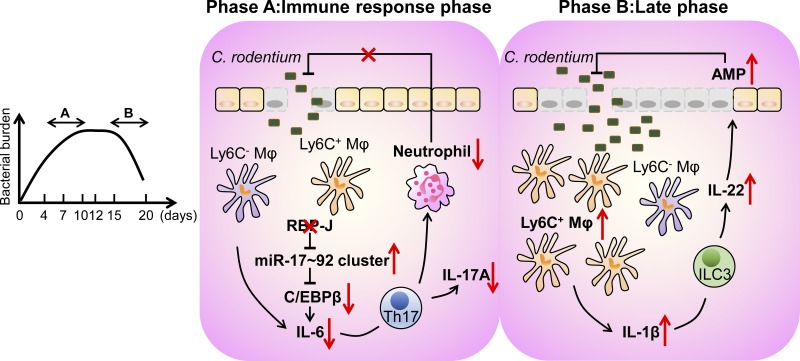
Transcription factor RBP-J in intestinal macrophages dynamically controls antibacterial immunity by interfacing with T cells and ILCs. Macrophage-intrinsic RBP-J supports intestinal Th17 cell responses via IL-6 production, whereas RBP-J deficiency unexpectedly reveals a compensatory role for ILC3 late during infection.
Abstract
Macrophages play pleiotropic roles in maintaining the balance between immune tolerance and inflammatory responses in the gut. Here, we identified transcription factor RBP-J as a crucial regulator of colonic macrophage–mediated immune responses against the enteric pathogen Citrobacter rodentium. In the immune response phase, RBP-J promoted pathogen clearance by enhancing intestinal macrophage-elicited Th17 cell immune responses, which was achieved by maintenance of C/EBPβ-dependent IL-6 production by overcoming miRNA-17∼92–mediated suppressive effects. RBP-J deficiency–associated phenotypes could be genetically corrected by further deleting miRNA-17∼92 in macrophages. In the late phase, noneradicated pathogens in RBP-J KO mice recruited abundant IL-1β–expressing CD64+Ly6C+ colonic macrophages and thereby promoted persistence of ILC3-derived IL-22 to compensate for the impaired innate and adaptive immune responses, leading to ultimate clearance of pathogens. These results demonstrated that colonic macrophage–intrinsic RBP-J dynamically orchestrates intestinal immunity against pathogen infections by interfacing with key immune cells of T and innate lymphoid cell lineages.
Graphical Abstract
Introduction
The intestine is the largest mucosal surface of the body, and it is continually exposed to various nonself agents such as dietary antigens, commensal bacteria, and pathogens (Mowat and Agace, 2014). Mononuclear phagocytes (MPs), consisting of macrophages and dendritic cells (DCs), are sentinels of the intestine and essential for maintenance of intestinal homeostasis as well as eliciting competent innate and adaptive immune responses against enteric pathogens (Cerovic et al., 2014; Varol et al., 2010). Distinct from many other tissue-resident macrophages, intestinal macrophages are mainly derived from circulating Ly6Chi monocytes and require constitutive replenishment throughout adult life (Bain et al., 2014; Ginhoux and Jung, 2014), although recent reports also suggest non–bone marrow origin of certain subsets of this population (Shaw et al., 2018). In addition, they can be distinguished from DCs by expression of the macrophage-specific marker Fcγ receptor 1 (CD64), which positively correlates with CX3C chemokine receptor 1 (CX3CR1) expression and negatively correlates with Ly6C expression (Tamoutounour et al., 2012; Zigmond and Jung, 2013). Under the steady state, mature intestinal macrophages play a central role in the maintenance of gut homeostasis by expressing high levels of anti-inflammatory genes such as Il10 and thus are hyposensitive to activation by inflammatory signals (Rivollier et al., 2012). During the initial phase of intestinal inflammation elicited by enteric infections, circulating Ly6Chi monocytes are recruited to the inflamed sites, resulting in the accumulation of pro-inflammatory macrophages that exert their functions by producing effector cytokines such as IL-6 and IL-23 (Zigmond et al., 2012). Despite of recent advancements characterizing intestinal macrophage ontogeny, the key molecular players and signaling networks that govern functionality of intestinal macrophages remain poorly defined.
Citrobacter rodentium is a natural mouse Gram-negative enteric bacterial pathogen widely used to mimic human infections of enterohemorragic Escherichia coli and enteropathogenic E. coli. Infecting mice with C. rodentium causes attaching and effacing mechanisms of epithelial cells and induces infiltration of inflammatory cells, having the advantages of representing physiological host–pathogen interactions (Crepin et al., 2016; Mundy et al., 2005). The infection of C. rodentium progresses through three distinct stages, including a colonization phase during the first 4 d post-infection (p.i.), a subsequent immune response phase, and a final convalescent phase after day 12. The immune responses to C. rodentium are composed of innate immune reactions (days 4–7) and adaptive immune responses (days 7–11) involving various immune cells such as innate lymphoid cells (ILCs), CD4+ T cells, and B cells producing C. rodentium–specific antibodies (Bry and Brenner, 2004; Bry et al., 2006; Collins et al., 2014; Maaser et al., 2004; Simmons et al., 2003; Zheng et al., 2008). Intestinal MPs are critical in maintaining a proper balance between tolerogenic reactions and pro-inflammatory immune responses by producing mediators, including pro- and anti-inflammatory cytokines, which are required for T helper cell differentiation in response to C. rodentium (Atarashi et al., 2008; Denning et al., 2007; Schreiber et al., 2013). In addition to cross-talking with T cells, via production of IL-1β and IL-23, intestinal MPs also support functionality of IL-22–producing ILC3 to sustain the expression of antimicrobial peptides (AMPs) such as lectins of the Reg3 family (Longman et al., 2014; Manta et al., 2013; Seo et al., 2015). However, the distinct roles of macrophages and DCs in intestinal immunity against C. rodentium remain controversial due to their partially overlapping surface markers and cellular functions.
Notch signaling in the gut is critical in formation of intestinal barrier and maintenance of homeostasis. Abnormal Notch pathway activities have been observed under various disease conditions of the intestine (Noah and Shroyer, 2013). The Notch signaling pathway is pivotal to a process known as lateral inhibition that results in specification of distinct lineages of intestinal epithelial cells (IECs) and requires transcriptional activities mediated by recombinant recognition sequence binding at the Jκ site (RBP-J; also named CSL), the master nuclear mediator of canonical Notch signaling. In the intestinal immune system, Notch–RBP-J signaling regulates the terminal differentiation of intestinal DCs and consequently controls T cell priming and ILC3 activation, consistent with a role of Notch in the development of certain DC subsets observed in other organs (Caton et al., 2007; Ishifune et al., 2014; Lewis et al., 2011; Satpathy et al., 2013). Thus, previous reports regarding Notch signaling in the intestinal system, such as those concerning IECs and DCs, mainly revealed its role in developmental processes, in line with the canonical function of this pathway in cell fate decisions. Recently, in addition to its well-characterized function in development, increasing evidence suggests a role for the Notch–RBP-J signaling pathway in regulating functional polarization and activation of mature immune cells such as macrophages (Foldi et al., 2016; Hu et al., 2008; Shang et al., 2016; Wang et al., 2010; Xu et al., 2012). However, the role of Notch–RBP-J signaling in tissue-specific resident macrophages in vivo such as colonic macrophages is largely unknown. Interestingly, RBP-J signaling has been reported to regulate miRNAs in nonimmune processes such as bone remodeling and neural stem cell differentiation (Gao et al., 2017; Miller et al., 2016). miRNAs are short noncoding RNAs that posttranscriptionally regulate gene expression via direct binding to 3′ untranslated regions (UTRs) of specific mRNAs (O’Connell et al., 2012). miRNAs play diverse roles in the immune system, yet whether miRNAs are targeted by Notch–RBP-J signaling in the context of immune regulation remains elusive.
Using a combination of genetic, genomic, immunological, and imaging approaches, we comprehensively analyzed the role of RBP-J in colonic macrophages during the course of host defense against the enteric pathogen C. rodentium. Using loss-of-function analyses, we described a highly dynamic manner by which RBP-J was involved in orchestration of colonic macrophage–mediated intestinal immune responses by interfacing with multiple key cells types such as T helper 17 (Th17) cells and ILC3. In the immune response phase of infection, RBP-J in colonic macrophages drove IL-6–dependent Th17 cell immune responses to eliminate bacterial pathogens. Interestingly, RBP-J deficiency led to manifestation of an unexpected role for ILC3 during the late phase of infection, which was crucial for pathogen eradication. Together, these findings revealed a critical role for RBP-J in colonic macrophage–mediated communication with T cells and ILCs to dynamically regulate the host defense program against enteric pathogens.
Results
RBP-J expression in colonic macrophages promotes C. rodentium clearance
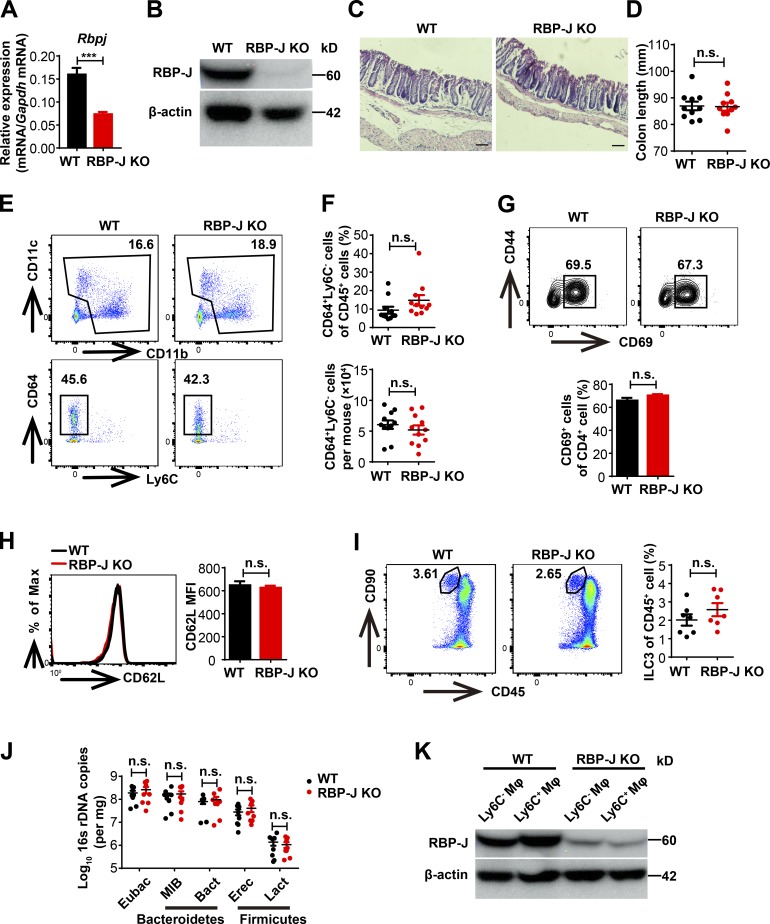
To identify the role of RBP-J in colonic macrophages in vivo, we characterized phenotypes of mice with RBP-J specifically deleted in the myeloid compartment (Rbpjfl/fl Lyz2-Cre, referred hereafter as RBP-J KO). Efficient deletion of Rbpj in sorted colonic macrophages as the CD11b+SiglecF−CD64+Ly6C− population was shown by quantitative real-time PCR (qPCR) and immunoblotting (Fig. S1, A and B). Histological assessments and colon length measurements revealed that under homeostasis, RBP-J KO mice displayed normal colonic architectures and did not exhibit signs of spontaneous colitis (Fig. S1, C and D). The absolute number of colonic macrophages (CD64+Ly6C−) as well as the percentages among lamina propria (LP) leukocytes did not differ between WT controls (Rbpj+/+ Lyz2-Cre) and RBP-J KO mice under the resting state (Fig. S1, E and F). T cell phenotypes and ILC3 population exhibited no differences between WT and RBP-J KO mice in the homeostatic condition (Fig. S1, G–I). In addition, the abundances of commensal bacteria displayed no differences between cohoused or separately housed WT and RBP-J KO mice (Fig. S1 J). The above results indicated that in contrast to colonic DCs, RBP-J was not required for the development of colonic macrophages or the manifestation of their homeostatic functions.
Figure S1.
WT and RBP-J KO mice display similar colonic architectures and abundance of commensal bacteria in homeostasis. (A and B) qPCR (A) and immunoblotting analysis (B) of RBP-J expression in sorted LP CD64+Ly6C− colonic macrophages from WT (Lyz2-Cre) and RBP-J KO (Rbpjfl/fl Lyz2-Cre) mice under the resting state. Data are pooled from two independent experiments (A); n = 3 in each group. (C) Histological analysis of distal colonic tissues by H&E staining under resting state (scale bars represent 50 µm). (D) Measurements of colon lengths in resting state. Data are pooled from three independent experiments; n ≥ 3 in each group. (E–I) LP mononuclear cell populations from 6–8-wk-old uninfected WT and RBP-J KO mice were determined by FACS. Data are pooled from two (G–I) or three (F) independent experiments; n ≥ 3 in each group. (E and F) Representative FACS plots (E) and cumulative data (F) quantitating percentages and cell numbers of CD64+Ly6C− colonic macrophages. (G–I) Representative FACS plots and cumulative data quantitating percentages of CD69+ cells out of CD4+ T cells (G), CD62L expression on CD4+ T cells (H), and ILC3 (CD45midCD3−Thy-1+) populations (I). (J) qPCR analysis of specific bacterial 16s rDNA copies in colon from cohoused WT and RBP-J KO mice in resting state. Data are pooled from three independent experiments; n ≥ 3 in each group. (K) Immunoblotting analysis of RBP-J protein in sorted LP CD64+Ly6C− and CD64+Ly6C+ colonic macrophages at day 5 p.i. Data are shown as mean ± SEM; n.s., not significant; ***, P < 0.001; two-tailed Mann-Whitney test (J) or two-tailed Student’s unpaired t test (other panels). Each symbol in D, F, I, and J represents an individual mouse.
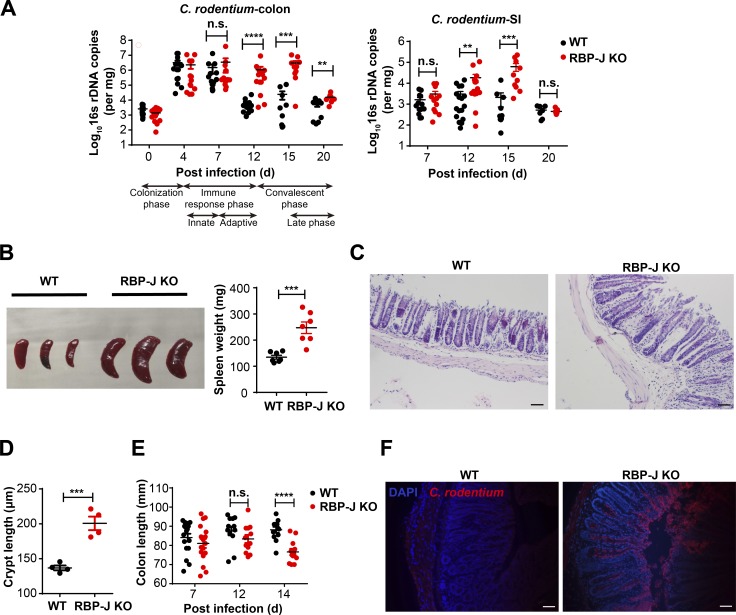
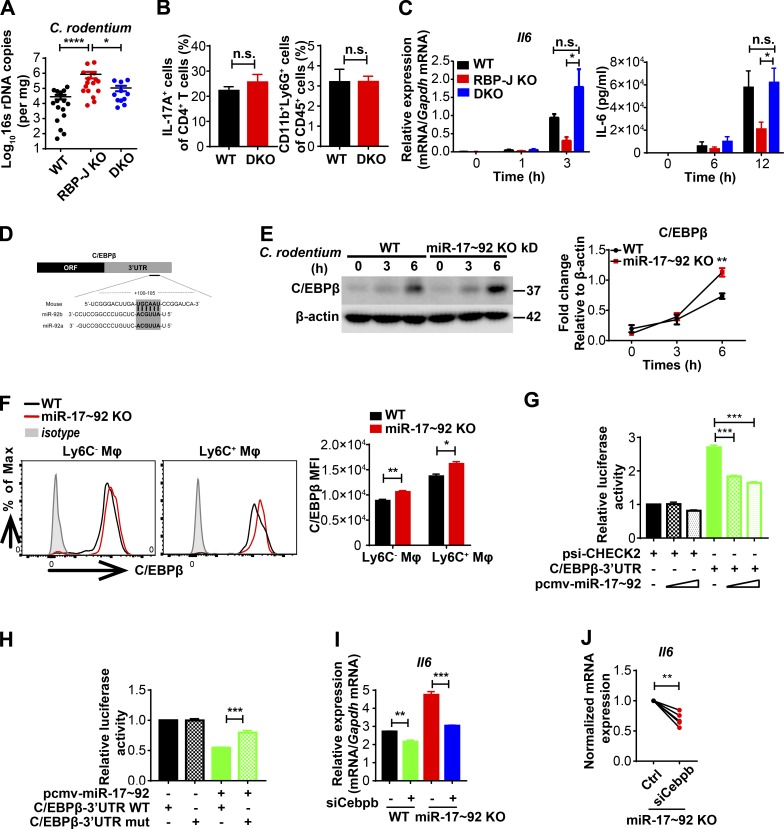
Next, we challenged WT and RBP-J KO mice with C. rodentium by oral gavage and monitored bacterial burdens in colon contents for up to 20 d. Similar to the observations in the uninfected condition, RBP-J protein in sorted colonic macrophages was efficiently deleted in RBP-J KO mice p.i. (Fig. S1 K). In WT mice, consistent with a previous report (Crepin et al., 2016), C. rodentium loads peaked at day 7 p.i., declined over time, and were nearly cleared after 3 wk (Fig. 1 A). The initial kinetics and peak of C. rodentium loads in RBP-J KO mice were comparable to those in WT mice. Interestingly, RBP-J KO mice harbored significantly higher bacterial burdens than WT mice at days 12 and 15 p.i. during the convalescent phase in colon and small intestine (Fig. 1 A). RBP-J KO mice developed severe systemic and colonic inflammation at day 14 p.i. evidenced by splenomegaly, worsened epithelial hyperplasia, increased mononuclear cell infiltration, and shortened colon lengths (Fig. 1, B–E). Infection with a green fluorescent C. rodentium strain (C. rodentium–GFP) visually demonstrated that more C. rodentium persisted in colonic lumen of RBP-J KO mice compared with WT mice and that translocation of pathogens into the colonic LP was evident in RBP-J KO, but not WT animals at day 12 p.i. (Fig. 1 F). During the late phase, C. rodentium was eventually expelled from RBP-J KO mice around day 20 p.i (Fig. 1 A), suggesting additional noncanonical mechanisms to compensate for the compromised immunity leading to bacterial clearance between days 15 and 20 p.i., which was further explored in latter parts of this study. In summary, these results identified RBP-J as a critical regulator in colonic macrophage–mediated immune responses against C. rodentium infection.
Figure 1.
Myeloid-specific RBP-J deficiency compromises host defense against C. rodentium. 6–8-wk-old WT (Lyz2-Cre) and RBP-J KO (Rbpjfl/fl Lyz2-Cre) mice were orally inoculated with 2 × 109 CFUs of C. rodentium, and tissues were harvested at the indicated time points p.i. (A) qPCR analysis of 16s rDNA copies to determine fecal bacterial burdens in colon (left) and small intestine (SI; right) at the indicated p.i. days. (B) Representative splenomegaly (left) and spleen weights (right) at day 14 p.i. (C and D) Histopathology (C) and crypt lengths (D) in the distal colon at day 14 p.i. (H&E; scale bars represent 50 µm). (E) Measurements of colon lengths at the indicated p.i. days. (F) Visualization of C. rodentium (red) and DAPI (blue) in the distal colon at day 12 p.i. (scale bars represent 50 µm). Data are representative of two independent experiments (B [left], C, D, and F) or pooled from two (B [right]) or four independent experiments (A and E); n ≥ 3 in each group. Data are shown as mean ± SEM; n.s., not significant; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; two-tailed Mann-Whitney test (A) or two-tailed Student’s unpaired t test (all other panels). Each symbol represents an individual mouse.
RBP-J in colonic macrophages promotes colonic Th17 cell immune responses during C. rodentium infection
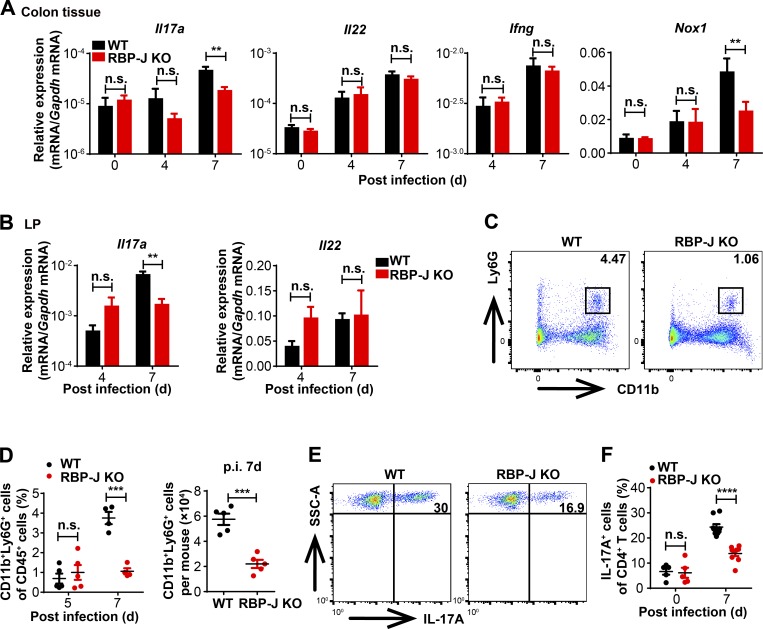
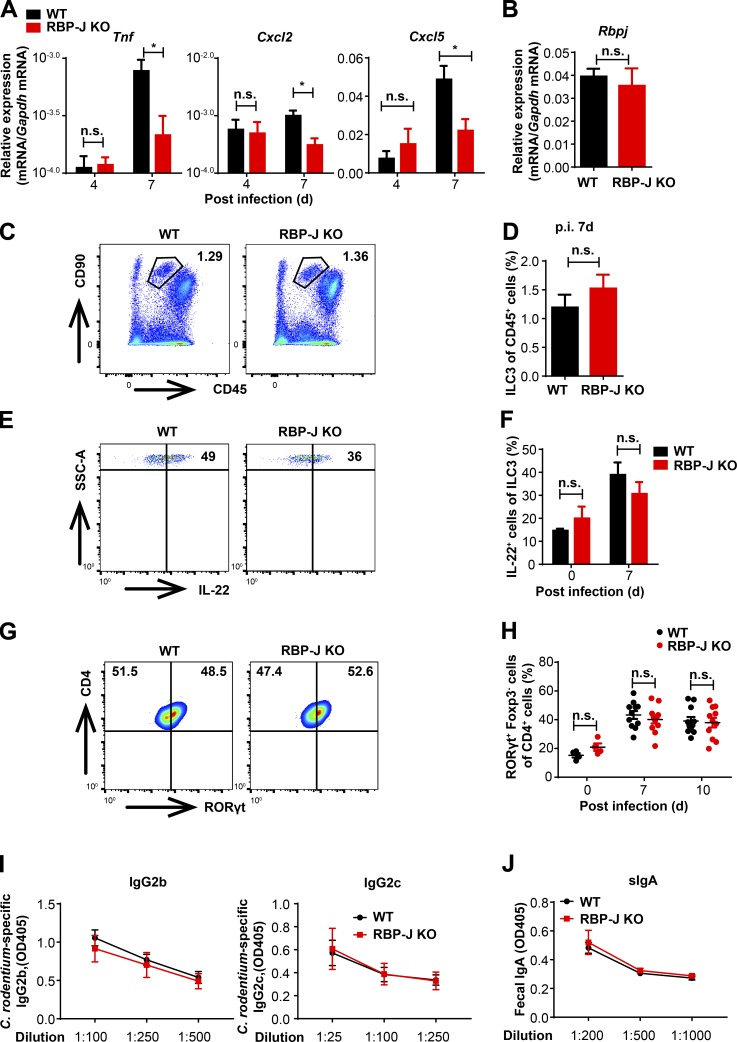
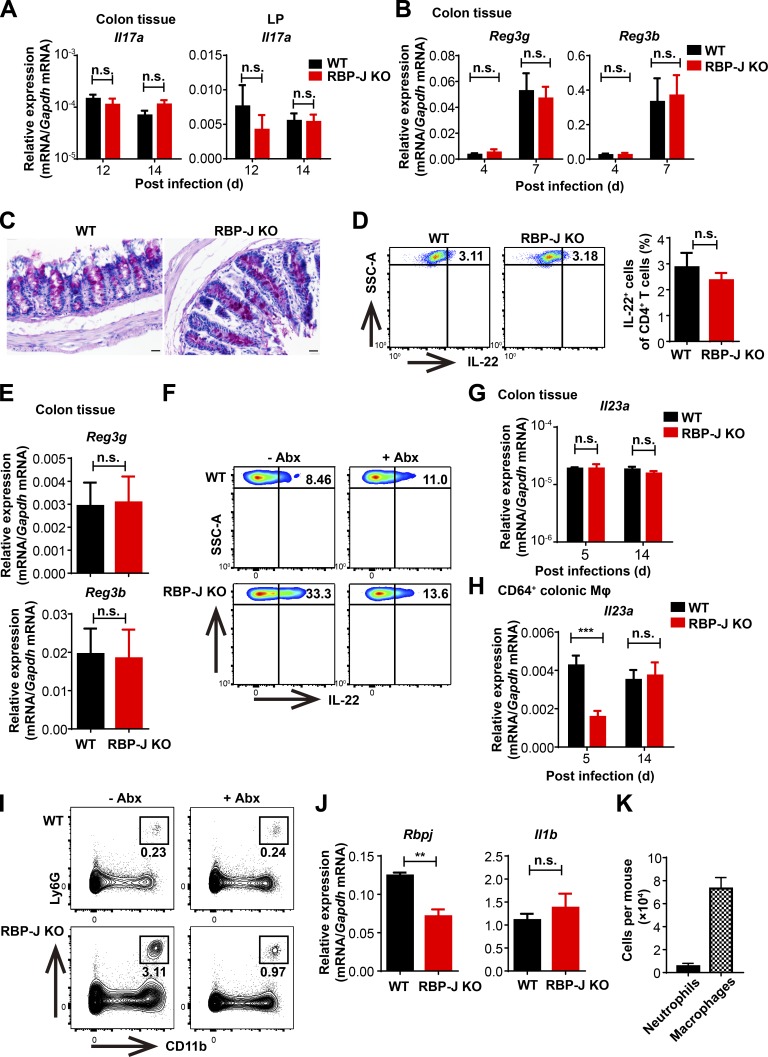
Next, we wished to characterize the cellular mechanisms underlying regulation of host defense by RBP-J in the immune response phase. IL-17A, IL-22, and IFN-γ play essential roles in modulating bacterial colonization and colonic inflammation during C. rodentium infection (Collins et al., 2014; Rubino et al., 2012). We found that compared with WT mice, colon homogenates as well as LP mononuclear cells from RBP-J KO mice expressed reduced levels of Il17a but comparable levels of Il22 and Ifng during the adaptive immune response phase at day 7 p.i. (Fig. 2, A and B). In addition, expression of an IL-17–dependent gene, Nox1, was decreased in RBP-J KO mice at day 7 p.i. (Fig. 2 A; Kumar et al., 2016). One of the critical indicators for IL-17 activities is the recruitment of neutrophils that aids in the elimination of pathogens (Ouyang et al., 2008; Xu and Cao, 2010). In line with the above-observed impaired IL-17–driven responses, recruitment of Ly6G+ neutrophils in LP was significantly diminished in RBP-J KO mice at day 7 p.i. (Fig. 2, C and D). Compromised neutrophil recruitment in RBP-J KO mice correlated with reduced expression of genes encoding neutrophilic chemokines Cxcl2 and Cxcl5 and genes encoding prototypical inflammatory cytokines such as Tnf (Fig. S2 A). Given that IL-17A is mainly produced by CD4+ T cells and ILCs (Korn et al., 2009; Sonnenberg and Artis, 2015), we monitored LP IL-17A–producing CD4+ T cells and ILC3 during the course of infection and found that frequency of IL-17A–producing LP CD4+ T cells was significantly reduced at day 7 p.i. in RBP-J KO mice, yet Rbpj expression in T cells was not affected by myeloid-specific deletion as expected (Fig. S2 B and Fig. 2, E and F). Nevertheless, neither the population of ILC3 (CD45midCD3−Thy-1+) nor the production of IL-22 by ILC3 displayed notable differences between WT and RBP-J KO mice during the adaptive immune response phase (Fig. S2, C–F), identifying IL-17A–producing CD4+ T cells, but not ILC3, as targets of RBP-J–mediated regulation. Intriguingly, Th17 cells defined by RORγt+ positivity were present at comparable percentages in infected WT and RBP-J KO mice (Fig. S2, G and H), implying that RBP-J expression in colonic macrophages was dispensable for initial priming of naive T cells. Moreover, serum anti–C. rodentium IgG titers and fecal secretory IgA titers at day 12 p.i. did not differ between WT and RBP-J KO mice (Fig. S2, I and J), indicating that humoral immune responses were likely not affected. Taken together, these data suggested that RBP-J expression in colonic macrophages cross-regulated the function of IL-17A–secreting effector T cells to promote adaptive immune responses against C. rodentium infection.
Figure 2.
RBP-J KO mice display impaired colonic Th17 cell immune responses upon C. rodentium infection. WT and RBP-J KO mice were orally inoculated with 2 × 109 CFUs of C. rodentium, and tissues and LP were harvested at the indicated time points p.i. (A and B) qPCR analysis of the indicated mRNAs in colon tissues (A) and LP mononuclear cells (B). (C and D) Colonic LP CD11b+Ly6G+ neutrophils were determined by flow cytometry analyses (FACS). Representative FACS plots at day 7 p.i. (C) and cumulative data of cell ratio (D, [left]) and absolute numbers (D, [right]) at the indicated p.i. days are shown. (E and F) Colonic LP mononuclear cells at day 7 p.i. were treated with PMA and ionomycin in vitro for 4–5 h, and representative FACS plots of IL-17A production in LP CD3+CD4+ T cells are shown. Data are pooled from two independent experiments (A, B, D, and F); n ≥ 3 in each group. Data are shown as mean ± SEM; n.s., not significant; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; two-tailed Student’s unpaired t test. Each symbol in D and F represents an individual mouse. SSC-A, side scatter area.
Figure S2.
RBP-J KO mice display normal activation of ILC3 and humoral immune responses during the adaptive immune response phase against C. rodentium infection. (A) 6–8-wk-old WT and RBP-J KO mice were orally inoculated with 2 × 109 CFUs of C. rodentium, and colons were harvested at the indicated time points p.i. qPCR analysis of the indicated mRNAs in colon tissues is shown. Data are pooled from two independent experiments; n = 3 in each group. (B) qPCR of Rbpj in sorted LP CD4+ T cells from uninfected mice. Data are pooled from two independent experiments; n = 3 in each group. (C–H) Representative FACS plots of colonic LP ILC3 (CD45midCD3−Thy-1+), IL-22–producing ILC3 and Th17 cells (CD3+CD4+RORγt+) at day 7 p.i. (C, E, and G) and cumulative data (D, F, and H). Data are pooled from two independent experiments; n ≥ 3 in each group. (I and J) Serum anti–C. rodentium immunoglobulin G2b (IgG2b) and IgG2c titers (I) and fecal secretory IgA (sIgA) titers (J) at day 12 p.i. were measured by ELISA. Data are pooled from two independent experiments; n = 3 in each group. Data are shown as mean ± SEM; two-tailed Student’s unpaired t test. n.s., not significant; SSC-A, side scatter area.
RBP-J facilitates macrophage-Th17 cell cross-talk by targeting IL-6
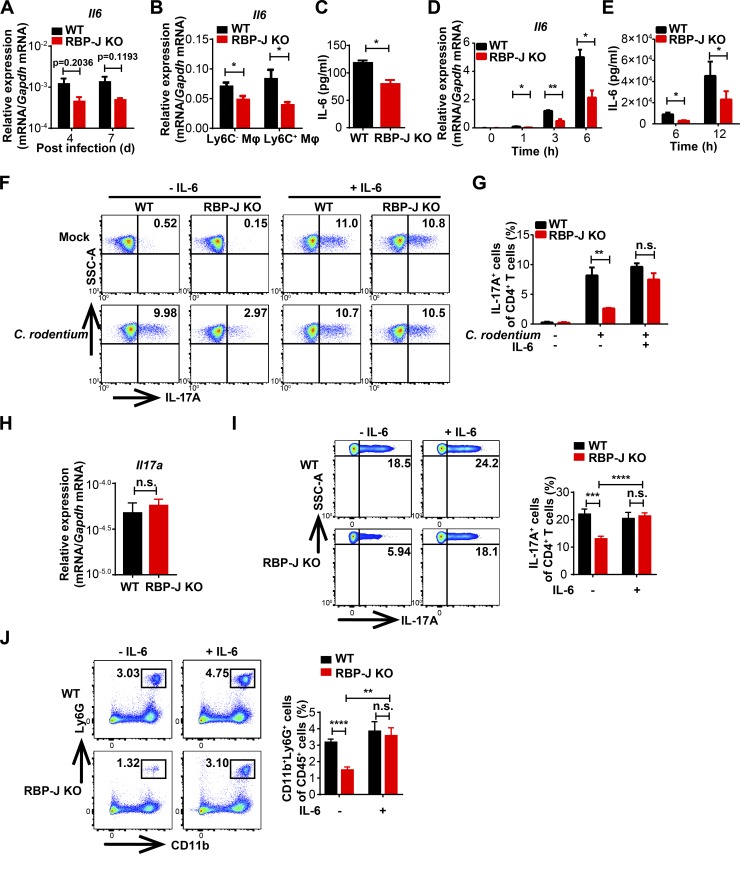
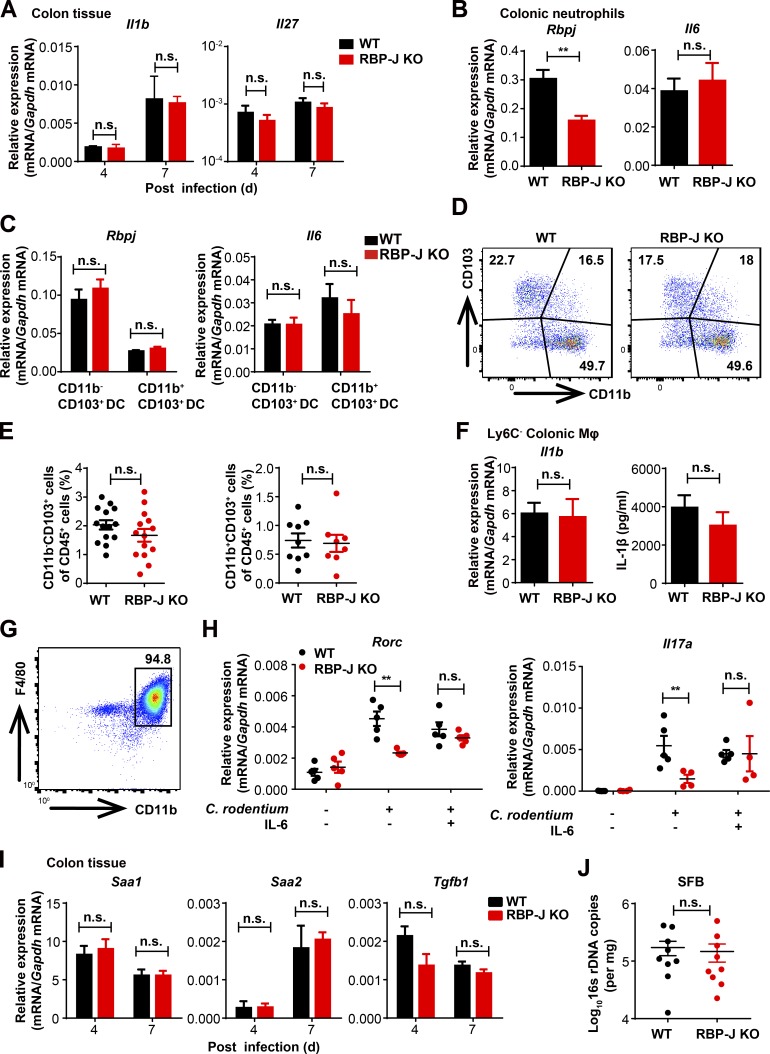
We next explored mechanisms by which RBP-J regulated colonic macrophage–mediated Th17 cell immune responses. Upon infections with enteric pathogens, cytokines such as IL-6 and IL-1β produced by colonic macrophages and DCs drive differentiation and activation of Th17 cells (Denning et al., 2007; Ivanov et al., 2006; Korn et al., 2009; Schreiber et al., 2013). We first examined cytokine expression in colon homogenates and found that Il6, but not Il1b and Il27, displayed a trend toward decreased expression in infected RBP-J KO mice (Fig. 3 A and Fig. S3 A). Analyses of sorted immune cell populations showed that expression of IL-6 was decreased in RBP-J–deficient colonic macrophages, but not in RBP-J–deficient neutrophils (Fig. 3, B and C; and Fig. S3 B). Consistent with the previously described deletion patterns of Lyz2-Cre mice (Jakubzick et al., 2008), Rbpj was not deleted in colonic DC populations (Fig. S3 C). As a result, DC populations and expression of Il6 in DCs were not affected by Lyz2-Cre–driven RBP-J deficiency (Fig. S3, C–E). Of note, no significant differences were observed between WT and RBP-J KO macrophages regarding IL-1β levels (Fig. S3 F). Consistent with the in vivo data, upon treatment with heat-killed C. rodentium, expression levels of IL-6 were decreased in RBP-J–deficient peritoneal macrophages (Fig. 3, D and E; and Fig. S3 G). Moreover, we mimicked the potential colonic macrophage-Th17 cell interaction using supernatants from macrophages stimulated with heat-killed C. rodentium. The production of IL-17A in T cells cultured with RBP-J KO macrophage supernatants was significantly decreased compared with WT supernatants (Fig. 3, F and G). Diminished IL-17A expression resulted from RBP-J–deficient macrophages was rescued by addition of recombinant IL-6 (Fig. 3, F and G). Consistently, the subdued mRNA levels of Il17a and Rorc in T cells in the RBP-J KO group were also rescued to levels comparable to those in the WT group by supplying exogenous IL-6 (Fig. S3 H). To further corroborate a causal role for IL-6 in vivo, recombinant IL-6 was injected intraperitoneally into infected mice at days 3 and 5 p.i. The impaired IL-17A production by CD4+ T cells and compromised neutrophil recruitment associated with RBP-J deficiency were rescued by IL-6 administration in vivo to the levels comparable with those observed in WT mice (Fig. 3, H–J). Expression of genes encoding other Th17 cell regulatory factors such as Saa1, Saa2, and Tgfb1 (Sano et al., 2015) in colon homogenates as well as the abundance of segmented filamentous bacteria displayed no significant differences between WT and RBP-J KO mice at days 4 and 7 p.i. (Fig. S3, I and J). Together, these results implied that RBP-J promoted IL-6 production in colonic macrophages to support local activation of Th17 cells upon C. rodentium infection.
Figure 3.
Macrophage-intrinsic RBP-J promotes Th17 cell immune responses via sustaining IL-6 expression. (A–C) WT and RBP-J KO mice were orally inoculated with 2 × 109 CFUs of C. rodentium. Colonic LP mononuclear cells were isolated at day 5 p.i. Data are pooled from two independent experiments; n = 3 in each group. (A) qPCR analysis of Il6 in colon tissues at the indicated p.i. days. (B) qPCR analysis of Il6 in sorted CD64+Ly6C− and CD64+Ly6C+ colonic macrophages. (C) IL-6 levels in the supernatants of sorted CD64+Ly6C− colonic macrophages (2 × 106 cells per ml) were measured by ELISA. (D and E) IL-6 levels of peritoneal macrophages stimulated with heat-killed C. rodentium (MOI = 1) were determined by qPCR (D) and ELISA (E). Data are pooled from three independent experiments. (F and G) Naive CD4+ T cells were sorted and cultured with the supernatants of peritoneal macrophages that were not treated (mock) or stimulated with heat-killed C. rodentium (MOI = 1) and TGF-β, without (left) or with (right) IL-6 for 72 h. FACS (F) and cumulative data (G) of IL-17A–expressing CD4+ T cells are shown. Data are pooled from three independent experiments. (H–J) 6–8-wk-old WT and RBP-J KO mice were intraperitoneally injected with recombinant IL-6 at days 3 and 5 p.i. and sacrificed at day 7 p.i. Data are pooled from two independent experiments, n ≥ 4 in each group. (H) qPCR analysis of Il17a in colon tissues. (I and J) Representative FACS plots (left) and cumulative data (right) of IL-17A production in LP CD3+CD4+ T cells (I) and colonic LP neutrophils (J). Data are shown as mean ± SEM; n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; two-tailed Student’s paired t test (D and E) or Student’s unpaired t test (other panels).
Figure S3.
RBP-J KO mice display normal functions of DCs and neutrophils. (A) qPCR analysis of Il1b and Il27 in colon tissues from WT and RBP-J KO mice at the indicated p.i. days. Data are pooled from two independent experiments, n = 3 in each group. (B–F) WT and RBP-J KO mice were orally inoculated with 2 × 109 CFUs of C. rodentium, and colonic LP mononuclear cells were isolated at day 5 p.i. Data are pooled from two (B, C, and F) or three (E) independent experiments. (B and C) qPCR analysis of Rbpj and Il6 in sorted CD11b+Ly6G+ colonic neutrophils (B) and CD11b−CD103+ and CD11b+CD103+ colonic DCs (C). (D and E) Representative FACS plots (D) and cumulative data (E) quantitating LP CD11c+MHCII+ DC subpopulations. (F) IL-1β levels in sorted CD64+Ly6C− colonic macrophages were determined by qPCR analysis (left) and ELISA (right). (G) FACS analysis of replated peritoneal cells intraperitoneally administrated with thioglycollate. (H) qPCR analysis of Rorc (left) and Il17a (right) mRNA expression in cultured CD4+ T cells performed as in Fig. 3 F. Data are pooled from three independent experiments. (I) qPCR analysis of indicated mRNAs in colon tissues from infected WT and RBP-J KO mice. Data are pooled from two independent experiments; n = 3 in each group. (J) qPCR analysis of segmented filamentous bacteria (SFB) in colon from WT and RBP-J KO mice at day 7 p.i. Data are pooled from two independent experiments; n ≥ 3 in each group. Data are shown as mean ± SEM; n.s., not significant; **, P < 0.01; two-tailed Mann-Whitney test (J) or two-tailed Student’s unpaired t test (other panels). Each symbol in J represents an individual mouse.
RBP-J inhibits the miR-17∼92 cluster to promote IL-6 production
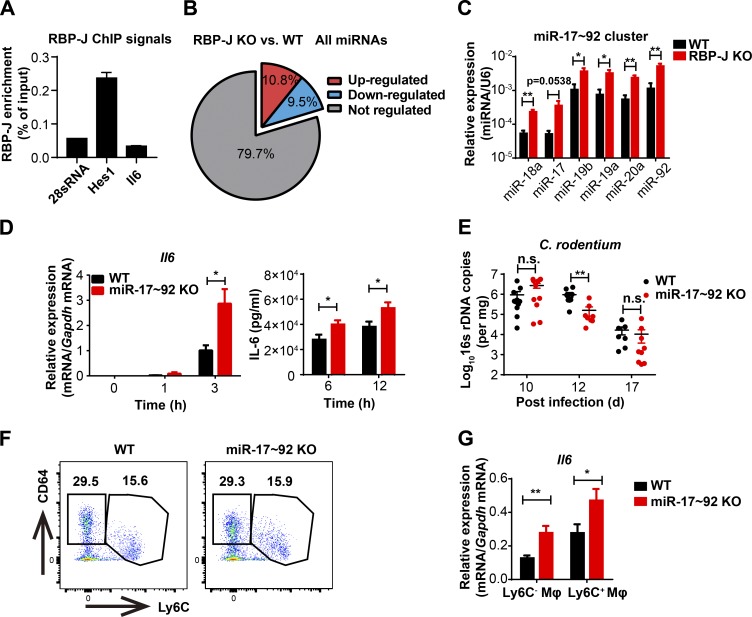
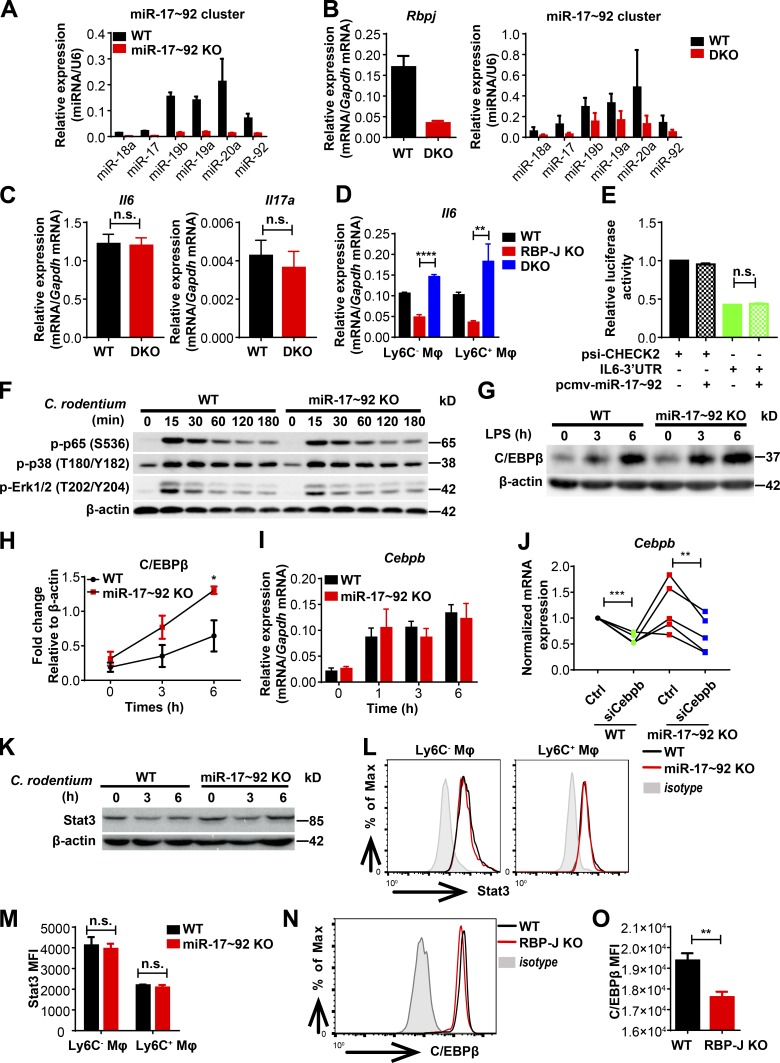
Next, we investigated the mechanisms by which RBP-J promoted the production of IL-6 in macrophages. Chromatin immunoprecipitation (ChIP) analysis with a RBP-J–specific antibody showed that binding of RBP-J with a putative site on the endogenous Il6 promoter was minimal relative to signals on a positive control gene (Fig. 4 A), suggesting that RBP-J may not directly regulate Il6 expression. Thus, we postulated that RBP-J promoted IL-6 production via indirect mechanisms. Given the precedence of RBP-J–mediated posttranscriptional regulation (Xu et al., 2012), we hypothesized that in colonic macrophages, RBP-J may exert regulatory functions by modulating expression of certain miRNAs. Small RNA sequencing (RNA-seq) analysis of RBP-J KO macrophages identified a number of RBP-J–regulated miRNAs (Fig. 4 B and Table S1). Validation experiments revealed that the miR-17∼92 cluster, consisting of six individual mature miRNAs generated from a single transcript, was drastically up-regulated in RBP-J KO colonic macrophages (Fig. 4 C), uncovering RBP-J as an endogenous suppressor of the miR-17∼92 cluster. To assess the role of miR-17∼92 cluster in macrophages, we generated miR-17∼92 myeloid-specific conditional knockout mice of the miR-17∼92fl/fl Lyz2-Cre genotype (referred hereafter as miR-17∼92 KO; Fig. S4 A). Stimulation of miR-17∼92 KO macrophages with heat-killed C. rodentium in vitro resulted in heightened expression of IL-6 (Fig. 4 D), indicating that the miR-17∼92 cluster served as a negative regulator of IL-6 production. To clarify the role of the miR-17∼92 cluster in colonic macrophages in vivo, WT and miR-17∼92 KO mice were infected with C. rodentium. In contrast to the observations in RBP-J KO mice, the kinetics of C. rodentium burdens showed that miR-17∼92 KO mice were more protective to C. rodentium infection than WT mice (Fig. 4 E). Despite similar compositions of colonic macrophage subsets (Fig. 4 F), miR-17∼92–deficient colonic macrophages exhibited elevated IL-6 production relative to WT controls (Fig. 4 G), suggesting that the miR-17∼92 cluster regulated effector functions instead of populations of colonic macrophages. These results raised the possibility that RBP-J regulated IL-6–dependent host responses by targeting the miR-17∼92 cluster.
Figure 4.
RBP-J promotes IL-6 production by releasing miR-17∼92 cluster–imposed inhibition. (A) RBP-J occupancy at the promoters of Hes1 and Il6 was analyzed by ChIP-qPCR in WT BMDMs. Data are from one representative experiment of two independent experiments. (B) A pie graph showing the percentages of RBP-J–up-regulated and down-regulated miRNAs among all miRNAs. (C) qPCR analysis of the miR-17∼92 cluster in sorted LP CD64+Ly6C− colonic macrophages. Data are pooled from two independent experiments. (D) WT (Lyz2-Cre) and miR-17∼92 KO (miR-17∼92fl/fl Lyz2-Cre) BMDMs were stimulated with heat-killed C. rodentium (MOI = 0.5) at the indicated time points. IL-6 levels were determined by qPCR (left) and ELISA (right). Data are pooled from three independent experiments. (E–G) WT and miR-17∼92 KO mice were orally inoculated with 4 × 109 CFUs of C. rodentium, and colonic LP mononuclear cells were isolated at day 5 p.i. (E) qPCR analysis of 16s rDNA copies to determine fecal bacterial burdens in colon. Data are pooled from two independent experiments; n ≥ 3 in each group. (F) Representative FACS plots of LP colonic macrophage subsets. (G) qPCR analysis of Il6 in sorted CD64+Ly6C− and CD64+Ly6C+ colonic macrophages. Data are pooled from two independent experiments; n = 3 in each group. Data are shown as mean ± SEM; n.s., not significant; *, P < 0.05; **, P < 0.01; two-tailed Student’s unpaired t test (C and G), two-tailed Student’s paired t test (D), or two-tailed Mann-Whitney test (E). Each symbol in E represents an individual mouse.
Figure S4.
RBP-J promotes IL-6 production by inhibition of the miR-17∼92–C/EBPβ axis. (A) qPCR analysis of the miR-17∼92 cluster in WT (Lyz2-Cre) and miR-17∼92 KO (miR-17∼92fl/fl Lyz2-Cre) BMDMs. (B) qPCR analysis of Rbpj mRNA and the miR-17∼92 cluster in WT (Lyz2-Cre) and DKO (Rbpjfl/fl miR-17∼92fl/fl Lyz2-Cre) BMDMs. (C) WT and DKO mice were orally inoculated with 2 × 109 CFUs of C. rodentium. qPCR analysis of Il6 (left) and Il17a (right) in LP mononuclear cells at day 7 p.i. Data are pooled from two independent experiments; n = 3 in each group. (D) qPCR analysis of Il6 in sorted LP colonic macrophage subsets from WT, RBP-J KO, and DKO mice at day 5 p.i. Data are pooled from two independent experiments; n = 3 in each group. (E) A luciferase reporter plasmid (psi-CHECK2) containing the IL-6 3′ UTR and an expression plasmid encoding miR-17∼92 cluster (pCMV-miR-17∼92) were cotransfected into HEK293T cells. Luciferase activities were determined and normalized to the condition with empty reporter plasmid alone. Data are pooled from two independent experiments. (F) Immunoblotting analysis of the indicated proteins in whole-cell lysates of BMDMs treated with heat-killed C. rodentium (MOI = 0.5) for various times (top lanes). (G and H) Immunoblotting analysis of C/EBPβ protein in whole-cell lysates of BMDMs treated with LPS (10 ng/ml). Data from one representative experiment (G) and densitometrical quantitation of cumulative data from three independent experiments (H) are shown. (I) qPCR analysis of Cebpb mRNA in BMDMs stimulated with heat-killed C. rodentium (MOI = 0.5). Data are pooled from three independent experiments. (J) qPCR analysis of Cebpb in BMDMs transfected with siCebpb or control siRNA. Data are pooled from three independent experiments. (K) Immunoblotting analysis of Stat3 in whole-cell lysates of BMDMs treated with heat-killed C. rodentium (MOI = 0.5) for various times (top lanes). (L and M) Representative FACS plots (L) and cumulative mean fluorescence intensity (MFI; M) of Stat3 expression in WT and miR-17∼92 KO colonic macrophages at day 5 p.i. Black lines represent WT mice, red lines represent miR-17∼92 KO mice, and shaded curves represent isotype control. Data are pooled from two independent experiments; n ≥ 3 in each group. (N and O) Representative FACS plots (N) and cumulative MFI (O) of C/EBPβ expression in WT and RBP-J KO colonic macrophages. Black lines represent WT mice, red lines represent RBP-J KO mice, and shaded curves represent isotype control. Data are pooled from two independent experiments; n = 3 in each group. Data are shown as mean ± SEM; n.s., not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001; two-tailed Student’s paired t test (J) or two-tailed Student’s unpaired t test (other panels).
RBP-J enhances IL-6 production by inhibition of the miR-17∼92–C/EBPβ axis
Next, we wished to provide genetic evidence for the miR-17∼92 cluster in RBP-J–mediated host defense process and generated Rbpjfl/fl miR-17∼92fl/fl Lyz2-Cre mice (referred hereafter as DKO; Fig. S4 B). Upon C. rodentium challenge, DKO mice were more resistant to infection than RBP-J KO mice as assessed by bacterial burdens (Fig. 5 A), suggesting that the compromised immune phenotypes in RBP-J KO mice could be rescued, at least partially, by further deleting the miR-17∼92 cluster. Moreover, no significant differences were detected between DKO and WT mice regarding the expression of Il6 and Il17a in LP mononuclear cells, the frequency of IL-17A–producing CD4+ T cells, and the recruitment of neutrophils at day 7 p.i. (Fig. S4 C and Fig. 5 B). Deletion of the miR-17∼92 cluster rescued RBP-J deficiency–associated defective Il6 expression in vivo in CD64+Ly6C− and CD64+Ly6C+ colonic macrophage subsets (Fig. S4 D). Consistently, defective expression of IL-6 in RBP-J KO macrophages upon in vitro stimulation with heat-killed C. rodentium was reversed in the DKO cells (Fig. 5 C). These data indicated that RBP-J promoted IL-6–dependent Th17 cell immune responses by suppressing the miR-17∼92 cluster in vivo and in vitro.
Figure 5.
RBP-J regulates host responses to C. rodentium by targeting the miR-17∼92–C/EBPβ axis. (A and B) WT, RBP-J KO, and DKO (Rbpjfl/fl miR-17∼92fl/fl Lyz2-Cre) mice were orally inoculated with 2 × 109 CFUs of C. rodentium. (A) qPCR analysis of 16s rDNA copies to determine fecal bacterial burdens in colon at day 14 p.i. Data are pooled from three independent experiments; n ≥ 3 in each group. (B) Colonic LP mononuclear cells were isolated at day 7 p.i. Cumulative data of IL-17A production in CD3+CD4+ T cells (left) and LP neutrophils (right) are shown. Data are pooled from two independent experiments; n = 3 in each group. (C) BMDMs were stimulated with heat-killed C. rodentium (MOI = 0.5). IL-6 levels were determined by qPCR (left) and ELISA (right). Data are pooled from three independent experiments. (D) Schematic illustration of the predicted targeting sites for the miR-17∼92 cluster within the 3′ UTR of Cebpb mRNA. (E) Immunoblotting analysis (left) and densitometrical quantitation of cumulative data from three independent experiments (right) of C/EBPβ in whole-cell lysates of BMDMs treated with heat-killed C. rodentium (MOI = 0.5) for various times (top lanes). (F) Representative FACS plots (left) and cumulative MFI (right) of C/EBPβ expression in colonic macrophages at day 5 p.i. Data are pooled from two independent experiments; n = 3 in each group. (G and H) Luciferase reporter plasmid (psi-CHECK2) containing the C/EBPβ 3′ UTR (G) or 3′ UTR mutant (H) and various doses of pCMV-miR17∼92 plasmids were cotransferred into HEK293T cells. Luciferase activities were determined and normalized to the condition with empty reporter plasmid alone. Data are pooled from three independent experiments. (I and J) qPCR analysis of Il6 in BMDMs transfected with siCebpb or control siRNA with LPS stimulation for 3 h. Representative data of one experiment (I; mean and SEM of triplicate measurements) and cumulative data from three independent experiments (J). Data are shown as mean ± SEM; n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; two-tailed Mann-Whitney test (A), two-tailed Student’s paired t test (C and J), or two-tailed Student’s unpaired t test (other panels). Each symbol in A represents an individual mouse.
Next, we wished to identify the mechanisms underlying the inhibitory effects of miR-17∼92 cluster on IL-6 expression. TargetScan (v7.0; Agarwal et al., 2015) analysis indicated that Il6 was likely not a direct target of the miR-17∼92 cluster, as the miR-17∼92 cluster “seed region” could not be identified in the Il6 3′ UTR (data not shown). Overexpression of the miR-17∼92 cluster did not affect IL-6–3′ UTR reporter–driven luciferase activity, supporting the bioinformatics analysis that the miR-17∼92 cluster may not regulate Il6 directly (Fig. S4 E). Moreover, miR-17∼92 cluster deficiency did not obviously affect the activation of canonical signaling pathways leading to Il6 induction (Fig. S4 F). Interestingly, CCAAT enhancer binding protein β (C/EBPβ; also termed NF-IL6), a transcription factor essential for Il6 expression (Akira et al., 1990), was informatically identified as a direct target of the miR-17∼92 cluster (Fig. 5 D). Upon C. rodentium or LPS stimulation, miR-17∼92 cluster deficiency resulted in heightened protein levels of C/EBPβ without altering Cebpb mRNA expression (Fig. 5 E and Fig. S4, G–I), indicating that the miR-17∼92 cluster inhibits C/EBPβ expression at the posttranscriptional level. Consistent with in vitro findings, miR-17∼92-deficient colonic macrophages exhibited enhanced C/EBPβ expression in vivo upon C. rodentium infection (Fig. 5 F). Luciferase activities associated with C/EBPβ 3′ UTR were attenuated upon overexpression of the miR-17∼92 cluster, and such attenuation was abolished when the putative miR-17∼92 binding site at the C/EBPβ 3′ UTR was mutated (Fig. 5, G and H), supporting that C/EBPβ is a direct target of the miR-17∼92 cluster. To evaluate whether changes in C/EBPβ were causally related to miR-17∼92 cluster–regulated Il6 expression, we knocked down Cebpb in miR-17∼92 KO bone marrow–derived macrophages (BMDMs) using RNA interference (Fig. S4 J). Upon LPS stimulation, Cebpb knockdown resulted in reduced Il6 expression in miR-17∼92–deficient cells (Fig. 5, I and J), implying that miR-17∼92 cluster deficiency–associated Cebpb up-regulation contributed to the IL-6 overproduction phenotype. In addition, although Stat3 has been implicated as a direct target of miR-17 (Zhang et al., 2011), no significant differences in Stat3 protein levels were detected after comparing WT and miR-17∼92–deficient BMDMs and colonic macrophages (Fig. S4, K–M). In contrast to the heightened expression of C/EBPβ in miR-17∼92 KO cells, RBP-J–deficient colonic macrophages exhibited decreased protein levels of C/EBPβ (Fig. S4, N and O). Taken together, these results suggested that RBP-J promoted IL-6 expression by targeting the miR-17∼92–C/EBPβ axis in vivo and in vitro and genetically implicated RBP-J–controlled miRNAs in colonic macrophage–mediated immune defense.
RBP-J KO mice mount high abundances of ILC3-derived IL-22 during the late phase of infection
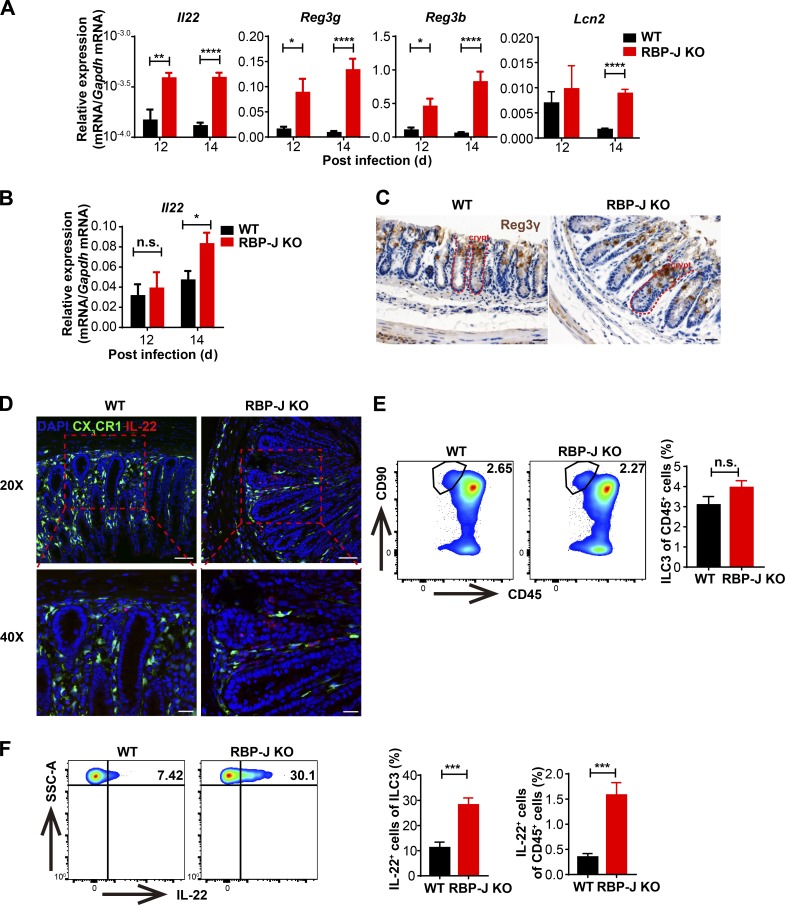
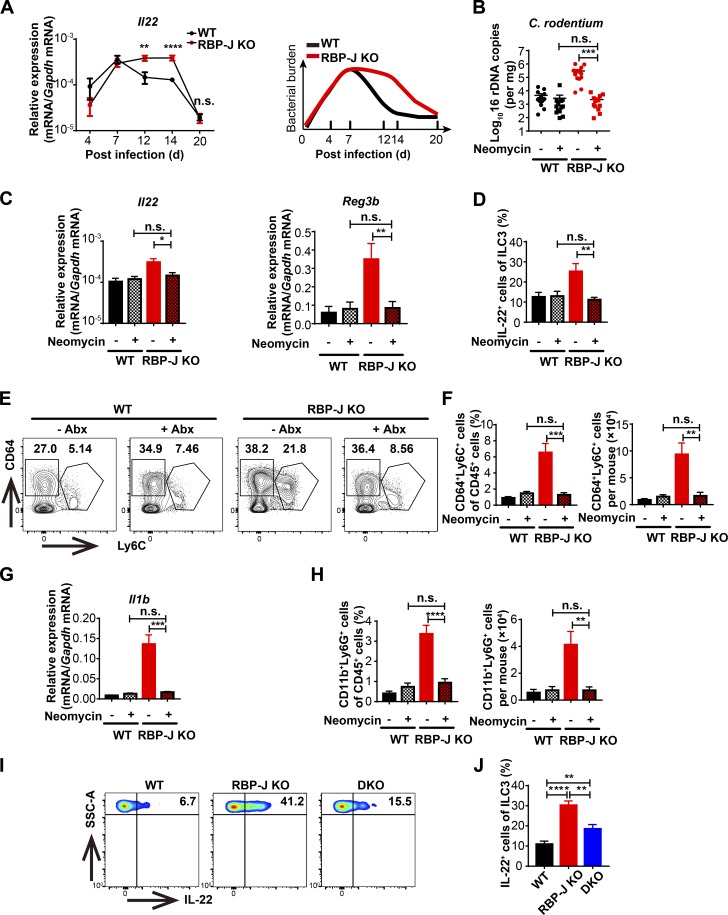
The above data showed that although the clearance of C. rodentium in RBP-J KO mice was markedly impaired, bacteria were eventually cleared at day 20 p.i. (Fig. 1 A), which was beyond the typical course of adaptive immune responses and plausibly mediated by alternative mechanisms. To determine how RBP-J KO mice with impaired innate and adaptive immune responses cleared C. rodentium in the late phase of infection, we evaluated expression of some key factors involved in the host defense. Intriguingly, Il22, but not Il17a expression in colon tissues and LP mononuclear cells was markedly elevated in RBP-J KO mice at days 12 and 14 p.i. (Fig. 6, A and B; and Fig. S5 A). Given that IL-22 promotes epithelial cells to secret AMPs to aid in the eradication of pathogens, we measured expression levels of common AMPs and found that Reg3g, Reg3b, and Lcn2 mRNA levels were increased in RBP-J KO colon tissues after day 12, but not before day 7 p.i. (Fig. 6 A and Fig. S5 B). In addition, histological and immune-histochemical analyses showed that at day 14 p.i., Reg3γ protein was increased in colon tissue from RBP-J KO mice that displayed goblet cell hyperplasia as assessed by Periodic acid–Schiff+ staining (Fig. 6 C and Fig. S5 C). These data suggested that noneradicated C. rodentium in RBP-J KO mice were likely cleared by IL-22–induced AMPs during the late phase of infection.
Figure 6.
RBP-J KO mice display heightened levels of intestinal ILC3-derived IL-22 during the late phase of infection. WT and RBP-J KO mice were orally inoculated with 2 × 109 CFUs of C. rodentium. (A and B) qPCR of Il22 and AMP mRNAs in colon tissues (A) and LP mononuclear cells (B). Data are pooled from two or three independent experiments; n ≥ 3 in each group. (C) Immunohistochemical analysis of Reg3γ protein levels at day 14 p.i. (scale bars represent 20 µm). (D) Immunofluorescence staining for IL-22 (red) and DAPI (blue) in the distal colon from WT (CX3CR1gfp/+ Lyz2-Cre) and RBP-J KO (CX3CR1gfp/+ Rbpjfl/fl Lyz2-Cre) mice at day 14 p.i. Scale bars represent 50 µm (top panels) and 20 µm (bottom panels). (E and F) Representative FACS plots (left) and cumulative data (right) of colonic LP ILC3 (CD45midCD3−Thy-1+) populations (E) and IL-22 production in ILC3 (F) at day 14 p.i. Data are pooled from three independent experiments; n ≥ 3 in each group. Data are shown as mean ± SEM; n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; two-tailed Student’s unpaired t test. SSC-A, side scatter area.
Figure S5.
Sustained IL-22 production by ILC3 in RBP-J KO mice is secondary to persistence of pathogens during the late phase of infection. WT and RBP-J KO mice were orally inoculated with 2 × 109 CFUs of C. rodentium. (A) qPCR analysis of Il17a in colon tissues (left) and LP mononuclear cells (right) at the indicated p.i. days. Data are pooled from two independent experiments; n ≥ 3 in each group. (B) qPCR analysis of Reg3g and Reg3b in colon tissues at the indicated p.i. days. Data are pooled from two independent experiments; n ≥ 3 in each group. (C) Periodic acid–Schiff staining in colons at day 14 p.i. (scale bars represent 20 µm). (D) Representative FACS plots (left) and cumulative data (right) of IL-22 production in LP CD4+ T cells (gated by CD3+CD4+) at day 14 p.i. Data are pooled from two independent experiments; n ≥ 3 in each group. (E) qPCR analysis of Reg3g and Reg3b in colon tissues at day 20 p.i. Data are pooled from two independent experiments; n = 3 in each group. (F) WT and RBP-J KO mice were orally inoculated with 2 × 109 CFUs of C. rodentium and were given neomycin sulfate individually at day 12 p.i. Representative FACS plots of IL-22–producing LP ILC3 (gated by CD45midCD3−Thy-1+) with or without neomycin treatment at day 14 p.i. (G and H) qPCR analysis of Il23a in colon tissues (G) and sorted CD64+ macrophages (H) at the indicated p.i. days. Data are pooled from two independent experiments; n = 3 in each group. (I) Representative FACS plots of LP neutrophils from WT and RBP-J KO mice with or without neomycin treatment at day 14 p.i. (J) qPCR analysis of Rbpj and Il1b in sorted LP neutrophils at day 14 p.i. Data are pooled from two independent experiments; n = 3 in each group. (K) Absolute numbers of neutrophils (CD11b+Ly6G+) and macrophages (CD11b+CD64+SiglecF−) in WT mice at day 14 p.i. Data are pooled from two independent experiments; n = 3 in each group. Data are shown as mean ± SEM; n.s., not significant; **, P < 0.01; two-tailed Student’s unpaired t test.
To identify the cellular source of sustained IL-22 in RBP-J KO mice, colon sections from day 14 p.i. WT and RBP-J KO mice on the CX3CR1-GFP reporter background (CX3CR1gfp/+ Rbpjfl/fl Lyz2-Cre) were stained with an anti-IL-22 antibody. In colonic LP, RBP-J KO mice exhibited enhanced numbers of IL-22–expressing cells, yet these IL-22+ cells did not colocalize with CX3CR1+ cells, indicating that IL-22 was not secreted by CX3CR1+ cells (Fig. 6 D). Next, we characterized CD45midCD3−Thy-1+ ILC3 population at day 14 p.i. and found that despite similarity in the ILC3 populations (Fig. 6 E), ILC3 in RBP-J KO mice produced significantly higher levels of IL-22 than those in WT mice (Fig. 6 F). Of note, the percentage of IL-22–producing CD4+ T cells showed no differences between WT and RBP-J KO mice (Fig. S5 D), suggesting that ILC3, but not CD4+ T cells were responsible for heightened IL-22 production in RBP-J KO mice. Overall, these results illustrated that colonic macrophages dynamically coordinated ILC3 effector functions during the late phase of infection in an RBP-J–regulated manner.
Sustained IL-22 production in RBP-J KO mice is dependent on CD64+Ly6C+ colonic macrophages secondary to noneradicated pathogens
To uncover the factors that contributed to prolonged production of IL-22 by ILC3, we monitored the kinetics of Il22 mRNA in colon tissues during the course of infection and found that fluctuations of Il22 and AMPs (Fig. 6 A and Fig. S5, B and E) tightly correlated with changes of bacterial burdens (Fig. 7 A) in WT and RBP-J KO mice. Thus, we speculated that the high abundances of IL-22 and AMPs in the convalescent phase of infection were sustained by noneradicated pathogens in RBP-J KO mice. To test this hypothesis, infected mice were treated with neomycin at day 12 p.i. to deplete noneradicated C. rodentium. Antibiotics treatment significantly reduced C. rodentium loads in RBP-J KO mice (Fig. 7 B) and subsequently inhibited expressions of IL-22 and AMPs in colon tissues to levels comparable with those observed in WT mice (Fig. 7 C). Consistently, ILC3 from neomycin-treated RBP-J KO mice showed diminished capacity to produce IL-22 compared with untreated RBP-J KO mice (Fig. S5 F and Fig. 7 D). To investigate the connections between IL-22–producing ILC3 and pathogens, we characterized the colonic macrophage subsets and found that neomycin treatment of RBP-J KO mice led to markedly diminished population of CD64+Ly6C+ macrophages along with decreased expression of Il1b, which is well known to support ILC3 activation (Fig. 7, E–G; Seo et al., 2015). In addition to IL-1β, IL-23 has also been implicated in ILC3 activation (Longman et al., 2014), whereas no significant differences in Il23a expression in colon tissues (Fig. S5 G) and sorted colonic macrophages (Fig. S5 H) were found between WT and RBP-J KO mice in the late phase of infection. Thus, these data implied that it was likely IL-1β not IL-23 that induced ILC3 hyperactivation in RBP-J KO mice. Furthermore, neomycin treatment also reversed excessive recruitment of neutrophils in RBP-J KO mice (Fig. 7 H and Fig. S5 I), yet the expression of Il1b was similar between WT and RBP-J–deficient neutrophils (Fig. S5 J). In addition, the comparison of the absolute numbers of neutrophils and colonic macrophages suggested that neutrophils were likely not the main source of IL-1β contributing to prolonged ILC3 activation (Fig. S5 K). These data implied that heightened immune responses late during the infection course were secondary to noneradicated pathogens.
Figure 7.
Persistent IL-22 activities manifest by RBP-J deficiency are dependent on noneradicated pathogens-elicited CD64+Ly6C+ colonic macrophages. (A) qPCR of Il22 in colon tissues (left) and schematic illustration of bacterial burdens in colon (right) from WT and RBP-J KO mice at the indicated p.i. days. Data are pooled from three independent experiments; n ≥ 3 in each group. (B–H) 6–8-wk-old mice were orally inoculated with 2 × 109 CFUs of C. rodentium and given neomycin sulfate individually at day 12 p.i., and mice were sacrificed at day 14 p.i. Data are pooled from three independent experiments; n ≥ 3 in each group. (B) qPCR analysis of 16s rDNA copies to determine fecal bacterial burdens in colon with or without neomycin treatment. (C) qPCR of Il22 and AMPs in colon tissues with or without neomycin treatment. (D) FACS cumulative data of IL-22–producing LP ILC3 (gated by CD45midCD3−Thy-1+) with or without neomycin treatment. (E and F) Representative FACS plots (E) and cumulative data (F) of CD64+Ly6C+ colonic macrophages with or without neomycin treatment. (G) qPCR of Il1b in colon tissues with or without neomycin treatment. (H) FACS cumulative data of LP neutrophils with or without neomycin treatment. (I and J) 6–8-wk-old mice were orally inoculated with 2 × 109 CFUs of C. rodentium. Representative FACS plots (I) and cumulative data (J) of IL-22 production in colonic LP ILC3 (gated by CD45midCD3−Thy-1+) at day 14 p.i. Data are pooled from three independent experiments; n ≥ 3 in each group. Data are shown as mean ± SEM; n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; two-tailed Mann-Whitney test (B) or two-tailed Student’s unpaired t test (other panels). Each symbol in B represents an individual mouse. SSC-A, side scatter area.
Next, we analyzed IL-22–producing ILC3 in RBP-J miR-17∼92 DKO mice that exhibited relatively normal immune responses against C. rodentium and thus harbored lower bacterial loads than RBP-J KO mice (Fig. 5, A and B). Closely correlated with bacterial loads, DKO mice displayed decreased frequency of IL-22-secreting ILC3 compared with RBP-J KO mice (Fig. 7, I and J). These results indicated that ILC3-elicited IL-22, sustained by CD64+Ly6C+ colonic macrophages secondary to noneradicated pathogens, was crucial to clearance of C. rodentium in the late phase of infection in RBP-J KO mice. In summary, these results uncovered an unexpected role of late ILC3-IL-22-AMPs during infection to compensate for the impaired innate and adaptive immune responses, which led to ultimate clearance of pathogens.
Discussion
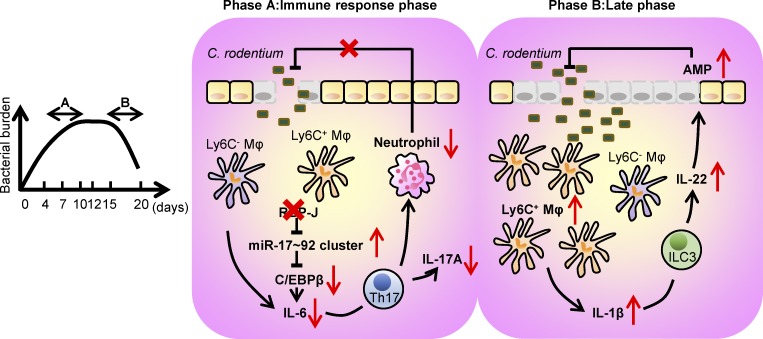
Maintenance of gut homeostasis requires a complex network involving the presence of commensal microbiota, proper barrier functions of IECs, and effective cross-talk between innate and adaptive immune cells. Macrophage, as a major innate sentinel residing in intestinal LP, acts as a critical player mediating immune responses to lumen-derived antigens. In this study, we identified RBP-J as a pivotal molecule in colonic macrophage–coordinated immune responses against C. rodentium (Fig. 8). These experiments provided genetic evidence that RBP-J in colonic macrophages facilitated effective clearance of C. rodentium by enhancing IL-6 production and thereby promoting Th17 cell responses through targeting the miR-17∼92-C/EBPβ molecular circuit. In the late phase of infection, delayed clearance of pathogens in RBP-J KO mice resulted in the prolonged activation of the IL-22–AMPs axis induced by noneradicated C. rodentium–recruited CD64+Ly6C+ colonic macrophages. This study uncovered the orchestration of intestinal immune responses by colonic macrophages at each stage of enteric bacterial infection, illustrated the cellular and molecular components of host–pathogen interaction related to the pathogenesis of infection-induced colitis, and dissected the regulatory mechanisms of RBP-J–mediated effects in colonic macrophages in vivo.
Figure 8.
A schematic model for the dynamical regulation of colonic macrophage–mediated immune responses against enteric bacterial infections by RBP-J. RBP-J is a crucial regulator in colonic macrophage–mediated immune responses against the enteric pathogen C. rodentium. In the immune response phase (phase A), RBP-J KO mice exhibit compromised clearance of pathogens due to diminished IL-6–induced Th17 immune responses secondary to up-regulation of the miRNA-17∼92 cluster and subsequent suppression of C/EBPβ protein expression. In the late phase (phase B), RBP-J KO mice eventually clear C. rodentium owing to late persistence of pathogen-dependent ILC3-derived IL-22 activated by CD64+Ly6C+ colonic macrophages.
Macrophages play important roles in antimicrobial defense during infection and influence the effector programs of other immune cells. In our study, RBP-J expression in macrophages was indispensable for eliciting competent innate immune responses against C. rodentium infection by controlling the production of key Th17 cell–promoting cytokines. Mechanistically, RBP-J facilitated the production of IL-6 via inhibition of the miR-17∼92 cluster. In the immune system, the miR-17∼92 cluster has been shown to promote lymphomagenesis by targeting PTEN and Bim in lymphocytes (Xiao et al., 2008), yet the function of the miR-17∼92 cluster in myeloid cells was unknown. Here, our data implicate that the miR-17∼92 cluster acts as a homeostatic inhibitor of inflammatory cytokine expression via a previously unappreciated target transcription factor, C/EBPβ, which is supported by multiple lines of experimental evidence, including reporter assays with 3′ UTR mutants. Nevertheless, it remains a major challenge to determine the exact targets of miRNAs, and we cannot exclude the involvement of other targets at this point. Importantly, compromised immune phenotypes resulted from RBP-J deficiency could be genetically corrected by further deleting the miR-17∼92 cluster, pinpointing this specific miRNA cluster as a molecular and functional target of RBP-J in colonic macrophages. These findings depicted a miRNA-centric indirect regulatory loop by which RBP-J regulated colonic macrophage effector functions to maintain the balance between inflammation and pathogen elimination in vivo. Small RNA-seq analysis in BMDMs identified a number of RBP-J–regulated miRNAs, including the miR-106a∼363 cluster, the miR-106b∼25 cluster, and the miR-181 family, whose immune functions demand future in-depth investigations (Table S1). In addition, further studies are required to decipher the molecular mechanism of such regulation by RBP-J.
Extensive literature has documented that IL-22–producing ILC3 is essential for innate immunity to enteric infections through induction of AMPs and maintenance of epithelial cell barrier function (Sonnenberg and Artis, 2015). However, the role of ILC3 in the late phase of infection remains to be determined. Due to the impaired immune responses, RBP-J KO mice displayed delayed clearance of pathogens, leading to aggravated C. rodentium–induced colitis in the convalescent phase of infection. We found that the fluctuations of Il22 in colon tissues were tightly correlated with C. rodentium burdens in feces during the course of infection and that RBP-J KO mice displayed prolonged production of IL-22 by ILC3 and subsequently AMPs by epithelial cells. ILCs that lack pattern-recognition receptors do not have the ability to recognize pathogen-associated molecular patterns directly (Robinette et al., 2015). Several lines of evidence including antibiotics treatment and experiments with DKO animals collectively supported that the sustained effector functions of ILC3 in RBP-J KO mice were secondary to excessive presence of CD64+Ly6C+ colonic macrophages as a result of noneradicated pathogens. These findings uncovered an unexpected role for ILC3–IL-22 in the late phase of infection, which was revealed under the conditions of defective innate and adaptive immunity. Late ILC3 responses functionally compensated for the otherwise compromised host defense and expanded the previous concept of ILC3 involvement in the innate phase of protection to the late phase.
Colonic MPs extend dendrites between IECs to uptake intestinal antigens in the gut lumen, among which colonic macrophages represent nonmigratory populations and do not traffic to draining lymph nodes to directly mediate T cell priming, a feature that is distinct from canonical CD103+ DCs (Schulz et al., 2009). Some reports have suggested that colonic macrophages have the capacity to direct T cell polarization by presenting antigens and producing mediators, including pro- and anti-inflammatory cytokines (Atarashi et al., 2008; Bauché et al., 2018; Denning et al., 2007; Panea et al., 2015; Schreiber et al., 2013). However, the exact contributions of distinct colonic macrophages and DC subsets in T cell activation are still controversial. Therefore, genetic approaches taking advantage of cell type–specific gene deletion as well as utilization of reporter mice, as performed in the current study, may aid in better understanding of functionality of intestinal antigen-presenting cells. Leaving the DC compartment intact using Lyz2-Cre, we found that colonic macrophages were the dominant source of certain pro-inflammatory cytokines such as IL-6 during enteric infections, which facilitated local activation and maintenance of Th17 cells. Despite normal populations and functions of CD11b−CD103+ and CD11b+CD103− colonic DCs, RBP-J deficiency in macrophages led to impaired IL-17A production of effector T cells (Fig. 2, E and F) without affecting normal Th17 cell differentiation as assessed by RORγt positivity (Fig. S2, G and H). The above lines of evidence are consistent with a model whereby DCs and macrophages play different, and perhaps sequential, roles in intestinal Th17 cell responses. Upon enteric bacterial infections, DCs instruct initial Th17 cell differentiation, and subsequently, macrophages act locally in the intestinal microenvironment to facilitate full-fledged Th17 cell effector functions, which is consistent with the minimal migratory capacity of colonic macrophages (Zigmond and Jung, 2013). Taken together, this study implies that colonic macrophages are indispensable for mounting optimal adaptive immunity and play nonredundant roles with DCs in host defense against C. rodentium.
Materials and methods
Mice
CX3CR1gfp/gfp mice (JAX stock 005582; Jung et al., 2000) and miR-17∼92fl/fl mice (JAX stock 008458; Ventura et al., 2008) were purchased from the Jackson Laboratory. Mice with a myeloid-specific deletion of the Rbpj were generated by crossing Rbpjfl/fl animals to animals with a Lyz2-Cre on the C57/BL6 background as described previously (Hu et al., 2008). miR-17∼92fl/fl Lyz2-Cre mice were generated by crossing miR-17∼92fl/fl with Lyz2-Cre mice. Rbpjfl/fl Lyz2-Cre mice were crossed to CX3CR1gfp/gfp mice to obtain Rbpjfl/fl Lyz2-Cre CX3CR1gfp/+ mice. Rbpjfl/fl Lyz2-Cre mice were crossed to miR-17∼92fl/fl Lyz2-Cre mice to obtain Rbpjfl/fl miR-17∼92fl/fl Lyz2-Cre mice. Gender- and age-matched mice with Rbpj+/+Lyz2-Cre, miR-17∼92+/+Lyz2-Cre, Rbpj+/+ Lyz2-Cre CX3CR1gfp/+, and Rbpj+/+ miR-17∼92+/+ Lyz2-Cre genotypes were bred in-house and used as controls. Unless otherwise specified, mice were cohoused after weaning for 3–4 wk, used at 6–8 wk of age, and separated p.i. according to their genotypes. All mice were on C57/BL6 background and kept under specific pathogen–free conditions at the Experimental Animal Facility, Tsinghua University. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committees at Tsinghua University.
Reagents, bacteria, and cell lines
4% sterile thioglycollate was purchased from Sigma-Aldrich. Cell culture–grade LPS (E. coli O111:B4) was purchased from Sigma-Aldrich. C. rodentium strain DBS100 (ATCC51459; American Type Culture Collection) and C. rodentium–GFP (a generous gift from Dr. Elizabeth Hartland, Hudson Institute of Medical Research, Monash University, Melbourne, Australia) were used to inoculate mice through oral gavage. For the colony-formation assays, C. rodentium was plated at serial dilutions onto MacConkey agar plates, and the number of CFUs was determined after overnight incubation at 37°C. For preparation of heat-killed C. rodentium, the pathogen was then harvested and washed twice with ice-cold PBS and heat inactivated at 60°C for 30 min.
HEK293T cells were cultured in DMEM supplemented with 10% FBS. All cells were cultured at 37°C with 5% CO2.
C. rodentium infection and depletion
C. rodentium was grown overnight in LB broth with shaking at 37°C. Mice were infected by oral gavage with 2 × 109 or 4 × 109 CFUs of C. rodentium in a total volume of 200 µl. Feces were collected for bacterial DNA analysis or CFU counts. C. rodentium was depleted by feeding mice with neomycin sulfate (1 mg/ml; Sigma-Aldrich) in a total volume of 250 µl individually at day 12 p.i. by oral administration.
Construction of GFP-expressing C. rodentium strain
GFP-expressing C. rodentium (C. rodentium–GFP) was kindly provided by Dr. Elizabeth Hartland. GFP was cloned into EcoRV–BamHI sites located in the Tet gene in pACYC184 vector driven from the Tet promoter, and the construct was transformed into C. rodentium. The primers used for cloning and strain construction are provided in Table S2.
Isolation of bacterial DNA
For isolation of lumen bacterial DNA, the intestinal tract was excised and the distal 5 cm of the colon was isolated. The luminal contents were collected by flushing with 1 ml sterile PBS, weighed, and homogenized. The luminal bacterial DNA was immediately isolated with the Stool Genomic DNA Kit (CWBIO) according to the manufacturer’s instructions.
Immunohistochemistry
Colon sections were stained with H&E for assessment of intestinal architecture and Periodic acid–Schiff, anti-Reg3g (AP5606c; Abgent) for immunohistochemistry. Slides were washed three times with 0.1% TBS-Tween before incubation with secondary antibodies. Stained slides were washed again in PBS and stained with DAB (TIANGEN) in conjunction with a hematoxylin counterstain (Solarbio). After dehydration, sections were mounted in neutral balsam.
Immunofluorescence histology
Colons were washed and fixed overnight at 4°C in a solution of 1% paraformadehyde in PBS. The tissues were incubated in a solution of 30% sucrose in PBS and the mixture of 30% sucrose and OCT compound 4583 (Sakura Finetek) separately at 4°C overnight. The samples were then embedded in OCT, frozen in a bath of ethanol cooled with liquid nitrogen and stocked at −80°C. Frozen samples were cut at 10-µm thickness and collected onto slides. Slides were dried at 50°C for 30 min and fixed in 1% paraformaldehyde for 10 min and processed for staining. The tissues were permeabilized in PBS/0.5% Triton X-100/0.3 M glycine at 37°C for 30 min and blocked in PBS/5% goat serum at room temperature for 1 h. The tissues were then incubated with indicated primary antibodies diluted (1:1,000) in PBS/5% goat serum at 4°C overnight (anti-green fluorescent PR, A11122 [Invitrogen]; and IL-22 antibody, NB100-737 [Novus]), and washed in PBS/0.2% Tween-20 at room temperature for 30 min three times. The tissues were incubated with Alexa dye-conjugated secondary antibodies (goat anti-rabbit IgG-TR, 1:200, sc-2780; Santa Cruz) and DAPI (1:200) in PBS/0.5% BSA at room temperature for 2 h and washed in PBS/0.2% Tween-20 at room temperature for 1 h five times before mounting with SlowFade Diamond Antifade Mountant (Life Technologies).
IL-6 administration in vivo
IL-6 (Peprotech) was intraperitoneally injected at days 3 and 5 after C. rodentium infection at a dose of 200 ng per mouse. Mice were then sacrificed at day 7 p.i. for experimental assessments.
Preparation of colonic LP mononuclear cells
Mice were sacrificed and colons were removed and placed in ice-cold calcium and magnesium-free HBSS. The colons were cut open longitudinally, thoroughly washed in ice-cold HBSS, and cut into 1.5-cm pieces. Colons were incubated twice in HBSS containing 10 mM Hepes, 10 mM EDTA (Promega), and 1 mM dithiothreitol (Fermentas) for 20 min at 37°C to remove epithelial cells and mucus. For isolation of LP mononuclear cells, the tissues were then digested in RPMI 1640 (with calcium and magnesium) containing 5% heat-inactivated FBS, 1 mg/ml Collagenase IV (Sigma-Aldrich), 1 mg/ml Dispase (Roche), and 100 µg/ml DNaseI (Sigma-Aldrich) for 75 min at 37°C with slow rotation after washing with HBSS containing 10 mM Hepes. The digested tissues were homogenized by vigorous shaking, passed through a 70-µm cell strainer, and resuspended in 40% Percoll (GE Healthcare) solution before gradient density centrifugation at 2,500 rpm for 20 min without brake at room temperature. Red blood cells were lysed using ammonium-chloride-potassium lysing buffer.
Cell isolation
For isolation of the thioglycollate-elicited peritoneal macrophages, mice were injected intraperitoneally with 1 ml of 4% sterile thioglycollate, and peritoneal cells were collected by washing with PBS 3.5 d after injection. Nonadherent cells were removed 12 h later, whereas adherent cells were replated in DMEM with 10% FBS and allowed to recover overnight.
Mouse BMDMs were obtained as described previously (Xu et al., 2012) and maintained in DMEM supplemented with 10% FBS and 10% supernatants of L929 mouse fibroblasts as conditioned medium providing macrophage colony-stimulating factor. After 5 d of culture, floating cells were discarded, and attached macrophages were plated in multiple well plates overnight prior to stimulation.
For isolation of splenic T cells, organs were mashed through a 70-µm cell strainer, and red blood cells were lysed using ammonium-chloride-potassium lysing buffer.
In vitro T cell differentiation
Splenic naive T cells (CD4+CD62LhiCD25−CD44lo) were isolated by positive selection using CD4 (L3T4) MACS (Miltenyi Biotech) and FACS. Cells were cultured in 48-well plates bound with anti-CD3 (6 µg/ml) and anti-CD28 (6 µg/ml, both from Sungene) for 3 d in the presence of 0.5 ng/ml recombinant human TGF-β (R&D Systems), with or without 20 ng/ml IL-6 (Peprotech).
Flow cytometry
Nonspecific antibody binding was blocked with anti-CD16/CD32 antibody before surface staining.
Fluorescence-conjugated mAb against CD45 (30-F11), IAb (AF6-120.1), Ly6C (HK1.4), CD64 (X54-5/7.1), CD103 (2E7), CX3CR1 (SA011F11), CD4 (GK1.5), CD44 (IM7), CD62L (MEL-14), CD69 (H1.2F3), and CD90.2 (30-H12) were purchased from BioLegend. Fluorescence-conjugated mAb against CD11b (M1/70), CD11c (N418), F4/80 (BM8), CD3 (17A2), IL-17a (eBio17B7), IL-22 (1H8PWSR), Foxp3 (FJK-16s), RORγt (AFKJS-9), and STAT3 (232209) were purchased from eBioscience. Fluorescence-conjugated mAb against SiglecF (E50-2440), Ly6G (RB6-8C5), and CD25 (7D4) were purchased from BD Biosciences. Isotype-matched antibodies (BioLegend) were used for control staining. All antibodies were used in 1:400 dilutions in 2 × 106 cells per 50 µl except CD64 (used in 1:200 dilution). Staining of surface molecules with fluorescently labeled antibodies was performed on ice for 30 min in the dark.
For measurement of intracellular cytokine expression, cells were isolated ex vivo and stimulated with PMA (50 ng/ml) and ionomycin (500 ng/ml; both from Sigma-Aldrich) for 4–5 h in the presence of Golgistop (BD Biosciences). Dead cells were excluded from the analysis by using Fixable Viability Dye eFluor 506 (eBioscience). Cells were stained with antibodies to surface antigens and then fixed and permeabilized with the Fixation and Permeabilization kit (BD Biosciences) according to the manufacturer’s instructions, followed by staining with antibodies.
For analysis of transcription factor, cells were stained with antibodies to surface antigens, fixed and permeabilized with the Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer’s instructions, and stained with antibodies or isotype control diluted in permeabilization buffer separately at room temperature for 30 min. For analysis of C/EBPβ, after permeabilization, cells were blocked in PBS/5% donkey serum at room temperature for 30 min and stained with anti-CEBPβ (1:1,000, ab32358; Abcam) or anti-rabbit IgG isotype (1:1,000, ab172730; Abcam) and then donkey anti-rabbit IgG-FITC (1:200, sc-2090; Santa Cruz) diluted in permeabilization buffer separately at room temperature for 30 min.
Flow cytometry analysis was performed on FACSFortessa or FACSAria III flow cytometer (BD Biosciences) and analyzed with FlowJo software (Tree Star).
Measurement of cytokines and antigen-specific Igs
Concentrations of IL-6 and IL-1β (BD Biosciences) in culture supernatants were measured with ELISAs according to the manufacturer’s recommendations.
C. rodentium–specific Ig amounts in sera and fecal were measured with SBA Clonotyping System (Southern Biotech) according to the manufacturer’s instructions. C. rodentium was grown overnight in LB broth with shaking at 37°C. After centrifugation, bacterial pellets were resuspended in sterile PBS and sonicated on ice. The supernatant was collected by removing the pellet after centrifugation at 13,000 rpm for 5 min and used to coat ELISA plates at the concentration of 5 µg/ml. Feces were collected from individual mice and then weighed and homogenized in sterile PBS at the concentration of 50 mg/ml. After centrifugation at 13,000 rpm for 5 min, the supernatants were collected and frozen at −80°C.
qPCR
RNA from tissues and cultured cells was extracted with Total RNA purification kit (GeneMark), and RNA from sorted cells was extracted with Trizol reagent (Invitrogen). Complementary DNA was synthesized from 1 mg total RNA by M-MLV reverse transcription (Takara). qPCR was performed using FastSYBR mixture (CWBIO) with specific primers (Table S2) on a real-time PCR system (StepOnePlus; Applied Biosystems). The comparative threshold cycle method and an internal control (Gapdh) were used to normalize the expression of target genes.
To measure mature miRNA, cDNA was prepared from total RNA with the TaqMan microRNA Reverse Transcription Kit (Applied Biosystems). qPCR was performed with TaqMan microRNA assays according to the manufacturer’s recommendations (Applied Biosystems). U6 small nuclear RNA was used as internal control to normalize the expression of miRNAs.
The abundance of specific intestinal bacterial groups was measured by qPCR with FastSYBR mixture (CWBIO) and universal 16s rDNA primers (Table S2). Bacterial abundance was determined using standard curves with reference to cloned bacterial DNA corresponding to a short segment of the 16s rRNA gene that was amplified using conserved specific primers. It should be noted that qPCR measures 16s rRNA gene copies per sample, not the actual bacterial numbers or CFUs.
ChIP assays
For RBP-J ChIP assays, BMDMs from WT mice were used. Approximately 15–20 × 106 cells were fixed by 1.0% methanol-free formaldehyde for 10 min at room temperature. The ChIP assay was performed using the SimpleChIP enzymatic ChIP kit (Cell Signaling Technology [CST]) according to the manufacturer’s instructions. The DNA–protein complexes were immunoprecipitated using 5.0 µl of RBP-J antibody from CST. The bound DNA fragments were subjected to qPCR with SYBR Green reagents. The primer sequences are listed in Table S2.
Small RNA-seq analysis
Total RNA was isolated, and the small RNA fractions were enriched with the mirVana miRNA Isolation Kit (Life Technologies) according to the manufacturer’s instructions. miRNA libraries were constructed per the Illumina TruSeq Small RNA Library preparation kit. High-throughput sequencing was performed using the Illumina HiSeq 1500. miRNA-seq reads were aligned to the mouse miRNA sequences in the miRBase database (release 21) using miRDeep2. Mature miRNA values were normalized by library size (corresponding to counts per million [cpm] mapped miRNA reads). miRNAs with cpm values <5 in all conditions were eliminated from further analysis. RBP-J–regulated miRNAs were defined as cpm values of miRNAs in RBP-J KO BMDMs versus those in WT cells >1.2 (up-regulated) or <0.6 (down-regulated) with P values < 0.05.
Western blotting
Whole-cell lysates were separated by 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane by electroblotting. After blocking with 5% fat-free milk, the membranes were incubated at 4°C overnight with the following primary antibodies: anti-CEBPβ (1:1,000, ab32358; Abcam), β-Actin (1:1,000, ac026; ABclonal Technology), phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204) rabbit mAb (1:1,000, 4370; CST), Phospho-p38 MAPK (Thr180/Tyr182) rabbit mAb (1:1,000, 9215; CST), Phospho-NF-κB p65 (Ser536) rabbit mAb (1:1,000, 3033; CST), RBPSU XP rabbit mAb (1:1,000, 5313; CST), and STAT3 mouse mAb (1:1,000, 9139; CST). The membranes were then washed and incubated with goat anti-rabbit IgG (H&L)-HRP conjugated antibody (1:10,000, BE0101; EASYBIO) or goat anti-mouse IgG antibody (1:10,000, BE0102; EASYBIO). Proteins were visualized with SuperSignal West Pico Chemiluminescent Substrate (34080; Thermo Scientific).
Luciferase reporter assays
We constructed a luciferase reporter plasmid with Cebpb 3′ UTR WT containing predicted miR-92–binding sites and mutant that deleted the putative 6-nt target sequence pairing with the seed region of the miR-17∼92 cluster with the psiCHECK2 vector (Promega). HEK293T cells were cotransfected with the Cebpb luciferase reporter plasmid and an expression plasmid (pCMV-miR-17∼92) encoding mouse miR-17∼92 cluster using FuGENE HD Transfection Reagent (Promega). After 24-h transfection, cell lysates were prepared and analyzed using Dual-Luciferase Report Assay System (Promega). The Renilla firefly luciferase activity was normalized by the firefly luciferase activity, and expression is presented as Renilla firefly luciferase/firefly luciferase activity ratio.
RNA-mediated interference
siRNA specifically targeting mouse Cebpb and nontargeting control siRNA were purchased from GenePharma Company. siRNA was transfected into BMDMs using INVI DNA RNA Transfection Reagent according to the manufacturer’s instructions (Invigentech). Cells were used 48 h after transfection.
Statistical analyses
Statistical analysis, excluding microbiome, was performed using Prism 6.0 (GraphPad Software). Statistical analyses were performed with averages of biological replicates. All results are shown as mean and SEM. Student’s unpaired t tests, Student’s paired t tests, and Mann-Whitney tests were used to statistically analyze the results. Statistical significance was defined as P < 0.05.
Data and software availability
Small RNA-seq data are deposited in the Genome Expression Omnibus under accession no. GSE103220.
Online supplemental material
Fig. S1 shows that WT and RBP-J KO mice display similar colonic architectures and abundance of commensal bacteria in homeostasis. Fig. S2 shows that RBP-J KO mice display normal activation of ILC3 and humoral immune responses during the adaptive immune response phase against C. rodentium infection. Fig. S3 shows that RBP-J KO mice display normal functions of DCs and neutrophils. Fig. S4 shows that RBP-J promotes IL-6 production by inhibition of the miR-17∼92–C/EBPβ axis. Fig. S5 shows that sustained IL-22 production by ILC3 in RBP-J KO mice is secondary to persistence of pathogens during the late phase of infection. Table S1 lists RBP-J–suppressed miRNAs. Table S2 lists the primers and oligonucleotides used in this study.
Supplementary Material
lists RBP-J–suppressed miRNAs.
lists primers and oligonucleotides used in this study.
Acknowledgments
We thank Elizabeth Hartland for providing C. rodentium with GFP. We are thankful for the support of the animal core facility at Tsinghua University.
This research was supported by the National Natural Science Foundation of China (grants 31725010 and 31821003 to X. Hu), the Ministry of Science and Technology of China (National Key Research Project 2015CB943201 to X. Hu), UK Royal Society/National Natural Science Foundation of China (Newton Advanced Fellowship 81661130161), Tsinghua-Peking Center for Life Sciences (X. Hu), and Institute for Immunology at Tsinghua University (X. Hu).
Author contributions: L. Kang designed research, performed experiments, analyzed data, and wrote the manuscript; X. Zhang performed miRNA-related experiments; L. Ji performed some FACS experiments and prepared mice for experiments; T. Kou performed plasmid construction experiments; S.M. Smith performed small RNA-seq; B. Zhao provided suggestions to the study; X. Guo provided key reagents in experimentation; I. Pineda-Torra and L. Wu provided valuable advice on the project; and X. Hu conceptualized the project, designed research, supervised experiments, and wrote the manuscript.
References
- Agarwal V., Bell G.W., Nam J.W., and Bartel D.P.. 2015. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 4:e05005 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., Nakajima T., Hirano T., and Kishimoto T.. 1990. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9:1897–1906. 10.1002/j.1460-2075.1990.tb08316.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Nishimura J., Shima T., Umesaki Y., Yamamoto M., Onoue M., Yagita H., Ishii N., Evans R., Honda K., and Takeda K.. 2008. ATP drives lamina propria T(H)17 cell differentiation. Nature. 455:808–812. 10.1038/nature07240 [DOI] [PubMed] [Google Scholar]
- Bain C.C., Bravo-Blas A., Scott C.L., Perdiguero E.G., Geissmann F., Henri S., Malissen B., Osborne L.C., Artis D., and Mowat A.M.. 2014. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15:929–937. 10.1038/ni.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauché D., Joyce-Shaikh B., Jain R., Grein J., Ku K.S., Blumenschein W.M., Ganal-Vonarburg S.C., Wilson D.C., McClanahan T.K., Malefyt R.W., et al. . 2018. LAG3+ Regulatory T Cells Restrain Interleukin-23-Producing CX3CR1+ Gut-Resident Macrophages during Group 3 Innate Lymphoid Cell-Driven Colitis. Immunity. 49:342–352.e5. 10.1016/j.immuni.2018.07.007 [DOI] [PubMed] [Google Scholar]
- Bry L., and Brenner M.B.. 2004. Critical role of T cell-dependent serum antibody, but not the gut-associated lymphoid tissue, for surviving acute mucosal infection with Citrobacter rodentium, an attaching and effacing pathogen. J. Immunol. 172:433–441. 10.4049/jimmunol.172.1.433 [DOI] [PubMed] [Google Scholar]
- Bry L., Brigl M., and Brenner M.B.. 2006. CD4+-T-cell effector functions and costimulatory requirements essential for surviving mucosal infection with Citrobacter rodentium. Infect. Immun. 74:673–681. 10.1128/IAI.74.1.673-681.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., and Reizis B.. 2007. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 204:1653–1664. 10.1084/jem.20062648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerovic V., Bain C.C., Mowat A.M., and Milling S.W.. 2014. Intestinal macrophages and dendritic cells: what’s the difference? Trends Immunol. 35:270–277. 10.1016/j.it.2014.04.003 [DOI] [PubMed] [Google Scholar]
- Collins J.W., Keeney K.M., Crepin V.F., Rathinam V.A., Fitzgerald K.A., Finlay B.B., and Frankel G.. 2014. Citrobacter rodentium: infection, inflammation and the microbiota. Nat. Rev. Microbiol. 12:612–623. 10.1038/nrmicro3315 [DOI] [PubMed] [Google Scholar]
- Crepin V.F., Collins J.W., Habibzay M., and Frankel G.. 2016. Citrobacter rodentium mouse model of bacterial infection. Nat. Protoc. 11:1851–1876. 10.1038/nprot.2016.100 [DOI] [PubMed] [Google Scholar]
- Denning T.L., Wang Y.C., Patel S.R., Williams I.R., and Pulendran B.. 2007. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat. Immunol. 8:1086–1094. 10.1038/ni1511 [DOI] [PubMed] [Google Scholar]
- Foldi J., Shang Y., Zhao B., Ivashkiv L.B., and Hu X.. 2016. RBP-J is required for M2 macrophage polarization in response to chitin and mediates expression of a subset of M2 genes. Protein Cell. 7:201–209. 10.1007/s13238-016-0248-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Zhang Y.F., Zhang Z.P., Fu L.A., Cao X.L., Zhang Y.Z., Guo C.J., Yan X.C., Yang Q.C., Hu Y.Y., et al. . 2017. miR-342-5p Regulates Neural Stem Cell Proliferation and Differentiation Downstream to Notch Signaling in Mice. Stem Cell Reports. 8:1032–1045. 10.1016/j.stemcr.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., and Jung S.. 2014. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14:392–404. 10.1038/nri3671 [DOI] [PubMed] [Google Scholar]
- Hu X., Chung A.Y., Wu I., Foldi J., Chen J., Ji J.D., Tateya T., Kang Y.J., Han J., Gessler M., et al. . 2008. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 29:691–703. 10.1016/j.immuni.2008.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishifune C., Maruyama S., Sasaki Y., Yagita H., Hozumi K., Tomita T., Kishihara K., and Yasutomo K.. 2014. Differentiation of CD11c+ CX3CR1+ cells in the small intestine requires Notch signaling. Proc. Natl. Acad. Sci. USA. 111:5986–5991. 10.1073/pnas.1401671111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., and Littman D.R.. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Jakubzick C., Bogunovic M., Bonito A.J., Kuan E.L., Merad M., and Randolph G.J.. 2008. Lymph-migrating, tissue-derived dendritic cells are minor constituents within steady-state lymph nodes. J. Exp. Med. 205:2839–2850. 10.1084/jem.20081430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., and Littman D.R.. 2000. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114. 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M., and Kuchroo V.K.. 2009. IL-17 and Th17 Cells. Annu. Rev. Immunol. 27:485–517. 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- Kumar P., Monin L., Castillo P., Elsegeiny W., Horne W., Eddens T., Vikram A., Good M., Schoenborn A.A., Bibby K., et al. . 2016. Intestinal Interleukin-17 Receptor Signaling Mediates Reciprocal Control of the Gut Microbiota and Autoimmune Inflammation. Immunity. 44:659–671. 10.1016/j.immuni.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K.L., Caton M.L., Bogunovic M., Greter M., Grajkowska L.T., Ng D., Klinakis A., Charo I.F., Jung S., Gommerman J.L., et al. . 2011. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 35:780–791. 10.1016/j.immuni.2011.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman R.S., Diehl G.E., Victorio D.A., Huh J.R., Galan C., Miraldi E.R., Swaminath A., Bonneau R., Scherl E.J., and Littman D.R.. 2014. CX3CR1+ mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J. Exp. Med. 211:1571–1583. 10.1084/jem.20140678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaser C., Housley M.P., Iimura M., Smith J.R., Vallance B.A., Finlay B.B., Schreiber J.R., Varki N.M., Kagnoff M.F., and Eckmann L.. 2004. Clearance of Citrobacter rodentium requires B cells but not secretory immunoglobulin A (IgA) or IgM antibodies. Infect. Immun. 72:3315–3324. 10.1128/IAI.72.6.3315-3324.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manta C., Heupel E., Radulovic K., Rossini V., Garbi N., Riedel C.U., and Niess J.H.. 2013. CX(3)CR1(+) macrophages support IL-22 production by innate lymphoid cells during infection with Citrobacter rodentium. Mucosal Immunol. 6:177–188. 10.1038/mi.2012.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.H., Smith S.M., Elguindy M., Zhang T., Xiang J.Z., Hu X., Ivashkiv L.B., and Zhao B.. 2016. RBP-J-Regulated miR-182 Promotes TNF-α-Induced Osteoclastogenesis. J. Immunol. 196:4977–4986. 10.4049/jimmunol.1502044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat A.M., and Agace W.W.. 2014. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 14:667–685. 10.1038/nri3738 [DOI] [PubMed] [Google Scholar]
- Mundy R., MacDonald T.T., Dougan G., Frankel G., and Wiles S.. 2005. Citrobacter rodentium of mice and man. Cell. Microbiol. 7:1697–1706. 10.1111/j.1462-5822.2005.00625.x [DOI] [PubMed] [Google Scholar]
- Noah T.K., and Shroyer N.F.. 2013. Notch in the intestine: regulation of homeostasis and pathogenesis. Annu. Rev. Physiol. 75:263–288. 10.1146/annurev-physiol-030212-183741 [DOI] [PubMed] [Google Scholar]
- O’Connell R.M., Rao D.S., and Baltimore D.. 2012. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 30:295–312. 10.1146/annurev-immunol-020711-075013 [DOI] [PubMed] [Google Scholar]
- Ouyang W., Kolls J.K., and Zheng Y.. 2008. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 28:454–467. 10.1016/j.immuni.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panea C., Farkas A.M., Goto Y., Abdollahi-Roodsaz S., Lee C., Koscsó B., Gowda K., Hohl T.M., Bogunovic M., and Ivanov I.I.. 2015. Intestinal Monocyte-Derived Macrophages Control Commensal-Specific Th17 Responses. Cell Reports. 12:1314–1324. 10.1016/j.celrep.2015.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivollier A., He J., Kole A., Valatas V., and Kelsall B.L.. 2012. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 209:139–155. 10.1084/jem.20101387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette M.L., Fuchs A., Cortez V.S., Lee J.S., Wang Y., Durum S.K., Gilfillan S., and Colonna M.; Immunological Genome Consortium . 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16:306–317. 10.1038/ni.3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino S.J., Geddes K., and Girardin S.E.. 2012. Innate IL-17 and IL-22 responses to enteric bacterial pathogens. Trends Immunol. 33:112–118. 10.1016/j.it.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Sano T., Huang W., Hall J.A., Yang Y., Chen A., Gavzy S.J., Lee J.Y., Ziel J.W., Miraldi E.R., Domingos A.I., et al. . 2015. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell. 163:381–393. 10.1016/j.cell.2015.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A.T., Briseño C.G., Lee J.S., Ng D., Manieri N.A., Kc W., Wu X., Thomas S.R., Lee W.L., Turkoz M., et al. . 2013. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14:937–948. 10.1038/ni.2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber H.A., Loschko J., Karssemeijer R.A., Escolano A., Meredith M.M., Mucida D., Guermonprez P., and Nussenzweig M.C.. 2013. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J. Exp. Med. 210:2025–2039. 10.1084/jem.20130903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz O., Jaensson E., Persson E.K., Liu X., Worbs T., Agace W.W., and Pabst O.. 2009. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 206:3101–3114. 10.1084/jem.20091925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S.U., Kuffa P., Kitamoto S., Nagao-Kitamoto H., Rousseau J., Kim Y.G., Núñez G., and Kamada N.. 2015. Intestinal macrophages arising from CCR2(+) monocytes control pathogen infection by activating innate lymphoid cells. Nat. Commun. 6:8010 10.1038/ncomms9010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Smith S., and Hu X.. 2016. Role of Notch signaling in regulating innate immunity and inflammation in health and disease. Protein Cell. 7:159–174. 10.1007/s13238-016-0250-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw T.N., Houston S.A., Wemyss K., Bridgeman H.M., Barbera T.A., Zangerle-Murray T., Strangward P., Ridley A.J.L., Wang P., Tamoutounour S., et al. . 2018. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J. Exp. Med. 215:1507–1518. 10.1084/jem.20180019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons C.P., Clare S., Ghaem-Maghami M., Uren T.K., Rankin J., Huett A., Goldin R., Lewis D.J., MacDonald T.T., Strugnell R.A., et al. . 2003. Central role for B lymphocytes and CD4+ T cells in immunity to infection by the attaching and effacing pathogen Citrobacter rodentium. Infect. Immun. 71:5077–5086. 10.1128/IAI.71.9.5077-5086.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg G.F., and Artis D.. 2015. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat. Med. 21:698–708. 10.1038/nm.3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoutounour S., Henri S., Lelouard H., de Bovis B., de Haar C., van der Woude C.J., Woltman A.M., Reyal Y., Bonnet D., Sichien D., et al. . 2012. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur. J. Immunol. 42:3150–3166. 10.1002/eji.201242847 [DOI] [PubMed] [Google Scholar]
- Varol C., Zigmond E., and Jung S.. 2010. Securing the immune tightrope: mononuclear phagocytes in the intestinal lamina propria. Nat. Rev. Immunol. 10:415–426. 10.1038/nri2778 [DOI] [PubMed] [Google Scholar]
- Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J., Newman J., Bronson R.T., Crowley D., Stone J.R., et al. . 2008. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 132:875–886. 10.1016/j.cell.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.C., He F., Feng F., Liu X.W., Dong G.Y., Qin H.Y., Hu X.B., Zheng M.H., Liang L., Feng L., et al. . 2010. Notch signaling determines the M1 versus M2 polarization of macrophages in antitumor immune responses. Cancer Res. 70:4840–4849. 10.1158/0008-5472.CAN-10-0269 [DOI] [PubMed] [Google Scholar]
- Xiao C., Srinivasan L., Calado D.P., Patterson H.C., Zhang B., Wang J., Henderson J.M., Kutok J.L., and Rajewsky K.. 2008. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 9:405–414. 10.1038/ni1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., and Cao X.. 2010. Interleukin-17 and its expanding biological functions. Cell. Mol. Immunol. 7:164–174. 10.1038/cmi.2010.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhu J., Smith S., Foldi J., Zhao B., Chung A.Y., Outtz H., Kitajewski J., Shi C., Weber S., et al. . 2012. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat. Immunol. 13:642–650. 10.1038/ni.2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Liu Q., Mi S., Liang X., Zhang Z., Su X., Liu J., Chen Y., Wang M., Zhang Y., et al. . 2011. Both miR-17-5p and miR-20a alleviate suppressive potential of myeloid-derived suppressor cells by modulating STAT3 expression. J. Immunol. 186:4716–4724. 10.4049/jimmunol.1002989 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., and Ouyang W.. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14:282–289. 10.1038/nm1720 [DOI] [PubMed] [Google Scholar]
- Zigmond E., and Jung S.. 2013. Intestinal macrophages: well educated exceptions from the rule. Trends Immunol. 34:162–168. 10.1016/j.it.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Zigmond E., Varol C., Farache J., Elmaliah E., Satpathy A.T., Friedlander G., Mack M., Shpigel N., Boneca I.G., Murphy K.M., et al. . 2012. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 37:1076–1090. 10.1016/j.immuni.2012.08.026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists RBP-J–suppressed miRNAs.
lists primers and oligonucleotides used in this study.
Data Availability Statement
Small RNA-seq data are deposited in the Genome Expression Omnibus under accession no. GSE103220.