Abstract
SUMO is a ubiquitin-like protein that covalently binds to lysine residues of target proteins and regulates many biological processes such as protein subcellular localization or stability, transcription, DNA repair, innate immunity, or antiviral defense. SUMO has a critical role in the signaling pathway governing type I interferon (IFN) production, and among the SUMOylation substrates are many IFN-induced proteins. The overall effect of IFN is increasing global SUMOylation, pointing to SUMO as part of the antiviral stress response. Viral agents have developed different mechanisms to counteract the antiviral activities exerted by SUMO, and some viruses have evolved to exploit the host SUMOylation machinery to modify their own proteins. The exploitation of SUMO has been mainly linked to nuclear replicating viruses due to the predominant nuclear localization of SUMO proteins and enzymes involved in SUMOylation. However, SUMOylation of numerous viral proteins encoded by RNA viruses replicating at the cytoplasm has been lately described. Whether nuclear localization of these viral proteins is required for their SUMOylation is unclear. Here, we summarize the studies on exploitation of SUMOylation by cytoplasmic RNA viruses and discuss about the requirement for nuclear localization of their proteins.
Keywords: SUMO, RNA viruses, Interferon
SUMO Conjugation
SUMOylation consists in the covalent attachment of the small ubiquitin-like modifier (SUMO) proteins to specific lysine residues of a target protein. This conjugation is an enzymatic process that occurs in cascade. The precursor SUMO proteins are proteolytically processed to the mature forms, exhibiting their C-terminal diglycine motifs, by sentrin/SUMO-specific proteases (SENPs). The resulting mature forms of SUMO are then activated by the SUMO-activating E1 enzyme (SAE1/SAE2). Subsequently, the SUMO molecule is transferred to the E2-conjugating enzyme Ubc9, the only E2 SUMOylation enzyme discovered to date. Finally, the carboxyl group of the glycine residues at the SUMO carboxy terminus forms an isopeptide linkage with the ε-amino group of a lysine residue on the target protein, a step that can be facilitated by SUMO E3 ligases. SUMOylation can be then reversed by SENPs. Usually, the target lysine for SUMO is located in the consensus sequence ψKxE (where ψ is a hydrophobic residue and x any residue) in the target protein, although SUMO can be also conjugated to lysine residues located in non-consensus sequences. Four different SUMO isoforms have been described in mammals (SUMO1 to SUMO4). SUMO1 protein is found mainly conjugated to targets due to its limited quantity and only shares 50% identity with SUMO2/3. SUMO2 and SUMO3 share 97% sequence identity and often are referred to as SUMO2/3, and its conjugation is stress-inducible [1]. SUMO4 remains poorly characterized and its physiological relevance is still unclear [2, 3]. The main function of SUMO proteins is to regulate protein–protein interaction, which ultimately results in changes in protein subcellular localization or stability, transcription, DNA repair, innate immunity, or antiviral defense. Interestingly, many SUMO-modified proteins contain SUMO-interacting motifs (SIMs), allowing the noncovalent interaction of SUMO substrates with SUMO or non-SUMOylated proteins [4, 5].
Regulation of the Type I Interferon (IFN) Production by SUMO
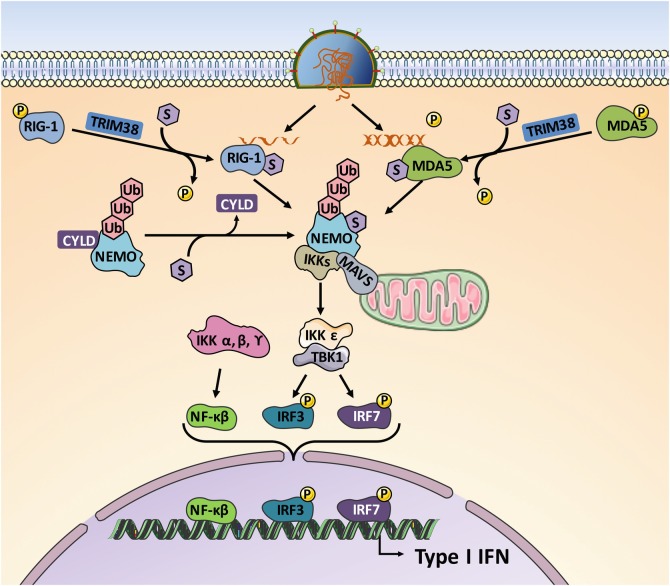
The first line of defense against viruses is the induction of type I interferons (IFNs). During RNA virus infection, the viral genomic RNA and replication intermediates produced by the virus are recognized as pathogen-associated molecular pattern (PAMP) by pattern-recognition receptor (PRR) proteins such as Toll-like receptors, retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetic and physiology 2 (LGP2), initiating a signaling cascade that results in the production of type I IFN. SUMO interacts with different intermediates along this signaling cascade modulating the transcriptional transactivation of type I IFN (Fig. 11.1).
Fig. 11.1.
Transactivation of type I IFN upon RNA virus infection by SUMO. Detection of the viral RNA by RIG-I and MDA5 led to activation of IFN production. SUMO, by conjugating MDA5 and RIG-I, inhibits their phosphorylation and induces their stabilization [6]. Conjugation of SUMO to NEMO inhibits its interaction with the deubiquitinase CYLD strengthening the activation of IKK and inducing an increase of type I IFN production [7]
SUMOylation of MDA5 and RIG-I, promoted by the SUMO E3 ligase TRIM38, has been shown to be required for its dephosphorylation and activation upon virus infection [6]. Once RIG-I is activated, it recruits the inhibitor of kB kinase (IKK) α, β, and ε through its interaction with MAVS, leading to the activation of NF-kβ, interferon regulatory factor (IRF)3, and IRF7, and consequently resulting in IFNβ and NF-kβ promoter activation. Frequently, the activation of both NF-kβ and IRF3/7 pathways by RIG-I in response to virus infection requires the NF-kβ essential modulator (NEMO), an adaptor protein that organizes the assembly of IKKs into activated high-molecular-weight complexes. Conjugation of SUMO2/3 to NEMO, a process downmodulated by SENP6, inhibits its interaction with the deubiquitinase CYLD strengthening the activation of IKK [7]. The noncanonical IKK kinase TANK-binding kinase 1 (TBK1), which mediates the activation of IRF3, has also been shown to be a pivotal player in antiviral innate immunity, and its modification by SUMO supports its antiviral function, likely through contributing to adaptor binding for the transduction of antiviral signaling [8].
In addition to its role as a modulator of the RIG-I phosphorylation, SUMO1 conjugation to RIG-I also inhibits its K48-linked polyubiquitination and degradation [6] and promotes its interaction with MAVS [9], inducing activation of the IFNβ promoter. In addition, upregulation of IFN expression has been observed as a result of the modification of the virus sensor MDA5 by SUMO1 after Ubc9 or PIAS2β overexpression [10].
SUMOylation can also have a negative impact on IFN production. SUMO1 can inactivate NF-kB by modifying IkBα and inhibiting its degradation [11]. In addition, SUMOylation of IRF3/7 in response to Toll-like receptor (TLR) and RIG-I activation can act as a transcriptional repressor of IRF3 and IRF7, a mechanism that has been postulated to be part of the negative feedback loop of normal IFN signaling [12]. However, conjugation of SUMO to IRF3 not necessarily inhibits IRF3. Ran and colleagues found that SUMO can conjugate to K87 in IRF3, a lysine residue involved in IRF3 K48-ubiquitin conjugation and degradation [13]. Consequently, I IFN downmodulation of IRF3 SUMOylation induced by SENP2 promotes its ubiquitination, negatively regulating the virus-triggered type induction and cellular antiviral response [13]. Whereas PIAS1 has been reported to work as E3 SUMO ligase for both IRF3 and IRF7 [14], the repression of the ability of IRF7 to induce type I IFN transcription by SUMOylation has been shown to be promoted by TRIM28 [15]. Together, these studies show that SUMO plays an important role in the type I IFN expression in response to RNA virus infection.
SUMO and Type I IFN Responses
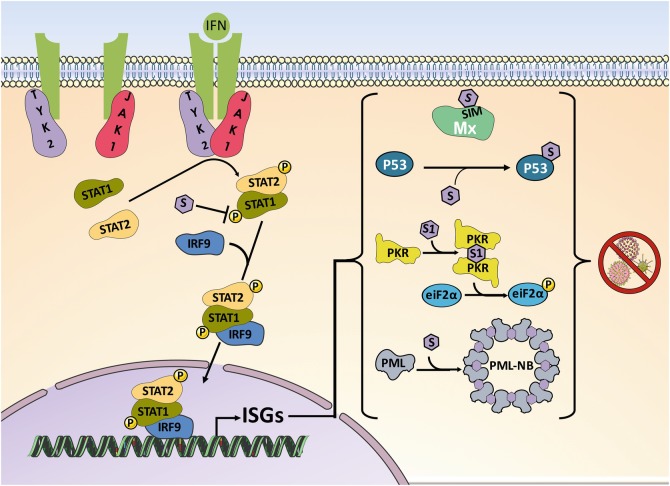
IFN binding to specific receptors at the cell surface activates the receptor-associated protein tyrosine kinases Janus kinase 1(JAK1) and tyrosine kinase 2 (TYK2), which phosphorylate the signal transducer and activator of transcription 1 (STAT1) and STAT2. These two proteins form dimers and associate with IRF9 to form a transcriptionally active IFN-stimulated gene factor 3 (ISGF3). ISGF3 binds to IFN-stimulated response elements (ISREs) leading to transcription of IFN-stimulated genes (ISGs). This canonical type I IFN signaling can be modulated by SUMO (Fig. 11.2).
Fig. 11.2.
Effect of SUMO on type I IFN response. Activation of STAT1 in response to IFN is downmodulated by SUMO conjugation. However, SUMO does not alter the IFNα signaling, likely because STAT2 is not affected by SUMO and it can compensate for the reduction in STAT1 phosphorylation [16–20]. Conjugation of SUMO to p53, PML, and MxA promotes their antiviral activities. The regulation of the antiviral activity of PKR by SUMO may depend on the SUMO isoform [21–23]
SUMO has been shown to interact with STAT1 in a covalent manner, and this interaction has been proposed to inhibit STAT1 activity, since a STAT1 SUMOylation mutant is hyperphosphorylated and exhibits increased DNA binding on STAT1 responsive gene promoters [16–18], and phosphorylation of STAT1 induced by IFNβ treatment inhibits its SUMOylation [19]. In addition, SUMO overexpression has been shown to reduce the IFN-induced STAT1 phosphorylation and downmodulate IFNγ transcriptional responses [20]. However, SUMO does not alter IFNα signaling, likely because STAT2 is not affected by SUMO and it can compensate for the reduction in STAT1 phosphorylation [20].
IFN establishes an antiviral state in cells by inducing a number of proteins with antiviral activity. Interestingly, many of these IFN-induced proteins are SUMO substrates and their activities are regulated by SUMOylation and/or noncovalent interaction with SUMO.
The GTPases Mx
exert antiviral activity against a variety of RNA and DNA viruses. MxA protein has been reported to interact with SUMO1, SAE2, and Ubc9 [24, 25]. In addition, a recruitment of Mx to PML-NBs and its modification by SUMO2/3 have also been reported [24]. Interestingly, noncovalent SUMO interaction but not SUMOylation has been found to be essential for the antiviral effect exerted by MxA on vesicular stomatitis virus (VSV), since Ubc9 depletion does not affect the protective effect of MxA on VSV replication [26].
The Tumor Suppressor Protein p53
can interfere with the replication of several viruses, and activation of p53 has been proposed as a potential therapy against viral infections [27]. IFN treatment induces the transactivation of p53 [28], but it also induces its SUMOylation, modification that contributes to the antiviral functions of IFN [29]. In addition, SUMOylation of p53 induced by IFN has also been shown to contribute to senescence induction, a process that helps to control virus replication [29, 30].
The Double-Stranded RNA (dsRNA)-Dependent Protein Kinase (PKR)
is an IFN-inducible protein that can phosphorylate the alpha subunit of the protein synthesis initiation factor eIF2α, resulting in a shut-off of protein translation, apoptosis induction, and inhibition of virus replication [31]. PKR is regulated by covalent and noncovalent interaction with SUMO [21–23]. SUMOylation, as well as noncovalent SUMO interaction, is required for efficient PKR–dsRNA interaction, PKR dimerization, phosphorylation of eIF-2α, and inhibition of VSV replication [21, 22]. Although virus infection promotes the modification of PKR by SUMO1 and SUMO2/3, recently, a specific role of SUMO3 counteracting PKR activation and stability upon virus infection has been proposed [23].
PML
is an IFN-induced protein that plays a key role in the establishment of innate antiviral responses. Thus, PML KO mice are more sensitive than WT to infection with different viruses such as lymphocytic choriomeningitis virus (LCMV), encephalomyocarditis virus (EMCV), or VSV, and cells from these mice are also more sensitive to infection with rabies virus [32–34]. In addition, overexpression of specific PML isoforms confers resistance to some virus infections. Thus, PMLIII protects against human foamy virus (HFV) [35], poliovirus [36], VSV, or influenza A virus [37], and PMLIV confers resistance to rabies virus and EMCV infection [34]. PML interacts with SUMO in a covalent and noncovalent manner. SUMOylation of PML is required for PML-NB formation and for promoting SUMOylation of specific substrates recruited on PML-NBs [38–40]. Moreover, modification of PMLIV by SUMO positively regulates IFNβ synthesis through IRF3 activation, and it is required for protecting cells from virus infection [41]. PML-III and PML-IV also recruit Ubc9 to PML-NBs, and this recruitment has been shown to be essential for the increase in global SUMOylation induced by IFN [42]. This is not the unique mechanism that contributes to the increase in global SUMOylation upon IFN treatment. Among the IFN-regulated proteins is Lin28B, a RNA binding protein that represses the expression of miRNAs from let-7 family [43–47] which targets SUMO transcripts [48].
The IFN-induced global SUMOylation, as well as the SUMOylation of viral restriction factors induced by IFN, is likely part of the cellular antiviral defense mechanisms of the host since some viruses have developed strategies to inhibit those processes. In addition, some viruses have evolved to hijack the SUMOylation machinery of the cell to regulate their own proteins.
SUMO and Ubc9 proteins are mainly located at the cell nucleus. Therefore, for many years it has been suggested that SUMOylation was relevant mainly for infection of nuclear replicating viruses. In agreement with this hypothesis, exploitation of the SUMO pathway has been reported to be a common strategy of DNA viruses and of those few RNA virus replicating in the nucleus, such as influenza virus or human immunodeficiency virus (HIV)-1 [49, 50]. Lately, numerous reports demonstrate that the SUMO pathway is also relevant for cytoplasmic RNA viruses.
SUMO and RNA viruses
Reoviridae
Viruses of the Reoviridae family are non-enveloped icosahedral virus of around 60–80 nm that contain from 9 to 12 segments of linear double-stranded RNA. Although reovirus replication occurs in the cytoplasm, several viral proteins have been detected in the cell nucleus. Thus, the nonstructural sigma 1 protein, a determinant of reovirus virulence, contains a functional nuclear localization signal (NLS) [51], and it can be detected in the nucleus during reovirus infection [52, 53]. The avian reovirus core protein sigma A accumulates in the nucleoplasm of mammalian cells or in the nucleolus and cytoplasm of avian cells [54], and the reovirus minor capsid protein mu 2 of specific strains localizes to nuclear speckles [55].
The Reoviridae family is constituted by several genera, whose members have a varied host range. Rotavirus is a genus in the Reoviridae family that is recognized as the single most important cause of severe gastroenteritis in infants of a wide range of mammals. Rotavirus genome is constituted by 11 dsRNA segments encoding six structural (VP1–4, VP6, and VP7) and six nonstructural (NSP1–6) proteins. Early stages of viral assembly and viral RNA replication take place in virus-induced inclusion bodies called viroplasms localized in the infected cell cytoplasm. Viroplasms are formed by VP1, VP2, VP3, VP6, NSP2, and NSP5. Five out of six of the viroplasms components, VP1, VP2, NSP2, VP6, and NSP5 proteins, are modified by SUMO; and three of them, VP1, VP2, and NSP2 proteins, also interact in a noncovalent manner with SUMO. SUMOylation of NSP5 has been shown to be essential for the formation of viroplasm-like structures (VLS) generated by overexpression of VP2 (VLS-VP2i). In addition, upregulation of SUMOylation positively modulates rotavirus replication and viral protein production, whereas interference of Ubc9 produces a marked decrease in the synthesis of viral proteins and virus titer [56]. The extensive exploitation of the SUMOylation machinery by rotavirus evokes to that described for influenza virus [57]. However, in contrast to influenza virus, rotavirus conducts its life cycle in the cytoplasm and, so far, nuclear localization of rotavirus proteins has not been reported.
Members of the Orthoreovirus genus have been evaluated as putative anticancer agents and, as reported for rotavirus, Ubc9 also contributes to their efficient replication. Ubc9 has been shown to interact with the outer fiber protein VP55 from the grass carp reovirus (GCRV)-104 or the type II GCRV, with sigma C from avian reovirus (ARV) and with sigma 1 from mammalian reovirus (MRV) by using yeast two-hybrid system. Furthermore, a positive correlation between Ubc9 levels and (GCRV)-104 replication has been reported [58]. Therefore, SUMOylation has been proposed as a tool to improve the therapeutic efficacy of oncolytic reoviruses. Although it has been hypothesized that SUMOylation of the outer fiber proteins may increase tropism for host cells, so far, no SUMOylation of orthoreovirus proteins has been demonstrated.
Paramyxoviridae
Paramyxoviridae family is constituted by single-stranded negative-sense RNA genome viruses. Paramyxovirus replication takes place in the cytoplasm; however, several paramyxoviral proteins have been detected in the nucleus in infected cells. So far, there is just one example of exploitation of SUMOylation machinery by paramyxovirus.
Parainfluenza virus 5 (PIV5) is a prototypic member of the Rubulavirus genus. The genome of PIV5 encodes eight known proteins. After evaluation of the putative SUMOylation of four proteins of the virus (the two components of the viral RNA-dependent RNA polymerase L and P, the nucleocapsid NP protein, and the nonstructural V protein), only the P protein was found to be SUMOylated and only by SUMO1 and not by SUMO2/3 [59]. Analysis of a recombinant PIV5 containing a mutant of the P protein in the SUMOylation motif revealed a reduction in the titer, viral RNA synthesis, and protein expression relative to the WT PIV5. Therefore, SUMOylation was proposed to regulate PIV5 gene expression through the regulation of viral RNA transcription [59]. Whether the P protein of PIV5 can enter into the nucleus is unknown.
Pneumoviridae
Pneumoviridae is a family of large enveloped single-stranded negative-sense RNA virus. One of the members of the family that affects humans is the respiratory syncytial virus (RSV). Although RSV is a cytoplasmic virus able to replicate in enucleated cells, its matrix (M) protein has been found inside the nucleus of infected cells [60].
Different reports reveal a relationship between RSV and the host cell SUMOylation machinery. Thus, an association between a cluster of host proteins involved in the SUMOylation process and the phosphoprotein (P) of the virus has been found after integration of proteome and transcriptome datasets [61]. In addition, SUMO seems to play a role in the pathogenesis of the virus [62]. The cellular damage and lung inflammation caused by RSV infection are associated with the generation of reactive oxygen species (ROS) and oxidative stress, via degradation of the transcription factor NF-E2-related factor (NRF2), which occurs in a SUMO-specific E3 ubiquitin-ligase-RING finger protein (RNF4)-dependent manner [62]. However, so far, no SUMOylation of pneumovirus proteins has been demonstrated.
Nodaviridae
Nodavirus are non-enveloped viruses with a genome consisting in two molecules of single-stranded positive-sense RNA. The family includes two genera, Alphanodavirus and Betanodavirus. Viruses belonging to the first genus infect insects, whereas betanodaviruses infect fishes. Nodavirus replicate in the host cell cytoplasm. However, the capsid protein of nodavirus contains a nuclear localization signal that targets the viral capsid to the nucleus of infected cells [63]. Infection with the betanodavirus red-spotted grouper nervous necrosis virus (RGNNV) induces the transcriptional upregulation of SUMO3, and SUMO3 overexpression increases IFN and ISRE promoter activity, suggesting that SUMOylation may play a role in the immune response of fish to viral infection [64]. However, SUMO3 has been shown to enhance RGNNV replication in vitro [64], suggesting that RGNNV is able to exploit the host SUMOylation machinery. So far, no SUMOylation of nodavirus proteins has been demonstrated.
Picornaviridae
Picornaviridae is a family of non-enveloped viruses with single-stranded positive-sense RNA genome containing usually a single open reading frame encoding a precursor protein that can be processed by viral proteinases. It contains more than 30 genera and more than 75 species. Although the replication of picornaviruses occurs in the cytoplasm, some viral 3C proteases have been found to enter the nucleus using a NLS present in the RNA-dependent RNA polymerase (3D), in order to prevent host transcription and cap-dependent translation to provide cellular resources for viral replication [65]. In addition to 3C and 3D, EMCV nonstructural 2A and 3B proteins have been also found in the cell nucleus [66].
The Enterovirus 71 (EV71) belongs to the Enterovirus genera within the Picornaviridae family, and it is the common cause of hand, foot, and mouth disease. The polymerase 3D of EV71 is modified by SUMO1 and SUMO3, and this modification promotes its K63-linked ubiquitination enhancing the stability of the viral protein. Consequently, SUMOylation is required for 3D polymerase activity and virus replication, and this may explain why SUMO1 overexpression enhances viral replication [67]. Interestingly, a SUMO-dependent ubiquitination of the protease 3C has also been described. SUMO modification of 3C has been proposed to be a host cell defense mechanism against virus replication and apoptosis induction during EV71 infection because SUMO-dependent ubiquitination of 3C was associated with reduced protease activity of 3C in vitro, increased degradation, decreased apoptosis induction and virus replication, and reduced EV71-elicited neurovirulence in a mouse model [68].
A connection between the human enteroviruses Coxsackievirus B5 (CVB5) or CVB3 and the host cell SUMOylation system has also been proposed. CVB5 infection induces redistribution of SUMO1 and Ubc9 all over the cell, which has led to the hypothesis that CVB5 might interfere with, and perhaps destabilize, protein SUMOylation [69]. In addition, CVB3 infection has been shown to induce the degradation of p53, likely through the promotion of the SUMOylation of the proteasome regulator REG to promote viral replication [70]. However, so far, no SUMOylation of CVB5 or CVB3 proteins has been demonstrated.
Another picornavirus that exploits the host cell SUMOylation machinery to modulate the activity of restriction factors is EMCV, the prototype of the Cardiovirus genus. EMCV, likely through the protease 3C, induces proteasome and SUMO-dependent degradation of PMLIII [33].
Bunyaviridae
Bunyaviridae is a large family of enveloped single-stranded RNA viruses. It is constituted by 5 genera: Phlebovirus, Nairovirus, Hantavirus, Orthobunyavirus, and Tospovirus. Although bunyavirus replication occurs in the cytoplasm, virus production is blocked in enucleated cells. Therefore, the role of the nucleus is unclear.
So far, a role of SUMO in the viral life cycle of viruses belonging to this family has only been reported for Hantavirus. The hantaviruses genome consists of three RNA segments encoding the nucleocapsid protein NP, two surface glycoproteins G1 and G2, and the viral polymerase L [71]. Interaction between the nucleocapsid protein of different hantaviruses, including Hantaan virus (HTVN), Seoul virus (SEOV), Tula virus (TULV), Puumala virus, and Andes virus (ANDV), and SUMO1 or SUMOylation pathway proteins, such as Ubc9, PIAS1, PIASxβ, or RanGAP, has been reported [72–74]. However, so far, no SUMOylation of NP was observed and the function of these interactions in the viral life cycle remains unknown.
Coronaviridae
Coronaviridae is a family of enveloped single-stranded positive-sense RNA viruses. It has two subfamilies, Coronavirinae and Torovirinae. The entire replication cycle takes place in the cytoplasm. However, the multifunctional nucleocapsid N protein of coronavirus can be detected in the nucleolus [75, 76], although this ability varies between N proteins of different coronaviruses. The severe acute respiratory syndrome-related coronavirus (SARS-Cov) N protein can directly interact with human Ubc9 [77] and is modified by SUMO [78]. Analysis of an N protein SUMOylation mutant revealed that SUMOylation promotes N homo-oligodimerization and nucleolar localization and plays certain roles in the N-protein-mediated interference of host cell division.
Arteriviridae
Arteriviridae is a family of enveloped, single-stranded positive-sense RNA virus that includes only the Arterivirus genus. One of the members of the family is the porcine reproductive and respiratory syndrome virus (PRRSV). The genome of PRRSV contains several overlapping open reading frames. The ORF1a and 1b encode the viral nonstructural proteins that play important roles in replication and transcription of the viral genome, in modulation of the host innate immunity, and in pathogenesis and virulence. The remaining ORFs encode the structural proteins of PRRSV including the viral nucleocapsid N protein. The N protein is one of the most abundant PRRSV proteins, highly immunogenic in pigs and with important roles in PRRSV replication and immune evasion. PRRSV replicates in the cytoplasm of infected cells. However, the N protein localizes specifically in the nucleus and nucleolus of virus-infected cells, localization that modulates the pathogenesis of the virus in pigs [79].
Wang and collaborators found many conserved lysine residues in various proteins of PRRSV that were predicted to be SUMOylated using in silico prediction analysis of the amino acid sequences of PRRSV structural and nonstructural proteins [80]. Furthermore, the authors demonstrated that the PRRSV N protein can be modified by SUMO and that the nonstructural PRRSV proteins NSP1β, NSP4, NSP9, NSP10, and N colocalize and interact with Ubc9 [80]. Altogether these data suggest that the virus can exploit the cell host SUMOylation pathway. However, the overall effect of SUMO on PRRSV seems to be negative, as knockdown of Ubc9 promotes virus replication and overexpression of Ubc9 inhibits viral genomic replication [80]. The role of PRRSV N protein SUMOylation, the consequences of the interaction between Ubc9 and the viral proteins, or the molecular mechanisms by which Ubc9 modulates PRRSV replication are still unknown.
Filoviridae
Filoviridae is constituted by enveloped virus containing a non-segmented negative-strand RNA genome. This family contains some of the deadliest pathogens known to date. There are three Filoviridae genera: Ebolavirus, Marburgvirus, and Cuevavirus. So far, only one of the virus of the family, Ebola virus (EBOV), has been probed to exploit the SUMOylation pathway.
EBOV encode seven structural proteins: the nucleoprotein NP, the polymerase cofactor VP35, the matrix protein VP40, the transcription activator VP30, the minor matrix protein VP24, the glycoprotein GP, and the RNA-dependent RNA polymerase L. The GP transcript can be edited giving rise to four alternative forms of gene products. In addition to serve as structural components, EBOV proteins play multiple roles in the virus life cycle.
SUMO has been proposed to be critical for EBOV life cycle, and modulation of the interaction between SUMOylation pathway components and viral proteins may represent a novel target for therapeutics to block EBOV infection. VP35 has been reported to interact with Ubc9 and PIAS1 and induce a PIAS1-mediated SUMOylation of IRF7 and IRF3, a mechanism through which EBOV VP35 disrupts antiviral responses [14]. In addition, EBOV hijacks the cellular SUMOylation system in order to modify its own proteins. Thus, the multifunctional VP40 protein, involved in regulating virus budding, nucleocapsid recruitment, virus structure and stability, and viral genome replication and transcription, is modified by SUMO [81]. Interestingly, SUMO was also found to be included into the viral particles formed by VP40. One consequence of VP40 SUMOylation is the regulation of its ubiquitination [81]. Although EBOV replication takes place in the cytoplasm of the infected cells, EBOV VP40 protein has been detected in the cell nucleus [82].
Flaviviridae
Flaviviridae is a family of small enveloped viruses with non-segmented positive-sense RNA genomes. Flaviviridae genome encodes a single polyprotein that is cleaved into three structural proteins (capsid (C), premembrane (prM), and envelope (E)), and seven nonstructural (NS) proteins. Although flavivirus replication occurs in the cytoplasm, some viral proteins translocate to the nucleus during infection contributing to viral replication [83]. The Flaviviridae family includes four genera: Flavivirus, Pestivirus, Hepacivirus, and Pegivirus.
The Flavivirus genus consists mainly of arthropod-borne viruses and includes important human pathogens such as dengue virus (DENV), Zika virus (ZIKV), or yellow fever virus (YFV). Several studies indicate that SUMO plays an important role in flavivirus replication and suggest that SUMO inhibitors may be a broad spectrum anti-flaviviral strategy. Thus, treatment with the SUMOylation inhibitor 2-D08 has been reported to significantly reduce the replication of ZIKV in vitro in different cell types and in a dose-response manner [84]. How SUMOylation favors ZIKV replication is not clear. The ZIKV RNA-dependent RNA polymerase NS5 has a putative SIM domain that is highly conserved among Zika strains that is required for the NS5-mediated suppression of type I IFN signaling [84]. However, functionality of the SIM domain in the interaction between ZIKV NS5 and Ubc9 or SUMO or SUMOylation of the viral protein has not been demonstrated.
2-D08 treatment, as well as Ubc9 interference, also reduces replication of DENV [84, 85]. Several DENV proteins might interact with the SUMOylation machinery. Ubc9 has been predicted to bind to different DENV proteins by using a computational approach [86]. Furthermore, interaction between Ubc9 and the nonstructural proteins NS2B, NS4B, and NS5, as well as with the envelope protein E, was probed using a yeast two-hybrid assay [87, 88]. However, after evaluation of the putative SUMO1 modification of ten different DENV proteins, only SUMOylation of NS5 was demonstrated [85]. An intact SIM domain in NS5 was shown to be required for its SUMOylation, which protected DENV NS5 against proteasome degradation, supporting virus replication [84]. Interestingly, NS5 protein contains two functional NLS and is one of the DENV proteins that can be found inside the nucleus, where it interferes with cellular splicing [89]. Although Ubc9 was required for efficient replication of DENV, a study on type 2 dengue virus (DV-2) showed that overexpression of Ubc9 reduces the viral plaque formation in mammalian cells, likely due to the involvement of Ubc9 in the host defense system to prevent virus propagation [87].
SUMOylation inhibitor treatment also significantly inhibited the replication of other flavivirus such as Japanese encephalitis virus (JEV), West Nile virus (WNV), or YFV. In addition, in silico analysis of 78 representative flaviviruses revealed the presence of a putative SIM domain at the NS5 protein in 92.3% of the analyzed viruses [84]. Interestingly, the authors found that only the insect-specific flavivirus lacked putative SIMs. Therefore, they proposed that it can be an evolutionarily conserved modification process among flaviviruses to enhance virus replication and suppress host antiviral response.
The SUMO system has been shown to be also required for the replication of hepatitis C virus (HCV), the best characterized member of the Hepacivirus genus of the Flaviviridae family with several proteins containing functional NLSs and nuclear export signals (NESs) including the core, NS2, NS3, and NS5 proteins [90]. HCV infection upregulates SUMO1 expression, a host factor essential for HCV replication [91]. Ubc9 has also been shown to be required for HCV RNA replication [92]. The requirement of the host cell SUMOylation machinery for HCV replication is probably linked to the SUMOylation-dependent stability of NS5A [92] and to the SUMO-mediated regulation of the expression of the viral core protein [91]. The importance of NS5 SUMOylation for HCV was also supported by the constant reversion of the SUMO-defective mutant K348R of HCV NS5 to the WT sequence during virus replication [92]. In addition, SUMO1 is also required for lipid droplet accumulation, organelles that work as a platform for virus replication, assembly, and production [91].
In contrast to the essential role of SUMO1 and Ubc9 in HCV replication, PIAS2-mediated SUMOylation has been identified as a restriction factor against HCV replication [93]. Thus, an enhancement of HCV core, NS3, and NS5A protein expression in infected cells, as well as of viral assembly and budding efficiency, was observed after knockdown of PIAS2 [93]. In contrast, exogenous overexpression of PIAS2 decreased HCV core, NS3, and NS5A expression and the viral assembly and budding efficiency. When expressed together with SUMO1, PIAS2 induced the degradation of HCV core, NS3, and NS5A proteins expressed from individual plasmids and after treatment with the proteasome inhibitor MG132, PIAS2 interacts with and enhances SUMOylation of the core protein. Therefore, PIAS2 has been proposed to mediate the degradation of the core, NS5A, and NS3 proteins through a ubiquitin-independent proteasomal pathway [93].
Interestingly, SUMO1 may have a role in the promotion of HCV replication by alcohol exposure. Thus, alcohol treatment has been shown to induce upregulation in PIASγ expression which induces autophagy, at least partially through promoting the accumulation of SUMO1 conjugated proteins, and consequently promotes HCV replication [94].
SUMO has also been demonstrated to be relevant for members of the Flaviviridae family not pathogenic for humans such as the classical swine fever virus (CSFV). CSFV belongs to the genus Pestivirus within Flaviviridae family. It has three structural components, including the core protein. Core protein of CSFV interacts with SUMO1 and Ubc9, and mutant viruses with deletions in the putative SUMOylation sites of the core protein show attenuated phenotypes and limited spreading within infected swine, suggesting that SUMOylation pathway may contribute to curtailing viral clearance [95]. Only mutation for both SUMO1 and Ubc9 interaction led to complete attenuation. However, whether the core protein is indeed modified by SUMO or whether the attenuation is related with reduced SUMOylation is not known.
Rhabdoviridae
Viruses from the Rhabdoviridae family are single-stranded negative-sense RNA viruses. The prototypes of this family are VSV and rabies virus (RABV). Although VSV infection does not induce global SUMOylation [26, 49], it induces the SUMOylation of specific proteins, promoting their antiviral activity, such as the tensin homolog deleted for chromosome 10 (PTEN) [96], the double-stranded RNA-dependent protein kinase (PKR) [22], p53 [29], and MxA [26]. Based on these reports, an upregulation of SUMOylation will likely be negative for VSV replication. In fact, although SUMO expression reduced IFN synthesis upon VSV infection, stable expression of SUMO1 or SUMO3 resulted in resistance to VSV infection [26]. However, whether the control of VSV replication by SUMO overexpression is mediated by SUMOylation is not so clear, since the synthesis of VSV proteins is not affected by Ubc9 knockdown [26].
Whether SUMOylation of p53, PKR, or PTEN protects against RABV infection is unknown. What it has been reported is that SUMO3 overexpression has a positive effect on RABV infection, likely due to the inactivation of IRF3 caused by its SUMOylation in response to RABV infection [26].
Conclusions
Type I interferon induces the transcription of multiple genes leading to immunomodulatory effects. Among these IFN upregulated genes, there are SUMO proteins as well as multiple targets of SUMO, suggesting that SUMO may play a role in immunity. Viruses have evolved to exploit the SUMOylation machinery of the cell to improve their replication. Many viral proteins are SUMOylated or influence the SUMOylation of cellular proteins. Initially, most of the viral proteins evaluated as putative SUMO substrates were nuclear proteins of DNA viruses or RNA viruses replicating in the nucleus because SUMO and SUMOylation pathway components are localized mainly at the cell nucleus. However, in recent years numerous studies have demonstrated that cytoplasmic RNA viruses also exploit the cellular SUMO pathway. Although SUMOylation may contribute to the control of virus replication, SUMO seems to have an overall positive effect in most of the cytoplasmic RNA viruses. Thus, SUMO proteins or components of the SUMOylation pathway such as Ubc9 are required for the replication of some picornavirus, reovirus, nodavirus, and flavivirus and play a role in pnemovirus or flavivirus pathogenesis. In addition, SUMO conjugates to viral proteins from reovirus, paramyxovirus, picornavirus, coronavirus, arterivirus, filovirus, and flavivirus, and only a few examples of a negative effect of SUMO have been reported for members of the Flaviviridae, Arteriviridae, and Rhabdoviridae families. Interestingly, many of the viral proteins from cytoplasmic RNA viruses that are SUMO substrates can be detected at the cell nucleus during the viral cell cycle. Additional information will be required in order to determine whether there is a relationship between these two events.
Acknowledgments
Funding at the laboratory of CR is provided by the Ministry of Science, Innovation and Universities (MICINN) and FEDER (BFU-2017-88880-P) and by GRC GI-2119 (Xunta de Galicia). SV is a predoctoral fellow funded by Xunta de Galicia. AEM is a recipient of a FPI fellowship from the Spanish MICINN. JGS laboratory is supported by grants from Secretaría Nacional de Ciencia, Tecnología e Innovación de Panamá (SENACYT) (FID017-2016), Ministerio de Economía y Finanzas (009044.060), and Dirección de Investigación, Universidad Interamericana de Panamá (DI-UIP633800). JMP and JGS are members of Sistema Nacional de Investigación (SNI from SENACYT).
Contributor Information
Rosa Barrio, Email: rbarrio@cicbiogune.es.
James D. Sutherland, Email: jsutherland@cicbiogune.es
Manuel S. Rodriguez, Email: Manuel.RODRIGUEZ@itav.fr
Carmen Rivas, Email: mcarmen.rivas@usc.es.
References
- 1.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 2.Guo D, Han J, Adam BL, Colburn NH, Wang MH, Dong Z, et al. Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochem Biophys Res Commun. 2005;337(4):1308–1318. doi: 10.1016/j.bbrc.2005.09.191. [DOI] [PubMed] [Google Scholar]
- 3.Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337(2):517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 4.Minty A, Dumont X, Kaghad M, Caput D. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem. 2000;275(46):36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 5.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101(40):14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu MM, Liao CY, Yang Q, Xie XQ, Shu HB. Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J Exp Med. 2017;214(4):973–989. doi: 10.1084/jem.20161015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Chen W, Wang Q, Li L, Wang C. Negative regulation of TLR inflammatory signaling by the SUMO-deconjugating enzyme SENP6. PLoS Pathog. 2013;9(6):e1003480. doi: 10.1371/journal.ppat.1003480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saul VV, Niedenthal R, Pich A, Weber F, Schmitz ML. SUMO modification of TBK1 at the adaptor-binding C-terminal coiled-coil domain contributes to its antiviral activity. Biochim Biophys Acta. 2014;1853(1):136–143. doi: 10.1016/j.bbamcr.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Mi Z, Fu J, Xiong Y, Tang H. SUMOylation of RIG-I positively regulates the type I interferon signaling. Protein Cell. 2010;1(3):275–283. doi: 10.1007/s13238-010-0030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu J, Xiong Y, Xu Y, Cheng G, Tang H. MDA5 is SUMOylated by PIAS2beta in the upregulation of type I interferon signaling. Mol Immunol. 2011;48(4):415–422. doi: 10.1016/j.molimm.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2(2):233–239. doi: 10.1016/S1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 12.Kubota T, Matsuoka M, Chang TH, Tailor P, Sasaki T, Tashiro M, et al. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J Biol Chem. 2008;283(37):25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ran Y, Liu TT, Zhou Q, Li S, Mao AP, Li Y, et al. SENP2 negatively regulates cellular antiviral response by deSUMOylating IRF3 and conditioning it for ubiquitination and degradation. J Mol Cell Biol. 2011;3(5):283–292. doi: 10.1093/jmcb/mjr020. [DOI] [PubMed] [Google Scholar]
- 14.Chang TH, Kubota T, Matsuoka M, Jones S, Bradfute SB, Bray M, et al. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009;5(6):e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang Q, Deng H, Li X, Wu X, Tang Q, Chang TH, et al. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J Immunol. 2011;187(9):4754–4763. doi: 10.4049/jimmunol.1101704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Begitt A, Droescher M, Knobeloch KP, Vinkemeier U. SUMO conjugation of STAT1 protects cells from hyperresponsiveness to IFNgamma. Blood. 2011;118(4):1002–1007. doi: 10.1182/blood-2011-04-347930. [DOI] [PubMed] [Google Scholar]
- 17.Gronholm J, Vanhatupa S, Ungureanu D, Valiaho J, Laitinen T, Valjakka J, et al. Structure-function analysis indicates that sumoylation modulates DNA-binding activity of STAT1. BMC Biochem. 2012;13:20. doi: 10.1186/1471-2091-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ungureanu D, Vanhatupa S, Gronholm J, Palvimo JJ, Silvennoinen O. SUMO-1 conjugation selectively modulates STAT1-mediated gene responses. Blood. 2005;106(1):224–226. doi: 10.1182/blood-2004-11-4514. [DOI] [PubMed] [Google Scholar]
- 19.Zimnik S, Gaestel M, Niedenthal R. Mutually exclusive STAT1 modifications identified by Ubc9/substrate dimerization-dependent SUMOylation. Nucleic Acids Res. 2009;37(4):e30. doi: 10.1093/nar/gkp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maarifi G, Maroui MA, Dutrieux J, Dianoux L, Nisole S, Chelbi-Alix MK. Small Ubiquitin-like Modifier Alters IFN Response. J Immunol. 2015;195(5):2312–2324. doi: 10.4049/jimmunol.1500035. [DOI] [PubMed] [Google Scholar]
- 21.de la Cruz-Herrera CF, Baz-Martinez M, Motiam AE, Vidal S, Collado M, Vidal A, et al. Phosphorylable tyrosine residue 162 in the double-stranded RNA-dependent kinase PKR modulates its interaction with SUMO. Sci Rep. 2017;7(1):14055. doi: 10.1038/s41598-017-12777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de la Cruz-Herrera CF, Campagna M, Garcia MA, Marcos-Villar L, Lang V, Baz-Martinez M, et al. Activation of the double-stranded RNA-dependent protein kinase PKR by small ubiquitin-like modifier (SUMO) J Biol Chem. 2014;289(38):26357–26367. doi: 10.1074/jbc.M114.560961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maarifi G, El Asmi F, Maroui MA, Dianoux L, Chelbi-Alix MK. Differential effects of SUMO1 and SUMO3 on PKR activation and stability. Sci Rep. 2018;8(1):1277. doi: 10.1038/s41598-018-19683-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelhardt OG, Ullrich E, Kochs G, Haller O. Interferon-induced antiviral Mx1 GTPase is associated with components of the SUMO-1 system and promyelocytic leukemia protein nuclear bodies. Exp Cell Res. 2001;271(2):286–295. doi: 10.1006/excr.2001.5380. [DOI] [PubMed] [Google Scholar]
- 25.Brantis-de-Carvalho CE, Maarifi G, Goncalves Boldrin PE, Zanelli CF, Nisole S, Chelbi-Alix MK, et al. MxA interacts with and is modified by the SUMOylation machinery. Exp Cell Res. 2015;330(1):151–163. doi: 10.1016/j.yexcr.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 26.Maarifi G, Hannoun Z, Geoffroy MC, El Asmi F, Zarrouk K, Nisole S, et al. MxA mediates SUMO-induced resistance to vesicular stomatitis virus. J Virol. 2016;90(14):6598–6610. doi: 10.1128/JVI.00722-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aloni-Grinstein R, Charni-Natan M, Solomon H, Rotter V. p53 and the viral connection: back into the future (double dagger) Cancers (Basel) 2018;10(6):178. doi: 10.3390/cancers10060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, et al. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature. 2003;424(6948):516–523. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- 29.Marcos-Villar L, Perez-Giron JV, Vilas JM, Soto A, de la Cruz-Hererra CF, Lang V, et al. SUMOylation of p53 mediates interferon activities. Cell Cycle. 2013;12(17):2809–2816. doi: 10.4161/cc.25868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baz-Martinez M, Da Silva-Alvarez S, Rodriguez E, Guerra J, El Motiam A, Vidal A, et al. Cell senescence is an antiviral defense mechanism. Sci Rep. 2016;6:37007. doi: 10.1038/srep37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, et al. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70(4):1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonilla WV, Pinschewer DD, Klenerman P, Rousson V, Gaboli M, Pandolfi PP, et al. Effects of promyelocytic leukemia protein on virus-host balance. J Virol. 2002;76(8):3810–3818. doi: 10.1128/JVI.76.8.3810-3818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El McHichi B, Regad T, Maroui MA, Rodriguez MS, Aminev A, Gerbaud S, et al. SUMOylation promotes PML degradation during encephalomyocarditis virus infection. J Virol. 2010;84(22):11634–11645. doi: 10.1128/JVI.01321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maroui MA, Pampin M, Chelbi-Alix MK. Promyelocytic leukemia isoform IV confers resistance to encephalomyocarditis virus via the sequestration of 3D polymerase in nuclear bodies. J Virol. 2011;85(24):13164–13173. doi: 10.1128/JVI.05808-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Regad T, Saib A, Lallemand-Breitenbach V, Pandolfi PP. de The H, Chelbi-Alix MK. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 2001;20(13):3495–3505. doi: 10.1093/emboj/20.13.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pampin M, Simonin Y, Blondel B, Percherancier Y, Chelbi-Alix MK. Cross talk between PML and p53 during poliovirus infection: implications for antiviral defense. J Virol. 2006;80(17):8582–8592. doi: 10.1128/JVI.00031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chelbi-Alix MK, Quignon F, Pelicano L, Koken MH. de The H. resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J Virol. 1998;72(2):1043–1051. doi: 10.1128/JVI.72.2.1043-1051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(5):a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24(3):331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, et al. Dynamics of component exchange at PML nuclear bodies. J Cell Sci. 2008;121(Pt 16):2731–2743. doi: 10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]
- 41.El Asmi F, Maroui MA, Dutrieux J, Blondel D, Nisole S, Chelbi-Alix MK. Implication of PMLIV in both intrinsic and innate immunity. PLoS Pathog. 2014;10(2):e1003975. doi: 10.1371/journal.ppat.1003975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maroui MA, Maarifi G, McManus FP, Lamoliatte F, Thibault P, Chelbi-Alix MK. Promyelocytic leukemia protein (PML) requirement for interferon-induced global cellular SUMOylation. Mol Cell Proteomics. 2018;17(6):1196–1208. doi: 10.1074/mcp.RA117.000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32(2):276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 44.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14(8):1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283(31):21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 46.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 47.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahin U, Ferhi O, Carnec X, Zamborlini A, Peres L, Jollivet F, et al. Interferon controls SUMO availability via the Lin28 and let-7 axis to impede virus replication. Nat Commun. 2014;5:4187. doi: 10.1038/ncomms5187. [DOI] [PubMed] [Google Scholar]
- 49.Domingues P, Golebiowski F, Tatham MH, Lopes AM, Taggart A, Hay RT, et al. Global reprogramming of host SUMOylation during influenza virus infection. Cell Rep. 2015;13(7):1467–1480. doi: 10.1016/j.celrep.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Everett RD, Boutell C, Hale BG. Interplay between viruses and host sumoylation pathways. Nat Rev Microbiol. 2013;11(6):400–411. doi: 10.1038/nrmicro3015. [DOI] [PubMed] [Google Scholar]
- 51.Hoyt CC, Bouchard RJ, Tyler KL. Novel nuclear herniations induced by nuclear localization of a viral protein. J Virol. 2004;78(12):6360–6369. doi: 10.1128/JVI.78.12.6360-6369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belli BA, Samuel CE. Biosynthesis of reovirus-specified polypeptides: expression of reovirus S1-encoded sigma 1NS protein in transfected and infected cells as measured with serotype specific polyclonal antibody. Virology. 1991;185(2):698–709. doi: 10.1016/0042-6822(91)90541-I. [DOI] [PubMed] [Google Scholar]
- 53.Rodgers SE, Connolly JL, Chappell JD, Dermody TS. Reovirus growth in cell culture does not require the full complement of viral proteins: identification of a sigma1s-null mutant. J Virol. 1998;72(11):8597–8604. doi: 10.1128/JVI.72.11.8597-8604.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vazquez-Iglesias L, Lostale-Seijo I, Martinez-Costas J, Benavente J. Different intracellular distribution of avian reovirus core protein sigmaA in cells of avian and mammalian origin. Virology. 2012;432(2):495–504. doi: 10.1016/j.virol.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Rivera-Serrano EE, Fritch EJ, Scholl EH, Sherry B (2017) A cytoplasmic RNA virus alters the function of the cell splicing protein SRSF2. J Virol 91(7). 10.1128/JVI.02488-16 [DOI] [PMC free article] [PubMed]
- 56.Campagna M, Marcos-Villar L, Arnoldi F, de la Cruz-Herrera CF, Gallego P, Gonzalez-Santamaria J, et al. Rotavirus viroplasm proteins interact with the cellular SUMOylation system: implications for viroplasm-like structure formation. J Virol. 2013;87(2):807–817. doi: 10.1128/JVI.01578-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pal S, Santos A, Rosas JM, Ortiz-Guzman J, Rosas-Acosta G. Influenza a virus interacts extensively with the cellular SUMOylation system during infection. Virus Res. 2011;158(1–2):12–27. doi: 10.1016/j.virusres.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 58.Yu F, Wang H, Wang L, Lu L. Orthoreovirus outer-fiber proteins are substrates for SUMO-conjugating enzyme Ubc9. Oncotarget. 2016;7(48):79814–79827. doi: 10.18632/oncotarget.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun D, Xu P, He B. Sumoylation of the P protein at K254 plays an important role in growth of parainfluenza virus 5. J Virol. 2011;85(19):10261–10268. doi: 10.1128/JVI.00389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghildyal R, Baulch-Brown C, Mills J, Meanger J. The matrix protein of human respiratory syncytial virus localises to the nucleus of infected cells and inhibits transcription. Arch Virol. 2003;148(7):1419–1429. doi: 10.1007/s00705-003-0112-y. [DOI] [PubMed] [Google Scholar]
- 61.Dapat C, Oshitani H. Novel insights into human respiratory syncytial virus-host factor interactions through integrated proteomics and transcriptomics analysis. Expert Rev Anti-Infect Ther. 2016;14(3):285–297. doi: 10.1586/14787210.2016.1141676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komaravelli N, Ansar M, Garofalo RP, Casola A. Respiratory syncytial virus induces NRF2 degradation through a promyelocytic leukemia protein - ring finger protein 4 dependent pathway. Free Radic Biol Med. 2017;113:494–504. doi: 10.1016/j.freeradbiomed.2017.10.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanapi UF, Yong CY, Goh ZH, Alitheen NB, Yeap SK, Tan WS. Tracking the virus-like particles of Macrobrachium rosenbergii nodavirus in insect cells. PeerJ. 2017;5:e2947. doi: 10.7717/peerj.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei J, Li C, Zhang X, Fan L, Wei S, Qin Q. Fish SUMO3 functions as a critical antiviral molecule against iridovirus and nodavirus. Fish Shellfish Immunol. 2019;86:1088–1095. doi: 10.1016/j.fsi.2018.12.059. [DOI] [PubMed] [Google Scholar]
- 65.Weng KF, Li ML, Hung CT, Shih SR. Enterovirus 71 3C protease cleaves a novel target CstF-64 and inhibits cellular polyadenylation. PLoS Pathog. 2009;5(9):e1000593. doi: 10.1371/journal.ppat.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aminev AG, Amineva SP, Palmenberg AC. Encephalomyocarditis virus (EMCV) proteins 2A and 3BCD localize to nuclei and inhibit cellular mRNA transcription but not rRNA transcription. Virus Res. 2003;95(1–2):59–73. doi: 10.1016/S0168-1702(03)00163-1. [DOI] [PubMed] [Google Scholar]
- 67.Liu Y, Zheng Z, Shu B, Meng J, Zhang Y, Zheng C, et al. SUMO modification stabilizes Enterovirus 71 polymerase 3D to facilitate viral replication. J Virol. 2016;90(23):10472–10485. doi: 10.1128/JVI.01756-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen SC, Chang LY, Wang YW, Chen YC, Weng KF, Shih SR, et al. Sumoylation-promoted enterovirus 71 3C degradation correlates with a reduction in viral replication and cell apoptosis. J Biol Chem. 2011;286(36):31373–31384. doi: 10.1074/jbc.M111.254896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gomes R, Guerra-Sa R, Arruda E. Coxsackievirus B5 induced apoptosis of HeLa cells: effects on p53 and SUMO. Virology. 2009;396(2):256–263. doi: 10.1016/j.virol.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 70.Gao G, Wong J, Zhang J, Mao I, Shravah J, Wu Y, et al. Proteasome activator REGgamma enhances coxsackieviral infection by facilitating p53 degradation. J Virol. 2010;84(21):11056–11066. doi: 10.1128/JVI.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Plyusnin A, Vapalahti O, Vaheri A. Hantaviruses: genome structure, expression and evolution. J Gen Virol. 1996;77(Pt 11):2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- 72.Kaukinen P, Vaheri A, Plyusnin A. Non-covalent interaction between nucleocapsid protein of Tula hantavirus and small ubiquitin-related modifier-1, SUMO-1. Virus Res. 2003;92(1):37–45. doi: 10.1016/S0168-1702(02)00312-X. [DOI] [PubMed] [Google Scholar]
- 73.Lee BH, Yoshimatsu K, Maeda A, Ochiai K, Morimatsu M, Araki K, et al. Association of the nucleocapsid protein of the Seoul and Hantaan hantaviruses with small ubiquitin-like modifier-1-related molecules. Virus Res. 2003;98(1):83–91. doi: 10.1016/j.virusres.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 74.Maeda A, Lee BH, Yoshimatsu K, Saijo M, Kurane I, Arikawa J, et al. The intracellular association of the nucleocapsid protein (NP) of hantaan virus (HTNV) with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9) Virology. 2003;305(2):288–297. doi: 10.1006/viro.2002.1767. [DOI] [PubMed] [Google Scholar]
- 75.Hiscox JA, Wurm T, Wilson L, Britton P, Cavanagh D, Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J Virol. 2001;75(1):506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wurm T, Chen H, Hodgson T, Britton P, Brooks G, Hiscox JA. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J Virol. 2001;75(19):9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fan Z, Zhuo Y, Tan X, Zhou Z, Yuan J, Qiang B, et al. SARS-CoV nucleocapsid protein binds to hUbc9, a ubiquitin conjugating enzyme of the sumoylation system. J Med Virol. 2006;78(11):1365–1373. doi: 10.1002/jmv.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li FQ, Xiao H, Tam JP, Liu DX. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579(11):2387–2396. doi: 10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee C, Hodgins DC, Calvert JG, Welch SK, Jolie R, Yoo D. The nuclear localization signal of the PRRS virus nucleocapsid protein viral replication in vitro and antibody response in vivo. Adv Exp Med Biol. 2006;581:145–148. doi: 10.1007/978-0-387-33012-9_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang C, Zeng N, Liu S, Miao Q, Zhou L, Ge X, et al. Interaction of porcine reproductive and respiratory syndrome virus proteins with SUMO-conjugating enzyme reveals the SUMOylation of nucleocapsid protein. PLoS One. 2017;12(12):e0189191. doi: 10.1371/journal.pone.0189191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baz-Martinez M, El Motiam A, Ruibal P, Condezo GN, de la Cruz-Herrera CF, Lang V, et al. Regulation of Ebola virus VP40 matrix protein by SUMO. Sci Rep. 2016;6:37258. doi: 10.1038/srep37258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nanbo A, Watanabe S, Halfmann P, Kawaoka Y. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci Rep. 2013;3:1206. doi: 10.1038/srep01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lopez-Denman AJ, Mackenzie JM. The IMPORTance of the nucleus during Flavivirus replication. Viruses. 2017;9(1):14. doi: 10.3390/v9010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu Z, Chu H, Wen L, Yuan S, Chik KK, Yuen TT, et al. Targeting SUMO modification of the non-structural protein 5 of Zika virus as a host-targeting antiviral strategy. Int J Mol Sci. 2019;20(2):E392. doi: 10.3390/ijms20020392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Su CI, Tseng CH, Yu CY, Lai MMC. SUMO modification stabilizes dengue virus nonstructural protein 5 to support virus replication. J Virol. 2016;90(9):4308–4319. doi: 10.1128/JVI.00223-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doolittle JM, Gomez SM. Mapping protein interactions between dengue virus and its human and insect hosts. PLoS Negl Trop Dis. 2011;5(2):e954. doi: 10.1371/journal.pntd.0000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chiu MW, Shih HM, Yang TH, Yang YL. The type 2 dengue virus envelope protein interacts with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9) J Biomed Sci. 2007;14(3):429–444. doi: 10.1007/s11373-007-9151-9. [DOI] [PubMed] [Google Scholar]
- 88.Khadka S, Vangeloff AD, Zhang C, Siddavatam P, Heaton NS, Wang L, et al. A physical interaction network of dengue virus and human proteins. Mol Cell Proteomics. 2011;10(12):M111012187. doi: 10.1074/mcp.M111.012187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, et al. The dengue virus NS5 protein intrudes in the cellular Spliceosome and modulates splicing. PLoS Pathog. 2016;12(8):e1005841. doi: 10.1371/journal.ppat.1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bonamassa B, Ciccarese F, Antonio VD, Contarini A, Palu G, Alvisi G. Hepatitis C virus and host cell nuclear transport machinery: a clandestine affair. Front Microbiol. 2015;6:619. doi: 10.3389/fmicb.2015.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Akil A, Wedeh G, Zahid Mustafa M, Gassama-Diagne A. SUMO1 depletion prevents lipid droplet accumulation and HCV replication. Arch Virol. 2016;161(1):141–148. doi: 10.1007/s00705-015-2628-3. [DOI] [PubMed] [Google Scholar]
- 92.Lee HS, Lim YS, Park EM, Baek SH, Hwang SB. SUMOylation of nonstructural 5A protein regulates hepatitis C virus replication. J Viral Hepat. 2014;21(10):e108–e117. doi: 10.1111/jvh.12241. [DOI] [PubMed] [Google Scholar]
- 93.Guo J, Chen D, Gao X, Hu X, Zhou Y, Wu C, et al. Protein inhibitor of activated STAT2 restricts HCV replication by modulating viral proteins degradation. Viruses. 2017;9(10):285. doi: 10.3390/v9100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ran M, Chen H, Liang B, Liao W, Jiang J, Huang J, et al. Alcohol-induced autophagy via upregulation of PIASy promotes HCV replication in human hepatoma cells. Cell Death Dis. 2018;9(9):898. doi: 10.1038/s41419-018-0845-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gladue DP, Holinka LG, Fernandez-Sainz IJ, Prarat MV, O’Donell V, Vepkhvadze N, et al. Effects of the interactions of classical swine fever virus Core protein with proteins of the SUMOylation pathway on virulence in swine. Virology. 2010;407(1):129–136. doi: 10.1016/j.virol.2010.07.040. [DOI] [PubMed] [Google Scholar]
- 96.Gonzalez-Santamaria J, Campagna M, Ortega-Molina A, Marcos-Villar L, de la Cruz-Herrera CF, Gonzalez D, et al. Regulation of the tumor suppressor PTEN by SUMO. Cell Death Dis. 2012;3:e393. doi: 10.1038/cddis.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]