ATC TTG |
317 |
| cpsA | F- GCA GTA CAG CAG TTT GTT GGA CTG ACC R – GAA TAT TTT CAT TAT CAG TCC CAG TC |
160 |
Repetitive-element PCR fingerprinting (REP-PCR)
REP-PCR was performed using the primer (5′-GCG CCG ICA TGC GGC ATT-3′) as described previously [26]. Amplification was performed in 1X PCR buffer, 2.5mM of MgCl2, 125 µM of each dNTP, 1 µM of primer, 1.5 U of Taq DNA polymerase (Promega, USA) and 10 µl of DNA template to a total of 25-µl reaction volume. PCR was performed at initial denaturation of 94°C for 4 min followed by 35 cycles of 94°C for 4 min, 42°C for 1 min, and 68°C for 8 min, with final extension of 72°C for 8 min. The PCR products were electrophoresed on a 2% agarose gel for 5 h at 100 V. The banding patterns of amplified DNA were analyzed using BioNumerics 7.0 (Applied Math, Kortrijk, Belgium). All the PCR fingerprint profiles were assigned arbitrary designations and the quantitative differences among the profiles were defined by the Dice coefficient; F. Cluster analysis was carried out according to the unweighted pair group with arithmetic averages (UPGMA) using a position tolerance of 0.15.
Antimicrobial susceptibility testing
Minimum inhibitory concentration (MIC) of the S. pneumoniae isolates were tested by broth microdilution tests according to CLSI guidelines [27]. In brief, the isolates were grown overnight on blood agar plates and suspended in cation-adjusted Mueller-Hinton broth (CAMHB) supplemented with 5% lysed horse blood to a 0.5 McFarland density, representing approximately 1 × 108 CFU/ml. The suspension was diluted to the final cell concentration of 1 × 106 CFU/ml. Subsequently, 1:1 ratio of cell suspension and penicillin was prepared, with the final concentration of 1 × 105 CFU/ml, in each well. The plate was incubated overnight before MIC determination. S. pneumoniae ATCC® 49619™ strain was used as QC strain and the breakpoint of penicillin was interpreted according to CLSI guideline.
DNA sequencing of pbp1a, pbp2b, and pbp2x genes
Ten strains of S. pneumoniae (three strains with oxacillin zone diameter ≤19 mm and seven randomly selected bacterial isolates which were susceptible to penicillin) were subjected to PCR and DNA sequencing for identification of mutations in pbp1a, pbp2b, and pbp2x genes. Three sets of primers were used to amplify pbp1a (1197 bp), pbp2b (1317bp), and pbp2x (1148 bp) genes as described by Zhou et al. [13]. The 25 µl PCR reaction mixture contained 1X PCR buffer, 1.5 mM of MgCl2, 50 µM of each dNTP, 0.4 µM of each primer, 1.25 U of Taq DNA polymerase (Promega, USA) and 8 µl of DNA template. PCR reaction was programmed to initial denaturation of 94°C for 5 min, 30 cycles of 94°C for 30 sec, 57°C for 30 sec, and 72°C for 1 min, with the final extension of 72°C for 7 min. The PCR products were then purified using MEGAquick-spinTM Total Fragment DNA Purification Kit (Intron Biotechnology) and sequenced by Integrated DNA Technologies, Inc. (Coralville, IA, USA). Sequence results were analyzed by the Basic Local Alignment Search Tool (BLAST) online with R6 as reference strain (GenBank accession no.: NC_003098).
Results
Serotyping of S. pneumoniae isolates
In accordance with the previous study, the serotyping revealed 13 S. pneumoniae serotypes among the isolates, with predominantly serotypes 19 F (26.3%), 6A/B (23.2%), and 23F (10.5%) (Table 2) [21]. Additional multiplex PCR assays and conventional PCR were performed on all NT isolates. According to the results obtained, 40% (6/15) of the NT isolates were successfully typed as 3 less common serotypes. Using the previously described scheme with modifications, serotypes 9N/L (n = 1), 17A (n = 4), and 23B (n = 1) were detected.
Table 2.
Serotype distribution of S. pneumoniae isolates obtained from UMMC, Malaysia, 2013 to 2015.
| Serotype | No. of isolates, n | % of Total |
|---|---|---|
| 3a,b | 2 | 2.1 |
| 6A/Ba,b,c | 22 | 23.2 |
| 19Aa,b | 4 | 4.2 |
| 19Fa,b | 25 | 26.3 |
| 23Fa,b | 10 | 10.5 |
| 6C | 6 | 6.3 |
| 9N/Lb,c | 1 | 1.1 |
| 11A/D/Fb,c | 4 | 4.2 |
| 15A/F | 1 | 1.1 |
| 17A | 4 | 4.2 |
| 23A | 4 | 4.2 |
| 23B | 1 | 1.1 |
| 34 | 2 | 2.1 |
| NTd | 9 | 9.5 |
| Total | 95 | 100 |
aSerotypes that have been included in PCV13.
bSerotypes that have been included in PPSV23 (conly serotypes 6B, 9N, and 11A are included)
dNT, non-typeable.
REP-PCR
The REP-PCR generated profiles with 8 to 12 bands with sizes ranging from 0.2 kb to 0.8 kb. For all the types obtained, the genetic similarity coefficient ranged from 70% to 100% as shown in Figure 1. REP-PCR had typed the 95 S. pneumoniae strains into 51 profiles and were coded REP-1 to REP-51. A total of five major clusters were observed (C1–C5) at the similarity of 80%. Cluster C1 had grouped 10 strains of serotype 19F together whereas all other clusters consisted a mixture of all the serotypes detected in this study.
Figure 1.
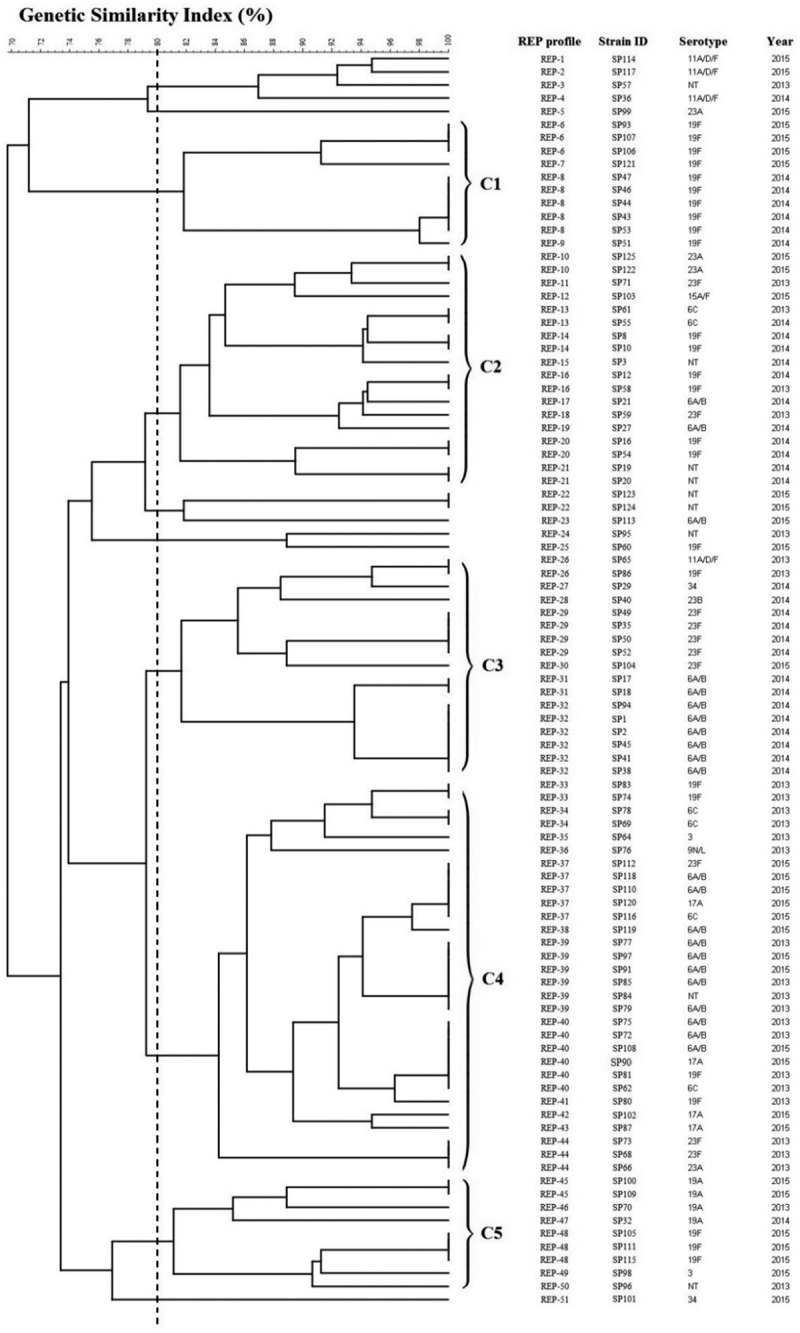
Genotypic relatedness of 95 S. pneumoniae isolates from 2013 to 2015, based on REP profile.
Although the isolates were closely related based on the dendrogram generated, several lineages were identified. For instance, serotype 19F was further subtyped into 12 profiles, with 3 clusters based on 80% similarity (Figure 2). The REP-PCR subtypes within serotype 19F differed from one another by minor bands in the high-molecular-weight region of the image (not shown). On the other hand, serotype 6A/B was further classified into two clusters with nine subtypes (Figure 3(a)) while for 23F, it was further profiled into two major clusters with six subtypes (Figure 3(b)).
Figure 2.
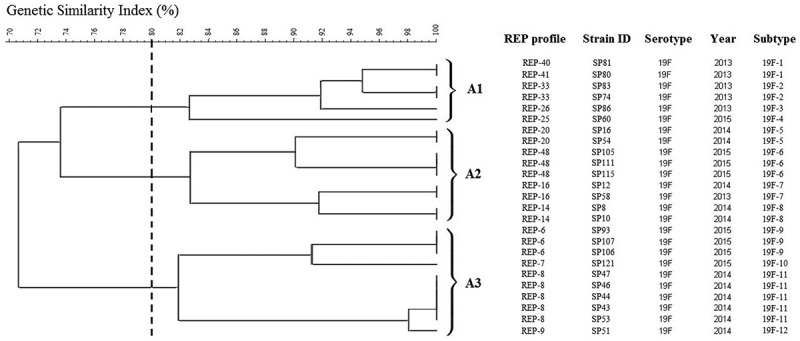
Genotypic relatedness of 25 serotype 19F S. pneumoniae isolates from 2013 to 2015 based on REP profile.
Figure 3.
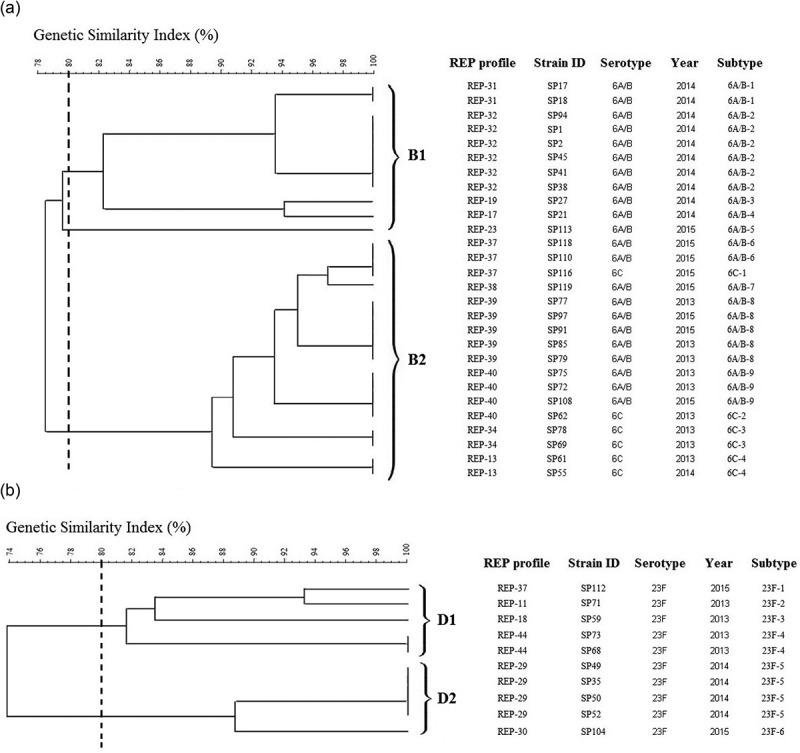
Genotypic relatedness of serotypes 6A/B, 6C, and 23F S. pneumoniae isolates from 2013 to 2015 based on REP profile. (a) Twenty-two serotypes 6A/B & 6C S. pneumoniae isolates; (b) 10 serotype 23F S. pneumoniae isolates.
Antimicrobial susceptibility testing and gene sequencing
The susceptibility data of the isolates were obtained from the Microbiology Diagnostic laboratory, UMMC. All but three isolates of S. pneumoniae were susceptible to penicillin based on ODD test. The susceptibility of three isolates, SP2, SP3, and SP47, were further confirmed using broth microdilution and all three strains were recorded to have MIC of 4 µg/ml, which represents penicillin-intermediate based on CLSI breakpoints (Table 3).
Table 3.
Antimicrobial susceptibility of 3 S. pneumoniae isolates.
| Strain no. | Serotypes | Penicillin MIC, µg/ml |
|---|---|---|
| SP2 | 6A/B | 4 |
| SP3 | NT | 4 |
| SP47 | 19F | 4 |
Sequence analysis of pbp1a, pbp2b, and pbp2x were performed on the three isolates and seven randomly selected susceptible strains (Table 4). Compared to the R6 reference strain, in pbp1a, two out of three intermediate isolates had a mutation at Thr371 in the STMK motif and Pro432→Thr. No mutations were observed in 557KTG motif for all three isolates. In pbp2b, no mutations were observed in the 385SVVK motif and 614KTG motif. However, all three isolates contained a Thr338→Ala mutation in the SSN motif. For pbp2x, all three strains contained a Thr338→Ala alteration in the STMK motif and Leu546→Val substitution in the KSG motif, while no mutations were found in the HSSN motif. All other seven representative strains had highly similar sequence the R6 strain.
Table 4.
Amino acid alterations of three conserved motifs of pbp1a, pbp2b, and pbp2x in 11 S. pneumoniae isolates.
| Strain no. | No. (%) of altered amino acids |
Changes in amino acids of conserved motifs forming or surrounding active pbp binding site |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
pbp1a 314–608 |
pbp2b 277–625 |
pbp2x 254–608 |
pbp1a | pbp2b | pbp2x | |||||||
| R6 | ─ | ─ | ─ | STMK | SRNVP | KTG | SVVK | SSNT | KTG | STMK | HSSN | LKSG |
| 20 | 1 (0.3) | 3 (0.9) | 1 (0.3) | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ |
| 36 | 1 (0.3) | 1 (0.3) | 0 (0.0) | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ |
| 46 | 1 (0.3) | 3 (0.9) | 2 (0.6) | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ |
| 69 | 16 (5.6) | 33 (9.5) | 32 (9.0) | ─ | ─ | ─ | ─ | ---A | ─ | ─ | L--- | V--- |
| 87 | 1 (0.3) | 3 (0.9) | 1 (0.3) | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ |
| 95 | 3 (1.0) | 3 (0.9) | 0 (0.0) | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ |
| 102 | 1 (0.3) | 1 (0.3) | 1 (0.3) | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ | ─ |
| 2a | 16 (5.6) | 29 (8.3) | 46 (13.0) | ─ | ─ | ─ | ─ | ---A | ─ | -A– | ─ | V--- |
| 3a | 40 (13.9) | 56 (16.1) | 40 (11.3) | -A– | ---T | ─ | ─ | ---A | ─ | -A– | ─ | V--- |
| 47a | 40 (13.9) | 18 (5.2) | 39 (11.2) | -A– | ---T | ─ | ─ | ---A | ─ | -A– | ─ | V--- |
aPenicillin-intermediate according to CLSI breakpoints.
Discussion
In Malaysia, pneumonia is the third leading cause of mortality in infant and ranked the second in toddler in 2016. However, surveillance data of pneumococcal disease in Malaysia is scarce and most of the study was performed before the implementation of PVC13 conjugate vaccine. There is an urgent need to study the pneumococcal serotype distribution in children in Malaysia. Our study has included S. pneumoniae isolates obtained from pediatric patients showing signs of LRTI. The limitation that we encountered during the study was the inability to confirm the colonizing or invasive state of the S. pneumoniae isolates, as the isolates were only obtained from nasopharyngeal specimens. Further, not all of the pediatric patients fulfilled the strict definition of pneumonia, despite showing signs of LRTI. Nonetheless, the serotype, genetic, and antimicrobial resistance data that we report are worth a note to the health-care providers, as colonizers may turn pathogenic and cause disease in susceptible individuals. Although our study represents a single-center based study, the data obtained may provide the first insight into current trend of S. pneumoniae serotypes causing LRTI or colonizing hospitalized children with respiratory diseases in our local community.
In this study, surprisingly, we found that the serotypes that included in PCV7 and PCV13 vaccines (Table 2) remain persistent despite of the introduction of the vaccines and this phenomenon is consistent with the previous study [28]. Elimination of disease-causing serotypes might not be achieved as the vaccine is not included in the routine national childhood immunization schedule in most of the Asian region. This may also be due to the high cost of conjugate vaccines that are available in private markets [28,29].
Furthermore, there was also slight increase of new serotypes observed for the non-vaccine serotypes (NVTs), namely 9N/L and 23B, whereby serotype 9N was first reported in Malaysia in 2014 [30]. This finding is somehow consistent with the post-PCV surveillance studies reported [31,32] and one serotype 9N/L were detected in this study. Furthermore, a healthy cohort study in Hong Kong revealed that 89.3% of the isolates from nasopharyngeal carriage were non-PCV13 serotypes [33]. Similar phenomenon was observed in the Netherlands, where serotypes 6C and 23B are currently prevalent, followed by serotypes 11A, 15B, and 23A, which are less invasive [34].
Capsular switching is a common occurrence in S. pneumoniae isolates whereby it involves a horizontal recombination of capsular DNAs via transformation, for instance, change of a serotype of a single clone happens by alteration or exchange of its cps locus. Genotyping is vital to closely monitor the clonal distribution of the pneumococcal population in Malaysia and to predict the impact of pneumococcal vaccine in Malaysia. As for most of the genotyping method, REP-PCR fingerprinting has been reported to have high discriminatory power, reliable, rapid, and reproducible [35,36].
As shown in Figure 1, cluster analysis based on REP data showed genetic diversity in 95 S. pneumoniae strains and closely related strains were able to be differentiated. This study reflected the pneumococcal causing strains isolated in UMMC were less likely to be related. Referring to the predominant serotypes, isolates were heterogeneous, only few variants accounting for most or all of the isolates within the same serotype (Figures 2 and 3). Strains that belonged to the same clone having the same fingerprint pattern, possibly indicating strong clonal spread is happening among children within the community. Similar finding was reported in Hungary in 2015 whereby most of the 19A strains collected from 40 nurseries were clonal with only a few small clusters, providing evidence of clonal spread in the community [37].
Interestingly, some REP clusters shared similar isolated years. For instance, the serotype 23F isolates which isolated in 2013 formed a distinct cluster (D1, Figure 3(b)) while isolates from 2014 formed another cluster (D2, Figure 3(b)). For serotype 6A/B, B1 cluster comprised isolates obtained from the isolation year 2014 while another larger cluster (B2) comprised of isolates from 2013 and 2015 (Figure 3(a)). These results suggesting that an endemic persistence is happening whereby a clone can reappear at different times and is in agreement with a previous study [38]. We also found a similar REP profile for REP-36 and REP-37 which represents serotype 9N/L and 23F, respectively. This could be attributed to the capsular switching phenomenon occurred which is parallel to the previous finding in 2013 whereby serotype change was observed from 9N to 23F, possibly due to a potential recombination import within dexB and aliA gene [39].
Based on the cluster analysis, three penicillin-intermediate strains, SP2, SP3, and SP47, were grouped into three different clusters, which reported to be serotype 6A/B, NT, and 19F, respectively. These findings are in agreement with the previous finding by Yu et al. in 2019 whereby penicillin-resistance strains mostly belonged to serotype 19F and 6A/B. In addition to that, Nguyen et al. also reported serotype 19F strains that were resistant to beta-lactam antimicrobials were commonly found in children in Vietnam [40,41]. Another previous study based on global populations in 2013 also revealed penicillin resistance strains have been shown to associate with specific pneumococcal serotypes, particularly 6A, 6B, 9V, 14, 15A, 19F, and 23F and the highest rate of penicillin resistance were in Africa [42].
Previous literature has suggested that pbp1a, pbp2b, and pbp2x are the major contributing factors of penicillin resistance. The conserved motifs, SXXK, SXN, and KTSG were altered, hence lowering the binding affinity toward penicillin [12,13]. In our study, most of the mutations found in all three pbp1a, pbp2b, and pbp2x genes are globally similar to those described previously, namely single substitution in conserved SSN motif of pbp2b as well as substitution of STMK motif and LKSG motif in pbp2x. As published, these substitutions have been the predominant motifs in causing intermediate penicillin resistance [43]. Based on the sequencing result of pbp1a, pbp2b, and pbp2x, two out of three intermediate strains shared a similar pattern of amino acid alteration closed to the conserved motifs. Identical mutation patterns were published by Diawara et al. previously. The absence of amino acid substitution in the conserved SVVK and KTG motif in this study is identical to previous report, hence suggesting that these two motifs are not involved in penicillin resistance development [44]. On the other hand, no mutations were observed in pbp1a gene of SP2 isolate, and this probably due to pbp1a only involves in developing high penicillin-resistance and it can only be mediated by the presence of mutation on pbp2b and pbp2x [43].
None of the strains in this study were resistant toward penicillin, which is consistent with recent findings in Malaysia [30]. The rate is lower than that reported by the Malaysian National Surveillance of Antimicrobial Resistance (NSAR). In fact, NSAR reported an average of 1.3% of PRSP in Malaysia from 2013 to 2015 [45]. The lower resistance rate obtained might be possibly due to the sample collection area that only focused on one medical center and the sample size of the study was relatively small, within a limited time frame.
In conclusion, this study provides an insight into the genotypic and phenotypic features of the respiratory isolates of S. pneumoniae in Malaysia. The use of vaccine in the country is highly recommended as the VTs are among the most dominant serotypes in our study. It is believed to be able to provide significant protection toward children <2 years of age and creates a herd immunity for the community. However, continued surveillance with larger geographical area is necessary in order to monitor the NVT trends caused by vaccine-induced selection pressure in the future.
Funding Statement
This study was supported by University of Malaya Research Grant [UMRG: RP026B-14HTM]; and Post Graduate Research Grant [PPP: PG159-2015B].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Wahl B, O’Brien KL, Greenbaum A, et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000–15. Lancet Glob Health. 2018;6(7):e744–e757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Berger S, Goekeri C, Gupta SK, et al. Delay in antibiotic therapy results in fatal disease outcome in murine pneumococcal pneumonia. Crit Care. 2018;22:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Weiser JN. The pneumococcus: why a commensal misbehaves. J Mol Med. 2010;88:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Park IH, Pritchard DG, Cartee R, et al. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J Clin Microbiol. 2007;45(4):1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu C, Xiong X, Xu W, et al. Serotypes and patterns of antibiotic resistance in strains causing invasive pneumococcal disease in children less than 5 years of age. PloS One. 2013;8(1):e54254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Makwana A, Ladhani SN, Kapatai G, et al. Rapid spread of pneumococcal nonvaccine serotype 7C previously associated with vaccine serotype 19F, England and Wales. Emerg Infect Dis. 2018;24(10):1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].van der Linden M, Falkenhorst G, Perniciaro S, et al. Effects of infant pneumococcal conjugate vaccination on serotype distribution in invasive pneumococcal disease among children and adults in Germany. PloS One. 2015;10(7):e0131494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brueggemann AB, Peto TE, Crook DW, et al. Temporal and geographic stability of the serogroup-specific invasive disease potential of Streptococcus pneumoniae in children. J Infect Dis. 2004;190(7):1203–1211. [DOI] [PubMed] [Google Scholar]
- [9].Department of Statistics Malaysia. Statistics on causes of death Malaysia. Malaysia: The Office of Chief Statistician; 2017. Available from: https://www.dosm.gov.my/v1/index.php?r=column/cthemeByCat&cat=401&bul_id=Y3psYUI2VjU0ZzRhZU1kcVFMMThGUT09&menu_id=L0pheU43NWJwRWVSZklWdzQ4TlhUUT09 [Google Scholar]
- [10].Shiow CT. Stopping Pneumococcus. 2018. Available from: https://www.thestar.com.my/lifestyle/health/2018/04/30/stopping-pneumococcus [Google Scholar]
- [11].Kosowska K, Jacobs M, Bajaksouzian S, et al. Alterations of penicillin-binding proteins 1A, 2X, and 2B in Streptococcus pneumoniae isolates for which amoxicillin MICs are higher than penicillin MICs. Antimicrob Agents Chemother. 2004;48(10):4020–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhanel GG, Wang X, Nichol K, et al. Molecular characterisation of Canadian paediatric multidrug-resistant Streptococcus pneumoniae from 1998–2004. Int J Antimicrob Agents. 2006;28(5):465–471. [DOI] [PubMed] [Google Scholar]
- [13].Zhou X, Liu J, Zhang Z, et al. Molecular characteristics of penicillin-binding protein 2b, 2x and 1a sequences in Streptococcus pneumoniae isolates causing invasive diseases among children in Northeast China. Eur J Clin Microbiol Infect Dis. 2016;35(4):633–645. [DOI] [PubMed] [Google Scholar]
- [14].Albrich WC, Monnet DL, Harbarth S. Antibiotic selection pressure and resistance in Streptococcus pneumoniae and Streptococcus pyogenes. Emerg Infect Dis. 2004;10(3):514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Appelbaum PC. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clinl Infect Dis. 1992;15(1):77–83. [DOI] [PubMed] [Google Scholar]
- [16].Chiba N, Morozumi M, Shouji M, et al. Changes in capsule and drug resistance of pneumococci after introduction of PCV7, Japan, 2010–2013. Emerg Infect Dis. 2014;20(7):1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Obolski U, Lourenço J, Thompson C, et al. Vaccination can drive an increase in frequencies of antibiotic resistance among nonvaccine serotypes of Streptococcus pneumoniae. Proc Nat Acad Sci. 2018;115(12):3102–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Le C-F, Palanisamy NK, Yusof MYM, et al. Capsular serotype and antibiotic resistance of Streptococcus pneumoniae isolates in Malaysia. PloS One. 2011;6(5):e19547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dube FS, van Mens SP, Robberts L, et al. Comparison of a real-time multiplex PCR and sequetyping assay for pneumococcal serotyping. PloS One. 2015;10(9):e0137349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144(11):3049–3060. [DOI] [PubMed] [Google Scholar]
- [21].Subramaniam P, Jabar KA, Kee BP, et al. Serotypes & penicillin susceptibility of Streptococcus pneumoniae isolated from children admitted to a tertiary teaching hospital in Malaysia. Indian J Med Res. 2018;148(2):225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dias CA, Teixeira LM, Carvalho M, et al. Sequential multiplex PCR for determining capsular serotypes of pneumococci recovered from Brazilian children. J Med Microbiol. 2007;56(9):1185–1188. [DOI] [PubMed] [Google Scholar]
- [23].Hanieh S, Hamaluba M, Kelly DF, et al. Streptococcus pneumoniae carriage prevalence in Nepal: evaluation of a method for delayed transport of samples from remote regions and implications for vaccine implementation. PloS One. 2014;9(6):e98739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kong F, Wang W, Tao J, et al. A molecular-capsular-type prediction system for 90 Streptococcus pneumoniae serotypes using partial cpsA–cpsB sequencing and wzy- or wzx-specific PCR. J Med Microbiol. 2005;54(4):351–356. [DOI] [PubMed] [Google Scholar]
- [25].Miernyk K, DeByle C, Harker-Jones M, et al. Serotyping of Streptococcus pneumoniae isolates from nasopharyngeal samples: use of an algorithm combining microbiologic, serologic, and sequential multiplex PCR techniques. J Clin Microbiol. 2011;49(9):3209–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Navia MM, Capitano L, Ruiz J, et al. Typing and characterization of mechanisms of resistance of Shigella spp. isolated from feces of children under 5 years of age from Ifakara, Tanzania. J Clin Microbiol. 1999;37(10):3113–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].CLSI . Performance standards for antimicrobial susceptibility testing. 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- [28].Jauneikaite E, Jefferies J, Hibberd M, et al. Prevalence of Streptococcus pneumoniae serotypes causing invasive and non-invasive disease in South East Asia: a review. Vaccine. 2012;30(24):3503–3514. [DOI] [PubMed] [Google Scholar]
- [29].Lin T-Y, Shah NK, Brooks D, et al. Summary of invasive pneumococcal disease burden among children in the Asia-Pacific region. Vaccine. 2010;28(48):7589–7605. [DOI] [PubMed] [Google Scholar]
- [30].Jefferies JM, Yusof MYM, Sekaran SD, et al. Novel clones of Streptococcus pneumoniae causing invasive disease in Malaysia. PloS One. 2014;9(6):e97912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ladhani SN, Collins S, Djennad A, et al. Rapid increase in non-vaccine serotypes causing invasive pneumococcal disease in England and Wales, 2000–17: a prospective national observational cohort study. Lancet Infect Dis. 2018;18(4):441–451. [DOI] [PubMed] [Google Scholar]
- [32].Liyanapathirana V, Nelson EAS, Ang I, et al. Emergence of serogroup 15 Streptococcus pneumoniae of diverse genetic backgrounds following the introduction of pneumococcal conjugate vaccines in Hong Kong. Diagn Microbiol Infect Dis. 2015;81(1):66–70. [DOI] [PubMed] [Google Scholar]
- [33].Chan K, Ip M, Chong P, et al. Nasopharyngeal colonisation and antimicrobial resistance of Streptococcus pneumoniae in Hong Kong children younger than 2 years. Hong Kong Med J. 2018;24(5 Supplement 6):4–7. [PubMed] [Google Scholar]
- [34].Vissers M, Wijmenga-Monsuur AJ, Knol MJ, et al. Increased carriage of non-vaccine serotypes with low invasive disease potential four years after switching to the 10-valent pneumococcal conjugate vaccine in The Netherlands. PloS One. 2018;13(3):e0194823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rodriguez M, Hogan PG, Satola SW, et al. Discriminatory indices of typing methods for epidemiologic analysis of contemporary Staphylococcus aureus strains. Medicine (Baltimore). 2015;94:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tang Y-W. Progress toward rapid and accurate Staphylococcus aureus strain typing. Clin Chem. 2009;55(12):2074–2076. [DOI] [PubMed] [Google Scholar]
- [37].Tóthpál A, Laub K, Kardos S, et al. Epidemiological analysis of pneumococcal serotype 19A in healthy children following PCV7 vaccination. Epidemiol Infect. 2016;144(7):1563–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Villalón P, Valdezate S, Medina-Pascual MJ, et al. Clonal diversity of nosocomial epidemic Acinetobacter baumannii strains isolated in Spain. J Clin Microbiol. 2011;49(3):875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wyres KL, Lambertsen LM, Croucher NJ, et al. Pneumococcal capsular switching: a historical perspective. J Infect Dis. 2012;207(3):439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nguyen HAT, Fujii H, Vu HTT, et al. An alarmingly high nasal carriage rate of Streptococcus pneumoniae serotype 19F non-susceptible to multiple beta-lactam antimicrobials among Vietnamese children. BMC Infect Dis. 2019;19(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yu -Y-Y, Xie X-H, Ren L, et al. Epidemiological characteristics of nasopharyngeal Streptococcus pneumoniae strains among children with pneumonia in Chongqing, China. Sci Rep. 2019;9(1):3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hackel M, Lascols C, Bouchillon S, et al. Serotype prevalence and antibiotic resistance in Streptococcus pneumoniae clinical isolates among global populations. Vaccine. 2013;31(42):4881–4887. [DOI] [PubMed] [Google Scholar]
- [43].Hakenbeck R, Brückner R, Denapaite D, et al. Molecular mechanisms of β-lactam resistance in Streptococcus pneumoniae. Future Microbiol. 2012;7(3):395–410. [DOI] [PubMed] [Google Scholar]
- [44].Diawara I, Nayme K, Katfy K, et al. Analysis of amino acid motif of penicillin-binding proteins 1a, 2b, and 2x in invasive Streptococcus pneumoniae nonsusceptible to penicillin isolated from pediatric patients in Casablanca, Morocco. BMC Res Notes. 2018;11(1):632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Institute for Medical Research (IMR). National Antibiotic Resistance Surveillance Report 2016. Kuala Lumpur: Ministry of Health Malaysia; 2016. [Google Scholar]


