Abstract
Objective.
Intracellular patch-clamp electrophysiology, one of the most ubiquitous, high-fidelity techniques in biophysics, remains laborious and low-throughput. While previous efforts have succeeded at automating some steps of the technique, here we demonstrate a robotic ‘PatcherBot’ system that can perform many patch-clamp recordings sequentially, fully unattended.
Approach.
Comprehensive automation is accomplished by outfitting the robot with machine vision, and cleaning pipettes instead of manually exchanging them.
Main results.
the PatcherBot can obtain data at a rate of 16 cells per hour and work with no human intervention for up to 3 h. We demonstrate the broad applicability and scalability of this system by performing hundreds of recordings in tissue culture cells and mouse brain slices with no human supervision. Using the PatcherBot, we also discovered that pipette cleaning can be improved by a factor of three.
Significance.
The system is potentially transformative for applications that depend on many high-quality measurements of single cells, such as drug screening, protein functional characterization, and multimodal cell type investigations.
Keywords: patch-clamp, robotics, machine vision
Introduction
The ability to profile individual cells has been transformative for investigating intact and diseased tissue, screening drugs, and studying cell types [1–4]. Existing microscopy-based assays typically analyze cells based on fluorescence; however, for many drug screening and cell profiling applications, electrical properties of cells must also be measured [5, 6]. The highest-fidelity technique to capture the electrical activity of single cells is whole-cell patch clamp recording which can report current and voltage fluctuations at a spatiotemporal resolution beyond the capability of other techniques [7]. The whole-cell configuration also uniquely offers direct molecular access to the cell membrane and cytosol, enabling fine control of the intracellular milieu. Among other uses, it is a gold-standard technique for screening and validating novel indicators and effectors of electrical activity such as opsins and fluorescent sensors [8–13]. However, the tradeoff for superior signal quality is low throughput since patch-clamp is notoriously laborious and time-consuming, precluding its widespread use for high-throughput screening [14].
Patch-clamp is low-throughput primarily due to its manual nature, in contrast to microscopy- and cytometry-based assays. It requires the use of micromanipulators to steer pipettes to individual cells of interest, pipette suction (typically, by mouth) to form a seal with the cell membrane, and pipette replacement after every recording. Further, when studying cells in intact tissue such as brain slices, additional care must be taken to avoid neighboring cells and compensate for tissue deformation. With significant practice, investigators become adept at these tasks but performing patch-clamp recordings is low-throughput (on the order of 10 recorded cells per day [15]), and highly variable in success rate (~30%–90% of attempts are successful) even for experienced users [16–18].
Parts of the patch-clamp technique have been automated by us and other groups but fully unsupervised operation has heretofore not been achieved [19]. Dissociated cells can be studied with planar patch-clamp systems that are fully automated but cannot be used for adherent cells or cells in tissue [20]. For these preparations, a computer-assisted system was previously developed to automate pressure control and aspects of actuator positioning to enable up to 12 cells to be patch-clamped simultaneously [21]. Similarly, our group has previously developed variations of the Autopatcher and ImagePatcher, robotic systems for performing blind and image-guided recordings, respectively [16, 22, 23]. However, these solutions and others [24–27] only automate several steps of a typical experiment. While these automation technologies do save time, they still cannot work unsupervised for more than a few minutes.
Accordingly, the main contribution of this paper is the development of the PatcherBot, a fully automated patch-clamp electrophysiology system that enables the user to walk away from the experiment after choosing target cells. In contrast to previous methods which have automated portions of a single trial, the PatcherBot performs multiple patch-clamp trials sequentially. This system is enabled by two advances. First, pipette cleaning [28] circumvents the need to manually swap pipettes between attempts. Second, machine vision techniques for cell detection, tracking, and pipette identification enable the automation of the most difficult manual aspects of patch-clamp recording. We have previously performed pilot trials of the system in cultured cells [28]. Here, we demonstrate its usability and quantify its throughput in a variety of electrophysiology experiments. The throughput improvement and commoditization offered by the PatcherBot could thus enable the application of single-cell electrophysiology to fields requiring large-scale data collection such as drug screening, cell characterization, and neuronal connectivity profiling.
Materials and methods
PatcherBot hardware
The robotic system was based on a conventional electrophysiology setup (SliceScope Pro 3000, Scientifica Ltd), comprising two motorized PatchStar micromanipulators mounted on a motorized stage. Samples (cultured cells and brain slices) were imaged using a 40 × objective (LUMPLFL40XW/IR, NA 0.8, Olympus) on a motorized focus drive, illuminated under differential interference contrast (DIC) with an infrared light-emitting diode (Scientifica), and captured with a Rolera Bolt camera (QImaging). Köhler illumination was set up and routinely checked to ensure consistent illumination. A peristaltic pump (120S/DV, Watson-Marlow) was used to perfuse cells and slices with buffer solution. Recordings were acquired using the Multiclamp 700b amplifier (Molecular Devices) and digitized to a USB-6221 OEM data acquisition board (National Instruments). We performed two main hardware modifications to the conventional Scientifica electrophysiology workstation to enable full automation. First, we built a custom two-channel pipette pressure controller. For each pipette, pressure was controlled by a ±10 psi regulator (QPV1TBNEEN10P10PSGAXL, ProportionAir) using an analog (0–10 V) control signal. The control signal for each regulator was generated by a microcontroller (Arduino Uno, Arduino) via a digital-to-analog converter (MAX539, Maxim Integrated). Individual pressure regulators for each pipette were necessary to ensure that different pressures could be maintained on each pipette, e.g. if one pipette is forming a seal (atmospheric or negative pressure), and another is approaching a neuron (positive pressure). The second modification was a custom-machined electrophysiology chamber (supplementary figure 1 (stacks.iop.org/JNE/16/046003/mmedia)) with small side chambers for cleaning and rinsing solutions. The chamber could be used for up to four manipulators.
PatcherBot software
A finite state machine architecture was implemented to repeatedly patch-clamp user-selected cells (supplementary figure 2). The overall steps in the block diagram were unchanged between HEK cell preparations and brain slice preparations; however, specific details of the ‘calibration’, ‘descend pipette’, ‘approach cell’ and ‘establish seal’ states necessitated adjustments. The software (written in LabVIEW, National Instruments) interfaces with the MATLAB-based cell tracking algorithm [29], communicates with the stage, manipulators, and pipette pressure controller with a serial interface, and communicates with the amplifier using an ActiveX interface.
Initialization
Upon software initialization, the user is asked to load the pipette and perform an initial calibration (calibrate, supplementary figure 2). To perform the calibration, the user first centers the pipette under the objective and the software zeros the coordinate systems of the manipulator and stage. At this time, it also stores an image of the pipette to be used for machine vision. Then, the user defines a ‘HOME’ position for the pipette. It serves as an intermediate position for the pipette between the sample chamber and the clean/rinse baths. The position must be sufficiently far from the sample to avoid pipette collisions with the objective. The user then identifies the positions of the clean/rinse baths. These steps are necessary for every pipette due to inter-pipette differences in taper and tip geometry. The pipette is subsequently returned to the ‘HOME’ position.
The user then chooses a ‘parking plane’, an XY plane normal to the slice surface (or cover slip for HEK cells) defined by z = zpark where zpark is ideally 20–50 µm above the tissue surface (or cover slip). Pipettes will ‘park’ on this plane prior to patching in order to perform the pipette refocusing routine (supplementary figure 4(c)). Finally, the user picks any number of cells, up to 10 per pipette when using Alconox as the cleaning agent (pick cells, supplementary figure 2). Selected cells are outlined with a red circle and an index number (supplementary figure 3). When a cell is picked, the program stores the coordinates of each cell as well as an image of the cell (200 × 200 pixels) for later machine vision use. Overall, the initialization procedure takes ~4 min for single-manipulator operation and ~8 min for dual-manipulator operation.
Unattended operation
Once the PatcherBot is initialized and loaded with cell locations, the remaining procedure is fully automated. First, a user-picked cell is brought into focus (focus on cell i, supplementary figure 2) and a recalibration is performed (recalibrate, supplementary figures 2 and 4(a)). When the stage focuses on a target cell, there is error (0–10 µm) that is caused by one or more of the following factors: (1) drift of tissue slice or the cover slip, (2) mechanical drift in the optical system, (3) imprecision in the stage actuation and (4) inaccuracies in the manual calibration. The error is corrected by finding the true cell position using normalized 2D cross-correlation (NI Vision Development Module, IMAQ Find Pattern) and moving the stage to that position (supplementary figure 4(b)). If the cell is not found in the field of view, the focusing plane is changed in 2 µm step increments over 20 µm. When the cell is found, the coordinate system of the stage and every active pipette undergo a translation by (xfound − xorig, yfound − yorig, zfound − zorig) where (xfound, yfound, zfound) are the coordinates of the detected cell and (xorig, yorig, zorig) are the original coordinates where the cell was expected to be. After the coordinate transform, the stage is focused on the true position of the cell. If the cell is not found in any plane, the stage returns to the original expected coordinates of the cell (xorig, yorig, zorig).
The stage subsequently focuses on (xfound, yfound, zpark), directly above the cell, in the ‘parking plane’ the user selected earlier. The pipette moves to those coordinates with some error (typically, 0–20 µm away from true location). This is due to two major factors: (1) micromanipulator drift or low repeatability and (2) calibration-based errors. The calibration-based errors stem from unavoidable angular inaccuracies of the initial manual calibration step. To correct for the error, we use the same refocusing technique as in the cell refocusing step (supplementary figure 4(c)). Briefly, the pipette tip is found using machine vision, moved to the true (xfound, yfound, zpark) location, and the coordinate system of the manipulator is adjusted to reflect the true pipette position. If the pipette is not found in any plane, it returns to the original coordinates and the software assumes that it is already centered.
In HEK cell experiments, typically performed on a monolayer of cells adherent to a glass cover slip, the pipette subsequently descends to 15 µm above the target cell (descend pipette, supplementary figure 2), at which point, a ‘blind’ cell approach algorithm is engaged (supplementary figure 5(a)). The pipette moves down in 1 µm steps until the targeted cell is encountered, based on a three-step resistance-based threshold, similar to the original Autopatcher algorithm [22]. In slice experiments, the pipette instead first moves in the XY plane above the slice such that it can approach the cell on a trajectory parallel to the pipette axis. The trajectory is planned such that the pipette arrives 15 µm above the target cell in the brain slice. As the pipette arrives, it deforms the surrounding brain tissue both due to mechanical insertion and due to pressurized intracellular solution being ejected from the pipette. In previous patch-clamp automation efforts, any such deformation would result in a failed attempt because the algorithm had no way of determining in which direction the cell moved. Here, the PatcherBot engages the cell tracker to observe cell boundaries in real time [29] (supplementary figure 5(b)). The cell tracker is comprised of a prefiltering stage to reduce interference due to the surrounding tissue, followed by deconvolution via a custom dynamic filtering algorithm and a thresholding step to accurately identify membrane boundaries. The cell tracker outputs the detected centroid of the neuron and moves the pipette towards the centroid (in x and y only) in 1 µm steps until the pipette tip is found to be within 2 µm (in x and y) of the cell centroid. Once the pipette is aligned with the cell in X and Y, it moves down in 1 µm steps until the neuron is detected with a resistance-based method. If the cell centroid moves at any time, the algorithm re-aligns the pipette with the cell centroid.
After the target cell is detected, sealing and break-in algorithms are engaged (establish seal, break in, supplementary figure 2). To establish a gigaseal, suction is applied with the pipette pressure controller based on the slope of the resistance trace over time (sampled at 2 Hz). Small resistance slopes (<2.5 MΩ s−1 in HEK cells, <0.25 MΩ s−1 in slices) indicate a slow or failing seal, thus more suction is applied in 5 mBar increments up to a maximum (−70 mBar). Large slopes (>40 MΩ s−1) indicate successful seal formation and suction is reduced in 5 mBar increments. For slopes between these extremes, suction is maintained. Once the measured resistance reaches 1 GΩ, the algorithm waits (5 s) and proceeds to the break-in state (break in, supplementary figure 2). Break-in is accomplished by short pulses of suction (100–1000 ms, −400 mBar). A break-in is considered successful when the measured resistance drops to under 800 MΩ and the holding current remains low (> −200 pA at −50 mV in HEK cells, −70 mV in slices). The whole-cell electrophysiology state (ephys, supplementary figure 2) consists of a voltage clamp protocol where cell parameters (access resistance, membrane resistance, holding current) are measured as well as a current clamp protocol (0 pA for 1 s, −300 to +300 pA step for 1 s, 0 pA for 1 s).
Pipette cleaning is performed as previously described [28] (clean pipette, supplementary figure 2) except due to the long unattended operation time, we needed a way to account for fluid evaporation (measured to be ~23 µl h−1). To do this, we relied on the observation that pipette transient capacitive current increases consistently (10 s of pA) when it touches fluid. Thus, the algorithm would descend the pipette into the clean and rinse bath at the user-provided XY coordinates until detecting a sudden increase in the transient current.
Dual manipulator operation
We made two changes to the single-manipulator patcherBot algorithm to enable it to be used for dual patching. First, a ‘pick cell’ state was added in which the algorithm decides which cell to target and which manipulator to use. This is needed because in single-manipulator trials, cells are simply patch-clamped in the order in which they were picked by the user, but for two manipulators, this could cause the pipettes to collide. The ‘pick cell’ state ensures that each manipulator is assigned to the cell closest to its home position from an array of un-patched cells. The second addition is ‘microscope reservation’ feature which ensures that each manipulator can ‘reserve’ the microscope stage and imaging system for the pick cell, calibration, pipette descent, and cell approach states. It is essential that each manipulator has complete control over the microscope during these steps since they rely on camera output. If a manipulator is ready for the ‘pick cell’ stage, but the microscope is reserved by the other manipulator, it must wait until the microscope is unreserved.
Culture and brain slice preparation
Human embryonic kidney (HEK293T) cells (American Type Culture Collection, Manassas, VA) were cultured as previously described [28]. Briefly, cells were cultured and passaged regularly in accordance with the manufacturer’s instructions. For patch-clamp recording, cells were grown on glass coverslips (12 mm diameter, No.2, VWR), and used within one week of splitting. Cells were not transfected.
All animal procedures were in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the Georgia Institute of Technology. For brain slice experiments, male mice (C57BL/6, P31–P46, Charles River) were anesthetized with isofluorane, and the brain was quickly removed and mounted in agar (2% w/v). Coronal sections (300 µm thick) were then sliced on a compresstome (VF-300, Precisionary Instruments) while the brain was submerged in ice-cold sucrose solution containing (in mM) 40 NaCl, 4 KCl, 1.25 NaH2PO4·H2O, 7 MgCl2, 25 NaHCO3, 10 D-Gluocse, 0.5 CaCl2·2H2O, 150 Sucrose (pH 7.3–7.4, 300–310 mOsm). The slices were incubated at 37 °C for 1 h in neuronal artificial cerebro-spinal fluid (ACSF) consisting of (in mM) 124 NaCl, 2.5 KCl, 1.25 NaH2PO4·H2O, 1.3 MgCl2, 26 NaHCO3, 10 D-Gluocse, 2 CaCl2·2H2O, 1 L-Ascorbate·H2O (pH 7.3–7.4, 290–300 mOsm). Prior to recording, the slices were maintained at room temperature for at least 15 min (22 °C–25 °C). The sucrose solution and neuronal ACSF were bubbled with 95% O2/5% CO2. Recordings were performed in mouse primary area V1, hippocampal area CA3, and thalamus.
Patch-clamp recording
Borosilicate pipettes were pulled on the day of the experiment using a horizontal puller (P-97, Sutter Instruments) to a resistance of 4–8 MΩ. For HEK cells, the intracellular solution was composed of (in mM): 120 KCl, 2 MgCl2, 10 EGTA, 10 HEPES (pH: 7.2–7.3, 290–300 mOsm) and recordings were performed at room temperature with constant superfusion of ACSF: (in mM) 161 NaCl, 10 HEPES, 6 D-Glucose, 3 KCl, 1 MgCl2, 1.5 CaCl2 (pH: 7.4). For neurons in brain slices, the intracellular solution was composed of (in mM) 135 K-Gluconate, 10 HEPES, 4 KCl, 1 EGTA, 0.3 Na-GTP, 4 Mg-ATP, 10 Na2-phosphocreatine (pH: 7.2–7.3, 290–300 mOsm). Recordings were performed at room temperature with constant superfusion of oxygenated neuronal ACSF.
Performance benchmarking
To evaluate PatcherBot performance, the user chose 10 cells for single-manipulator HEK cell studies, between 3 and 10 for brain slice studies, between 18 and 24 for dual-manipulator HEK cell studies, and up to 30 for single-manipulator HEK cell studies with Tergazyme. An attempt was considered successful upon the successful completion of the break-in state (see Unattended operation).
For machine vision benchmarking, a pipette or cell refocusing attempt was considered successful if there was sufficient similarity between one image of the pipette taken in focus in the beginning of the experiment and after refocusing. Image similarity was quantified using the IMAQ Find Pattern function in NI LabVIEW, which outputs a similarity score. The similarity score is the result of a normalized 2D cross-correlation algorithm, scaled to a value between 0 (no similarity) and 1000 (exact match). The similarity score threshold was empirically determined based on lighting conditions and set to between 700 and 900. In brain slices, cell area was measured manually after the experiment by outlining boundaries in saved cell images. Image contrast was found by calculating the range (min to max) of image histograms.
Throughput modeling
We created a computational model to extrapolate throughput for PatcherBot systems with more than two manipulators. Success rates and trial durations of pilot single-manipulaotr and two-manipulator PatcherBot experiments were used to set default model parameters. For HEK293 cells, the whole-cell success rate was set to 77% to be consistent with our empirical observations for a single-manipulator PatcherBot. In the simulations, unsuccessful attempts lasted 3.5 min while successful attempts lasted one minute longer to mimic running a whole-cell recording. Other time considerations included the 1 min microscope reservation time per manipulator and the 1 min of cleaning after each attempt. After ten cleans, the pipette would have to be replaced and re-calibrated which we modeled with a 4 min reloading time. Any setup time prior to starting the first attempt was not included in the model. We simulated 8 h of PatcherBot runtime, and computed an average hourly throughput rate. Monte Carlo simulations were run 100 000 times for each manipulator number using MATLAB (Mathworks) on a personal computer.
Statistics
The success rate of the PatcherBot as a function of (1) number of performed cleans, (2) lateral displacement, (3) cell area, (4) image quality, and (5) cell depth was assessed using a logistic regression model (MATLAB function mnrfit). An odds ratio (OR) was calculated for each category. An odds ratio can be interpreted as the odds of success improving given a 1-unit increase in the category; for example, a cell area OR of 1.5 indicates that the likelihood of the formation of a successful gigaseal or whole-cell increases by 1.5 with every 1 µm2 added to the cell area. Odds ratios are reported with 95% confidence intervals (CI). To assess the effect of the number of performed cleans on recording characteristics, we used a linear mixed effects model (MATLAB function fitlme). The response variables were Ra and TGS and the predictor variable was the reuse number. Analyses and statistics were computed using MATLAB (Mathworks). Data are presented as mean ± standard deviation unless otherwise stated. Data normality was assessed using the Anderson–Darling test. Probability values P < 0.05 were considered to be significant.
Results
System overview
The PatcherBot (figure 1(a), supplementary figure 1, supplementary video 1) was built by augmenting standard patch-clamp electrophysiology system with a custom-made pipette cleaning chamber and pressure control hardware [30]. Pipette cleaning was performed automatically using Alconox (Alconox Inc) as the detergent, as previously demonstrated [28]. For robotic control, a software package was written to interface with three-axis manipulators, XY stage, Z focus drive, pressure control system, amplifier, and camera.
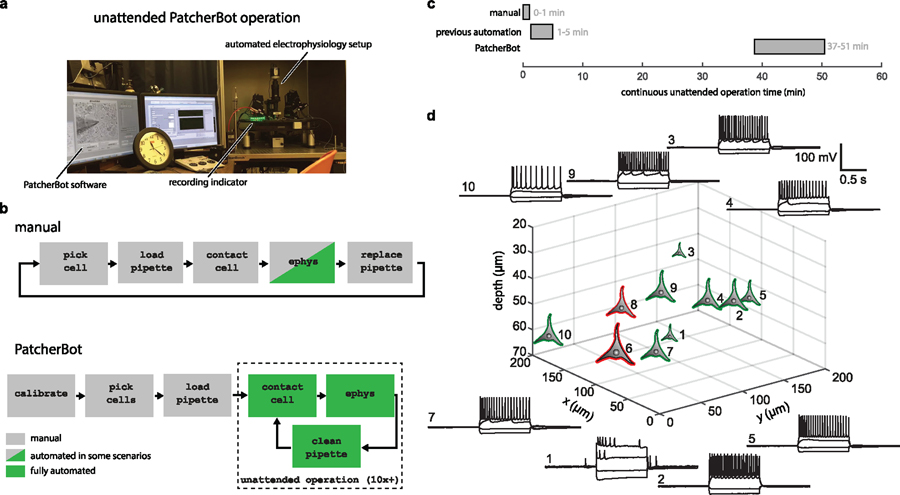
Figure 1.
PatcherBot system and operation. (a) Experimental setup: PatcherBot is built on a conventional Scientifica SliceScope electrophysiology system. Software performs unattended single-cell electrophysiology. Recording indicator lights up upon establishing a whole-cell configuration. (b) Simplified workflow of patch-clamp experiments. In manual experiments, only the electrophysiology (ephys) component is automated in some types of experiments. In the PatcherBot, a calibration state is added to enable unattended operation. See detailed block diagram (supplementary figure 2). (c) Comparison of approximate unattended operation time of the PatcherBot and conventional manual experiments as well as previous patch-clamp automation techniques. Recordings are assumed to be short (1 min) for all three modalities. Manual and previous automation timing information was taken from [21] (supplementary table 1). (d) Representative whole-cell recordings in brain slices obtained using the PatcherBot. Green neuron symbols represent successful whole-cell recordings; red symbols represent failed attempts. Cells are shown in a coordinate system that depicts their centroid location in the slice. The (0, 0, 0) point corresponds to the location where manual calibration was performed. Numbers represent order in which cells were chosen and patched (1, 2, 3, …). Cell 1 had low spike amplitude, likely due to an unhealthy cell or incomplete break-in.
PatcherBot software only requires user input for several minutes to perform basic calibration steps and choose cells (figure 1(b), supplementary figure 2). First, a manual calibration step is performed to align manipulator and stage coordinate systems. Second, the user selects cells of interest using the graphical user interface (supplementary figure 3). Subsequently, all steps of the patch-clamp procedure are done automatically, conferring a ~50-fold increase in unattended operation time over manual experiments and a ~10-fold increase over previous automation efforts (figure 1(c), supplementary table 1). At the end of the unattended experiment, the user can collect and process the data from all successfully recorded cells (figure 1(d)).
Single-cell electrophysiology in cultured cells
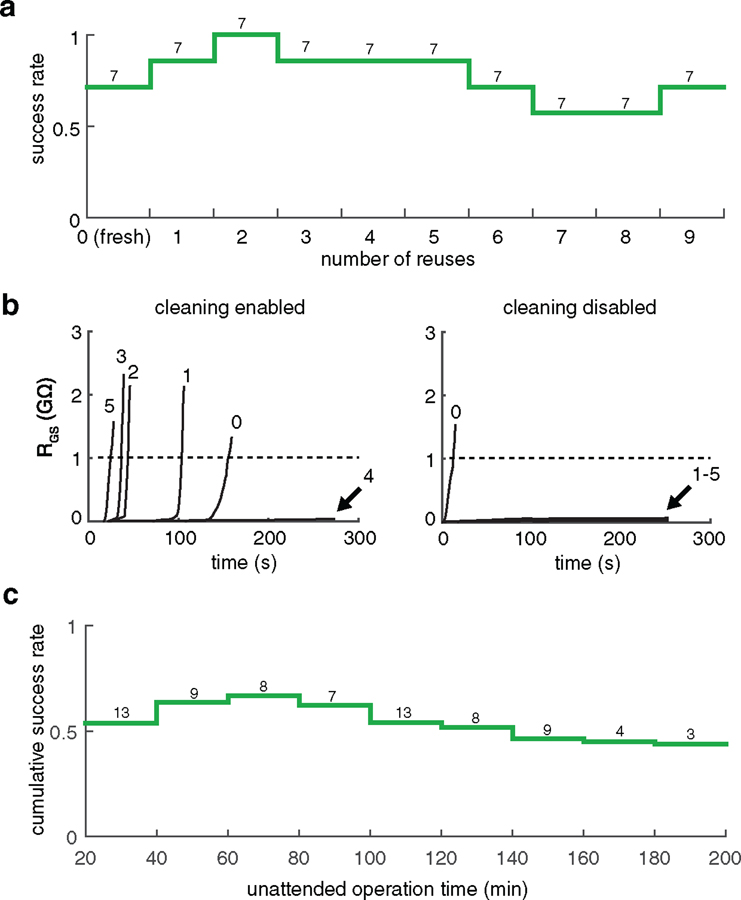
We first validated PatcherBot performance in HEK cells cultured on glass coverslips. After the user installed each pipette, performed the initial calibration, and selected cells, the PatcherBot was engaged with no further experimenter intervention. It achieved an overall success rate of 77% (n = 54 whole-cell recordings/70 attempts, 7 pipettes). This is comparable to the 60%–70% manual success rates often reported in similar experiments [15, 31], although differences in preparations preclude direct comparisons. The number of performed cleans was not a significant factor in determining whether a particular trial will be successful, suggesting that there was no degradation in performance over the ten attempts per pipette (figure 2(a)). In a separate experiment, we confirmed that disabling pipette cleaning prevented successful recordings (figure 2(b)).
Figure 2.
PatcherBot performance in cultured HEK cells. (a) Whole-cell success rate of pipettes over ten trials (nine reuses) in HEK cells. Reuses did not significantly decrease likelihood of whole-cell recording (odds ratio, OR = 1.17, CI: 0.13–1.18, P = 0.12). Numbers above columns indicate number of trials performed at each reuse number. (b) Seal resistances attained with and without cleaning. When cleaning is enabled (left), a pipette can achieve successive gigaseals (RGS ⩾ 1GΩ) on multiple cells. When cleaning is disabled (right) by draining Alconox from the clean bath, a gigaseal is obtained with a fresh pipette (0) but not on any other attempt (1–5). (c) Cumulative success rate in long-hold experiments. Increased unattended operation time contributed to slight diminishment in success rate (OR = 1.02, CI: 1.01–1.03, P = 0.0019, n = 74 attempts). Alconox was used as the cleaning agent.
We then demonstrated that the PatcherBot is suitable for drug discovery and characterization experiments. Drug studies often require long (5–30 min) patch-clamp recording experiments to allow for multiple drug application and washout steps and to study drug kinetics [32, 33]; we therefore programmed the PatcherBot to perform 20 min long recordings. It obtained whole-cell recordings for 98–182 min with an overall success rate of 43% (n = 32 whole-cell recordings/74 attempts, 7 pipettes, figure 2(c), supplementary video 2). The success rate was slightly diminished over ~1.5 h, perhaps due to cell health deterioration outside the culture medium.
To demonstrate how the PatcherBot could be used as a tool for high-throughput screening, we ran it for four hours and measured the throughput. The overall success rate was 67% (n = 4 four-hour experiments, n = 136 whole-cell recordings/202 attempts, 27 pipettes, supplementary video 1). During the four-hour experiments, the only user intervention was replacing the pipette and performing a recalibration approximately once every hour. Cells were held for short periods of time (1 min) during which a brief electrophysiological survey of a cell was performed. Based on these experiments, a user can expect on average 34 whole-cell recordings in 51 attempts over a span of 4 h.
Machine vision for targeting cells in intact tissue
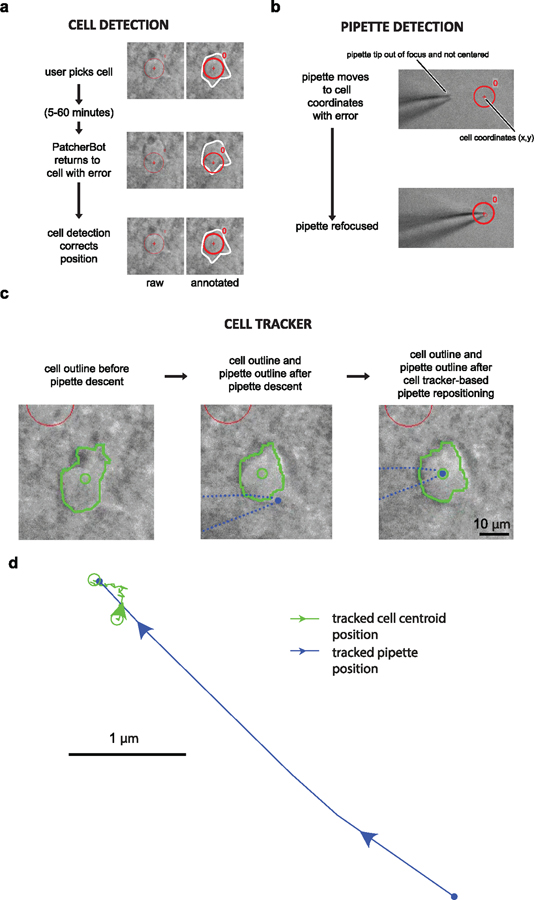
Brain slice recording tends to be more difficult than recordings in cultured cells, and thus could benefit more from full automation. Navigating pipettes to cells of interest in the 3D, heterogeneous environment of brain tissue is challenging even for trained operators. Computer-initiated navigation to target cells without feedback is error-prone due to the accumulation of mechanical errors in patch-clamp actuators. The PatcherBot corrects for these errors and cell motion in real time using machine vision (figure 3, [29]). Actuator errors (micromanipulator and stage) were mitigated by performing pipette auto-focus and cell auto-focus routines (99% and 79% success rate, respectively, with ~1 µm accuracy, n = 161 attempts, supplementary figures 4(b) and (c)). Real-time cell tracking compensated for cell displacement during the final approach of the pipette (supplementary figure 5, supplementary video 3). The two auto-focus routines and cell tracking were subsequently benchmarked together.
Figure 3.
Machine vision elements in brain slices. (a) Sequence of events for cell detection. After the user initially picks the cell (top panels), the system comes back to patch-clamp it after 5–60 min with some inaccuracy (middle panel). The cell detection procedure re-centers the cell (bottom panel). Images on the left show raw screen capture frames, images on the right are annotated for clarity. Red circle and crosshair show the expected cell location; white outline shows actual cell boundary. Circle diameter: 10 µm. (b) Before (top) and after (bottom) the pipette detection state. The algorithm successfully refocused on the pipette after it entered the field of view off-center. (c) Sample cell tracking results. See supplementary video 3 (cell tracker ON) for real-time video of this trial. Cell boundaries (green outline) are automatically tracked and the centroid (green circle) is computed. Pipette tip location on the screen is estimated from the manipulator position (blue dot). Left: cell boundary before pipette descent. Center: cell position and pipette position after pipette descent. Right: cell position and pipette position after trajectory adjustment. Red circle on top left of images is a different cell. (d) Cell centroid position (green) and pipette position (blue) during the ‘approach cell’ state. The pipette moves laterally towards the tracked cell centroid. Same attempt as in (c) and supplementary video 3.
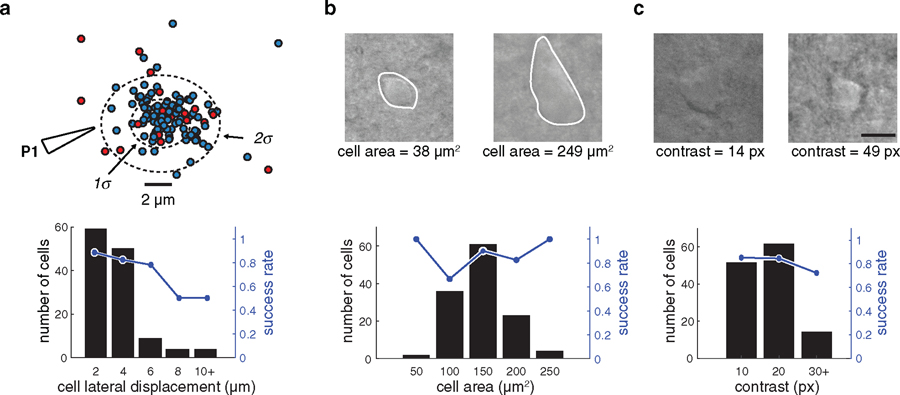
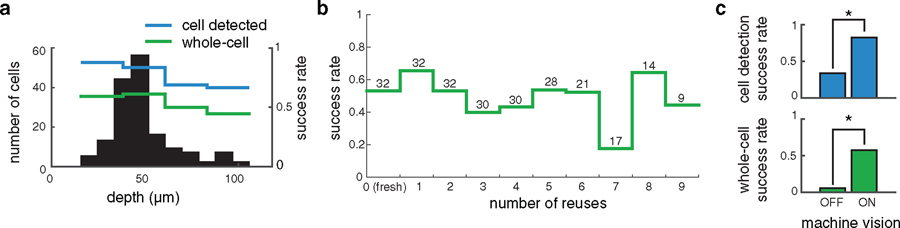
Brain tissue is deformed by pipette entry, which, if uncompensated, could lead to the PatcherBot missing the target cell. Machine vision successfully compensated for the motion in 133 cells/161 attempts (83%, supplementary figure 2) where a successful compensation was defined as the algorithm reaching the ‘establish seal’ state with the target cell. Overall, neurons in brain tissue moved by a magnitude of 2.5 ± 1.9 µm (n = 126 trials) in the XY plane in response to pipette descent. Large cell displacements (⩾4 µm) could not be fully compensated with machine vision and led to decreased success rate (figure 4(a)). Cells tended to displace in the direction of pipette approach, likely due to the pressure applied by the pipette on the tissue during the descent state. Depth, but not cell size was correlated with the magnitude of the displacement (depth versus displacement: r = 0.35, P = 0.0001; cell size versus displacement: r = −0.11, P = 0.20; Pearson’s r), indicating that cells deeper in the tissue tended to be moved more by the pipette than shallower cells.
Figure 4.
Benchmarking machine vision performance in brain slices. (a) Top: scatterplot of cell displacement (in XY plane) after pipette descent. Blue dots denote successful cell detection (algorithm reaching the ‘establish seal’ state but not necessarily whole-cell), red circles denote failed detection (pipette missing the cell). ‘P1’ shows the approximate direction of pipette 1 entering tissue. The 1σ and 2σ dotted ellipses represent 1 and 2 standard deviations of the displacements, respectively. Bottom: distribution of cell displacement. Most (87%) cells exhibited displacements of ⩽4 µm. Larger cell displacements in tissue significantly decreased the likelihood of successful cell detection (OR = 1.3, CI: 1.05–1.72, P = 0.0187). (b) Top: sample cell images and outlines of cells (white) used to calculate cell area. Bottom: distribution of cell areas. The size of the target cell did not significantly affect cell detection (OR = 0.99, CI: 0.97–1.00, P = 0.11). (c) Top: sample cell images showing poor (left) and good (right) image quality. Image quality was defined as the number of unique pixel values in the image. Bottom: distribution of contrast values. Image contrast did not significantly affect cell detection (OR = 1.06, CI: 0.99–1.13, P = 0.07, n = 126 attempts).
PatcherBot performance in brain slices
We subsequently evaluated the ability of the PatcherBot to attain successful recordings in various regions of brain slices. The PatcherBot functioned unattended (15–48 min operation time, supplementary video 4) and achieved a success rate in cortical areas similar to the success rate in subcortical areas (cortical whole-cell success rate: 104 cells/205 attempts = 51%; subcortical whole-cell success rate: 17 cells/40 attempts = 43%, P = 0.39, Fisher’s Exact Test). Thalamic recordings exhibited expected characteristics of neurons associated with that region such as bursting, sag current, and after-hyperpolarization bursts [34–36] in response to current injections (supplementary figure 6). Success rate was not significantly affected by targeted cell size (figure 4(b)), imaging contrast (figure 4(c)), or depth (figure 5(a)), suggesting that our automation approach can be readily applied to cells in many brain regions. As in the HEK cell preparation, no deterioration of success rate over subsequent pipette reuses was observed (figure 5(b)). On the other hand, as expected, when all machine vision is disabled, the success rates of cell detection and whole-cell recording decrease (figure 5(c)).
Figure 5.
PatcherBot performance in brain slices. (a) Success rates of cell detection and whole-cell recordings as a function of cell depth in slices. Data from cortical and sub-cortical experiments were combined. The depth of the target cell did not significantly impact cell detection likelihood (OR = 1.00, CI: 0.98–1.03, P = 0.8, n = 126 attempts) and whole-cell recording likelihood (OR = 1.00, CI: 0.98–1.03, P = 0.65, n = 126 attempts). (b) Whole-cell success rate in brain slices. Reuses did not significantly decrease likelihood of whole-cell recording (OR = 1.07, CI: 0.97–1.17, P = 0.19, n = 245 attempts). (c) Success rate of cell detection and whole-cell recordings with and without machine vision (machine vision off: n = 18 attempts; machine vision on: n = 161 attempts). Cell detection (top): P = 2.4 × 10−5, whole-cell (bottom): P = 1.7 × 10−5, Fisher’s exact test. Alconox was used as the cleaning agent.
Multi-manipulator PatcherBot
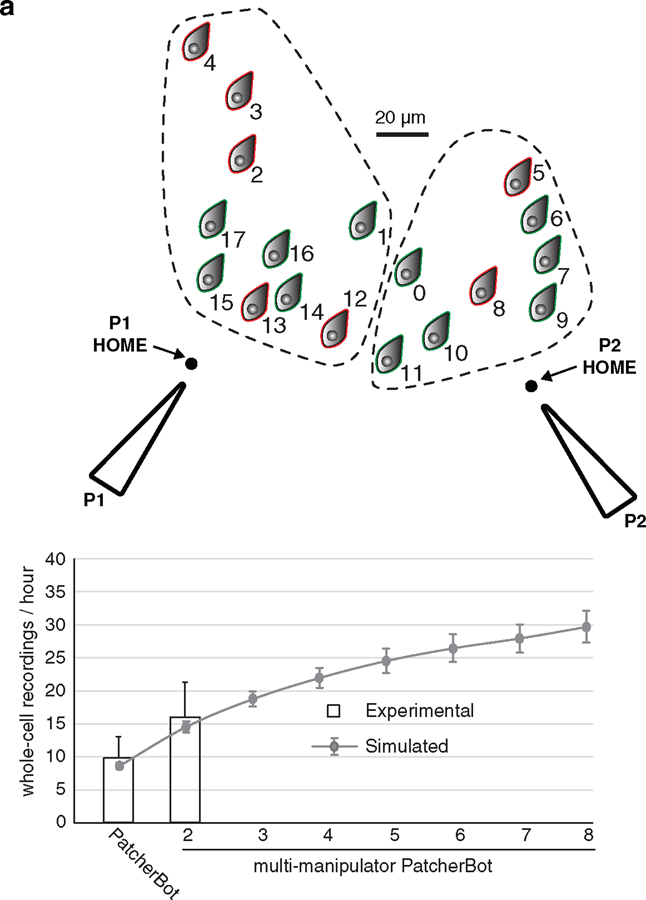
Unlike a human operator, the PatcherBot can command many manipulators, pressure channels, and electrophysiology controls simultaneously; thus, it can theoretically surpass the throughput of even the most experienced investigator. To showcase this, we selected up to 24 cells and programmed two manipulators to perform patch-clamp trials as quickly as possible to maximize throughput (figure 6(a), supplementary videos 5 and 6). The dual-manipulator PatcherBot operated unattended for 38–49 min, achieving a success rate of 62% (n = 83 whole-cell recordings/134 attempts, 7 pipettes per manipulator). Thus, by adding a second manipulator, the average throughput increased from 13 attempts h−1 to 25 attempts h−1. The throughput increase was less than two-fold as would be expected from a perfectly parallelized process because (1) manipulator speed was decreased from 3.4 mm s−1 to 0.4 mm s−1 to reduce vibration and (2) microscope optics that are needed for machine vision had to be shared between the manipulators (supplementary figure 7).
Figure 6.
Multi-patch electrophysiology. (a) Representative two-manipulator PatcherBot experiment in cultured HEK cells. Left dashed line encircles cells patch-clamped by pipette 1 (P1); right dashed line encircles cells patch-clamped by pipette 2 (P2). Cells are shown in an XY coordinate system because they are effectively in the same z plane on the coverslip. Green cell symbols represent successful whole-cell recordings; red symbols represent failed attempts. Pipettes not to scale. (b) Experimental and simulated results of multi-patch system scalability. From simulations, the largest increases in whole-cell recordings/hour are predicted in going from 1 to 2 manipulators (5.7 cells h−1), 2 to 3 (4.1 cells h−1) and 3 to 4 (3.1 cells h−1).
To determine the optimal number of manipulators that should be used to attain maximum throughput, we created a computational model. The model predicted that going from one manipulator to two will result in the most drastic increase in throughput, with progressively smaller increases as more manipulators are added (figure 6(b)). Two factors contributed to diminishing returns on throughput when adding more manipulators: (1) manipulators waiting for the microscope optics will contribute to a bottleneck and (2) a limit of ten cleans per pipette [28] means a significant amount of time is spent manually replacing pipettes, especially for many manipulators.
Improvements to pipette cleaning
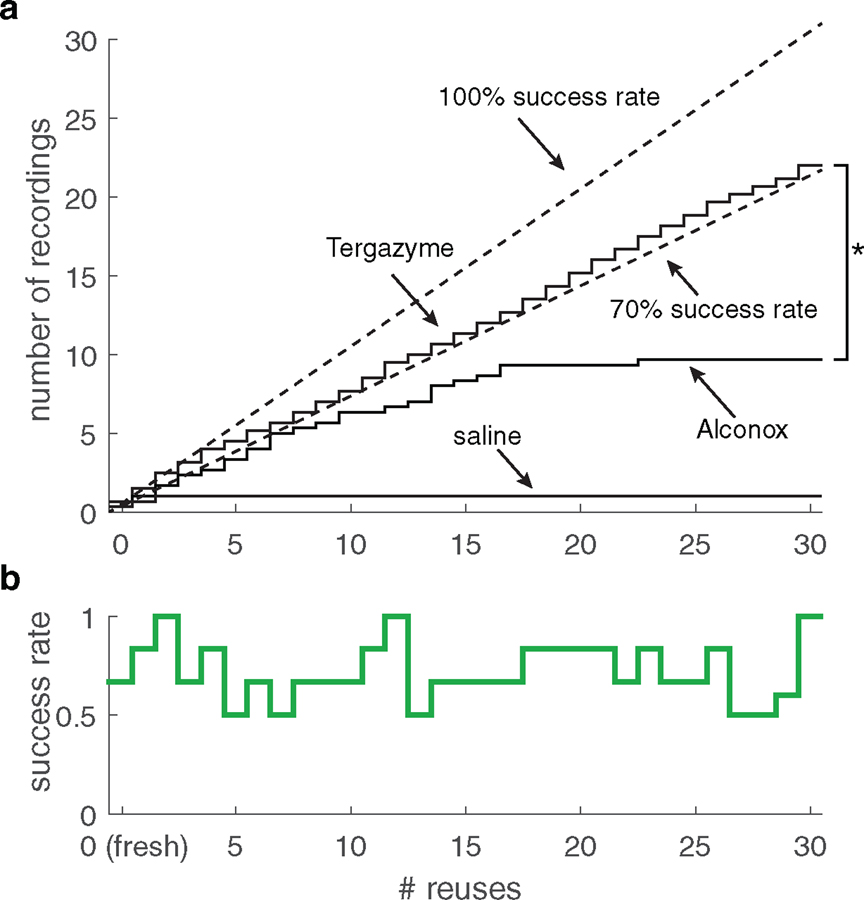
The PatcherBot enabled us to screen candidate detergents in an attempt to find one that can reliably clean pipettes better than Alconox. When cleaning pipettes with Alconox, the PatcherBot can only patch-clamp ~10 cells without recording quality degrading [28], which is the main bottleneck preventing even longer unsupervised operation. We suspected that residual proteins adsorbed to the pipette tip were responsible for recording quality degradation after many cleans. Tergazyme (Alconox Inc) is an inexpensive glassware detergent similar to Alconox with an additional protease enzyme Subtilisin Carlsberg (from the bacterium Bacillus licheniformis) which removes proteins adsorbed to glass [37]. Demonstrating the superiority of a candidate cleaning solution over Alconox requires hundreds of recordings to reach statistical significance so we used the PatcherBot for these experiments. In experiments where the user was blinded to the cleaning solution being used, Tergazyme-cleaned pipettes outperformed Alconox-cleaned pipettes after 30 cleaning cycles (31 attempted cells, figure 7(a)). We found no relationship between the number of cleans and the patch-clamp success rate or quality parameters in Tergazyme-cleaned pipettes (figure 7(b), supplementary figure 7). With Tergazyme as the cleaning agent, the PatcherBot ran unattended for 109–120 min.
Figure 7.
Using the PatcherBot to validate Tergazyme as an improved cleaning solution for pipettes. (a) Cumulative success rate of Alconox and Tergazyme over 31 pipette uses in cultured HEK cells. Tergazyme overall success rate: 71.7% (n = 132 whole-cell recordings/184 attempts, 6 pipettes), Alconox overall success rate: 39.0% (n = 29 whole-cell recordings/75 attempts, 3 pipettes), saline overall success rate: 6.4% (n = 3 whole-cell recordings/47 attempts, 3 pipettes). *P = 2.6 × 10−5 (Kolmogorov–Smirnov test). (b) Whole-cell success rate over 31 uses with Tergazyme as the cleaning agent using Tergazyme data from (a). Reuses did not significantly decrease likelihood of whole-cell recording (OR = 1.00, CI: 0.97–1.04, p = 0.89, n = 184 attempts).
Discussion
We have created a system for walk-away automation of patch-clamp recordings. Due to its ability to perform multiple trials sequentially, it provides an approximately ten-fold increase in unattended operation time compared to previous automation efforts. We demonstrate the scalability of the system to multiple pipettes and show that it can work in cell cultures as well as brain slices.
The PatcherBot can be extended beyond patch-clamp to be broadly useful for single-cell in situ studies requiring high throughput. For example, it could be used for single cell harvesting [38] or automated cell labeling using plasmids [39, 40] and viruses [41]. These techniques are powerful due to their ability to target single cells in much higher specificity than bulk transfection, but they still suffer from low throughput and success rate. Automation could open the door to large-scale studies of cell morphology, gene expression, and activity in neurons [42, 43]. Since the system does not require fluorescently tagged cells for targeting, it could also be used to study human brain tissue samples.
The approach presented here is a significant step towards turning high-quality single-cell electrophysiology into an easy-to-use device analogous to a plate reader. By largely removing the trained human from the loop, electrophysiology setups can be redesigned to mimic automated microscopes that function as a ‘black box’ to the user. Further, miniaturization efforts in actuators, amplifiers, and microscopes are paving the way towards substantially reduced space needs and costs [44–46].
We anticipate that the PatcherBot will have impact in the pharmaceutical and proteomic domains. For drug development, the PatcherBot could be used to screen ion channel modulators to characterize specific and non-specific effects. For proteomics, it could be used for electrophysiological screening of fluorescent indicators and optogenetic actuators. In these and other applications involving functional screening, the PatcherBot could expedite, commoditize and standardize the process. The low degree of human involvement makes it possible for one trained user to operate many PatcherBot systems simultaneously, increasing throughput even further.
We anticipate that integration with existing automated microscopy hardware and software will be sufficient to enable the PatcherBot to function as a screening tool. For drug screening, the software can be extended to control dispensing syringe pumps, valves, or injection devices. To perform functional screening of ion-conducting opsins [13, 47–49] or voltage indicators [10, 38, 50–52], the PatcherBot software can be programmed to initiate light stimulation and time-series fluorescence imaging. Preliminary studies indicate that pipette cleaning does not alter cell pharmacokinetics [28] but further evaluation will be performed to quantify cross-contamination in cleaned pipettes. To improve data quality and throughput for screening, we foresee the need for additional software refinement that will include (1) mimicking best practices from trained electrophysiologists using machine learning, (2) checking recording quality in real time and applying remedies if quality degrades, (3) automatically selecting cells based on health or fluorescence, and (4) performing auto-calibration. These refinements could greatly add to the usefulness of the PatcherBot as a screening tool, enabling its use in core facilities and high-throughput screening pipelines.
Large-scale multimodal cell profiling experiments in brain tissue have been highly impactful but are labor-intensive and therefore rare [53–55]. To our knowledge, the PatcherBot is the first system to perform repeated fully automated probing of non-fluorescently labeled cells in living brain tissue. In this study, we only demonstrated the ability of the PatcherBot to record electrical activity but we foresee that it could also soon be augmented to study gene expression and morphology in the same cell [42]. Several developments would be needed to make this a reality. First, our algorithm does not presently monitor recording quality; a more nuanced break-in and quality control algorithm would be developed to ensure that access resistance is low throughout the experiment. To study single-cell gene expression, reused pipettes would need to be tested extensively for genetic cross-contamination. To study morphology, pipettes would also be filled with a dye or small molecule that would diffuse into cells to stain them. We found that cell health degradation over time (noticeable after ~1 h in our preparations) is a biological constraint that limits unattended operation time, but we envision this can be mitigated with environmental control of humidity, temperature and CO2.
We demonstrated the PatcherBot working with only one or two manipulators but adding more could improve throughput well beyond the capabilities of even the most skilled human experimenters. Human operators can only patch-clamp one cell at a time while the PatcherBot can control many manipulators, pressure lines, and electrical command signals simultaneously, giving it the ability to record from cells in a highly parallel fashion. Our simulations suggest that for short, individual recordings, using four or six manipulators may be optimal, as the increase in throughput in adding one more manipulator in those cases is marginal. Meanwhile, for sampling neuronal connections, adding manipulators exponentially increases the number of testable pairs but demands even more attention and skill from the user [21, 56]. Scaled-up versions of the PatcherBot could greatly simplify and expedite large connectivity studies [53, 57, 58]; however, more sophisticated algorithms than those presented here are necessary to patch-clamp a cell in a brain slice without disturbing another existing recording. Beyond neuronal connections, electrical interactions between neurons and astrocytes are not presently well-understood; the multi-pipette PatcherBot could also be used to comprehensively characterize those relationships.
Supplementary Material
Acknowledgments
IK acknowledges funding from the Georgia Tech Neural Engineering Center Seed Grant. JL and CJR acknowledge funding from NSF Grant CCF-1409422, the James S McDonnell Foundation Grant 220020399, and DSO National Laboratories of Singapore. CRF acknowledges the NIH BRAIN Initiative Grant (NEI and NIMH 1-U01-MH106027–01), NIH 1-R01-NS102727–02, NIH Single Cell Grant 1-R01-EY023173–01, and NIH Computational Neuroscience Training grant (5-T90-DA032466–02).
Footnotes
Supplementary material for this article is available online
Competing interests
IK, WAS and CRF are inventors on a US patent application 15/232,770 related to pipette cleaning technology, licensed to Sensapex. IK and MCY have consulting agreements with Neuromatic Devices which manufactures pipette pressure control systems. JL and CJR are inventors on a US patent application 16/116,192 related to cell membrane tracking in tissue.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- [1].Zeisel A et al. 2015. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq Science 347 1138–42 [DOI] [PubMed] [Google Scholar]
- [2].Saadatpour A, Lai S, Guo G and Yuan G-C 2015. SinCgle-cell analysis in cancer genomics Trends Genet 31 576–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Krutzik PO, Crane JM, Clutter MR and Nolan GP 2008. High-content single-cell drug screening with phosphospecific flow cytometry Nat. Chem. Biol 4 132–42 [DOI] [PubMed] [Google Scholar]
- [4].Paşca SP et al. 2011. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome Nat. Med 17 1657–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Denyer J, Worley J, Cox B, Allenby G and Banks M 1998. HTS approaches to voltage-gated ion channel drug discovery Drug Discov. Today 3 323–32 [Google Scholar]
- [6].Bagal SK et al. 2013. Ion channels as therapeutic targets: a drug discovery perspective J. Med. Chem 56 593–624 [DOI] [PubMed] [Google Scholar]
- [7].Hamill OP, Marty A, Neher E, Sakmann B and Sigworth FJ 1981. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches Pflüg. Arch 391 85–100 [DOI] [PubMed] [Google Scholar]
- [8].Lee D, Hyun JH, Jung K, Hannan P and Kwon H-B 2017. A calcium- and light-gated switch to induce gene expression in activated neurons Nat. Biotechnol 35 858–63 [DOI] [PubMed] [Google Scholar]
- [9].Wang W et al. 2017. A light- and calcium-gated transcription factor for imaging and manipulating activated neurons Nat. Biotechnol 35 864–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hochbaum DR et al. 2014. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins Nat. Methods 11 825–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen T-W et al. 2013. Ultra-sensitive fluorescent proteins for imaging neuronal activity Nature 499 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Boyden ES, Zhang F, Bamberg E, Nagel G and Deisseroth K 2005. Millisecond-timescale, genetically targeted optical control of neural activity Nat. Neurosci 8 1263–8 [DOI] [PubMed] [Google Scholar]
- [13].Berndt A, Yizhar O, Gunaydin LA, Hegemann P and Deisseroth K 2009. Bi-stable neural state switches Nat. Neurosci 12 229–34 [DOI] [PubMed] [Google Scholar]
- [14].Park J. et al. Screening fluorescent voltage indicators with spontaneously spiking HEK cells. PLoS One. 2013;8:e85221. doi: 10.1371/journal.pone.0085221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Milligan CJ et al. 2009. Robotic multiwell planar patch-clamp for native and primary mammalian cells Nat. Protoc 4 244–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kolb I et al. 2016. Integration of autopatching with automated pipette and cell detection in vitro J. Neurophysiol 116 1564–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stuart GJ, Dodt HU and Sakmann B 1993. Patch-clamp recordings from the soma and dendrites of neurons in brain slices using infrared video microscopy Pflüg. Arch 423 511–8 [DOI] [PubMed] [Google Scholar]
- [18].Blanton MG, Lo Turco JJ and Kriegstein AR 1989. Whole cell recording from neurons in slices of reptilian and mammalian cerebral cortex. J. Neurosci. Methods 30 203–10 [DOI] [PubMed] [Google Scholar]
- [19].Annecchino LA and Schultz SR 2018. Progress in automating patch clamp cellular physiology Brain Neurosci. Adv 2 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fertig N, Blick RH and Behrends JC 2002. Whole cell patch clamp recording performed on a planar glass chip Biophys. J 82 3056–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Perin R and Markram HA 2013. Computer-assisted multi-electrode patch-clamp system J. Vis. Exp (80) e50630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kodandaramaiah SB, Franzesi GT, Chow BY, Boyden ES and Forest CR 2017. Automated whole-cell patch-clamp electrophysiology of neurons in vivo Nat. Methods 9 585–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Suk H-J et al. 2017. Closed-loop real-time imaging enables fully automated cell-targeted patch-clamp neural recording in vivo Neuron 95 1037–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Suter BA. et al. Ephus: multipurpose data acquisition software for neuroscience experiments. Frontiers Neural Circ. 2010;4:100. doi: 10.3389/fncir.2010.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Campagnola L, Kratz MB and Manis PB 2014. ACQ4: an open-source software platform for data acquisition and analysis in neurophysiology research Frontiers Neuroinformatics 8 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu Q and Chubykin AA 2017. Application of automated image-guided patch clamp for the study of neurons in brain slices JoVE J. Vis. Exp (125) e56010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Annecchino LA et al. 2017. Robotic automation of in vivo two-photon targeted whole-cell patch-clamp electrophysiology Neuron 95 1048–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kolb I. et al. Cleaning patch-clamp pipettes for immediate reuse. Sci. Rep. 2016;6:35001. doi: 10.1038/srep35001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lee J, Kolb I, Forest CR and Rozell CJ 2018. Cell membrane tracking in living brain tissue using differential interference contrast microscopy IEEE Trans. Image Process 27 1847–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kodandaramaiah SB et al. 2016. Assembly and operation of the autopatcher for automated intracellular neural recording in vivo Nat. Protoc 11 634–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yang R, Fang Y, Yang J and Lai KWC 2017. Design and analysis of electrical resistance feedback for automated patch clamp on adherent cells IEEE Trans. Autom. Sci. Eng 14 844–54 [Google Scholar]
- [32].Connor M, Bagley EE, Chieng BC and Christie MJ 2015. β-Arrestin-2 knockout prevents development of cellular µ-opioid receptor tolerance but does not affect opioid-withdrawal-related adaptations in single PAG neurons Br. J. Pharmacol 172 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Venkatesh N, Lamp ST and Weiss JN 1991. Sulfonylureas, ATP-sensitive K+ channels, and cellular K+ loss during hypoxia, ischemia, and metabolic inhibition in mammalian ventricle Circ. Res 69 623–37 [DOI] [PubMed] [Google Scholar]
- [34].Pape H-C and McCormick DA 1989. Noradrenaline and serotonin selectively modulate thalamic burst firing by enhancing a hyperpolarization-activated cation current Nature 340 715–8 [DOI] [PubMed] [Google Scholar]
- [35].Pape HC 1996. Queer current and pacemaker: the hyperpolarization-activated cation current in neurons Annu. Rev. Physiol 58 299–327 [DOI] [PubMed] [Google Scholar]
- [36].Leist M. et al. Two types of interneurons in the mouse lateral geniculate nucleus are characterized by different h-current density. Sci. Rep. 2016;6:24904. doi: 10.1038/srep24904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Alconox Inc 2017. Tergazyme enzyme active powdered detergent (https://alconox.com/resources/standarddocuments/sds/sds_tergazyme_english_ghs.pdf)
- [38].Piatkevich KD. et al. A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol. 2018;14:352. doi: 10.1038/s41589-018-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rancz EA et al. 2011. Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics Nat. Neurosci 14 527–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Steinmeyer JD and Yanik MF 2012. High-throughput single-cell manipulation in brain tissue PLoS One 7 e35603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schubert R et al. 2017. Virus stamping for targeted single-cell infection in vitro and in vivo Nat. Biotechnol 36 81–8 [DOI] [PubMed] [Google Scholar]
- [42].Cadwell CR. et al. Multimodal profiling of single-cell morphology, electrophysiology, and gene expression using Patch-seq. Nat. Protoc. 2017;12:2531. doi: 10.1038/nprot.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Economo MN. et al. A platform for brain-wide imaging and reconstruction of individual neurons. eLife. 5:e10566. doi: 10.7554/eLife.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Harrison RR et al. 2015. Microchip amplifier for in vitro, in vivo, and automated whole cell patch-clamp recording J. Neurophysiol 113 1275–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kolb I et al. 2014. Linear micro-actuation system for patch-clamp recording Proc. 29th Ann. Meeting of the American Society for Precision Engineering (Boston, MA) Nov 2014 [Google Scholar]
- [46].Ghosh KK et al. 2011. Miniaturized integration of a fluorescence microscope Nat. Methods 8 871–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kato HE. et al. Structural mechanisms of selectivity and gating in anion channelrhodopsins. Nature. 2018;561:349. doi: 10.1038/s41586-018-0504-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Klapoetke NC et al. 2014. Independent optical excitation of distinct neural populations Nat. Methods 11 338–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chuong AS et al. 2014. Noninvasive optical inhibition with a red-shifted microbial rhodopsin Nat. Neurosci 17 1123–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gong Y et al. 2015. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor Science 350 1361–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Xu F, Shi D-Q, Lau P-M, Lin MZ and Bi G-Q 2018. Excitation wavelength optimization improves photostability of ASAP-family GEVIs Mol. Brain 11 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Abdelfattah AS et al. 2016. A bright and fast red fluorescent protein voltage indicator that reports neuronal activity in organotypic brain slices J. Neurosci 36 2458–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Jiang X. et al. Principles of connectivity among morphologically defined cell types in adult neocortex. Science. 2015;350:aac9462. doi: 10.1126/science.aac9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zeng H et al. 2012. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures Cell 149 483–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tasic B et al. 2016. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics Nat. Neurosci 19 335–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wang G et al. 2015. An optogenetics- and imaging-assisted simultaneous multiple patch-clamp recording system for decoding complex neural circuits Nat. Protoc 10 397–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Perin R, Berger TK and Markram H 2011. A synaptic organizing principle for cortical neuronal groups Proc. Natl Acad. Sci 108 5419–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Song S, Sjöström PJ, Reigl M, Nelson S and Chklovskii DB 2005. Highly nonrandom features of synaptic connectivity in local cortical circuits PLoS Biol 3 e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.