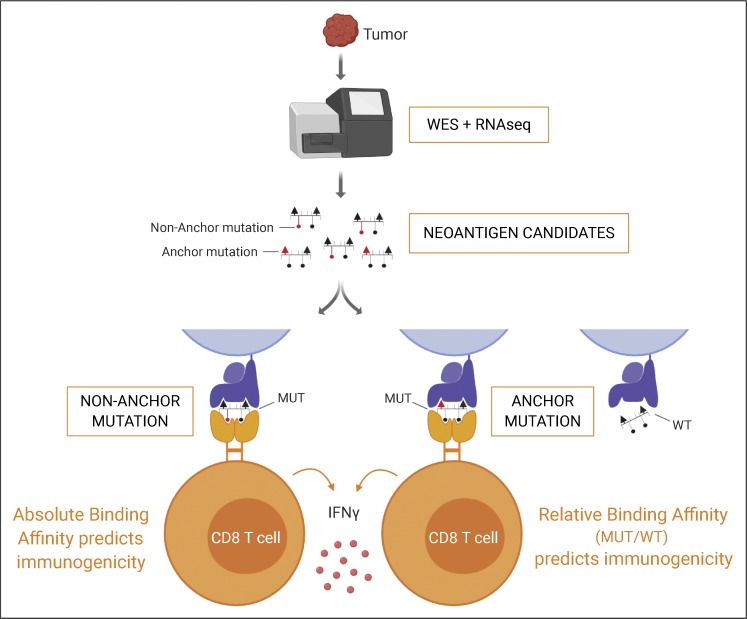
Cancer mutations create neoantigens that are of great interest for immunotherapy, but their accurate identification remains a challenge. Capietto et al. show that mutation position is an important determinant for immunogenicity, and incorporating this feature into prediction algorithms improves their identification.
Abstract
Tumor-specific mutations can generate neoantigens that drive CD8 T cell responses against cancer. Next-generation sequencing and computational methods have been successfully applied to identify mutations and predict neoantigens. However, only a small fraction of predicted neoantigens are immunogenic. Currently, predicted peptide binding affinity for MHC-I is often the major criterion for prioritizing neoantigens, although little progress has been made toward understanding the precise functional relationship between affinity and immunogenicity. We therefore systematically assessed the immunogenicity of peptides containing single amino acid mutations in mouse tumor models and divided them into two classes of immunogenic mutations. The first comprises mutations at a nonanchor residue, for which we find that the predicted absolute binding affinity is predictive of immunogenicity. The second involves mutations at an anchor residue; here, predicted relative affinity (compared with the WT counterpart) is a better predictor. Incorporating these features into an immunogenicity model significantly improves neoantigen ranking. Importantly, these properties of neoantigens are also predictive in human datasets, suggesting that they can be used to prioritize neoantigens for individualized neoantigen-specific immunotherapies.
Graphical Abstract
Introduction
Tumor-specific mutations can generate neoantigens if peptides containing the mutation are presented on the surface of tumor cells by MHC class I (MHC-I) molecules and are recognized by T cells as neoepitopes. These neoepitopes have the potential to be highly immunogenic, because they have never been seen by the immune system and thus never subjected to central tolerance.
Recent studies have shown that neoepitopes drive effective CD8 T cell responses in cancer patients. Neoantigen-specific CD8 T cells expand in response to checkpoint blockade immunotherapies (Rizvi et al., 2015; Snyder et al., 2014), and high neoantigen or mutation burden is generally predictive of response to checkpoint blockade (Hellmann et al., 2018; Le et al., 2015; Nathanson et al., 2017; Rizvi et al., 2015; Snyder et al., 2014; Van Allen et al., 2015). Finally, adoptive transfer of tumor infiltrating T lymphocytes recognizing neoantigens has been shown to mediate tumor regression in patients (Tran et al., 2014, 2016; Zacharakis et al., 2018). These studies present a strong rationale for developing immunotherapies targeting neoantigens, including vaccines.
The vast majority of somatic mutations in cancer are unique to each patient and thus require the design of an individualized vaccine tailored for each patient. Recent advances in next-generation sequencing have enabled the rapid identification of somatic mutations and make such individualized therapies feasible. One challenge, however, is the accurate identification of immunogenic neoantigens. Only a small fraction of mutations are processed into peptides, presented on MHC-I, and recognized as foreign by the immune system (Capietto et al., 2017). Computational predictions have been successfully used to identify neoantigens that elicit T cell responses in preclinical models (Duan et al., 2014; Gubin et al., 2014; Yadav et al., 2014), as well as in human vaccination studies (Carreno et al., 2015; Ott et al., 2017; Sahin et al., 2017). To a great extent, these computational methods predict neoantigens based on the predicted binding affinity (BA) of the mutated peptides to MHC-I molecules. Since the BA of a peptide to MHC-I plays an important role in determining its presentation to CD8 T cells, the approach is conceptually attractive. In fact, early studies demonstrated that viral peptides with high affinity to MHC-I tend to be more immunogenic (Sette et al., 1994a,b).
The current success rate of neoepitope predictions remains poor, however, primarily because methods do not accurately take into account a variety of other features that can influence the functional presentation of mutant peptides, such as intracellular protease activity, antigen expression level, and T cell receptor repertoire. Studies have also suggested that peptide/MHC-I stability may be a better predictor of immunogenicity than peptide affinity (Harndahl et al., 2012; Strønen et al., 2016; van der Burg et al., 1996).
Another important determinant of immunogenic potential is the binding of the presented peptides to the TCR. The affinity of the TCR for the peptide/MHC-I complex has been shown to impact immunogenicity, and several studies have identified that immunogenic microbial peptides are enriched for residues with certain physicochemical properties at TCR contact residues (Calis et al., 2013; Chowell et al., 2015). In addition, it has recently been proposed that mutant peptides with sequence homology to immunogenic microbial epitopes may be recognized by cross-reactive T cells and therefore are more likely immunogenic (Balachandran et al., 2017; Łuksza et al., 2017; Snyder et al., 2014). Finally, the MHC-I peptide must be recognized as foreign by the immune system. The majority of cancer mutations result from a single amino acid substitution that may not always be sufficient for the immune system to distinguish the mutant peptide from its WT (self) counterpart. Indeed, our earlier study suggested that mere presentation of mutated peptides was insufficient for immunogenicity, and that peptides with mutations predicted to be in contact with the TCR, and thus more recognizably different from the WT counterpart, are more likely to be immunogenic (Yadav et al., 2014). Alternatively, mutations that increase peptide BA to MHC-I in comparison to the WT counterpart may be more likely to be immunogenic if they result in a bona fide presentation of novel epitopes not previously seen by the immune system. Duan et al. (2014) identified the differential agretopic index, i.e., the difference in predicted MHC-I BA between WT and corresponding mutant peptides, as a superior predictor of antigenicity compared with the predicted absolute affinity of a candidate neoepitope. Other more recent studies have also suggested that neoepitopes with increased affinity for MHC-I in comparison to the WT counterpart may be more immunogenic (Balachandran et al., 2017; Łuksza et al., 2017). In this study, we aimed to further characterize the properties of immunogenic neoepitopes and found that it is possible to significantly refine the predictive power of peptide BA data by incorporating a more precise view of how individual mutations may or may not alter interactions with MHC-I molecules.
Results
Assessing immunogenicity of neoantigen candidates in mouse models
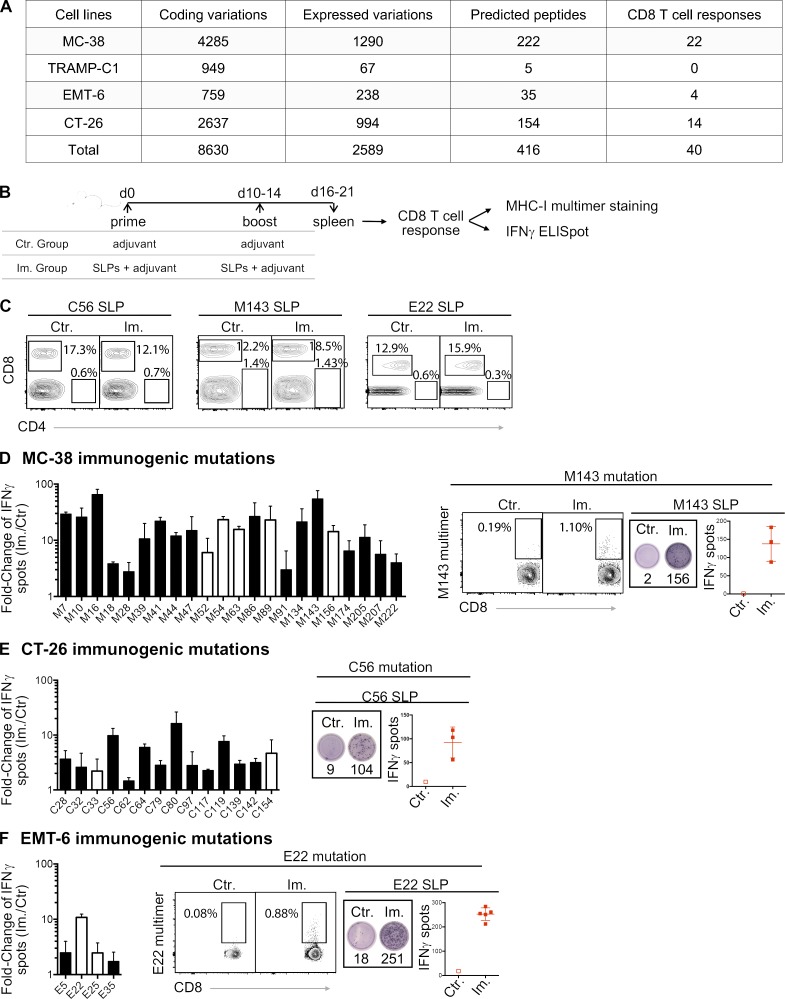
We performed whole-exome sequencing and RNA sequencing (RNA-seq) on four mouse tumor cell lines (MC-38, TRAMP-C1, EMT-6, and CT-26) to identify single nucleotide variants with evidence of expression of the mutated alleles (Fig. 1 A and Data S1). We then selected 416 mutations across the four cell lines that may generate neoantigen candidates based on scanning all 8–11-mer peptides containing a mutated residue and predicting their BA to the MHC-I alleles of the mouse strain from which the cell lines were derived (Fig. 1 A). We identified a total of 409 unique long peptides that harbored the mutation in the middle across the four mouse tumor cell lines and tested their immunogenicity by immunizing healthy mice with synthetic long peptides (SLPs) in combination with two adjuvants, polyinosinic:polycytidylic acid (poly(I:C)) and agonistic anti-CD40 antibody (Fig. 1 B). These 24-mer SLPs require internalization and proteasome-dependent processing by dendritic cells for efficient presentation on MHC-I to T cells (Rosalia et al., 2013). Antigenicity was assessed by IFN-gamma release following in vitro restimulation of CD8 T cells (CD4-depleted splenocytes; Fig. 1 C) with the mutant SLP by ELISpot assay and/or MHC-I/peptide multimer staining of CD8 T cells (Fig. 1 B). Neoantigens from 40 mutations out of the 409 predicted candidates induced CD8 T cell responses (Fig. 1, A and D–F). These results confirmed that a large majority of candidate neoantigens, as predicted by BA alone, are not antigenic with this vaccine, consistent with previous observations (Gubin et al., 2014; Kreiter et al., 2015; Ott et al., 2017; Sahin et al., 2017; Yadav et al., 2014).
Figure 1.
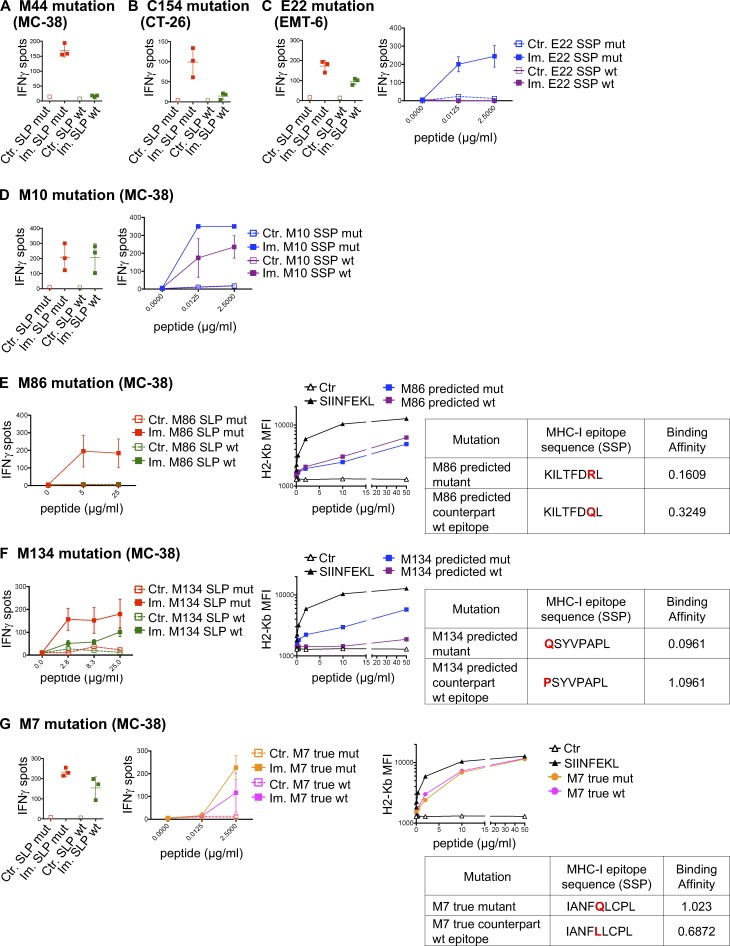
MHC-I immunogenicity of neoantigen candidates. (A) Number of exome and transcript variations, predicted peptides, and detected CD8 T cell responses for four mouse tumor cell lines (MC-38, TRAMP-C1, EMT-6, and CT-26). (B) Schema depicting the study protocol. Naive mice were immunized with mutant predicted SLPs + adjuvant (Im., immunized; adjuvants: anti-CD40 antibody and poly(I:C)) or adjuvant only (Ctr, control) on day 0 and days 10–14. CD8 T cell responses were assessed in spleen 6–7 d following the last immunization, either by IFN-gamma ELISpot (after CD4 T cell depletion) or MHC-I multimer staining. (C) Representative flow cytometry graphs from purity analysis of CD4 depletion from experiments assessing CD8 T cell responses shown in D–F. Purity of the depletion was ≥98%. (D–F) Representative data (from one experiment, repeated 3–11 independent times) of detected MHC-I immunogenic mutations are shown for each mouse model (D-MC-38, E-CT-26, and F-EMT-6). Anchor (white) and nonanchor (black) mutations are shown for each tumor model. Mean (± SD) fold-change of IFN-gamma spot numbers from CD8 T cells (5 × 105 CD4-depleted splenocytes/well) between control (Ctr., n = 1) and immunized (Im., n = 3–5) mice after overnight stimulation with the mutant predicted SLP (25 µg/ml). Representative data and pictures of mutant-specific MHC-I multimer staining and/or IFN-gamma ELISpot for one mutation are shown for each mouse model. Each image is a representative well from an ELISpot plate.
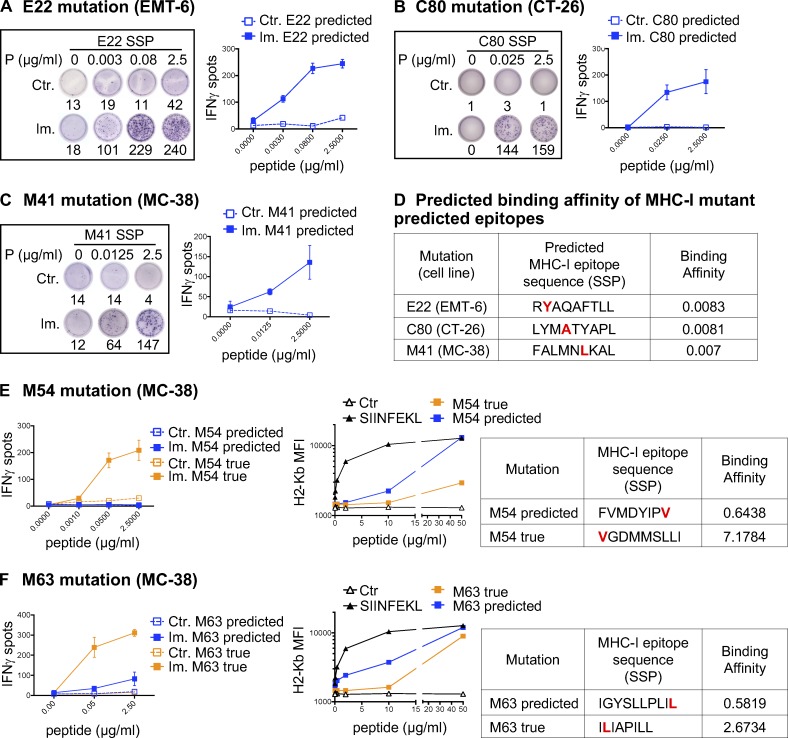
For 39 of 40 antigenic mutations, we assessed the immunogenic potential of the predicted optimal neoepitopes (i.e., of the subsequence of the mutant SLP that achieved the optimal binding prediction) by using synthetic short peptides (SSPs) in an in vitro T cell restimulation assay and/or by T cell staining with MHC-I/peptide multimers (Fig. 2, A–C; and Table S1). In some cases, our data suggest that multiple epitopes containing the same mutation may be antigenic (Fig. S1 and not depicted). This was not unexpected, as for each mutation, we selected the predicted neoepitope with the highest BA among the multiple epitopes within the SLP that are predicted to bind MHC-I and be presented (Fig. 2 D).
Figure 2.
Validation of MHC-I optimal predicted mutant epitopes. (A–C) Representative pictures of ELISpot wells and mean (± SD) IFN-gamma spot numbers from CD8 T cells (5 × 105 CD4-depleted splenocytes/well) from control (Ctr., n = 1) or immunized (Im., n = 3) mice (in vivo protocol as in Fig. 1 B) after stimulation with the predicted optimal mutant SSPs (A-E22 mutation, EMT-6; B-C80 mutation, CT-26; C-M41 mutation, MC-38). Each image is a representative well from an ELISpot plate. (D) Sequence and BA (percentile rank) of the predicted optimal mutant epitopes in A–C (E22, H-2Kd; C80, H-2Kd; M41, H-2Db). (E and F) Mean (± SD) IFN-gamma spot numbers from CD8 T cells (5 × 105 CD4-depleted splenocytes/well) from control (Ctr., n = 1) or immunized (Im., n = 3) mice (in vivo protocol as in Fig. 1 B) after stimulation with predicted and true mutant optimal epitopes (MC-38; E-M54 mutation; F-M63 mutation). Binding assay of the optimal and true SSPs to H-2Kb on Tap-1 KO EL-4 cells is shown for M54 and M63, as well as the sequence and BA (percentile rank) of the predicted optimal mutant epitopes. Each experiment was performed twice independently.
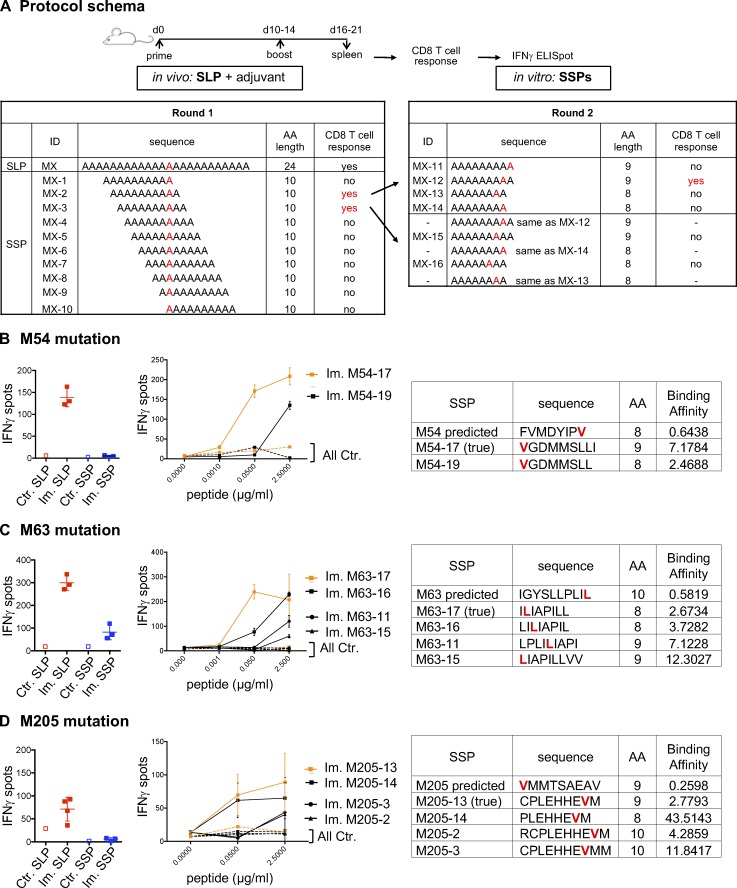
Figure S1.
Immunogenicity of predicted MHC-I optimal mutant epitopes. (A) CD8 T cells (5 × 105 CD4-depleted splenocytes/well) from control (Ctr., n = 1) or immunized (Im., n = 3) mice with mutant SLPs were isolated following the in vivo protocol as shown in Fig. 1 B and in vitro cultured with SLPs or SSPs to assess IFN-gamma release by ELISpot assay. To determine alternative neoepitopes of the poorly or nonimmunogenic predicted neoepitope candidate, IFN-gamma release was measured by neoantigen-specific CD8 T cells generated on SLP vaccines and restimulated in vitro with overlapping 10-, 9-, or 8-mer peptides containing the mutation. (B–D) Representative data of IFN-gamma spots (mean ± SD) from MC-38 mutations (B, M54; C, M63; D, M205) comparing the predicted mutant SLP (25 µg/ml) and optimal SSP (2.5 µg/ml) are shown, as well as the dose response (mean ± SEM to various concentrations) to 10-, 9-, or 8-mer immunogenic alternate optimal mutant peptides. Sequence, length, and predicted BA (H-2Kb for M54 and M63; H-2Db for M205) of each peptide (mutation in red) are shown. Each experiment was independently repeated twice.
In 43.6% of the 39 cases, neoantigen-specific CD8 T cells generated upon vaccination with SLPs did not recognize (13 neoantigens) or poorly recognized (4 neoantigens) the best predicted neoepitope candidate (Fig. 2, E and F; and Table S1). For 10 mutations where the predicted optimal binder was poorly antigenic or not at all, we made overlapping peptide libraries from the SLP sequence to determine alternative neoepitopes and tested 10-, 9-, and 8-mer neoepitope candidates for each of these mutations (Fig. S1 A and not depicted; see Materials and methods for details). Where we successfully identified the alternative neoepitope eliciting the strongest CD8 T cell response in the ELISpot assay, we refer to it as the “true neoepitope” (Fig. 2, E and F; and Fig. S1). Surprisingly, all of the true neoepitopes from these mutations were predicted to be weak MHC-I binders (i.e., their predicted percentile rank was >2 for all alleles; Fig. 2, E and F; and Fig. S1). In line with a recent study (Ebrahimi-Nik et al., 2019), these results clearly show that peptide binding ability to MHC-I is an insufficient predictor of immunogenicity.
We then experimentally assessed the MHC-I binding of the true neoepitopes by measuring the peptide’s ability to stabilize MHC-I on the surface of transporter associated with antigen processing (TAP)–deficient EL-4 cells. As a positive control, we used the well characterized H-2Kb binding peptide, SIINFEKL, derived from ovalbumin (Fig. 2, E and F). All true neoepitopes tested were found to bind H-2Kb, although their measured binding affinities were either lower than or comparable to those of the initially predicted neoepitopes (Fig. 2, E and F). As we were able to assess only the binding to H-2Kb, we cannot exclude the possibility that these true neoepitopes might show a higher BA to H-2Db.
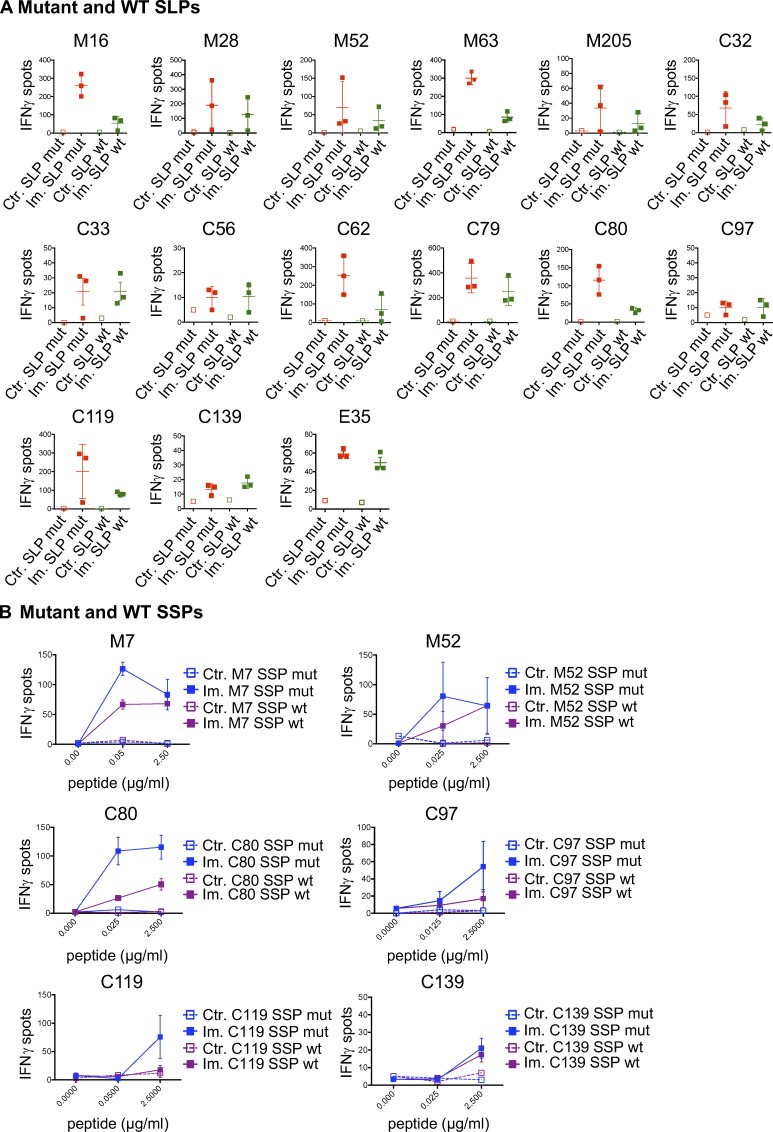
We next assessed the cross-reactivity of the neoantigen-specific CD8 T cells to the WT counterpart of the predicted neoepitope. Mice were immunized against mutant SLPs, and neoantigen-specific CD8 T cells were in vitro restimulated with either mutant or WT SLPs or SSPs. The 40 immunogenic mutations were evaluated, and in 45% (18 of 40) of the cases, we observed IFN-gamma release by neoantigen-specific CD8 T cells stimulated with both the mutated and WT counterpart peptides, demonstrating some degree of cross-reactivity of the neoantigen-induced TCRs to WT peptides (Fig. 3, Table S1, and Fig. S2). While it was a surprisingly high rate of cross-reactivity, recognition of the WT counterpart by the neoantigen-specific TCR was generally weaker (Fig. 3, Table S1, and Fig. S2) and independent of the predicted binding affinities (Table S2).
Figure 3.
Cross-reactivity between MHC-I mutant and WT counterpart neoantigens. (A–G) Representative data of mean ± SD IFN-gamma spot numbers from CD8 T cells (5 × 105 CD4-depleted splenocytes/well) from control mice (Ctr., n = 1) or mice immunized with mutant SLPs (Im., n = 3) as depicted in Fig. 1 B, after in vitro restimulation with the mutant SLP or its WT counterpart (25 µg/ml; A-M44 mutation, MC-38; B-C154 mutation, CT-26; C-E22 mutation, EMT-6; D-M10 mutation, MC-38; E-M86 mutation, MC-38; F-M134 mutation, MC38; and G-M7 mutation, MC-38) or with the predicted mutant optimal epitope SSP or its WT counterpart (C, D, and G). Binding assay of the optimal mutant and its WT counterpart SSPs to H-2Kb on Tap-1 KO EL-4 cells is shown, as well as the sequence and the BA (percentile rank) of the predicted optimal epitopes (E–G). Each experiment was performed twice independently.
Figure S2.
Cross-reactive mutant MHC-I neoantigen-specific T cells. (A and B) CD8 T cells (5 × 105 CD4-depleted splenocytes/well) from control (Ctr., n = 1) or immunized (Im., n = 3) mice with mutant SLPs were isolated following the in vivo protocol shown in Fig. 1 B and in vitro cultured with mutant or WT counterpart SLPs (A; 25 µg/ml) or SSPs (B; 0–2.5 µg/ml) to assess cross-reactivity through IFN-gamma release by ELISpot assay. Representative data of IFN-gamma spots (mean ± SEM) from MC-38 mutations (M#), CT-26 (C#), and EMT-6 (E#) are shown. Each experiment was independently repeated twice.
The position of the mutation defines MHC-I affinity of a peptide as a determinant of immunogenicity
We next characterized computationally predictable properties of the immunogenic mutations. Previous work had suggested that predicted MHC-I BA of peptides correlates with their immunogenicity potential (Sette et al., 1994b). In our study, we found that the predicted absolute BA (calculated as a percentile rank using NetMHCpan 4.0) of immunogenic neoepitopes was stronger than that of the nonimmunogenic candidates (Fig. 4 A), with mean predicted BA values of 0.2 for immunogenic neoepitopes versus 0.5 for nonimmunogenic. Note that since we preselected candidate neoantigens for antigenicity testing based on strong binding affinities, the mean BA values in such a comparison are conditional on that selection. We also obtained affinity and immunogenicity data for 81 neoepitopes identified in human studies (Data S1), including a compilation of reports of spontaneous immunity to neoepitopes in cancer patients (Fritsch et al., 2014), human vaccination studies (Carreno et al., 2015), and studies testing predicted neoepitopes for immunogenicity in cancer patients or healthy donors (Cohen et al., 2015; Strønen et al., 2016). We observed a similar trend in the human dataset, in which the absolute affinity of human neoepitopes was significantly higher than that of the nonimmunogenic candidates, with mean predicted BA values of 0.4 and 1.3, respectively (Fig. 4 A).
Figure 4.
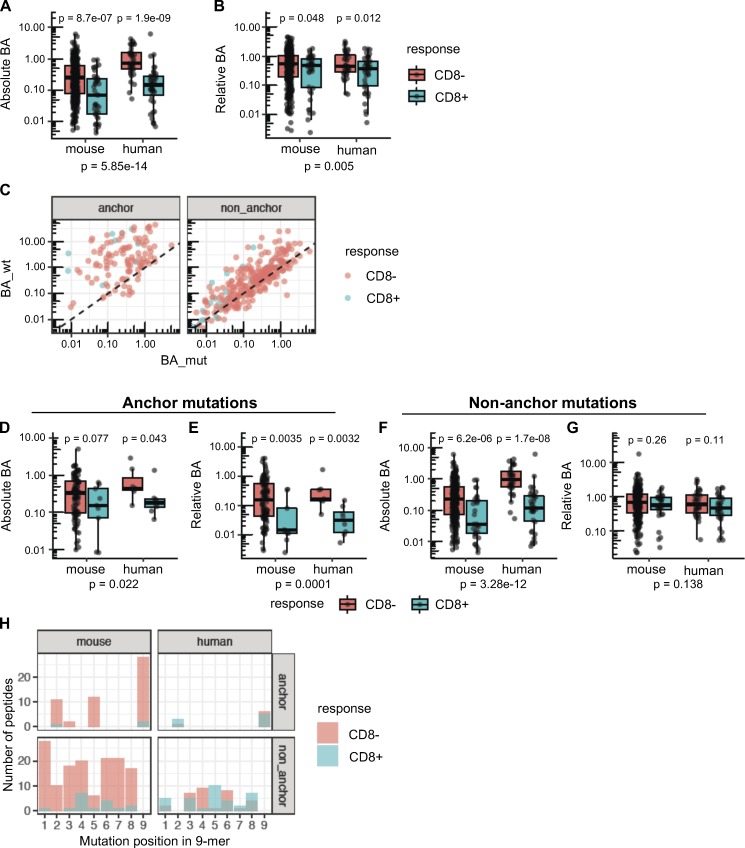
Position of the mutation determines the importance of absolute or relative affinity for immunogenicity of neoepitopes. (A and B) Absolute BA (A) and relative BA (B) of candidate neoepitopes are shown for the neoantigens that induced CD8 T cell responses (blue) and for the neoantigens that were nonimmunogenic (red). A candidate neoepitope for a given mutation was identified as the peptide that had the best BA value of all 8–11-mer mutant peptides across all MHC-I alleles. P values in the plots are shown based on a t test on log-transformed values. P values in the subtitles are meta-analysis P values calculated using Fisher’s method. No distinction of anchor or nonanchor mutation was made here. Number of data points: 369 (mouse, CD8−), 40 (mouse, CD8+), 39 (human, CD8−), and 42 (human, CD8+). (C) Predicted BA values of mouse candidate neoepitopes and their WT counterparts are shown as scatter plots (Spearman correlation coefficient 0.81 and 0.32 for nonanchor and anchor mutations, respectively). Both anchor and nonanchor mutated peptides contain cases that show CD8 responses. Both anchor and nonanchor scatter plots show significant correlation in predicted BA between the mutant peptides and their WT counterparts (Spearman correlation test P value of 5.16 × 10−4 and 6.39 × 10−70 for anchor [n = 113] and nonanchor [n = 296] cases, respectively). (D–G) Predicted absolute BA of the neoepitope (D) and the predicted relative BA of the neoepitope to its WT counterpart (E) are shown, specifically for neoepitopes mutated at anchor residues. Number of data points: 104 (mouse, CD8−), 9 (mouse, CD8+), 7 (human, CD8−), and 8 (human, CD8+). Similarly, F and G show the predicted absolute BA and predicted relative BA at nonanchor mutations. Number of data points: 265 (mouse, CD8−), 31 (mouse, CD8+), 32 (human, CD8−) and 34 (human, CD8+). P values in the plots are based on t test, while the P values in the subtitles are meta-analysis P values calculated using Fisher’s method. (H) Number of peptides mutated at each position. Only 9-mer peptides are shown to retain comparability of positions across peptides, as a majority of the predicted neoepitopes were 9-mers. Peptide counts of immunogenic and nonimmunogenic peptides are shown as overlapping bar plots.
More recently, it has been proposed that differential MHC-I BA between the mutant epitope and its WT counterpart is a superior predictor of antigenicity than the absolute affinity of a candidate neoepitope (Balachandran et al., 2017; Duan et al., 2014; Ghorani et al., 2018; Rech et al., 2018). Using our systematic dataset, we defined relative BA as the ratio of the BA values of the mutant epitope and the corresponding WT epitope to the same HLA allele. Relative BA showed a weaker difference between immunogenic and nonimmunogenic neoepitope candidates compared with absolute affinity, when all the candidates were evaluated (Fig. 4 B).
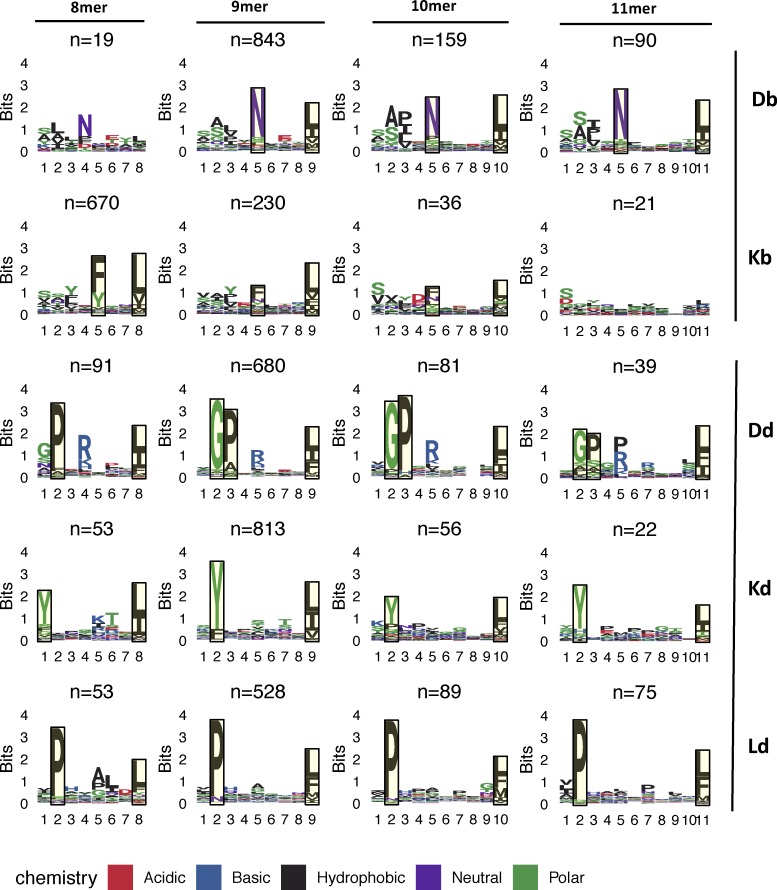
We then reasoned that relative BA might be particularly relevant among the subset of neoepitopes with a mutation at the anchor residue, while absolute BA, which is the MHC-I affinity of a mutant peptide independent of its WT counterpart, should always be a predictor of immunogenicity. To enable anchor residue identification for the mouse alleles from our cell lines across different peptide lengths, we first determined the MHC peptidome eluted from MC-38 and EMT-6 mouse tumor cell lines to further specify the MHC binding motifs for H-2Kb, H-2Db, H-2Kd, H-2Ld, and H-2Dd alleles (Fig. 5). We then determined the anchor positions for different peptide lengths for each allele (see Materials and methods for details) and classified the mutations as either anchor or nonanchor mutations. Immunogenic peptides were identified at similar rates in both anchor or nonanchor mutated peptides, at 10.5% (31/296) and 8% (9/113), respectively (Fig. 4 C; Fisher’s exact test P value of 0.58). The strength of the T cell response, measured by IFN-gamma spot count, also did not appear to differ between the two groups (Fig. 1, D–F). We then compared absolute and relative BA values of immunogenic versus nonimmunogenic neoepitope candidates, looking at anchor mutation and nonanchor mutation cases separately. Surprisingly, in the case of anchor mutations among the mouse dataset, immunogenic neoepitope candidates did not have higher absolute affinities than nonimmunogenic neoepitope candidates (Fig. 4 D); however, both mouse and human datasets exhibited significantly higher relative affinities (lower relative BA values; Fig. 4 E) of immunogenic neoepitope candidates. On the other hand, absolute BA was highly predictive of antigenicity when the mutations were at nonanchor positions in the neoepitope candidates for both mouse and human datasets (Fig. 4 F), and consistent with our expectation, relative BA was not predictive of antigenicity when a nonanchor residue was mutated (Fig. 4 G).
Figure 5.
MHC-I peptide binding motifs. H-2′b′ and H-2′d′ allele motifs facetted by allele and peptide length are shown. Peptides identified from peptide elution experiments were clustered using GibbsCluster v2.0, using the largest cluster (default parameters and trash cluster option) that yielded the maximum total KLD, with the exceptions of the Db 8-mer and Kb 11-mer motifs. The H-2Db 8-mer motif was manually selected from a smaller cluster of a three-cluster solution that did not have maximum KLD but was similar to the other k-mer motifs from that allele. The H-2Kb 11-mer motif was derived from the full set of original peptides due to the small number of peptides. Putative anchor positions were selected for each allele and peptide length based on the information content at the residues and are shown in the highlighted blocks. Anchor positions were not defined for H-2Db 8-mer peptides as well as H-2Kb 11-mer peptides due to the low number of eluted peptides.
The majority of immunogenic nonanchor mutations are at positions predicted to be in contact with the TCR
The TCR-MHC-I/peptide interaction is another essential component of immunogenicity. Our earlier work suggested that presentation of mutated peptides was not sufficient for immunogenicity, and that peptides with mutations predicted to be in contact with the TCR were more likely to be immunogenic (Yadav et al., 2014). Amino acids at positions 4–6 in MHC-I–bound peptides have been found to be more important for immunogenicity, likely because they interact directly with the TCR (Calis et al., 2013; Glanville et al., 2017). Among the 9-mer neoepitopes identified, nonanchor mutations were observed at several positions in the peptides, with a majority at position 4 in the mouse neoepitopes. Among the 9-mer neoepitopes identified in the human data, a majority of mutations were at position 5 (Fig. 4 H). These results support our previous findings that the position of nonanchor mutations is a potential determinant of immunogenicity.
Physicochemical properties are not consistent predictors of immunogenicity
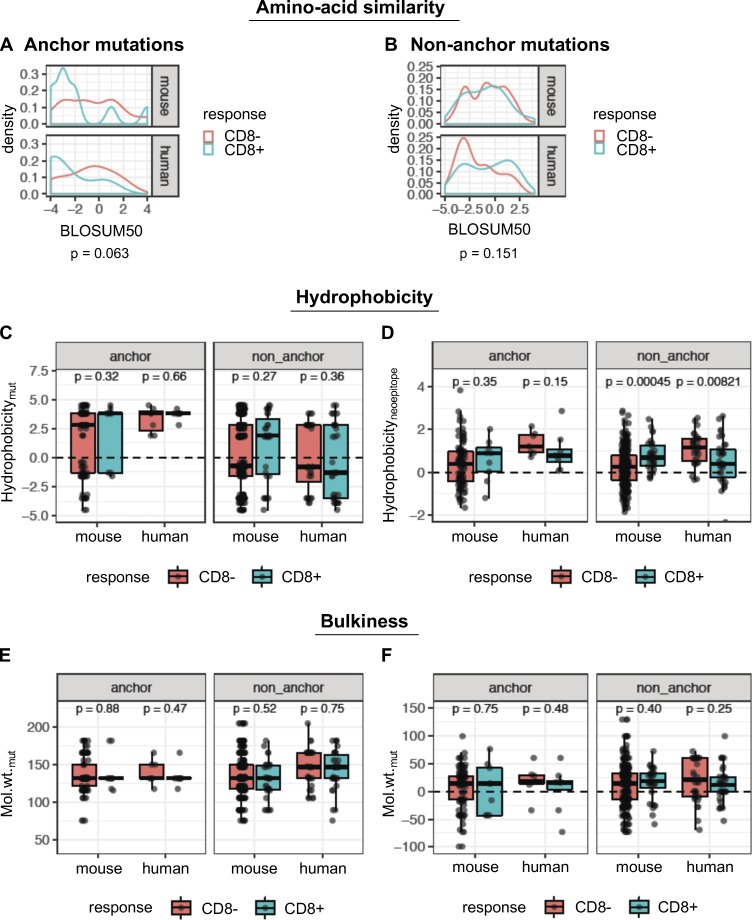
To further investigate peptide-intrinsic properties of mutant neoepitopes, we reasoned that nonconservative amino acid substitutions should create a higher likelihood of immunogenicity. Using BLOSUM50 as a measure of amino acid similarity, we found that immunogenic peptides exhibited a trend toward a higher degree of dissimilarity between the mutant and WT amino acid residues at anchor positions, although this was not statistically significant (Fig. 6 A). Unexpectedly, there was no evidence that the BLOSUM50 score was associated with immunogenicity in the case of mutations at nonanchor residues (Fig. 6 B).
Figure 6.
Physicochemical properties are not consistent predictors of immunogenicity. (A and B) Density plots of distributions of BLOSUM50 scores for the single amino acid substitutions, in immunogenic (blue) or nonimmunogenic (red) neoepitope candidates for anchor (A) and nonanchor (B) mutations. (C) The GRAVY hydrophobicity index of the mutant amino acid of the predicted neoepitope was compared between immunogenic and nonimmunogenic peptides. (D) The mean hydrophobicity index across all amino acids of the predicted neoepitope was also compared. Wilcoxon test was used to compare the hydrophobicity values between immunogenic and nonimmunogenic peptides. (E) Molecular weight of the mutant amino acid is shown for immunogenic and nonimmunogenic predicted neoepitopes. (F) The shift in molecular weight due to mutation is shown for the peptides. Wilcoxon test was used to compare the values between immunogenic and nonimmunogenic peptides. Wilcoxon rank sum test was used to compare the metrics (BLOSUM scores, hydrophobicity, etc.) between immunogenic and nonimmunogenic neoepitope candidates, separately for mouse and human cohorts. Species-specific values from this analysis are shown in the box plots. For the BLOSUM scores, the species-specific P values were combined into a single P value using Fisher’s method. Number of data points for anchor mutations: 104 (mouse, CD8−), 9 (mouse, CD8+), 7 (human, CD8−), and 8 (human, CD8+). Number of data points for nonanchor mutations: 265 (mouse, CD8−), 31 (mouse, CD8+), 32 (human, CD8−), and 34 (human, CD8+).
We also examined the contribution of hydrophobicity and bulkiness of the mutated amino acid to immunogenicity. Both properties have been previously shown to impact binding to the TCR (Chowell et al., 2015). No consistent differences were observed between immunogenic and nonimmunogenic peptides, for either the residue-specific hydrophobicity at the mutant position (Fig. 6 C) or mean hydrophobicity across the peptide (Fig. 6 D). In fact, for nonanchor mutations, mean hydrophobicity of the peptide showed opposite trends between the mouse and human data. Similarly, neither the molecular weight of the mutant amino acid (Fig. 6 E) nor the change in molecular weight due to the mutation (Fig. 6 F) was significantly different between immunogenic and nonimmunogenic cases.
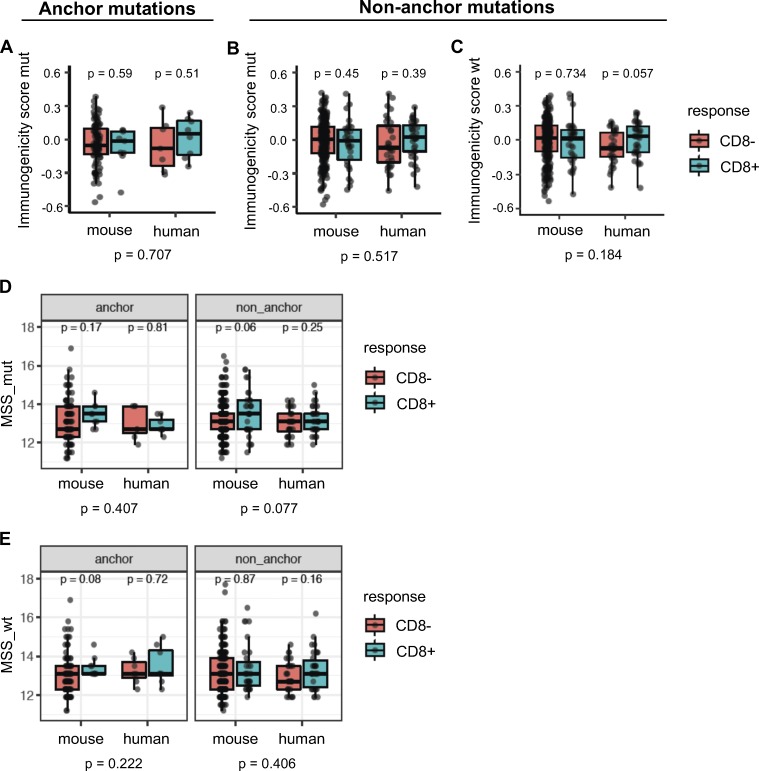
Another approach to evaluate the propensity of TCRs to bind peptides presented by MHC-I is to examine the similarity of the peptide candidates to immunogenic microbial peptides (Balachandran et al., 2017; Calis et al., 2013). We first used the method developed by Calis et al. (2013) that assesses whether the nonanchor amino acids in the peptide candidates are preferentially enriched in immunogenic microbial peptides. Both Calis immunogenicity scores (CISs) of neoepitopes or their WT counterparts were not predictive of their immunogenicity (Fig. 7, A–C). Next, we used a method similar to the one developed by Balachandran et al. (2017) that considers sequence similarity of the entire MHC-I peptide, including anchor positions, to immunogenic microbial peptides. We determined microbial similarity score (MSS) of an amino acid sequence as the highest basic local alignment search tool (BLAST) score after aligning the peptide to a set of 2,329 immunogenic microbial peptides (Balachandran et al., 2017). MSS of the mutant or WT counterpart peptides were not predictive of immunogenicity in either the mouse or human data for both anchor and nonanchor mutations (Fig. 7, D and E).
Figure 7.
Similarity to immunogenic microbial peptides is not a consistent predictor of immunogenicity. The Calis model was used to score candidate neoepitopes or their WT counterparts for similarity of nonanchor amino acids with the nonanchor amino acid content of immunogenic microbial peptides. (A) When the mutation was at an anchor position, immunogenic and nonimmunogenic peptides did not differ in their immunogenicity scores from the Calis model. (B and C) For nonanchor mutations, median scores of immunogenic peptides in the human dataset were higher than nonimmunogenic peptides for both the mutant peptides (B) and their WT counterparts (C), but the trend was not statistically significant. (D and E) Each neoepitope or its WT counterpart was aligned to a set of immunogenic pathogenic peptides using BLAST, and the highest BLAST score was used as an MSS (MSS_mut, for neoepitopes, and MSS_wt, for their WT counterparts). P values shown at the bottom are meta-analysis P values across mouse and human datasets. P values within the plots (species-specific P values) were based on a Wilcoxon test, while the P values across the species were based on Fisher’s test using the species-specific P values. Numbers of data points are the same as in Fig. 6.
Predictors using mutation position context improve neoantigen ranking
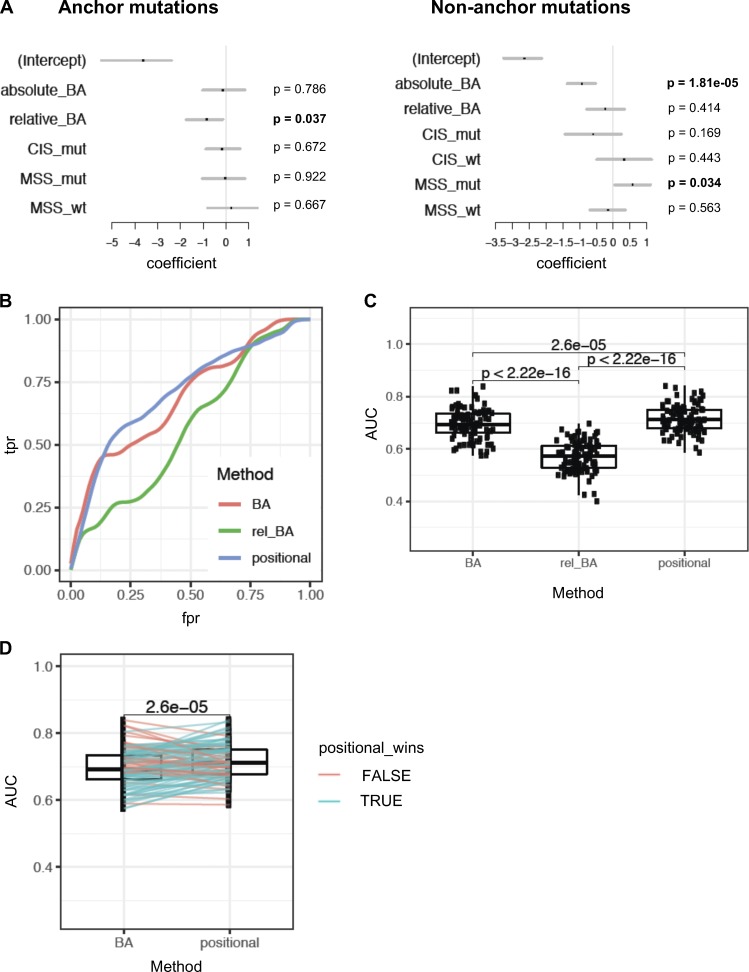
To assess which features are the most important predictors of immunogenicity when considered together, we used multivariate logistic regression to model the likelihood of immunogenicity. Since anchor and nonanchor mutations represent functionally distinct modalities, we generated separate models for each using the mouse vaccination data. We included in our analysis the following features: absolute BA, relative BA, CIS (CIS_mut and CIS_wt), and MSS (MSS_mut and MSS_wt). In accordance with our earlier univariate observations, relative BA was significantly predictive only for anchor mutations, while absolute BA was predictive for nonanchor mutations (Fig. 8 A). Among the scores that evaluate similarity to microbial peptides, only MSS_mut was significant for nonanchor mutations, although not for anchor mutations (Fig. 8 A).
Figure 8.
Positional model ranks neoantigens better than absolute or relative affinity alone. (A) Multivariate models were used to assess the significance of absolute BA of the mutant peptide (absolute_BA), relative affinity (relative_BA), CIS for mutant and WT peptides (CIS_mut and CIS_wt, respectively), and MSS (MSS_mut and MSS_wt). The analysis was done separately for neoepitope candidates with anchor mutations and for those without anchor mutations. Since CIS is identical for the mutant and WT peptides when the mutation is at an anchor, only CIS_mut was considered for the anchor mutation case. The set of these two models is referred to as a positional model. P values are mentioned for each separated model. (B) Performance assessment of the positional model, compared with models that use absolute or relative affinity alone without considering position of mutations with respect to anchor. Receiver-operator characteristic curves (true positive rate vs. false positive rate, or equivalently, sensitivity vs. [1 − specificity]) are shown based on 100 bootstrap iterations. A representative curve is shown for each method, using a Loess fit of 100 bootstrap iterations. (C) AUC of these curves across 100 bootstrap iterations were compared between the three methods (using paired t test). (D) Performance assessment of the positional model relative to predicted absolute BA was assessed by 100 bootstrap iterations. At each iteration, performance of ranking by the positional model and by predicted absolute BA alone was assessed on the held-out data to obtain AUC values. Pairwise comparison of the AUC values from the two methods is shown, and a paired t test was done to assess the difference. Blue lines indicate the iterations where the AUC of the positional model was higher than absolute BA on the held-out data; red lines indicate the cases where it was lower.
We next examined if we could use the predictive features of immunogenic neoepitopes for improving their prioritization. Given that the predictive utility of relative BA versus absolute BA depends on the positioning of the altered amino acid, we incorporated both into an approach that explicitly models anchor versus nonanchor position. Specifically, we used logistic regression to model the likelihood of immunogenicity based on relative BA for anchor mutations, but based on absolute BA and MSS_mut for nonanchor mutations. Two separate “positional models” were derived, one for anchor and the other one for nonanchor mutations. To assess the performance of the positional model in the mouse dataset without overfitting, we used subsampling via 100 bootstrap iterations. In each iteration, we trained the model on a random subset of the mouse data and tested the performance of this trained model on the held-out data. The positional model showed significant improvement in performance in predicting immunogenic peptides, compared with models that used absolute BA alone or relative BA alone (Fig. 8, B–D). Thus, using our mouse vaccination data, we show that ranking of predicted neoepitopes to prioritize immunogenic candidates can be improved by taking into account the anchor versus nonanchor positioning of the mutation. Since the predictive properties of neoantigens in the mouse data were also predictive in human data, such an approach may also be beneficial when training on human data to achieve improved immunogenicity predictors, once a sufficient amount of human immunogenicity data become available from ongoing vaccine studies.
Discussion
Induction of CD8 T cell responses by a tumor-specific mutated peptide depends on the success of several steps in the antigen presentation pathway in both dendritic cells and tumor cells. These steps include peptide processing and translocation into the ER, binding of the peptide to MHC-I and stabilization of the complex at the cell surface, and finally, the presence of T cells in the repertoire that have appropriate affinity toward the neoepitope but not to the WT peptide counterpart (Capietto et al., 2017). Current strategies to identify neoantigens mainly focus on the peptide/MHC-I BA step, and although they significantly enrich for immunogenic neoepitopes, a majority of the predicted candidates are false positives (Carreno et al., 2015; Ott et al., 2017; Sahin et al., 2017). Mass spectrometry (MS) remains the only method that allows direct identification of peptides presented on MHC-I and is currently being used in conjunction with machine learning tools to further define the rules underlying antigen presentation and to continue improving the binding prediction models (Abelin et al., 2017; Schumacher et al., 2017).
Characterization of neoepitope properties that determine immunogenicity has been challenging because of the paucity of immunogenicity data, both for naturally occurring T cell responses and those elicited by vaccination. Although absolute MHC-I BA clearly correlates with immunogenicity (Carreno et al., 2015; Fritsch et al., 2014), other studies suggest that relative affinity may be more important and that mutations that increase BA to MHC-I in comparison to the WT counterpart are more likely to be immunogenic, since they may result in a novel epitope (Balachandran et al., 2017; Duan et al., 2014).
In this study, we generated a large-scale immunogenicity data resource, and we employed it to better characterize the properties of immunogenic neoepitopes. We identified >2,500 expressed mutations from four mouse tumor models, selected 416 candidate neoantigens based on MHC-I BA prediction, and tested 409 candidates for immunogenicity by vaccinating healthy mice with a peptide-based vaccine platform. While there may be some variations between responses induced by different vaccine platforms, rules of neoantigen immunogenicity should remain unchanged (Kreiter et al., 2015). We identified two groups of immunogenic neoantigens: those with high predicted absolute affinity, and those with high predicted relative affinity compared with the WT counterpart. We showed that both of these properties are relevant for immunogenicity, but in two different functional contexts. In the first case, where a nonanchor residue is mutated, absolute affinity is a strong predictor of immunogenicity, but neoepitopes and their WT counterparts typically have comparable affinity. Interestingly, the majority of neoepitopes with nonanchor mutations carried the mutation at central positions, which have been shown to interact with TCRs. These results are consistent with our previous observation that a direct interaction between the mutated residue and the TCR reduces the probability of cross-reactivity between mutant peptide and the WT counterpart, and the established tolerance against the mutant peptide (Yadav et al., 2014).
In the second case, where the mutation is at an anchor residue, increased affinity relative to the corresponding WT peptide is predictive of immunogenicity. We speculate that neoepitopes with higher relative affinity are more prone to be recognized as “non-self,” or foreign, since the WT peptide (self) counterpart is likely poorly presented to T cells. While it does not seem that absolute affinity is as important when anchor residues are mutated, our analysis may underestimate the role of the absolute affinity for anchor residues, since neoepitope candidates were preselected based on a cutoff of predicted absolute affinity. Future vaccination studies using candidates with a wider range of predicted affinities may help to clarify this point. Importantly, we did not observe differences in either the frequency of immunogenic mutations between these two groups or differences in their immunogenic strength as measured by the magnitude of the T cell responses. However, the majority of the immunogenic mutations were at nonanchor positions, possibly for the simple reason that there are fewer anchor positions (typically only two per peptide compared with six to nine nonanchor positions). Our data show that both absolute affinity and relative affinity are predictors of neoepitope immunogenicity, although they are best used for ranking neoepitopes when taking into account the position of the mutated amino acids. Furthermore, this behavior was validated with a human dataset. Although the number of data points is low in the human datasets, especially for anchor mutations, the results were consistent with the mouse data. Notably, Fritsch et al. (2014) have also observed that neoepitopes can originate from both anchor and nonanchor residue mutations, wherein the anchor positioning interpretation was based on conservation of BA. However, the predictive value of neoepitope properties could not be tested in their study due to lack of nonimmunogenic peptide data (Fritsch et al., 2014). Here, we formally identified predictive properties of neoepitopes when the mutation is at an anchor versus a nonanchor residue by comparing neoepitopes to nonimmunogenic neoepitope candidates. Furthermore, we identified the anchor positions in mouse alleles using MS to identify anchors independently of binding predictions.
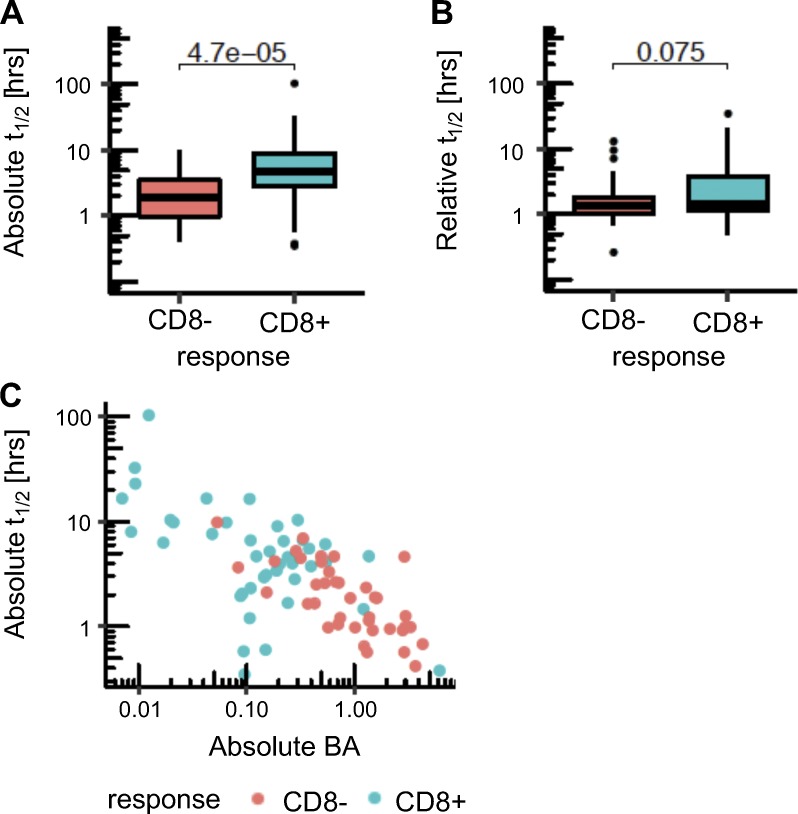
Several studies have emphasized that peptide/MHC-I complex stability provides additional information beyond affinity for immunogenicity potential (Harndahl et al., 2012; Jørgensen et al., 2014; Rasmussen et al., 2016). We used a stability prediction method developed for human data, NetMHSstabpan (Rasmussen et al., 2016), and determined the absolute and relative stability of the mutant peptides in the human dataset (Fig. S3, A and B). We found a higher predicted half-life for the immunogenic neoepitopes (9 h on average) compared with the nonimmunogenic cases (mean of 2.4 h). However, stability was highly correlated with affinity (Fig. S3 C), and we cannot make a definitive conclusion about the utility of stability predictions. It is also notable that previous affinity-balanced comparisons of stability for immunogenic and nonimmunogenic candidates yielded only a modest 3% improvement in reduction of false positives (Jørgensen et al., 2014). A limitation of the evaluation of stability is the sparsity of data to train stability prediction. Additional data will be needed to fully assess the contribution of stability of the peptide/MHC complex.
Figure S3.
Comparison of peptide/MHC stability between immunogenic and nonimmunogenic neoepitope candidates. (A and B) Absolute (A) and relative (B) stability values as predicted by NetMHCstabpan are shown for the neoepitopes that induced CD8 T cell responses (blue) and for the neoepitope candidates that were nonimmunogenic (red) in the human data. P values in the plots are shown based on a t test. (C) Correlation between BA and stability for the human cohort is shown. Spearman’s ρ value is −0.64 (P value = 2.11 × 10−10 for correlation test).
TCR affinity for the MHC-I/peptide complex modulates TCR signaling strength and the magnitude of the T cell response. It has been proposed that immunogenic peptides have biochemical properties that favor interaction with TCRs. We used multiple approaches to explore this question and were unable to find evidence to support it. Using the BLOSUM50 score, we found a trend for higher dissimilarity between the mutant amino acid and the WT counterpart for immunogenic neoantigens only for mutations at anchor positions (Fig. 6, A and B), i.e., at precisely those positions predicted not to interact with the TCR. For the neoepitopes with nonanchor mutations, this difference was not predictive of immunogenicity. Thus, in the case of nonanchor mutations, the residues facing the TCR may have similar properties between the mutant and WT peptides. This observation is consistent with the current model that to ensure a wide breadth of the T cell repertoire, T cells recognizing self-antigens with low avidity can escape negative selection in the thymus and cross-react with mutant/foreign peptides (Birnbaum et al., 2014; Nelson et al., 2015; Sandberg et al., 2000; Yu et al., 2015). In further support of this idea, we observed cross-reactivity of T cells to the WT counterpart peptides in 45% of the cases evaluated, with a mainly weaker T cell response against the WT peptide compared with the neoantigens. This high rate of cross-reactivity with WT counterparts suggests that vaccination against neoantigens still carries a potential risk of inducing autoimmunity. However, it is not clear whether the weaker responses to WT counterparts would translate into effective T cell responses. Further investigation is needed to understand the relevance of the observed cross reactivity to autoimmunity.
We also observed a particularly strong bias toward high predicted MHC-I BA for immunogenic peptides containing nonanchor mutations. The high predicted affinity of the peptides to MHC-I may contribute to increasing T cell avidity by presenting more peptides at the cell surface, thereby compensating for lower affinity of the TCR to the peptides. When we assessed the immunogenic potential of nonanchor mutations by measuring their similarity to microbial peptides, we found that immunogenic mutant peptides tended to be more similar to immunogenic microbial peptides (Fig. 7 D), at least in the case of the mouse dataset. However, this trend of similarity was not observed with human immunogenicity data, which was less extensive. Since this similarity metric depends on how comprehensive the database of immunogenic pathogenic peptides is, alternative approaches to capture this similarity may be needed.
Our study provides a basis for better understanding the immunogenicity of neoepitopes, and it highlights the utility of preclinical models for generating immunogenicity data. We found that both absolute and relative affinities are predictors of immunogenicity of mouse neoepitopes in a position-dependent manner, and we confirmed this result with a human dataset. Furthermore, using a simple logistic regression-based model, we showed that this insight can be used to modestly, but significantly, improve the ranking of predicted neoepitopes. Thus, modeling immunogenicity prediction by taking into account biophysical properties in the physiological contexts where those properties are relevant can improve predictions, as would be the expectation. With the generation of even larger-scale immunogenicity data resources in the future, advanced modeling that takes context-dependent properties into account can be achieved. Furthermore, coupled with advances in our understanding of peptide processing, our results may lead to better prioritization of neoepitope candidates and thus more effective immunotherapies, including personalized cancer vaccination approaches.
Materials and methods
Sequencing, read mapping, and variant calling
Next-generation sequencing and data processing of MC-38, TRAMP-C1, CT-26, and EMT-6 cancer cell lines were performed as previously described (Yadav et al., 2014). Briefly, exome capture was performed using the SureSelectXT mouse exon kit (Agilent). Exome capture libraries were then sequenced on a HiSeq 2000 (Illumina) using the HiSeq sequencing Kit (200 cycles). 60 M (2 × 75 bp) exome reads were sequenced from each cell line. Reads were mapped to the mouse genome (NCBI build 37 or mm9) using GSNAP (Wu and Nacu, 2010). Only uniquely mapped reads were retained for further analysis. Exome sequencing–based variants were called using GATK (DePristo et al., 2011) and annotated for effects on transcripts using the variant effector predictor tool (McLaren et al., 2010). RNA-seq–based variants from the reference mouse genome were called using these criteria: variant allele should be supported by at least two reads, variant allele frequency was ≥4%, and variant allele was not strand-biased (based on read alignment to the genome, Fisher’s exact test, P < 0.05). A vast majority of the variants were supported by exactly two reads, and these accounted for a majority of variants <20% variant allele frequency. As an ad hoc filter, RNA-seq variants with exactly two-read support or <20% variant allele frequency were filtered out.
Identification of candidate neoepitopes
For each somatic mutation that passed our filters, we defined a long peptide (25-mer) neoantigen candidate that contains the mutation at the center. We used NetMHC v3.4 to predict MHC-I BA of all 8–11-mer mutant peptides from the 25-mer peptide sequences to all MHC-I alleles of the mouse strain the cell line was derived from. We then selected the peptide-MHC pair with the lowest half-maximal inhibition (IC50) value, and we call such a mutant peptide that has the best score a candidate neoepitope. We selected 416 mutations that encoded candidate neoepitopes at IC50 ≤500 nM. At the time of peptide synthesis in this study, we used the IC50 metric from NetMHC v3.4 for predicting BA. However, subsequent to peptide synthesis, a newer version of NetMHC, NetMHCpan 4.0 was published, and we used NetMHCpan 4.0 for further analysis of the peptide sequences in this study. Moreover, percentile rank values performed better than IC50 values from NetMHCpan 4.0 in prioritizing immunogenic peptides, so we used percentile rank values instead of IC50.
Selection of peptides for immunogenicity testing
Of 416 mutated sequences, three were duplicated across the four cell lines. Four other mutations, which resulted in responses in ELISpot assays that contained both CD4 and CD8 T cells, were not tested for CD8 responses and were also removed from consideration. Thus, only 409 predicted neoantigen candidates were tested for immunogenicity. We injected 24-aa-long SLPs with the last amino acid being removed from the 25-mer predicted peptide sequence to facilitate peptide synthesis. In cases where the optimal predicted epitope was poorly antigenic or not at all, we first synthesized overlapping peptide libraries of the SLP sequence with 10-aa length (all including the mutation) and tested their immunogenicity through ELISpot assay (see below for the ELISpot method details) with neoantigen-specific CD8 T cells generated upon vaccination with SLP. For each 10-mer peptide inducing a CD8 T cell response, we then synthesized 9- and 8-aa-long peptides and tested them in a similar manner to the 10-mer peptides.
Mice immunization with SLPs
All animal studies were reviewed and approved by Genentech’s Institutional Animal Care and Use Committee. Age-matched 8–10-wk-old female C57BL/6 or BALB/c mice (The Jackson Laboratory) were injected intraperitoneally with either 100 µg mutant predicted SLP each (1 to 5 SLPs per mouse) in combination with adjuvant (50 µg anti-CD40 Ab clone FGK45 and 100 µg poly(I:C); InvivoGen) in PBS or adjuvant alone. Mice were immunized on day 0 and between days 10 and 14. Splenocytes were used for detection of antigen-specific CD8 T cells and/or IFN-gamma release 6–8 d following the last injection (see ELISpot and multimer staining method sections).
ELISpot
5 × 105 splenocytes or CD4-depleted splenocytes (using the manufacturer’s procedure with CD4 L3T4 beads and LS columns from Miltenyi) were cultured overnight at 37°C in RPMI 1640 containing 10% FBS (1% penicillin/streptomycin, 1% Hepes, 1% GlutaMAX, and 1% sodium pyruvate) in a 96-well mouse IFN-gamma ELISpot plate (R&D Systems). CD4-depleted splenocytes (containing dendritic cells) were used to determine the CD8 T cell response in an ELISpot assay, and only CD8 T cells with <2% of CD4 T cells remaining after depletion were considered in the analysis (Fig. 1 C). For stimulation, each mutant predicted 24-mer SLPs (25 µg/ml), optimal mutant, or WT counterpart MHC-I epitopes (8–11 mer SSPs) were added into the culture at various concentrations (as noted in graphs) and compared with mice immunized with adjuvant alone after stimulation in vitro with the same concentrations of specific peptides. For analysis, IFN-gamma spots were counted using the manufacturer’s procedure (R&D Systems mouse IFN-gamma kit) and an automatic ELISpot reader (AID). All samples were tested in technical duplicate and/or 3–5 biological replicates in each individual experiment, and 3–11 experiments were performed depending on the mutation. One screening study was outsourced to WuXi (China). Peptide synthesis was performed by New England Peptide, Anaspec, or Genscript (75% purity).
Multimer staining and flow cytometry
To identify peptide-specific T cells, cells were stained with PE-conjugated peptide–MHC-I dextramers (MHC-I–peptide complex; Immudex) for 20 min followed by staining with cell surface markers CD3, CD4, B220 (BD Biosciences), and CD8 (BioLegend). The purity of the CD4 depletion was evaluated by staining the depleted cells with CD3, CD4 (BD Biosciences), and CD8 (BioLegend) surface antibodies. Only samples with ≥98% purity were used for analysis. Samples were acquired on BD FACSCanto II and analyzed using FlowJo software (v10).
Tap1 and 2 knockout EL-4 cells
The EL4 parental cell line and CRISPR subclones were cultured in RPMI 1640 supplemented with 100 U penicillin, 100 U streptomycin, and 0.29 mg/ml glutamine at 37°C, 5% CO2. sgRNAs were designed using CRISPR3 (Callow et al., 2018; Mali et al., 2013) and cloned into a plasmid enabling coexpression of each sgRNA, SpCas9, and mCherry. Mouse Tap1 CRISPR plasmid constructs (mTap1-1 targeting 5′-CAGCGCTGGATTACTGTAC-3′, mTap1-2 targeting 5′-AAGAAGAGACGTCTCTACC-3′, and mTap1-3 targeting 5′-CGCTGGAGTTTGCAAGTGA-3′) and mouse Tap2 CRISPR plasmid constructs (mTap2-1 targeting 5′-CACAGCACTCCAAGTCGCA-3′, mTap2-2 targeting 5′-GTCGTGTAATTGACATCCT-3′, and mTap2-3 targeting 5′-CGTATCCGCAGGTTGATCC-3′) were nucleofected into the EL4 cell line using the Cell Line Nucleofector Kit L (Amaxa, VCA-1005), and single-cell subclones were sorted by FACS onto 96-well plates. Subclones were screened by flow cytometry for decreased surface MHC-I expression using the Alexa Fluor 647 anti-mouse H-2Kb/H-2Db antibody (clone 28-8-6, BioLegend, 114612).
For testing the relative affinities of H-2Kb peptides, subclone mTap1-2H, targeted by the mTap1-2 CRISPR guide RNA, was seeded at 2 × 105 cells per well on 96-well plates containing 10 µg/ml β2-microglobulin with peptides at various concentrations in a total volume of 50 µl culture medium and subsequently incubated for 16 h at 26°C, 5% CO2. The surface expression of H-2Kb, quantified as median fluorescence intensity, was determined by flow cytometry using the Alexa Fluor 647 anti-mouse H-2Kb antibody (clone AF6-88.5, BioLegend, 116512).
MHC-I peptide isolation and MS
MHC-I peptide profiling was conducted for the H-2Db and H-2Kb alleles of the C57BL/6-derived murine colon adenocarcinoma cell line MC-38 (Kerafast) as described previously (Yadav et al., 2014). MHC-I peptide profiling was conducted for the H-2Dd, H-2Kd, and H-2Ld alleles of the BALB/c-derived murine mammary carcinoma cell line EMT-6 (ATCC) as described previously (Chong et al., 2018) with the following modifications. Approximately 200 million cells were lysed, and MHC-I molecules were immunoprecipitated using three different antibodies covalently coupled to protein A cartridges (Agilent) using dimethyl pimelimidate (Sigma-Aldrich) to extract H-2Dd, Kd, and Ld-specific peptides. Antibodies were derived from hybridoma clones 34-4-20S (H-2Dd specific), K9-18 (H-2Kd specific), and 30-5-7S (H-2Ld specific) and purified before use. MHC-I complexes were bound to antibody resin using the AssayMAP Bravo liquid handler (Agilent), washed with 50 column volumes (CV) of TBS followed by 20 CV of 25 mM Tris, pH 8.0, and eluted using 20 CV of 1% acetic acid. Peptides were isolated and desalted via C18 cartridge using the AssayMAP Bravo (Agilent) before MS analysis.
Peptides were loaded onto a fused-silica Picofrit column (inner diameter = 100 µm, tip = 15 µm, length = 250 mm; NewObjective), packed with 1.7 µm Acquity C18 (130Å, Waters), and separated by ultra-performance liquid chromatography (Ultimate3000, Thermo Fisher Scientific). The gradient used solvent A (2% acetonitrile in water with 0.1% formic acid) and solvent B (98% acetonitrile in water with 0.1% formic acid) and increased from 10% to 40% B over 30 min at a flow rate of 500 nl/min. The eluted peptides were analyzed by Top10 data-dependent acquisition in an Orbitrap Fusion Lumos hybrid mass spectrometer (Thermo Fisher Scientific) equipped with a nano-electrospray ionization source with spray voltage set at 2,000 V. Mass spectral data were acquired using Orbitrap MS scans (R = 60,000 at m/z 400), followed by Orbitrap MS/MS scans (R = 15,000 at m/z 400) subjected to high-energy collision-induced dissociation at 27% collision energy. The mass range for selection of singly, doubly, or triply charged precursors was set at 250-1,250 m/z, with a maximum injection time of 120 ms and AGC target values of 1E6 (MS) and 1E5 (MS/MS).
MS data analysis and binding motif identification
Tandem MS results were submitted for protein database searching using PEAKS (v8.5, Bioinfors). Raw files were converted into mzXML peak lists using ReAdW (v4.3.1), which were then filtered by mass (500–1,500 daltons), charge (+1, 2, or 3), and retention time (10–45 min). The data were searched against a Uniprot-derived Mus musculus database (UP000000589, downloaded March 12, 2019, 22,286 genes) with no enzyme specificity, variable methionine oxidation (+15.99 daltons), variable deamidation (+0.98 daltons), 20 ppm precursor ion mass tolerance, and 0.02 daltons fragment ion mass tolerance. A contaminant database derived from the Contaminant Repository for Affinity Purification (v2012-01-01) was also used to remove nonspecific identifications. Search results were filtered using a linear discriminant algorithm to an estimated peptide false discovery rate of 1%.
H-2′b′ allele peptides were identified from three separate experiments with MC-38 cell line, and H-2′d′ allele peptides were identified from a single experiment with EMT-6 cell line. For each given allele and each peptide length, ranging from 8 to 11 aa long, peptides were used as input for GibbsCluster 2.0 Server with default parameters and the trash cluster option. Clustering solutions with maximum total Kullback–Leibler divergence (KLD) were chosen. In cases where the clustering solution yielded multiple groups, the peptides belonging to the larger group were selected to generate the final motif. The Db 8-mer and Kb 11-mer motifs were the only exceptions. The Db 8-mer motif was derived from the smaller group from a three-cluster solution that did not represent the maximum KLD, but was similar to the other k-mer motifs from that allele. The Kb 11-mer motif was derived from the full set of original peptides, as the number of peptides was too low to perform meaningful clustering. Final motif images were generated using R v3.5.1 and ggseqlogo v0.1.
Anchors (Fig. 5) were defined for each allele and each peptide length as the two residues with the highest information content, with the exception of Dd 9-, 10-, and 11-mer motifs, which had positions 2 and 3 with high information content besides the C-terminal residue, resulting in three anchors. The Dd 8-mer motif seemed to have a distinct behavior at the A-pocket compared with the other peptide lengths and was assigned two anchor positions.
Evaluation of immunogenicity
For each long peptide evaluated for CD8 T cell response by ELISpot, positivity of the assay was declared by using a likelihood ratio test between two nested linear models, one containing the treatment fixed effect and the other without this effect. When multiple experiments were conducted on the same mouse, and/or across multiple days, the model included mouse identity or day as a random effect. Spot count data were log-transformed, and a pseudo-count of 0.1 was added. The R package lme4 (Bates et al., 2015) was used to fit the linear mixed effects model. Inference was made by comparing the two nested models using the function lme4::anova.merMod. P values were corrected for multiple testing using the Benjamini–Hochberg method. Positive responses were declared as the ones with adjusted P value ≤0.05, median spot count of ≥10 after vaccination, and a positive effect size of ≥2 (i.e., doubling of spot counts after treatment).
In some cases, we manually overrode the statistical call, when we had reasonable supporting criteria to do so. These mutations were statistically borderline-negative but were manually called as positive based on additional experiments: C117, fold-change was borderline (1.94), but we tested both SLP and SSP and the results indicated a CD8 response; C64, borderline P value (0.1) for SLP, but SSP also showed a response.
Calculation of peptide properties
Several measures of physicochemical properties of peptides have been used in this study. They are described as follows: (a) IC50 values for peptide-MHC BA, as predicted by NetMHC-3.4 (IEDB tools v2.13, predict_binding.py with method as “ann”; tool available at https://downloads.iedb.org/tools/mhci/). (b) BA of peptide to MHC, as percentile rank scores, based on NetMHCpan 4.0 (IEDB tools v2.19, netMHCpan executable script from the suite). This is the same as absolute BA. (c) Relative BA: ratio of the BA values of candidate neoepitope to the BA value of the WT counterpart, for binding the optimal predicted MFHC allele. (d) Absolute t1/2: half-life of peptide/MHC complex in hours, predicted by NetMHCstabpan (IEDB tools v2.17, predict_binding.py script with method “netmhcstabpan”). (e) Relative t1/2: ratio of the t1/2 value of candidate neoepitope to the t1/2 value of the WT counterpart, for binding the optimal predicted MHC allele. (f) CIS: immunogenicity scores were calculated using IEDB tools (IEDB_Immunogenicity-1.0, script predict_immunogenicity.py, available at https://downloads.iedb.org/tools/immunogenicity/). The method masks anchor residues and calculates an immunogenicity score based on enrichment of nonanchor residues in known immunogenic peptides versus nonimmunogenic peptides, mostly from pathogenic peptides. The mask positions were provided based on our anchor inferences as described in MS data analysis and binding motif identification. (g) BLOSUM50 matrix was used to calculate BLOSUM scores for substitution of the WT amino acid with the mutant amino acid. (h) MSS: a similarity score of a query peptide to a database of immunogenic peptides from pathogens (Balachandran et al., 2017) was calculated by taking the best BLAST alignment score of the query peptide to all immunogenic peptides in the database. This is a slightly different score from the one used by Balachandran et al. (2017) that takes the sum of exponentials of each alignment score between the query peptide and the peptides from the database. (i) Hydrophobicity, molecular weight: GRAVY Hydrophobicity index and molecular weight of amino acids or of peptides was calculated using the R package Peptides (Osorio et al., 2015).
Considering that a TCR recognizes an epitope and the allele together, we computed relative BA keeping the allele constant. For an anchor mutation, if the same allele presented the WT counterpart peptide, the TCR facing residues will be the same between the mutant and the WT peptide. We hypothesized that this would result in tolerance to the presented mutant peptide, unless the BA of the WT peptide to the same allele is low. On the other hand, if the mutant peptide and its WT counterpart are presented by different alleles, it is unlikely that the binding of the WT peptide would impact the immunogenicity of the mutant peptide via tolerance mechanisms since the alleles would be different. Therefore, we do not consider such scenarios in our predictions.
Model development and assessment
For inference on various predictors of immunogenicity for both mouse and human data, separate multivariate logistic models were made for anchor and nonanchor mutations (Fig. 8). The model formula used was
Two different models were made this way, for anchor and nonanchor mutations, for mouse data. BLOSUM50 was omitted, as it correlates with affinity predictions. Forest plots were made to represent the coefficients and 95% confidence intervals, using forestplot R package (Gordon and Lumley, 2017). The intercepts in the two models represent the odds of immunogenicity of a predicted neoepitope that has low absolute BA, equal affinity to its WT counterpart, and low CIS and MSS values (absolute BA of 1, relative BA of 1, and CIS and MSS values of 0 for mutant peptide and its WT counterpart).
Model performance assessment was done on the training and the test data using the R package ROCR (Sing et al., 2005). Bootstrap performance assessment was done on the mouse model by iterating 100 times over the mouse vaccination data. At each iteration, approximately two-thirds of the data was used to train the models separately for anchor and nonanchor mutations. To ensure a reasonable number of data points in each category (anchor and nonanchor mutations), the sampling was done separately for the neoepitopes in each of these categories. At each iteration, all 113 anchor data points and 296 nonanchor data points were sampled with replacement. As a result, across 100 iterations, the held-out data amounted to 137–162 data points across the two categories, which is approximately one-third of the total number of data points of 409. At each iteration, two models were trained on the sampled data, one for each of the categories. The trained model was used to predict the outcome in the held-out data. AUC (area under the curve) values were determined for these predictions. AUC values were recorded for each iteration of bootstrap, and later on, used to compare the methods using a paired t test.
Online supplemental material
Fig. S1 shows the immunogenicity of predicted MHC-I optimal mutant epitopes. Fig. S2 shows cross-reactive mutant MHC-I neoantigen-specific T cells. Fig. S3 is a comparison of peptide/MHC stability between immunogenic and nonimmunogenic neoepitope candidates. Table S1 lists MHC-I immunogenic mutations for MC-38, CT-26, and EMT-6 mouse tumor cell lines. Table S2 shows the independence of neoepitope-specific TCR cross-reactivity and predicted BA of mutant neoepitopes or their WT counterparts. Data S1 lists neoantigen candidates in four mouse tumor cell lines.
Supplementary Material
lists MHC-I immunogenic mutations for MC-38, CT-26, and EMT-6 mouse tumor cell lines.
shows the independence of neoepitope-specific TCR cross-reactivity and predicted BA of mutant neoepitopes or their WT counterparts.
lists neoantigens candidates in four mouse tumor cell lines.
Acknowledgments
We acknowledge technical support from BioRender with the graphical abstract.
Author contributions: A.-H. Capietto planned and performed experiments, analyzed and interpreted data, and wrote the manuscript. S. Jhunjhunwala analyzed and interpreted whole-exome sequencing and RNA-seq data, predicted neoantigens, performed statistical data analysis, and wrote the manuscript. P. Lupardus analyzed the amino acid composition of immunogenic peptides. J. Wong generated cell lines and performed BA studies. T. Nozawa and Z. Fan performed vaccinations. L. Hänsch, J. Cevallos, Y. Chestnut, A. Fernandez, M. Singh, and Z. Fan performed ELISpot assay and tetramer staining. C.C. de la Cruz coordinated outsourced work. B. Haley designed CRISPR constructs. S.B. Pollock and Q.T. Phung performed and analyzed MS data. N. Lounsbury analyzed and interpreted the MHC peptide binding motifs. J.R. Lill oversaw the MS analysis. L. Taraborrelli grew cells for MS assay. I. Mellman and R. Bourgon assisted with data analysis and preparation of the manuscript. L. Delamarre oversaw all the work performed, planned experiments, interpreted data, and wrote the manuscript.
References
- Abelin J.G., Keskin D.B., Sarkizova S., Hartigan C.R., Zhang W., Sidney J., Stevens J., Lane W., Zhang G.L., Eisenhaure T.M., et al. 2017. Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity. 46:315–326. 10.1016/j.immuni.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran V.P., Luksza M., Zhao J.N., Makarov V., Moral J.A., Remark R., Herbst B., Askan G., Bhanot U., Senbabaoglu Y., et al. 2017. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 551:512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B., and Walker S.. 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67:1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Birnbaum M.E., Mendoza J.L., Sethi D.K., Dong S., Glanville J., Dobbins J., Ozkan E., Davis M.M., Wucherpfennig K.W., and Garcia K.C.. 2014. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 157:1073–1087. 10.1016/j.cell.2014.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calis J.J., Maybeno M., Greenbaum J.A., Weiskopf D., De Silva A.D., Sette A., Keşmir C., and Peters B.. 2013. Properties of MHC class I presented peptides that enhance immunogenicity. PLOS Comput. Biol. 9:e1003266 10.1371/journal.pcbi.1003266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow M.G., Watanabe C., Wickliffe K.E., Bainer R., Kummerfield S., Weng J., Cuellar T., Janakiraman V., Chen H., Chih B., et al. 2018. CRISPR whole-genome screening identifies new necroptosis regulators and RIPK1 alternative splicing. Cell Death Dis. 9:261 10.1038/s41419-018-0301-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capietto A.H., Jhunjhunwala S., and Delamarre L.. 2017. Characterizing neoantigens for personalized cancer immunotherapy. Curr. Opin. Immunol. 46:58–65. 10.1016/j.coi.2017.04.007 [DOI] [PubMed] [Google Scholar]
- Carreno B.M., Magrini V., Becker-Hapak M., Kaabinejadian S., Hundal J., Petti A.A., Ly A., Lie W.R., Hildebrand W.H., Mardis E.R., and Linette G.P.. 2015. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 348:803–808. 10.1126/science.aaa3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong C., Marino F., Pak H., Racle J., Daniel R.T., Müller M., Gfeller D., Coukos G., and Bassani-Sternberg M.. 2018. High-throughput and Sensitive Immunopeptidomics Platform Reveals Profound Interferonγ-Mediated Remodeling of the Human Leukocyte Antigen (HLA) Ligandome. Mol. Cell. Proteomics. 17:533–548. 10.1074/mcp.TIR117.000383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowell D., Krishna S., Becker P.D., Cocita C., Shu J., Tan X., Greenberg P.D., Klavinskis L.S., Blattman J.N., and Anderson K.S.. 2015. TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc. Natl. Acad. Sci. USA. 112:E1754–E1762. 10.1073/pnas.1500973112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen C.J., Gartner J.J., Horovitz-Fried M., Shamalov K., Trebska-McGowan K., Bliskovsky V.V., Parkhurst M.R., Ankri C., Prickett T.D., Crystal J.S., et al. 2015. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J. Clin. Invest. 125:3981–3991. 10.1172/JCI82416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43:491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan F., Duitama J., Al Seesi S., Ayres C.M., Corcelli S.A., Pawashe A.P., Blanchard T., McMahon D., Sidney J., Sette A., et al. 2014. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J. Exp. Med. 211:2231–2248. 10.1084/jem.20141308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Nik H., Michaux J., Corwin W.L., Keller G.L., Shcheglova T., Pak H., Coukos G., Baker B.M., Mandoiu I.I., Bassani-Sternberg M., and Srivastava P.K.. 2019. Mass spectrometry driven exploration reveals nuances of neoepitope-driven tumor rejection. JCI Insight. 5:129152 10.1172/jci.insight.129152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch E.F., Rajasagi M., Ott P.A., Brusic V., Hacohen N., and Wu C.J.. 2014. HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol. Res. 2:522–529. 10.1158/2326-6066.CIR-13-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorani E., Rosenthal R., McGranahan N., Reading J.L., Lynch M., Peggs K.S., Swanton C., and Quezada S.A.. 2018. Differential binding affinity of mutated peptides for MHC class I is a predictor of survival in advanced lung cancer and melanoma. Ann. Oncol. 29:271–279. 10.1093/annonc/mdx687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville J., Huang H., Nau A., Hatton O., Wagar L.E., Rubelt F., Ji X., Han A., Krams S.M., Pettus C., et al. 2017. Identifying specificity groups in the T cell receptor repertoire. Nature. 547:94–98. 10.1038/nature22976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M., and Lumley T.. 2017. forestplot: Advanced forest plot using ‘grid’ graphics. R package version 1.7.2. https://CRAN.R-project.org/package=forestplot
- Gubin M.M., Zhang X., Schuster H., Caron E., Ward J.P., Noguchi T., Ivanova Y., Hundal J., Arthur C.D., Krebber W.J., et al. 2014. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 515:577–581. 10.1038/nature13988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harndahl M., Rasmussen M., Roder G., Dalgaard Pedersen I., Sørensen M., Nielsen M., and Buus S.. 2012. Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur. J. Immunol. 42:1405–1416. 10.1002/eji.201141774 [DOI] [PubMed] [Google Scholar]
- Hellmann M.D., Nathanson T., Rizvi H., Creelan B.C., Sanchez-Vega F., Ahuja A., Ni A., Novik J.B., Mangarin L.M.B., Abu-Akeel M., et al. 2018. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. 33:843–852.e4. 10.1016/j.ccell.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen K.W., Rasmussen M., Buus S., and Nielsen M.. 2014. NetMHCstab - predicting stability of peptide-MHC-I complexes; impacts for cytotoxic T lymphocyte epitope discovery. Immunology. 141:18–26. 10.1111/imm.12160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiter S., Vormehr M., van de Roemer N., Diken M., Löwer M., Diekmann J., Boegel S., Schrörs B., Vascotto F., Castle J.C., et al. 2015. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 520:692–696. 10.1038/nature14426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D., et al. 2015. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 372:2509–2520. 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łuksza M., Riaz N., Makarov V., Balachandran V.P., Hellmann M.D., Solovyov A., Rizvi N.A., Merghoub T., Levine A.J., Chan T.A., et al. 2017. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 551:517–520. 10.1038/nature24473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., and Church G.M.. 2013. RNA-guided human genome engineering via Cas9. Science. 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., and Cunningham F.. 2010. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 26:2069–2070. 10.1093/bioinformatics/btq330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson T., Ahuja A., Rubinsteyn A., Aksoy B.A., Hellmann M.D., Miao D., Van Allen E., Merghoub T., Wolchok J.D., Snyder A., and Hammerbacher J.. 2017. Somatic Mutations and Neoepitope Homology in Melanomas Treated with CTLA-4 Blockade. Cancer Immunol. Res. 5:84–91. 10.1158/2326-6066.CIR-16-0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R.W., Beisang D., Tubo N.J., Dileepan T., Wiesner D.L., Nielsen K., Wüthrich M., Klein B.S., Kotov D.I., Spanier J.A., et al. 2015. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity. 42:95–107. 10.1016/j.immuni.2014.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio V., Rondón-Villarreal P., and Torres R.. 2015. Peptides: A package for data mining of antimicrobial peptides. R J. 7:4–14. 10.32614/RJ-2015-001 [DOI] [Google Scholar]
- Ott P.A., Hu Z., Keskin D.B., Shukla S.A., Sun J., Bozym D.J., Zhang W., Luoma A., Giobbie-Hurder A., Peter L., et al. 2017. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 547:217–221. 10.1038/nature22991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M., Fenoy E., Harndahl M., Kristensen A.B., Nielsen I.K., Nielsen M., and Buus S.. 2016. Pan-Specific Prediction of Peptide-MHC Class I Complex Stability, a Correlate of T Cell Immunogenicity. J. Immunol. 197:1517–1524. 10.4049/jimmunol.1600582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech A.J., Balli D., Mantero A., Ishwaran H., Nathanson K.L., Stanger B.Z., and Vonderheide R.H.. 2018. Tumor Immunity and Survival as a Function of Alternative Neopeptides in Human Cancer. Cancer Immunol. Res. 6:276–287. 10.1158/2326-6066.CIR-17-0559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S., et al. 2015. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 348:124–128. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosalia R.A., Quakkelaar E.D., Redeker A., Khan S., Camps M., Drijfhout J.W., Silva A.L., Jiskoot W., van Hall T., van Veelen P.A., et al. 2013. Dendritic cells process synthetic long peptides better than whole protein, improving antigen presentation and T-cell activation. Eur. J. Immunol. 43:2554–2565. 10.1002/eji.201343324 [DOI] [PubMed] [Google Scholar]
- Sahin U., Derhovanessian E., Miller M., Kloke B.P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B., et al. 2017. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 547:222–226. 10.1038/nature23003 [DOI] [PubMed] [Google Scholar]
- Sandberg J.K., Franksson L., Sundbäck J., Michaelsson J., Petersson M., Achour A., Wallin R.P., Sherman N.E., Bergman T., Jörnvall H., et al. 2000. T cell tolerance based on avidity thresholds rather than complete deletion allows maintenance of maximal repertoire diversity. J. Immunol. 165:25–33. 10.4049/jimmunol.165.1.25 [DOI] [PubMed] [Google Scholar]
- Schumacher F.R., Delamarre L., Jhunjhunwala S., Modrusan Z., Phung Q.T., Elias J.E., and Lill J.R.. 2017. Building proteomic tool boxes to monitor MHC class I and class II peptides. Proteomics. 17:1600061 10.1002/pmic.201600061 [DOI] [PubMed] [Google Scholar]
- Sette A., Sidney J., del Guercio M.F., Southwood S., Ruppert J., Dahlberg C., Grey H.M., and Kubo R.T.. 1994a Peptide binding to the most frequent HLA-A class I alleles measured by quantitative molecular binding assays. Mol. Immunol. 31:813–822. 10.1016/0161-5890(94)90019-1 [DOI] [PubMed] [Google Scholar]
- Sette A., Vitiello A., Reherman B., Fowler P., Nayersina R., Kast W.M., Melief C.J., Oseroff C., Yuan L., Ruppert J., et al. 1994b The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 153:5586–5592. [PubMed] [Google Scholar]
- Sing T., Sander O., Beerenwinkel N., and Lengauer T.. 2005. ROCR: visualizing classifier performance in R. Bioinformatics. 21:3940–3941. 10.1093/bioinformatics/bti623 [DOI] [PubMed] [Google Scholar]
- Snyder A., Makarov V., Merghoub T., Yuan J., Zaretsky J.M., Desrichard A., Walsh L.A., Postow M.A., Wong P., Ho T.S., et al. 2014. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371:2189–2199. 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strønen E., Toebes M., Kelderman S., van Buuren M.M., Yang W., van Rooij N., Donia M., Böschen M.L., Lund-Johansen F., Olweus J., and Schumacher T.N.. 2016. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 352:1337–1341. 10.1126/science.aaf2288 [DOI] [PubMed] [Google Scholar]
- Tran E., Robbins P.F., Lu Y.C., Prickett T.D., Gartner J.J., Jia L., Pasetto A., Zheng Z., Ray S., Groh E.M., et al. 2016. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N. Engl. J. Med. 375:2255–2262. 10.1056/NEJMoa1609279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E., Turcotte S., Gros A., Robbins P.F., Lu Y.C., Dudley M.E., Wunderlich J.R., Somerville R.P., Hogan K., Hinrichs C.S., et al. 2014. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 344:641–645. 10.1126/science.1251102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen E.M., Miao D., Schilling B., Shukla S.A., Blank C., Zimmer L., Sucker A., Hillen U., Foppen M.H.G., Goldinger S.M., et al. 2015. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 350:207–211. 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg S.H., Visseren M.J., Brandt R.M., Kast W.M., and Melief C.J.. 1996. Immunogenicity of peptides bound to MHC class I molecules depends on the MHC-peptide complex stability. J. Immunol. 156:3308–3314. [PubMed] [Google Scholar]
- Wu T.D., and Nacu S.. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 26:873–881. 10.1093/bioinformatics/btq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M., Jhunjhunwala S., Phung Q.T., Lupardus P., Tanguay J., Bumbaca S., Franci C., Cheung T.K., Fritsche J., Weinschenk T., et al. 2014. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 515:572–576. 10.1038/nature14001 [DOI] [PubMed] [Google Scholar]
- Yu W., Jiang N., Ebert P.J., Kidd B.A., Müller S., Lund P.J., Juang J., Adachi K., Tse T., Birnbaum M.E., et al. 2015. Clonal Deletion Prunes but Does Not Eliminate Self-Specific αβ CD8(+) T Lymphocytes. Immunity. 42:929–941. 10.1016/j.immuni.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharakis N., Chinnasamy H., Black M., Xu H., Lu Y.C., Zheng Z., Pasetto A., Langhan M., Shelton T., Prickett T., et al. 2018. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med. 24:724–730. 10.1038/s41591-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists MHC-I immunogenic mutations for MC-38, CT-26, and EMT-6 mouse tumor cell lines.
shows the independence of neoepitope-specific TCR cross-reactivity and predicted BA of mutant neoepitopes or their WT counterparts.
lists neoantigens candidates in four mouse tumor cell lines.