Abstract
Alphavirus nsP3 proteins contains long, intrinsically disordered, hypervariable domains, HVD, which serve as hubs for interaction with many cellular proteins. Here, we have deciphered the mechanism and function of HVD interaction with host factors in alphavirus replication. Using NMR spectroscopy, we show that CHIKV HVD contains two SH3 domain-binding sites. Using an innovative chemical shift perturbation signature approach, we demonstrate that CD2AP interaction with HVD is mediated by its SH3-A and SH3-C domains, and this leaves the SH3-B domain available for interaction with other cellular factor(s). This cooperative interaction with two SH3 domains increases binding affinity to CD2AP and possibly induces long-range allosteric affects in HVD. Our data demonstrate that BIN1, CD2AP and SH3KBP1 play redundant roles in initiation of CHIKV replication. Point mutations in both CHIKV HVD binding sites abolish its interaction with all three proteins, CD2AP, BIN1 and SH3KBP1. This results in strong inhibition of viral replication initiation.
1. Introduction
Alphaviruses are a group of important human and animal pathogens, which are widely distributed on all continents (Strauss and Strauss, 1994; Weaver et al., 2012). In natural conditions, alphaviruses are transmitted by mosquito vectors between amplifying vertebrate hosts. The Old World (OW) alphaviruses are generally less pathogenic than the New World (NW) representatives. In humans, they induce diseases characterized by rash, fever and arthritis (Weaver and Lecuit, 2015). Recently, one of the OW alphaviruses, chikungunya virus (CHIKV), has attracted attention because of the severity of the caused disease and rapid distribution of the virus to new areas (Rezza and Weaver, 2019). Currently, it is already circulating in both hemispheres (Weaver and Lecuit, 2015). In humans, CHIKV induces severe polyarthritis characterized by excruciating joint pain that can continue for years. Despite the public health threat, neither licensed vaccines nor therapeutic means against CHIKV infection are available.
CHIKV genome (G RNA) encodes only a handful of proteins. The nonstructural proteins, nsP1, nsP2, nsP3 and nsP4, are translated directly from G RNA as precursor polyproteins P123 and P1234 (Strauss and Strauss, 1994). Their gradual processing by nsP2-associated protease activity results in formation of viral replication complexes (vRCs) that demonstrate different, processing-dependent template specificities. As for other alphaviruses, CHIKV nsP1, nsP2 and nsP4 exhibit enzymatic functions that are required for RNA synthesis. The nsP3 protein is also an integral and indispensable component of vRCs, but it appears not to be directly involved in synthesis and posttranscriptional modifications of virus-specific RNAs. This protein contains conserved N-terminal macro and alphavirus unique (AUD) domains, and a C-terminal, ~200-aa-long, hypervariable domain (HVD) (Rupp et al., 2015). Alphavirus HVDs were proposed to be intrinsically disordered domains (IDPs) that contain combinations of short linear motifs mediating their interactions with several host proteins (Foy et al., 2013; Frolov et al., 2017; Kim et al., 2016; Meshram et al., 2018). Thus, HVDs serve as hubs for recruitment of cellular factors that are required for vRC formation and function (Kim et al., 2016). The important characteristic of these interactions with host proteins is the high level of redundancy and involvement of not a single but multiple protein family members and even multiple families of cellular proteins, such as FXR and G3BP families. This likely allows alphaviruses to replicate in varieties of cell types and tissues, in which expression of each particular protein may vary.
In the previous studies, we and others have identified a number of proteins that interacted with CHIKV nsP3 HVD in vertebrate and mosquito cells (Frolov et al., 2017; Kim et al., 2016; Meshram et al., 2018; Mutso et al., 2018; Neuvonen et al., 2011). These HVD-specific host factors included members of the G3BP and NAP1 families, and several Src homology 3 (SH3) domain-containing proteins, such as CD2AP, SH3KBP1 and BIN1. We have demonstrated that G3BP family members play an indispensable role in vRC formation by recruiting viral G RNA into the site of vRC assembly at the plasma membrane (Kim et al., 2016). However, the presence of G3BP-binding sites only in HVD was insufficient to support CHIKV replication (Meshram et al., 2018). These data strongly suggested that interaction with other cellular partners, including SH3 domain-containing proteins, is also a critical determinant of CHIKV replication. Understanding of HVD interactions with host factors may lead to development of new therapeutic means against CHIK fever, but they need to be fully characterized.
In this study, we applied NMR spectroscopy to define CHIKV HVD interaction with CD2AP. We have assigned the backbone resonances for the entire 199-aa-long CHIKV HVD and then identified the specific amino acids that directly interact with individual SH3 domains and their natural combination in CD2AP. Our data demonstrate that individual CD2AP SH3 domains bind to two different motifs in CHIKV HVD, although with different affinity. Importantly, in the N-terminal fragment of CD2AP that contains all SH3 domains, only two of three SH3 domains interact with specific sites in CHIKV HVD. This cooperative binding has strong positive effect on the affinity of CD2AP-HVD interaction. Strikingly, we found that binding of SH3 domains also induces long-range allosteric perturbation in the HVD peptide that mediates interaction with other host factors, G3BP1 and G3BP2. Point mutations in the binding sites of CHIKV HVD sites abolished interactions with CD2AP and other SH3 domain-containing cellular proteins, BIN1 and SH3KBP1. The same point mutations in HVD-coding sequence in CHIKV genome had profound negative effect on CHIKV infectivity and replication. The negative effect was particularly strong in the human fibroblasts, which are the primary target of CHIKV infection in vivo.
2. Materials and methods
Protein purification
CHIKV HVD-coding sequence (aa 325–523) was amplified from the infectious cDNA clone of CHIKV 181–25 and cloned into pE-SUMOpro-3 plasmid (LifeSensors Inc.). Sequence encoding Twin-Strep-tag (GSWSHPQFEKGGGSGGGSGGGSWSHPQFEK) was introduced into C-terminus of the fragment by PCR. The resulting plasmid was used to transform the E. coli strain Rosetta 2(DE3)pLacI (Novagen). The 15N- or 15N- and 13C-labelled proteins were produced by growing bacteria in M9 minimal media supplemented with [15N]NH4Cl and d-[13C6]glucose (Cambridge Isotope Laboratories). Protein expression was induced by 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) after cell reached the density of 1–1.2 OD600. Cells continued to grow for 16–18 h at 25° C. Freshly prepared or frozen pellets were lysed in Emulsiflex B15 (Avestin). The recombinant CHIKV HVD was purified on a HisTrap HP column (GE Healthcare). After cleavage of SUMO tag, CHIKV HVD was additionally purified on StrepTactin Sepharose column (GE Healthcare). Size exclusion chromatography on Superdex 200 10/30 column (GE Healthcare) in phosphate buffer [50 mM N2HPO4 pH 6.8, 200 mM NaCl, 2 mM tris(hydroxypropyl)phosphine (TCEP)] was used as a final purification step. The purified CHIKV HVD was concentrated to ~0.3–0.5 mM.
Nucleotide sequences of all CD2AP SH3 domain-containing constructs were codon-optimized for expression in E. coli, and synthetic DNA fragments were obtained from Integrated DNA Technologies. They were cloned into pE-SUMOpro-3 plasmid (LifeSensors Inc.). The resulting sequences encoded His-tags at the N-termini. The aa sequences of all expressed proteins are presented in Supplementary Fig. 13. Plasmids were transformed into E. coli BL21 Star (DE3) (Thermo Fisher), and proteins were produced in M9 media. The expression was induced by 1 mM IPTG after cells reached the density of 1–1.2 OD600. Then cells continued to grow at 25°C for 16–18 h. Freshly prepared or frozen cell pellets were lysed as described above. The recombinant proteins were first purified on a HisTrap HP column (GE Healthcare), followed by buffer exchanged using HiPrep 26/10 desalting column (GE Healthcare). Purified proteins were concentrated to ~1 mM.
The protein’s purity and identity were confirmed by SDS-PAGE and mass spectrometry. In the CD2AP derivatives, the first methionine was present in less than 50% of purified products. Protein concentrations were determined on 280 nm using an extinction coefficients of 22,760 M−1cm−1 (CHIKV HVD), 9,530 M−1cm−1 (SH3-A), 12,660 M−1 cm−1 (SH3-B), 13,940 M−1 cm−1 (SH3-C) and 37,410 M−1 cm−1 (SH3-All), which were determined using ProteinCalculator v3.4 (http://protcalc.sourceforge.net/).
NMR spectroscopy
All NMR experiments were performed in the buffer containing 25 mM Na-phosphate pH 6.8, 50 mM NaCl, 1 mM TCEP, 0.02% NaN3 and 10 (v/v) % D2O.
Targeted acquisition (TA) (Isaksson et al., 2013; Jaravine and Orekhov, 2006; Jaravine et al., 2008; Unnerstale et al., 2016)
Two sets of NMR experiments were recorded. The first was done at 25° C on a 700MHz Bruker AVANCE III-HD spectrometer equipped with 5mm CP-TCI probe. The second was performed at 20° C on an 800MHz Bruker AVANCE III-HD spectrometer equipped with 3mm CP-TCI probe. At both temperatures, the TA procedure was implemented using the non-uniformly sampled best-TROSY experiments (Favier and Brutscher, 2011): 3D HNCO, 3D HNCOCA, 3D HNCA, 3D HNCACO, 3D HNCOCACB and 3D HNCACB. The latter two experiments were optimized for detection of cross peaks of 13Cβ nuclei, and all these spectra were recorded with a relaxation delay of 0.2 s. The experimental parameters for the 3D experiments are summarised in Supplementary Table 1.
5D HACACONH with XLSY approach
The 5D HACACONH spectrum was recorded at 20° C on an 800MHz Bruker AVANCE III-HD spectrometer equipped with 3mm CP-TCI probe and reconstructed using XLSY approach (Pustovalova et al., 2018). The experimental set included 300 hyper-complex non-uniformly sampled (NUS) points and ten 2D radial sampled (RS) projections: four orthogonal projections (1HNi/15Ni, 1HNi/13COi−1, 1HNi/13Cαi-1, 1HNi/1Hαi-1); six diagonal projections including: one projection with simultaneous evolution in all four indirect spectral dimensions (i.e. all dimensions except for 1HNi), all four projections with simultaneous evolution of three out of four indirect dimensions, and one projection with evolutions in 13Cαi-1 and 1Hαi-1 directions. The data were acquired with 8 scans on the time domain grid of 128×128×128×128 complex points with spectral width/acquisition time of 26 ppm/60 ms for 15N; 6 ppm/106 ms for 13CO; 50 ppm/13 ms for 13Cα and 3.5 ppm/46 ms for 1Hα. Additionally, we recorded a 3% NUS 3D HNCO spectrum with the same spectral width and acquisition times as in the corresponding dimensions of the 5D HACACONH. The 3D HNCO spectrum was reconstructed using the IST-VE Compressed Sensing algorithm3,5 in the MddNMR software (Kazimierczuk and Orekhov, 2011; Orekhov and Jaravine, 2011), and then used for processing of the 5D HACACONH spectrum as an independent 3D projection. The direct dimension of the 5D experiment was processed in TopSpin3.5 software (Bruker BioSpin), and the amide region (8.6–7.5 1H ppm, 160 points) was extracted. Further processing was performed using a home-built script (Pustovalova et al., 2018) in MATLAB software. All the RS and NUS data, which were acquired as hyper-complex time domain points with quadrature detection for all indirect spectral dimensions, were converted to 5D complex virtual echo representation (Mayzel et al., 2014). This resulted in 4 orthogonal and 26 independent diagonal 2D projections of the 5D spectrum. For each of 160 points in the direct spectral dimension, the final experimental data consisted of 4681 complex points of NUS data. The final 5D HACACONH spectrum reconstructed by the XLSY procedure contained 29150 points statistically significant non-zero intensities. A peak list for the 5D spectrum was obtained using a home-built script in MATLAB software and manually verified.
Assignment of CHIKV-HVD
Clean peak lists provided by the TA procedure were mostly automatically assigned by FLYA software (Schmidt and Guntert, 2012). The peak lists and results of the automatic assignment for two temperatures 20°C and 25°C were further verified manually using CARA software (Keller, 2004) versus high-resolution 3D spectra that were also provided by the TA. These spectra acquired at two temperatures were the key for resolving ambiguities and obtaining the full spectrum assignment. The peak list from the 5D HACACONH spectrum was used as an independent reference and control for the curated TA assignment as well as a source of 1Hα chemical shifts. Additional validation was performed through selective type MUSIC experiments (Schubert et al., 2001) allowing to distinguish groups of amino acid types and their consecutive residues.
Binding experiments were performed by recording 2D best-TROSY (1H −15N) spectra on Bruker Avance III spectrometer equipped with a cryo-enhanced QCI-P probe and operating at 600MHz. The used parameters were as follows: relaxation delay of 0.3 s, 400 t1 increments and NS=16. The control 1D spectra were used to assess the purity and stability of the samples. 0.1mM of TSP was added as a reference for chemical shift. All NMR spectra were processed by TopSpin3.5 software. Unlabeled SH3-A, SH3-B, SH3-C and SH3-All proteins were titrated into uniformly 15N-labelled CHIKV HVD sample at 25°C.
Analysis of the spectra was performed using CcpNmr software package. The magnitude of the combine chemical shift deviations (Δδ(H+N)) was calculated using equation 1 (Ortega-Roldan et al., 2009):
| (1) |
in which the proton (Δδ(H)) and the nitrogen (Δδ(N)) chemical shift perturbations (CSP) were defined as a difference in chemical shift between the equilibrium mixture of the bound (δ1Hbound and δ15Nbound, correspondingly) and the free states (δ1Hfree and δ15Nfree, correspondingly) induced by the different proteins (eq. 2).
| (2) |
The normalizing coefficient of value 6.51 in eq. 1 between proton (Δδ(H)) and the nitrogen (Δδ(N)) CSP is adopted from elsewhere (Ortega-Roldan et al., 2009).
KD values were calculated by fitting equation (3) for both KD and Δδmax using Δδ(H+N) for the residues described in the text for each site (Williamson, 2013).
| (3) |
Δδobs is Δδ(H+N), [P]t is total protein concentration, [L]t is total ligand concentration.
Cell cultures
The BHK-21 cells were kindly provided by Paul Olivo (Washington University, St. Louis, MO). The NIH 3T3, BJ-5ta, MRC-5, HFF-1 and HeLa cells were obtained from the American Tissue Culture Collection (Manassas, VA). BHK-21, NIH 3T3, HEK 293 and HeLa cells were maintained in alpha minimum essential medium supplemented with 10% fetal bovine serum (FBS) and vitamins. BJ-5ta, MRC-5 and HFF-1 cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% FBS.
Plasmid constructs
Plasmid encoding infectious cDNA of CHIKV 181–25 containing GFP under control of subgenomic promoter was described elsewhere (Kim et al., 2016). Mutations indicated in Fig. 6 were introduced using standard recombinant DNA techniques. Plasmids encoding cDNA of VEEV replicons containing Flag-GFP and Flag-GFP fused with CHIKV HVD were described elsewhere (Frolov et al., 2017). The sequence of mutant CHIKV HVD (mHVD) was introduced using standard cloning techniques. Nucleotide sequences of the plasmids are available from the authors upon request.
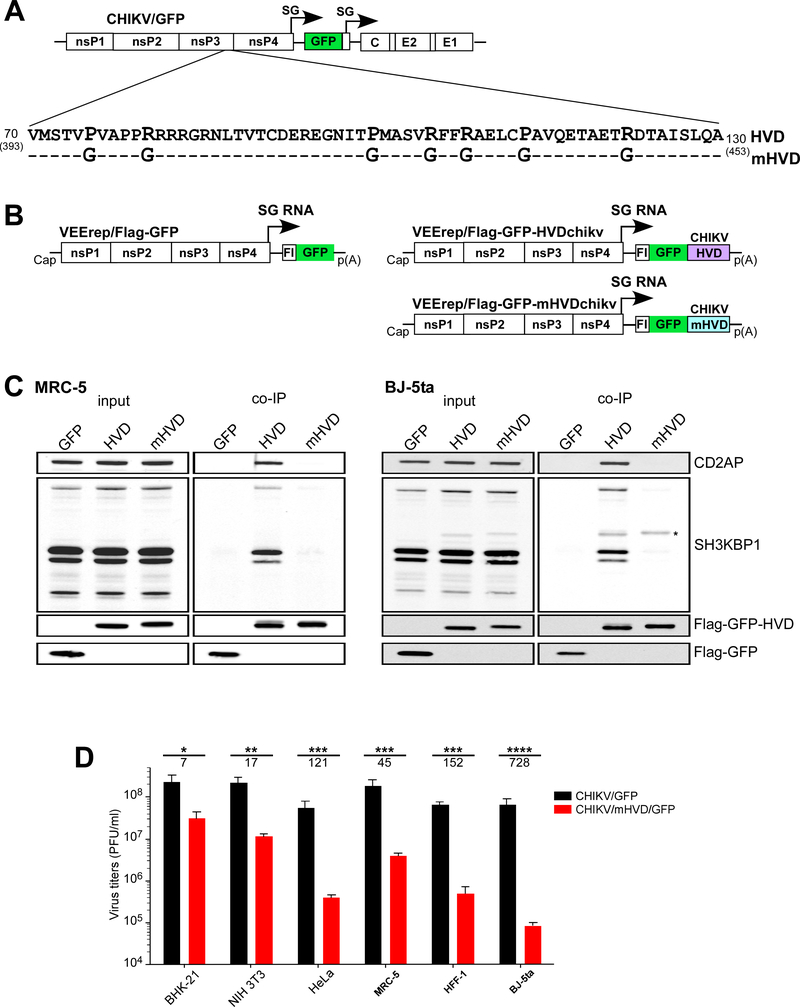
Fig. 6. Mutation is Site 1 and Site 2 of CHIKV HVD abolish binding of CD2AP and SH3KBP1 and strongly inhibit viral replication.
(A) The schematic presentation of CHIKV/GFP genome and mutations introduced into CD2AP-binding sites of CHIKV HVD. Dashes indicate identical aa. (B) The schematic presentation of the designed VEEV replicons, which encoded Flag-GFP alone, and wt and mutant HVDs fused with Flag-GFP. (C) Human MRC-5 and BJ-5ta cells were infected with VEEV replicons encoding Flag-GFP, or Flag-GFP fused with either wt or mutant CHIKV HVD. Protein complexes were isolated using magnetic beads with anti-Flag MAbs and further analyzed by Western blot using indicated antibodies. An asterisk marks the nonspecific staining of Flag-GFP-HVDs. (D) Equal numbers of indicated cells in the wells of 6-well plate were infected with the indicated viruses at an MOI of 0.04 PFU/cell. After incubation for 15 h at 37° C, media were harvested, and viral titers were determined by plaque assay on BHK-21 cells. Significances of differences among the values were determined by two-tailed unpaired t-test (*, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001; n=3). Data are presented as mean with SD (n=3). Numbers under asterisks indicate average fold differences between the titers.
Western blot analysis
Equal numbers of different cells were lysed directly in protein gel loading buffer. Proteins were separated in 4–12% NuPAGE gels (ThermoFisher) and transferred to nitrocellulose membrane. The following antibodies were used for detection of cellular proteins: anti-CD2AP rabbit polyclonal antibodies (sc-9137, SantaCruz), anti-SH3KBP1 monoclonal antibodies (sc-166862, SantaCruz), anti-BIN1 rabbit polyclonal antibodies (13679S, Cell Signaling) and anti-GAPDH monoclonal antibodies (66031–1-Ig, Proteintech). Secondary Abs labeled with IRDye 680RD or IRDye 800CW infrared dyes were acquired from LI-COR Biosciences. Membranes were imaged and analyzed on Odyssey Imaging System (LI-COR Biosciences).
Co-immunoprecipitation and Western blot analysis
MRC-5, Bj-5ta and NIH 3T3 cells (2×106) were infected with packaged VEEV replicons expressing Flag-GFP, Flag-GFP-HVDchikv and Flag-GFP-mHVDchikv at an MOI 20 inf.u/cell. Cells were harvested at 3–4 h post infection (p.i.). Protein complexes were isolated from post-nuclear fraction using magnetic beads loaded with anti-Flag MAbs (Sigma) as previously described (Kim et al., 2016). The lysates and eluates were separated by electrophoresis in 4–12% NuPAGE gels (ThermoFisher) followed by transfer to nitrocellulose membrane. The following antibodies were used for detection of cellular proteins: anti-CD2AP monoclonal antibodies (sc-9137, SantaCruz), anti-SH3KBP1 monoclonal antibodies (sc-166862, SantaCruz), anti-Flag monoclonal antibodies (F1804, Sigma). Secondary Abs labeled with IRDye 680RD or IRDye 800CW infrared dyes were acquired from LI-COR Biosciences. Membranes were imaged and analyzed on Odyssey Imaging System (LI-COR Biosciences).
RNA transcriptions
Plasmids were purified by ultracentrifugation in CsCl gradients and linearized by Not I digestion. Viral G RNAs were synthesized by using SP6 RNA polymerase in the presence of cap analog (New England Biolabs) according to the manufacturer’s recommendations (Invitrogen). The yields of the transcripts were analyzed by agarose gel electrophoresis under nondenaturing conditions. Aliquots of the reaction mixtures were used without additional purification for electroporation of BHK-21 cells in the previously described conditions. Viruses were harvested at 24 h post electroporation, and titers were determined by plaque assay on BHK-21 cells. Packaging of VEEV-based replicons into infectious virions was performed as described elsewhere (Meshram et al., 2018).
Viral infections
5×105 cells were seeded in the wells of 6-well Costar plates in the corresponding media. After 4 h-long incubation at 37°C, cells were infected at MOIs indicated in the figure legends in 0.2 ml PBS supplemented with 1% FBS. After incubation for 1 h at 37°C, cells were washed 3 times with 1 ml PBS and then incubated in the corresponding media for the times indicated in figure legends. Viral titers in the harvested samples were determined by plaque assay on BHK-21 cells (Akhrymuk et al., 2018).
RT-qPCR
Viral genomic RNAs were isolated from 100 μl aliquots using Direct-Zol RNA kit according to the manufacturer’s instructions (Zymo Research). These RNA samples were used for cDNA synthesis with Superscript IV reverse transcriptase (Invitrogen) using reverse primer 5’-TTCAAGAGCAGCGAATCTACC-3’. cDNAs were further used for qPCR analysis using above reverse primer and the direct primer 5’-GCAAGTCAGCCATTATCAAGAAC-3’. The qPCRs were performed using SsoFast EvaGreen supermix (Bio-Rad) in a CFX96 real-time PCR detection system (Bio-Rad). The specificities of the qPCR products were confirmed by analyzing their melting temperatures.
Viral infectivity
Different cell lines were seeded into the wells of 6-well Costar plates at the concentration of 5×105 cells/well. They were infected at the same time with serial 10-fold dilutions of viral stocks for 1 h at 37°C. Cells were covered with 0.5% agarose supplemented with DMEM and 3% FBS. Foci of GFP-positive cells were imaged at 48 h p.i., then incubation continued, and plaques were stained with crystal violet at 70 h p.i. to determine viral titers specific to each cell line (PFU/ml). Since the designed mutant was unable to form plaques on most of human cell lines, the cell-specific titers were determined in GFP-positive foci-forming or infectious units (inf.u/ml).
Data availability
The final chemical shift assignments of the backbone 1HN, 15N, 13CO, 13Cα, 13Cβ atoms of CHIKV HVD at 20°C have been deposited in the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu/) under BMRB accession code 27868. Other data are available from corresponding authors upon reasonable request.
3. Results
Human fibroblasts preferentially express CD2AP and SH3KBP1
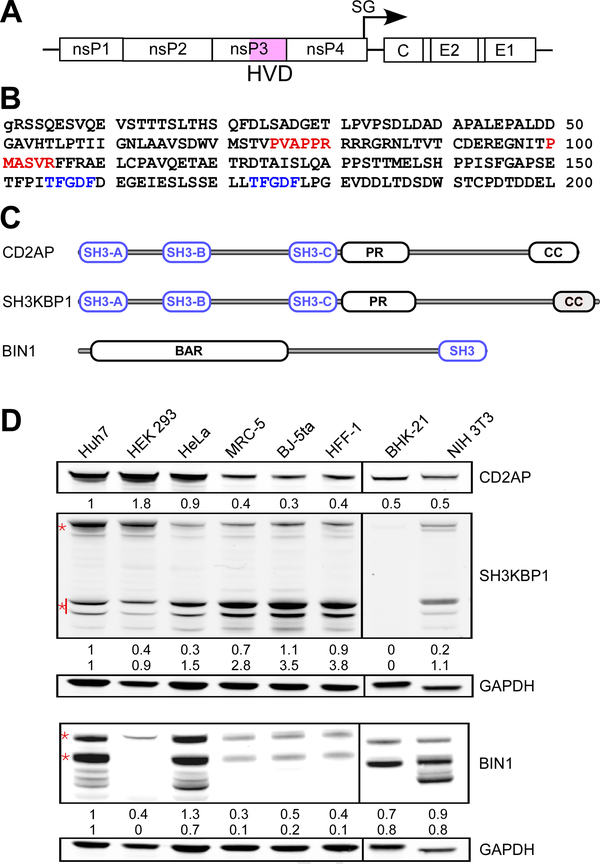
The 199-aa-long hypervariable domain (HVD) of CHIKV nsP3 is is located at the C-terminus of nsP3 (Figs. 1A and B). Previously, it has been shown to interact with at least three cellular SH3 domain-containing proteins CD2AP, BIN1 and SH3KBP1 (Frolov et al., 2017; Meshram et al., 2018). The latter host factors differ in the number of encoded SH3 domains, their sequences and location (Fig. 1C). In initial experiments, we compared the levels of expression of these proteins in standard, commonly used cell lines, such as hamster BHK-21, mouse NIH 3T3 and human HeLa, HEK 293 and Huh7 cells, and in human fibroblasts, which included MRC-5, BJ-5ta and HFF-1 cells. The latter cell lines were investigated because dermal fibroblasts are known to be the primary target of CHIKV replication during initial rounds of in vivo infection (Schilte et al., 2010). We found that in these cell lines, the SH3 domain-containing proteins were present at very different concentrations (Fig. 1D). Multiple BIN1 isoforms have been detected in both rodent, BHK-21 and NIH 3T3, and human, Huh7 and HeLa, cells. BIN1 expression in HEK 293 cells was barely detectable, and its expression in all human fibroblasts, MRC-5, HFF-1 and BJ-5ta cells, was also very low. CD2AP and SH3KBP1 proteins were represented more universally in the analyzed cells, particularly in all the cell lines of human fibroblasts. Since these cells are highly susceptible to CHIKV infection and are relevant to CHIKV pathogenesis, we focused our further experiments on analysis of CHIKV nsP3 HVD interaction with CD2AP. SH3KBP1 is a close homolog of CD2AP (Fig. 1C), and the generated CD2AP-based results are likely applicable to this protein as well.
Fig. 1. Differential expression of CD2AP, SH3KBP1 and BIN1 in the cells susceptible to CHIKV infection.
(A) The schematic presentation of CHIKV genome. (B) The aa sequence of CHIKV 181–25 nsP3 HVD, which corresponds to aa 325–523 of the full-length nsP3 protein. The previously predicted motifs that could interact with SH3 domain-containing proteins are depicted in red. Two sites of G3BP1/2 binding are depicted in blue. (C) The schematic presentation of the domain structures of CD2AP, SH3KBP1 and BIN1. (D) Western blot analysis of CD2AP, SH3KBP1 and BIN1 expression in different cell lines. Of all isoforms, the bands, which were used for quantitative analysis of protein expression, are marked by red asterisks.
All CD2AP SH3 domains can interact with two sites in CHIKV HVD
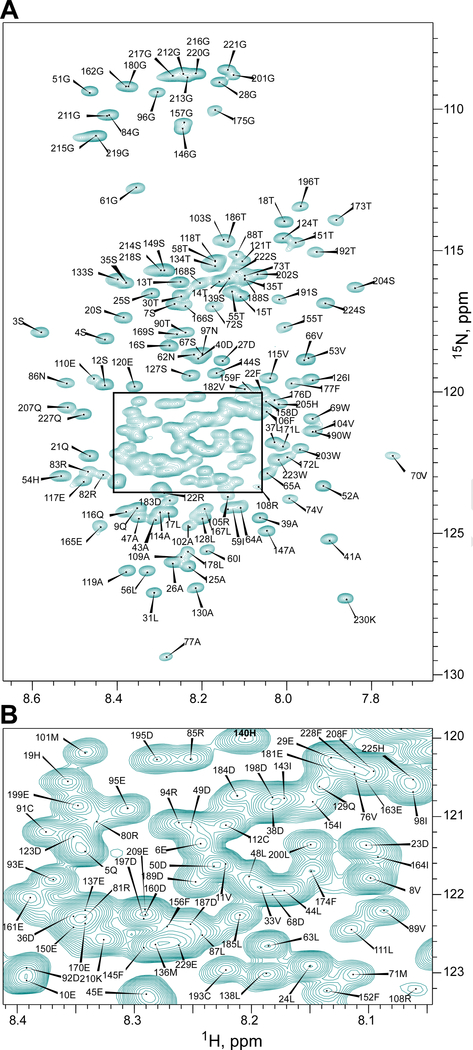
We have previously performed a preliminary backbone assignment of 199-aa-long CHIKV HVD and demonstrated that it is an intrinsically disordered protein domain (IDP) (Meshram et al., 2018). In this work, application of targeted acquisition (TA) (Isaksson et al., 2013; Jaravine and Orekhov, 2006; Jaravine et al., 2008; Unnerstale et al., 2016), extra-large spectroscopy (XLSY) (Pustovalova et al., 2018), and manual verification led to almost complete assignment of the backbone resonances of CHIKV HVD: 97% of 1HN, 97% of 15N, 98% 13Cα, 96% of 13Cβ, 98% of 13CO and 73% of 1Hα signals (Fig. 2 and Fig. S-1, supplementary data). The assignment data unambiguously proved that CHIKV HVD is disordered in solution (Supplementary Fig. 2) and allowed us to characterize in detail its interactions with the SH3-binding domains of CD2AP protein.
Fig. 2. 1H-15N-TROSY-HSQC spectrum of the CHIKV-HVD with assignment of the cross peaks that belong to the amide groups.
(A) The area between 1H: 8.65–7.65 ppm and 15N: 130–107 ppm that covers all observed cross peaks. (B) The magnified overlapped spectrum region indicated in panel (A). The cross peaks were labeled according to the numbers of the aa in CHIKV-HVD in the sequence presented in Fig. 1B. Spectrum was acquired on a 800MHz Bruker AVANCE III-HD spectrometer at 20° C.
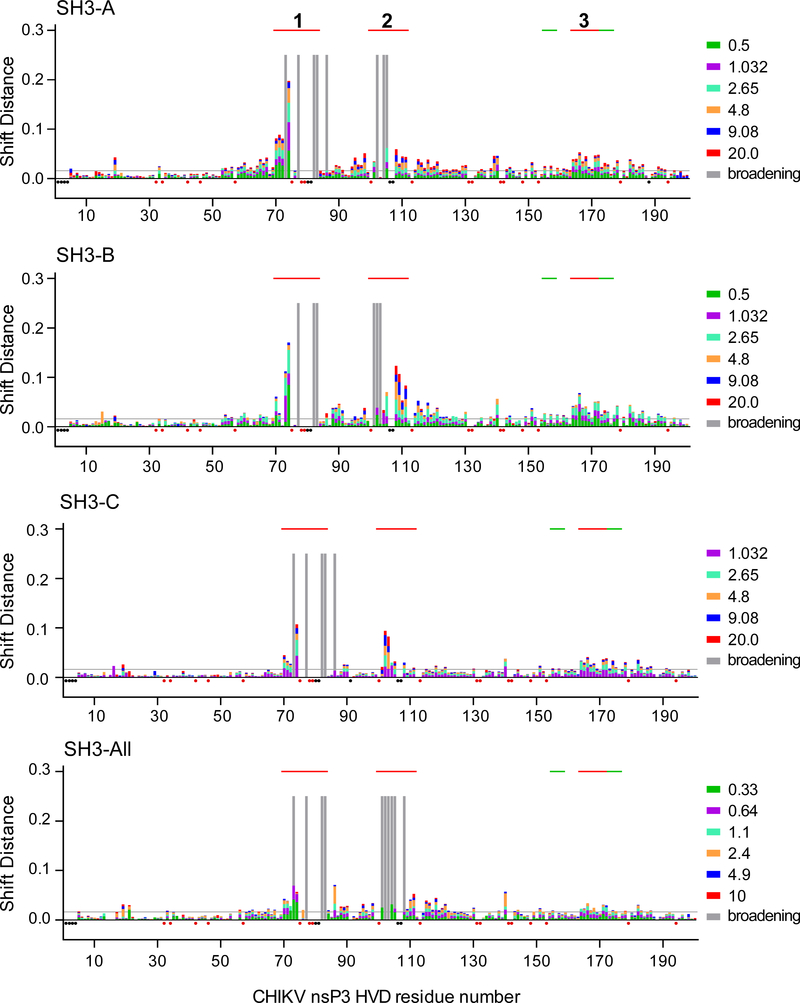
CD2AP contains three SH3 domains that are separated by long disordered linkers (Fig. 1C and Supplementary Fig. 3). To further understand CD2AP binding to CHIKV HVD, we first analyzed interactions of individual SH3 domains with the full-length HVD. Mapping of intermolecular interfaces of protein complexes was done by assessing chemical shift perturbations (CSP) in the 1H-15N best-TROSY spectra of the 15N-labeled CHIKV HVD in the presence of different concentrations of unlabeled individual SH3 domains (Supplementary Figs S4–6). All of the domains unambiguously demonstrated interactions with CHIKV HVD, albeit the induced specific changes in the CHIKV HVD NMR spectra were different (Fig. 3). The most significant changes in the chemical shift (2 standard deviations, 2σ) were found in three sites: aa 70–83 (Site 1), aa 101–111 (Site 2) and aa 164–172 (Site 3).
Fig. 3. CHIKV HVD interaction with individual SH3 domains of CD2AP and their combination in SH3-All.
Weighted chemical shift perturbations of the amide bonds (1H and 15N nuclei) in the 15N-labeled CHIKV HVD are presented as superimposed bars on the CHIKV HVD sequence. The lists of the titration ratios (SH3:HVD) are indicated at the right side of the panels. The amino acids that showed broadening of cross peaks during titrations are indicated by grey bars. The unassigned resonances and prolines are indicated by black and red dots, respectively. The thresholds of interacting/non-interacting aa are indicated as grey lines at the level of two standard deviations (2σo). Solid red lines on the tops of the panels indicate the positions of binding sites 1 and 2 and allosteric site 3. The green lines show the positions of G3BP-binding sites.
All three SH3 domains had the strongest effects on the CSP specific to Site 1 (aa 70–83, Fig. 3) that contained the 75PVAPPR80 sequence. The latter peptide resembles both canonical (PxxPxR) and non-canonical (PxxxPR) SH3 domain-binding motifs (Saksela and Permi, 2012). It also matches the consensus motifs that were described for CD2AP SH3-A and SH3-B domains (PT(V,I,L)P(A,F,V)xxR) and for SH3-C domain (PT(I,V)P(A)A(Q,T,P)P(A)R) (Rouka et al., 2015). The addition of any of three SH3 domains to CHIKV HVD at 0.2:1 ratio led to immediate resonances broadening in the spectrum of several residues in Site 1 (77A, 82R and 83R), and their corresponding cross peaks fell below the detection limit (marked as gray bars in Fig. 3). Noteworthy, these cross peaks remained exchange-broadened even in the presence of larger excess of SH3 protein (20:1 ratio). Similar absence of cross peaks for a bound complex has been previously described for several other IDPs (Jensen et al., 2011; Schneider et al., 2015; Schwarten et al., 2013). These studies suggested that interacting kinetics can be characterized by more than two conformations and, as a result, the bound state exhibits additional dynamics on the μs-ms timescale. Residues 80R and 81R were excluded from the analysis due to their location in a crowded part of the spectra. Based on the data, we concluded that the motif 75PVAPPRRRR83 functions as the core binding site, or docking site, for all individual CD2AP-specific SH3 domains.
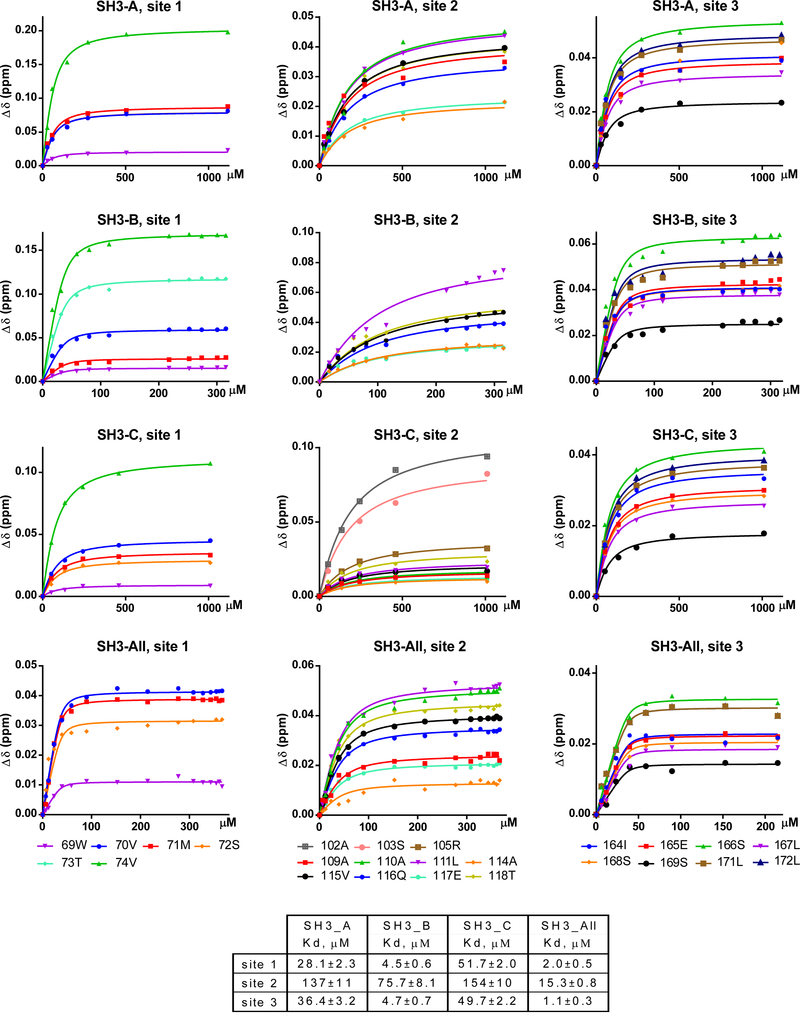
The cross peaks for CHIKV HVD residues in the N-terminal fragment of Site 1 (70V, 71M, 72S and 74V) exhibited significant shifts with increasing concentrations of SH3 domains. However, these changes were dramatically different for each tested domain, suggesting variations in kinetics and affinities of interactions (Fig. 4A and Supplementary Fig. 8). Consequently, KD values, which were determined by global fitting, greatly differed (Fig. 5). The SH3-B domain demonstrated the strongest binding with KD of 4.5±0.6 μM. The SH3-A and SH3-C exhibited lower affinities with KD of 28.1±2.3 and 51.7±2.0 μM, respectively.
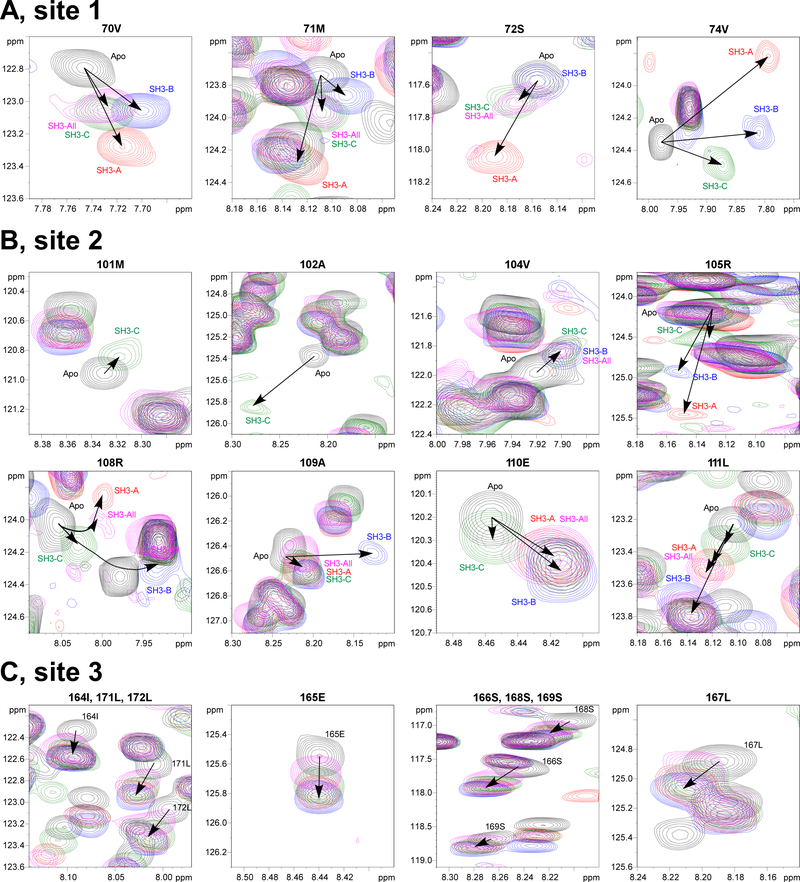
Fig. 4. Different CHIKV HVD/SH3 complexes demonstrate specific CSP signatures.
Superposition of the expanded 1H-15N best-TROSY spectra of the apo 15N-CHIKV HVD (black cross peaks) with the corresponding spectra of 15N-CHIKV HVD, titrated with unlabelled SH3 domains. The SH3-A-, SH3-B-, SH3-C- and SH3-All-specific cross peaks are indicated in red, blue, green and pink, respectively. (A) The N-terminal element of Site 1, represented by aa 70–74; (B) The N- and C-terminal elements of Site 2, represented by aa 101–105 and 108–111, respectively; and (C) Site 3, represented by aa 164–172. Arrows indicate the directions of the chemical shift perturbations in the presence of the SH3 domains at increasing concentrations. Several cross peaks of these sequences are not shown because they are either located in a crowed area of the spectrum or the intensity decreased below the detection limit.
Fig. 5. Individual CD2AP SH3 domains and SH3-All have different affinities to Sites 1 and 2 in CHIKV HVD.
Experimental titration and globally fitted curves of (1H, 15N) weighted CSPs extracted from 1H-15N best-TROSY spectra of the 15N-CHIKV HVD in the titration experiments with different concentrations of unlabelled domains SH3-A, SH3-B, SH3-C and SH3-All.
Changes in CSPs (Δδ(H+N)) with increasing concentrations of the individual SH3 domains depend on the underlying exchange rate (kex) between free and bound states of the proteins and on the relation between kex and the CS difference between free- and bound-state signals (Δδ(H+N)). In addition, the maximum difference (Δδ(H+N)) is determined by the local structural rearrangements of the interacting aa of IDP with SH3 domains and by the internal structural properties of the interacting complex. Thus, the interplay between kex and (Δδ(H+N)) may provide unique CSP signatures in 2D plane of 1H- 15N spectra of the individual aa of the IDP, which are involved in the dynamical interaction. Keeping this in mind, we attempted to map the CSP signature for each complex in 2D plane of 1H- 15N spectrum of HVD for the resonances 70–74 of Site 1.
Interaction of SH3-C domain with Site 1 of CHIKV HVD induced linear movement of all the 1H–15N correlation peaks corresponding to aa 70–74, towards the cross peaks of the bound state (Fig. 4 and Supplementary Fig. 8). This was suggestive that these residues were in fast exchange regime, i.e. kex is higher than the chemical shift difference between the free state and the bound state signals (kex ≫ Δδ(H+N)). This pattern correlated with the lower affinity of SH3-C to Site 1 (Fig. 5). Formation of SH3-A and SH3-B complexes with CHIKV HVD in the Site 1 followed different routes. In the presence of these proteins, residue 74V exhibited the largest CSP, possibly due to close proximity to aromatic spins of the SH3 domains. The increase in concentration of SH3-A or SH3-B domains resulted in broadening of the resonances and a reduction of their intensities half way through the titration (Supplementary Fig. 8). This was an indication of intermediate exchange regime (kex ≈Δδ(H+N)) of 74V. For the N-terminal aa 70–72, the shielding induced by binding of the SH3 domains was reduced and resulted in smaller CSP difference between free and bound states. Therefore, the CSP for those cross peaks of HVD/SH3-A and HVD/SH3-B complexes were in the fast exchange regime similar to CHIKV HVD/SH3-C complex. Importantly, both the directions of CSPs and the maximum values, Δδ(H+N) of the cross peaks 70–74 were specific for each applied SH3 domain (Fig. 4). These parameters did not depend on exchange regime and likely reflected structural signatures of different SH3-HVD complexes. Therefore, we proposed that they could be used as a CSP signature for the identification of binding of particular SH3 domain in the context of combination of SH3s in the natural CD2AP aa sequence.
The second binding site in CHIKV HVD that exhibited significant CSP was identified between aa 101–118 (Fig. 3). This peptide contains a non-canonical SH3 domain-binding motif 100PMASVR105, which demonstrates similarity with previously described CD2AP SH3-binding site (Rouka et al., 2015). As was found for Site 1, the resonances of the residues in the docking site (aa 101–105) for SH3-A and SH3-B complexes were broadening with increasing concentration of the SH3 domains. The signals also became undetectable at saturation point (Supplementary Fig. 9). For the SH3-C complex, the corresponding 1H–15N correlation peaks moved linearly towards the chemical shifts of the bound state. The cross peaks in all three complexes under saturated conditions were only detected starting from residue 105. This allowed an unambiguous definition of the directions of CSPs and their max values (Δδ(H+N)) (Fig. 4 and Supplementary Fig. 10). Residues 106 and 107 are located in a crowded region and thus, were not analyzed.
Residues 108–118 exhibited large CSP for SH3-B and SH3-A, but not for SH3-C domains. Moreover, aa 108R, 109A and 110E in the complexes with SH3-B and SH3-A, CSP trajectories demonstrated presence of curvatures. This was suggestive of the existence of either heterogeneity in binding or the possibility of existence of two or more binding events in Site 2 (Zuiderweg, 2002). Similar observation has been previously made for CIN85A (SH3KBP1) binding to Cbl-b-derived peptide, and simultaneous interaction of two domains with overlapping binding sites (type I and type II) was proposed (Ababou et al., 2009). This possibility is under investigation. Nevertheless, we were able to clearly distinguish unique CSP signatures for interactions of each SH3 domain with the Site 2 (Fig. 4 and Supplementary Fig. 10).
We determined apparent KD values for all complexes in Site 2 (Fig. 5). Compared to Site 1, all SH3 domains exhibited 3-to-15-fold lower binding affinity to Site 2. The SH3-B domain had the highest affinity (KD 75.7±8.1 μM), but it was ~15-fold lower than that to Site 1 (KD 4.5±0.6 μM). The interactions of SH3-A and SH3-C domains with Site 2 were even weaker, and had KDs of 137±11 and 154±10 μM, respectively.
Binding of SH3 domains to Site 1 induces long-range allosteric perturbation
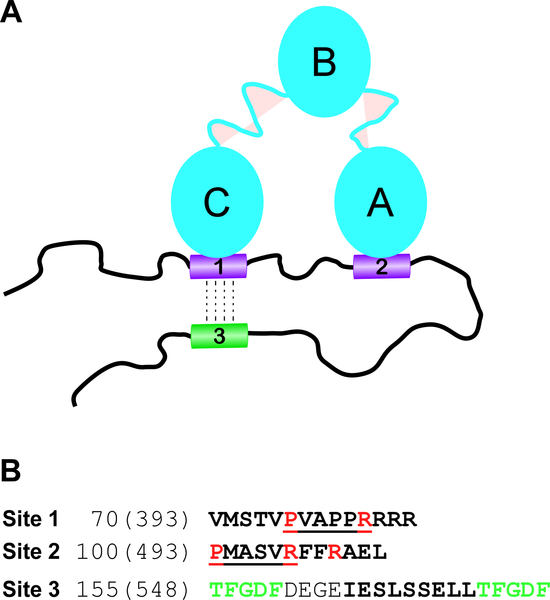
In addition to Sites 1 and 2, in the presence of individual SH3 domains at increasing concentrations, the cross peaks for residues 164–172 of CHIKV HVD (Site 3) were also clearly shifting (Fig. 4C and Supplementary Fig. 11). At saturating concentrations of the SH3 domains, the maximum CSP values of all residues in this site were above the threshold, defined as 2σo deviations (Fig. 3). The distinguishing characteristic of the CSP signature of the Site 3 was that neither the direction of trajectories nor the maximum CSP values for each residue depended on the type of the SH3 domain. Interestingly, for each used SH3 domain, the estimated by global fitting apparent KD values were similar to those determined for Site 1 (Fig. 5). These data suggests that Site 3 does not directly bind SH3 domains, and the detected CSPs were rather the result of allosteric responses to the binding of any SH3 domain to Site 1. This hypothesis is in part supported by other data. (i) We have previously demonstrated that deletion of Site 3-containing fragment in CHIKV HVD did not abolished CD2AP/HVD interaction (Meshram et al., 2018). (ii) Site 3 does not contain prolines and aa sequences exhibiting similarity with known SH3 domain-binding motifs. (iii) Site 3 is positioned between two binding sites of cellular G3BP proteins (Schulte et al., 2016). This connecting peptide has been shown to form an α-helix upon binding the NTF2 domain of G3BP1 to the upstream- and downstream-located motifs in SFV HVD (Schulte et al., 2016). However, further studies are needed to provide mechanistic understanding of how binding of SH3 domains to Site 1 and, potentially to Site 2, can induce the allosteric response in Site 3, which is located 100 aa downstream in disordered HVD and what the exact nature of this potential allosteric response.
In conclusion, we found that all three SH3 domains of CD2AP are capable of binding to each of two proline-rich motifs in CHIKV HVD. Importantly, we have identified the CSP signatures for binding of individual domains to each site. In addition, interactions of the Site 1 with any SH3 domain seems to induce an allosteric response in Site 3 located in the C-terminus of HVD.
CD2AP interacts with CHIKV HVD through cooperative binding of SH3-A and SH3-C domains
Next, we characterized binding of CHIKV HVD to the entire N-terminal domain of CD2AP (SH3-All, Fig. 1C) that contains all three SH3 domains connected by natural linker aa sequences. As for individual SH3 domains, mapping of the intermolecular interfaces of protein complexes was performed by analyzing CSP in the 1H-15N best-TROSY spectra of the N-labelled CHIKV HVD in the presence of different concentrations of unlabeled SH3-All protein. A striking observation was that despite the high molecular weight of the complex (MW 63 kD), the intensities and line widths of the cross peaks of residues outside the identified SH3 domain-binding motifs remained unchanged. Thus, the HVD fragments that are not involved in interactions retained high degree of disorder and flexibility (Fig. 3). The CSP plot demonstrated that SH3-All interacted with the same sites of CHIKV HVD as did individual SH3 domains (Fig. 4).
Residues 77A, 82R and 83R of the docking motif of Site 1 were already broadened at HVD to SH3-All ratio of 1.0:0.1, and comparison of the cross peaks for residues 70V, 71M and 72S showed striking similarities in trajectories of these cross peaks (CSP signatures) for HVD/SH3-All and HVD/SH3-C complexes (Fig. 4A and Supplementary Fig. 8). This strongly suggested that Site 1 predominantly interacts with SH3-C. Interactions with other SH3 domains are not detected. Thus, despite that all SH3 domains of SH3-All could potentially bind in the HVD-specific Site 1, only the SH3-C domain functioned as an interacting partner in this context.
Interaction of SH3-All with the Site 2 also revealed interesting characteristics. As described above, each individual CD2AP-specific SH3 domain could bind to this site, albeit with low affinities (Fig. 5). However, in the experiments with SH3-All, the Site 2-specific signals of residues 101 to 105 were already not detected at the first point of titration, at ratio 0.1:1 (Fig. 4 and Supplementary Fig. 9). This was an indication of a stronger affinity of SH3-All, compared to those of the individual domains. For aa 108R-111L, the CSP trajectories and their maximums were identical to the CSP signatures observed for binding of the SH3-A domain to CHIKV HVD (Fig. 4 and Supplementary Fig. 10). Thus, we concluded that SH3-A predominantly mediates a high affinity binding of SH3-All to CHIKV HVD in Site 2. Interestingly, the collected data also provided no evidence of SH3-B/HVD interaction, despite that it was clearly detected in the experiments with individual SH3-B. Further validation of the lack of interaction between SH3-B domain and HVD in the context of SH3-All requires analysis of 15N-labeled SH3-All binding to HVD. This work is currently in progress.
The CSP signatures of Site 3, which we proposed to be an allosteric site, were identical in the experiments with SH3-All and individual SH3 domains (Fig. 4C and Supplementary Fig. 11). Taken together, these results additionally support our hypothesis that shifts of the Site 3-specific cross peaks are rather allosterically induced by SH3-All binding to Site 1 than by direct binding of SH3-All to Site 3.
Importantly, in the study of SH3-All interactions with HVD, multiple changes in the cross peaks in the best-TROSY HVD spectrum became clearly detectable already at an HVD:SH3-All ratio of 1.0:0.1. The increase in ratio to 1.0:2.5 led to full saturation of the complex, and additional increase did not cause further changes in the spectra. This was a strong indication that the binding affinity of the SH3-All to CHIKV HVD is higher than those detected in the experiments with individual SH3 domains. The global fitting analysis performed on all residues involved in fast exchange regime for both Site 1 and Site 2 did not generate a good fit suggesting that these sites still bind individually. The analysis of binding in each site independently generated the apparent KD values of 2.0±0.5 μM and 15.3±0.8 μM, respectively. These KD values were more than 10-fold lower than those obtained in the binding of the individual domains. The decrease in KD values for each site indicated cooperativity between these two binding events despite that these domains are connected through long disordered peptide linkers.
Thus, using CSP signature-based approach, we found that CD2AP cooperatively binds to two sites of CHIKV HVD by SH3-A and SH3-C domains. SH3-B domain is not involved in binding and may potentially make complex with another cellular protein(s), which remains to be identified. Presence of multiple SH3 domains in the CD2AP strongly increases the affinity of its binding to CHIKV HVD.
Mutations in SH3 domain-binding sites of CHIKV HVD strongly affect CHIKV replication in vitro
We used the obtained structural data to inactivate both of the HVD-specific, SH3 domain-binding sites. A few point mutations were introduced into docking sites of binding Site 1 (P75G and R80G) and Site 2 (P100G and R105G) of CHIKV HVD (Fig. 6A). To ensure complete inhibition of CD2AP/HVD binding, we introduced two additional point mutation in C-terminal part of Site 2 (P113G and R122G), which could potentially form a weak third binding site. First, sequences encoding both wt and mutant CHIKV HVDs were cloned as Flag-GFP fusions under control of subgenomic promoter into Venezuelan equine encephalitis virus (VEEV) replicon (Fig. 6B) (Meshram et al., 2018). These packaged replicons VEErep/Flag-GFP-HVDchikv, VEErep/Flag-GFP-mHVDchikv and control VEErep/Flag-GFP were used to infect MRC-5 and BJ-5ta human fibroblasts. CHIKV HVD-specific complexes were isolated using Flag-specific MAbs and analyzed by Western blot. The introduced mutations blocked CD2AP binding to CHIKV HVD in both cell lines (Fig. 6C). Importantly, mutant CHIKV HVD also did not bind other SH3 domain-containing proteins, such as SH3KBP1 and BIN1 (Fig. 6C and Supplementary Fig. 12). Thus, introduced mutations completely abrogated interactions of CHIKV HVD with all previously identified SH3 domain-containing host factors.
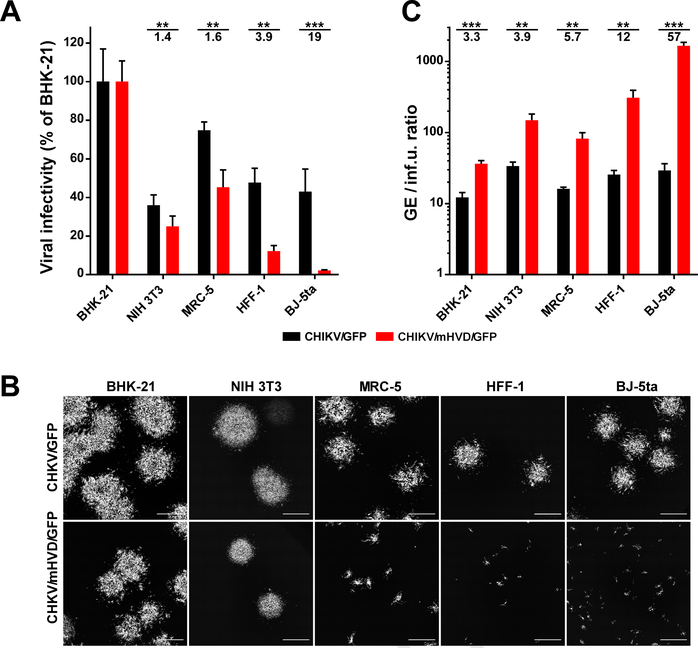
We also introduced the above mutations into infectious cDNA of CHIKV strain 181–25 (Gorchakov et al., 2012; Levitt et al., 1986) (Fig. 6A). Additional GFP gene was cloned into cDNAs of the mutant (CHIKV/mHVD/GFP) and parental virus (CHIKV/GFP) under control of duplicated subgenomic promoter, and GFP expression was used for monitoring the infection spread. The stocks of CHIKV/GFP and CHIKV/mHVD/GFP were generated by electroporation of the in vitro-synthesized RNAs into BHK-21 cells and used to infect rodent and human cell lines at the same multiplicity of infection (MOI). In all of the used cell lines, titers of the harvested mutant virus were 1-to-4 orders of magnitude lower than those of parental CHIKV/GFP (Fig. 6D). The differences were particularly evident in the used cell lines of human origin, which included HeLa, MRC-5, HFF-1 and BJ-5ta cells. Since both CHIKV/GFP and CHIKV/mHVD/GFP had exactly the same envelope, the alteration in infection spread could not result from changes in glycoprotein interaction with the receptor and other steps of viral entry. The most plausible explanation was that the mutations had negative effect on either vRC formation or its function in viral RNA replication or both.
To further dissect the effect of the mutations, the same samples of the parental CHIKV/GFP and its CHIKV/mHVD/GFP derivative were used for plaque assay on BHK-21, NIH 3T3, MRC-5, HFF-1 and BJ-5ta cells. For each cell line, we compared the efficiencies of plaque formation (Figs. 7A and B) and the ratio of concentrations of genome equivalents to concentrations of viral infectious units (GE/inf.u.) in the same stocks of viruses rescued by electroporation of the in vitro-synthesized RNAs into BHK-21 cells (Fig. 7C). In HFF-1 and BJ-5ta cells, CHIKV/mHVD/GFP was unable to produce distinguishable plaques, and titers were determined by counting small GFP-positive foci (Fig. 7B). CHIKV/GFP demonstrated similar GE:inf.u. ratio on all of the cells, ranging from 10:1 to 30:1 (Fig. 7C). CHIKV/mHVD/GFP was less infectious and, depending on the cell line, the GE-IU ratio was between 30:1 in the most susceptible BHK-21 cells and ~1650:1 in BJ-5ta cells. Accordingly, in all of the cell lines, the sizes of GFP-positive foci formed by CHIKV/mHVD/GFP under agarose cover at day 2 post infection, were dramatically smaller than those of CHIKV/GFP (Fig. 7B).
Fig. 7. Mutations in CD2AP-binding sites of CHIKV HVD strongly affect viral infectivity.
(A) The RNA electroporation-derived stocks of CHIKV/GFP and CHIKVmut/GFP were synchronously titrated on the indicated cell lines as described in Materials and Methods. Infectious viral titers were normalized to those determined on BHK-21 cells. (B) In the titration experiments, images of GFP-positive foci formed by viruses on both cell lines were taken at 40 h p.i. directly under agarose cover. (C) Ratio of viral concentrations in the same stocks determined in GE/ml and inf.u/ml on the indicated cell lines. Since CHIKVmut/GFP was unable to form plaques on most of cell lines, for uniform presentation, infectious titers were used as infectious units per ml (inf.u/ml) instead of PFU/ml. Significances of differences among the values were determined by two-tailed unpaired t-test (*, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001; n=3). Data are presented as mean with SD (n=3). Numbers under asterisks indicate average fold differences.
Taken together, the data demonstrated that the mutations introduced into HVD caused strong negative effect on the early step(s) of viral replication in human cells, and thus, had profound negative effect on viral infectivity. The detected differences in infectivities were likely determined by the efficiency of formation of early vRCs and recruitment of viral G RNA before it could be degraded after release from the incoming virions.
4. Discussion
In recent years, biology of IDPs has become a rapidly developing field. To date, many disordered protein sequences have been shown to contain combinations of short linear motifs that mediate interactions with other proteins factors during formation of multicomponent complexes. Viral proteins are not an exception. Many viruses encode IDPs, but their functions in virus replication and interactions with host are poorly understood (Feuerstein et al., 2012; Kim et al., 2016; Meshram et al., 2018). Alphavirus nsP3 proteins contain long, C-terminal, intrinsically disordered HVDs that were recently proposed to serve as hubs for recruiting cellular proteins during vRC assembly (Kim et al., 2016; Meshram et al., 2018). Importantly, combinations of the HVD-binding cellular proteins were found to be specific for geographically separated alphavirus species, and this strongly suggests critical role of HVD evolution in viral adaptation to different hosts and/or mosquito vectors. The distinguishing characteristic of alphavirus HVDs is that interacting host factors have either redundant or additive functions. Therefore, single knockout (KO) of cellular genes that encode HVD-binding proteins often has no effect on viral replication (Kim et al., 2016). Similarly, the effects of HVD-specific mutations, which alter interaction with particular host factors, on viral replication can be difficult to detect. However, CHIKV is more sensitive than other studied alphaviruses to both HVD modifications and KO of cellular genes that encode HVD-interacting proteins (Kim et al., 2016). Mutations in CHIKV nsP3 HVD that block binding of cellular G3BP proteins or other HVD-interacting cellular partners either make virus nonviable or strongly affect its replication rates in a cell-dependent manner (Kim et al., 2016; Meshram et al., 2018).
CD2AP is one of the CHIKV HVD-interacting host factors that was reproducibly co-isolated from human and other cells (Frolov et al., 2017; Meshram et al., 2018; Mutso et al., 2018). The latter protein contains SH3 domains and belongs to a very large family of the SH3 domain-containing proteins (Mayer, 2001). This family is involved in a wide range of processes in cell biology (Kaneko et al., 2008). The distinguishing characteristics of interactions mediated by SH3 domains are a relatively low affinity and binding specificity. However, in numerous co-IP experiments, only CD2AP, closely related SH3KBP1, and BIN1, but not other SH3 domain family members, were identified as CHIKV HVD-binding partners (Frolov et al., 2017; Meshram et al., 2018; Mutso et al., 2018). This was an indication of their specific use by CHIKV. Moreover, despite having exceptionally low level of identity, all alphavirus HVDs contain proline-rich motifs, suggesting that binding of the SH3 domain-containing cellular proteins is likely common for all of the members of the genus. Thus, it appears to be a critical determinant of viral replication and one of potential targets for drug development.
CD2AP has three SH3 domains and the pattern of their interaction with CHIKV HVD and binding sites remained uncharacterized. Due to low affinity and promiscuous binding of SH3 domains with proline-rich motifs, it is impossible to predict the pattern of interaction of the latter protein with CHIKV HVD. To date, two major classes of SH3 binding motifs have been identified (Teyra et al., 2017). However, there is a great number of exceptions from the consensus sequence, and a wide variety of other motifs can mediate such interaction. Therefore, to unambiguously identify the CD2AP-binding aa sequences in CHIKV HVD, we applied NMR spectroscopy. All three individual CD2AP SH3 domains were interacting with two same HVD motifs with low affinity. Site 1 has previously shown high affinity interaction with BIN1 (Neuvonen et al., 2011; Tossavainen et al., 2016). Site 2 was predicted to bind CD2AP using mutagenesis approach (Mutso et al., 2018). However, our experimental data demonstrate that both sites interact with any CD2AP SH3 domain, and Site 1 has higher affinity than Site 2. Thus, the data obtained in the experiments with individual CD2AP SH3 domains suggested a possibility of existence of multiple binding patterns for the natural combination of SH3 domains in CD2AP. Based on the higher affinity of SH3-B domain binding, it was also expected to be the main interacting partner of CHIKV HVD. Surprisingly, this is not the case. We found that directions and shifts of cross peaks outside the docking domains (CSP signatures) were specific for each site and each individual SH3 domain. Thus, every interaction displayed unique CSP signature, and they were clearly preserved in assays of SH3-ALL interaction with HVD. This was likely because in CD2AP, the domains were separated by long disordered aa linkers. According to these signatures, SH3-C domain in SH3-All exclusively interacts with Site 1, and SH3-A binds to Site 2. The detected strong increase in KD for each site suggests cooperativity in SH3-C and SH3-A interaction in the context of SH3-All. Importantly, despite the highest binding affinity as an individual domain, in the context of SH3-All, SH3-B demonstrated no detectable interaction with CHIKV HVD. The affinities of SH3-All to HVD sites 1 and 2 remained several orders lower than that previously determined for BIN1 SH3 domain (Tossavainen et al., 2016). This explains the previous data on preferential binding of BIN1 with CHIKV HVD in NIH 3T3 cells, where both BIN1 and CD2AP present in well-detectable concentration (Frolov et al., 2017).
A common approach for analysis of the SH3 domains’ interaction with cellular partners is characterization of their individual binding with short target peptides. Our data show that this approach is not applicable for analysis of proteins that contain multiple SH3 domains and partners having multiple binding sites. CD2AP is an adaptor protein involved in interaction with numerous cellular proteins, and our approach utilizing CSP signature will be valuable for characterization of complex multivalent interactions of this protein with cellular partners.
Another intriguing outcome of this work is identification of a potential long-range allosteric effect in Site 3 upon formation of complexes of the SH3 domains with Site 1 of CHIKV HVD. The importance of allosteric interactions in the regulation of function of disordered proteins and proteins with large disordered regions has been theoretically predicted (Hilser and Thompson, 2007; Wodak et al., 2019) but, thus far, no model amendable to structural study has been identified. Interestingly, the indicated Site 3 is located in the fragment that determines interaction of CHIKV HVD with G3BP proteins, and this interaction is indispensable for CHIKV RNA replication. More studies are needed to understand the nature of the allosteric interaction and whether it has biological significance. The schematic working model of proposed CD2AP-CHIKV HVD interaction is presented in Fig. 8.
Fig. 8. Schematic model of CHIKV HVD-CD2AP complex.
(A) CD2AP and CHIKV HVD form bivalent complex. SH3-C and SH3-A domains of CD2AP bind to Site 1 and 3 of CHIKV HVD respectively. Such bivalent interaction strongly increases affinity of CD2AP interaction with CHIKV HVD. Site 3 undergoes an allosteric modification upon interaction of any SH3 domain with Site 1. This modification may regulate binding of G3BP1 and G3BP2 to the motifs that flank Site 3. (B) Amino acid sequences of binding sites 1 and 2 and allosteric site 3 of CHIKV nsP3 HVD. The docking motifs in Sites 1 and 2 are underlined. The amino acids that were mutated in this work are depicted in red. The allosteric Site 3 is marked by bold font and flanking G3BP binding sites are depicted in green.
The role of the SH3 domain-containing proteins in CHIKV and other OW alphaviruses remains poorly understood, and published data are very contradictory. The BIN1-binding site was structurally characterized for CHIKV Semliki Forest virus (Tossavainen et al., 2016). Mutations in this site did not significantly affect CHIKV replication in mammalian cells (NIH 3T3 and Vero), but strongly reduced replication in mosquito cells (Goertz et al., 2018; Meshram et al., 2018). Interestingly, deletion of this site made CHIKV strain 37997 nonviable in one study and had no effect on of replication of CHIKV strain LR2006-OPY1 in another (Goertz et al., 2018; Mutso et al., 2018). The mutation in Site 2 also had marginal effect on CHIKV replication (Mutso et al., 2018). These data suggested that BIN1/CD2AP/SH3KBP1 have the same role in CHIKV replication and use of specific protein depends on cell type and specific strain.
Identification of two SH3 domain-binding sites in CHIKV HVD allowed us to design a viral mutant, which no longer interacted with any of the three SH3 domain-containing proteins, CD2AP, BIN1 and SH3KBP1. A very point few mutations had strong negative effect on viral replication rates, and it was more prominent in human than in rodent cells. Importantly, CHIKV with mutated HVD remained capable of replication. This was an indication that other host binding partners, such as NAP1 family members (Frolov et al., 2017; Meshram et al., 2018), also supported some level of viral replication. Since it was cell specific, these factors may have different impact on CHIKV infection in mouse and human cells or can be present in these cells at different concentrations. The detected profound increases in GE/inf.u ratios of the mutant on human cell lines imply that the HVD-specific mutations strongly affected viral ability to initiate replication and to form primary vRCs on very few molecules of CHIKV genome delivered into the cells.
The accumulated data support our hypothesis that all three SH3 domain-containing proteins, CD2AP, SH3KBP1 and BIN1, that demonstrate an ability to interact with CHIKV HVD play redundant role in establishing viral replication. However, the exact nature of this common function remains to be established. CD2AP and SH3KBP1 are adaptor proteins involved in assembly of numerous signaling complexes involved in regulation of actin assembly, endocytosis, signal transduction etc. (Dikic, 2002; Kaneko et al., 2008; Kowanetz et al., 2004; Tang and Brieher, 2013). BIN1, in turn, is implicated in regulation of endocytosis, membrane recycling and cytoskeleton regulation (Prokic et al., 2014). However, these functions in regulation of cell biology do not provide information about the roles of the proteins in CHIKV replication. Moreover, to date, HVDs of all of the studied alphaviruses demonstrate interaction with one or more SH3 domain-containing host factors (Frolov et al., 2017; Kim et al., 2016; Meshram et al., 2018). Therefore, characterization of the common functions of these bindings will elucidate new critical aspects of alphavirus-host interactions.
Supplementary Material
Highlights.
Hypervariable domain of CHIKV nsP3 (HVD) contains two SH3 domain-binding motifs
SH3-A and SH3-C domains of CD2AP cooperatively mediate its interaction with CHIKV HVD
Chemical shift perturbation signature can be applied effectively for identification of interacting domains in multidomain proteins
Mutations in both motifs of CHIKV HVD inhibit initiation of virus replication
CD2AP, SH3KBP1 and BIN1 play redundant roles in CHIKV replication
Acknowledgements
This work was supported by National Institute of Allergy and Infectious Diseases grants R01AI118867 and R01AI133159 to E.I.F., Swedish Foundation for Strategic Research grant ITM17-0218 to P.A. and Swedish Research Council research grant 2015-04614 to V.Y.O.
Footnotes
Conflict of Interest
The authors have no conflict of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ababou A, Pfuhl M, Ladbury JE, 2009. Novel insights into the mechanisms of CIN85 SH3 domains binding to Cbl proteins: solution-based investigations and in vivo implications. J Mol Biol 387, 1120–1136. [DOI] [PubMed] [Google Scholar]
- Akhrymuk I, Lukash T, Frolov I, Frolova EI, 2018. Novel mutations in nsP2 abolish chikungunya virus-induced transcriptional shutoff and make virus less cytopathic without affecting its replication rates. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, 2002. CIN85/CMS family of adaptor molecules. FEBS Lett 529, 110–115. [DOI] [PubMed] [Google Scholar]
- Favier A, Brutscher B, 2011. Recovering lost magnetization: polarization enhancement in biomolecular NMR. J Biomol NMR 49, 9–15. [DOI] [PubMed] [Google Scholar]
- Feuerstein S, Solyom Z, Aladag A, Favier A, Schwarten M, Hoffmann S, Willbold D, Brutscher B, 2012. Transient structure and SH3 interaction sites in an intrinsically disordered fragment of the hepatitis C virus protein NS5A. J Mol Biol 420, 310–323. [DOI] [PubMed] [Google Scholar]
- Foy NJ, Akhrymuk M, Akhrymuk I, Atasheva S, Bopda-Waffo A, Frolov I, Frolova EI, 2013. Hypervariable domains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes. J Virol 87, 1997–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Kim DY, Akhrymuk M, Mobley JA, Frolova EI, 2017. Hypervariable Domain of Eastern Equine Encephalitis Virus nsP3 Redundantly Utilizes Multiple Cellular Proteins for Replication Complex Assembly. J Virol 91, e00371–00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goertz GP, Lingemann M, Geertsema C, Abma-Henkens MHC, Vogels CBF, Koenraadt CJM, van Oers MM, Pijlman GP, 2018. Conserved motifs in the hypervariable domain of chikungunya virus nsP3 required for transmission by Aedes aegypti mosquitoes. PLoS Negl Trop Dis 12, e0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Wang E, Leal G, Forrester NL, Plante K, Rossi SL, Partidos CD, Adams AP, Seymour RL, Weger J, Borland EM, Sherman MB, Powers AM, Osorio JE, Weaver SC, 2012. Attenuation of Chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. J Virol 86, 6084–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilser VJ, Thompson EB, 2007. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci U S A 104, 8311–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaksson L, Mayzel M, Saline M, Pedersen A, Rosenlow J, Brutscher B, Karlsson BG, Orekhov VY, 2013. Highly efficient NMR assignment of intrinsically disordered proteins: application to B- and T cell receptor domains. PLoS One 8, e62947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaravine VA, Orekhov VY, 2006. Targeted acquisition for real-time NMR spectroscopy. J Am Chem Soc 128, 13421–13426. [DOI] [PubMed] [Google Scholar]
- Jaravine VA, Zhuravleva AV, Permi P, Ibraghimov I, Orekhov VY, 2008. Hyperdimensional NMR spectroscopy with nonlinear sampling. J Am Chem Soc 130, 3927–3936. [DOI] [PubMed] [Google Scholar]
- Jensen MR, Communie G, Ribeiro EA Jr., Martinez N, Desfosses A, Salmon L, Mollica L, Gabel F, Jamin M, Longhi S, Ruigrok RW, Blackledge M, 2011. Intrinsic disorder in measles virus nucleocapsids. Proc Natl Acad Sci U S A 108, 9839–9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Li L, Li SS, 2008. The SH3 domain--a family of versatile peptide- and protein-recognition module. Front Biosci 13, 4938–4952. [DOI] [PubMed] [Google Scholar]
- Kazimierczuk K, Orekhov VY, 2011. Accelerated NMR Spectroscopy by Using Compressed Sensing. Ang. Chem. Int. Ed 50, 5556–5559. [DOI] [PubMed] [Google Scholar]
- Keller RLJ, 2004. The Computer Aided Resonance Assignment Tutorial. CANTINA Verlag. [Google Scholar]
- Kim DY, Reynaud JM, Rasalouskaya A, Akhrymuk I, Mobley JA, Frolov I, Frolova EI, 2016. New World and Old World Alphaviruses Have Evolved to Exploit Different Components of Stress Granules, FXR and G3BP Proteins, for Assembly of Viral Replication Complexes. PLoS Pathog 12, e1005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowanetz K, Husnjak K, Holler D, Kowanetz M, Soubeyran P, Hirsch D, Schmidt MHH, Pavelic K, De Camilli P, Randazzo PA, Dikic I, 2004. CIN85 associates with multiple effectors controlling intracellular trafficking of epidermal growth factor receptors. Mol Biol Cell 15, 3155–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt NH, Ramsburg HH, Hasty SE, Repik PM, Cole FE Jr., Lupton HW, 1986. Development of an attenuated strain of chikungunya virus for use in vaccine production. Vaccine 4, 157–162. [DOI] [PubMed] [Google Scholar]
- Mayer BJ, 2001. SH3 domains: complexity in moderation. J Cell Sci 114, 1253–1263. [DOI] [PubMed] [Google Scholar]
- Mayzel M, Kazimierczuk K, Orekhov VY, 2014. The causality principle in the reconstruction of sparse NMR spectra. Chem. Commun. (Camb.) 50, 8947–8950. [DOI] [PubMed] [Google Scholar]
- Meshram CD, Agback P, Shiliaev N, Urakova N, Mobley JA, Agback T, Frolova EI, Frolov I, 2018. Multiple Host Factors Interact with Hypervariable Domain of Chikungunya Virus nsP3 and Determine Viral Replication in Cell-Specific Mode. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutso M, Morro AM, Smedberg C, Kasvandik S, Aquilimeba M, Teppor M, Tarve L, Lulla A, Lulla V, Saul S, Thaa B, McInerney GM, Merits A, Varjak M, 2018. Mutation of CD2AP and SH3KBP1 Binding Motif in Alphavirus nsP3 Hypervariable Domain Results in Attenuated Virus. Viruses 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvonen M, Kazlauskas A, Martikainen M, Hinkkanen A, Ahola T, Saksela K, 2011. SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication. PLoS Pathog 7, e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orekhov VY, Jaravine VA, 2011. Analysis of non-uniformly sampled spectra with multi-dimensional decomposition. Prog. Nucl. Magn. Reson. Spectrosc. 59, 271–292. [DOI] [PubMed] [Google Scholar]
- Ortega-Roldan JL, Jensen MR, Brutscher B, Azuaga AI, Blackledge M, van Nuland NA, 2009. Accurate characterization of weak macromolecular interactions by titration of NMR residual dipolar couplings: application to the CD2AP SH3-C:ubiquitin complex. Nucleic Acids Res 37, e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokic I, Cowling BS, Laporte J, 2014. Amphiphysin 2 (BIN1) in physiology and diseases. J Mol Med (Berl) 92, 453–463. [DOI] [PubMed] [Google Scholar]
- Pustovalova Y, Mayzel M, Orekhov VY, 2018. XLSY: Extra-Large NMR Spectroscopy. Angew Chem Int Ed Engl 57, 14043–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezza G, Weaver SC, 2019. Chikungunya as a paradigm for emerging viral diseases: Evaluating disease impact and hurdles to vaccine development. PLoS Negl Trop Dis 13, e0006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouka E, Simister PC, Janning M, Kumbrink J, Konstantinou T, Muniz JR, Joshi D, O’Reilly N, Volkmer R, Ritter B, Knapp S, von Delft F, Kirsch KH, Feller SM, 2015. Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3). J Biol Chem 290, 25275–25292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW, 2015. Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol 96, 2483–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksela K, Permi P, 2012. SH3 domain ligand binding: What’s the consensus and where’s the specificity? FEBS Lett 586, 2609–2614. [DOI] [PubMed] [Google Scholar]
- Schilte C, Couderc T, Chretien F, Sourisseau M, Gangneux N, Guivel-Benhassine F, Kraxner A, Tschopp J, Higgs S, Michault A, Arenzana-Seisdedos F, Colonna M, Peduto L, Schwartz O, Lecuit M, Albert ML, 2010. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J Exp Med 207, 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E, Guntert P, 2012. A New Algorithm for Reliable and General NMR Resonance Assignment. J Am Chem Soc 134, 12817–12829. [DOI] [PubMed] [Google Scholar]
- Schneider R, Maurin D, Communie G, Kragelj J, Hansen DF, Ruigrok RW, Jensen MR, Blackledge M, 2015. Visualizing the molecular recognition trajectory of an intrinsically disordered protein using multinuclear relaxation dispersion NMR. J Am Chem Soc 137, 1220–1229. [DOI] [PubMed] [Google Scholar]
- Schubert M, Oschkinat H, Schmieder P, 2001. MUSIC, selective pulses, and tuned delays: amino acid type-selective (1)H-(15)N correlations, II. J Magn Reson 148, 61–72. [DOI] [PubMed] [Google Scholar]
- Schulte T, Liu L, Panas MD, Thaa B, Dickson N, Gotte B, Achour A, McInerney GM, 2016. Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication. Open biology 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarten M, Solyom Z, Feuerstein S, Aladag A, Hoffmann S, Willbold D, Brutscher B, 2013. Interaction of nonstructural protein 5A of the hepatitis C virus with Src homology 3 domains using noncanonical binding sites. Biochemistry 52, 6160–6168. [DOI] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG, 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58, 491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang VW, Brieher WM, 2013. FSGS3/CD2AP is a barbed-end capping protein that stabilizes actin and strengthens adherens junctions. J Cell Biol 203, 815–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyra J, Huang H, Jain S, Guan X, Dong A, Liu Y, Tempel W, Min J, Tong Y, Kim PM, Bader GD, Sidhu SS, 2017. Comprehensive Analysis of the Human SH3 Domain Family Reveals a Wide Variety of Non-canonical Specificities. Structure 25, 1598–1610 e1593. [DOI] [PubMed] [Google Scholar]
- Tossavainen H, Aitio O, Hellman M, Saksela K, Permi P, 2016. Structural Basis of the High Affinity Interaction between the Alphavirus Nonstructural Protein-3 (nsP3) and the SH3 Domain of Amphiphysin-2. J Biol Chem 291, 16307–16317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnerstale S, Nowakowski M, Baraznenok V, Stenberg G, Lindberg J, Mayzel M, Orekhov V, Agback T, 2016. Backbone Assignment of the MALT1 Paracaspase by Solution NMR. PLoS One 11, e0146496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Lecuit M, 2015. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med 372, 1231–1239. [DOI] [PubMed] [Google Scholar]
- Weaver SC, Winegar R, Manger ID, Forrester NL, 2012. Alphaviruses: population genetics and determinants of emergence. Antiviral Res 94, 242–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MP, 2013. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc 73, 1–16. [DOI] [PubMed] [Google Scholar]
- Wodak SJ, Paci E, Dokholyan NV, Berezovsky IN, Horovitz A, Li J, Hilser VJ, Bahar I, Karanicolas J, Stock G, Hamm P, Stote RH, Eberhardt J, Chebaro Y, Dejaegere A, Cecchini M, Changeux JP, Bolhuis PG, Vreede J, Faccioli P, Orioli S, Ravasio R, Yan L, Brito C, Wyart M, Gkeka P, Rivalta I, Palermo G, McCammon JA, Panecka-Hofman J, Wade RC, Di Pizio A, Niv MY, Nussinov R, Tsai CJ, Jang H, Padhorny D, Kozakov D, McLeish T, 2019. Allostery in Its Many Disguises: From Theory to Applications. Structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuiderweg ER, 2002. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry 41, 1–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The final chemical shift assignments of the backbone 1HN, 15N, 13CO, 13Cα, 13Cβ atoms of CHIKV HVD at 20°C have been deposited in the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu/) under BMRB accession code 27868. Other data are available from corresponding authors upon reasonable request.