ABSTRACT
The present study was conducted to investigate the antimicrobial susceptibility profiles of Salmonella serotypes, especially fluoroquinolone-resistant strains, recovered from clinical samples in Iran. A full electronic search using related keywords was conducted in Persian and English languages in ISI Web of Knowledge, PubMed, Scopus, Google Scholar and the Scientific Information Database (SID) search engines to find papers published between 1983 and 1 July 2019. According to the inclusion and exclusion criteria, 46 eligible articles were selected for the final analysis out of the initial 13,186 studies retrieved. The pooled prevalence of quinolone-resistant Salmonella serotypes in clinical specimens in Iran was 2.9% to ciprofloxacin and 48.1% to nalidixic acid. Additional data on antibiotic resistance was as follows: 54.3% to tetracycline, 50.6% to ceftizoxime, 50.2% to streptomycin, 37.9% to ampicillin, 36.5% to kanamycin, 33.5% to trimethoprim-sulfamethoxazole, 27.2% to chloramphenicol, 19.1% to cephalothin, 8.8% to ceftriaxone, 7.6% to cefotaxime, 7.4% to aztreonam, 7.2% to gentamicin, 7% to cefepime, 6.8% to ceftazidime, 5.8% to cefixime, 2.7% to imipenem and 2.2% to meropenem. Findings of the present study showed a rising trend of resistance to the drugs of choice for the treatment of Salmonella infections, i.e. ampicillin, chloramphenicol and trimethoprim-sulfamethoxazole in Iran. However, ciprofloxacin, third-generation cephalosporins and carbapenems are still effective antibiotics especially against multi-drug resistant strains in Iran.
KEYWORDS: Antibiotic resistance, Salmonella, fluoroquinolone, Iran
Introduction
The genus Salmonella belongs to the family Enterobacteriaceae and includes two main species, i.e. Salmonella enterica and Salmonella bongori. This genus has around 2,600 unique serotypes, which are characterized as Gram-negative, facultative anaerobe, rod-shaped and motile with peritrichous flagella [1–3]. Salmonella serotypes are also known as enteric bacteria and cause zoonotic diseases that vary in severity from a local infection called gastroenteritis to systemic infections such as septicemia, paratyphoid fever and enteric fever (typhoid fever) [1,2,4]. Additionally, asymptomatic colonization of Salmonella serotypes adapted to humans in the gallbladder can establish human chronic carriers, which along with oral ingestion of contaminated water and food products such as poultry, eggs and dairy products are considered as the major dissemination routes for human diseases [2,4]. Individuals younger than 5 and older than 60 years as well as immunocompromised patients are more susceptible to Salmonella infections [2,4]. On the other hand, Salmonella infections are important in both developed and developing countries in terms of hospitalization as well as public health and economic impacts [5,6]. However, the efficacy of antibiotic treatment for Salmonella infections has been challenged by the emergence of antibiotic-resistant, especially multidrug-resistant (MDR), Salmonella serotypes [5]. Antibiotic therapy is not needed for Salmonella-induced gastroenteritis while for invasive Salmonella infections, ampicillin, chloramphenicol and trimethoprim-sulfamethoxazole are used as the first-line treatments [1,2]. However, emerging MDR Salmonella species have changed the treatment regimen toward using fluoroquinolones and third-generation cephalosporins [1]. Nonetheless, the prevalence of fluoroquinolone-resistant Salmonella species is growing according to the World Health Organization (WHO) reports, warning that these species may become a great threat to human health [7]. The prevalence of antibiotic resistance of Salmonella serotypes has been studied sporadically in different cities of Iran but there has been no comprehensive study in this regard. Therefore, the present systematic review and meta-analysis were conducted to determine the antimicrobial susceptibility profiles of Salmonella serotypes, especially fluoroquinolone-resistant serotypes, recovered from clinical samples in Iran.
Methods
Search strategy
This systematic review and meta-analysis were performed based on the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines [8]. Two authors searched both international and national databases including the Information Sciences Institute (ISI) Web of Knowledge, PubMed, Scopus, Google Scholar and the Scientific Information Database (SID) to find studies published between 1983 and 1 July 2019. Eligible studies were peer-reviewed scientific articles addressing antimicrobial susceptibility profiles of Salmonella serotypes, and published in English or Persian languages. Additionally, the references of included studies were manually searched to find missing studies. The search terms along with connectors (AND/OR) were ‘drug resistan*’ OR ‘antibiotic resistan*’ OR ‘antimicrobial resistan*’ AND ‘Salmonella’ AND ‘clinical sample’ AND ‘Iran’.
Study selection and quality assessment
The identified studies were further assessed in terms of eligibility for inclusion. We included studies reporting the prevalence of resistance, studies evaluating Salmonella serotypes isolated from clinical samples and studies limited to Iran. We excluded articles which had insufficient information, non-original articles, and data from other countries or on non-clinical samples. We only chose one of the articles with the same first author and the same time period of study. The Joanna Briggs Institute (JBI) critical appraisal checklist for studies reporting prevalence data was used for the quality assessment of the included studies [9]. Articles were considered as a high-quality study when received more than 5 scores, medium-quality with 4–5 scores and low-quality with lower than 4 scores. We also excluded articles with quality scores lower than 4.
Data extraction and analysis
Important details of studies were extracted from articles that met the inclusion criteria (Table 1). These details included first author surnames, score of quality assessment, province of study, period of study, age group, sample size, type of tested samples, important Salmonella serotypes, antibiotic susceptibility testing method, number of Salmonella serotypes resistant to different antibiotics, number of Salmonella serotypes producing extended-spectrum β-lactamases (ESBLs) and number of multidrug-resistant Salmonella serotypes. Collected primary data on antibiotic resistance from eligible articles was transferred to Comprehensive Meta-Analysis (CMA) software (Biostat, Englewood, NJ) and used for calculating microbial resistance profiles for each antibiotic. Data synthesis was done and expressed as a percentage and 95% confidence intervals (95% CIs) based on random- or fixed-effects models. The CMA software was also applied to assess two characteristics in the included studies, i.e. the existence of heterogeneity using I2 statistic and Chi-square test (significance defined at p < 0.1), as well as publication bias using the funnel plots. I2 values of 25%, 50% and 75% were considered as low, moderate and high levels of heterogeneity, respectively. At a low heterogeneity, i.e. I2 < 25%, a fixed-effects model was used for meta-analysis. The existence of visual asymmetry in funnel plots was considered as a sign of potential publication bias.
Table 1.
Extracted information from eligible studies included in the meta-analysis.
| Antibiotic resistance (n) (%) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author (Ref) | Quality score |
Province | Year | Age group | Sample origin |
Strain (n) |
Salmonella serotypes | AST | AMP | CHL | TMP-SMX | CIP | NAL | CAZ | CRO |
| Farahani [10] | 7 | Different cities | NA | NA | Stool | 36 | Enteritidis | Disk diffusion | NA | 32 (91.2) |
NA | 8 (23.5) |
32 (88.2) |
1 (2.9) |
NA |
| Soltan Dallal [11] | 7 | Different cities | 2012-2013 | <60 | Stool | 74 | NA | Disk diffusion | 17 (23) |
NA | NA | 74 (100) |
65 (87.8) |
NA | 18 (24.3) |
| Saboohi [12] | 6 | Different cities | 2008-2010 | NA | Stool Blood Abscess Urine BM SF |
85 | NA | Disk diffusion | 12 (14.1) |
NA | NA | 0 (0) |
49 | 9 (10.5) |
6 (7) |
| Iranshahi [13] | 7 | Different cities | 2007-2008 | <65 | Stool Blood BM | 53 | NA | Disk diffusion | NA | NA | NA | 0 0 |
28 (52.8) |
NA | NA |
| Sepehri Rad [14] | 7 | NA | 2008-2010 | NA | Stool Blood Ascites Abscess Urine BM SF |
83 | Typhi Paratyphi Enteritidis |
Disk diffusion | 66 (79) |
18 (21) |
12 (14) |
4 (4) |
47 (56) |
9 (10) |
6 (7) |
| Amir Mozafari [15] | 5 | NA | 2005-2006 | NA | Stool | 45 | Typhi Paratyphi Enteritidis Typhimurium |
Disk diffusion | NA | NA | NA | NA | 11 (24.4) |
NA | NA |
| Bialvaei [16] | 7 | East Azerbaijan | 2009-2013 | <70 | Stool | 91 | Enteritidis Typhimurium |
Disk diffusion | 78 (85.7) |
56 (61.5) |
85 (93.4) |
NA | NA | NA | NA |
| Aminshahidi [17] | 7 | Fars | 2014-2015 | <18 | Stool | 14 | NA | Disk diffusion | 2 (14.2) |
NA | 2 (14.2) |
0 (0) |
NA | 1 (7.1) |
NA |
| Anvarinejad [18] | 8 | Fars | 2008 – 2014 | NA | Blood | 19 | NA | Disk diffusion | 3 (15.7) |
4 (21) |
3 (15.7) |
0 (0) |
6 (31.5) |
0 (0) |
0 (0) |
| Abdollahi [19] | 7 | Fars | NA | NA | Stool | 96 | Typhi Paratyphi Enteritidis Typhimurium |
Disk diffusion | 47 (49) |
17 (18) |
24 (25) |
0 (0) |
23 (24) |
NA | NA |
| Yousefi-Mashouf [20] | 6 | Hamadan | 2001-2004 | <68 | Stool Blood Urine |
296 | Typhi Paratyphi Enteritidis Typhimurium |
Disk diffusion | 214 (72.2) |
105 (35.4) |
94 (31.7) |
8 (2.7) |
NA | NA | NA |
| Afzali [21] | 7 | Isfahan | 2000 − 2001 | NA | Stool | 66 | NA | Disk diffusion | NA | 1 (1.5) |
29 (43.9) |
7 (10.5) |
25 (37.9) |
NA | NA |
| Soltan Dallal [22] | 7 | Mazandaran | 2013-2014 | NA | Stool | 4 | Enteritidis | Disk diffusion | 3 | 0 (0) |
4 (100) |
0 (0) |
4 (100) |
0 (0) |
NA |
| Eshaghi Zadeh [23] | 7 | Tehran | 2016-2017 | <14 | Stool | 30 | Typhi Paratyphi Enteritidis Typhimurium |
Disk diffusion | NA | 2 (6.7) |
8 (26.7) |
5 (16.7) |
16 (53.3) |
1 (3.3) |
1 (3.3) |
| Fardsanei [24] | 7 | Tehran | 2015-2016 | NA | Stool | 44 | Enteritidis | Disk diffusion | NA | 1 (2.3) |
8 (18.2) |
40 (90.9) |
34 (77.3) |
4 (9.1) |
3 (6.8) |
| Ranjbar [25] | 8 | Tehran | 2015-2016 | NA | Stool | 138 | NA | Disk diffusion | 11 (7.9) |
NA | NA | 0 (0) |
NA | 40 (28.9) |
40 (28.9) |
| Ranjbar [26] | 7 | Tehran | 2015 | NA | Stool Blood Urine |
21 | Typhimurium | Disk diffusion | 12 (57) |
14 (67) |
3 (14) |
0 (0) |
2 (9) |
0 (0) |
0 (0) |
| Abaspour shoushtari [27] | 6 | Tehran | 2015 | NA | Stool | 60 | NA | Disk diffusion | 50 (83.3) |
38 (38.3) |
60 (100) |
NA | NA | NA | 27 (45) |
| Najafi [28] | 7 | Tehran | 2015 | NA | Stool Blood CSF Urine |
48 | Enteritidis Typhimurium |
Disk diffusion | 5 | 15 (31.2) |
0 (0) |
NA | NA | NA | 5 |
| Amiri [29] | 7 | Tehran | 2015 | NA | Stool | 60 | Typhimurium | Disk diffusion | NA | NA | 13 (21.7) |
0 (0) |
42 (70) |
NA | NA |
| Malehmir [30] | 6 | Tehran | 2014-2015 | NA | NA | 138 | NA | Disk diffusion | NA | NA | NA | 0 (0) |
92 (66.6) |
NA | NA |
| Amini [31] | 6 | Tehran | 2014 | NA | Stool | 46 | Typhimurium | Disk diffusion | 38 (82.6) |
37 (80.4) |
20 (43.1) |
NA | NA | 3 (6.5) |
4 (8.6) |
| Mirjafari Tafti [32] | 8 | Tehran | 2012 − 2014 | <60 | Stool | 83 | Enteritidis | Disk diffusion | 47 (56.6) |
38 (45.7) |
71 (85.5) |
2 (2.4) |
10 (12) |
NA | 2 (2.4) |
| Salimian Rizi [33] | 6 | Tehran | 2012 − 2013 | NA | Stool Blood | 110 | NA | Disk diffusion | 27 (24.5) |
30 (27.3) |
70 (63.6) |
0 (0) |
52 (47.3) |
7 (6.4) |
7 (6.4) |
| Farahani [34] | 8 | Tehran | 2012-2016 | <10 | Stool | 371 | NA | Disk diffusion | 45 (12.1) |
NA | 84 (22.6) |
NA | 230 (61.9) |
NA | NA |
| Soltan Dallal [35] | 6 | Tehran | 2011 | <10 | Stool | 13 | Typhi Paratyphi |
Disk diffusion | 10 (76.9) |
1 (7.6) |
1 (7.6) |
NA | 1 (7.6) |
NA | NA |
| Bakhshi [36] | 8 | Tehran | 2009-2012 | <5 | Stool | 50 | Enteritidis | Disk diffusion | NA | NA | 19 (38) |
3 (6) |
26 (52) |
NA | NA |
| Firoozeh [37] | 5 | Tehran | 2009-2010 | NA | NA | 58 | Paratyphi Enteritidis Typhimurium |
Disk diffusion | 13 (22.4) |
10 (17.2) |
12 (20.6) |
1 (1.8) |
43 (74.1) |
7 (12.1) |
3 (6.9) |
| Ranjbar [38] | 8 | Tehran | 2008-2010 | NA | Stool Blood Urine |
38 | NA | Disk diffusion | 1 (2.6) |
2 (5.2) |
12 (31.5) |
0 (0) |
36 (94.7) |
2 (5.2) |
4 (10.5) |
| Tajbakhsh [39] | 7 | Tehran | 2008-2010 | NA | Stool | 202 | Enteritidis Typhimurium |
Disk diffusion | 29 (14.3) |
27 (13.3) |
70 (34.6) |
0 (0) |
90 (44.5) |
9 (4.4) |
9 (4.4) |
| Hamidian [40] | 7 | Tehran | 2008-2009 | NA | Stool | 174 | Typhi Paratyphi Enteritidis Typhimurium |
Disk diffusion | NA | NA | NA | 0 (0) |
89 (51.1) |
NA | NA |
| Rajaei [41] | 7 | Tehran | 2008-2009 | NA | Stool Blood Ascites Abscess Urine BM SF |
84 | Typhi Paratyphi Typhimurium |
Disk diffusion | 6 (7.1) |
23 (27.4) |
25 (29.8) |
1 (1.2) |
54 (64.3) |
2 (2.4) |
NA |
| Eshraghi [42] | 5 | Tehran | 2008 | NA | Stool | 14 | Enteritidis Paratyphi |
Disk diffusion | NA | 0 (0) |
3 (21.4) |
0 (0) |
10 (71.4) |
0 (0) |
0 (0) |
| Hamidian [43] | 8 | Tehran | 2007-2008 | NA | Stool | 129 | Typhi Paratyphi Enteritidis |
Disk diffusion | 20 (15.5) |
19 (14.7) |
47 (36.4) |
0 (0) |
59 (45.7) |
NA | NA |
| Ranjbar [44] | 8 | Tehran | 2007-2008 | <12 | Stool Blood Urine |
139 | Enteritidis Typhimurium | Disk diffusion | 22 (15.8) |
19 (13.7) |
30 (21.6) |
0 (0) |
85 (61.2) |
6 (4.3) |
6 (4.3) |
| Tajbakhsh [45] | 8 | Tehran | 2007-2008 | NA | Stool | 71 | Typhi Paratyphi Enteritidis |
Disk diffusion | 10 (14) |
8 (11) |
13 (18) |
0 | 16 (22) |
0 (0) |
NA |
| Naghoni [46] | 8 | Tehran | 2006-2008 | NA | NA | 138 | Enteritidis Typhimurium |
Disk diffusion | 22 (15.9) |
18 (13) |
28 (20.3) |
NA | 89 (64.5) |
6 (4.3) |
6 (4.3) |
| Irajian [47] | 6 | Tehran | 2007 | NA | Stool | 50 | Typhi Paratyphi |
Disk diffusion | 13 (26) |
23 (46) |
32 (64) |
0 | 31 (62) |
1 (2) |
NA |
| Morshed [48] | 5 | Tehran | 2005-2007 | NA | Stool | 9 | Enteritidis | Disk diffusion | 3 (33.3) |
1 (11.1) |
1 (11.1) |
0 (0) |
7 (77.8) |
0 (0) |
0 (0) |
| Pourakbari [49] | 7 | Tehran | 2001-2005 | NA | Blood | 42 | NA | Disk diffusion | 23 (54.7) |
11 (26) |
9 (21.4) |
NA | NA | 15 (35.7) |
32 (76.1) |
| Bahrmand [50] | 6 | Tehran | 1994 | NA | Stool Blood | 33 | Typhi | Disk diffusion | 29 (89.3) |
22 (67.9) |
22 (67.9) |
0 (0) |
2 (7.1) |
NA | NA |
| Velayati [51] | 8 | Tehran | 1986 | <5 | Stool | 56 | Enteritidis Typhimurium |
Disk diffusion | 52 (92.8) |
49 (87.6) |
NA | NA | 1 (1.7) |
NA | NA |
| Farhoudi-Moghaddam [52] | 8 | Tehran | 1983-1986 | <5 | NA | 508 | Typhi Typhimurium |
Disk diffusion | 434 (85.4) |
420 (82.7) |
374 (73.6) |
0 (0) |
14 (2.7) |
NA | NA |
| Araghinezhad [53] | 7 | Tehran | NA | NA | Stool | 60 | NA | Disk diffusion | NA | NA | 10 (16.6) |
NA | NA | NA | 0 (0) |
| Bakhshi [54] | 6 | Tehran | NA | NA | Stool | 36 | NA | Disk diffusion | 9 (25) |
1 (2.8) |
10 (27.8) |
4 (11.1) |
NA | NA | NA |
| Amini [55] | 5 | Tehran | NA | <5 | NA | 11 | Enteritidis | Disk diffusion | 1 (9) |
3 (27.3) |
0 (0) |
NA | NA | NA | 1 (9) |
| Author (Ref) | Antibiotic resistance (n) (%) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | ZOX | FEP | CFM | CEF | TET | GEN | MEM | IPM | STR | ATM | KAN | ESBLs | MDR | |
| Farahani [10] | NA | NA | NA | NA | NA | NA | 4 (11.2) |
1 (2.9) |
5 (14.7) |
NA | NA | 17 (47.1) |
NA | NA |
| Soltan Dallal [11] | 28 (37.8) |
NA | NA | NA | NA | 43 (58.1) |
61 (82.4) |
NA | NA | NA | NA | NA | NA | NA |
| Saboohi [12] | 9 (10.5) |
NA | 5 (5.8) |
6 (7) |
NA | NA | NA | NA | NA | NA | NA | NA | 2 (2.3) |
NA |
| Iranshahi [13] | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Sepehri Rad [14] | 6 (7) |
NA | 6 (7) |
NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Amir Mozafari [15] | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Bialvaei [16] | 35 (38.4) |
35 (38.4) |
NA | NA | NA | 82 (91.1) |
20 (21.9) |
NA | NA | NA | NA | NA | 29 (31.8) | NA |
| Aminshahidi [17] | 2 (14.2) |
NA | NA | NA | NA | NA | 0 (0) |
0 (0) |
NA | NA | NA | NA | 1 (7.2) |
NA |
| Anvarinejad [18] | 0 (0) |
NA | 0 (0) |
0 (0) |
NA | 2 (10.5) |
0 (0) |
0 (0) |
0 (0) |
NA | 0 (0) |
NA | 0 (0) |
2 (10.5) |
| Abdollahi [19] | 5 (5) |
NA | NA | NA | NA | NA | NA | NA | 0 (0) |
NA | NA | NA | 5 (5.2) |
NA |
| Yousefi-Mashouf [20] | 177 (59.7) |
125 (42.2) |
NA | NA | NA | NA | 39 (13.1) |
NA | NA | NA | NA | NA | NA | NA |
| Afzali [21] | NA | 59 (89.4) |
NA | NA | 55 (83.3) |
53 (80.3) |
NA | NA | NA | NA | NA | NA | NA | NA |
| Soltan Dallal [22] | 0 (0) |
NA | NA | NA | NA | 4 (100) |
NA | NA | NA | NA | NA | NA | NA | NA |
| Eshaghi Zadeh [23] | 1 (3.3) |
NA | NA | NA | NA | 11 (36.7) |
NA | 0 (0) |
0 (0) |
12 (40) |
NA | NA | NA | NA |
| Fardsanei [24] | 3 (6.8) |
NA | 5 (11.4) |
NA | NA | 8 (18.2) |
NA | NA | 0 (0) |
18 (40.9) |
NA | NA | NA | NA |
| Ranjbar [25] | 6 (4.3) |
NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 40 (28.9) |
NA |
| Ranjbar [26] | 0 (0) |
NA | NA | NA | NA | 17 (81) |
0 (0) |
NA | 0 (0) |
5 (24) |
NA | 3 (14) |
NA | 1 (4.7) |
| Abaspour shoushtari [27] | NA | NA | NA | NA | NA | 28 (46.7) |
60 (100) |
NA | 60 (100) |
43 (71.7) |
NA | NA | NA | NA |
| Najafi [28] | NA | NA | NA | NA | NA | 27 (56.2) |
0 (0) |
NA | 0 (0) |
0 (0) |
NA | NA | NA | NA |
| Amiri [29] | NA | NA | NA | NA | NA | NA | NA | 0 (0) |
0 (0) |
NA | NA | NA | NA | NA |
| Malehmir [30] | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Amini [31] | NA | NA | NA | NA | 37 (80.4) |
32 (69.5) |
4 (8.6) |
NA | NA | NA | NA | NA | 1 (2.1) |
NA |
| Mirjafari Tafti [32] | NA | NA | NA | 3 (3.6) |
NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Salimian Rizi [33] | 3 (2.7) |
NA | NA | NA | NA | 37 (33.6) |
1 (0.9) |
NA | 0 (0) |
NA | 6 (5.5) |
NA | 4 (3.6) |
3 (2.7) |
| Farahani [34] | 25 (6.7) |
NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 63 (17) |
NA |
| Soltan Dallal [35] | NA | 0 (0) |
NA | NA | 3 (23) |
NA | 1 (7.6) |
NA | NA | NA | NA | NA | NA | 1 (5) |
| Bakhshi [36] | NA | NA | NA | NA | NA | 31 (50) |
1 (2) |
NA | NA | 26 (52) |
NA | NA | NA | NA |
| Firoozeh [37] | 2 (3.4) |
NA | NA | 4 (6.9) |
1 (1.8) |
ND | 4 (6.9) |
NA | 0 (0) |
39 (67.3) |
5 (8.6) |
13 (22.4) |
NA | 6 (10.3) |
| Ranjbar [38] | 4 (10.5) |
NA | NA | NA | NA | 34 (89.4) |
0 (0) |
NA | NA | 29 (77.1) |
NA | 24 (63) |
NA | NA |
| Tajbakhsh [39] | 10 (4.9) |
NA | NA | NA | NA | 80 (39.6) |
1 (0.4) |
NA | NA | NA | NA | NA | 7 (3.4) |
8 (3.9) |
| Hamidian [40] | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Rajaei [41] | NA | NA | NA | NA | NA | NA | NA | NA | NA | 25 (29.8) |
NA | NA | NA | NA |
| Eshraghi [42] | 0 (0) |
NA | NA | NA | NA | 4 (28.6) |
0 (0) |
0 (0) |
0 (0) |
1 (7.1) |
NA | NA | NA | NA |
| Hamidian [43] | NA | NA | NA | NA | 8 (6.2) |
56 (43.4) |
0 (0) |
NA | NA | NA | 11 (8.5) |
NA | 3 (2.3) |
9 (6.9) |
| Ranjbar [44] | 6 (4.3) |
NA | NA | NA | 6 (4.3) |
72 (51.8) |
0 (0) |
NA | 0 (0) |
59 (42.8) |
ND | 31 (22.3) |
6 (3.2) |
NA |
| Tajbakhsh [45] | 0 (0) |
NA | NA | NA | 0 (0) |
18 (25) |
0 (0) |
NA | 0 (0) |
NA | NA | 10 (14) |
NA | NA |
| Naghoni [46] | 6 (4.3) |
NA | NA | NA | 6 (4.3) |
70 (50.7) |
NA | NA | NA | 59 (42.7) |
NA | 31 (22.5) |
NA | NA |
| Irajian [47] | NA | NA | NA | NA | NA | NA | 14 (28) |
NA | NA | NA | NA | 17 (34) |
1 (2) |
6 (12) |
| Morshed [48] | NA | NA | NA | 0 (0) |
1 (11.1) |
3 (33.3) |
0 (0) |
NA | 0 (0) |
3 (33.3) |
NA | 2 (22.2) |
NA | NA |
| Pourakbari [49] | NA | NA | NA | NA | 21 (50) |
NA | 6 (14.2) |
NA | NA | NA | NA | 13 (30.9) |
NA | NA |
| Bahrmand [50] | NA | NA | NA | NA | NA | 20 (60.7) |
NA | NA | NA | 27 (82.1) |
NA | 4 (10.7) |
NA | 15 (45.4) |
| Velayati [51] | NA | NA | NA | NA | 31 (55.3) |
44 (78.5) |
50 (89.2) |
NA | NA | 53 (94.6) |
NA | 53 (94.6) |
NA | NA |
| Farhoudi-Moghaddam [52] | NA | NA | NA | NA | 80 | 406 (80) |
0 (0) |
NA | NA | 390 (76.8) |
NA | 412 (81.1) |
NA | NA |
| Araghinezhad [53] | NA | NA | 0 (0) |
NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Bakhshi [54] | NA | NA | NA | NA | NA | 17 (47.2) | NA | NA | NA | 17 (47.2) |
NA | NA | NA | NA |
| Amini [55] | NA | NA | NA | NA | NA | 0 (0) |
0 (0) |
NA | 0 (0) |
0 (0) |
NA | NA | NA | NA |
Abbreviations: AMP-ampicillin; CHL-chloramphenicol; TMP/SMX-trimethoprim-sulfamethoxazole; CIP-ciprofloxacin; NAL-nalidixic acid; CAZ-ceftazidime; CRO-ceftriaxone; CTX-cefotaxime; ZOX-ceftizoxime; FEP-cefepime; CFM-cefixime; CEF-cephalothin; TET-tetracycline; GEN-gentamicin; MEM-meropenem; IPM-imipenem; STR-streptomycin; ATM-aztreonam; KAN-kanamycin; AST-antimicrobial susceptibility testing; ESBLs-extended-spectrum β-lactamases; MDR-multidrug-resistant (combined resistance to ampicillin, chloramphenicol and trimethoprim-sulfamethoxazole); BM-bone marrow; SF-synovial fluid; CSF-cerebrospinal fluid; NA-data not available.
Finally, we assessed antimicrobial resistance trends of Salmonella serotypes to important antibiotics, i.e. ciprofloxacin, nalidixic acid, ampicillin, chloramphenicol and trimethoprim-sulfamethoxazole and third-generation cephalosporins in Iran from 1983 to 2019.
Results
Study characteristics
As shown in Figure 1, a total of 46 unique studies out of 13,186 records were included in this meta-analysis after screening titles, abstracts and full texts of eligible studies presenting data on the antibiotic resistance of Salmonella serotypes in Iran. Briefly, 12,081 records were initially excluded because of being duplicate studies obtained from different databases. Then, 735 duplicates, non-original and non-relevant articles were excluded through the evaluation of titles and abstracts. Among 370 studies identified for full-text screening, 185 duplicates and 39 articles with inadequate data were excluded along with 100 articles reporting antibiotic resistance in non-clinical samples. The included studies, 11 in Persian and 35 in English, were reported from different provinces of Iran and received quality scores between 5 and 8 (Table 1). Disk diffusion was the most commonly used method for antimicrobial susceptibility testing in the included studies. As shown in Table 1, Salmonella serotypes were isolated from all age groups, i.e. pediatric, juvenile and adult patients.
Figure 1.
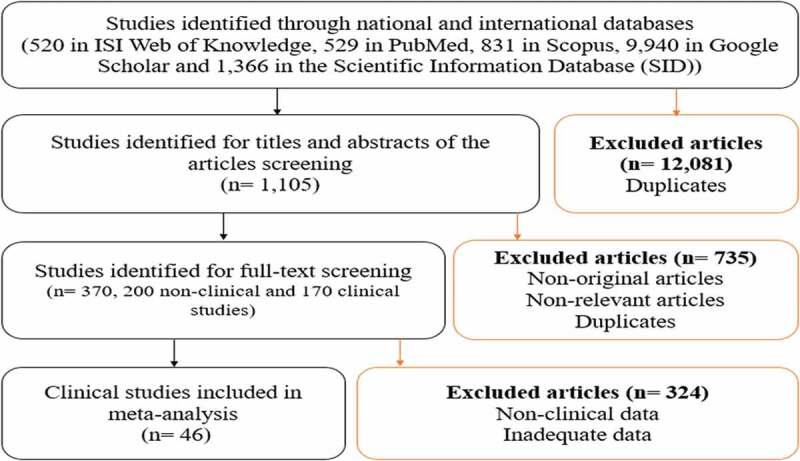
Systematic review flowchart.
Quinolone-resistant Salmonella serotypes
Thirty-four and 35 studies evaluated antibiotic resistance rates of Salmonella serotypes against ciprofloxacin (Figure 2(a)) and nalidixic acid, respectively. The level of heterogeneity among the studies was high (>75%), hence a random-effects model was used to calculate the weighted average. Additionally, in publication bias evaluation, we observed a visual asymmetry of the funnel plot (Figure 2(b)). Overall resistance prevalence of quinolone-resistant Salmonella serotypes isolated from clinical specimens in Iran was as follows: 2.9% (95% CI: 1.4–6; I2 = 84.4%; Q = 212.7; p = 0.00) to ciprofloxacin and 48.1% (95% CI: 39.9–56.4; I2 = 92.8%; Q = 475.6; p = 0.00) to nalidixic acid. As shown in Figure 3(a) and Table 2, we also evaluated the trends of antimicrobial resistance during 12-year intervals. From 1983 to 2019, the resistance trend of Salmonella serotypes to ciprofloxacin and nalidixic acid in Iran was increasing with a gentle and fast slope, respectively.
Figure 2.
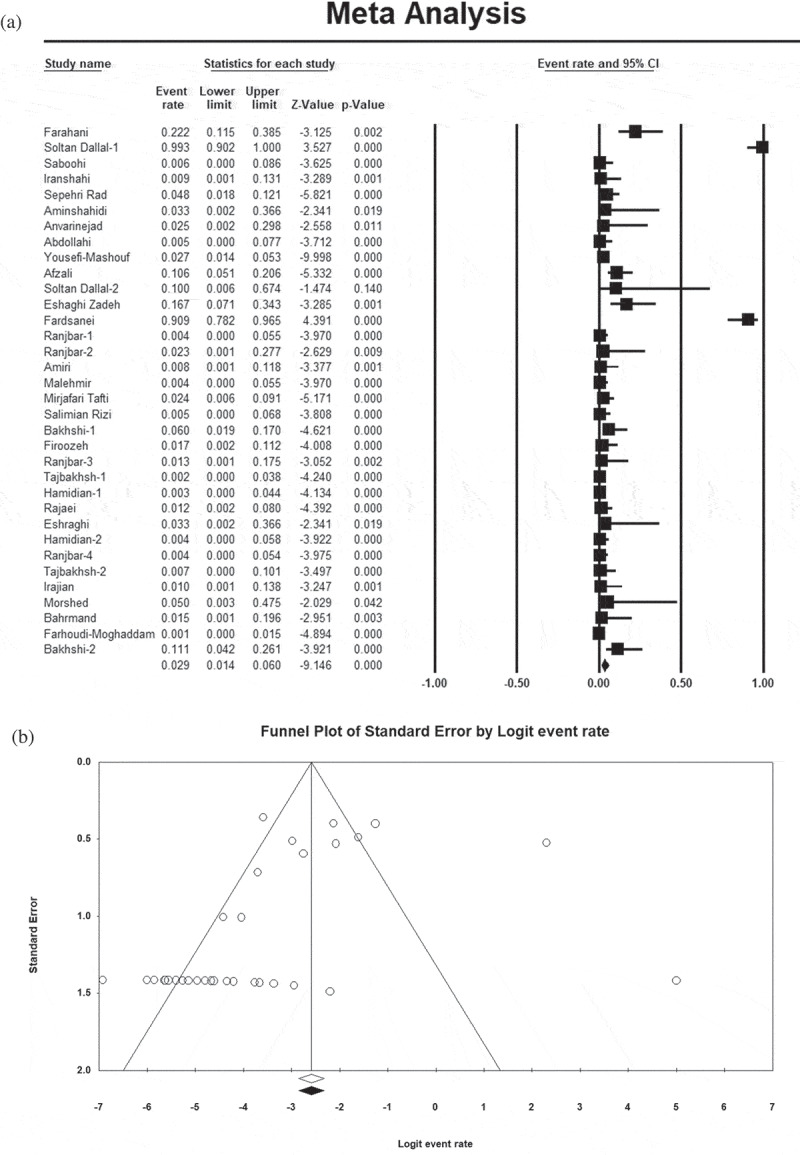
Forest plot (a) and funnel plot (b) showing the prevalence of antibiotic resistance of Salmonella serotypes to ciprofloxacin.
Figure 3.
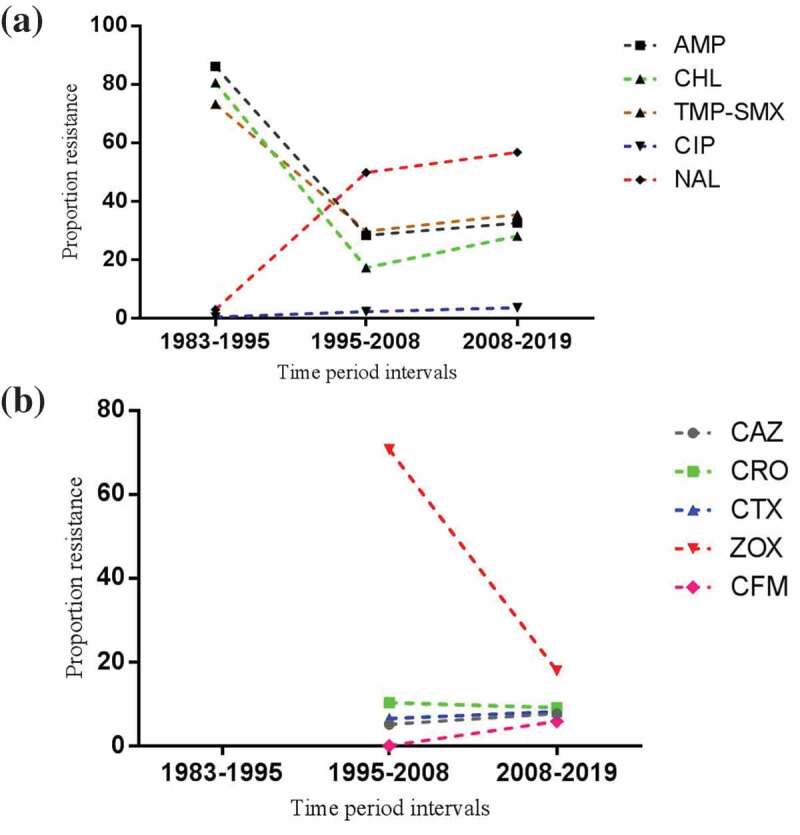
Antimicrobial resistance trends of Salmonella serotypes to different drugs in Iran over time. (a) ciprofloxacin, nalidixic acid, ampicillin, chloramphenicol and trimethoprim-sulfamethoxazole trends and (b) third-generation cephalosporins trends.
Table 2.
Proportion of Salmonella serotypes resistant to therapeutic antibiotics during a 12-year intervals.
| Proportion of resistant isolates (%) (95% CIs) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Year | Strain (n) | AMP | CHL | TMP-SMX | CIP | NAL | CAZ | CRO | CTX | ZOX | CFM |
| 1983-1995 | 597 | 86.1 (83–88.6) |
80.5 (69.7–88.1) |
73.2 (69.3–76.7) |
0.4 (0–5.2) |
2.9 (1.8–4.7) |
NA | NA | NA | NA | NA |
| 1995-2008 | 1052 | 28.3 (12.8–51.7) |
17.2 (10.5–26.8) |
29.7 (22.3–38.2) |
2.2 (0.9–5.4) |
49.8 (39.3–60.4) |
5.1 (1.5–16.1) |
10.3 (1.2–52.6) |
6.5 (0.7–40.3) |
70.7 (18–96.4) |
0 |
| 2008-2019 | 2341 | 32.5 (21–46.5) |
28 (18.5–40) |
35.4 (25.1–47.2) |
3.5 (1.1–11) |
56.7 (48.6–64.4) |
7.7 (4.6–12.5) |
9.1 (5.3–15.2) |
8.2 (4.5–14.5) |
17.9 (1.5–76) |
5.8 (3.4–9.6) |
Abbreviations: AMP-ampicillin; CHL-chloramphenicol; TMP/SMX-trimethoprim-sulfamethoxazole; CIP-ciprofloxacin; NAL-nalidixic acid; CAZ-ceftazidime; CRO-ceftriaxone; CTX-cefotaxime; ZOX-ceftizoxime; CFM-cefixime; NA-data not available.
Salmonella serotypes resistance profiles to the first-line treatments for invasive infections
Meta-analyses with random-effects models were used to assess Salmonella serotypes resistance profiles to ampicillin (I2 = 96.9%; Q = 1035.6; p = 0.00), chloramphenicol (I2 = 95.3%; Q = 716.2; p = 0.00) and trimethoprim-sulfamethoxazole (I2 = 93.8%; Q = 582.7; p = 0.00). In Iran, 37.9% (95% CI: 26.2–51.3) of Salmonella serotypes were resistant to ampicillin, 33.5% (95% CI: 26–42) to trimethoprim-sulfamethoxazole and 27.2% (95% CI: 18.7–37.8) to chloramphenicol were resistant. There were signs of publication bias in the included studies evaluating the resistance of Salmonella serotypes to each of the three above-mentioned antibiotics. As shown in Figure 3(a), the susceptibility of Salmonella serotypes to the first-line antibiotics increased from 1983 to 2008 but showed a decreasing trend from 2008 to 2019. Additionally, the prevalence of MDR serotypes of Salmonella was 9% (95% CI: 4.3–18; I2 = 83.4%; Q = 48.4; p = 0.00) in Iran.
Salmonella serotypes resistance profiles to the third-generation cephalosporins
Antibiotic resistance profiles of Salmonella serotypes to the third-generation cephalosporins were as follows: 50.6% (95% CI: 26.5–74.4; I2 = 92.7%; Q = 41.1; p = 0.00) to ceftizoxime, 8.8% (95% CI: 5.1–14.9; I2 = 89.1%; Q = 202.4; p = 0.00) to ceftriaxone, 7.6% (95% CI: 3.8–14.6; I2 = 95%; Q = 425.6; p = 0.00) to cefotaxime, 6.8% (95% CI: 4.3–10.7; I2 = 79.2%; Q = 106; p = 0.00) to ceftazidime and 5.8% (95% CI: 3.4–9.5; I2 = 0.0%; Q = 1.4; p = 0.83) to cefixime. Apart from cefixime, the prevalence of antibiotic resistance was pooled using random-effects models. The trends of antimicrobial resistance to ceftizoxime was decreasing, while it was almost constant for ceftazidime, ceftriaxone, cefotaxime and cefixime over the time period from 1995 to 2019 (Figure 3(b)). Additionally, the prevalence of ESBLs producing Salmonella serotypes was 6.5% (95% CI: 3.5–11.7; I2 = 89.4%; Q = 113.8; p = 0.00) in Iran.
Other Salmonella serotypes resistance profiles
Resistance to other antibiotics were as follows: 54.3% (95% CI: 45–63.3; I2 = 91.9%; Q = 310.1; p = 0.00) to tetracycline, 50.2% (95% CI: 38.4–62; I2 = 91.6%; Q = 202.7; p = 0.00) to streptomycin, 36.5% (95% CI: 20–56.9; I2 = 96.5%; Q = 344.1; p = 0.00) to kanamycin, 19.1% (95% CI: 8.2–38.6; I2 = 95.6%; Q = 253.8; p = 0.00) to cephalothin, 7.4% (95% CI: 4.9–10.9; I2 = 0.0%; Q = 1.5; p = 0.66) to aztreonam, 7.2% (95% CI: 3.4–14.5; I2 = 91.4%; Q = 292.7; p = 0.00) to gentamicin, 7% (95% CI: 4.4–11; I2 = 9.3%; Q = 4.4; p = 0.35) to cefepime, 2.7% (95% CI: 0.9–8.4; I2 = 71.9%; Q = 53.4; p = 0.00) to imipenem and 2.2% (95% CI: 0.8–6.2; I2 = 0.0%; Q = 0.7; p = 0.97) to meropenem.
Discussion
Recently, it has been reported that the prevalence of Salmonella strains resistant to antimicrobial agents, especially quinolone-resistant Salmonella serotypes, is increasing. This increasing prevalence poses a serious public health concern in both developed and developing countries [1,56]. Therefore, obtaining epidemiological information on drug resistance can help physicians and health-care professionals choosing proper antimicrobial agents and avoid treatment failure. Ciprofloxacin is a known fluoroquinolone antibiotic in the treatment of life-threatening Salmonella infections [57]. However, according to the WHO report, Salmonella serotypes are becoming increasingly drug-resistant bacteria and fluoroquinolone-resistant Salmonella serotypes have been placed in the high-priority category in terms of the urgency of the need to new antibiotics [7]. Resistance rate to ciprofloxacin in Salmonella strains in the present meta-analysis was low (2.9%) (Figure 2(a)). Our findings showed higher rates of resistance compared with those reported from Korea, France, the United States, Greece, Turkey (0%) and Thailand (0.3%) while the rates were lower compared with China (9.2%) [57–60]. On the other hand, tracking the antibiotic resistance trends of Salmonella serotypes during successive years is important for sustaining treatment regimens and preventing treatment failure. The trend of ciprofloxacin resistance in Salmonella serotypes in Iran showed a rather mild increase from 1983 to 2019 (0.4% to 3.5%) (Table 2 and Figure 3(a)). It shows that ciprofloxacin can still be used as an effective antibiotic against infections due to Salmonella serotypes in Iran. Contrary to ciprofloxacin, resistance rate to another quinolone, i.e. nalidixic acid was increasing during the monitored years (2.9% to 56.7%). Overall resistance to nalidixic acid in Iran was high (48.1%), which is similar to Korea (43.3%) and China (56%) [58,60]. Differences in results can be attributed to different Salmonella serotypes and regional variations. The main mechanisms involved in resistance to fluoroquinolones in Salmonella strains include mutations in the DNA gyrase genes, efflux pumping and maybe alterations in the expression of outer membrane proteins or lipopolysaccharides [59]. In the present study, Salmonella serotypes displayed a higher level of resistance to conventional antibiotics used as the first-line treatments for Salmonella-induced enteric fever infection, i.e. ampicillin (37.9%), chloramphenicol (27.2%) and trimethoprim-sulfamethoxazole (33.5%) compared with newer agents, i.e. fluoroquinolones and extended-spectrum cephalosporins. However, the trend of resistance of Salmonella serotypes to these antibiotics was variable from 1983 to 2019 in Iran (Figure 3(a)). On the other hand, frequency of MDR strains, combined resistance to ampicillin, chloramphenicol and trimethoprim-sulfamethoxazole, was low in Iran (9%). However, the prevalence of MDR strains is variable worldwide. This is due to the widespread use of the mentioned antibiotics that has caused these drugs to become obsolete in some regions [56]. Given the results of this study, continuing these antibiotics in Iran can lead to a similar outcome. Resistance rates to the above-mentioned three drugs in Iran were much higher than those reported for the United States, Greece, Turkey and Italy [59]. Extended-spectrum cephalosporins are another class of antibiotics, which can be used in severe infections when ciprofloxacin is contraindicated [57]. Fortunately, the resistance of Salmonella serotypes against both classes of antibiotics, i.e. fluoroquinolones and extended-spectrum cephalosporins, was low in Iran, except for ceftizoxime (50.6%) (6.8% to ceftazidime, 8.8% to ceftriaxone, 7.6% to cefotaxime and 5.8% to cefixime). Furthermore, the frequency of ESBLs which confer Salmonella serotypes resistance to the third-generation cephalosporins was low in Iran (6.5%). On the other hand, the trend of resistance of Salmonella serotypes to these drugs in Iran was not worrisome (Figure 3(b)). However, given the ability of Salmonella serotypes to establish zoonotic infections as well as the human chronic carriers, overuse of fluoroquinolones and extended-spectrum cephalosporins in both clinical settings and animal industry can lead to the spread of antimicrobial resistance [59]. In addition to foods of animal products, which can act as the primary source of antimicrobial-resistant Salmonella infection, the bacterium is able to acquire resistance genes from other enteric pathogens through transferable plasmids, transposons, and integrons [59]. Therefore, it is necessary to apply strategies to decrease drug-resistant Salmonella infections, such as stopping the use of antimicrobial agents in food animal industries and continuous monitoring of drug resistance of foodborne Salmonella in both clinical and non-clinical specimens via routine susceptibility testing. In addition to the third-generation cephalosporins, the use of azithromycin has been recommended as the treatment of choice against infections caused by MDR and fluoroquinolone-resistant Salmonella serotypes [56]. In addition to azithromycin, carbapenems and tigecycline are drugs of choice for the treatment of Salmonella infections resistant to classical first-line antibiotics, fluoroquinolones and third-generation cephalosporins [61]. In accordance with the reported results from Korea (0%) [58], the prevalence of imipenem-resistant Salmonella serotypes was low in Iran (2.7%). Our results showed that meropenem resistance rate was also low in Iran (2.2%). However, there was not enough information on azithromycin- and tigecycline-resistant Salmonella serotypes in Iran.
Conclusion
Findings of the present study showed a rising trend of resistance to the drugs of choice for the treatment of Salmonella infections, i.e. ampicillin, chloramphenicol and trimethoprim-sulfamethoxazole in Iran. Therefore, to prevent the emergence and spread of MDR strains in Iran, the following measures are recommended: prudent use of antibiotics, performing continuous antimicrobial susceptibility testing, using effective antibiotics with low bacterial resistance rates such as ciprofloxacin, third-generation cephalosporins and carbapenems, and testing bacterial resistance to other effective antibiotics such as azithromycin and tigecycline. Additionally, there is a need for additional comprehensive systematic reviews and meta-analyses in Iran to obtain information on the prevalence of resistant Salmonella isolates in non-clinical samples. This information will help reducing the spread of resistance from animal to human pathogens.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Almashhadany DA. Occurrence and antimicrobial susceptibility of Salmonella isolates from grilled chicken meat sold at retail outlets in Erbil City, Kurdistan region, Iraq. Ital J Food Saf. 2019;8(2):8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Carroll KC, Butel JS, Morse SA.. Jawetz Melnick & Adelbergs medical microbiology. 27th ed. Pennsylvania: McGraw Hill Professional; 2016. p. 239–242. [Google Scholar]
- [3].Murray PR, Rosenthal KS, Pfaller MA. Medical microbiology. 8th ed. UK: Elsevier Health Sciences; 2015. p. 259–260. [Google Scholar]
- [4].Ruby T, McLaughlin L, Gopinath S, et al. Salmonella’s long-term relationship with its host. FEMS Microbiol Rev. 2012;36(3):600–615. [DOI] [PubMed] [Google Scholar]
- [5].Eng SK, Pusparajah P, Ab Mutalib NS, et al. Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci. 2015;8(3):284–293. [Google Scholar]
- [6].Ailes E, Budge P, Shankar M, et al. Economic and health impacts associated with a Salmonella typhimurium drinking water outbreak− Alamosa, CO, 2008. PloS One. 2013;8(3):e57439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].World Health Organization . WHO publishes list of bacteria for which new antibiotics are urgently needed. Geneva: WHO; 2017. [Google Scholar]
- [8].Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6(7):e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Munn Z, Moola S, Lisy K, et al. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and incidence data. Int J Evid Based Healthc. 2015;13:147–153. [DOI] [PubMed] [Google Scholar]
- [10].Farahani RK, Ehsani P, Ebrahimi-Rad M, et al. Molecular detection, virulence genes, biofilm formation, and antibiotic resistance of Salmonella enterica serotype Enteritidis isolated from poultry and clinical samples. Jundishapur J Microbiol. 2018;11(10):e69504. [Google Scholar]
- [11].Soltan Dallal MM, Motalebi S, Masoomi Asl H, et al. Burden of Food-Related Illness Caused by Resistant Salmonella spp. and Shigella spp.: Harbingers of Multistate Outbreaks in 2012 and 2013. Int J Enteric Pathog. 2015;3(4):1–4. [Google Scholar]
- [12].Saboohi R, Rajaei B, Rad NS, et al. Molecular detection and association of qnrA, qnrB, qnrS and blaCMY resistance genes among clinical isolates of Salmonella spp. in Iran. Adv Microbiol. 2014;4(01):63–68. [Google Scholar]
- [13].Iranshahi N, Ranjbar R, Siadat SD, et al. Evaluation of nalidixic acid susceptibility testing for screening of clinical strains of Salmonella with decreased susceptibility to ciprofloxacin. Iran J Med Microbiol. 2009;2(3 and 4):39–45. [Article in Persian]. [Google Scholar]
- [14].Sepehri Rad N, Razavi MR, Siadat SD, et al. Evaluation of antibiotic resistance to fluoroquinolones and third generation cephalosporins in Iranian clinical isolates of salmonella spp. Int J Mol Clin Microbiol. 2012;2:194–198. [Google Scholar]
- [15].Amir Mozafari N, Forouhesh Tehrani H, Niakani M. Nalidixic acid resistance rate in typhoidal and non-typhoidal Salmonella isolated from hospitalized patients during one year period (2005-2006). RJMS. 2007;14(56):43–51. [Article in Persian]. [Google Scholar]
- [16].Bialvaei AZ, Pourlak T, Aghamali M, et al. The prevalence of CTX-M-15 extended-spectrum β-lactamases among Salmonella spp. and Shigella spp. isolated from three Iranian hospitals. Eur J Microbiol Immunol. 2017;7(2):133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Aminshahidi M, Arastehfar A, Pouladfar G, et al. Diarrheagenic Escherichia coli and Shigella with high rate of extended-spectrum Beta-lactamase production: two predominant etiological agents of acute diarrhea in Shiraz, Iran. Microb Drug Resist. 2017;23(8):1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Anvarinejad M, Pouladfar GR, Pourabbas B, et al. Detection of Salmonella spp. with the BACTEC 9240 automated blood culture system in 2008-2014 in Southern Iran (Shiraz): biogrouping, MIC, and antimicrobial susceptibility profiles of isolates. Jundishapur J Microbiol. 2016;9(4):e26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Abdollahi A, Najafipour S, Kouhpayeh SA, et al. Salmonella enterica: serotyping, drug resistance & extended spectrum of β-Lactamase (ESBLs). J Fasa Univ Med Sci. 2011;1(1):38–44. [Article in Persian]. [Google Scholar]
- [20].Yousefi-Mashouf R, Moshtaghi AA. Frequency of typhoidal and non-typhoidal Salmonella species and detection of their drugs resistance patterns. J Res Health Sci. 2007;7(1):49–56. [PubMed] [Google Scholar]
- [21].Afzali H, Taghavi Ardekani A, Rasa H. Evaluation of antibiotic sensitivity of Shigella, Salmonella, and Vibrio cholera in patients with acute diarrhea referred to reference laboratory of Kashan University of Medical Sciences from 2000 to 2001. KAUMS J (FEYZ). 2001;5(3):47–58. [Article in Persian]. [Google Scholar]
- [22].Soltan Dallal MM, Khalilian M, Masoumi Asl H, et al. Molecular epidemiology and antimicrobial resistance of Salmonella spp. isolated from resident patients in Mazandaran Province, Northern Iran. J Food Qual Hazards Control. 2016;3(4):146–151. [Google Scholar]
- [23].Eshaghi SZ, Fahimi H, Fardsanei F, et al. Antimicrobial resistance and presence of cass 1 integrons among different serotypes of Salmonella spp. recovered from children with diarrhea in Tehran, Iran. Infect Disord Drug Targets. 2019;19:1–7. [DOI] [PubMed] [Google Scholar]
- [24].Fardsanei F, Dallal MM, Douraghi M, et al. Antimicrobial resistance, virulence genes and genetic relatedness of Salmonella enterica serotype Enteritidis isolates recovered from human gastroenteritis in Tehran, Iran. J Glob Antimicrob Resist. 2018;12:220–226. [DOI] [PubMed] [Google Scholar]
- [25].Ranjbar R, Ardashiri M, Samadi S, et al. Distribution of extended-spectrum β-lactamases (ESBLs) among Salmonella serogroups isolated from pediatric patients. Iran J Microbiol. 2018;10(5):294–299. [PMC free article] [PubMed] [Google Scholar]
- [26].Ranjbar R, Elhaghi P, Shokoohizadeh L. Multilocus sequence typing of the clinical isolates of Salmonella enterica serovar Typhimurium in Tehran hospitals. Iran J Med Sci. 2017;42(5):443–448. [PMC free article] [PubMed] [Google Scholar]
- [27].AbaspourShoushtari F. Investigating classes of integrons in Salmonella infantis isolated from clinical samples and their antibiotic resistance profile. J Ilam Univ Med Sci. 2018;25(6):97–105. [Article in Persian]. [Google Scholar]
- [28].Najafi MR, Parviz M, Amini K. Detection of cmlA/tetR, bla PSE-1, bla TEM and sip B in the Salmonella strains by multiplex-PCR method and their antibiotic resistance pattern. Med Sci. 2017;27(2):119–125. [Article in Persian]. [Google Scholar]
- [29].Amiri S, Moradli G. Molecular identification of virulence genes (agfA and mgtC) in Salmonella Typhimurium strains isolated from children with gastroenteritis using multiplex PCR method and determination of their antibiotic susceptibility pattern. J Babol Univ Med Sci. 2016;18(10):40–45. [Article in Persian]. [Google Scholar]
- [30].Malehmir S, Ranjbar R, Harzandi N. The molecular study of antibiotic resistance to quinolones in Salmonella enterica strains isolated in Tehran, Iran. Open Microbiol J. 2017;11:189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Amini K, Mobasseri P, Mokhtari A. Detection of blaPSE and blaTEM genes encoding B-Lactamase in clinical samples of Salmonella Typhimurium by multiplex PCR. Iran J Med Microbiol. 2016;10(3):73–78. [Article in Persian]. [Google Scholar]
- [32].Mirjafari Tafti ZS, Rahbar M, Eslami P, et al. A survey of the epidemiology and antibiotic resistance patterns of enteropathogens isolates in an Iranian hospital. Int J Enteric Pathog. 2016;4(1):1–4. [Google Scholar]
- [33].Rizi KS, Peerayeh SN, Bakhshi B, et al. Prevalence of the blaCTX-M-1 group and their transferability in resistant clinical isolates of Salmonella serogroups from several hospitals of Tehran. Iran J Microbiol. 2015;7(4):203–207. [PMC free article] [PubMed] [Google Scholar]
- [34].Farahani NN, Jazi FM, Nikmanesh B, et al. Prevalence and antibiotic susceptibility patterns of Salmonella and Shigella species isolated from pediatric diarrhea in Tehran. Arch Pediatr Infect Dis. 2018;6(4):e57328. [Google Scholar]
- [35].Soltan Dallal MM, Rastegar Lari A, Sharifi Yazdi MK. Pattern of serotyping and antibiotic resistance of Salmonella in children with diarrhea. J Gorgan Uni Med Sci. 2014;16(1):100–105. [Article in Persian]. [Google Scholar]
- [36].Bakhshi B, Dehghan-Mouriaabadi A, Kiani P. Heterogeneity of multidrug-resistant Salmonella enterica isolates with increasing frequency of resistance to ciprofloxacin during a 4-year period in Iran. Microb Drug Resist. 2018;24(4):479–488. [DOI] [PubMed] [Google Scholar]
- [37].Firoozeh F, Shahcheraghi FE, Salehi TZ, et al. Antimicrobial resistance profile and presence of class I integrongs among Salmonella enterica serovars isolated from human clinical specimens in Tehran, Iran. Iran J Microbiol. 2011;3(3):112–117. [PMC free article] [PubMed] [Google Scholar]
- [38].Ranjbar R, Rahmati H, Shokoohizadeh L. Detection of common clones of Salmonella enterica serotype Infantis from human sources in Tehran hospitals. Gastroenterol Hepatol Bed Bench. 2018;11(1):54–59. [PMC free article] [PubMed] [Google Scholar]
- [39].Tajbakhsh M, Avini MY, Alikhajeh J, et al. Emergence of bla CTX-M-15, bla TEM-169 and bla PER-1 extended-spectrum β-lactamase genes among different Salmonella enterica serovars from human faecal samples. Infect Dis. 2016;48(7):550–556. [DOI] [PubMed] [Google Scholar]
- [40].Hamidian M, Tajbakhsh M, Tohidpour A, et al. Detection of novel gyrA mutations in nalidixic acid-resistant isolates of Salmonella enterica from patients with diarrhoea. Int J Antimicrob Agents. 2011;37(4):360–364. [DOI] [PubMed] [Google Scholar]
- [41].Rajaei B, Siadat SD, Rad NS, et al. Molecular detection of antimicrobial resistance gene cassettes associated with class 2 integron in Salmonella serovars isolated in Iran. Br Microbiol Res J. 2014;4(1):132–141. [Google Scholar]
- [42].Eshraghi S, Dalall MM, Fardsanei F, et al. Salmonella Enteritidis and antibiotic resistance patterns: a study on 1950 children with diarrhea. Tehran Univ Med J. 2010;67(12):882–886. [Article in Persian]. [Google Scholar]
- [43].Hamidian M, Tajbakhsh M, Walther-Rasmussen J, et al. Emergence of extended-spectrum beta-lactamases in clinical isolates of Salmonella enterica in Tehran, Iran. Jpn J Infect Dis. 2009;62(5):368–371. [PubMed] [Google Scholar]
- [44].Ranjbar R, Giammanco GM, Farshad S, et al. Serotypes, antibiotic resistance, and class 1 integrons in Salmonella isolates from pediatric cases of enteritis in Tehran, Iran. Foodborne Pathog Dis. 2011;8(4):547–553. [DOI] [PubMed] [Google Scholar]
- [45].Tajbakhsh M, Hendriksen RS, Nochi Z, et al. Antimicrobial resistance in Salmonella spp. recovered from patients admitted to six different hospitals in Tehran, Iran from 2007 to 2008. Folia Microbiol. 2012;57(2):91–97. [DOI] [PubMed] [Google Scholar]
- [46].Naghoni A, Ranjbar R, Tabaraie B, et al. High prevalence of integron-mediated resistance in clinical isolates of Salmonella enterica. Jpn J Infect Dis. 2010;63(6):417–421. [PubMed] [Google Scholar]
- [47].Irajian G, Ranjbar R, Jazayeri Moghadas A. Detection of extended spectrum beta lactamase producing Salmonella spp. and multidrug resistance pattern. Iran J Pathol. 2009;4(3):128–132. [Google Scholar]
- [48].Morshed R, Peighambari SM. Drug resistance, plasmid profile and random amplified polymorphic DNA analysis of Iranian isolates of Salmonella Enteritidis. New Microbiol. 2010;33(1):47–56. [PubMed] [Google Scholar]
- [49].Pourakbari B, Sadr A, Ashtiani MT, et al. Five-year evaluation of the antimicrobial susceptibility patterns of bacteria causing bloodstream infections in Iran. J Infect Dev Ctries. 2012;6(02):120–125. [DOI] [PubMed] [Google Scholar]
- [50].Bahrmand AR, Velayati AA. Antimicrobial resistance pattern and plasmid profile of Salmonella Typhi isolated from an outbreak in Tehran province. Scand J Infect Dis. 1997;29(3):265–269. [DOI] [PubMed] [Google Scholar]
- [51].Velayati AA, Ghazi Saidi K, Taravati MR. A study of Salmonella, Shigella and enteropathogenic Escherichia coli serotypes in acute gastroenteritis children under the age of five. Med J Islam Repub Iran. 1987;1(1):22–31. [Google Scholar]
- [52].Farhoudi-Moghaddam AA, Katouli M, Jafari A, et al. Antimicrobial drug resistance and resistance factor transfer among clinical isolates of salmonellae in Iran. Scand J Infect Dis. 1990;22(2):197–203. [DOI] [PubMed] [Google Scholar]
- [53].Aghdasi-Araghinezhad R, Amini K. Study of antibiotic resistance pattern and incidence of pathogenic genes of mgtC, spi4R, agfA, invE/A and ttrC in Salmonella Infantis isolated from clinical specimens. Feyz J Kashan Univ Med Sci. 2017;21:442–449. [Google Scholar]
- [54].Bakhshi B, Eftekhari N, Pourshafie MR. Genetic elements associated with antimicrobial resistance among intestinal bacteria. Jundishapur J Microbiol. 2014;7(5):e9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Amini K. Prevalence of antibiotic resistance genes in Salmonella Enteritidis isolated from animal and human and determining their antibiotic resistance patterns. J Comp Pathobiol. 2016;12(4):1733–1740. [Article in Persian]. [Google Scholar]
- [56].Britto CD, Wong VK, Dougan G, et al. A systematic review of antimicrobial resistance in Salmonella enterica serovar Typhi, the etiological agent of typhoid. PLOS Negl Trop Dis. 2018;12(10):e0006779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Weill FX, Guesnier F, Guibert V, et al. Multidrug resistance in Salmonella enterica serotype Typhimurium from humans in France (1993 to 2003). J Clin Microbiol. 2006;44(3):700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yoon KB, Song BJ, Shin MY, et al. Antibiotic resistance patterns and serotypes of Salmonella spp. isolated at Jeollanam-do in Korea. Osong Public Health Res Perspect. 2017;8(3):211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Su LH, Chiu CH, Chu C, et al. Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin Infect Dis. 2004;39(4):546–551. [DOI] [PubMed] [Google Scholar]
- [60].Qu M, Lv B, Zhang X, et al. Prevalence and antibiotic resistance of bacterial pathogens isolated from childhood diarrhea in Beijing, China (2010–2014). Gut Pathog. 2016;8(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Dyson ZA, Klemm EJ, Palmer S, et al. Antibiotic resistance and typhoid. Clin Infect Dis. 2019;68(Supplement_2):S165–S170. [DOI] [PMC free article] [PubMed] [Google Scholar]


