Dear Editor,
Brassinosteroid (BR) regulates diverse physiological and developmental processes in plants (Choudhary et al., 2012). BR binds to the receptor kinase BRI1 to trigger a phosphorylation/dephosphorylation cascade, which includes BRI1 phosphorylation of Brassinosteroid Signaling Kinase 1 (BSK1) and Constitutive Differential Growth 1 (CDG1), CDG1 phosphorylation of BSU1, and BSU1 dephosphorylation of BIN2 (a GSK3-like kinase) (Kim et al., 2011; Kim et al., 2009; Wang et al., 2014). Almost all the downstream BR signaling components are encoded by multiple genes of a family.
The Arabidopsis BSU1 family (BSUf; BSU1, BSL1, BSL2, and BSL3) is classified into a unique phosphatase family, the PPKL (Protein Phosphatases with Kelch-Like repeat domains), which is not found in animals (Kutuzov and Andreeva, 2002; Mora-Garcia et al., 2004). BSUf members contain an N-terminal Kelch-repeat domain and a C-terminal phosphatase domain that is closely related to Protein Phosphatase 1. The knockdown of BSL2 and BSL3 by artificial micro-RNA in the bsu1bsl1 mutant (amiBSL2,3/bsu1bsl1) causes pleiotropic defects, including severe dwarfism similar to strong BR-deficient or BR-insensitive mutants and abnormal stomatal development (Kim et al., 2009; Kim et al., 2012; Youn et al., 2013). A recent study suggested that BSUf members exhibit specific/non-overlapping functions in addition to BR signaling, although all four members are involved in BR signaling (Maselli et al., 2014). However, the biochemical characteristics underlying the functional redundancy and diversity of PPKL family members are largely unknown.
To identify proteins that interact with BSU1, we performed mass spectrometry (MS) analysis of proteins co-immunoprecipitated with BSU1-YFP from transgenic Arabidopsis overexpressing BSU1-YFP. Unexpectedly, MS analysis revealed high sequence coverage of three BSU1 homologs as well as the BSU1-YFP bait, suggesting that BSU1 interacts with other BSUf members in plant cells (Supplemental Figure 1A). To further investigate these interactions, transgenic plants co-expressing two different BSUf members, each fused to a different tag (myc or YFP), were generated. BSU1-myc was co-immunoprecipitated by anti-YFP antibody from protein extracts of transgenic plants co-expressing BSL1-YFP and BSU1-myc, or BSL2-YFP and BSU1-myc (Figure 1A and Supplemental Figure 1B). BSU1-BSU1 interaction was also confirmed with BSU1-YFP expressed from the native BSU1 promoter in transgenic Arabidopsis plants (Supplemental Figure 2). Similarly, BSL1-myc was co-immunoprecipitated with BSL1-YFP and with BSL2-YFP in protein extracts from the corresponding double transgenic plants (Figure 1B and Supplemental Figure 1C).
Figure 1. BR Signaling is Mediated by Oligomerized BSU1 Family Members.
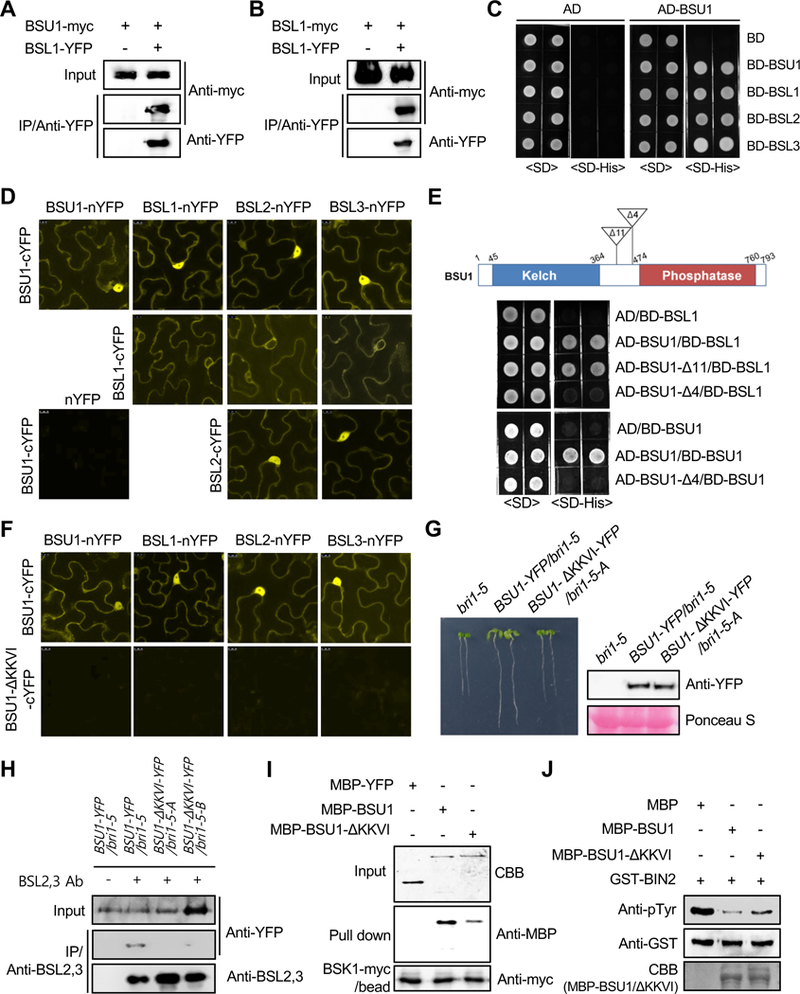
(A-B) Co-immunoprecipitation assays from double transgenic Arabidopsis plants co-expressing the indicated constructs. Immunoprecipitations were performed from protein extracts of double transgenic Arabidopsis plants with anti-YFP antibodies. Associated proteins were immunoblotted with anti-myc and anti-YFP antibodies.
(C) Yeast two-hybrid assay to examine the interactions between BSU1 and BSUf members. BSUf members were expressed as fusion proteins with the DNA binding domain (BD) in yeast cells expressing BSU1 fused to the activation domain (AD). Yeast cells were grown on synthetic dropout (SD) or SD-Histidine (His) medium.
(D) BiFC assay between BSUf members. The indicated constructs were co-transformed into tobacco epidermal cells. Scale bars, 10 μm.
(E) Yeast two-hybrid assays using the two BSU1 deletion constructs. BSU1-Δ11 harbors a deletion of 11 amino acids (437 to 447) while BSU1-Δ4 (BSU1-ΔKKVI) harbors a deletion of 4 amino acids (454 to 457).
(F) The ΔKKVI mutation abolishes the interaction between BSUf members in BiFC assays. Scale bars, 10 μm.
(G) Phenotypes of 7-d-old bri1–5 mutants overexpressing either BSU1-YFP or BSU1-ΔKKVI-YFP. Immunoblot analysis shows the levels of protein in the Arabidopsis lines.
(H) Oligomerization of BSU1-ΔKKVI-YFP with BSL2,3 in two independent 35S-BSU1-ΔKKVI-YFP/bri1–5 plants (-A and -B). Protein extracts of Arabidopsis plants expressing 35S-BSU1-YFP or 35S-BSU1-ΔKKVI-YFP were immunoprecipitated with anti-BSL2,3 antibodies. Associated proteins were detected by immunoblotting with anti-YFP antibodies.
(I) Pull down assays to investigate BSK1 binding to BSU1 and BSU1-ΔKKVI. MBP-YFP, MBP-BSU1, and MBP-BSU1-ΔKKVI were pulled down with immobilized BSK1-myc. Bound proteins were detected by immunoblotting with anti-MBP antibodies.
(J) Comparison of the phosphatase activities of BSU1 and BSU1-ΔKKVI. GST-BIN2 was incubated with MBP, MBP-BSU1, or MBP-BSU1-ΔKKVI. Bound proteins were analyzed by immunoblotting with anti-pTyr antibodies and anti-GST antibodies.
The interactions between BSUf members were further analyzed to determine which of these interactions are direct. In vitro pull-down and yeast two-hybrid assays showed that all BSUf members interact with BSU1 (Supplemental Figure 3 and Figure 1C). Next, Bimolecular Fluorescence Complementation (BiFC) was used to investigate these interactions. In tobacco epidermal cells co-expressing a BSUf member fused to the N-terminal half of YFP (nYFP) and BSU1 fused to the C-terminal half of YFP (cYFP), strong fluorescence was detected (Figure 1D). In addition, other combinations between BSUf members also yielded fluorescence signals in the BiFC assay. In contrast, no fluorescence was detected in tobacco cells co-expressing nYFP and BSU1-cYFP. Subcellular localization of full length YFP-fusion proteins of the BSUf members revealed that three members localized in both the nucleus and cytoplasm. However, BSL1-YFP localized exclusively to the cytoplasm but not in the nucleus (Supplemental Figure 4). In BiFC assays, the fluorescence signals resulting from BSL1 binding to BSL1, BSL2 or BSL3 were excluded from the nucleus and were relatively weak, whereas the other interaction signals were detected both in the nucleus and in the cytoplasm (Figure 1D). In contrast, the BiFC signal caused by the interaction between BSU1 and BSL1 was observed in both the nucleus and in the cytoplasm. This pattern corresponds to the subcellular localization of BSU1-YFP (Supplemental Figure 4). These findings indicate that all BSUf members except BSU1 are retained in cytoplasm when they are associated with BSL1. On the other hand, BSUf complexes containing BSU1 localize to both the nucleus and the cytoplasm.
Chemical crosslinking was performed with Bis-sulfosuccinimidyl Suberate (BS3) treatment to analyze the oligomeric status of MBP-BSU1 (monomer, 145 kDa). SDS-PAGE analysis revealed that the molecular weight of BS3-treated MBP-BSU1 oligomers was greater than the largest 500 kDa protein marker (Supplemental Figure 5). Gel filtration chromatography indicated that MBP-BSU1 tetramers are likely to be dominant, whereas dimeric forms of MBP-BSU1 were not detected (Supplemental Figure 6A). In cross-linking of BSU1-myc obtained from transgenic plant, BSU1-myc tetramers were also predominant (Supplemental Figure 7). However, although our cross-linking and size exclusion chromatography experiments are consistent with BSU1 tetramers in vitro, the oligomeric state of the native proteins in vivo cannot be assessed with certainty.
We next aimed to determine the critical region mediating BSUf member oligomerization. BSU1 binding to truncated BSU1 or BSL1 fragments was examined. It revealed that the C-terminal region of BSU1 or BSL1 mediates the interaction with BSU1 (Supplemental Figure 8 and 9). In addition to the phosphatase domain, two other conserved regions are also present in the C-terminal regions of all BSUf members (Supplemental Figure 8). Thus, we focused on these two conserved sequences and performed yeast two-hybrid assays with deletion constructs for those regions. The ΔKKVI mutation (which lacks residues 454 to 457) in BSU1 abolished homo-dimerization as well as hetero-dimerization of BSU1. In contrast, the BSU1-Δ11 mutant, which lacks residues 437 to 447, interacted with BSL1 as well as BSU1 (Figure 1E). In vitro pull down assays also showed that the interactions of His-BSU1 and MBP-BSUf members were greatly reduced by the ΔKKVI mutation (Supplemental Figure 10). Consistently, no oligomeric form of MBP-BSU1-ΔKKVI was observed in the gel filtration chromatography (Supplemental Figure 6B). Moreover, no BiFC fluorescence signal was detected in tobacco cells co-expressing BSU1-ΔKKVI-cYFP and one of the BSUf members fused to nYFP (Figure 1F) although BSU1-ΔKKVI-YFP showed the same pattern of subcellular localization as BSU1-YFP in tobacco epidermal cells (Supplemental Figure 11A). Co-immunoprecipitation assay further confirmed that homo-oligomerization between BSU1-myc and BSU1-YFP was abolished by the ΔKKVI mutation (Supplemental Figure 11B). Taken together, our results indicate that a short conserved sequence within the middle region of the BSUf members is essential for their oligomerization. KKVI residues of BSU1 do not belong to core catalytic domain of plant protein phosphatases and the ΔKKVI mutation did not affect phosphatase activity of BSU1 in phosphatase assay (Supplemental Figure 12), suggesting that non-oligomeric BSU1-ΔKKVI protein is suitable for examining the biological role of BSU1 oligomerization.
To investigate the functional role of BSUf oligomerization in BR signaling, we generated transgenic bri1–5 lines overexpressing either BSU1-YFP or BSU1-ΔKKVI-YFP and then compared their phenotypes. The dwarf phenotype of the bri1–5 mutants was suppressed by overexpression of BSU1-YFP. However, a similar level of BSU1-ΔKKVI-YFP failed to suppress the dwarf phenotype of the bri1–5 mutant (Figure 1G and Supplemental Figure 13A). Moreover, unlike BSU1-YFP, BR-regulated gene expression in the bri1–5 mutant was not rescued by BSU1-ΔKKVI expression (Supplemental Figure 13B). We further screened a BSU1-ΔKKVI-YFP/bri1–5 plant which exhibits the phenotypic suppression comparable to that of BSU1-YFP/bri1–5. Immunoblot analysis indicated that approximately 7 times more BSU1-ΔKKVI than wild-type BSU1 is required to suppress the growth defect of the bri1–5 mutant to the same extent (Supplemental Figure 14). In those two independent BSU1-ΔKKVI-YFP/bri1–5 plants, co-immunoprecipitation of BSU1-ΔKKVI-YFP with BSL2,3 was greatly reduced regardless of their expression level (Figure 1H), demonstrating that the partial suppression of bri1–5 phenotype by BSU1-ΔKKVI-YFP is due to residue activity of the non-oligomeric form of BSU1.
BSUf members are known to act downstream of BSK1 and CDG1, but upstream of the GSK3-like kinase BIN2 (Kim et al., 2011). To address the mechanism by which BSUf oligomerization positively modulates BR signaling, the interactions of BSK1 and CDG1 with BSU1-ΔKKVI were investigated. The pull-down assay using BSK1-myc immunoprecipitated from transgenic BSK1-myc plants revealed that a ΔKKVI mutation in MBP-BSU1 greatly reduced the binding affinity to BSK1-myc (Figure 1I). Similarly, the interaction of GST-BSK1 with MBP-BSU1 was much stronger than that with MBP-BSU1-ΔKKVI (Supplemental Figure 15). Interestingly, however, oligomerization of BSU1 did not alter BSU1 interaction with either GST-CDG1 or GST-CDL1 (CDG1-Like 1, Supplemental Figure 16). We next investigated whether the oligomerization of BSUf members affects their phosphatase activity on BIN2. In vitro assays showed that the ΔKKVI mutation reduced BSU1-mediated dephosphorylation of BIN2 Tyr200, suggesting that BSU1 oligomers are more efficient in BIN2 dephosphorylation compared with BSU1 monomers (Figure 1J).
To determine whether BR signaling regulates BSUf oligomerization, co-immunoprecipitation assays were performed after BR treatment. However, BR treatment did not alter the amount of either BSU1-myc or BSL1-myc that was co-immunoprecipitated with BSL2-YFP in protein extracts from BSU1-myc/BSL2-YFP or BSL1-myc/BSL2-YFP double transgenic plants (Supplemental Figure 17A and 17B). In addition, co-immunoprecipitation using anti-BSL2,3 antibodies revealed that BR treatment did not alter the amount of BSU1-myc or BSU1-YFP co-immunoprecipitated with endogenous BSL2,3 (Supplemental Figure 17C and 17D). BSUf members appear to remain in equilibrium between monomers and oligomers regardless of the on/off status of BR signaling. Instead of altering the oligomeric status of BSUf members, upstream BR signaling components such as BSK1 may signal preferentially through BSUf oligomers.
In conclusion, our study provides evidence that the PPKL BSUf members form homo- and hetero-oligomeric association, which is required for effective BR signaling and regulation of subcellular localization. Our findings shed light on the molecular mechanisms underlying the functional overlap and diversity of this unique family of phosphatases.
Supplementary Material
ACKNOWLEDGMENTS
FUNDING
This work was supported by grants from the Next-Generation BioGreen 21 Program (SSAC, PJ01114901 to T.W.K, PJ01113201 to S.K.K), Rural Development Administration and by a Basic Science Research Program grant (2012R1A1A1011986 to T.W.K) from the National Research Foundation of Korea (NRF), which is funded by the Ministry of Science, ICT & Future Planning, Republic of Korea. This research was also supported by a grant from NIH (R01GM066258) to Z.Y.W.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information is available at Molecular Plant Online.
No conflict of interest declared.
REFERENCES
- Choudhary SP, Yu JQ, Yamaguchi-Shinozaki K, Shinozaki K, and Tran LS (2012). Benefits of brassinosteroid crosstalk. Trends Plant Sci 17, 594–605. [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan S, Burlingame AL, and Wang ZY (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, and Wang ZY (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11, 1254–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Michniewicz M, Bergmann DC, and Wang ZY (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutuzov MA, and Andreeva AV (2002). Protein Ser/Thr phosphatases with kelch-like repeat domains. Cell Signal 14, 745–750. [DOI] [PubMed] [Google Scholar]
- Maselli GA, Slamovits CH, Bianchi JI, Vilarrasa-Blasi J, Cano-Delgado AI, and Mora-Garcia S (2014). Revisiting the evolutionary history and roles of protein phosphatases with Kelch-like domains in plants. Plant Physiol 164, 1527–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H, and Chory J (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18, 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Bai MY, and Wang ZY (2014). The brassinosteroid signaling network-a paradigm of signal integration. Curr Opin Plant Biol 21C, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JH, Kim TW, Kim EJ, Bu S, Kim SK, Wang ZY, and Kim TW (2013). Structural and functional characterization of Arabidopsis GSK3-like kinase AtSK12. Mol Cells 36, 564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


