Hyper-activation of NLRP3 inflammasomes contributes to the development of endotoxemia, but the molecular mechanisms are poorly defined. Tang et al. demonstrate that sequential ubiquitination of NLRP3 is crucial to keep NLRP3 inflammasomes in check and limits endotoxemia.
Abstract
Aberrant NLRP3 inflammasome activation contributes to the development of endotoxemia. The importance of negative regulation of NLRP3 inflammasomes remains poorly understood. Here, we show that the E3 ubiquitin ligase Cbl-b is essential for preventing endotoxemia induced by a sub-lethal dose of LPS via a caspase-11/NLRP3–dependent manner. Further studies show that NLRP3 undergoes both K63- and K48-linked polyubiquitination. Cbl-b binds to the K63-ubiquitin chains attached to the NLRP3 leucine-rich repeat domain (LRR) via its ubiquitin-associated region (UBA) and then targets NLRP3 at K496 for K48-linked ubiquitination and proteasome-mediated degradation. We also identify RNF125 as an additional E3 ubiquitin ligase that initiates K63-linked ubiquitination of the NLRP3 LRR domain. Therefore, NLRP3 is sequentially ubiquitinated by K63- and K48-linked ubiquitination, thus keeping the NLRP3 inflammasomes in check and restraining endotoxemia.
Introduction
Sepsis, a systemic inflammatory response syndrome (SIRS) in patients following infection or injury, causes millions of deaths globally each year (Angus et al., 2001; Deutschman and Tracey, 2014; Hutchins et al., 2014; Martin et al., 2003). Sepsis caused by gram-negative bacteria is thought to be largely due to the host's response to LPS or endotoxins (Bosmann and Ward, 2013). IL-1, particularly IL-1β, is one of several pro-inflammatory cytokines produced during SIRS that serves to initiate the host inflammatory response and to integrate nonspecific immunity. Many of IL-1’s effects are beneficial in times of stress, but when produced for extended periods of time or in excessive quantities, IL-1β contributes to host morbidity and mortality (Bosmann and Ward, 2013; Pruitt et al., 1995; Wiersinga et al., 2014). In addition, pyroptosis mediated by caspase-11 (Casp-11; Kayagaki et al., 2015; Shi et al., 2015), which has been shown to be a direct cytosolic receptor for LPS (Shi et al., 2014), also contributes to the mortality of high-dose LPS-induced endotoxemia (Kayagaki et al., 2011). The release of IL-1β and the induction of pyroptosis during sepsis are tightly controlled by a multi-protein complex, termed the nucleotide-binding oligomerization domain (NOD)–, leucine-rich repeat (LRR)–, and pyrin domain-containing protein 3 (NLRP3) inflammasome. This complex consists of NLRP3, apoptosis-associated speck-like protein (ASC), and Casp-1 (Davis et al., 2011; Martinon et al., 2009; Martinon and Tschopp, 2004), or in some instances a noncanonical inflammasome involving Casp-11/gasdermin D (GSDMD; Kayagaki et al., 2015; Shi et al., 2015) rather than Casp-1. Nlrp3−/−, Casp11−/−, and Casp1−/− mice are protected from LPS-induced endotoxemia under various conditions (Kayagaki et al., 2011; Li et al., 1995; Mao et al., 2013; Sarkar et al., 2006; Wang et al., 1998), while a naturally occurring polymorphism for human Casp-12, a putative regulator of Casp-1, or mice deficient for Casp-12 have been linked to sepsis in both humans and mice (Saleh et al., 2006; Saleh et al., 2004). Moreover, NLRP3 polymorphisms have been linked to sepsis morbidity (Zhang et al., 2011). Thus, NLRP3 inflammasome activation appears to be a prerequisite for a competent immune response during endotoxemia. However, the mechanisms for negatively regulating the activation of the NLRP3 inflammasomes during endotoxemia are largely unknown.
Casitas-B-lineage lymphoma protein-b (Cbl-b) is a RING finger E3 ubiquitin ligase that plays a crucial role in T cell activation, tolerance induction, and differentiation (Bachmaier et al., 2000; Chiang et al., 2000; Guo et al., 2012; Harada et al., 2010; Heissmeyer et al., 2004; Jeon et al., 2004; Li et al., 2004; Qiao et al., 2008; Qiao et al., 2014; Qiao et al., 2013; Zhang et al., 2002), but little is known about Cbl-b’s role in the innate immune response. NLRP3 undergoes polyubiquitination when BRCC3, a deubiquitinating enzyme, is inhibited (Py et al., 2013), suggesting that ubiquitination is one of the major mechanisms regulating NLRP3 inflammasome activity. It was reported that tripartite motif 31 (TRIM31) may ubiquitinate NLRP3 and induce NLRP3 degradation during LPS priming (Song et al., 2016), suggesting that TRIM31 may mainly affect the expression of NLRP3 during the priming phase. Therefore, the E3 ubiquitin ligase(s) specifically responsible for NLRP3 ubiquitination induced by NLRP3 inflammasome activators and the biological relevance of NLRP3 ubiquitination have not been fully characterized. Interestingly, although Cbl-b does not inhibit the signaling derived from TLRs (Xiao et al., 2016), we show here that it specifically inhibits IL-1β production by macrophages induced by canonical and noncanonical NLRP3 inflammasome stimuli. Therefore, we hypothesized that Cbl-b may be the E3 ubiquitin ligase that negatively regulates NLRP3.
In this study, we show that the ubiquitin-associated region (UBA) of Cbl-b binds to the K63 ubiquitin chains that attach to the NLRP3 LRR domain upon NLRP3 inflammasome stimulation. Cbl-b then targets NLRP3 for K48-linked polyubiquitination. We also found that RNF125, a RING finger E3 ubiquitin ligase, initially induces K63-linked polyubiquitination of NLRP3 within the LRR domain, which is required for the recruitment of Cbl-b. Therefore, our data collectively indicate that NLRP3 undergoes sequential K63- and K48-linked polyubiquitination mediated by RNF125 and Cbl-b, respectively, which is essential for controlling its activation and ultimately endotoxemia and polymicrobial sepsis.
Results
Cbl-b selectively dampens NLRP3 inflammasomes in vitro
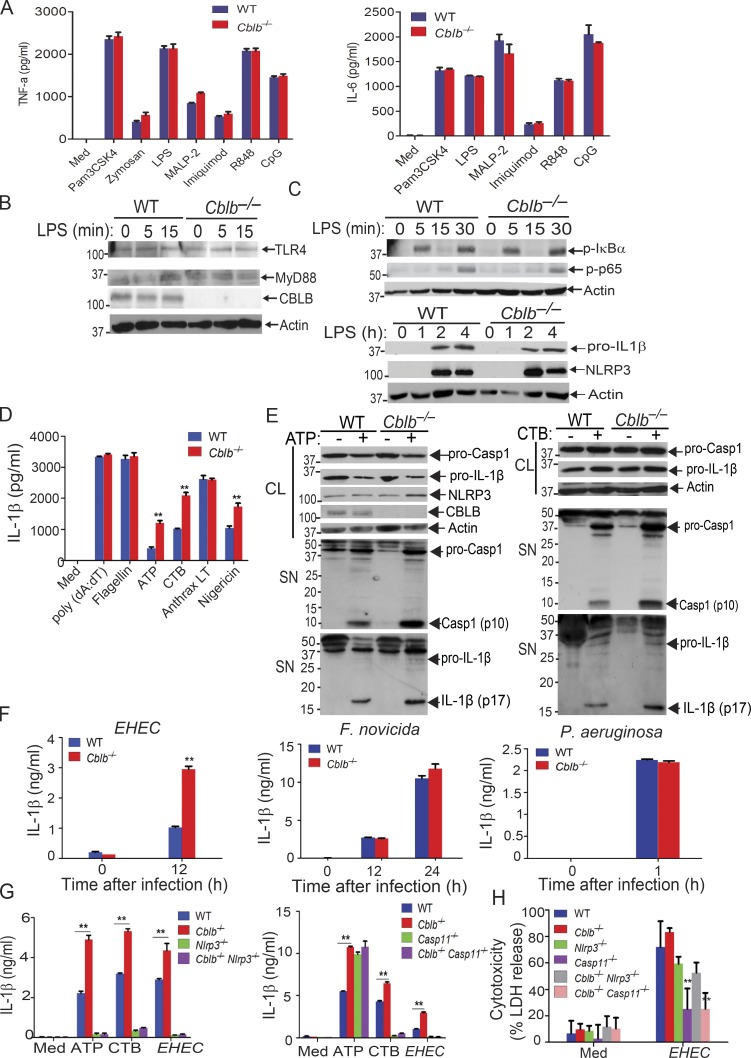
There are conflicting published views about the involvement of Cbl-b in TLR4 or MyD88-mediated signaling in monocytes or macrophages (Bachmaier et al., 2007; Han et al., 2010; Yee and Hamerman, 2013). We therefore revisited whether Cbl-b plays a significant role in the production of pro-inflammatory cytokines in response to TLR ligands. We stimulated WT and Cblb−/− bone marrow–derived macrophages (BMDMs) with various TLR ligands and measured the production of TNF-α and IL-6 in cell culture supernatants. Consistent with recent published findings (Xiao et al., 2016; Yee and Hamerman, 2013), we detected no difference in the production of these pro-inflammatory cytokines by BMDMs generated from WT and Cblb−/− mice (Fig. 1 A). We failed to observe any reduction in TLR4 or MyD88 degradation in BMDMs lacking Cbl-b in response to LPS stimulation (Fig. 1 B). Furthermore, phosphorylation of IκBα and p65 and induction of NLRP3 and pro–IL-1β were comparable between WT and Cblb−/− BMDMs in response to LPS stimulation (Fig. 1 C). Taken together, our data suggest that Cbl-b does not regulate TLR signaling.
Figure 1.
Cbl-b selectively inhibits canonical and noncanonical NLRP3 inflammasomes. (A) TNF-α and IL-6 production by BMDMs from WT and Cblb−/− mice stimulated with TLR ligands for 24 h. Med, medium; CpG, CpG oligodeoxynucleotides. (B) Immunoblot analysis of TLR4, MyD88, and CBLB of WT and Cblb−/− BMDMs stimulated with LPS (500 ng/ml) at indicated time points. (C) Immunoblot analysis of phospho-IκBα, phospho-p65, NLRP3, pro–IL-1β, and NLRP3 of WT and Cblb−/− BMDMs stimulated with LPS at various time points. (D) IL-1β production by LPS-primed BMDMs stimulated with ATP (2.5 mM) for 30 min, nigericin (20 µM) for 3 h, CTB (40 µg/ml) for 16 h, anthrax LT (500 µg/ml) for 6 h, poly(dA:dT) (1 µg/106 cells) for 6 h, and flagellin (6.25 µg/106 cells) for 4 h. (E) Immunoblots of pro–Casp-1, pro–IL-1β, NLRP3, and CBLB expression in cell extracts and Casp-1 activation and IL-1β maturation in the supernatants of LPS-primed WT and Cblb−/− BMDMs stimulated with ATP (2.5 mM) for 30 min and CTB (40 µg/ml) for 16 h. CL, cell lysate; SN, supernatants. (F) IL-1β production by LPS-primed WT and Cblb−/− BMDMs after infection with EHEC (MOI = 25:1), P. aeruginosa (MOI = 30:1), and F. novicida (MOI = 100:1) for the indicated periods of time. (G) IL-1β production by LPS-primed BMDMs from WT, Cblb−/−, Nlrp3−/−, Cblb−/−Nlrp3−/−, Casp11−/−, and Cblb−/−Casp11−/− mice stimulated with ATP (2.5 mM) for 30 min or CTB (40 µg/ml) for 16 h or infected with EHEC (MOI = 25:1) for 12 h. (H) Lactate dehydrogenase (LDH) release from LPS-primed BMDMs from WT, Cblb−/−, Nlrp3−/−, Cblb−/−Nlrp3−/−, Casp11−/−, and Cblb−/−Casp11−/− mice infected with EHEC (MOI = 25:1) for 12 h. Data are shown as mean ± SD. Data are representative of three independent experiments. **, P < 0.01; Student’s t test. p, phospho.
To determine whether Cbl-b regulates canonical and noncanonical NLRP3 inflammasomes, we stimulated LPS-primed BMDMs from WT and Cblb−/− mice with either (1) ATP and nigericin, which activate the NLRP3 inflammasome (Davis et al., 2011; Martinon et al., 2009; Rathinam et al., 2012a; Tschopp and Schroder, 2010); (2) cholera toxin B (CTB), which activates the noncanonical NLRP3 inflammasome (Kayagaki et al., 2011); (3) anthrax lethal toxin (LT), which activates the NLRP1 inflammasome (Pétrilli et al., 2007; Rathinam et al., 2012a); (4) poly(dA:dT), which activates the absent in melanoma 2 (AIM2) inflammasome (Rathinam et al., 2010; Rathinam et al., 2012a); or (5) flagellin, which activates the NLRC4 inflammasome (Pétrilli et al., 2007; Rathinam et al., 2012a). We found no difference in IL-1β production by WT and Cblb−/− BMDMs via stimulation through anthrax LT, flagellin, and poly(dA:dT); however, ATP, nigericin, and CTB induced significantly higher production of IL-1β by Cblb−/− BMDMs compared with their WT controls (Fig. 1 D). The increased IL-1β production by Cblb−/− BMDMs upon ATP or CTB stimulation correlated with the heightened generation of active Casp-1 p10 and mature IL-1β p17 as revealed by immunoblotting of the supernatants collected from the cultures (Fig. 1 E). To further determine whether Cbl-b regulates NLRP3 inflammasome activation triggered by microbes, we infected WT and Cblb−/− BMDMs with either enterohemorrhagic Escherichia coli strain 700927 (hereafter called EHEC), which activates the noncanonical NLRP3 inflammasome (Rathinam et al., 2012b), Pseudomonas aeruginosa, which triggers the NLRC4 inflammasome (Sutterwala et al., 2007), or Francisella novicida, which activates the AIM2 inflammasome (Rathinam et al., 2010). Cblb−/− BMDMs only produced significantly more mature IL-1β than WT BMDMs upon infection with EHEC but not P. aeruginosa and F. novicida (Fig. 1 F).
To define whether Cbl-b regulates both the canonical and noncanonical NLRP3 inflammasomes, we introduced NLRP3 deficiency and Casp-11 deficiency into Cblb−/− background and generated Cblb−/−Nlrp3−/− and Cblb−/−Casp11−/− mice. We measured IL-1β production and pyroptosis by BMDMs from WT, Cblb−/−, Nlrp3−/−, Cblb−/−Nlrp3−/−, Casp11−/−, and Cblb−/−Casp11−/− mice upon LPS-priming and ATP, CTB, or EHEC stimulation. NLRP3 deficiency abrogated hyper–IL-1β production by Cblb−/− BMDMs upon stimulation with ATP and CTB or infection with EHEC (Fig. 1 G, left panel). In contrast, loss of Casp-11 only diminished hyper–IL-1β production induced by CTB and EHEC but not ATP by Cblb−/− BMDMs (Fig. 1 G, right panel). Pyroptosis induced by EHEC was comparable between WT and Cblb−/− BMDMs, and deficiency for Casp-11, but not NLRP3, markedly inhibited pyroptosis (Fig. 1 H). Therefore, our data support the notion that Cbl-b regulates Casp-11– and NLRP3-dependent IL-1β production upon canonical and noncanonical inflammasome stimuli but does not appear to regulate NLRP3-independent but Casp-11–dependent pyroptosis induced by EHEC.
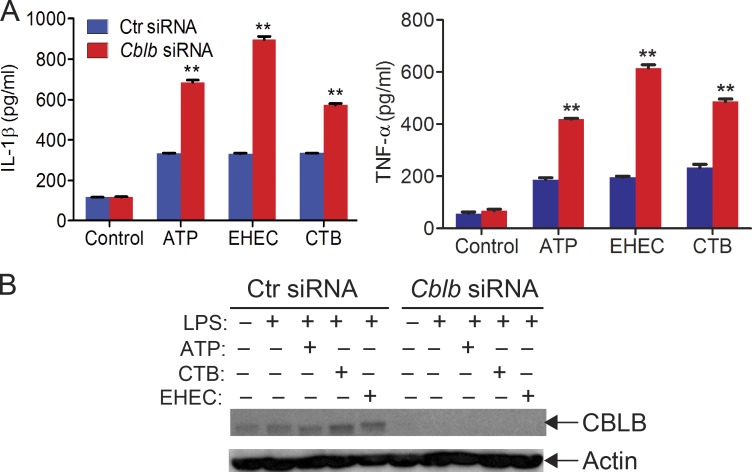
To determine whether Cbl-b has a similar effect on NLRP3 inflammasome activation in human macrophages, we generated human monocyte–derived macrophages (MDMs; Rajaram et al., 2011) and transfected MDMs with CBLB-specific siRNA or scrambled siRNA by nucleofection. Knocking down CBLB in MDMs resulted in heightened IL-1β production upon LPS priming and ATP stimulation (Fig. S1), similar to the data obtained by using mouse BMDMs. These results suggest that the role of Cbl-b in NLRP3 inflammasome in mouse macrophages is translatable to human macrophages. These findings also suggest that Cbl-b regulates both canonical and noncanonical NLRP3 inflammasomes, which are mediated by Casp-1 and Casp-11, respectively (Kayagaki et al., 2011; Rathinam et al., 2012b).
Figure S1.
Silencing the CBLB gene in human macrophages leads to heightened IL-1β and TNF-α production upon LPS priming and stimulation with ATP, CTB, and EHEC. Related to Fig. 1. (A) ELISA analysis of IL-1β and TNF-α production by human MDMs transfected with control siRNA or CBLB-specific siRNA (200 nM) by using Lonza nucleofector reagent before priming with LPS (100 ng/ml) and stimulation with ATP (2.5 nM, 30 min), CTB (20 ng/ml, 6 h), and EHEC (MOI = 25:1, 8 h). Data are shown as mean ± SD. **, P < 0.01; Student’s t test. (B) Immunoblot analysis of CBLB in MDMs treated with control siRNA or CBLB-specific siRNA. Actin was used as a loading control. Data are representative of three independent experiments.
Cbl-b inhibits a sub-lethal dose of LPS-induced endotoxemia via a Casp-11/NLRP3–dependent manner
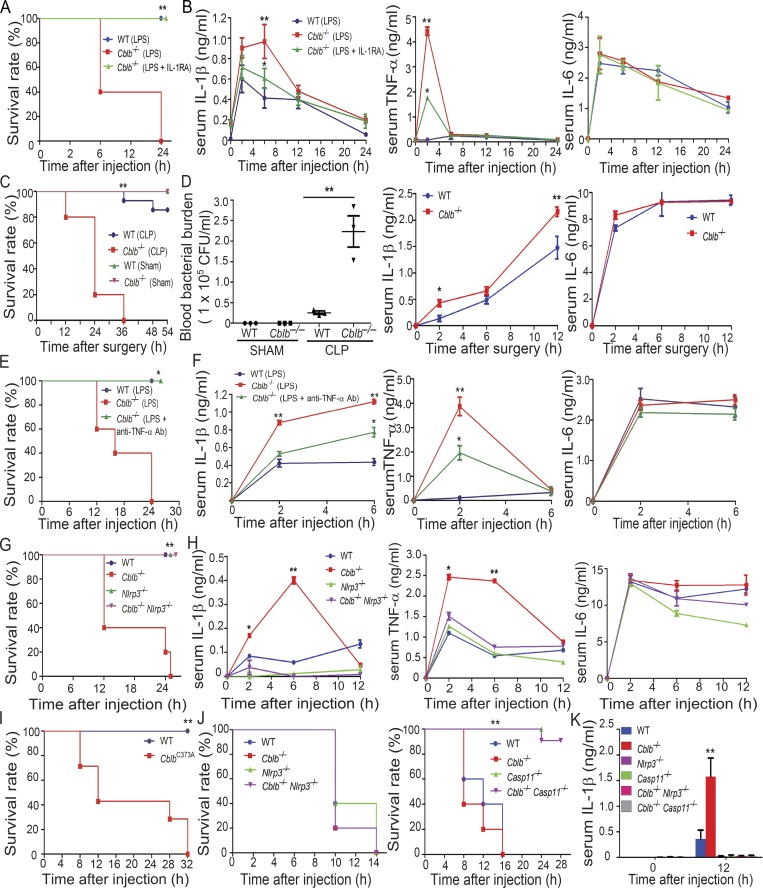
Lethality to LPS is mediated by the pro-inflammatory cytokine IL-1β secreted by myeloid cells (Fantuzzi and Dinarello, 1996; Kang et al., 2013; Scheibel et al., 2010) or by Casp-11 (Broz and Monack, 2013; Kayagaki et al., 2011; Rathinam et al., 2012b) and can be independent of TLR4 at very high doses (Hagar et al., 2013; Kayagaki et al., 2013). To test whether Cbl-b regulates NLRP3 inflammasomes in vivo, we challenged WT and Cblb−/− mice with a sub-lethal dose of LPS (5 mg/kg; E. coli 0111:B4; Sigma-Aldrich) by intraperitoneal injection. Although WT mice all survived 24 h after LPS injection, all Cblb−/− mice died (Fig. 2 A). Serum TNF-α was significantly higher in the Cblb−/− mice than the WT mice at 2 h, and IL-1β was increased at 2–6 h after injection in Cblb−/− mice, whereas there was no difference in serum IL-6 levels in WT and Cblb−/− mice (Fig. 2 B). To determine the role of Cbl-b in polymicrobial sepsis, the cecal ligation and puncture (CLP)–induced sepsis model was used. Cblb−/− mice were found to be highly susceptible to sub-lethal CLP, which correlated with heightened blood bacterial burden and serum IL-1β but not IL-6 after CLP (Fig. 2, C and D).
Figure 2.
Loss of Cbl-b increases the susceptibility of mice to sub-lethal LPS-induced endotoxemia via a Casp-11/NLRP3–dependent manner. (A) Kaplan-Meier survival curve of WT and Cblb−/− mice injected with a sub-lethal dose of LPS (5 mg/kg) in the presence or absence of IL-1RA pretreatment. WT + LPS (n = 9), Cblb−/− + LPS (n = 10), and Cblb−/− + LPS/IL-1RA (25 mg/kg; n = 7). **, P < 0.01 (Cblb−/− + LPS vs. WT + LPS or Cblb−/− + LPS/IL-1RA); log-rank test. (B) ELISA of serum IL-1β, TNF-α, and IL-6 levels from WT and Cblb−/− mice injected with LPS (1 mg/kg) with or without IL-1RA pretreatment (n = 5). Data are shown as mean ± SD. **, P < 0.01 (WT + LPS vs. Cblb−/− + LPS); *, P < 0.05 (Cblb−/− + LPS vs. Cblb−/− + LPS/IL-1RA), Student’s t test. (C) Survival rate of WT and Cblb−/− mice undergoing sub-lethal CLP. CLP: WT (n = 8), Cblb−/− (n = 10); Sham: WT (n = 5), Cblb−/− (n = 5). **, P < 0.01 (WT CLP vs. Cblb−/− CLP); log-rank test. (D) Blood bacterial burden (CFU/ml) of WT (n = 5) and Cblb−/− mice (n = 5) undergoing sub-lethal CLP or Sham at 6 h after surgery. Data are represented as mean ± SEM. **, P < 0.01 (WT CLP vs. Cblb−/− CLP); by unpaired two-tailed Student’s t test. ELISA of serum IL-1β and IL-6 levels of WT and Cblb−/− mice undergoing sub-lethal CLP. Data are shown as mean ± SD. *, P < 0.05; **, P < 0.01; Student’s t test. (E) Kaplan-Meier survival rate of WT mice (n = 6) injected with LPS (5 mg/kg) or Cblb−/− mice (n = 6) pretreated with or without a neutralizing anti–TNF-α (50 µg/mouse) and then injected with LPS (5 mg/kg). *, P < 0.05; log-rank test. (F) ELISA of serum IL-1β, TNF-α, and IL-6 levels from WT mice (n = 6) injected with LPS (1 mg/kg) or Cblb−/− mice (n = 6) pretreated with or without a neutralizing anti–TNF-α (50 µg/mouse) and then injected with LPS (1 mg/kg). Data are shown as mean ± SD. *, P < 0.05; **, P < 0.01; Student’s t test. (G) Kaplan-Meier survival curve of WT, Cblb−/− and Nlrp3−/−, and Cblb−/−Nlrp3−/− mice injected with LPS (5 mg/kg; n = 5). **, P < 0.01 (Cblb−/− vs. Nlrp3−/− or Cblb−/−Nlrp3−/−); log-rank test. (H) ELISA of serum IL-1β, TNF-α, and IL-6 levels from WT, Cblb−/−, Nlrp3−/−, and Cblb−/−Nlrp3−/− mice injected with LPS (1 mg/kg) at 0, 2, 6, and 12 h (n = 5). Data are shown as mean ± SD. *, P < 0.05; **, P < 0.01 (Cblb−/−− vs. Nlrp3−/− or Cblb−/−Nlrp3−/−); Student’s t test. (I) Kaplan-Meier survival rate of WT (n = 5) and CblbC373A mice (n = 7) injected with LPS (5 mg/kg). **, P < 0.01; log-rank test. (J) Kaplan-Meier survival rate of WT, Cblb−/−, Nlrp3−/−−, Casp11−/− and Cblb−/−Nlrp3−/−, and Cblb−/−Casp11−/− mice (n = 5) injected with a lethal dose of LPS (54 mg/kg). **, P < 0.01 (Cblb−/− vs. Casp11−/− and Cblb−/−Casp11−/−); log-rank test. (K) Serum IL-1β levels detected after 20 mg/kg LPS injection in mice (n = 4) of the indicated genotypes at 12 h. Data are represented as mean ± SD. **, P < 0.01 (Cblb−/− vs. other groups); Student’s t test. Data are representative of three independent experiments (A–D and G, H, J, K) and representative of two independent experiments (E, F, and I). Ab, antibody.
To determine whether heightened IL-1β is the major cause of the LPS-induced lethality observed in Cblb−/− mice, we injected Cblb−/− mice with the IL-1R antagonist (IL-1RA) before LPS challenge. As shown in Fig. 2 A, blocking IL-1R prevented the death of Cblb−/− mice after LPS injection. This prevention was associated with attenuated serum levels of IL-1β and TNF-α but not IL-6 (Fig. 2 B), suggesting that there is a positive feedback between IL-1β and TNF-α and that both IL-1β and TNF-α contribute to the lethality of Cblb−/− mice. In support of this notion, blocking TNF-α using a neutralizing anti–TNF-α antibody also prevented the death of Cblb−/− mice, which correlated with a reduction of TNF-α and IL-1β but not IL-6 in the sera (Fig. 2, E and F). To further confirm these data, we used Cblb−/−Nlrp3−/− animals. NLRP3 deficiency completely rescued Cblb−/− mice from death induced by a sub-lethal dose of LPS (Fig. 2 G), which was associated with a dramatic reduction of IL-1β and TNF-α in the serum (Fig. 2 H). Therefore, our data indicate that Cbl-b inhibits NLRP3-dependent IL-1β generation in vivo and that increased TNF-α in the sera in Cblb−/− mice upon LPS challenge is likely secondary to IL-1β.
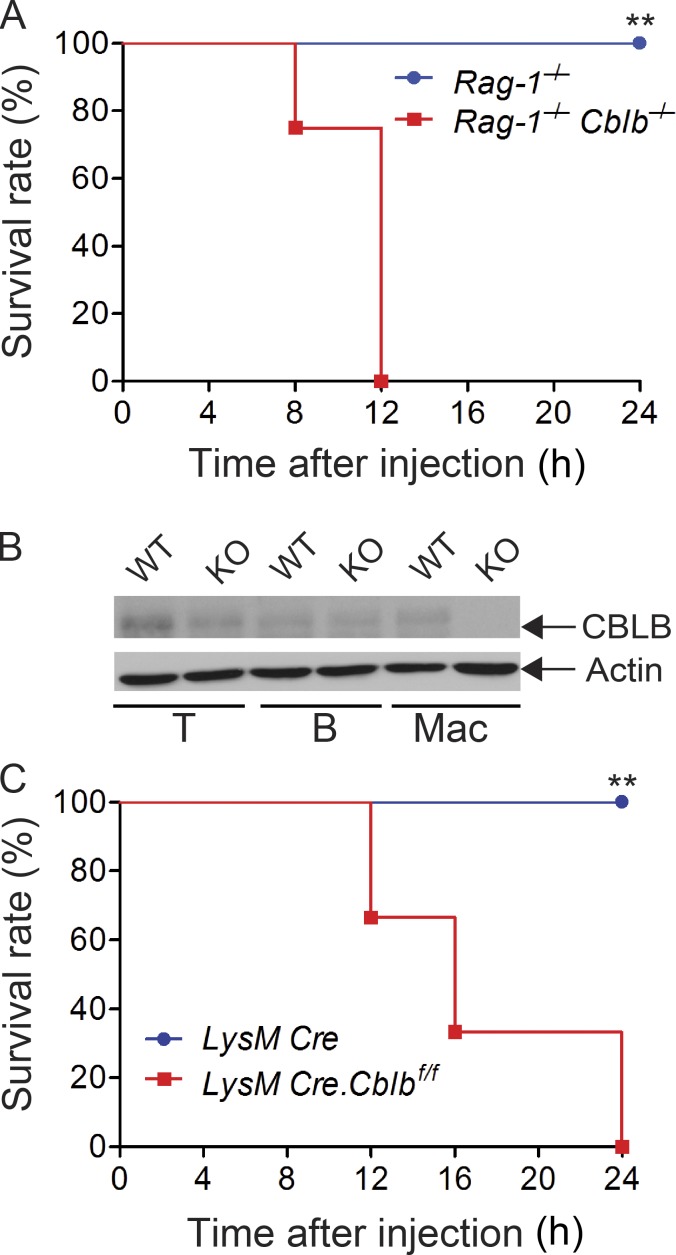
To exclude the potential confounding effect of the adaptive immune system on LPS-induced lethality in Cblb−/− mice, we generated Cblb−/−Rag1−/− mice, which do not have T and B cells. Injection of a sub-lethal dose of LPS into Rag1−/−Cblb−/− mice recapitulated the phenomenon observed in Cblb−/− mice (Fig. S2 A), suggesting that lethality to the typically sub-lethal dose of LPS is primarily mediated by IL-1β produced by innate immune cells. To further confirm this, we generated mice harboring loxP-flanked Cblb allele (Cblbf/f; Fig. S2 B) and crossed them with LysM Cre mice (Clausen et al., 1999) to obtain mice that specifically lack Cbl-b in the myeloid cell lineage (LysM Cre-Cblbf/f; Fig. S2 B). Indeed, a sub-lethal dose of LPS induced high mortality in LysM Cre-Cblbf/f mice (Fig. S2 C), thus recapitulating the phenomenon observed in Cblb−/− mice. A similar finding was observed in mice expressing an E3 ubiquitin ligase dead mutation (C373A; Fig. 2 I), suggesting that inhibition of LPS-induced endotoxemia by Cbl-b is dependent on its E3 ubiquitin ligase activity. Taken together, our data indicate that the E3 ubiquitin ligase activity of Cbl-b in myeloid cells inhibits NLRP3 inflammasome-mediated endotoxemia.
Figure S2.
Innate immune cells are essential for the hypersensitivity to a sub-lethal dose of LPS-induced endotoxemia in the absence of Cbl-b. Related to Fig. 2. (A) Survival rate of Rag1−/− and Rag1−/−Cblb−/− mice (n = 8 per group) after injection of LPS (2 mg/kg). **, P < 0.01; log-rank test. (B) Immunoblot analysis of lysates of T cells (T), B cells (B), and BMDMs (Mac) from LysM Cre (WT) and LysM Cre-Cblbf/f (KO) mice with antibodies against CBLB and actin. (C) Survival rate of LysM Cre and LysM Cre-Cblbf/f mice (n = 6 per group) after injection of LPS (5 mg/kg). **, P < 0.01; log-rank test. Data are representative of two independent experiments.
It was previously shown that lethal-dose LPS-induced lethality is dependent on pyroptosis induced by Casp-11 but not Casp-1 (Kayagaki et al., 2011). To determine whether Cbl-b also regulates lethality induced by a lethal dose of LPS, we injected WT, Cblb−/−, Casp11−/−, Nlrp3−/−, Cblb−/−Nlrp3−/−, and Cblb−/−Casp11−/− mice with a lethal dose (54 mg/kg) of LPS. All WT, Cblb−/−, Nlrp3−/−, and Cblb−/−Nlrp3−/− mice died within 14 h after injection at a similar rate (Fig. 2 J, left panel), whereas Casp11−/− and Cblb−/−Casp11−/− mice survived from the first 16 h of LPS injection (Fig. 2 J, right panel). Injection of LPS at 20 mg/kg to WT, Cblb−/−, Casp11−/−, Nlrp3−/−, Cblb−/−Casp11−/−, and Cblb−/−Nlrp3−/−mice showed that deficiency for both NLRP3 and Casp-11 abrogated hyper–IL-1β levels in the sera in Cblb−/− mice (Fig. 2 K). Collectively, these data suggest that Cbl-b regulates Casp-11/NLRP3–dependent inflammasome activation, which triggers IL-1β production, but it does not regulate NLRP3-independent, but Casp-11–dependent, noncanonical inflammasome activation, which mainly triggers pyroptosis.
Cbl-b associates with NLRP3 in macrophages upon NLRP3 inflammasome activation
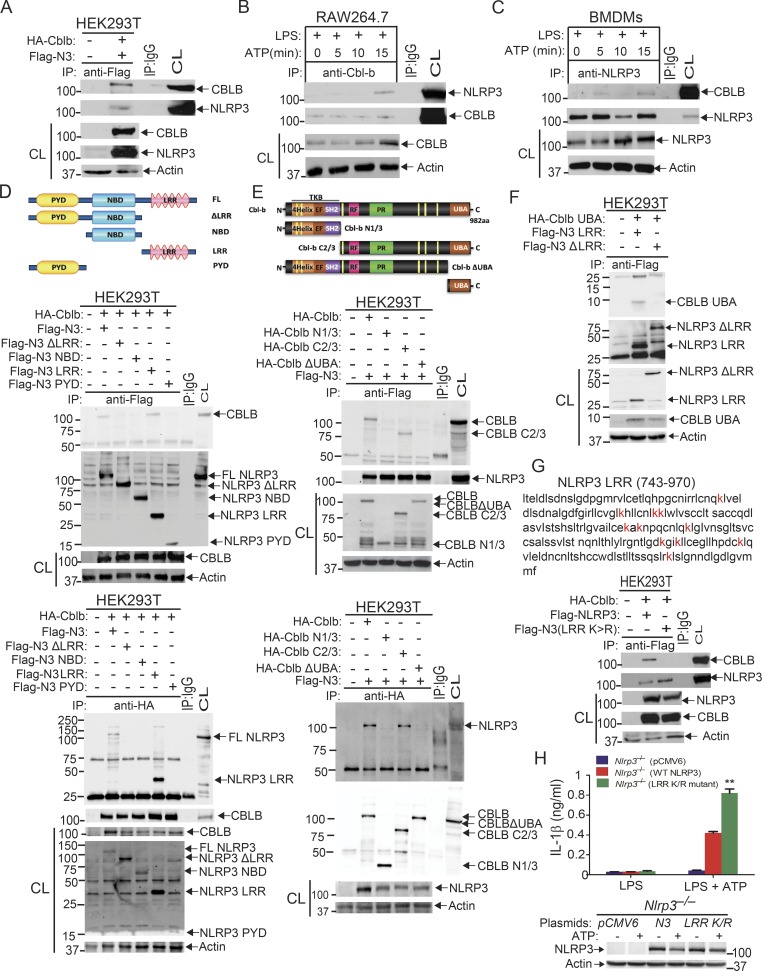
Recent studies indicate that NLRP3 inflammasomes are regulated by a protein ubiquitination–dependent mechanism (Py et al., 2013), but the E3 ubiquitin ligase(s) that ubiquitinates NLRP3 has not been fully characterized. Our data above collectively indicate that Cbl-b negatively inhibits NLRP3 inflammasomes. To determine whether Cbl-b ubiquitinates NLRP3, we first determined whether Cbl-b physically associates with NLRP3. We expressed hemagglutinin (HA)-tagged Cbl-b and Flag-tagged NLRP3 in HEK293T cells through transient transfection. HEK293T cells lack P2X receptors and have low phagocytic ability, making them relatively resistant to some NLRP3-activating stimuli like ATP and crystalline substances such as monosodium urate and alum (Gu et al., 2010; Subramanian et al., 2013). Therefore, nigericin, an ionophore that catalyzes an electroneutral potassium/proton exchange across lipid bilayers to induce NLRP3 activation, was used as the stimulus (Gu et al., 2010; Subramanian et al., 2013). Immunoprecipitation of Flag-tagged NLRP3 followed by Western blotting with anti-HA antibodies indicated that Cbl-b interacts with NLRP3 in HEK293T cells (Fig. 3 A). To determine whether the Cbl-b–NLRP3 association requires ASC, we performed a coimmunoprecipitation (Co-IP) assay using RAW264.7 cells, a mouse macrophage cell line that lacks ASC (Pelegrin et al., 2008). Upon LPS/ATP stimulation, Cbl-b associated with NLRP3 (Fig. 3 B), indicating that their association is independent of ASC. To verify that Cbl-b interacts with endogenous NLRP3, we performed a Co-IP using BMDMs derived from C57BL/6 mice. Again, upon LPS priming and ATP stimulation, Cbl-b bound to NLRP3 (Fig. 3 C), consistent with the data generated using RAW264.7 cells.
Figure 3.
Cbl-b UBA region is required to form a complex with the NLRP3 LRR domain. (A) Immunoblot analysis of Flag immunoprecipitates (IP) of lysates from HEK293T cells transfected with Flag-tagged NLRP3 and HA-tagged Cbl-b. CL, cell lysate. (B) Immunoblot analysis of Cbl-b immunoprecipitates of lysates of RAW264.7 cells primed with LPS (100 ng/ml) for 4 h and stimulated with ATP (2.5 mM) for indicated time points. (C) Immunoblot analysis of NLRP3 immunoprecipitates of lysates of BMDMs primed with LPS and stimulated with ATP. (D) Immunoblot analysis of Flag or HA immunoprecipitates of lysates of HEK293T cells transfected with HA-tagged Cbl-b and Flag-tagged NLRP3 or NLRP3 mutants (NLRP3 ΔLRR, NBD, LRR, and PYD). (E) Immunoblot analysis of Flag or HA immunoprecipitates of lysates of HEK293T cells transfected with HA-tagged Cbl-b, Cbl-b N1/3, Cbl-b C2/3, or Cbl-b ΔUBA and Flag-tagged NLRP3. (F) Immunoblot analysis of Flag immunoprecipitates of lysates of HEK293T cells transfected with HA-tagged Cbl-b UBA together with NLRP3 LRR or NLRP3ΔLRR in the presence of MG132 (10 µM, overnight). (G) Immunoblot analysis of Flag immunoprecipitates of lysates of HEK293T cells transfected with HA-tagged Cbl-b together with Flag-tagged NLRP3 or NLRP3 LRR K/R mutant. (H) IL-1β production by Nlrp3−/− BMDMs reconstituted with Flag-tagged NLRP3 or NLRP3 LRR K/R mutant plasmid, primed with LPS, and stimulated with ATP for 30 min. n = 3 mice per group, each with three repeated wells. Error bars are mean ± SD. **, P < 0.01 (Nlrp3−/− BMDMs transfected with WT NLRP3 vs. with NLRP3 LRR K/R mutant) group; Student’s t test. Data are representative of three independent experiments. Actin was used as a loading control. CL, cell lysates from transfected HEK293T cells, WT BMDMs, or RAW264.7 cells.
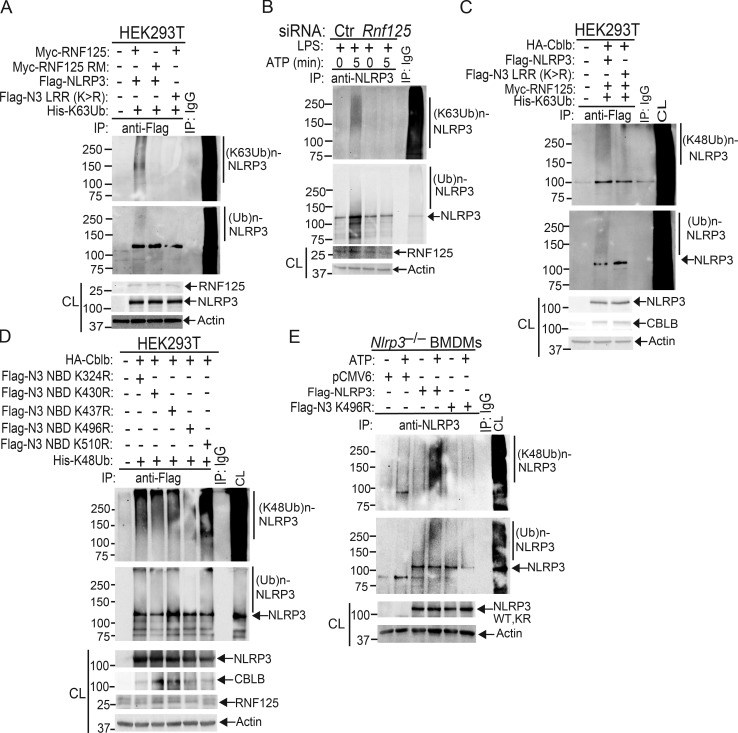
To define which domain of NLRP3 interacts with Cbl-b, we transfected HEK293T cells with Flag-tagged NLRP3 and its mutants (Fig. 3 D, upper panel). As shown in the middle and lower panels of Fig. 3 D, the interaction of NLRP3 and Cbl-b was dependent upon the LRR domain of NLRP3. Finally, we investigated which domain of Cbl-b binds to the NLRP3 LRR domain. By using a series of Cbl-b truncated fragments shown in Fig. 3 E, we found that deletion of the UBA region of Cbl-b abrogated the binding of Cbl-b to NLRP3, indicating that the UBA region of Cbl-b binds to NLRP3’s LRR domain. To confirm whether the Cbl-b UBA region directly binds to NLRP3’s LRR domain, we transfected HEK293T cells with Flag-tagged NLRP3 LRR or NLRP3ΔLRR together with HA-tagged Cbl-b UBA. Indeed, the Cbl-b UBA region directly bound to the NLRP3 LRR domain (Fig. 3 F). To determine whether Cbl-b UBA binds to the ubiquitin chains that attach to the NLRP3 LRR domain, we mutated all 11 lysine residues within the LRR domain. We found that the NLRP3 LRR K/R mutant failed to bind to Cbl-b (Fig. 3 G), therefore supporting the model that the Cbl-b UBA region binds to ubiquitin chains attached to the LRR domain of NLRP3. This notion is further supported by the fact that reconstituting Nlrp3−/− BMDMs with NLRP3 LRR K/R mutant led to heightened IL-1β production (Fig. 3 H).
Cbl-b targets NLRP3 for ubiquitination and proteasome-mediated degradation
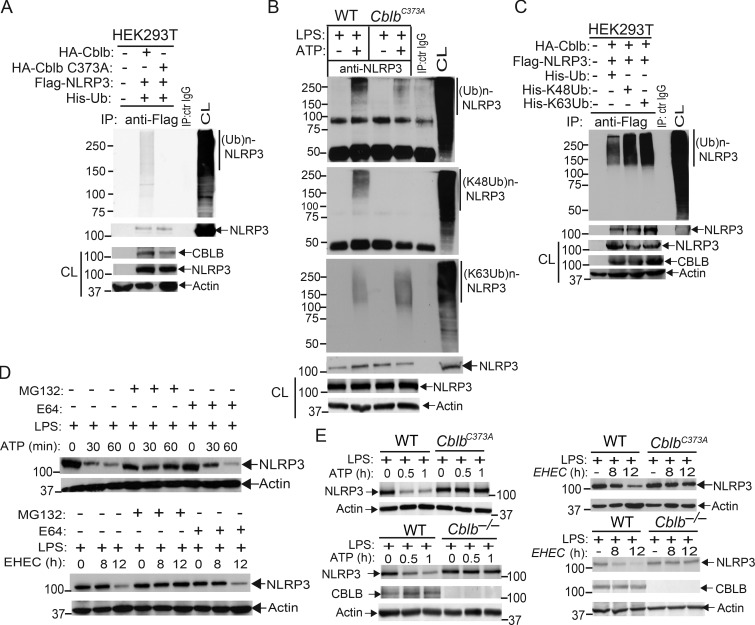
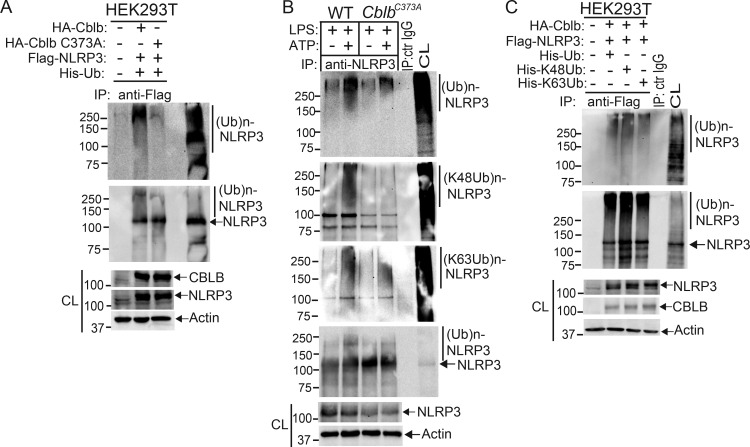
To assess whether Cbl-b targets NLRP3 for ubiquitination, we transfected HEK293T cells with Flag-tagged NLRP3, His-tagged ubiquitin, and HA-tagged Cbl-b or Cbl-b C373A mutant lacking E3 ubiquitin ligase activity. Indeed, Cbl-b but not Cbl-b C373A potentiated NLRP3 ubiquitination (Fig. 4 A and Fig. S3 A). To further confirm that Cbl-b is the E3 ubiquitin ligase for NLRP3, WT and CblbC373A BMDMs were primed with LPS and stimulated with ATP. NLRP3 ubiquitination was markedly reduced in BMDMs from CblbC373A mice (Fig. 4 B and Fig. S3 B). These data indicate that Cbl-b inhibits NLRP3 inflammasomes by ubiquitinating NLRP3.
Figure 4.
Cbl-b targets NLRP3 for K48-linked polyubiquitination. (A) Anti-His immunoblot analysis of Flag immunoprecipitates (IP) of lysates from HEK293T cells transfected with Flag-tagged NLRP3, His-tagged ubiquitin, and HA-tagged Cbl-b or Cbl-b C373A. CL, cell lysate; ctr, control. (B) Anti-ubiquitin immunoblot analysis of NLRP3 immunoprecipitates of lysates from WT and CblbC373A BMDMs primed with LPS (100 ng/ml, 4 h) and stimulated with ATP (2.5 mM, 5 min) and reprobed with anti–K48-ubiquitin and anti–K63-ubiquitin. (C) Anti-His immunoblot analysis of Flag immunoprecipitates of lysates from HEK293T cells transfected with Flag-tagged NLRP3, HA-tagged Cbl-b together with His-tagged ubiquitin, His-tagged K48 ubiquitin, or His-tagged K63 ubiquitin. (D) NLRP3 immunoblot analysis of lysates from LPS-primed WT BMDMs pretreated with E-64 (10 µM) and MG-132 (5 µM) for 30 min and then stimulated with ATP or infected with EHEC for indicated time points. (E) NLRP3 and Cbl-b immunoblot analysis of lysates from WT and CblbC373A or Cblb−/− BMDMs primed with 100 ng/ml LPS for 4 h and then stimulated with ATP or infected with EHEC for various time points. Data are representative of three independent experiments.
Figure S3.
Verification of Cbl-b as the E3 ubiquitin ligase that initiates K48-linked polyubiquitination of NLRP3. Related to Fig. 4. (A) Anti-His and anti-Flag immunoblot analysis of Flag immunoprecipitates (IP) of lysates from HEK293T cells transfected with Flag-tagged NLRP3, His-tagged ubiquitin, and HA-tagged Cbl-b or Cbl-b C373A under the denaturing condition by heating to 95°C for 10 min. CL, cell lysate. (B) Anti-ubiquitin and anti-NLRP3 immunoblot analysis of NLRP3 immunoprecipitates of lysates from WT and CblbC373A BMDMs primed with LPS (100 ng/ml, 4 h) and stimulated with ATP (2.5 mM, 5 min) and reprobed with anti–K48 ubiquitin and anti–K63 ubiquitin under the denaturing condition. (C) Anti-His and anti-Flag immunoblot analysis of Flag immunoprecipitates of lysates from HEK293T cells transfected with Flag-tagged NLRP3 and HA-tagged Cbl-b together with His-tagged ubiquitin, His-tagged K48 ubiquitin, or His-tagged K63 ubiquitin under the denaturing condition. Ctr, control. Data are representative of three independent experiments (A) and representative of two independent experiments (B and C).
Next, we determined whether NLRP3 undergoes K48- or K63-linked polyubiquitination by using K48- and K63-specific ubiquitin antibodies. Indeed, NLRP3 from WT BMDMs underwent both K48- and K63-linked polyubiquitination. Surprisingly, although K48-linked ubiquitination of NLRP3 was diminished in BMDMs expressing Cbl-b C373A, K63-linked ubiquitination of NLRP3 remained unaltered (Fig. 4 B and Fig. S3 B). To confirm this finding, we transfected HEK293T cells with HA-tagged Cbl-b and Flag-tagged NLRP3, together with His-tagged ubiquitin, K48 ubiquitin, or K63 ubiquitin. Consistent with a previous report (Py et al., 2013), NLRP3 underwent both K48- and K63-linked poyubiquitination (Fig. 4 C and Fig. S3 C). Our data suggest that Cbl-b mediates K48-linked polyubiquitination of NLRP3 and that the K63-linked ubiquitination of NLRP3 observed in HEK293T cells and CblbC373A BMDMs may be due to an additional endogenous E3 ubiquitin ligase.
To define the functional significance of NLRP3 ubiquitination, WT BMDMs were primed with LPS and stimulated with ATP or infected with EHEC for various time points in the presence or absence of MG-132 (proteasome inhibitor) or E-64 (lysosome inhibitor). NLRP3 underwent degradation upon ATP stimulation or EHEC infection, which was prevented by MG-132 but not E-64 (Fig. 4 D). These findings indicate that NLRP3 degradation is dependent on the proteasome. These observations are further supported by the fact that NLRP3 undergoes degradation in LPS-primed, ATP-stimulated, or EHEC-infected WT BMDMs and that this degradation was diminished in BMDMs expressing Cbl-b C373A mutant or lacking Cbl-b (Fig. 4 E). Collectively, these data support the notion that Cbl-b is the E3 ubiquitin ligase for NLRP3 and mediates the degradation of NLRP3.
Identification of RNF125 as a novel E3 ubiquitin ligase inducing K63-linked ubiquitination of the NLRP3 LRR domain that recruits Cbl-b
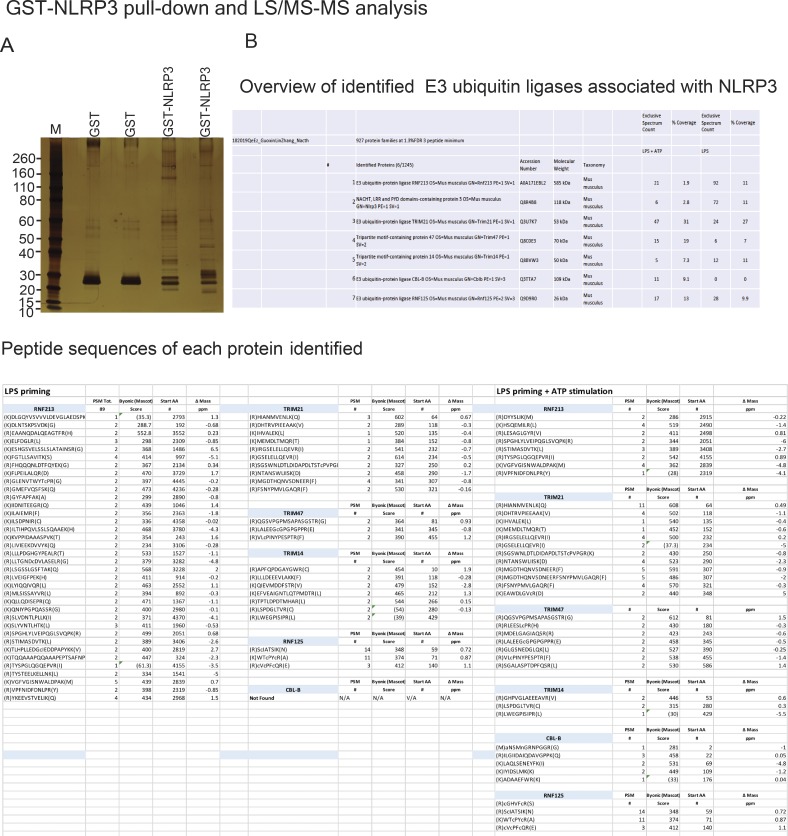
The attachment of ubiquitin chains to the NLRP3 LRR domain and their association with the Cbl-b UBA region suggests that an additional E3 ubiquitin ligase other than Cbl-b is involved in the initiation of NLRP3 K63-linked polyubiquitination. To identify the potential E3 ubiquitin ligase that initiates NLRP3 LRR ubiquitination, we performed a GST-NLRP3 pull-down assay coupled with mass spectrometry analysis. To this end, we primed WT BMDMs with LPS for 4 h and stimulated them with ATP for 5 min. The BMDMs primed with LPS but without ATP stimulation were used as a control. The BMDM lysates were incubated with GST-NLRP3 immobilized on glutathione-agarose beads. Complexes eluted from the beads were resolved by SDS-PAGE and analyzed by silver staining (Fig. S4 A). The proteins eluted from the beads were loaded on suspension trapping filters, captured as a fine dispersion, digested with trypsin, and subjected to liquid chromatography–mass spectrometry (LC-MS/MS) analysis of resulting peptides to identify the NLRP3-interacting proteins. We selected the potential E3 ubiquitin ligases based on whether they bound to NLRP3 upon LPS priming or LPS priming plus ATP stimulation with the probability >70%. In addition to Cbl-b, we identified several additional E3 ubiquitin ligases including TRIM213, TRIM21, TRIM47, Ring finger protein 125 (RNF125), and RNF213 also binding to NLRP3 (Fig. S4 B). Note that among these candidates, Cbl-b was the only E3 ubiquitin ligase that bound to NLRP3 upon ATP stimulation (Fig. S4 B).
Figure S4.
Identification of E3 ubiquitin ligases that bind to NLRP3. Related to Fig. 5. (A) Silver staining of GST or GST-NLRP3–binding proteins eluted from lysates of B6 BMDMs primed with LPS and stimulated with ATP for 5 min. (B) LC-MS/MS analysis of GST-NLRP3–binding E3 ubiquitin ligases eluted from lysates of B6 BMDMs primed with LPS and stimulated with ATP, captured, and trypsin digested. The top six hits were selected from three independent experiments.
Figure S5.
Verification of RNF125 as an additional E3 ubiquitin ligase to initiate K63-linked polyubiquitination of NLRP3 and confirmation of K496 as the ubiquitination site of NLRP3. Related to Figs. 5 and 6. (A) Anti-His and anti-Flag immunoblot analysis of Flag immunoprecipitates (IP) of lysates from Rnf125 gene–silenced HEK293T cells transfected with Flag-tagged NLRP3 or NLRP3 LRR (K>R), Myc-tagged RNF125, or Myc-RNF125 RM mutant together with His-tagged K63 ubiquitin under the denaturing condition. CL, cell lysate. (B) Anti-K63 ubiquitin and anti-NLRP3 immunoblot analysis of NLRP3 immunoprecipitates of lysates from WT BMDMs transfected with Rnf125 siRNA or a control (Ctr) siRNA, primed with LPS, and stimulated with ATP under the denaturing condition. (C) Anti–K48-ubiquitin and anti-Flag immunoblot of Flag immunoprecipitates of lysates from HEK293T cells transfected with Flag-tagged NLRP3 or NLRP3 LRR (K/R) mutant, HA-tagged Cbl-b, Myc-RNF125, and His-tagged K63 ubiquitin under the denaturing condition. (D) Anti-His and anti-Flag immunoblot analysis of Flag immunoprecipitates of lysates from HEK293T cells transfected with HA-tagged Cbl-b and His-tagged K48 ubiquitin together with Flag-tagged NLRP3 K/R mutants (K324R, K430R, K437R, K496R, and K510R) under the denaturing condition. (E) Anti–K48 ubiquitin and anti-NLRP3 immunoblot analysis of NLRP3 immunoprecipitates of lysates from Nlrp3−/− BMDMs reconstituted with pCMV6, Flag-tagged NLRP3, or NLRP3 K496R plasmid under the denaturing condition (upper panel). Anti-NLRP3 and anti-actin immunoblots of lysates from Nlrp3−/− BMDMs reconstituted with Flag-tagged NLRP3 or NLRP3 K496R plasmid, primed with LPS, and stimulated with ATP under the denaturing condition (lower panel). Data are representative of two independent experiments (A, B, C, and E) and representative of three independent experiments (D).
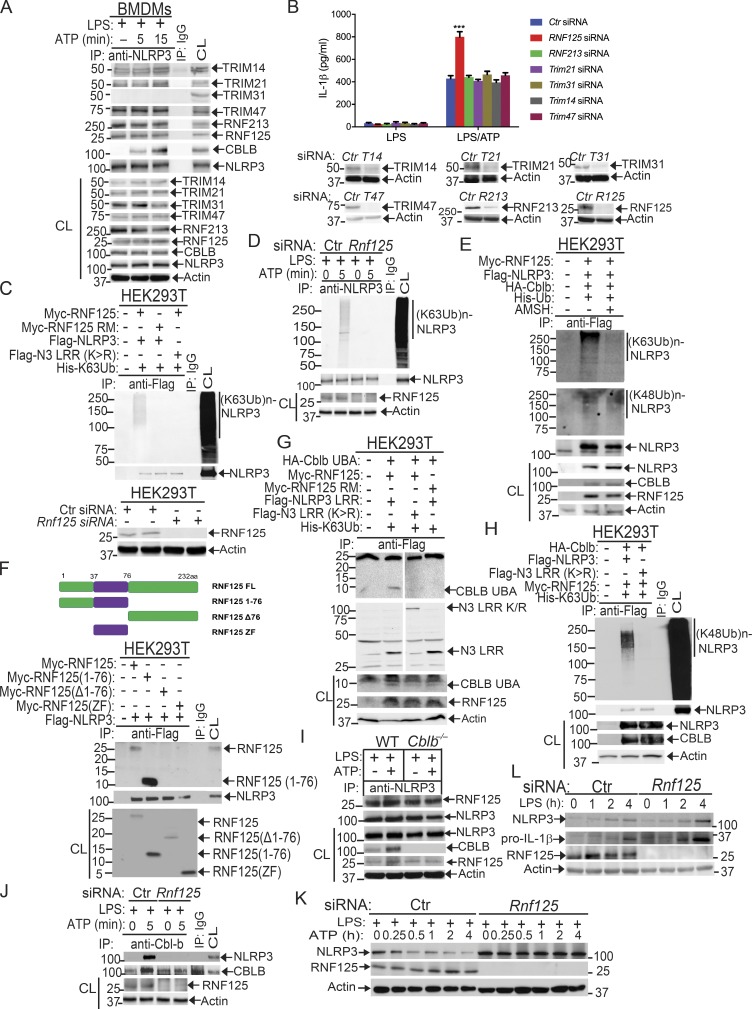
To further verify the association of these E3 ubiquitin ligases with NLRP3 and the role that they play in NLRP3 inflammasome activation, we performed Co-IP experiments, with the focus on the above E3 ubiquitin ligases identified by LC-MS/MS. We also included TRIM31 since it is reported that TRIM31 is a potential E3 ubiquitin ligase for NLRP3 (Song et al., 2016). These experiments revealed that TRIM14, TRIM21, TRIM47, RNF213, and RNF125 bound to NLRP3 in WT BMDMs upon LPS priming and ATP stimulation (Fig. 5 A). In contrast, we could not detect an interaction between NLRP3 and TRIM31 by LC-MS/MS and Co-IP assays at the time points that we tested (Fig. 5 A and Fig. S4 B). To examine whether any of these E3 ubiquitin ligases are involved in NLRP3 inflammasome activation, we silenced TRIM14, TRIM21, TRIM47, TRIM31, RNF213, and RNF125 in WT BMDMs by specific siRNA. We found that silencing RNF125, but not TRIM family members and RNF213, resulted in heightened IL-1β production upon LPS priming and ATP stimulation (Fig. 5 B). The failure to reproduce the TRIM31–NLRP3 interaction reported by Song et al. (2016) is currently unknown. Nevertheless, our data collectively indicate that RNF125 is the initial E3 ubiquitin ligase that induces NLRP3 K63-linked polyubiquitination within its LRR domain. We should also note that HEK293T cells expressed detectable RNF125 (Fig. 5 C), which may explain why NLRP3 underwent both K63- and K48-linked polyubiquitination without transfection with exogenous RNF125.
Figure 5.
Identification of RNF125 as an additional E3 ubiquitin ligase that initiates K63-linked polyubiquitination of NLRP3. (A) Immunoblot analysis of NLRP3 immunoprecipitates (IP) and total cell lysates (CL) from WT BMDMs primed with LPS and stimulated with ATP (2.5 mM) for 5 and 15 min with antibodies against TRIM14, TRIM21, TRIM31, TRIM47, RNF125, RNF213, CBLB, and NLRP3. (B) IL-1β production by BMDMs from WT mice (n = 3) that were transfected with siRNAs specific for Trim14, Trim21, Trim31, Trim47, Rnf125, and Rnf213, primed with LPS, and stimulated with ATP. Data are represented as mean ± SD.P < 0.001 (Rnf125 siRNA-transfected BMDMs vs. other siRNA-transfected BMDMs); Student's t test. Cell lysates from the above BMDMs were blotted with antibodies against TRIM14, TRIM21, TRIM31, TRIM47, RNF125, RNF213, and actin. Ctr, control. (C) Anti-His immunoblot analysis of Flag immunoprecipitates of lysates from Rnf125 gene-silenced HEK293T cells transfected with Flag-tagged NLRP3 or NLRP3 LRR (K>R), Myc-tagged RNF125, or Myc-RNF125 RM mutant together with His-tagged K63 ubiquitin. (D) Anti-K63 ubiquitin immunoblot analysis of NLRP3 immunoprecipitates of lysates from WT BMDMs transfected with Rnf125 siRNA or a control siRNA, primed with LPS, and stimulated with ATP. (E) Anti-K63 and K48 immunoblot analysis of Flag immunoprecipitates of lysates from HEK293T cells transfected with Flag-tagged NLRP3, HA-tagged Cbl-b, Myc-RNF125, and His-tagged ubiquitin treated with or without AMSH. (F) Anti-Myc immunoblot analysis of Flag immunoprecipitates of lysates from HEK293T cells transfected with Myc-tagged RNF125, RNF125 (1–76), RNF125 (Δ1-76), or RNF126 (ZF), together with Flag-tagged NLRP3. (G) Anti-HA immunoblot analysis of Flag immunoprecipitates of lysates from HEK293T cells transfected with Flag-tagged NLRP3 LRR or NLRP3 LRR (K/R) mutant, HA-tagged Cbl-b UBA, Myc-RNF125 or RNF125 RM, and His-tagged K63 ubiquitin in the presence of MG132 (10 µM, overnight). (H) Anti–K48-ubiquitin immunoblot of Flag immunoprecipitates of lysates from HEK293T cells transfected with Flag-tagged NLRP3 or NLRP3 LRR (K/R) mutant, HA-tagged Cbl-b, Myc-RNF125, and His-tagged K63 ubiquitin. (I) Anti-RNF125 immunoblot analysis of NLRP3 immunoprecipitates of lysates from WT and Cblb−/− BMDMs that were primed with LPS and stimulated with ATP. (J) Anti-NLRP3 immunoblot analysis of Cbl-b immunoprecipitates of lysates from WT BMDMs transfected with Rnf125 siRNA or a control siRNA, primed with LPS, and stimulated with ATP. (K) Anti-NLRP3 and anti-RNF125 immunoblot analysis of lysates from WT BMDMs transfected with Rnf125 siRNA or a control siRNA, primed with LPS, and stimulated with ATP. (L) Anti-NLRP3 and anti–pro-IL-1β immunoblot analysis of WT BMDMs transfected with Rnf125 siRNA or a control siRNA, primed with LPS for 1, 2, and 4 h. Data are representative of three independent experiments. Actin was used as a loading control.
To further determine whether RNF125 is the E3 ubiquitin ligase to induce K63-linked polyubiquitination of NLRP3 within its LRR domain, we knocked down endogenous RNF125 in HEK293T cells by Rnf125 siRNA and then transfected these cells with Myc-tagged RNF125 or RNF125 Ring mutant (RM), which contains C37R and A40M mutations within the RNF125 Ring finger domain (Kim et al., 2015), together with Flag-tagged NLRP3 or NLRP3 LRR K/R mutant in the presence of K63 ubiquitin. Indeed, RNF125 induced NLRP3 K63-linked polyubiquitination, and this ubiquitination was abrogated in HEK293T cells expressing RNF125 RM or an NLRP3 LRR K/R mutation (Fig. 5 C and Fig. S5 A). To further confirm these data, we silenced the Rnf125 gene in WT BMDMs by Rnf125 siRNA, primed them with LPS, and stimulated with ATP. Silencing Rnf125 abrogated K63-linked polyubiquitination of NLRP3 (Fig. 5 D and Fig. S5 B). To verify this result, we employed the K63-specific deubiquitinating enzyme AMSH (associated molecule with the SH3 domain of signal transducing adaptor molecule), which has been shown to specifically remove K63-ubiquitin chains (McCullough et al., 2004). We cotransfected HEK293T cells with HA-tagged Cbl-b, Flag-tagged NLRP3, Myc-tagged RNF125, and His-tagged ubiquitin. The cell lysates were immunoprecipitated with anti-Flag and then treated with or without the K63-specific deubiquitinating enzyme AMSH. As shown in Fig. 5 E, treating the Flag immunoprecipitates with AMSH abrogated K63- but not K48-linked polyubiquitination of NLRP3. These data indicate that RNF125 is the initiating E3 ubiquitin ligase by directing K63-linked polyubiquitination of the NLRP3 LRR domain. In support of these data, we found that RNF125 N-terminal 1–76 fragment interacted with NLRP3 (Fig. 5 F).
Since NLRP3 undergoes both K63- and K48-linked polyubiquitination, we hypothesized that RNF125 targets NLRP3 LRR domain for K63-linked polyubiquitination, which subsequently recruits Cbl-b to NLRP3 via its UBA domain. To test this, we transfected HEK293T cells with RNF125 or RNF125 RM, HA-tagged Cbl-b or Cbl-b UBA, Flag-tagged NLRP3 or NLRP3 LRR K/R mutant, and His-tagged K63 ubiquitin. We found that K63 ubiquitin chains were required for the binding of Cbl-b UBA to NLRP3 and for K48-linked polyubiquitination of NLRP3 (Fig. 5, G and H and Fig. S5 C). In further support of this notion, knocking down Rnf125 in WT BMDMs abrogated the interaction between Cbl-b and NLRP3, whereas RNF125–NLRP3 interaction remained intact in the absence of Cbl-b (Fig. 5, I and J). Furthermore, silencing the Rnf125 gene abrogated NLRP3 degradation (Fig. 5 K). Note that silencing the Rnf125 gene led to an increase in the expression of NLRP3 and pro–IL-1β during LPS priming (Fig. 5 L), suggesting that RNF125 inhibits the priming process. Taken together, our data collectively indicate that NLRP3 undergoes sequential K63- and K48-linked polyubiquitination mediated by RNF125 and Cbl-b, respectively.
Lysine 496 is the site for the attachment of K48-linked ubiquitin chains to NLRP3
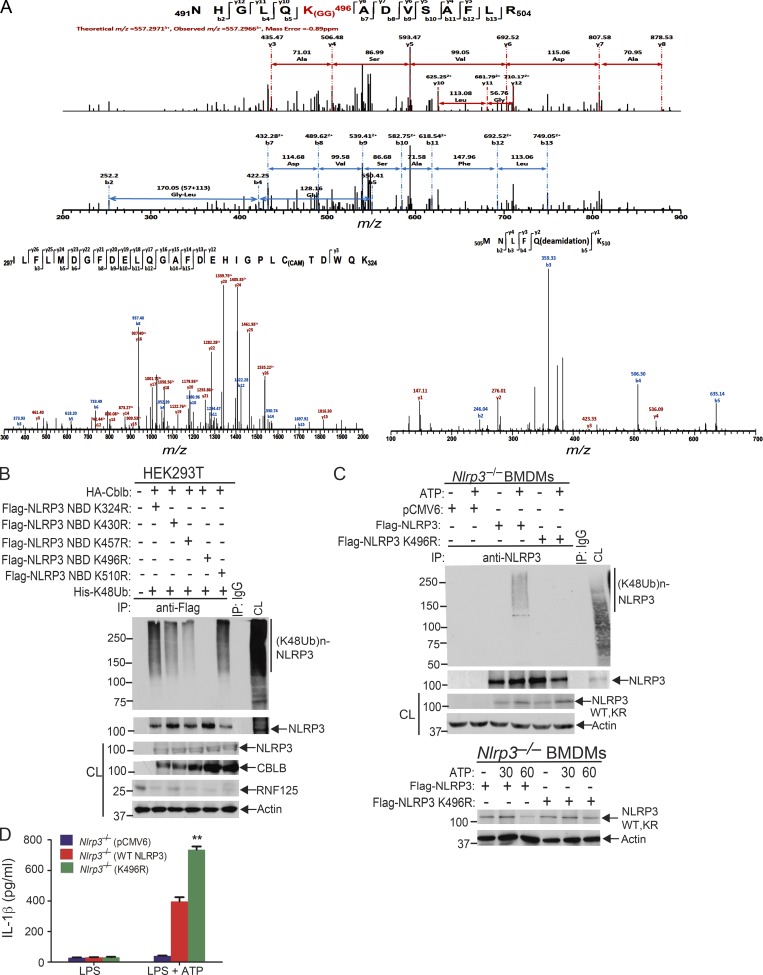
To identify which lysine residue(s) of NLRP3 is the potential ubiquitination site(s) mediated by Cbl-b, we transfected HEK293T cells with HA-tagged Cbl-b, Flag-tagged NLRP3, and His-tagged ubiquitin and then stimulated with nigericin and lysed in radioimmunoprecipitation assay (RIPA) buffer. The Flag-tagged NLRP3 was affinity purified using the FLAG M Purification Kit. The enriched fractions of NLRP3 and ubiquitinated NLRP3 were resolved on SDS-PAGE and were Coomassie blue stained. The bands corresponding to NLRP3 and ubiquitinated NLRP3 were cut into small slices in gel digested with trypsin and were processed for mass spectrometric analysis. Although we failed to identify any lysine residues within the NLRP3 LRR domain that were ubiquitinated, we did identify that 491NHGLQK(GG)496ADVSAFLR504 within the NLRP3 nucleotide-binding domain (NBD) was ubiquitinated (Fig. 6 A). The reason for the failure to identify the ubiquitination sites within the NLRP3 LRR domain might be due to the higher affinity of ubiquitin chains attached to K496 compared with the ubiquitin chains attached to lysine residues within the NLRP3 LRR domain. Alternatively, attachment of large ubiquitin chains to the NLRP3 LRR domain may also affect fragmentation. Furthermore, the protein mobility smear caused by polyubiquitin chains will reduce NLRP3 protein concentrations across the gel slices, which can adversely affect detection.
Figure 6.
Lysine 496 is the site for the attachment of K48-linked ubiquitin chains to NLRP3. (A) Mass spectrometric analysis of Flag-tagged NLRP3 showed that K496 [491NHGLQK(GG)496ADVSAFLR504] was ubiquitinated. (B) Anti-His immunoblot analysis of Flag immunoprecipitates (IP) of lysates from HEK293T cells transfected with HA-tagged Cbl-b and His-tagged K48 ubiquitin together with Flag-tagged NLRP3 lysine (K)/arginine (R) mutants (K324R, K430R, K437R, K496R, and K510R). CL, cell lysate. (C) Anti-K48 ubiquitin immunoblot analysis of NLRP3 immunoprecipitates of lysates from Nlrp3−/− BMDMs reconstituted with pCMV6, Flag-tagged NLRP3, or NLRP3 K496R plasmid (upper panel). Anti-NLRP3 and anti-actin immunoblots of lysates from Nlrp3−/− BMDMs reconstituted with Flag-tagged NLRP3 or NLRP3 K496R plasmid, primed with LPS, and stimulated with ATP (lower panel). (D) IL-1β production by BMDMs from Nlrp3−/− mice (n = 3) reconstituted with Flag-tagged NLRP3 or NLRP3 K496R plasmid, primed with LPS, and stimulated with ATP for 30 min. Data are represented as mean ± SD. **, P < 0.01 (Nlrp3−/− BMDMs transfected with WT NLRP3 vs. with NLRP3 K496R); Student’s t test. Data are representative of two independent experiments (A) and representative of three independent experiments (B–D).
Since the NLRP3 LRR domain underwent K63-linked polyubiquitination, we reasoned that Cbl-b mediates K48-linked polyubiquitination within the NBD domain of NLRP3. To confirm whether K496 is the ubiquitination site of NLRP3, we generated NLRP3 K496R mutant. We also used the UbPred, a random forest-based predictor of potential ubiquitination sites in proteins (http://www.ubpred.org) to predict the potential ubiquitination sites within the NLRP3 NBD domain. We identified four other lysine residues within the NBD domain with low to medium confidence (K324, K430, and K510) by the UbPred, of which K324 and K510 were also detected by LS-MS/MS analysis (Fig. 6 A). We also included K437, which is within the NLRP3 NBD domain, although UbPred shows this lysine residue with no confidence. We generated NLRP3 K324R, K430R, K437R, and K510R mutants and transfected HEK293T cells with HA-tagged Cbl-b, Flag-tagged NLRP3 or NLRP3 K/R mutants, and His-tagged K48 ubiquitin. Mutation of NLRP3 at K496R completely abrogated K48-linked polyubiquitination of NLRP3 induced by Cbl-b (Fig. 6 B and Fig. S5 D). Reconstituting Nlrp3−/− BMDMs with WT NLRP3, but not NLRP3 K496R, rescued K48-linked ubiquitination and subsequent degradation of NLRP3 (Fig. 6 C and Fig. S5 E), which correlated with heightened IL-1β production by Nlrp3−/− BMDMs reconstituted with NLRP3 K496R (Fig. 6 D). Therefore, our data collectively indicate that lysine 496 within the NLRP3 NBD domain is the site for K48-linked ubiquitin chain attachment.
In vivo silencing of Rnf125 increases the susceptibility of mice to septic shock induced by a sub-lethal dose of LPS
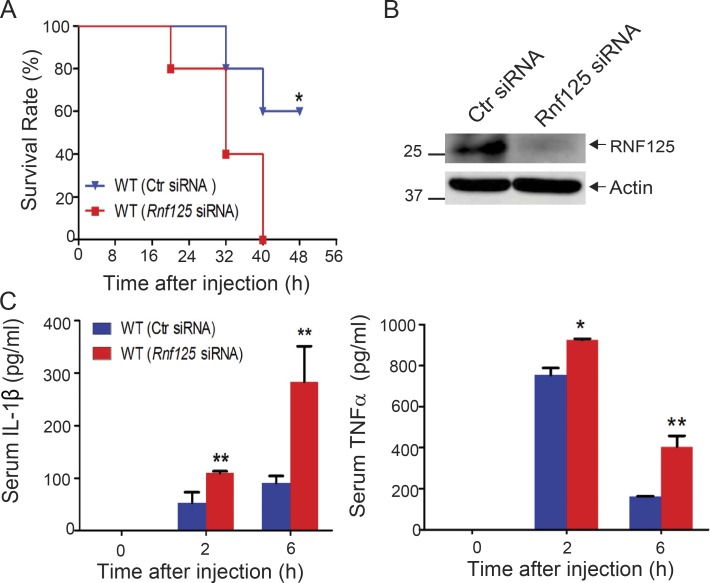
Since RNF125 is required for the initiation of K63-linked polyubiquitination of the NLRP3 LRR domain, which recruits Cbl-b, it is likely that RNF125-deficient mice should also be highly susceptible to LPS-induced endotoxemia as shown in Cblb−/− mice. Recently, we established a protocol that silences the Cblb gene in WT mice in vivo (Xiao et al., 2016). To test this, we silenced the Rnf125 gene by in vivo delivery of Rnf125-specific siRNA via tail vein injection. As expected, all the mice receiving Rnf125 siRNA died within 40 h after injection with a sub-lethal dose of LPS, whereas 60% of mice receiving control siRNA survived (Fig. 7 A), which correlated with heightened IL-1β and TNF-α in the sera (Fig. 7 C). These data indicate that the phenotype of mice with Rnf125 gene silencing recapitulated the one observed in Cblb−/− mice.
Figure 7.
Systemic in vivo delivery of Rnf125-specific siRNA renders mice susceptible to endotoxemia induced by a sub-lethal dose of LPS. (A) Kaplan-Meier survival curve of C57BL/6 mice treated with in vivo–grade Rnf125-specific siRNA or a nonsense siRNA (2 mg/kg/mouse) via tail vein injection for 24 h before LPS injection (5 mg/kg). *, P < 0.05 by log-rank test. (B) Immunoblot analysis for RNF125 in spleen cells from C57BL/6 mice that were treated with the control siRNA or Rnf125-specific siRNA. Actin was used as a loading control (Ctr). (C) Serum IL-1β and TNF-α levels from C57BL/6 mice (five mice per group) that were treated with the control siRNA or Rnf125-specific siRNA before LPS injection (1 mg/kg). Data are shown as mean ± SD. *, P < 0.05; **, P < 0.01; Student’s t test. Data are representative of three independent experiments (A and C) and representative of two independent experiments (B).
Discussion
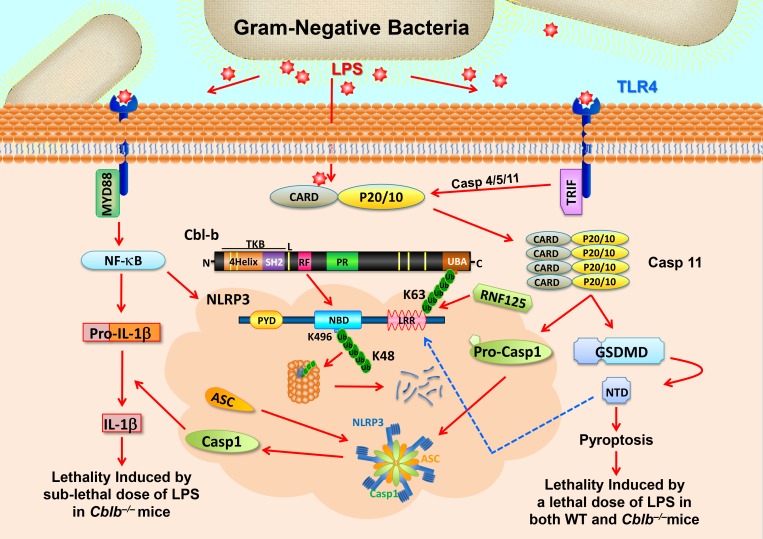
It has been shown that NLRP3 ubiquitination negatively regulates NLRP3 inflammasome in vitro (Py et al., 2013), but the E3 ubiquitin ligase or ligases that ubiquitinate NLRP3 are not fully defined. Furthermore, the biological relevance of in vivo NLRP3 ubiquitination remains to be established. Here, we first report that NLRP3 undergoes sequential K63- and K48-linked polyubiquitination, which is mediated by E3 ubiquitin ligases RNF125 and Cbl-b respectively (Figs. 4 and 5). Consistent with this, Cbl-b deficiency or inactivation leads to hyper-activation of canonical and noncanonical NLRP3 inflammasomes (Fig. 1, D–H). Mice deficient for Cbl-b are extremely sensitive to a sub-lethal dose of LPS, which is rescued by IL-1RA treatment or introduction of NLRP3 deficiency (Fig. 2, A and E). Furthermore, mice with Rnf125 gene silencing are also highly susceptible to a sub-lethal dose of LPS-induced endotoxemia (Fig. 7 A). Therefore, our data demonstrate that RNF125 and Cbl-b coordinately regulate NLRP3 inflammasome activation by targeting NLRP3 for ubiquitination in vitro and in vivo (Fig. 8).
Figure 8.
Schematic model for RNF125 and Cbl-b in LPS-induced endotoxemia. When mice receive a sub-lethal dose of LPS, LPS triggers (1) TLR4 to initiate a MyD88-dependent signaling pathway, which activates NF-κB and induces the expression of NLRP3 and pro–IL-1β and (2) TRIF-dependent pathway that induces the expression of Casp-11. LPS also undergoes endocytosis and binds its cytosolic sensor Casp-11 to induce oligomerizations of Casp-11, which cleaves GSDMD to liberate its N-terminal domain (NTD). NTD may trigger the activation of the NLRP3 inflammasome, leading to the release of IL-1β without inducing pyroptosis. RNF125 targets the NLRP3 LRR domain for K63-linked polyubiquitination. The K63 ubiquitin chains attached to the NLRP3 LRR domain recruit Cbl-b by binding its UBA region. Cbl-b then ubiquitinates NLRP3 at K496 within its NBD and targets NLRP3 to the proteasome for degradation, thus keeping the NLRP3 inflammasome in check. In the absence or inactivation of Cbl-b, mice are highly susceptible to LPS-induced endotoxemia. However, when mice receive a lethal dose of LPS exposure, LPS binds to Casp-11 and mainly triggers Casp-11–mediated pyroptosis via GSDMD. Cbl-b is unable to control this noncanonical inflammasome-induced lethality. CARD, caspase recruitment domain; PYD, pyrin domain; TKB; protein tyrosine kinase-binding; L, linker region; RF, Ring finger; P, peptide; PR, proline-rich region; Toll/IL‐1R domain–containing adaptor‐inducing IFN‐β. Red arrows indicate activation or induction, whereas the blue dashed arrow indicates possible activation.
It has been shown that inhibiting BRCC3, a deubiquitinating enzyme, results in NLRP3 ubiquitination, thus suppressing NLRP3 inflammasome activity (Py et al., 2013). However, the E3 ubiquitin ligases responsible for ubiquitinating NLRP3 are not fully characterized. It is reported that TRIM31 may be the E3 ubiquitin ligase for NLRP3 (Song et al., 2016), but the in vivo relevance of this report has not been established because of a lack of TRIM31 E3 ubiquitin ligase dead knock-in mice. Furthermore, loss of TRIM31 also only moderately slows the kinetics of NLRP3 degradation induced by LPS in the presence of cycloheximide (Song et al., 2016). In addition, none of the NLRP3 inflammasome stimuli were used to induce NLRP3 degradation (Song et al., 2016). Therefore, whether NLRP3 degradation observed in this study is induced by NLRP3 inflammasome stimuli is unknown. In our GST-NLRP3 pull-down assay coupled with LC-MS/MS analysis, we found that TRIM14, TRIM21, TRIM47, RNF125, and RNF213 but not TRIM31 associated with NLRP3 (Fig. 5 A, upper panel, and Fig. S4 B). Furthermore, silencing the Trim31 gene in WT BMDMs does not affect LPS-primed, ATP-stimulated production of IL-1β (Fig. 5 B). Therefore, TRIM31 may not be the E3 ubiquitin ligase that targets NLRP3 for ubiquitination.
Intriguingly, we found that Cbl-b binds to the NLRP3 LRR domain via its UBA region (Fig. 3, D–F), suggesting that Cbl-b binds to the ubiquitin chains that attach to the NLRP3 LRR domain. Consistent with this notion, mutating the 11 lysine residues to arginine within the LRR domain abrogates Cbl-b–NLRP3 interaction (Fig. 3 G). Reconstituting Nlrp3−/− BMDMs with the NLRP3 K/R mutant results in heightened IL-1β production in response to LPS priming and ATP stimulation (Fig. 3 H). The binding of Cbl-b to the ubiquitin chains attached to the NLRP3 LRR domain is critical for NLRP3 degradation in the proteasome because NLRP3 degradation is abrogated in macrophages expressing inactive Cbl-b upon ATP stimulation or EHEC infection (Fig. 4 E). Through a GST-NLRP3 pull-down assay coupled with LC-MS/MS and Co-IP experiments, we identified RNF125 as an initial E3 ubiquitin ligase to induce K63-linked NLRP3 ubiquitination within its LRR domain (Fig. 5 and Fig. S5, A–C). This results in the recruitment of Cbl-b via its UBA region (Figs. 3 and 5), enabling Cbl-b to target NLRP3 NBD for K48-linked polyubiquitination at K496 (Fig. 6 and Fig. S5, D and E) and proteasome-mediated degradation (Fig. 4). To our knowledge, our data are the first to identify a novel sequential ubiquitination event mediated by two E3 ubiquitin ligases, RNF125 and Cbl-b. Therefore, our data also indicate that ubiquitin chains can function as an adaptor to recruit E3 ubiquitin ligases to their substrates.
It was recently shown that Casp-11 is a cytosolic sensor for LPS (Shi et al., 2014) and Casp-11 mediates noncanonical inflammasome activation independently of TLR4 (Hagar et al., 2013; Kayagaki et al., 2013). Further analysis indicates that GSDMD, a substrate of Casp-11 and Casp-1, is a key player of pyroptosis (Kayagaki et al., 2015; Shi et al., 2015). It was reported that LPS-induced lethality in mice is mediated by Casp-11 because mice deficient for Casp-11 are rescued from death induced by a lethal dose of LPS (Kayagaki et al., 2011). We showed that mice lacking Cbl-b or expressing Cbl-b C373A die from an injection of a sub-lethal dose of LPS (5 mg/kg), which is rescued by deficiency of NLRP3 or by IL-1RA treatment, which blocks both IL-1α and IL-1β (Fig. 2, A, B, and G). These data suggest that Cblb−/− or CblbC373A mice are likely to die from aberrant production of IL-1β and subsequent TNF-α rather than pyroptosis. Indeed, blocking TNF-α by an anti–TNF-α neutralizing antibody also prevents LPS-induced mortality in Cblb−/− mice (Fig. 2 E). In support of this, a lethal dose of LPS-induced mortality in Cblb−/− mice is not rescued by NLRP3 deficiency but by Casp-11 deficiency (Fig. 2 J). These are further supported by our in vitro data that loss of Casp-11, but not NLRP3, impairs EHEC-triggered pyroptosis in WT and Cblb−/− macrophages (Fig. 1 H). Although it has been suggested that the cleavage of GSDMD by both Casp-1 and Casp-11 is a key executive event in pyroptosis, macrophages deficient for Casp-11, but not Casp-1, display defective pyroptosis triggered by cytosolic LPS (Napier et al., 2016). This suggests that noncanonical inflammasomes mainly trigger Casp-11–mediated pyroptosis independently of Casp-1. Our data are further supported by a recent report that GSDMD is required for IL-1 secretion in living macrophages without pyroptosis (Evavold et al., 2018). Collectively, our data suggest that gram-negative bacteria-induced sepsis may activate both canonical and noncanonical inflammasome pathways, while Cbl-b mainly regulates the release of IL-1β (Fig. 8). It is important to note that we did not observe any difference in IL-1β in the sera of WT and Cblb−/− mice infected with Candida albicans, suggesting that Cbl-b does not regulate Casp-8–mediated inflammasomes (Xiao et al., 2016).
Previous studies suggest that Cbl-b may target TLR4 or MyD88 for ubiquitination (Bachmaier et al., 2007; Han et al., 2010). However, TNF-α and IL-6 production by macrophages lacking Cbl-b stimulated with TLR ligands 1–9 is not increased (Fig. 1 A). Furthermore, LPS-induced expression of TLR4, MyD88, NLRP3, and pro–IL-1β and activation of NF-κB are also not augmented in macrophages lacking Cbl-b (Fig. 1, B and C). Therefore, our data collectively indicate that Cbl-b does not regulate TLR signaling. In support of these data, it was reported that Cblb−/− BMDMs are not hypersensitive to TLR stimulation (Yee and Hamerman, 2013). Although LPS-induced endotoxemia is mainly mediated by innate immune cells, T cells do respond to LPS via TLR4 (Zanin-Zhorov et al., 2007). However, Rag1−/−Cblb−/− mice injected with a sub-lethal dose of LPS (Fig. S2 A) recapitulate the phenotype observed in Cblb−/− mice, suggesting that Cbl-b deficiency in innate immune cells is responsible for the phenotype observed. Likewise, mice lacking Cbl-b in myeloid cells but not T cells are also highly sensitive to a sub-lethal dose of LPS (Fig. S2 C). Therefore, our data indicate that Cbl-b expression in innate immune cells is crucial for the suppression of LPS-induced endotoxemia.
In summary, we have identified RNF125 and Cbl-b as the key E3 ubiquitin ligases that keep NLRP3 inflammasomes in check by targeting NLRP3 for sequential K63- and K48-linked polyubiquitination. Thus, NLRP3 inflammasome-mediated endotoxemia is prevented in the presence of RNF125 and Cbl-b (Fig. 8).
Materials and methods
Mice
C57BL/6J, B6.129S7-Rag1tm1Mom/J (Rag1−/−), and B6.129S6-Nlrp3tm1Bhk/J (Nlrp3−/−) mice were purchased from the Jackson Laboratory. Cblb−/− mice were kindly provided by Dr. Josef M. Penninger (University of Toronto, Toronto, ON, Canada). Casp11−/− mice (Wang et al., 1998) were kindly provided by Dr. Junying Yuan (Harvard University, Cambridge, MA). Cblb−/− mice were crossed with Rag1−/−, Nlrp3−/−, and Casp11−/− mice to generate Cblb−/−Rag1−/−, Cblb−/−Nlrp3−/−, and Cblb−/−Casp11−/− mice. CblbC373A mice were described previously (Oksvold et al., 2008).
The mouse strain carrying the Cblbtm1a(KOMP)Wtsi allele was generated at The Ohio State University Genetically Engineered Mouse Modeling Core Facility by standard embryonic stem (ES) cell technology (Piovan et al., 2014). The ES clone EPD0703_2_B11 was acquired from the International Knockout Mouse Phenotyping Consortium (project #79117; http://www.mousephenotype.org/). Prior to microinjection, the identity of the targeted ES cells was verified by 5′ long-range PCR using a primer external to the targeting vector. Chimeric males were bred to C57BL/6 Albino females, and germline transmission was verified by PCR to detect the mutant together with the WT allele in the F1 heterozygous mice. Prior to utilization of the strain for experiments, mice were crossed to a Flipe ubiquitous strain (ACTB:FLPe B6J, JAX strain #005703; Rodríguez et al., 2000) to eliminate the lacZ/neo cassette and obtain the clean tm1c allele according to the breeding schemes recommended by the International Knockout Mouse Phenotyping Consortium. The LoxP flanked Cblb allele (Cblbf/f) was crossed to LysM Cre knock-in allele to specifically delete Cbl-b in myeloid cells including macrophages. All mice were 8–12 wk of age when used, and both male and female mice were used in this study. All animal experimentation involving LPS injection, CLP, and in vivo delivery of nonsense and Rnf125-specific siRNA was approved by the Institutional Animal Care and Use Committees of The Ohio State University and the University of Iowa.
Reagents
Antibodies against CBLB (G-1; sc-8006), HA (Y-11; sc-805), GST (sc-33613), Casp-1 (P10; M20; sc-514), TLR4 (sc-10741), and MyD88 (sc-11356) were purchased from Santa Cruz Biotechnology, Inc. Anti-NLRP3 (D4D8T; #15101 and 15101S), anti–IL-1β (3A6; #12242), anti–K48 linkage–specific polyubiquitin (#4289), anti–K63 linkage–specific polyubiquitin (#12930), anti–phospho-NF-κB p65 (S536; #3031), and anti–phospho-IκBα (5A5; #9246) were purchased from Cell Signaling Technology, Inc. Anti-ubiquitin (05–944) was purchased from EMD Millipore. Anti-Flag (M2; F3165 and F2555) and anti-actin (AC74; A2228) were obtained from Sigma-Aldrich. Anti-RNF125 (LS-B11326) was purchased from LSBio. Anti–c-Myc (9E10; MA1-980) was purchased from Invitrogen. Antibody against TRIM31 (OWL-A48647) was obtained from One World Lab. Antibodies against TRIM14 (PA5-50806), TRIM21 (PA5-52178), TRIM47 (PA5-50892), and RNF213 (PA5-51902) were obtained from Thermo Fisher Scientific. ELISA kits for mouse IL1β (432606), TNF-α (430906), and IL-6 (431306) were purchased from BioLegend. HRP-conjugated goat anti-rabbit IgG or rabbit anti-mouse IgG were purchased from Kirkegaard & Perry Laboratories. Protein G-sepharose was purchased from GE Healthcare. Human recombinant AMSH was purchased from Boston Biochem, Inc.
Bacteria
The growth conditions for EHEC (700927 strain), F. novicida (JSG1819), and P. aeruginosa (PAO1) were described previously (Bell et al., 2010; Dyszel et al., 2010; Wyckoff and Wozniak, 2001).
Plasmids and transfection
Flag-tagged NLRP3 plasmid and its truncated fragments were provided by Dr. Fabio Martinon (University of Lausanne, Lausanne, Switzerland). HA-tagged Cbl-b and its truncated fragments were described previously (Davies et al., 2004; Xiao et al., 2015). Flag-tagged RNF125 and its truncated fragments were from Dr. Ze’ev A. Ronai (Sanford-Burnham Medical Research Institute, La Jolla, CA), in which the Flag tag was replaced with a Myc tag at Mutagenex Inc. His-tagged WT ubiquitin and K48 and K63 ubiquitin were purchased from Boston Biochem. HEK293T cells were transfected with various plasmids by calcium precipitation (Qiao et al., 2014).
Generation of BMDMs and activation of inflammasomes
BM cells from WT and various knockout mouse strains were harvested from the femurs and tibias of mice. Cells were cultured in DMEM containing 10% FBS and 30% conditioned medium from L929 cells expressing M-CSF as described (Xiao et al., 2016). After 1 wk of culture, nonadherent cells were removed, and adherent cells were 80%–90% F4/80+CD11b+ as determined by flow cytometric analysis. LPS-primed BMDMs were infected with P. aeruginosa (multiplicity of infection [MOI] = 30:1), EHEC (MOI = 25:1), and F. novicida (MOI = 100:1) for 1.5 h and then cultured in 100 µg/ml gentamycin. Other stimulations included ATP (2.5 mM, 30 min), monosodium urate (200 µg/ml, 4 h), CTB (40 µg/ml, 16 h), nigericin (20 µM, 3 h), and anthrax LT (500 µg/ml, 6 h). Poly(dA:dT) (1 µg/106 cells, 6 h) and flagellin (6.25 µg/106 cells, 4 h) were transfected using Lipofectamine (Invitrogen).
Generation of MDMs, silencing of the Cblb gene, and activation of NLRP3 inflammasome
Human MDMs were generated as previously described (Rajaram et al., 2011). In brief, peripheral blood mononuclear cells from healthy donors were isolated from heparinized blood on Ficoll-sodium diatrizoate gradients and then cultured for 5 d in RPMI containing 20% autologous serum (2.0 × 106 mononuclear cells/ml) at 37°C. On day 5, human MDMs were transfected with control siRNA or Accell human CBLB siRNA (100 or 200 nM; Dharmacon RNA Technologies) by using Lonza nucleofector reagent and plated in RPMI 1640 containing 20% autologous serum. After 36 h, the MDMs were washed, primed with LPS, and stimulated with ATP or CTB or infected with EHEC. The protocol was approved by The Ohio State University Institutional Review Board.
ELISA and cell death assay
Cell culture supernatants and serum were assayed by ELISA for IL-1β, IL-6, and TNF-α. Cell death was measured by a lactate dehydrogenase assay kit from Sigma-Aldrich.
LPS-induced endotoxemia
Various groups of mice at 8–12 wk of age were injected i.p. with a sub-lethal dose of LPS (5 mg/kg; E. coli 0111:B4) and monitored for survival rate for 24 h. Some Cblb−/− mice were injected with IL-1RA (25 mg/kg) or TNF-α neutralization antibody (50 µg/mouse) before LPS injection. For serum pro-inflammatory cytokines, 1 mg/kg of LPS was injected i.p., and serum was collected every 4 h for 24 h. In some experiments, mice were injected i.p. with a lethal dose of LPS (54 mg/kg) and monitored every 4 h for 14–28 h. To determine the serum IL-1β levels, the mice were injected i.p. with 20 mg/kg of LPS, and serum was collected at 12 h after LPS injection.
Induction of polymicrobial sepsis by CLP
Polymicrobial sepsis was induced via CLP as previously described (Moreno et al., 2006). In brief, mice were anesthetized with aerosol isoflurane (3%–4% in 100% O2 flow at 2 liter/min). A 1-cm midline incision was made on the anterior abdomen. The cecum was ligated with a 2–0 vicryl suture at a point ∼1 cm from the cecal tip, and the cecum was punctured with a 21-gauge needle three times. The cecum was gently squeezed to express a small amount of fecal material and then returned to the central abdominal cavity. In sham-operated control animals, the cecum was isolated but neither ligated nor punctured. The abdominal incision was closed in two layers with 6–0 vicryl sutures. The muscle layer was closed with vicryl suture by applying simple running sutures, and the skin was closed with a 5–0 vicryl suture by applying simple interrupted sutures. The mice were monitored for survival rate for 14–28 h. Approximately 50–100 µl of blood was obtained via submandibular vein bleeding at the time of and 12 h after surgery.
Detection of NLRP3 inflammasome activation by Western blot analysis
BMDMs that were primed with LPS and stimulated with various inflammation activators or infected with various pathogens were lysed in RIPA buffer. The supernatants collected from the BMDM cultures were chloroform-methanol precipitated. The cell lysates or the supernatants were blotted with antibodies to Casp-1 p10 (1:1,000), IL-1β p17 (1:1,000), and actin (1:3,000).
Immunoprecipitation
For Co-IP assays, WT BMDMs were primed with LPS (100 ng/ml) for 4 h and stimulated with ATP (2.5 mM) for various times. Cells were lysed in 0.5% NP-40 lysis buffer. The cell lysates were immunoprecipitated with anti-NLRP3 (1:200) and blotted with anti-TRIM14 (1:500), anti-TRIM21 (1:500), anti-TRIM31 (1:500), anti-TRIM47 (1:500), anti-CBLB (1:500), anti-RNF213 (1:500), and anti-RNF125 (1:500). For Co-IP assays in HEK293T cells transfected with various HA-tagged Cbl-b or Cbl-b mutants or Flag-tagged NLRP3 or NLRP3 mutants, the cell lysates were immunoprecipitated with anti-Flag (1:200) and blotted with anti-HA (1:1,000) or, alternatively, immunoprecipitated with anti-HA and blotted with anti-Flag (1:1,000). To detect NLRP3 ubiquitination, BMDMs from WT and CblbC373A mice were primed with LPS (100 µg/ml) for 4 h and stimulated with ATP for 5 min and then lysed in RIPA buffer containing 2% SDS, sonicated, and diluted to 0.5% of SDS using RIPA buffer without SDS before immunoprecipitation in order to disrupt the proteins associated with NLRP3. In some experiments, a denaturing step was used by heating to 95°C for 10 min before sonication. The cell lysates were immunoprecipitated with anti-NLRP3 and blotted with anti-ubiquitin (1:1,000) or with anti–K48- or anti–K63-specific ubiquitin antibodies (1:1,000). To assess the protein stability of NLRP3, BMDMs from WT, Cblb−/−, or CblbC373A mice were primed with LPS and stimulated with ATP or CTB or infected with EHEC at indicated time points and blotted with antibodies against NLRP3 (1:1,000). To determine whether NLRP3 undergoes proteasome- or lysosome-mediated degradation, WT BMDMs were primed with LPS, pretreated with MG-132 (5 µM) or E-64 (10 µM) for 30 min, and then stimulated with ATP, CTB, or EHEC at various times and lysed. The cell lysates were blotted with anti-NLRP3.
Deubiquitination assay
To determine whether NLRP3 ubiquitination occurs sequentially by RNF125 and then Cbl-b, we transfected HEK293T cells with Myc-tagged RNF125, HA-tagged Cbl-b, Flag-tagged NLRP3, and His-tagged ubiquitin, lysed in RIPA buffer, and immunoprecipitated with anti-Flag. The Flag immunoprecipitates were treated with or without AMSH (200 nM) in deubiquitinating enzyme buffer (50 mM Tris-HCl, pH 7.2, 25 mM KCl, 5 mM MgCl2, and 1 mM dithiothreitol) at 37°C for 30 min (McCullough et al., 2004). The reaction was terminated by addition of SDS sample buffer. The ubiquitination and deubiquitination of NLRP3 was detected by immunoblotting with anti-K63 ubiquitin and anti-K48 ubiquitin antibodies, respectively.
Identification of E3 ubiquitin ligases that initiate K63-linked polyubiquitination of NLRP3
To identify potential NLRP3-binding E3 ubiquitin ligases, GST and GST–NLRP3 protein were incubated with lysates from WT BMDMs primed by LPS and stimulated by ATP for 5 min, followed by incubation with glutathione-sepharose beads.
Suspension trapping filters for cleanup and digestion
Homemade suspension trapping filters were fashioned from 8 × 3–mm punches of Munktell MK 360 Circles 9.0 cm (3600-0900) discs (Ahlstrom Laboratory Filters) M, stacked above two layers of C18 resin (Empore C18; 3M) in a 200-µl pipette tip (Zougman et al., 2014). This was filled with 100 mM TRIS-HCl (pH 7.4) in methanol. The sample was solubilized with SDS (4% wt/vol) and 20 mM dithiothreitol and incubated at 95°C for 5 min, then alkylated with chloroacetamide in the dark for 30 min. Phosphoric acid was added to 3%, and the sample was transferred to the trapping pipette. Proteins were captured on quartz fibers as a fine dispersion while mass spectrometry–incompatible materials passed through the tip. The stack was then rinsed and refilled with cold trypsin in 50 mM AmBiC and set to digest at 47°C for 2 h. Peptides were eluted with 300 μl of 50% ACN and 0.1% TFA, lyophilized and stored at −20°C.
LC-MS/MS
Full MS1 profile data were acquired on a Q-Exactive hf (Thermo Fisher Scientific) from 380–1,700 m/z at a resolution of 60,000 (Yu et al., 2015). The 10 most abundant precursors were selected with a mass window of 2 m/z thomson and subjected to higher-energy collisional dissociation at 37% activation efficiency. A 30-s dynamic exclusion improved selection of lower abundant ions. Fragmentation data were acquired in centroid at 30,000 resolution.
Initial spectral searches were performed with both Mascot version 2.6.2 (MatrixScience) and Byonic search engines (Protein Metrics ver. 2.8.2) against the 2/06/2016 UniprotKB and reverted entry database for mouse. With 5 and 10 ppm tolerance for precursor and fragments, respectively, searches allowed accepted fixed Cys mods of 57 D, as well as variable mods of 16 (M), 80 (S, T), and 114 (K) Th. Final discriminant scores were determined by Scaffold Q + S ver. 4.7 (Proteome Software) at 1.2% false discovery rate.
Identification of potential NLRP3 ubiquitination sites by mass spectrometry
NLRP3 protein was purified by a FLAG Purification Kit from HEK293T cells cotransfected with Flag-tagged NLRP3, HA-tagged Cbl-b, and His-tagged ubiquitin, which was treated with nigericin. The lysates were separated by SDS-PAGE. The bands corresponding to NLRP3 and ubiquitinated NLRP3 were cut, washed, and digested with trypsin overnight after reduction and alkylation. Peptides were extracted from the gel pieces and dried in a vacufuge, and peptides were resuspended in 20 μl of 50 mM acetic acid for LC-MS/MS analysis.
Capillary-liquid chromatography–nanospray tandem mass spectrometry (capillary–LC/MS/MS) performed on a Thermo Fisher Scientific LTQ orbitrap mass spectrometer equipped with a microspray source (Michrom Bioresources Inc.) operated in positive ion mode. Samples were separated on a capillary column (0.2 × 150 mm Magic C18AQ 3µ 200A; Bruker Daltonics) using an UltiMate 3000 HPLC system from Thermo Fisher Scientific. Mobile phase A was 50 mM acetic acid in water, and acetonitrile was used as mobile phase B. Flow rate was set at 2 µl/min. Tandem mass spectrometry data were acquired with a spray voltage of 2.2 kV, and a capillary temperature of 175°C was used. The scan sequence of the mass spectrometer was based on the preview mode data-dependent TopTen method. Sequence information from the tandem mass spectrometry data were processed by converting the raw files into a merged file (.mgf) using MS convert (ProteoWizard). The resulting mgf files were searched using Mascot Daemon by Matrix Science version 2.3.2. The fragment mass tolerance was set to 0.5 D. Considered variable modifications were Ubiquitinylation (K), oxidation (Met), deamidation (N and Q), and carbamidomethylation (Cys). Identified ubiquitinylated peptides were manually checked for validation.
Generation of NLRP3 LRR K/R mutants and RNF125 truncated fragments
Single and multiple lysine-to-arginine (K-to-R) encoding mutations of NLRP3 (NLRP3 K324R, K430R, K437R, K496R, 3 K570R, and NLRP3 LRR 11K/R) and Myc-tagged RNF125, N-terminal 1–76 fragment, zinc finger, and Δ1-76 were generated by site-directed mutagenesis at Mutagenex.
Nlrp3−/− BMDM reconstitution
Nlrp3−/− BMDMs were transfected with constructs expressing Flag-tagged NLRP3, NLRP3 LRR K/R, or NLRP3 K496R by Lipofectamine 2000 according to the manufacturer’s instructions.
In vitro knockdown experiments
Rnf125 Accell siRNA, Rnf213 Accell siRNA, Trim21 Accell siRNA, Trim31 Accell siRNA, Trim14 Accell siRNA, Trim47 Accell siRNA, or nonsense siRNA was obtained from Dharmacon. BMDMs or HEK293T cells (for Rnf125 siRNA) were plated in 12 wells and were transiently transfected with 2 µg of siRNAs plus 4 µl Lipofectamine 2000 according to the manufacturer’s instruction. 36 h later, cells were harvested. The cell lysates were blotted for specific antibodies against TRIM14, TRIM21, TRIM31, TRIM47, RNF125, and RNF213, respectively.
In vivo delivery of Rnf125 siRNA
WT mice were treated with Rnf125 Accell siRNA (2 mg/kg/mouse) or a nonsense siRNA intravenously. 24 h later, mice were i.p. injected with LPS (5 mg/kg) for the survival study. In a parallel experiment, the spleen cells from WT mice received Rnf125 Accell siRNA, or the control siRNA were collected at day 2 and lysed in RIPA buffer. The cell lysates were blotted with anti-RNF125 and anti-actin, respectively.
Data analysis and statistical analysis
ELISA data were analyzed by using the Student’s t test. Survival data were analyzed by using the Kaplan-Meier log-rank test. Differences were considered significant at P < 0.05. No animals were excluded from the analysis. Mice were allocated to experimental groups based on their genotypes and were randomized within their sex- and age-matched groups. No statistical method was used to predetermine sample size. It was assumed that normal variance occurred between the experimental groups.
Online supplemental material
Fig. S1 shows that silencing the CBLB gene in human macrophages leads to heightened IL-1β and TNF-α production upon LPS priming and ATP, EHEC, and CTB stimulation. Fig. S2 shows that Cblb−/−Rag1−/− and LysM Cre-Cblbf/f mice are hypersensitive to septic shock induced by a sub-lethal dose of LPS. Fig. S3 shows that Cbl-b targets NLRP3 for K48-linked polyubiquitination in RIPA buffer containing SDS under the denaturing condition. Fig. S4 shows the identification of potential E3 ubiquitin ligases that bind to NLRP3 by LC-MS/MS analysis. Fig. S5 shows the confirmation of RNF125 as the initial E3 ubiquitin ligase to target NLRP3 LRR for K63-linked polyubiquitination and verification of K496 as the ubiquitination site of NLRP3 under the denaturing condition.
Acknowledgments
We thank Dr. J.M. Penninger for providing Cblb−/− mice and Drs. Z. Ronai and F. Martinon for providing RNF125 and NLRP3 constructs. We also thank the Genetically Engineered Mouse Modeling Core and the Proteomics Shared Resource at The Ohio State University Comprehensive Cancer Center and the Proteomics Facility at the University of Iowa Carver College of Medicine for their assistance with generation of Cblbf/f mice and mass spectrometry experiments.
The University of Iowa Proteomic facility is supported by an endowment from the Carver Foundation and by a Howard Hughes Medical Institute grant to Dr. K. Campbell. This work was supported by the US National Institutes of Health (R01 AI090901, R01 AI121196, R01 AI123253, and R21 AI117547 to J. Zhang) and American Heart Association grant-in-aid (#16GRNT26990004; to J. Zhang).
Authors contributions: J. Tang and S. Tu performed most of experiments and analyzed the data; G. Lin, H. Guo, C. Yan, Q. Liu, L. Huang, N. Tang, and Y. Xiao performed some in vitro and in vivo experiments; M.R. Pope designed and supervised NLRP3-associated E3 ubiquitin ligase identification by mass spectrometry; M.V.S. Rajaram designed and performed human macrophage experiments and edited the manuscript; A.O. Amer provided Casp11−/− mice; B.M. Ahmer, J.S. Gunn., and D.J. Wozniak helped with experiments involving EHEC, F. novicida, and P. aeruginosa infections; L. Tao helped with data analysis; C. Vincenzo supervised the generation of Cblbf/f mice; L. Zhang performed the experiments to identify NLRP3 ubiquitination site by mass spectrometry; W.Y. Langdon provided CblbC373A knock-in mice and edited the manuscript; J.B. Torrelles analyzed data and edited the manuscript; S. Lipkowitz provided HA-tagged Cbl-b and Cbl-b mutants; J. Zhang conceived and supervised the research and analyzed data; and J. Tang and J. Zhang wrote the manuscript.
References
- Angus D.C., Linde-Zwirble W.T., Lidicker J., Clermont G., Carcillo J., and Pinsky M.R.. 2001. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 29:1303–1310. 10.1097/00003246-200107000-00002 [DOI] [PubMed] [Google Scholar]
- Bachmaier K., Krawczyk C., Kozieradzki I., Kong Y.Y., Sasaki T., Oliveira-dos-Santos A., Mariathasan S., Bouchard D., Wakeham A., Itie A., et al. 2000. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 403:211–216. 10.1038/35003228 [DOI] [PubMed] [Google Scholar]
- Bachmaier K., Toya S., Gao X., Triantafillou T., Garrean S., Park G.Y., Frey R.S., Vogel S., Minshall R., Christman J.W., et al. 2007. E3 ubiquitin ligase Cblb regulates the acute inflammatory response underlying lung injury. Nat. Med. 13:920–926. 10.1038/nm1607 [DOI] [PubMed] [Google Scholar]
- Bell B.L., Mohapatra N.P., and Gunn J.S.. 2010. Regulation of virulence gene transcripts by the Francisella novicida orphan response regulator PmrA: role of phosphorylation and evidence of MglA/SspA interaction. Infect. Immun. 78:2189–2198. 10.1128/IAI.00021-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosmann M., and Ward P.A.. 2013. The inflammatory response in sepsis. Trends Immunol. 34:129–136. 10.1016/j.it.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P., and Monack D.M.. 2013. Noncanonical inflammasomes: caspase-11 activation and effector mechanisms. PLoS Pathog. 9:e1003144 10.1371/journal.ppat.1003144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y.J., Kole H.K., Brown K., Naramura M., Fukuhara S., Hu R.J., Jang I.K., Gutkind J.S., Shevach E., and Gu H.. 2000. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 403:216–220. 10.1038/35003235 [DOI] [PubMed] [Google Scholar]
- Clausen B.E., Burkhardt C., Reith W., Renkawitz R., and Förster I.. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8:265–277. 10.1023/A:1008942828960 [DOI] [PubMed] [Google Scholar]
- Davies G.C., Ettenberg S.A., Coats A.O., Mussante M., Ravichandran S., Collins J., Nau M.M., and Lipkowitz S.. 2004. Cbl-b interacts with ubiquitinated proteins; differential functions of the UBA domains of c-Cbl and Cbl-b. Oncogene. 23:7104–7115. 10.1038/sj.onc.1207952 [DOI] [PubMed] [Google Scholar]
- Davis B.K., Wen H., and Ting J.P.. 2011. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29:707–735. 10.1146/annurev-immunol-031210-101405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschman C.S., and Tracey K.J.. 2014. Sepsis: current dogma and new perspectives. Immunity. 40:463–475. 10.1016/j.immuni.2014.04.001 [DOI] [PubMed] [Google Scholar]
- Dyszel J.L., Soares J.A., Swearingen M.C., Lindsay A., Smith J.N., and Ahmer B.M.. 2010. E. coli K-12 and EHEC genes regulated by SdiA. PLoS One. 5:e8946 10.1371/journal.pone.0008946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evavold C.L., Ruan J., Tan Y., Xia S., Wu H., and Kagan J.C.. 2018. The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion from Living Macrophages. Immunity. 48:35–44.e6. 10.1016/j.immuni.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantuzzi G., and Dinarello C.A.. 1996. The inflammatory response in interleukin-1 beta-deficient mice: comparison with other cytokine-related knock-out mice. J. Leukoc. Biol. 59:489–493. 10.1002/jlb.59.4.489 [DOI] [PubMed] [Google Scholar]
- Gu B.J., Saunders B.M., Jursik C., and Wiley J.S.. 2010. The P2X7-nonmuscle myosin membrane complex regulates phagocytosis of nonopsonized particles and bacteria by a pathway attenuated by extracellular ATP. Blood. 115:1621–1631. 10.1182/blood-2009-11-251744 [DOI] [PubMed] [Google Scholar]
- Guo H., Qiao G., Ying H., Li Z., Zhao Y., Liang Y., Yang L., Lipkowitz S., Penninger J.M., Langdon W.Y., and Zhang J.. 2012. E3 ubiquitin ligase Cbl-b regulates Pten via Nedd4 in T cells independently of its ubiquitin ligase activity. Cell Reports. 1:472–482. 10.1016/j.celrep.2012.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar J.A., Powell D.A., Aachoui Y., Ernst R.K., and Miao E.A.. 2013. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 341:1250–1253. 10.1126/science.1240988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Jin J., Xu S., Liu H., Li N., and Cao X.. 2010. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat. Immunol. 11:734–742. 10.1038/ni.1908 [DOI] [PubMed] [Google Scholar]
- Harada Y., Harada Y., Elly C., Ying G., Paik J.H., DePinho R.A., and Liu Y.C.. 2010. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J. Exp. Med. 207:1381–1391. 10.1084/jem.20100004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissmeyer V., Macián F., Im S.H., Varma R., Feske S., Venuprasad K., Gu H., Liu Y.C., Dustin M.L., and Rao A.. 2004. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 5:255–265. 10.1038/ni1047 [DOI] [PubMed] [Google Scholar]
- Hutchins N.A., Unsinger J., Hotchkiss R.S., and Ayala A.. 2014. The new normal: immunomodulatory agents against sepsis immune suppression. Trends Mol. Med. 20:224–233. 10.1016/j.molmed.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon M.S., Atfield A., Venuprasad K., Krawczyk C., Sarao R., Elly C., Yang C., Arya S., Bachmaier K., Su L., et al. 2004. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity. 21:167–177. 10.1016/j.immuni.2004.07.013 [DOI] [PubMed] [Google Scholar]
- Kang T.B., Yang S.H., Toth B., Kovalenko A., and Wallach D.. 2013. Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity. 38:27–40. 10.1016/j.immuni.2012.09.015 [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., et al. 2011. Non-canonical inflammasome activation targets caspase-11. Nature. 479:117–121. 10.1038/nature10558 [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Wong M.T., Stowe I.B., Ramani S.R., Gonzalez L.C., Akashi-Takamura S., Miyake K., Zhang J., Lee W.P., Muszyński A., et al. 2013. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 341:1246–1249. 10.1126/science.1240248 [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Stowe I.B., Lee B.L., O’Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T., et al. 2015. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 526:666–671. 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- Kim H., Frederick D.T., Levesque M.P., Cooper Z.A., Feng Y., Krepler C., Brill L., Samuels Y., Hayward N.K., Perlina A., et al. 2015. Downregulation of the Ubiquitin Ligase RNF125 Underlies Resistance of Melanoma Cells to BRAF Inhibitors via JAK1 Deregulation. Cell Reports. 11:1458–1473. 10.1016/j.celrep.2015.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Allen H., Banerjee S., Franklin S., Herzog L., Johnston C., McDowell J., Paskind M., Rodman L., Salfeld J., et al. 1995. Mice deficient in IL-1 β-converting enzyme are defective in production of mature IL-1 β and resistant to endotoxic shock. Cell. 80:401–411. 10.1016/0092-8674(95)90490-5 [DOI] [PubMed] [Google Scholar]
- Li D., Gál I., Vermes C., Alegre M.L., Chong A.S., Chen L., Shao Q., Adarichev V., Xu X., Koreny T., et al. 2004. Cutting edge: Cbl-b: one of the key molecules tuning CD28- and CTLA-4-mediated T cell costimulation. J. Immunol. 173:7135–7139. 10.4049/jimmunol.173.12.7135 [DOI] [PubMed] [Google Scholar]
- Mao K., Chen S., Chen M., Ma Y., Wang Y., Huang B., He Z., Zeng Y., Hu Y., Sun S., et al. 2013. Nitric oxide suppresses NLRP3 inflammasome activation and protects against LPS-induced septic shock. Cell Res. 23:201–212. 10.1038/cr.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.S., Mannino D.M., Eaton S., and Moss M.. 2003. The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 348:1546–1554. 10.1056/NEJMoa022139 [DOI] [PubMed] [Google Scholar]
- Martinon F., and Tschopp J.. 2004. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 117:561–574. 10.1016/j.cell.2004.05.004 [DOI] [PubMed] [Google Scholar]
- Martinon F., Mayor A., and Tschopp J.. 2009. The inflammasomes: guardians of the body. Annu. Rev. Immunol. 27:229–265. 10.1146/annurev.immunol.021908.132715 [DOI] [PubMed] [Google Scholar]
- McCullough J., Clague M.J., and Urbé S.. 2004. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166:487–492. 10.1083/jcb.200401141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S.E., Alves-Filho J.C., Alfaya T.M., da Silva J.S., Ferreira S.H., and Liew F.Y.. 2006. IL-12, but not IL-18, is critical to neutrophil activation and resistance to polymicrobial sepsis induced by cecal ligation and puncture. J. Immunol. 177:3218–3224. 10.4049/jimmunol.177.5.3218 [DOI] [PubMed] [Google Scholar]
- Napier B.A., Brubaker S.W., Sweeney T.E., Monette P., Rothmeier G.H., Gertsvolf N.A., Puschnik A., Carette J.E., Khatri P., and Monack D.M.. 2016. Complement pathway amplifies caspase-11-dependent cell death and endotoxin-induced sepsis severity. J. Exp. Med. 213:2365–2382. 10.1084/jem.20160027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksvold M.P., Dagger S.A., Thien C.B., and Langdon W.Y.. 2008. The Cbl-b RING finger domain has a limited role in regulating inflammatory cytokine production by IgE-activated mast cells. Mol. Immunol. 45:925–936. 10.1016/j.molimm.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Pelegrin P., Barroso-Gutierrez C., and Surprenant A.. 2008. P2X7 receptor differentially couples to distinct release pathways for IL-1beta in mouse macrophage. J. Immunol. 180:7147–7157. 10.4049/jimmunol.180.11.7147 [DOI] [PubMed] [Google Scholar]
- Pétrilli V., Dostert C., Muruve D.A., and Tschopp J.. 2007. The inflammasome: a danger sensing complex triggering innate immunity. Curr. Opin. Immunol. 19:615–622. 10.1016/j.coi.2007.09.002 [DOI] [PubMed] [Google Scholar]
- Piovan C., Amari F., Lovat F., Chen Q., and Coppola V.. 2014. Generation of mouse lines conditionally over-expressing microRNA using the Rosa26-Lox-Stop-Lox system. Methods Mol. Biol. 1194:203–224. 10.1007/978-1-4939-1215-5_11 [DOI] [PubMed] [Google Scholar]
- Pruitt J.H., Copeland E.M. III, and Moldawer L.L.. 1995. Interleukin-1 and interleukin-1 antagonism in sepsis, systemic inflammatory response syndrome, and septic shock. Shock. 3:235–251. 10.1097/00024382-199504000-00001 [DOI] [PubMed] [Google Scholar]
- Py B.F., Kim M.S., Vakifahmetoglu-Norberg H., and Yuan J.. 2013. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell. 49:331–338. 10.1016/j.molcel.2012.11.009 [DOI] [PubMed] [Google Scholar]
- Qiao G., Li Z., Molinero L., Alegre M.L., Ying H., Sun Z., Penninger J.M., and Zhang J.. 2008. T-cell receptor-induced NF-kappaB activation is negatively regulated by E3 ubiquitin ligase Cbl-b. Mol. Cell. Biol. 28:2470–2480. 10.1128/MCB.01505-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao G., Zhao Y., Li Z., Tang P.Q., Langdon W.Y., Yang T., and Zhang J.. 2013. T cell activation threshold regulated by E3 ubiquitin ligase Cbl-b determines fate of inducible regulatory T cells. J. Immunol. 191:632–639. 10.4049/jimmunol.1202068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao G., Ying H., Zhao Y., Liang Y., Guo H., Shen H., Li Z., Solway J., Tao E., Chiang Y.J., et al. 2014. E3 ubiquitin ligase Cbl-b suppresses proallergic T cell development and allergic airway inflammation. Cell Reports. 6:709–723. 10.1016/j.celrep.2014.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram M.V., Ni B., Morris J.D., Brooks M.N., Carlson T.K., Bakthavachalu B., Schoenberg D.R., Torrelles J.B., and Schlesinger L.S.. 2011. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc. Natl. Acad. Sci. USA. 108:17408–17413. 10.1073/pnas.1112660108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V.A., Jiang Z., Waggoner S.N., Sharma S., Cole L.E., Waggoner L., Vanaja S.K., Monks B.G., Ganesan S., Latz E., et al. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11:395–402. 10.1038/ni.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V.A., Vanaja S.K., and Fitzgerald K.A.. 2012a Regulation of inflammasome signaling. Nat. Immunol. 13:333–342. 10.1038/ni.2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V.A., Vanaja S.K., Waggoner L., Sokolovska A., Becker C., Stuart L.M., Leong J.M., and Fitzgerald K.A.. 2012b TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell. 150:606–619. 10.1016/j.cell.2012.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., and Dymecki S.M.. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140. 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Saleh M., Vaillancourt J.P., Graham R.K., Huyck M., Srinivasula S.M., Alnemri E.S., Steinberg M.H., Nolan V., Baldwin C.T., Hotchkiss R.S., et al. 2004. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 429:75–79. 10.1038/nature02451 [DOI] [PubMed] [Google Scholar]
- Saleh M., Mathison J.C., Wolinski M.K., Bensinger S.J., Fitzgerald P., Droin N., Ulevitch R.J., Green D.R., and Nicholson D.W.. 2006. Enhanced bacterial clearance and sepsis resistance in caspase-12-deficient mice. Nature. 440:1064–1068. 10.1038/nature04656 [DOI] [PubMed] [Google Scholar]
- Sarkar A., Hall M.W., Exline M., Hart J., Knatz N., Gatson N.T., and Wewers M.D.. 2006. Caspase-1 regulates Escherichia coli sepsis and splenic B cell apoptosis independently of interleukin-1beta and interleukin-18. Am. J. Respir. Crit. Care Med. 174:1003–1010. 10.1164/rccm.200604-546OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel M., Klein B., Merkle H., Schulz M., Fritsch R., Greten F.R., Arkan M.C., Schneider G., and Schmid R.M.. 2010. IkappaBbeta is an essential co-activator for LPS-induced IL-1beta transcription in vivo. J. Exp. Med. 207:2621–2630. 10.1084/jem.20100864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang Y., Gao W., Ding J., Li P., Hu L., and Shao F.. 2014. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 514:187–192. 10.1038/nature13683 [DOI] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F.. 2015. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 526:660–665. 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- Song H., Liu B., Huai W., Yu Z., Wang W., Zhao J., Han L., Jiang G., Zhang L., Gao C., and Zhao W.. 2016. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat. Commun. 7:13727 10.1038/ncomms13727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian N., Natarajan K., Clatworthy M.R., Wang Z., and Germain R.N.. 2013. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 153:348–361. 10.1016/j.cell.2013.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterwala F.S., Mijares L.A., Li L., Ogura Y., Kazmierczak B.I., and Flavell R.A.. 2007. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204:3235–3245. 10.1084/jem.20071239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J., and Schroder K.. 2010. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 10:210–215. 10.1038/nri2725 [DOI] [PubMed] [Google Scholar]
- Wang S., Miura M., Jung Y.K., Zhu H., Li E., and Yuan J.. 1998. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 92:501–509. 10.1016/S0092-8674(00)80943-5 [DOI] [PubMed] [Google Scholar]
- Wiersinga W.J., Leopold S.J., Cranendonk D.R., and van der Poll T.. 2014. Host innate immune responses to sepsis. Virulence. 5:36–44. 10.4161/viru.25436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff T.J., and Wozniak D.J.. 2001. Transcriptional analysis of genes involved in Pseudomonas aeruginosa biofilms. Methods Enzymol. 336:144–151. 10.1016/S0076-6879(01)36586-2 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Qiao G., Tang J., Tang R., Guo H., Warwar S., Langdon W.Y., Tao L., and Zhang J.. 2015. Protein Tyrosine Phosphatase SHP-1 Modulates T Cell Responses by Controlling Cbl-b Degradation. J. Immunol. 195:4218–4227. 10.4049/jimmunol.1501200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Tang J., Guo H., Zhao Y., Tang R., Ouyang S., Zeng Q., Rappleye C.A., Rajaram M.V., Schlesinger L.S., et al. 2016. Targeting CBLB as a potential therapeutic approach for disseminated candidiasis. Nat. Med. 22:906–914. 10.1038/nm.4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee N.K., and Hamerman J.A.. 2013. β(2) integrins inhibit TLR responses by regulating NF-κB pathway and p38 MAPK activation. Eur. J. Immunol. 43:779–792. 10.1002/eji.201242550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.L., Summers R.M., Li Y., Mohanty S.K., Subramanian M., and Pope R.M.. 2015. Rapid identification and quantitative validation of a caffeine-degrading pathway in Pseudomonas sp. CES. J. Proteome Res. 14:95–106. 10.1021/pr500751w [DOI] [PubMed] [Google Scholar]
- Zanin-Zhorov A., Tal-Lapidot G., Cahalon L., Cohen-Sfady M., Pevsner-Fischer M., Lider O., and Cohen I.R.. 2007. Cutting edge: T cells respond to lipopolysaccharide innately via TLR4 signaling. J. Immunol. 179:41–44. 10.4049/jimmunol.179.1.41 [DOI] [PubMed] [Google Scholar]
- Zhang J., Bárdos T., Li D., Gál I., Vermes C., Xu J., Mikecz K., Finnegan A., Lipkowitz S., and Glant T.T.. 2002. Cutting edge: regulation of T cell activation threshold by CD28 costimulation through targeting Cbl-b for ubiquitination. J. Immunol. 169:2236–2240. 10.4049/jimmunol.169.5.2236 [DOI] [PubMed] [Google Scholar]
- Zhang A.Q., Zeng L., Gu W., Zhang L.Y., Zhou J., Jiang D.P., Du D.Y., Hu P., Yang C., Yan J., et al. 2011. Clinical relevance of single nucleotide polymorphisms within the entire NLRP3 gene in patients with major blunt trauma. Crit. Care. 15:R280 10.1186/cc10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zougman A., Selby P.J., and Banks R.E.. 2014. Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics. 14:1006–1000. 10.1002/pmic.201300553 [DOI] [PubMed] [Google Scholar]