Cathryn R. Nagler discusses the opportunities of creating new microbiome-based drugs.
Abstract
Microbiome-modulating therapeutics have the potential to revolutionize medicine. Here, I discuss the challenges and opportunities inherent in creating new microbiome-based drugs.
32 million Americans currently suffer from food allergies. When I was a child, peanut butter and jelly was often a school-day lunch. By the time my children finished elementary school, peanuts were banned from their classrooms to protect students with life-threatening peanut allergies. The start of the 21st century brought striking increases in food allergies as well as other noncommunicable diseases, including obesity, inflammatory bowel disease, autism, and asthma, among others. What can account for this marked generational change? We (and others) have proposed that it is the absence of immunoregulatory signals formerly provided by protective intestinal bacteria (Prioult and Nagler-Anderson, 2005). Reintroducing these bacteria, or their metabolites, has the potential to create a new class of drugs to prevent or treat these increasingly prevalent diseases.
Identity crisis
We have learned a lot about the microbiome during the past 15 yr. We had greatly underestimated the quantity, complexity, and diversity of our microbial inhabitants because the majority are anaerobes which, until recently, were not readily culturable by standard microbiological methods. Current estimates suggest that resident bacteria equal or slightly outnumber the cells of their human hosts (Sender et al., 2016). Microbes populate the skin and all mucosal surfaces, but most are found in the gut, which contains trillions of bacteria (not to mention the less well-studied bacteriophages, viruses, fungi, and archaea that permeate these complex communities). Culture-independent sequencing methodologies opened the window to a vast microbial world we are only beginning to understand and explore. Many reports describing associations of commensal bacteria with human diseases initially focused on the changing abundance of various taxa in healthy and pathological states. Emerging evidence suggests that abundance may not be the most important variable; low abundance populations can have outsized effects (Fischbach et al., 2018). Moreover, the microbiota fluctuates in response to environmental factors, particularly diet, making it difficult to interpret human samples collected at a single point in time. Several long-term studies have concluded that the microbiome as a whole is generally stable within an individual over time, but certain taxa are more susceptible to change (Faith et al., 2013). Numerous investigators turned to mouse models, but many studies have drawn conclusions about the role of the microbiota based on experimental designs which have, more often than not, been inadequately controlled for baseline differences between the variables to be tested and those designated as controls (Stappenbeck and Virgin, 2016). Ironically, the controls that are in place in most academic mouse colonies may limit their utility as models of human diseases; recent work shows that “wildling” mice, genetically inbred strains colonized with the microbiota of wild mice, are better translational models for human disease than their specific pathogen–free counterparts (Rosshart et al., 2019). Therefore, despite abundant evidence that the microbiome profoundly influences health and disease, we still lack a clear definition of what constitutes health.
Missing microbes
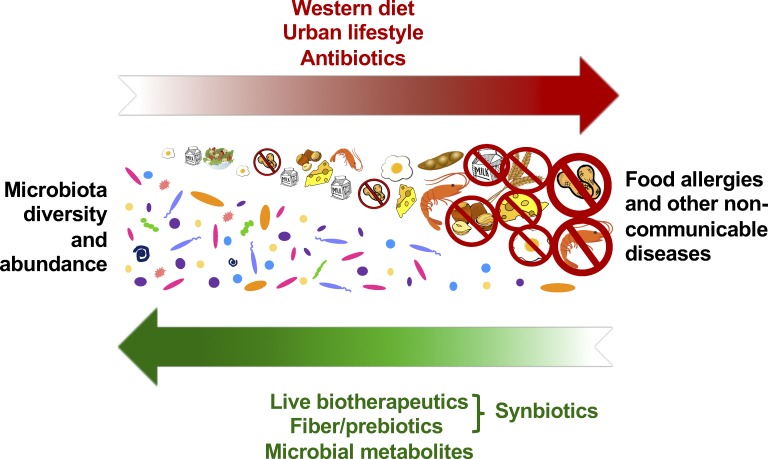
Examination of fecal samples from preindustrial societies clearly demonstrates a marked loss in diversity in our 21st century microbiome (Smits et al., 2017). Many lifestyle factors have contributed to reduced microbial diversity including the use (and abuse) of antibiotics and antimicrobials and living conditions that minimize exposure to the outside world and its attendant microbes. Diet also plays a critical and perhaps uniquely manipulable role. Intestinal bacteria ferment dietary fiber and produce metabolites, such as short chain fatty acids, which are critical to intestinal health. Declining fiber consumption (common in our fast-food society) can be directly tied to reduced microbial diversity in murine models (Sonnenburg et al., 2016). Immigration to the United States and transition to a low-fiber Western diet results in loss of specific taxa within a single generation (Vangay et al., 2018). The microbes are not only important in defining the microbiome and its impact on health, but also the metabolites that they produce. The short chain fatty acid butyrate, in particular, is an important energy source for colonic epithelial cells (Byndloss et al., 2018). Fiber-digesting, butyrate-producing bacteria, particularly those in the Clostridia class, induce an IL-22–dependent barrier protective response, which promotes homeostatic tolerance between bacteria and host (Stefka et al., 2014). These data suggest that, while reestablishment of the community structure of our 20th century microbiomes is unrealistic, it may be possible to target selected key taxa, their metabolites, and induced gene products for reintroduction to achieve at least partial restoration of function (Fig. 1).
Figure 1.
Decreased microbial diversity and abundance are causal to the increasing prevalence of food allergies and other noncommunicable diseases. Modern lifestyle factors including antibiotic use, urban housing, and Western diet have depleted populations of gut bacteria, such as butyrate-producing Clostridia, that perform barrier-protective functions critical to the maintenance of both physiological and immunological homeostasis. Reintroduction of selected bacterial taxa or their metabolites, particularly in the context of fiber and prebiotics, may restore mucosal function to prevent or treat disease.
Harnessing the microbiome’s potential
Purposeful consumption of bacteria is not a new phenomenon; probiotics have been used for thousands of years in foods like yogurt with claims of promoting health, although there is little evidence for therapeutic efficacy (NASPGHAN Nutrition Report Committee, 2006). The next generation of probiotics will exploit our growing knowledge of the obligate anaerobes that comprise the majority of our intestinal inhabitants. Transplantation of donor feces has been successful for the treatment of Clostridium difficile colitis (Bakken et al., 2011) but, given all that remains unknown about the microbiota, this approach must be used with great caution. The microbiome-modulating therapeutics of the future will be comprised of well-characterized anaerobic bacteria (or their metabolites) administered alone or in consortia. Whole genome sequencing will provide information on antibiotic susceptibility (and resistance) to allow for depletion of administered bacteria if needed. Turning oxygen-sensitive anaerobes into drugs is fraught with challenges, the most obvious of which is exposure to oxygen en route from the laboratory or pharmacy to the bedside. Some bacteria may be orally deliverable in pills as spores or in freeze-dried formulations. But will they colonize? It has generally been difficult to introduce new live biotherapeutics into a replete microbiota (whether healthy or dysbiotic). Pre-treatment with antibiotics to open a niche may be necessary but has inherent risks in an already compromised host. Another less commonly explored option is to use synbiotic formulations, combinations of bacteria and their preferred prebiotics, including fiber. Synbiotics address the difficulty of achieving colonization by increasing the competitive opportunities of the therapeutic rather than depleting resident microbes with antibiotics. This method also allows for controlled and intentional manipulation of the therapeutic bacteria’s metabolic activity in vivo, effectively using dietary prebiotics to maximize output of desired metabolites.
Direct administration of microbial metabolites is another promising avenue. The challenge in this case will be delivering the metabolite in bioactive form to target sites in the distal small intestine or colon, surviving the gauntlet of acidic pH and proteolytic enzymes that mark that journey. What is the physiological dose (or concentration) necessary for therapeutic efficacy? What frequency of administration will be necessary to maintain bioactivity? Will a single microbial metabolite be sufficient to induce protective effects? Is ileal/colonic absorption of the metabolite critical for efficacy or can systemic administration achieve the same effects? These are difficult questions that will need to be determined empirically, initially in animal models. It is possible, however, that the animal model findings may not be transferrable directly to treat human disease, and combination approaches may be required. Co-delivering microbes, prebiotics, and metabolites together may increase efficacy by enhancing colonization or stimulating multiple related metabolic programs. One approach we envision will combine administration of a microbiome-modulating bioactive polymer with a butyrate-producing live biotherapeutic that we have shown has a causal role in preventing allergic responses to food in a murine model (Feehley et al., 2019). To enhance colonization and bioactivity of this live biotherapeutic in vivo, we will identify optimal sources of both growth-promoting metabolites and the specific fibers that are most efficiently fermented to produce butyrate.
Although, as we have summarized, many challenges exist, drugging the microbiome presents enormous opportunities for new medicines. Clinical trials of microbiome-modulating therapeutics are already underway. There is good reason to hope that this new class of drugs will halt or reverse the progression of noncommunicable diseases, including food allergies, for future generations.
Acknowledgments
I thank L.A. Hesser, S. Cao, M. Mimee, and J.A. Hubbell for their helpful comments and edits to this viewpoint and L.A. Hesser for the figure artwork.
This work was supported by grants from the National Institute of Health (AI146099 and AI106302) and The Sunshine Charitable Foundation.
The author is the president and co-founder of ClostraBio, Inc.
References
- Bakken J.S., et al. . 2011. Clin. Gastroenterol. Hepatol. 10.1016/j.cgh.2011.08.014 [DOI] [Google Scholar]
- Byndloss M.X., et al. . 2018. Mucosal Immunol. 10.1038/s41385-018-0010-y [DOI] [PubMed] [Google Scholar]
- Faith J.J., et al. . 2013. Science. 1237439 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feehley T., et al. . 2019. Nat. Med. 10.1038/s41591-018-0324-z [DOI] [Google Scholar]
- Fischbach M.A. 2018. Cell. 10.1016/j.cell.2018.07.038 [DOI] [Google Scholar]
- NASPGHAN Nutrition Report Committee 2006. J. Pediatr. Gastroenterol. Nutr. 10.1097/01.mpg.0000239990.35517.bf [DOI] [Google Scholar]
- Prioult G., and Nagler-Anderson C.. 2005. Immunol. Rev. 10.1111/j.0105-2896.2005.00277.x [DOI] [PubMed] [Google Scholar]
- Rosshart S.P., et al. . 2019. Science. 10.1126/science.aaw4361 [DOI] [Google Scholar]
- Sender R., et al. . 2016. Cell. 10.1016/j.cell.2016.01.013 [DOI] [Google Scholar]
- Smits S.A., et al. . 2017. Science. 10.1126/science.aan4834 [DOI] [Google Scholar]
- Sonnenburg E.D., et al. . 2016. Nature. 10.1038/nature16504 [DOI] [Google Scholar]
- Stappenbeck T.S., and Virgin H.W.. 2016. Nature. 10.1038/nature18285 [DOI] [PubMed] [Google Scholar]
- Stefka A.T., et al. . 2014. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1412008111 [DOI] [Google Scholar]
- Vangay P., et al. . 2018. Cell. 10.1016/j.cell.2018.10.029 [DOI] [Google Scholar]