Abstract
Salmonella enterica serovar Infantis (S. Infantis) is one of the dominant serovars of the bacterial pathogen S. enterica. In recent years, the number of human infections caused by S. Infantis has been increasing in many countries, and often the emerging population harbors a unique virulence-resistant megaplasmid called plasmid of emerging S. Infantis (pESI). Here, we report the complete gap-free genome sequence of the S. Infantis Israeli emerging clone and compare its chromosome and pESI sequences with other complete S. Infantis genomes. We show a conserved presence of the Salmonella pathogenicity islands 1–6, 9, 11, 12, and CS54 and a common integration of five bacteriophages in the S. Infantis chromosome. In contrast, we found variable presence of additionally three chromosomally integrated phages and eight modular regions in pESI, which contribute to the genetic and phenotypic diversity (including antimicrobial resistance) of this ubiquitous foodborne pathogen.
Keywords: Salmonella Infantis, WGS, pESI, mobile genetic elements, HGT, virulence
Introduction
The abundant foodborne pathogen Salmonella enterica (S. enterica) is a Gram-negative, highly diverse bacterium that can infect and colonize a broad array of animal and human hosts. This single bacterial species comprises of >2,600 antigenically distinct serovars that can be classified according to their host-specificity and their occasioned disease (Gal-Mor 2019).
Non-typhoidal serovars (NTS) like Salmonella enterica serovar Typhimurium (S. Typhimurium) or S. enterica serovar Infantis (S. Infantis) are known to possess a wide host-specificity and are capable of infecting various animal species including reptiles, birds, and mammals. In immunocompetent humans, infection with NTS serovars normally provokes a self-limiting localized inflammation of the terminal ileum and colon, called gastroenteritis. The assessed annual global burden of gastroenteritis caused by NTS infections is 78.7 million incidents, resulting in 59,000 deaths (Havelaar et al. 2015).
Amongst >2,600 S. enterica serovars known to date, S. Infantis is one of the most prevalent serovars worldwide. In the United States, S. Infantis was ranked sixth, in the occurrence hierarchy (Crim et al. 2015) and in the European Union, S. Infantis was rated third in the prevalence order, following serovars Enteritidis and Typhimurium (ECDC 2014). Moreover, in recent years, S. Infantis is the most frequently reported S. enterica serovar from food-producing animals (mainly from the poultry production chain) and various food products in Europe (EFSA 2017).
Latterly, we demonstrated that serovar Infantis is largely associated with infections of infants younger than two years old and adheres better to host cells than the serovar Typhimurium. Nevertheless, in comparison to S. Typhimurium, S. Infantis was shown to be less invasive in humans and causes lower inflammation in the colitis mouse model. These differences were attributed to lower expression levels of the Salmonella pathogenicity island (SPI) 1 genes in S. Infantis compared with S. Typhimurium (Aviv et al. 2019).
In Israel, a rapid and clonal emergence of S. Infantis was reported in 2010, and from 2008 to 2015 S. Infantis was the most dominant serovar isolated from both human and poultry sources (Gal-Mor et al. 2010; Aviv et al. 2014). Noteworthy, the emergence of S. Infantis has been further reported in multiple countries around the world including Germany (Hauser et al. 2012), France, Belgium (Cloeckaert et al. 2007), Hungary (Nogrady et al. 2008), Russia (Bogomazova et al. 2020), Honduras (Liebana et al. 2004), Japan (Shahada et al. 2010), and Australia (Ross and Heuzenroeder 2008), indicating that S. Infantis is a globally emerging serovar and a primary source of poultry infection and human salmonellosis. A recent study addressing the genetic structure of the global S. Infantis population has shown that S. Infantis is a polyphyletic serovar and has evolved in three separate lineages, with specifically one dominant emerging lineage (Gymoese et al. 2019).
Previously, we have reported that the fast and clonal S. Infantis emergence was facilitated by lateral acquisition of a novel virulence-resistance megaplasmid, designated pESI (standing for plasmid of emerging S. Infantis) that contributes to multidrug resistance and enhanced pathogenicity of pESI-positive strains (Gal-Mor et al. 2010; Aviv et al. 2014). We specifically showed that pESI encodes several virulence factors, including the yersiniabactin—iron acquisition system, as well as the Klf and Ipf chaperon-usher fimbriae. Furthermore, this plasmid carries various mobile elements encoding antibiotic and mercury resistance genes and at least three independent toxin/antitoxin systems (MazEF/PemKI, CcdAB, and VagCD) (Aviv et al. 2014, 2016, 2017).
Subsequently, genetically related pESI-like plasmids were also found in additional emergent S. Infantis strains in Spain (Iriarte et al. 2017), Switzerland (Hindermann et al. 2017), Italy (Franco et al. 2015), Hungary (Szmolka et al. 2018), Japan (Yokoyama et al. 2015), the USA (Tate et al. 2017), and Russia (Bogomazova et al. 2020). These findings indicate worldwide dissemination of S. Infantis strains harboring pESI-like megaplasmids that play an important role in the evolution and epidemiology of globally emerging S. Infantis lineages.
Here, we report the complete and gap-free genome sequence of the emerging S. Infantis clone, represented by the 119944 Israel-isolated strain and present genomic analysis and comparison with other complete genomes of this serovars. Our results demonstrate a conserved distribution of 10 SPIs and five chromosomal prophages integrated into the genome of S. Infantis. Furthermore, we define core and variable regions in pESI and highlight the circulation of pESI-like plasmids among globally emerging S. Infantis strains.
Materials and Methods
Whole-Genome Sequencing
Genomic DNA from S. Infantis strain 119944, as a representative isolate of the emergent S. Infantis population in Israel (Gal-Mor et al. 2010) was isolated using the GenElute Bacterial Genomic DNA Kit (Sigma–Aldrich). Whole-genome sequencing that was performed at the Technion Genomic Center of the Israeli Institute of Technology (Haifa, Israel) has generated 16 × 106 paired‐ends shorts reads by an Illumina Genome Analyzer IIx platform (Illumina, Inc.) and 291,510 long reads using a MinION sequencer (Oxford Nanopore Technologies). The average MinION reads length was 11,538 bp and the N50 was 28,095 bp long. The quality of the short Illumina and the long MinIon reads (fastq files) was evaluated using FastQC (version 0.11.5) and NanoPlot tools, respectively.
Genome Assembly
Both the short (Illumina) and long (MinION) reads were combined for hybrid de novo assembly using the Unicycler (version 0.4.8-beta) pipeline (Wick et al. 2017a, 2017b). Unicycler assembler employed SPAdes (version 3.13) with error correction and automatic selection of k-mer length to produce short reads assembly graph (contigs). In the next step, the miniasm and Racon Unicycler’s modules were used for long reads and contigs assembly. The resulting assembly was then polished by pilon (version 1.22). The hybrid assembly of the S. Infantis 119944 genome resulted in two closed scaffolds corresponding to the chromosome (4,725,957 bp) and the pESI plasmid (285,081 bp), while the genome was covered 895×. The complete S. Infantis 119944 chromosome (accession number CP047881) and pESI (accession number CP047882) assembles were deposited in the NCBI database.
Bioinformatics Analyses
Genome comparison of the S. Infantis 119944 chromosome and plasmid was done against all S. Infantis complete genomes found at the NCBI database including FSIS1502916 (assembly number GCA_001931575.1); FARPER-219 (GCA_006402875.1); FSIS1502169 (GCA_001931555.1); N55391 (GCA_001931595.1); CVM44454 (GCA_001931615.1); 1326/28 (GCA_000953495.1); NCTC6703 (GCA_900478235.1); and CFSAN003307 (GCA_002863785.1). Sequences alignment was conducted using Mauve (Rissman et al. 2009) and compared by BRIG (Alikhan et al. 2011). Phages and their integration sites were identified using PHASTER (Arndt et al. 2016). SPIs sequences were downloaded from the pathogenicity islands database (PAIDB) (http://www.paidb.re.kr/browse_pais.php?m=p#Salmonella enterica) and BLASTed against the assembled S. Infantis 119944 genome.
Results and Discussion
To advance better understanding of the global epidemiology and genomics of S. Infantis we applied hybrid assembly while combining short reads from Illumina sequencing together with long reads from MinION platform. This approach allowed determining a complete gap-free genome sequence of S. Infantis isolate 119944 that was covered 895×. The complete genome of S. Infantis 119944 has a 53.2% GC content and composes of one circular 4,725,957 bp chromosome and a 285,081 bp plasmid, which we previously named pESI. Using the NCBI prokaryotic genome annotation pipeline (PGAP), we found that the S. Infantis 119944 genome encodes 4,853 genes, 4,612 proteins, 84 tRNA genes, and harbors122 pseudogenes.
To account for conserved and unique regions in the 119944 genome, the chromosome and the pESI plasmids were compared with eight S. Infantis complete genomes available at the NCBI database. Supplementary table 1, Supplementary Material online shows the main features and relevant metadata of the compered S. Infantis genomes. The S. Infantis genome size of these nine completely sequenced strains varied between 4,630,342 bp (strain NCTC6703) and 5,089,781 bp (FARPER-219) and harbored between none and two plasmids.
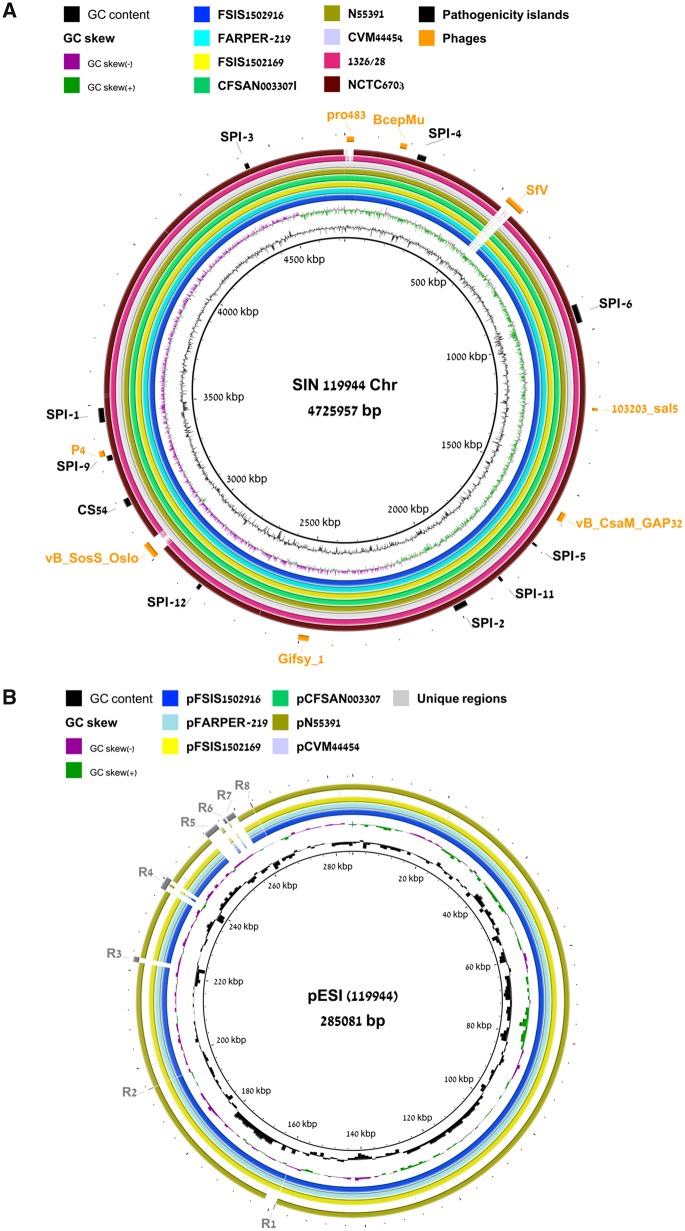
The distribution of SPIs was constant across S. Infantis 119944 and the other compared genomes, and all of them were found to carry intact SPIs-1–6, SPI-9, SPI-12, and CS54 (fig. 1A and table 1). In addition, all of these S. Infantis genomes harbor a 9 kb shorter version of SPI-11, instead of the 15.7 kb island, known in S. Choleraesuis SC-B67 (Jacobsen et al. 2011). Nonetheless, all of the SPI-11-associated virulence genes are present in the S. Infantis SPI-11, including pagC, pagD, msgA, envF, and the T3SS effector gene sopF.
Fig. 1.
—Chromosome and pESI sequence comparison between S. Infantis strains. (A) The chromosome sequence of strain 119944 was compared with the complete chromosome sequence of eight S. Infantis strains (FSIS1502916; FARPER-219; FSIS1502169; N55391; CVM44454; 1326/28; NCTC6703; and CFSAN003307 using the BRIG tool [Alikhan et al. 2011]). The genome coordinates of the S. Infantis 119944 strain are shown by the internal black ring and the GC content is indicated by the black plot. The distribution of SPIs and prophage across the genomes are shown by black and orange boxes, respectively. (B) The DNA sequence of plasmids from six S. Infantis strains that were found to harbor megaplasmids (pFSIS1502916, pFARPER-219, pFSIS1502169, pN55391, pCVM44454, and pCFSAN003307) were compare to the sequence of pESI in 119944 using BRIG. Eight variable regions (R1–R8) that were found to be uniquely present in pESI in 119944 are indicated by the gray boxes.
Table 1.
Distribution of PAIs and Prophages in the S. Infantis 119944 Genome
| Element | Name | Accession Number | Start | End | Length (bp) | Integrity | Unique (to 119944), Diverse or Conserved |
|---|---|---|---|---|---|---|---|
| Phage | pro483 | NC_028943 | 6,016 | 27,791 | 21,776 | Intact | Diverse |
| Phage | BcepMu | NC_005882 | 167,739 | 188,179 | 20,441 | Incomplete | Conserved |
| Phage | SfV | NC_003444 | 529,849 | 589,113 | 59,265 | Intact | Unique |
| Phage | 103203 sal5 | NC_031946 | 1,234,425 | 1,243,356 | 8,932 | Incomplete | Conserved |
| Phage | vB CsaM GAP32 | NC_019401 | 1,565,183 | 1,594,251 | 29,069 | Incomplete | Conserved |
| Phage | Gifsy 1 | NC_010392 | 2,471,266 | 2,503,308 | 32,043 | Intact | Conserved |
| Phage | vB SosS Oslo | NC_018279 | 3,002,249 | 3,050,660 | 48,412 | Intact | Diverse |
| Phage | P4 | NC_001609 | 3,342,337 | 3,360,947 | 18,611 | Incomplete | Conserved |
| PAIa | SPI-1 | NC_006905_P5 (43.5 kb) | 3,445,488 | 3,489,766 | 44,279 | Intact | Conserved |
| PAIa | SPI-2 | NC_006905_P3 (41.8 kb) | 1,971,307 | 2,013,190 | 41,884 | Intact | Conserved |
| PAIa | SPI-3 | NC_006905_P6 (12.8 kb) | 4,410,311 | 4,423,076 | 12,766 | Intact | Conserved |
| PAIa | SPI-4 | NC_006905_P7 (26.7 kb) | 227,272 | 253,975 | 26,704 | Intact | Conserved |
| PAIa | SPI-5 | NC_006905_P1 (5.7 kb) | 1,690,962 | 1,697,840 | 6,879 | Intact | Conserved |
| PAIa | SPI-6 | NC_003198_P1 (58.7 kb) | 913,174 | 971,088 | 57,915 | Intact | Conserved |
| PAIa | SPI-9 | NC_003198_P4 (15.7 kb) | 3,325,575 | 3,341,871 | 16,297 | Intact | Conserved |
| PAIa | SPI-11 | NC_006905_P2 (15.7 kb) | 1,839,408 | 1,848,476 | 9,069 | Incomplete | Conserved |
| PAIa | SPI-12 | NC_006905_P4 (11.1 kb) | 2,837,772 | 2,848,978 | 11,207 | Intact | Conserved |
| PAIa | CS54 | AF140550 (25.3 kb) | 3,173,887 | 3,199,139 | 25,253 | Intact | Conserved |
SPIs sequences for comparison were downloaded from http://www.paidb.re.kr/browse_pais.php?m=p#Salmonella enterica.
Salmonella enterica serovar Infantis 119944 genome was found to possess eight bacteriophages in its chromosome (fig. 1A and table 1). Five of which (Burkholderia cenocepacia phage BcepMu; Salmonella Phage 103203 sal5; Cronobacter phage vB CsaM GAP32; Gifsy 1; and Enterobacteria phage P4) were present in all of the compared S. Infantis genomes. In contrast, a 59.2 kb Enterobacteria SfV phage (accession number NC_003444.1, spanning positions 529849–589113) was found only in the 119944 genome. Similarly, the 21.7 kb Escherichia phage pro483 (NC_028943, covering positions 6016–27791) and the 48.4 kb Salmonella phage vB SosS Oslo (NC_001609, integrated between positions 3002249 and 3050660) were found in only a subset of the S. Infantis genomes. These results show that while SPIs distribution is conserved among the tested S. Infantis genomes, bacteriophages repertoire is diverse and contributes significantly to the genetic diversification of S. Infantis strains.
Next, we compared the genetic similarity between the corresponding S. Infantis plasmids. Six out of the eight compared S. Infantis strains were found to harbor megaplasmids with size ranging from 178.2 to 316.1 Mb, whereas two S. Infantis strains (1326/28 and NCTC6703) did not carry any plasmid. Among the six S. Infantis megaplasmids found, only five were actually pESI-related (fig. 1B), carried by S. Infantis strains that were isolated between 2014 and 2017. These results are consistent with the notion that pESI plasmids are associated with emerging (recent) S. Infantis strains and thus far were not identified in older isolates (Aviv et al. 2014).
Despite very high sequence similarity between pESI-related plasmids that were isolated from different geographically regions, the pESI of strain 119944 was found to contain eight unique regions ranging in size between 166 and 3,079 bp (indicated as unique regions R1–R8 in fig. 1B), which were not present in any of the other pESI-like plasmids included in this cohort. These are modular regions comprising insertion sequences elements, transposases or hypothetical proteins found in various plasmids. Interestingly, instead of region 8 (R8), all other pESI-like plasmids contain a different mobile element (possibly a transposon that carries transposases and IS 6-like insertion sequences), encoding arsenic resistance genes cluster as well as the blaCTX-M-65 gene (coding for an extended-spectrum β lactamases), which are lacking in the 119944 pESI.
Another recent study (Gymoese et al. 2019) that have used incomplete genomes of 105 S. Infantis isolates identified 16 strains harboring a conserved pESI-like plasmids of ∼280–283 kb. None of these plasmids contains the blaCTX-M-1 or blaCTX-M-65 genes as reported in pESI-like ESBL-positive plasmids (Franco et al. 2015; Hindermann et al. 2017; Tate et al. 2017). These differences highlight the modular nature of pESI and its genetic plasticity facilitated by its ability to “mix and match” mobile genetic elements and integrate them into a conserved pESI backbone. Moreover, because the ESBL-positive pESI-like plasmids were isolated from S. Infantis strains in USA (Tate et al. 2017), Peru (Vallejos-Sanchez et al. 2019), Switzerland (Hindermann et al. 2017), and Italy (Franco et al. 2015), it is highly possible that these pESI derivatives are globally disseminated.
In summary, we applied a state-of-the-art hybrid assembly approach and determined a gap-free complete sequence of a S. Infantis emerging clone that harbors the virulence-resistance megaplasmid pESI. By a conservative genomic comparison with other complete S. Infantis genomes, we defined core presence of ten SPIs and five prophages and identified conserved and variable regions in the pESI plasmid. We showed that the genetic and phenotypic diversity (especially antimicrobial resistance) of emerging S. Infantis strains is shaped by a varying repertoire of chromosomal prophages and integration of different mobile genetic elements into a conserved pESI backbone.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study at the Gal-Mor laboratory was supported by grant numbers: I-41-416.6-2018 from the German-Israeli Foundation for Scientific Research and Development (GIF); A128055 from the Research Cooperation Lower Saxony—Israel (The Volkswagen Foundation); and 2616/18 from the joint ISF-Broad Institute program. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Literature Cited
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA.. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12(1):402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D, et al. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44(W1):W16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv G, Rahav G, Gal-Mor O.. 2016. Horizontal transfer of the Salmonella enterica serovar Infantis resistance and virulence plasmid pESI to the gut microbiota of warm-blooded hosts. MBio 7(5):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv G, et al. 2014. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ Microbiol. 16(4):977–994. [DOI] [PubMed] [Google Scholar]
- Aviv G, et al. 2017. The plasmid-encoded Ipf and Klf fimbriae display different expression and varying roles in the virulence of Salmonella enterica serovar Infantis in mouse vs. avian hosts. PLoS Pathog. 13(8):e1006559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv G, et al. 2019. Differences in the expression of SPI-1 genes pathogenicity and epidemiology between the emerging Salmonella enterica serovar Infantis and the model Salmonella enterica serovar Typhimurium. J Infect Dis. 220(6):1071–1081. [DOI] [PubMed] [Google Scholar]
- Bogomazova AN, et al. 2020. Mega-plasmid found worldwide confers multiple antimicrobial resistance in Salmonella Infantis of broiler origin in Russia. Int J Food Microbiol. 319:108497. [DOI] [PubMed] [Google Scholar]
- Cloeckaert A, et al. 2007. Dissemination of an extended-spectrum-beta-lactamase blaTEM-52 gene-carrying IncI1 plasmid in various Salmonella enterica serovars isolated from poultry and humans in Belgium and France between 2001 and 2005. Antimicrob Agents Chemother. 51(5):1872–1875. [pii] 10.1128/AAC.01514-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crim SM, et al. 2015. Preliminary incidence and trends of infection with pathogens transmitted commonly through food – Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006–2014. MMWR Morb Mortal Wkly Rep. 64(18):495–499. [PMC free article] [PubMed]
- ECDC 2014. Annual epidemiological report 2014 food- and waterborne diseases and zoonoses. Stockholm (Sweden: ): European Centre for Disease Prevention and Control. [Google Scholar]
- EFSA 2017. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J 15. doi:10.2903/j.efsa.2017.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco A, et al. 2015. Emergence of a clonal lineage of multidrug-resistant ESBL-Producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One 10(12):e0144802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mor O. 2019. Persistent infection and long-term carriage of typhoidal and nontyphoidal Salmonellae. Clin Microbiol Rev. 32. doi: 10.1128/CMR.00088-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mor O, et al. 2010. Multidrug-resistant Salmonella enterica serovar Infantis. Emerg Infect Dis. 16(11):1754–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymoese P, et al. 2019. WGS based study of the population structure of Salmonella enterica serovar Infantis. BMC Genomics 20(1):870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser E, et al. 2012. Clonal dissemination of Salmonella enterica serovar Infantis in Germany. Foodborne Pathog Dis. 9(4):352–360. [DOI] [PubMed] [Google Scholar]
- Havelaar AH, et al. 2015. World Health Organization Global Estimates and Regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 12(12):e1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindermann D, et al. 2017. Salmonella enterica serovar Infantis from food and human infections, Switzerland, 2010–2015: poultry-related multidrug resistant clones and an emerging ESBL producing clonal lineage. Front Microbiol. 8:1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriarte A, et al. 2017. Draft genome sequence of Salmonella enterica subsp. enterica Serovar Infantis strain SPE101, isolated from a chronic human infection. Genome Announc. 5. doi:10.1128/genomeA.00679-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen A, Hendriksen RS, Aaresturp FM, Ussery DW, Friis C.. 2011. The Salmonella enterica pan-genome. Microb Ecol. 62(3):487–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebana E, et al. 2004. Pediatric infection due to multiresistant Salmonella enterica serotype Infantis in Honduras. J Clin Microbiol. 42(10):4885–4888. [pii] 10.1128/JCM.42.10.4885-4888.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogrady N, et al. 2008. Prevalence and characterization of Salmonella Infantis isolates originating from different points of the broiler chicken-human food chain in Hungary. Int J Food Microbiol. 127(1–2):162–167. [pii] 10.1016/j.ijfoodmicro.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Rissman AI, et al. 2009. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25(16):2071–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross IL, Heuzenroeder MW.. 2008. A comparison of three molecular typing methods for the discrimination of Salmonella enterica serovar Infantis. FEMS Immunol Med Microbiol. 53(3):375–384. [DOI] [PubMed] [Google Scholar]
- Shahada F, Sugiyama H, Chuma T, Sueyoshi M, Okamoto K.. 2010. Genetic analysis of multi-drug resistance and the clonal dissemination of beta-lactam resistance in Salmonella Infantis isolated from broilers. Vet Microbiol. 140(1–2):136–141. [DOI] [PubMed] [Google Scholar]
- Szmolka A, et al. 2018. Molecular epidemiology of the endemic multiresistance plasmid pSI54/04 of Salmonella Infantis in broiler and human population in Hungary. Food Microbiol. 71:25–31. [DOI] [PubMed] [Google Scholar]
- Tate H, et al. 2017. Comparative analysis of extended spectrum beta- lactamase CTX-M-65-producing Salmonella Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob Agents Chemother. 61(7):e00488–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejos-Sanchez K, et al. 2019. Whole-genome sequencing of a Salmonella enterica subsp. enterica Serovar Infantis strain isolated from broiler chicken in Peru. Microbiol Resour Announc. 8. doi:10.1128/MRA.00826-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE.. 2017a. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom. 3:e000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE.. 2017b. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 13(6):e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama E, et al. 2015. A novel subpopulation of Salmonella enterica serovar Infantis strains isolated from broiler chicken organs other than the gastrointestinal tract. Vet Microbiol. 175(2–4):312–318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.