Fate mapping of resident microglia and peripheral monocytes reveals that the myeloid cells that migrate to and associate with amyloid plaques in a mouse model of Alzheimer’s disease are exclusively derived from resident microglia.
Abstract
Alzheimer’s disease (AD) is accompanied by a robust inflammatory response mediated by plaque-associated myeloid cells of the brain. These cells exhibit altered gene expression profiles and serve as a barrier, preventing neuritic dystrophy. The origin of these cells has been controversial and is of therapeutic importance. Here, we genetically labeled different myeloid populations and unequivocally demonstrated that plaque-associated myeloid cells in the AD brain are derived exclusively from resident microglia, with no contribution from circulating peripheral monocytes.
Introduction
Microglia arise from yolk sac erythromyeloid progenitors that invade the neuroepithelium early in development (embryonic day 8.5; Ginhoux et al., 2010; Schulz et al., 2012), where they undergo self-renewal throughout life in mice (Askew et al., 2017; Tay et al., 2017) and humans (Askew and Gomez-Nicola, 2018; Réu et al., 2017). Under normal conditions, there is no contribution to the brain’s parenchymal myeloid cell population from the periphery (Kierdorf and Prinz, 2017); however, yolk sac–derived monocytes in the choroid plexus are gradually replaced by peripheral monocytes from the bone marrow (Goldmann et al., 2016). Microglia perform surveillance of the brain through continuous extension and retraction of processes, monitoring for tissue integrity, synapse function, and pathogens (Davalos et al., 2005; Nimmerjahn et al., 2005).
Alzheimer’s disease (AD) is typified by a robust myeloid cell–mediated inflammatory response that contributes to disease pathogenesis. In the AD brain, an early increase in β amyloid (Aβ) levels is followed by condensation into plaques within the brain parenchyma. A subset of myeloid cells migrate to and associate with these Aβ plaques, disrupting their homeostasis and exhibiting progressive changes in their gene expression. The conversion of plaque-associated cells from a “homeostatic” state to a “disease-associated microglial” or “microglial neurodegeneration” phenotype is characterized by distinctive gene expression signatures, including high levels of Trem2 expression (Keren-Shaul et al., 2017; Krasemann et al., 2017). Importantly, these plaque-associated myeloid cells effect a barrier function, enveloping the plaques with their processes and phagocytically remodeling the plaque, leading to its compaction, thereby preventing neuritic dystrophy (Yuan et al., 2016).
The ontogenic origin of plaque-associated myeloid cells has been controversial owing to ambiguity about whether they originate from yolk sac–derived resident brain microglia or, alternatively, peripheral blood–borne monocytes that infiltrate the AD brain and subsequently acquire certain features shared with resident microglia. This point is of some importance in designing therapeutic interventions targeting this population of cells. The suggestion that monocytes infiltrate the AD brain, as occurs in other central nervous system disorders, and have salutary effects on AD pathogenesis is not new (El Khoury and Luster, 2008; Simard and Rivest, 2006; Simard et al., 2006). El Khoury et al. (2007) reported peripheral myeloid cells infiltrated the AD brain in a CCL2-dependent manner, and loss of CCR2 was associated with fewer parenchymal microglia, increased plaque burden, and increased Aβ peptide levels. In contrast, Naert and Rivest (2012) reported that Ccr2 deletion had little effect on plaque burden and promoted an increase in the abundance of plaque-associated microglia exhibiting an anti-inflammatory phenotype. Attempts to resolve this conflict have been complicated by study design and technical considerations (Morganti et al., 2014).
We previously argued that the plaque-associated myeloid cells were likely derived from infiltrating peripheral “inflammatory” monocytes, owing to their sustained expression of markers that identify these cells in the blood, i.e., CD45hi and Ly6C, and low levels of the endogenous microglial marker P2RY12 (Jay et al., 2015; Korin et al., 2017). The interpretation of these data were confounded by evidence suggesting that microglial expression of these markers is altered in the context of AD, and therefore they may not be definitive markers of cell ontogeny. Here, we have used inducible genetic markers to definitively identify the ontogenic origin of plaque-associated myeloid cells in the AD brain. Fate mapping reveals that plaque-associated myeloid cells derive exclusively from the endogenous yolk sac–derived microglia.
Results and discussion
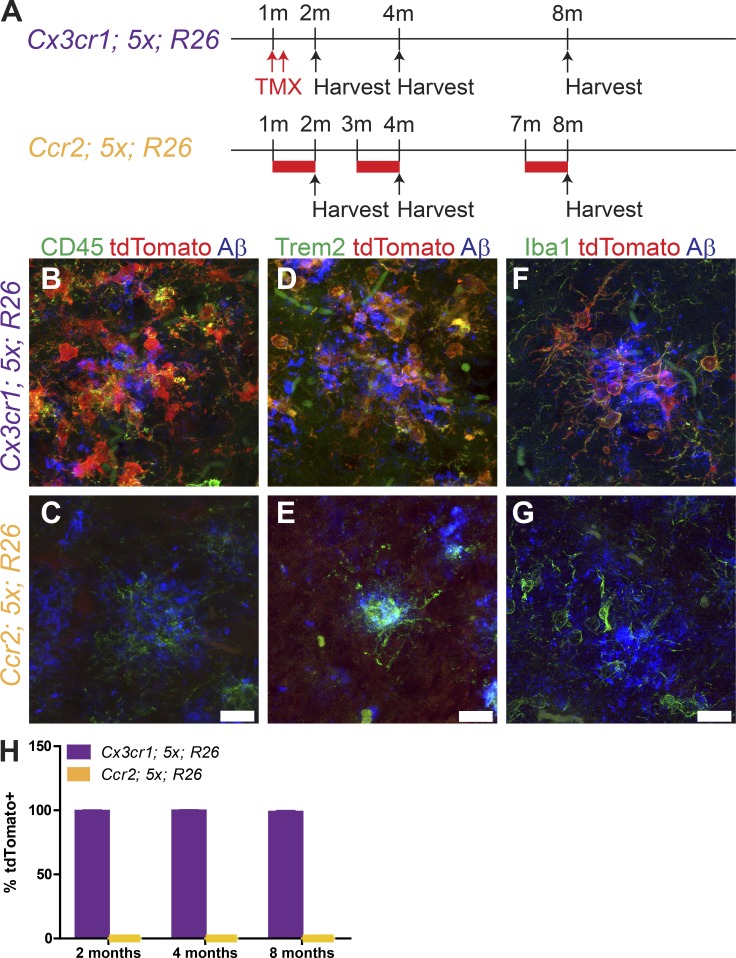
In an effort to identify the ontogeny of plaque-associated myeloid cells as either resident microglia-derived or peripheral monocytes that have infiltrated into the brain and migrated through the parenchyma to plaques, we used genetic labeling strategies to inducibly and irreversibly label each subset of myeloid cells (Fig. 1 A). Selective expression of the fluorescent marker tdTomato via tamoxifen-inducible Cx3cr1-CreERT2 (Yona et al., 2013) exclusively labels resident microglia in the brain, allowing for their identification apart from infiltrating blood-borne macrophages. Similarly, we employed an inducible Ccr2-driven Cre (Ccr2-CreERT2), which is highly expressed in circulating Ly6Chi monocytes (Croxford et al., 2015), to inducibly express tdTomato. These murine lines were crossed into the 5xFAD AD mouse model (Oakley et al., 2006), which exhibits rapid plaque development over 2–8 mo.
Figure 1.
Plaque-associated myeloid cells are resident, yolk sac–derived microglia. (A) Schematic for tamoxifen exposure to drive expression of tdTomato in Cx3cr1CreERT2; 5xFAD; ROSA26tdTomato (top, purple) or Ccr2CreERT2; 5xFAD; ROSA26tdTomato (bottom, yellow) mice. Tamoxifen treatment (TMX) is indicated by red arrows (subcutaneous injection) or red bars (tamoxifen-containing chow). m, month. (B–G) Representative confocal images of IHC for CD45 (B and C), TREM2 (D and E), and Iba1 (F and G) in the subiculum at 8 mo of age in Cx3cr1CreERT2; 5xFAD; ROSA26tdTomato (B, D, and F) and Ccr2CreERT2; 5xFAD; ROSA26tdTomato (C, E, and G) mice treated with tamoxifen. Scale bars, 15 µm (bars in C, E, and G apply to B, D, and F). (H) Quantification of the fraction of Iba1+ cells expressing tdTomato in the cortex, hippocampus, and thalamus in Cx3cr1CreERT2; 5xFAD; ROSA26tdTomato (purple bars) and Ccr2CreERT2; 5xFAD; ROSA26tdTomato (yellow bars) mice treated with tamoxifen. Data presented as mean ± SEM from six mice/genotype/age from three or more independent experiments.
The Cx3cr1-CreERT2; 5xFAD; ROSA26-tdTomato (Cx3cr1; 5x; R26) mice were treated with tamoxifen at 1 mo of age to induce tdTomato expression. The mice were sacrificed at 2, 4, or 8 mo of age (Fig. 1 A). We performed immunohistochemistry (IHC) for tdTomato plus CD45 or TREM2, two molecules whose high levels of expression are commonly associated with peripheral myeloid cells. We found that virtually all CD45- and TREM2-positive cells around plaques were tdTomato positive using the resident microglial driver Cx3cr1-CreERT2 (Fig. 1, B and D).
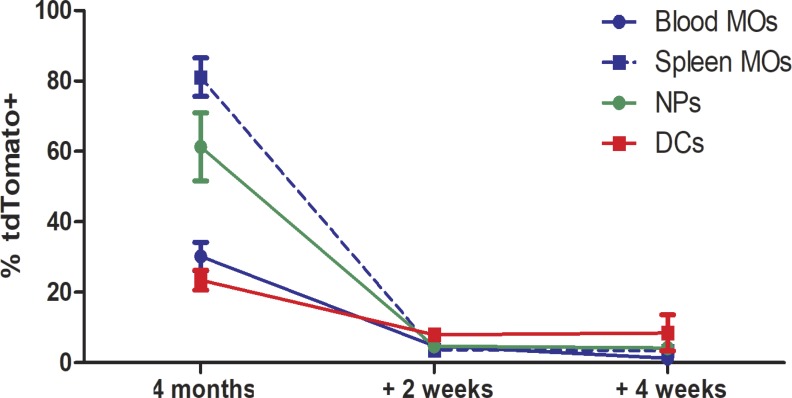
The Ccr2-CreERT2; 5xFAD; ROSA26-tdTomato (Ccr2; 5x; R26) mice were induced to express tdTomato by tamoxifen treatment for 28 d, starting at 1, 3, or 7 mo of age (Fig. 1 A). The sustained tamoxifen treatment was required for the efficient labeling of circulating Ccr2+ monocytes, owing to their brief lifetimes in the blood (Geissmann et al., 2003; Fig. S1), and over an interval in which rapid plaque growth is occurring. We were unable to detect any CD45+ and TREM2+ cells around plaques that were positive for tdTomato (Fig. 1, C and E).
Figure S1.
Kinetics of labeled peripheral monocytes using Ccr2-CreERT2. The fraction of monocytes (MOs; CD11b+CD115+), neutrophils (NPs; CD11b+Ly6G+), and dendritic cells (DCs; CD11b+CD11c+MHCII+) expressing tdTomato immediately after 5 d of tamoxifen-containing chow, or following a 2 or 4-wk return to normal chow. Data presented are mean ± SEM from eight mice/time point from three or more independent experiments.
To ensure we did not miss any Ccr2-lineage cells migrating into the brain, we performed IHC for Iba1, which detects both endogenous microglia and monocytes that have infiltrated the brain. We counted the fraction of Iba1+tdTomato+ cells throughout the cortex, hippocampus, and thalamus (Fig. 1, F and H) and found that >98% of the Iba1+ cells were tdTomato+ in Cx3cr1; 5x; R26 mice treated with tamoxifen, while none of the Iba1+ cells in Ccr2; 5x; R26 mice treated with tamoxifen were tdTomato+ (Fig. 1, G and H). These data demonstrate that parenchymal myeloid cells in the AD brain are resident, yolk sac–derived microglia, with no additional contribution from peripheral monocyte populations. Thus, the AD brain does not elicit the engraftment of peripheral myeloid cells.
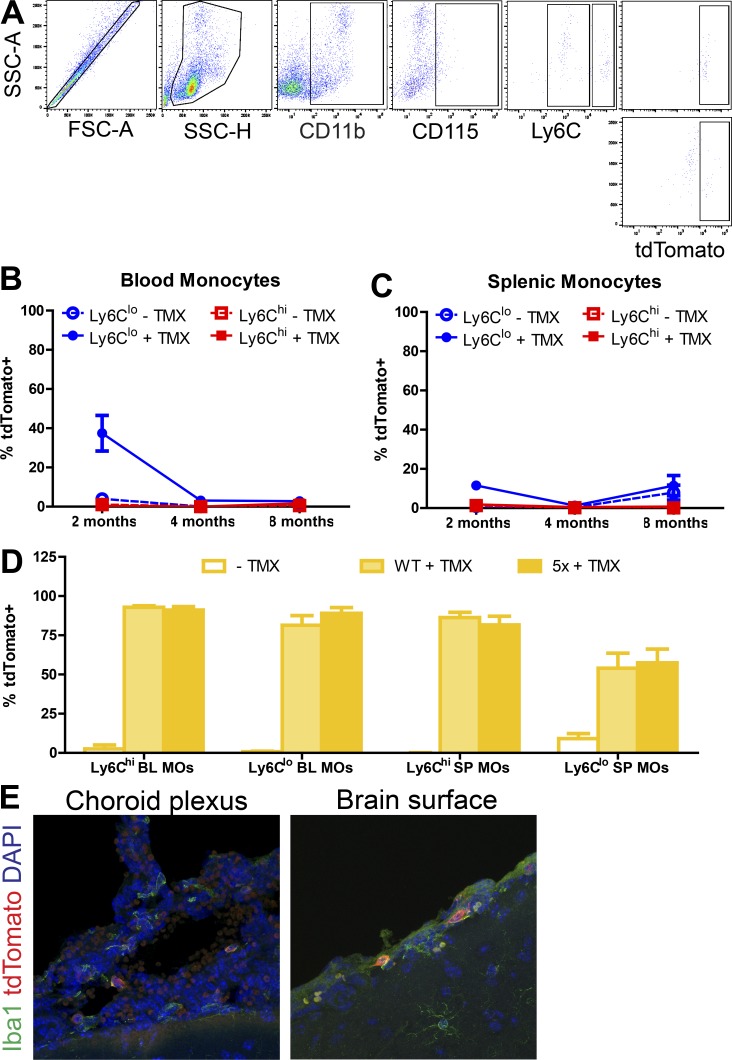
To confirm that the labeling of myeloid cells in the brain was reflective of actual labeling and not due to Cx3cr1+-labeled peripheral monocytes trafficking into the brain or a failure to efficiently label Ccr2-lineage cells, we performed flow cytometry to assess tdTomato expression in peripheral monocyte populations from the blood or spleen (Fig. 2 A) using previously described gating strategies (Croxford et al., 2015) to identify monocytes (CD11b+CD115+, either Ly6Chi or Ly6Clo).
Figure 2.
Labeling of peripheral monocyte populations following tamoxifen exposure. (A) Representative flow sorting scheme for tdTomato expression in Ly6Chi and Ly6Clo monocytes from the spleen. FSC, forward scatter; SSC, side scatter. (B and C) Fraction of Ly6Chi and Ly6Clo monocytes in the blood (B) or spleen (C) expressing tdTomato in Cx3cr1CreERT2; ROSA26tdTomato mice at 2, 4, or 8 mo of age following tamoxifen or control injections at 1 mo of age. Data presented as mean ± SEM from five or more mice/age/treatment from three or more independent experiments. (D) Fraction of Ly6Chi and Ly6Clo monocytes expressing tdTomato in Ccr2CreERT2; ROSA26tdTomato mice. Data presented as mean ± SEM from ≥15 mice/age/treatment from ≥3 independent experiments. TMX, tamoxifen. (E) Representative confocal images of IHC for Iba1 and tdTomato in the choroid plexus and on the brain surface in 8-mo-old Ccr2CreERT2; 5xFAD; ROSA26tdTomato mice treated with tamoxifen for 28 d. Scale bar, 15 µm.
Cx3cr1; 5x; R26 mice injected twice at 1 mo of age with tamoxifen exhibited some residual labeling of Ly6Clo monocytes in the blood (Fig. 2 B) and spleen (Fig. 2 C) at 2 mo of age, but it was absent by 4 and 8 mo of age. Ccr2; 5x; R26 mice on tamoxifen-containing chow for 28 d before harvest at 2, 4, or 8 mo of age showed robust tdTomato expression in both Ly6Chi and Ly6Clo monocytes in both the blood and spleen, irrespective of 5xFAD genotype (Fig. 2 D). Furthermore, lineage-tracing studies demonstrated that a fraction of monocytes at the brain interfaces (i.e., choroid plexus and meninges) consist of bone marrow–derived monocytes (Goldmann et al., 2016). Choroid plexus monocytes initially arise from yolk sac progenitors but are gradually replaced by bone marrow–derived monocytes in a CCR2-dependent manner. Iba+ cells in the choroid plexus and on the surface of the brain were assessed for tdTomato expression, and a subset were found to be tdTomato+ (Fig. 2 E), further validating labeling of peripheral monocytes.
The origins and nature of the plaque-associated myeloid cells in the AD brain have attracted considerable attention and engendered substantial controversy owing to the florid and distinctive phenotypes adopted by these cells, which differ dramatically from adjacent, uninvolved parenchymal microglia. Moreover, this discussion has been influenced by the recent revelation of the ontogenic origins of various macrophage populations associated with the brain, which contravened conventional understanding (Goldmann et al., 2016). The idea that peripheral monocytes or macrophages might enter the AD brain arose from earlier studies of other central nervous system disorders, most prominently multiple sclerosis, in which peripheral immune cells infiltrate the diseased brain and participate in disease pathogenesis (Kierdorf and Prinz, 2017).
An influential study (El Khoury et al., 2007) reported that knockout of the chemokine receptor CCR2 in a mouse model of AD resulted in fewer plaque-associated myeloid cells and accelerated disease progression and argued that bone marrow–derived monocytes enter the brain through the vasculature, migrate, and associate with amyloid plaques. The identification of the infiltrates as peripherally derived hinged largely on their expression of CD11b and CD45 (Korin et al., 2017; Mildner et al., 2011). However, recent work has refuted this assertion (Mrdjen et al., 2018). Transcriptomic studies (Keren-Shaul et al., 2017; Krasemann et al., 2017) have allowed delineation of microglia from macrophages and other immune cells, but have not been able to resolve the issue of ontogenic origin of the plaque-associated cells, owing to the rapid and diverse changes in myeloid cell gene expression in the brain microenvironment (Lund et al., 2018).
The issue of whether peripheral monocytes can enter the AD brain is of some significance, since therapeutically targeting these cells is of potential interest. Indeed, approaches to blocking their entry into the brain and treating circulating cells before their entry have been suggested. Alternative proposals include replacing endogenous microglia with peripheral macrophages (Cronk et al., 2018; Shemer et al., 2018) or using these cells to introduce molecules of interest into the brain (Böttcher et al., 2019).
We previously argued that plaque-associated myeloid cells were likely peripheral in origin (Jay et al., 2015), owing to their high levels of CD45 and Ly6C expression by IHC. This conclusion was challenged by Colonna and colleagues, who performed parabiosis experiments and were unable to detect any peripheral myeloid cells in the AD brain (Wang et al., 2016). While parabiosis models have the significant benefit over bone marrow grafts in determining myeloid cell origin, in that they do not rely on methods that may compromise the blood–brain barrier and therefore obscure the relative contribution of various cell types, these studies still represent an artificial system that could misrepresent the immune environment. We initiated our studies to provide an independent evaluation of this question using an alternative experimental approach. The present data definitively demonstrate that plaque-associated myeloid cells are exclusively brain-resident microglia in the absence of potentially confounding experimental conditions.
Materials and methods
Mice
5xFAD mice (Oakley et al., 2006; The Jackson Laboratory, MMRRC 34848-JAX) were mated to either homozygous CreERT2 Cx3cr1CreERT2 (Yona et al., 2013; Jackson 020940) or Ccr2CreERT2 (Croxford et al., 2015) mice provided by B. Becher to generate Cx3cr1CreERT2/+; 5xFAD or Ccr2CreERT2/+; 5xFAD mice, which were in turn mated to homozygous tdTomato ROSA26tdTomato mice, producing experimental Cx3cr1CreERT2/+; 5xFAD; R26tdTomato/+ and Cx3cr1CreERT2/+; wild type; R26tdTomato/+ mice or Ccr2CreERT2/+; 5xFAD; R26tdTomato/+ mice and Ccr2CreERT2/+; wild type; R26tdTomato/+ mice. Same-sex littermates were randomly assigned to control and experimental groups. Experimental mice were 2, 4, and 8 mo of age. All mice were maintained and experiments were performed in accordance with Institutional Animal Care and Use Committee guidelines at Case Western Reserve University.
Husbandry and housing conditions
Mice were ear-notched and genotyped by tail snipping at postnatal day 18 (P18), weaned at P21, and housed in cages with same-sex littermates under conditions approved by the Institutional Animal Care and Use Committee. Animals were genotyped using a PCR-based strategy suggested by the vendor for Cx3cr1CreERT2, 5xFAD, and ROSA26tdTomato. Primers for Ccr2CreERT2 are available on request.
Tamoxifen administration
Tamoxifen (Sigma-Aldrich) was dissolved in corn oil to 5% concentration and administered in 200-µl doses via two subcutaneous injections 48 h apart (5 mg/dose) to Cx3cr1CreERT2/+; 5xFAD; R26tdTomato/+ and Cx3cr1CreERT2/+; wild type; R26tdTomato/+ mice at 1 mo of age. Tamoxifen-containing chow (400 mg/kg citrate; Envigo, TD130860) was provided to Ccr2CreERT2/+; 5xFAD; R26tdTomato/+ mice and Ccr2CreERT2/+; wild type; R26tdTomato/+ mice for 28 d at 1, 3, or 7 mo of age, after which mice were euthanized. For kinetic labeling studies, 4-mo-old Ccr2CreERT2/+; 5xFAD; R26tdTomato/+ mice and Ccr2CreERT2/+; wild type; R26tdTomato/+ mice were given tamoxifen-containing chow for 5 d and then euthanized or put on normal chow until euthanasia 2 or 4 wk after ending tamoxifen-containing chow.
Tissue harvest and cell preparation
Mice were euthanized by cervical dislocation following isofluorane (Henry Schein) anesthesia. Blood was collected by cardiac puncture into sodium citrate (3.8% wt/vol) as an anticoagulant. RBC Lysis Buffer (BioLegend) was diluted to 1× and used 9:1 to lyse blood cells for 10 min. Cells were pelleted and washed in PBS and were then ready for staining with fluorescence cytometry antibodies. Spleens were dissected out and held in 10% FBS/PBS on ice until processing into single cells for fluorescence cytometry. Tissue was mechanically homogenized and passed through a 70-µm filter cell strainer, followed by resuspension in 1× RBC Lysis Buffer. Lysis was stopped by adding PBS, and cells were pelleted and washed for staining with fluorescence cytometry antibodies. Brains were dissected out and bisected along the midline, with one hemisphere immersion-fixed in 4% paraformaldehyde (PFA)/PBS for IHC and the other processed into single cells for fluorescence cytometry. Briefly, tissue was mechanically homogenized and passed through a 70-µm strainer, followed by 30/37/70% Percoll (GE Healthcare) gradient centrifugation in PBS (vol/vol; 20,000 g for 20 min at room temperature without brakes), removal of myelin with suction, and collection of cells from the 37/70% interface, and washed in PBS. Samples were then ready for staining with fluorescence cytometry antibodies.
Flow cytometry
Pooled portions of samples were used for unstained, single-stained control, and isotype controls, whereas individual samples were stained with a master mix of antibodies. All cell suspensions were incubated with FcR block (1:100; CD16/CD32, clone 92; eBioscience; 14-0161-85) before staining. Blood cells were stained using Alexa Fluor 700–conjugated CD11b clone M1/70 (1:200; Thermo Fisher Scientific, 56-0112-82), allophycocyanin (APC)-conjugated CD115 clone AFS98 (1:200; BioLegend, 135509), Alexa Fluor 488–conjugated Ly6C clone HK1.4 (1:200; Thermo Fisher Scientific, 53-5932-80), phycoerythrin cyanine (PEC) 5.5-conjugated Ly6G clone RB6-8C5 (1:200; eBioscience, 35-5931-82), PEC7-conjugated F4/80 clone BM8 (1:200; eBioscience, 25-4801-82), or PEC7-conjugated CD3 clone 17A2 (1:200; BioLegend, 100219), Alexa Fluor 488–conjugated CD4 clone GK1.5 (1:200; eBioscience, 53-0041-82), and APC-conjugated CD8b clone YTS156.7.7 (1:200; BioLegend, 126613) diluted in flow buffer (FB; 0.2% 0.5 M EDTA, 2% FBS). Spleen cells were stained using Alexa Fluor 700–conjugated CD11b clone M1/70 (1:200), APC-conjugated CD115 clone AFS98 (1:200), and Alexa Fluor 488–conjugated Ly6C clone HK1.4 (1:200), or APC-conjugated CD11b (1:200), Alexa Fluor 700–conjugated CD11c clone N418 (1:200; Thermo Fisher Scientific, 56-0114-80), and Alexa Fluor 488–conjugated MHCII clone M5/114.15.2 (1:200; BioLegend, 107615) diluted in FB. Brain cells were stained using Alexa Fluor 488–conjugated CD45 clone 30-F11 (1:200; BioLegend, 103122) and APC-conjugated CD11b clone M1/70 (1:200) in FB. After 30 min of surface staining on ice, cells were washed in FB, fixed in 4% PFA/FB, resuspended in FB, and filtered using Falcon tubes. Flow cytometry was performed using a BD FACSAria SORP (Becton Dickinson) and analyzed with FlowJo software (TreeStar). Standard, strict side scatter height versus area was used to discriminate doublets and gate singlets. Forward scatter area versus side scatter area was used to gate cells.
Tissue processing and IHC
Brains were harvested as described above. One hemisphere was postfixed overnight in 4% PFA, cryopreserved in 30% sucrose/PBS, embedded in optimal cutting temperature compound, and sectioned at 30 µm on a Leica CM 1950 cyostat. Sections were permeabilized with PBS containing 0.1% Triton X-100 and incubated in 1× Reveal Decloaker (Biocare Medical, RV1000) diluted in ddH2O for 10 min at 95–100°C for antigen retrieval, followed by 30-min cooling. Sections were then incubated in blocking buffer containing 5% normal goat serum and 0.3% Triton X-100 in PBS at room temperature for 1 h. Primary antibodies were diluted in blocking buffer, applied to sections, and incubated overnight at 4°C. Primary antibodies included rabbit anti-RFP (1:500; Rockland; 600-401-379), rat anti-CD45 clone IBL-3/16 (1:500; Bio-Rad; MCA1388); biotinylated sheep anti-TREM2 (1:200; R&D Systems; BAF1729), rat anti-Dectin1 clone R1-8g7 (1:50; InvivoGen; mabg-mdect), goat anti-Iba1 (1:500; Abcam; ab107159), mouse anti-Aβ clone 6E10 (1:1,000; BioLegend; 803001), and rat anti-MHCII (I-A/I-E; 1:100; Thermo Fisher Scientific; 14-5321-82). Primary antibodies were visualized with the appropriate Alexa Fluor 488–, 546–, and 647–conjugated secondary antibodies (1:1,000; Thermo Fisher Scientific) for 1 h at room temperature. Cell nuclei were counterstained with DAPI, and slices were mounted and coverslipped with ProLong Gold (Thermo Fisher Scientific; P36930).
Microscope image acquisition and quantification
IHC was performed on eight evenly spaced medial to lateral sections per animal. Confocal images were acquired at 63×, with 1-µm optical slices spanning the tissue depth on an LSM 800 microscope (Zeiss). Maximum projections were reconstructed using ImageJ (National Institutes of Health).
Quantification of IHC experiments was conducted by observers blinded to tamoxifen treatment and 5xFAD genotype. The number of Iba1+, CD45+, or MHCII+ cells positive for tdTomato were manually scored from every 12th sagittal section (8–10 sections/animal). Iba1+ cells were counted from three cortical regions (motor, somatosensory, and visual cortex), the hippocampus (subiculum, CA1, CA2, CA3), and thalamus at 40× magnification. Values were summed across sections for each animal to determine the fraction of each population that was tdTomato+. Data are graphed as the mean ± SEM.
Statistical analysis
Prism (GraphPad) was used for all statistical analyses. Grubb’s test with a cutoff of α = 0.05 was used to determine statistical outliers. Statistical significance was determined using a two-way ANOVA with Bonferroni post hoc analysis. Both male and female mice were used in this study, and all groups were sex-matched for each experiment. Each n represents a single biological replicate.
Online supplemental material
Fig. S1 illustrates the kinetics of labeled cell populations in Ccr2-CreERT2 mice following 5 d of tamoxifen treatment followed by a return to regular chow. These data show the short lifetime of these circulating cells in the blood.
Acknowledgments
Thanks to Brad Casali and Kathryn MacPherson for input during manuscript preparation.
This work was funded by the Alzheimer’s Association (BFG-15-364590 to G.E. Landreth), the National Institute on Aging (AG050597 and AG051495 to G.E. Landreth), and Northeast Ohio Medical University institutional funds (to E.G. Reed-Geaghan).
Author contributions: Conceptualization, E.G. Reed-Geaghan and G.E. Landreth; methodology, E.G. Reed-Geaghan, A. Croxford, B. Becher, and G.E. Landreth; investigation, E.G. Reed-Geaghan; writing (original draft), E.G. Reed-Geaghan and G.E. Landreth; writing (review and editing), E.G. Reed-Geaghan, A. Croxford, B. Becher, and G.E. Landreth; funding acquisition, E.G. Reed-Geaghan and G.E. Landreth.
References
- Askew K., and Gomez-Nicola D.. 2018. A story of birth and death: Insights into the formation and dynamics of the microglial population. Brain Behav. Immun. 69:9–17. 10.1016/j.bbi.2017.03.009 [DOI] [PubMed] [Google Scholar]
- Askew K., Li K., Olmos-Alonso A., Garcia-Moreno F., Liang Y., Richardson P., Tipton T., Chapman M.A., Riecken K., Beccari S., et al. 2017. Coupled Proliferation and Apoptosis Maintain the Rapid Turnover of Microglia in the Adult Brain. Cell Reports. 18:391–405. 10.1016/j.celrep.2016.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C., Schlickeiser S., Sneeboer M.A.M., Kunkel D., Knop A., Paza E., Fidzinski P., Kraus L., Snijders G.J.L., Kahn R.S., et al. NBB-Psy . 2019. Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat. Neurosci. 22:78–90. 10.1038/s41593-018-0290-2 [DOI] [PubMed] [Google Scholar]
- Cronk J.C., Filiano A.J., Louveau A., Marin I., Marsh R., Ji E., Goldman D.H., Smirnov I., Geraci N., Acton S., et al. 2018. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J. Exp. Med. 215:1627–1647. 10.1084/jem.20180247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxford A.L., Lanzinger M., Hartmann F.J., Schreiner B., Mair F., Pelczar P., Clausen B.E., Jung S., Greter M., and Becher B.. 2015. The Cytokine GM-CSF Drives the Inflammatory Signature of CCR2+ Monocytes and Licenses Autoimmunity. Immunity. 43:502–514. 10.1016/j.immuni.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Davalos D., Grutzendler J., Yang G., Kim J.V., Zuo Y., Jung S., Littman D.R., Dustin M.L., and Gan W.B.. 2005. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8:752–758. 10.1038/nn1472 [DOI] [PubMed] [Google Scholar]
- El Khoury J., and Luster A.D.. 2008. Mechanisms of microglia accumulation in Alzheimer’s disease: therapeutic implications. Trends Pharmacol. Sci. 29:626–632. 10.1016/j.tips.2008.08.004 [DOI] [PubMed] [Google Scholar]
- El Khoury J., Toft M., Hickman S.E., Means T.K., Terada K., Geula C., and Luster A.D.. 2007. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med. 13:432–438. 10.1038/nm1555 [DOI] [PubMed] [Google Scholar]
- Geissmann F., Jung S., and Littman D.R.. 2003. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 19:71–82. 10.1016/S1074-7613(03)00174-2 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R., et al. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 330:841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T., Wieghofer P., Jordão M.J., Prutek F., Hagemeyer N., Frenzel K., Amann L., Staszewski O., Kierdorf K., Krueger M., et al. 2016. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17:797–805. 10.1038/ni.3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay T.R., Miller C.M., Cheng P.J., Graham L.C., Bemiller S., Broihier M.L., Xu G., Margevicius D., Karlo J.C., Sousa G.L., et al. 2015. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J. Exp. Med. 212:287–295. 10.1084/jem.20142322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. 2017. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169:1276–1290.e17. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Kierdorf K., and Prinz M.. 2017. Microglia in steady state. J. Clin. Invest. 127:3201–3209. 10.1172/JCI90602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korin B., Ben-Shaanan T.L., Schiller M., Dubovik T., Azulay-Debby H., Boshnak N.T., Koren T., and Rolls A.. 2017. High-dimensional, single-cell characterization of the brain’s immune compartment. Nat. Neurosci. 20:1300–1309. 10.1038/nn.4610 [DOI] [PubMed] [Google Scholar]
- Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. 2017. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity. 47:566–581.e9. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund H., Pieber M., Parsa R., Han J., Grommisch D., Ewing E., Kular L., Needhamsen M., Espinosa A., Nilsson E., et al. 2018. Competitive repopulation of an empty microglial niche yields functionally distinct subsets of microglia-like cells. Nat Commun. 9:4845 10.1038/s41467-018-07295-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner A., Schlevogt B., Kierdorf K., Böttcher C., Erny D., Kummer M.P., Quinn M., Brück W., Bechmann I., Heneka M.T., et al. 2011. Distinct and non-redundant roles of microglia and myeloid subsets in mouse models of Alzheimer’s disease. J. Neurosci. 31:11159–11171. 10.1523/JNEUROSCI.6209-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morganti J.M., Jopson T.D., Liu S., Gupta N., and Rosi S.. 2014. Cranial irradiation alters the brain’s microenvironment and permits CCR2+ macrophage infiltration. PLoS One. 9:e93650 10.1371/journal.pone.0093650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrdjen D., Pavlovic A., Hartmann F.J., Schreiner B., Utz S.G., Leung B.P., Lelios I., Heppner F.L., Kipnis J., Merkler D., et al. 2018. High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity. 48:599 10.1016/j.immuni.2018.02.014 [DOI] [PubMed] [Google Scholar]
- Naert G., and Rivest S.. 2012. Hematopoietic CC-chemokine receptor 2 (CCR2) competent cells are protective for the cognitive impairments and amyloid pathology in a transgenic mouse model of Alzheimer’s disease. Mol. Med. 18:297–313. 10.2119/molmed.2011.00306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A., Kirchhoff F., and Helmchen F.. 2005. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 308:1314–1318. 10.1126/science.1110647 [DOI] [PubMed] [Google Scholar]
- Oakley H., Cole S.L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., et al. 2006. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26:10129–10140. 10.1523/JNEUROSCI.1202-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réu P., Khosravi A., Bernard S., Mold J.E., Salehpour M., Alkass K., Perl S., Tisdale J., Possnert G., Druid H., and Frisén J.. 2017. The Lifespan and Turnover of Microglia in the Human Brain. Cell Reports. 20:779–784. 10.1016/j.celrep.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W., et al. 2012. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 336:86–90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Shemer A., Grozovski J., Tay T.L., Tao J., Volaski A., Süß P., Ardura-Fabregat A., Gross-Vered M., Kim J.S., David E., et al. 2018. Engrafted parenchymal brain macrophages differ from microglia in transcriptome, chromatin landscape and response to challenge. Nat. Commun. 9:5206 10.1038/s41467-018-07548-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard A.R., and Rivest S.. 2006. Neuroprotective properties of the innate immune system and bone marrow stem cells in Alzheimer’s disease. Mol. Psychiatry. 11:327–335. 10.1038/sj.mp.4001809 [DOI] [PubMed] [Google Scholar]
- Simard A.R., Soulet D., Gowing G., Julien J.P., and Rivest S.. 2006. Bone marrow-derived microglia play a critical role in restricting senile plaque formation in Alzheimer’s disease. Neuron. 49:489–502. 10.1016/j.neuron.2006.01.022 [DOI] [PubMed] [Google Scholar]
- Tay T.L., Mai D., Dautzenberg J., Fernández-Klett F., Lin G., Sagar M., Datta M., Drougard A., Stempfl T., Ardura-Fabregat A., et al. 2017. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 20:793–803. 10.1038/nn.4547 [DOI] [PubMed] [Google Scholar]
- Wang Y., Ulland T.K., Ulrich J.D., Song W., Tzaferis J.A., Hole J.T., Yuan P., Mahan T.E., Shi Y., Gilfillan S., et al. 2016. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213:667–675. 10.1084/jem.20151948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., et al. 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 38:79–91. 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P., Condello C., Keene C.D., Wang Y., Bird T.D., Paul S.M., Luo W., Colonna M., Baddeley D., and Grutzendler J.. 2016. TREM2 Haplodeficiency in Mice and Humans Impairs the Microglia Barrier Function Leading to Decreased Amyloid Compaction and Severe Axonal Dystrophy. Neuron. 92:252–264. 10.1016/j.neuron.2016.09.016 [DOI] [PubMed] [Google Scholar]