Highlights
-
•
Consensus-based error-correction methods and their application in viral quasispecies.
-
•
Principles, advantages, flaws and their clinical applications of major NGS methods.
-
•
Discussion on both experimental design and computational analysis related to NGS.
-
•
Critical considerations to better unravel viral mutational landscape.
Keywords: Next-generation sequencing (NGS), Viruses, Quasispecies, RNA, Rare variants, Consensus-based error correction
Abstract
Next-generation sequencing (NGS) has revolutionized the scale and depth of biomedical sciences. Because of its unique ability for the detection of sub-clonal variants within genetically diverse populations, NGS has been successfully applied to analyze and quantify the exceptionally-high diversity within viral quasispecies, and many low-frequency drug- or vaccine-resistant mutations of therapeutic importance have been discovered. Although many works have intensively discussed the latest NGS approaches and applications in general, none of them has focused on applying NGS in viral quasispecies studies, mostly due to the limited ability of current NGS technologies to accurately detect and quantify rare viral variants. Here, we summarize several error-correction strategies that have been developed to enhance the detection accuracy of minority variants. We also discuss critical considerations for preparing a sequencing library from viral RNAs and for analyzing NGS data to unravel the mutational landscape.
1. Background
Developing low-frequency variants or mutations is a self-protective approach for various types of cells or organisms that is evolutionarily preservative to survive under stressful conditions through a variety of scales, from mitochondria, to tumor cells, to viruses (Andino and Domingo, 2015; He et al., 2010; Mwenifumbo and Marra, 2013; Salehi et al., 2015; Salk et al., 2018; Woo and Reifman, 2012). Viruses, particularly RNA viruses, possess a great capability to evolve and mutate in order to rapidly respond to host immune selection pressure. Consequently, they generate a population with a large number of variable but closely related genomes, also known as quasispecies (Andino and Domingo, 2015; Woo and Reifman, 2012). Accurate characterization of low-frequency variants could not only provide invaluable insights into molecular mechanisms but also aid clinical decision making (Andino and Domingo, 2015; Godoy et al., 2019; Parker and Chen, 2017; Pawlotsky, 2002; Suzuki et al., 2017). Minority variants in RNA viruses are often generated by error-prone replication (Domingo et al., 2012). Previous studies have shown that, among influenza, even those with a frequency below the detection limit of conventional surveillance methods, are evidently associated with antibody escape in vaccinated humans (Dinis et al., 2016) and could cause a large global public health burden (Chambers et al., 2015). However, enhancing the sensitivity and specificity for identifying minority variants still remains as one of the major challenges in virology. These viruses, including coronavirus (Li et al., 2020; Wu et al., 2020; Zhu et al., 2020), cytomegalovirus (CMV) (Sahoo et al., 2013), human immunodeficiency virus (HIV) (James et al., 2019; Kyeyune et al., 2016; Rawson et al., 2017), influenza virus (Chambers et al., 2015; Zaraket et al., 2010), hepatitis C virus (HCV) (Itakura et al., 2015), poliovirus (Acevedo et al., 2014), and others, have a superior ability to adapt to a new environment and emerge as drug- and vaccine-resistant mutants. Kyeyune et al. (2016) have shown that poor prognosis can be foreseen by the detection of drug-resistant mutations at a frequency of as low as 1 % in a human immunodeficiency virus (HIV)-infected patient.
Applications of next-generation sequencing (NGS) in basic and clinical virology research have grown rapidly over the past decade (Houldcroft et al., 2017), particularly for virus discovery (Datta et al., 2015) and diagnosis (Barzon et al., 2013, 2011; Capobianchi et al., 2013; Gardy and Loman, 2018; Kuroda et al., 2010; Prachayangprecha et al., 2014). Compared with conventional gold-standard Sanger sequencing, NGS provides considerably more sequencing reads for a lower cost and allows multiplexing of samples (Shendure et al., 2017). Although NGS technologies enable acquisition of a vast amount of sequencing data, high error rates from 0.1 % to 15 %, depending on platforms and applications, often impede the detection of rare mutations (Salk et al., 2018). To improve the accuracy of NGS for identifying low-frequency viral variants, a variety of error-correction approaches have been developed and applied to investigate viral quasispecies (Table 1 ). Several bioinformatics tools or pipelines for variant calling have been developed specifically for studying viral variants (Huber et al., 2017; McElroy et al., 2013; Verbist et al., 2015; Zagordi et al., 2010) and calculating the complexity of a quasispecies as well as measuring the genetic distance between two similar quasispecies (Marinier et al., 2019). Many more variants callers have been discussed and compared in dedicated reviews or methodological comparison papers (Hwang et al., 2015; Lee et al., 2020; Pereira et al., 2020). Implementing an existing variant-calling tool in NGS data analysis is relatively simple and saves additional costs for sample preparation. However, error corrections made by variant-calling tools have a low positive predictive value. They are not optimal for amplicon analysis because they are mostly based on the assumption that the error rate is randomly distributed (Posada-Cespedes et al., 2017). Therefore, another more innovative and accurate approach, named the consensus-based error-correction method, has become increasingly popular in NGS studies (Salk et al., 2018). Three major related approaches have currently been applied in virus quasispecies studies: Tag-based sequencing (Geller et al., 2016; Hauck et al., 2018; Jabara et al., 2011; Seifert et al., 2016), circular sequencing (CirSeq) (Acevedo et al., 2014), and intramolecular-ligated nanopore consensus sequencing (INC-Seq) (Li et al., 2016).
Table 1.
Comparison of various NGS approaches in virus quasispecies analysis.
| Principle | Strengths | Weaknesses | Error frequency |
|---|---|---|---|
|
Unique molecular identifiers (UID or Safe-SeqS): • Randomly generated UID • Allowing identification of every single reverse-transcribed viral RNA • Only mutations that exist in a majority of sequences with an identical UID considered as true variants |
• Preservation of minor variant frequency • Multiplexing possible |
• Incapable of correcting reverse transcription polymerase chain reaction (RT-PCR) errors • Risk of tag clashes when tag diversity is inadequate |
1.4 × 10−5 |
|
Duplex sequencing (DupSeq): • Molecular barcodes applied to each double-stranded DNA molecule • Simultaneously identifying the two complementary strands and distinguish them • True mutations present in a majority of sequences in each strand group and the complementary strand group |
• Multiplexing possible | • Incapable of correcting PCR errors that occur during reverse transcription. • Risk of tag clashes when tag diversity is inadequate • DupSeq cannot be applied directly to RNA templates, which can cause the loss of preservation of minor variant frequency of RNA viruses |
5 × 10−8 |
|
Circular sequencing (CirSeq): • Fragments of viral RNA followed by self-ligation into circularized RNAs for rolling circle amplification. The amplicon composed of many tandem repeats of the circularized RNA • Mutations present in most of repeats on the same molecule considered as true variants |
• No probe or primer design required • Preservation of minor variant frequency |
• A tendency towards G-to-A and C-to-T errors in the absence of uracil-DNA glycosylase and formamidopyrimidine-DNA glycosylase • Large amounts of viral RNA (>1 μg) required for library preparation • Very limited length of sequences that can be genotyped as tandem copies on short-read platforms |
7.6 × 10−6 |
|
Intramolecular-ligated nanopore consensus sequencing (INC-Seq): • Viral RNAs directly self-ligated into closed loops for rolling-circle amplification • Each amplicon composed of concatenated repeats of a starting viral molecule • Similar to the CirSeq but with many more copies of much longer fragments |
• Capability of extremely long-read sequencing (possible to identify multidrug-resistant variants in a single viral genome) • Multiplexing possible • Rapid and field-deployable • No probe or primer design required |
• High single-read error rates (about 1%–5%) • Requirement of high coverage to minimize the effect of sequencing errors |
3 × 10−2 |
In this mini-review, we mainly deliberate on the consensus-based error-correction approaches to characterize the population structure of single-strand RNA viruses. Although identification of novel viruses is also extremely important and challenging, it requires very different techniques and approaches (Houldcroft et al., 2017; Illingworth et al., 2017; McCrone and Lauring, 2016), which are beyond the scope of the current review. Along with an increasing number of applications in viral quasispecies research, it is important to evaluate various approaches used in NGS for improving the information–noise ratios of the obtained NGS data. Details of each method together with their major advantages and disadvantages are discussed and summarized in this work. Obtaining high-accuracy NGS in studies of viral quasispecies not only relies on error corrections of NGS data, but also depends on the well-tailored design of experimental and computational analysis approaches. Thus, we also briefly address the technical and analytical considerations when applying NGS to unravel the mutational landscape in viral quasispecies, particularly in comparative studies.
2. Approaches for enhancing the accuracy of NGS in virus quasispecies studies
In the last decade, several NGS approaches and platforms have been developed for viral whole-genome sequencing (WGS) and quasispecies studies in order to enhance infection control and disease management (Houldcroft et al., 2017). A majority of these studies have focused on specific short amplicons that can be sequenced on a short-read platform, such as Roche 454 (Sopena et al., 2018), Illumina (Sutar et al., 2019), or Ion Torrent technology (Goodwin et al., 2016). These amplicon strategies require a relatively simpler analysis workflow because only short regions of the viral genome are in focus. In comparison to single amplicon deep sequencing, WGS involves markedly more data processing procedures, such as de novo assembly and alignment within existing genome databases, but it can deliver a more complete view of the heterogeneity within viral populations, which is particularly important for the identification of novel viruses (Goodwin et al., 2016; Marston et al., 2013). Long-read sequencing has the advantage of directly obtaining information in repetitive sequences with a single read and consequently eliminating ambiguous information in those repetitive regions, but it still suffers from relatively high error rates (Amarasinghe et al., 2020). A high coverage could significantly reduce the error rates, but that entails a higher cost, relatively more computational power and longer times for analysis. To facilitate its comprehension, in this review we mainly concentrate on short-read approaches by discussing consensus-based error-correction methods (Table 1) for enhancing the accuracy of NGS data in virus quasispecies studies.
2.1. Tag-based sequencing
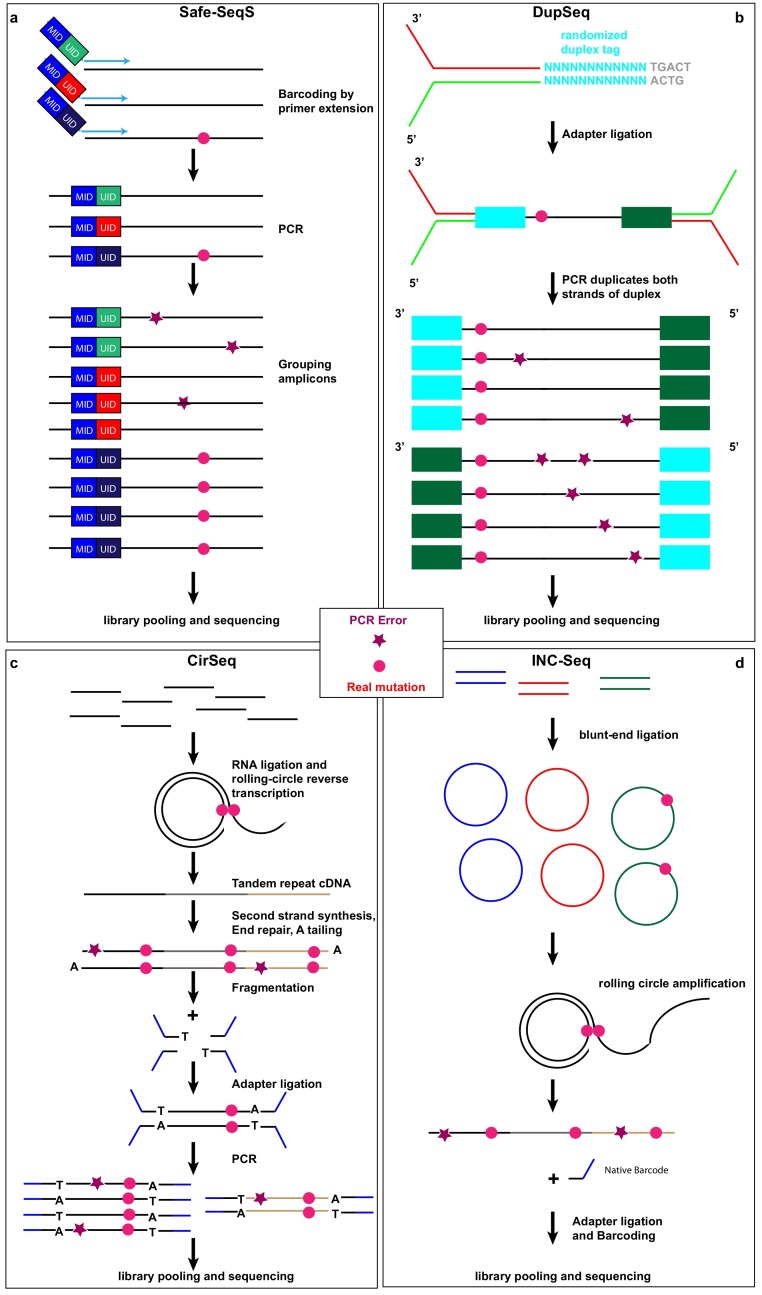
This is the most commonly used error-correction approach in short-read NGS platforms, in which a DNA library is typically amplified by polymerase chain reaction (PCR) before sequencing. Zhou et al. (2015), Hauck et al. (2018), Jabara et al.(2011), and Seifert et al. (2016) have applied randomly generated unique molecular identifiers (UIDs, also known as Safe-SeqS (Fig. 1 a), “molecular barcodes”, “primer IDs”, or “tags”). UIDs are linked to the primer for reverse transcription in order to label each single-stranded viral cDNA derived from a particular RNA molecule before PCR amplification. Each UID is passed on to all its derivative PCR copies, thus allowing the grouping of all sequence reads derived from the same viral RNA molecule template. Sequences with the same UID are then collapsed to a consensus sequence. Thus, each of these collapsed sequences correspond to one original viral RNA strand. Differences between sequences within a family of sequences with the same UID are due to technical substitution errors during PCR or sequencing and can be easily corrected (Hiatt et al., 2010; Kinde et al., 2011). Applying UIDs for error correction can decrease the sequencing error frequency to 1.4 × 10−5 (Fox et al., 2014).
Fig. 1.
Library preparation approaches of consensus-based error correction for investigating virus quasispecies. (a) Safe-SeqS uses primers linked to unique molecular identifiers (UIDs) and mouse identifiers (MIDs) for reverse transcription, which not only enables the recognition of every original viral RNA strand after PCR amplification, but also allows multiplexing of samples in the same sequencing run. (b) DupSeq applies randomized duplex tags to each double-stranded DNA molecule in a way that derivative PCR products of the two strands can be informatively related to each other but also distinguishable. Consensus wild-type or mutation sequences are reached only if the reads of each of the double strands show identical sequences. (c) CirSeq begins by circularizing of single-stranded DNA fragments without any exogenous molecular barcodes followed by rolling-circle amplification, fragmentation and sequencing. (d) INC-Seq also entails circularization single-stranded DNA fragments followed by rolling-circle amplification of the loop; however, the end product is a long DNA strand (>10Kb) comprising concatenated copies of one of the strands of the starting molecule to be sequenced on a long-read platform. For INC-Seq, only in-silico fragmentation is performed for analysis following sequencing. For CirSeq and INC-seq, the random fragmentation points of the starting molecules serve as endogenous UIDs for consensus-based error correction. For all above-mentioned four methods, after library preparation, pooling and sequencing, sequences originating from the same viral RNA strand of the same sample, are collapsed to a single consensus sequence. True mutations (pink circle) can be distinguished from PCR errors (purple star). Due to limited space, sequencing errors are not marked here.
Another tag-based error-correction approach, named duplex sequencing (DupSeq, Fig. 1b), has been applied to study the genetic variation of HCV (Geller et al., 2016). DupSeq utilizes special tags to label each double-stranded cDNA molecule derived from the same viral RNA after reverse transcription and subsequent complementary DNA synthesis by DNA polymerase so that derivative PCR copies of the two strands can be informatively related to each other but remain distinct (Schmitt et al., 2012). Consensuses are first generated for each single-strand group with the same tag and then compared to that of the complementary strand. Sequencing or PCR errors are extremely unlikely to take place at the same positions of the two DNA strands by chance. The double checking principle as indicated by the name of DupSeq can thus significantly reduce the sequencing error frequency down to 5 × 10−8 (Fox et al., 2014). However, compared with UID approaches, DupSeq cannot be directly applied to RNA templates. Before inserting tags, it requires additional reverse transcriptase PCR and second-strand PCR, which can significantly impact the low-frequency RNA templates in the samples (Head et al., 2014). Therefore, DupSeq might particularly suffer from the loss of preservation of variant frequency of RNA viruses. For both UID/Safe-SeqS and DupSeq error-correction approaches, mistakes that occur during reverse transcription, second-strand synthesis, and PCR recombination will escape correction (Zanini et al., 2017). Moreover, there is the risk of tag clash when the diversity of barcodes is too little to label each independent molecule. On the other hand, tags with too many random nucleotides could also directly contribute to PCR biases (Kou et al., 2016).
2.2. CirSeq
CirSeq (Fig. 1c) is another consensus sequencing method used in short-read NGS. In this case, viral RNAs are fragmented into very short pieces and self-ligated into many circularized RNAs that serve as templates for complementary DNA (cDNA) synthesis. CirSeq incorporates rolling-circle reverse transcription of circularized viral RNA to generate tandem repeat cDNA in order to enrich the target sequences (Acevedo et al., 2014; Whitfield and Andino, 2016). Thus, unlike the tag-based sequencing approach that requires exogenous barcodes to label each viral RNA or cDNA copy, CirSeq makes use of physically jointed copies of the sequence for consensus calling. True mutations can be distinguished from either amplification or sequencing errors by building a consensus sequence based on the linked copies to a single molecule. CirSeq, however, has a tendency towards G-to-A and C-to-T errors derived from base damage due to cytosine deamination; therefore, it is necessary to add in uracil-DNA glycosylase and formamidopyrimidine-DNA glycosylase during rolling circle amplification in order to eliminate such errors caused by DNA damage (Lou et al., 2013). The sequencing error frequency of CirSeq is about 7.6 × 10−6 (Fox et al., 2014). Because CirSeq is built on sequencing tandem repeats on the single-end Illumina sequencing platform, only short sequence fragments (<150 base pair (bp)) can be genotyped in this approach. Due to this in-built requirement, CirSeq thus particularly suffers from the constraint on short-length fragments and the inability to perform paired-end sequencing relative to the other major approaches. Moreover, CirSeq requires the input of large amounts of viral RNA (>1 μg) for library preparation (Whitfield and Andino, 2016).
2.3. INC-Seq
INC-Seq (Fig. 1d) is a direct consensus sequencing approach based on long-read nanopore sequencing, a platform developed by Oxford Nanopore Technologies (Li et al., 2016; Mikheyev and Tin, 2014). Akin to the CirSeq technique, INC-Seq begins by intramolecular circularizing of RNA molecules to form closed loops. Each RNA loop molecule further undergoes rolling-circle reverse transcription (RT)-PCR amplification to form a long cDNA product comprising concatenated repeats descended from the starting RNA molecule. After sequencing, the resultant reads consist of a long string of tandem copies similar to the results of the CirSeq technique but with many more copies of much longer fragments. True mutations are identified as the variants present in the majority of tandem repeats on the same single molecule, whereas technical substitution errors from RT-PCR or sequencing should not be found in a majority of repeats. The challenge is, however, that this approach has a high raw-read error rate of 5 %–20 % (Salk et al., 2018). Therefore, high coverage is required to reduce the impact of sequencing errors (Houldcroft et al., 2017).
It is worth noting that the aforementioned approaches are mainly applied to studies of single-strand RNA viruses, which tend to mutate much faster than double-strand RNA viruses and hence represent a major challenge in deciphering viral population structures. Characterization of variants of double-strand RNA viruses that only account for a small fraction of pathological viruses could benefit from particular approaches, such as DupSeq, which has been reported to detect ultralow-frequency variants from double-strand DNA samples (Kennedy et al., 2014; Schmitt et al., 2012).
3. Improving detection of rare variants in comparative studies
The genetic diversity of RNA viruses facilitates their adaptation to new environments and evasion of host immunity. Monitoring quasispecies evolution in infected hosts under treatment or after vaccination is important for the early detection of escape mutants. This analysis is complicated by the need not only to minimize technical sequencing artifacts, but also to enhance the comparability among different samples. Technical artifacts/biases correspond to systematic PCR or sequencing errors due to variability in sample processing and experimental design (Head et al., 2014). Artifacts, which cannot be otherwise eradicated, must be eliminated by experimental design. There are several ways of improving NGS data quality for comparing heterogeneous samples in a virus quasispecies study, including (1) sample and library preparation protocols that limit experimental biases, (2) single-molecule consensus sequencing that allows for the identification of true mutations, but exclusion of sequencing errors, (3) computational strategies for read normalization. We describe here mainly two of the strategies.
3.1. Sample and library preparation
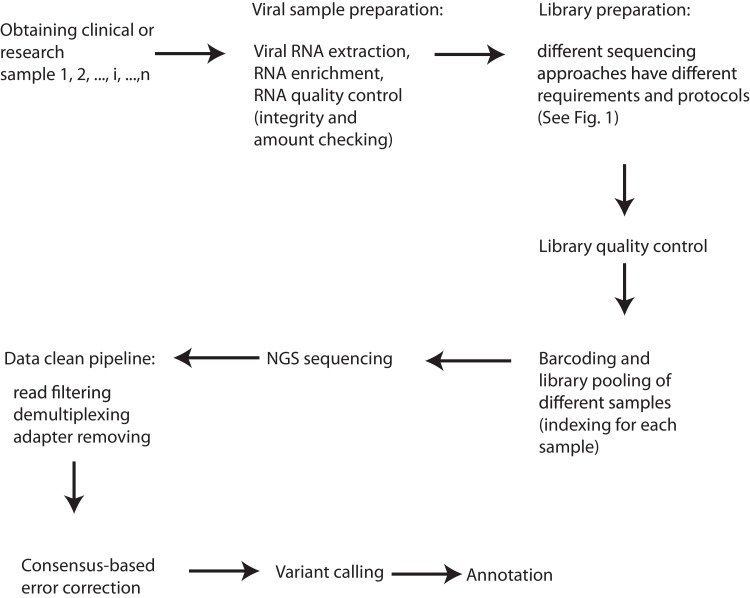
When it comes to sample and library preparation for analyzing complex populations such as virus quasispecies, it is especially critical to reduce sequencing biases to obtain a faithful picture of the analyzed samples (Acevedo and Andino, 2014; Chen et al., 2018; Forth and Hoper, 2019; Head et al., 2014; Verhoeven et al., 2018). Thus, even within the same experiment, only viral samples with similar RNA quantity and quality should be compared. Following RNA extraction and viral RNA enrichment from the host RNA (Forth and Hoper, 2019; Houldcroft et al., 2017; Sathiamoorthy et al., 2018; Singanallur et al., 2019), the first critical quality control (QC) step (Fig. 2 ) is to test both quantity and integrity of the starting viral RNA (Hauck et al., 2018; Ng et al., 2018; Yang et al., 2016). The necessity to control virus tire or genome copy numbers in comparative studies has been demonstrated in several studies as false positive variant calls become more evident with lower material inputs (Gallet et al., 2017; Illingworth et al., 2017; McCrone and Lauring, 2016). Ideally, in one comparative study, all the final concentrations of sequencing libraries should be identical in order to reduce false-positive calls. Furthermore, following the library preparation, the quantity of the library also should be examined (Ng et al., 2018) and in principle, only similar amount of libraries should be directly compared. Since fragmentation is required for the CirSeq approach (Fig. 1c), the fragment size distribution should be also analyzed at least in the CirSeq workflow to allow for sensible comparison (Acevedo and Andino, 2014; Lou et al., 2013).
Fig. 2.
A general experimental and computational workflow for improving NGS data quality of virus quasispecies studies. For the comparative studies or clinical samples, we start from different samples (1,2,i,…,n). One has to first go through different experimental steps, such as sample preparation, library preparation, library quality control, sample indexing, library pooling and sequencing. Then, computational steps are followed, such as data cleaning, consensus-based error correction, variant calling and annotation.
3.2. Barcoding technique and multiplex sequencing
In addition to sample and library QC, bias can be further reduced by pooling various indexed or barcoded samples. Molecular barcodes on short-read NGS platforms allow consensus-based error corrections and the detection of low-frequency variants down to ∼0.001 % mutations per base pair and the total error rate varies dependent on the length of the target sequences (Geller et al., 2016; Salk et al., 2018). In addition, multiplex sequencing is possible by assigning an additional barcode, e.g., mouse identifier (MID) (Fig. 1a), to the library of each sample (Hauck et al., 2018). Multiplexing is possible for all the aforementioned four sequencing methods (Salk et al., 2018). This can be done in comparative studies, in which the library of treated (e.g., vaccinated or infected) and control mice specifically tagged with a distinguishable barcode allows pooled samples into the same sequencing run (Fig. 1a), which reduces potential technical bias associated with run-to-run variability.
To further reduce substitution errors, one can implement one of the approaches in Fig. 1 to ensure the unique identification of every original viral RNA strand (or cDNA strand) after PCR amplification. After library preparation and deep sequencing, all sequences obtained are grouped by UIDs or by endogenous fragmentation points and compared for different type of errors. Sequences with the same UID or fragmentation point, i.e., originating from the same viral RNA strand, are collapsed into a single sequence. If within a family of grouped sequences there are differences between sequences due to late PCR errors or sequencing errors, these differences should be corrected using statistical approaches, such as cumulative binomial distribution, in order to assign and rank the probability of a correct base at each given position for various numbers of read copies. Thus, this tag-based error-correction approach eliminates most amplification biases and identifies the majority of substitution errors (Fig. 2).
Each NGS platform varies with its specific error profile that requires particular downstream computational and statistical handling. For instance, the Ion Torrent technology is prone to make insertion–deletion (indel) errors in homopolymeric stretches of DNA (Goodwin et al., 2016). Although the widely-used Illumina technology usually possesses an accuracy rate higher than 99.5 %, the platform displays a tendency towards substitution errors (Allhoff et al., 2013; Minoche et al., 2011). While indel errors may be a problem in the case of de novo sequencing, they can be easily identified and removed by comparison to the corresponding reference viral sequence as demonstrated by several works (Hauck et al., 2018; Song et al., 2017; Yeo et al., 2012).
Using this technique, the chance of the same sequencing error that occurs within sequences of the same UID family is extremely low. According to the estimations of Kinde et al. (2011), the PCR amplification errors are around 2.2 × 10−6 distinct alterations/bp. With such an extremely-low error rate, even with a read number of 7.8 × 105 in a sample, for the targeted sequence with a length of 165bp [e.g., the conserved long α-helix (LAH) domain of influenza hemagglutinin protein (Hauck et al., 2018)], the estimated total PCR amplification error is only around 280 reads (2.2 × 10−6 × 165 × 7.8 × 105). Therefore, the low-frequency viral variants [with a frequency higher than 1 % out of the entire population as demonstrated by Peng et al. (2015)] can be confidently considered as biologically mutated sequences rather than artifacts.
If without financial constraints and computational analysis limits, one should simply increase the sequencing coverage or depth to increase the confidence level. However, experimental cost, computational capacity and budget might always constrain us, which requires us to have optimal experimental design to balance between both experimental and computational cost and sequencing output. Estimation of the number of raw reads, or correspondingly the sequencing coverage/depth is not a trivial issue. Although there is a classic formula for us to estimate these numbers by: the sequencing coverage = the number of total reads × the read length/the length of target sequence or genome (Lander and Waterman, 1988). However, the estimation cannot work properly when the virus variant is very rare. Apparently, for the low-frequency variants, we need to increase the number of raw reads and the sequencing depth to increase the confidence and decrease the errors. However, except for empirical numbers obtained by different groups, no one knows exactly which sequencing depth or raw reads are needed to more accurately characterize viral quasispecies. For instance, as compellingly demonstrated by Griffith et al. (2015), the standard depth of 50x coverage can only detect 10 % of single nucleotide polymorphisms (SNPs) with minority variance (=<15 %) in tumor samples. They concluded that the coverage as high as 10,000x could be required to validate rare variants. We could foresee that characterization of viral quasispecies might encounter similar issues as did deciphering tumor clonal architecture. For more details about estimation of sequencing depth, please refer to the dedicated review published elsewhere (Sims et al., 2014).
4. Specific considerations in bioinformatics data analyses to detect low-frequency viral variants
General computational workflows to analyze and correct NGS data have been discussed elsewhere (Dolled-Filhart et al., 2013; Pabinger et al., 2014; Reinert et al., 2015; Salk et al., 2018; Treangen and Salzberg, 2011) and are beyond this review. In general, bioinformatics analysis of short NGS reads involves five main steps: (i) quality assessment of raw sequencing data, such as trimming, filtering, and others; (ii) read alignment; (iii) variant call (Fig. 2); (iv) annotation by comparing with knowledge databases; and (v) visualization of aligned reads and mutations (Pabinger et al., 2014; Posada-Cespedes et al., 2017). Several particular issues should be addressed in virus quasispecies studies. One of the key issues in computational analysis is related to variant calling. There are several popular variant callers. One example is MinVar (Huber et al., 2017) that is based on LoFreq (Wilm et al., 2012). The other variant callers include ShoRAH (Zagordi et al., 2011) and its extension (McElroy et al., 2013), SNVer (Wei et al., 2011), deepSNV (Gerstung et al., 2012), SAMtools (Li, 2011), GATK (McKenna et al., 2010), Ion-Torrent specific TVC and others. For comparison and evaluation of different methods, please refer to the dedicated reviews or comparative work (Hwang et al., 2015; Lee et al., 2020; Pereira et al., 2020). In short, each method has its own pros and cons. Investigation of viral diversity is very sensitive to the used variant calling methods (McCrone and Lauring, 2016). None of them alone can reliably identify authentic minority variants or mutations and therefore often a combination of several variant callers are required to reach better results (Leung et al., 2014). While UID-based barcoding NGS approaches can significantly improve the identification of low-frequency variants, the sampling bias on original templates that is introduced during PCR amplification remains challenging. It is also challenging to remove errors introduced in the UIDs during PCR amplification. In this context, Kou et al. tried to correct false UIDs to avoid the false identification of mutations (Kou et al., 2016). They clustered UIDs that differed only in one or two nucleotides into a single UID family. In addition, it has been observed that the first nucleotide of the sample UID and the last nucleotide of the UID are more error prone at least on some sequencing platforms (Brodin et al., 2015). Therefore, UIDs with minor differences are grouped into the same UID group family, and the positions of potential error bases should be taken into consideration.
Construction of consensus sequences is another critical step in analyzing UID-derived sequences. The consensus sequences are often constructed from the sequencing reads labelled with the same UIDs that have been retrieved for a minimal number of times, e.g., >= 3 times (Brodin et al., 2015). So far, most approaches treat multiple-sequence alignments in the same way irrespective of the UID family size. However, statistically, it is obvious that the higher the number of read copies for the given UID, the lower the probability that reads with identical bases occur by chance. This should be integrated into analysis pipelines to further refine error corrections. One could at least rank viral variants by confidence by including such approaches.
5. Concluding remarks and outlook
Dissecting viral population structures has important biomedical applications but is subject to a wide range of experimental and computational challenges. Improved NGS approaches provide the opportunity to better identify low-frequency, nevertheless clinically relevant viral variants (Houldcroft et al., 2017). Several major experimental methods applying consensus-based error correction to enhance data accuracy have been proposed and discussed, and more powerful instruments and creative approaches are under development. Consensus-based error-correction approaches can identify those error occurrences during PCR and sequencing. But these approaches require a relatively high sequencing coverage and are compromised by the impaired efficiency of the tag labeling. In the library preparation processes for NGS, which includes adapter ligation and multiple clean-up cycles, there exists usually an inevitable loss of the starting materials. This might result in the loss of preservation of minority-variant frequency, especially in the case of using viral or clinical samples that contain a limited amount of materials (Illingworth et al., 2017). To circumvent some of the labelling-related issues, a non-consensus-based error-correction approach (named overlapping paired-end read sequencing) has been shown to significantly scale down sequencing error frequency (5 × 10−4) and to improve the accuracy of rare-variant detection (Chen-Harris et al., 2013). For the overlapping paired-end read sequencing, as indicated by the name, each pair of read deriving from the same viral RNA should be exactly complementary. If not exactly complementary, the reads will be regarded as errors, therefore reducing false positive discovery of minority variants.
From a computational point of view, variant identification and sequence annotation are currently performed in separate steps. In the near future, both steps of variant call and viral protein structural annotation may be integrated into a single iterative analysis loop. For instance, with the advancement of the methods in protein structure prediction based on mutations, viral variants that might cause viral protein structural changes and reduce viral fitness in the host would be ranked lower. This may require more computational analysis power, including cloud computing (Langmead and Nellore, 2018).
In the context of translational virology, the current approaches for the diagnosis of viral infection need to be applied (Barzon et al., 2013, 2011; Capobianchi et al., 2013). All the clinical samples suffer from high ratios of host-to-viruses genetic inputs and a low amount of starting materials (Fernandez-Cassi et al., 2018). The first step is therefore to enrich and purify the viral materials from biopsies (Houldcroft et al., 2017). In order to compensate a relatively small number of starting templates, the number of PCR amplification cycles might need to be slightly increased, which might relatively compromise PCR-related errors. In routine clinical tests, measurement speed is another critical step, which often requires receiving results within hours rather than days (Capobianchi et al., 2013). This indicates the need for even higher throughput instruments compared with the current available machines. Computational analysis can also constitute a bottleneck to the analysis. Sequence assembly is particularly computationally intensive and demands much more computational power in clinical settings (Shendure et al., 2017). To address all these clinic-related challenges, there is still a long way to go even in consideration of the unparalleled high development pace of NGS or even third-generation sequencing approaches (Editorial, 2018; Lavezzo et al., 2016).
Author contributions
IL proposed and drafted the manuscript. F.H. conceptualized the framework and revised the manuscript. C.M. revised the manuscript.
Declaration of competing interest
None declared.
Acknowledgments
The work in Luxembourg was supported by the funding from the European Union’s Seventh Framework Programme for research, technological development and demonstration (FP7/2007-2013) under grant agreement Nr. 602437. Feng Q. He was partially supported by Luxembourg Fonds National de la Recherche (FNR) CORE Programme grant (CORE/14/BM/8231540/GeDES), FNR AFR-RIKEN bilateral program (TregBAR), PRIDE program grants (PRIDE/11012546/NEXTIMMUNE and PRIDE/10907093/CRITICS).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.197963.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Acevedo A., Andino R. Library preparation for highly accurate population sequencing of RNA viruses. Nat. Protoc. 2014;9(7):1760–1769. doi: 10.1038/nprot.2014.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo A., Brodsky L., Andino R. Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature. 2014;505(7485):686–690. doi: 10.1038/nature12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allhoff M., Schonhuth A., Martin M., Costa I.G., Rahmann S., Marschall T. Discovering motifs that induce sequencing errors. BMC Bioinf. 2013;14(Suppl 5):S1. doi: 10.1186/1471-2105-14-S5-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21(1):30. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R., Domingo E. Viral quasispecies. Virology. 2015;479-480:46–51. doi: 10.1016/j.virol.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzon L., Lavezzo E., Militello V., Toppo S., Palu G. Applications of next-generation sequencing technologies to diagnostic virology. Int. J. Mol. Sci. 2011;12(11):7861–7884. doi: 10.3390/ijms12117861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzon L., Lavezzo E., Costanzi G., Franchin E., Toppo S., Palu G. Next-generation sequencing technologies in diagnostic virology. J. Clin. Virol. 2013;58(2):346–350. doi: 10.1016/j.jcv.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Brodin J., Hedskog C., Heddini A., Benard E., Neher R.A., Mild M., Albert J. Challenges with using primer IDs to improve accuracy of next generation sequencing. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0119123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capobianchi M.R., Giombini E., Rozera G. Next-generation sequencing technology in clinical virology. Clin. Microbiol. Infect. 2013;19(1):15–22. doi: 10.1111/1469-0691.12056. [DOI] [PubMed] [Google Scholar]
- Chambers B.S., Parkhouse K., Ross T.M., Alby K., Hensley S.E. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014-2015 influenza season. Cell Rep. 2015;12(1):1–6. doi: 10.1016/j.celrep.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Qiu Y., Zhuang Q., Wang S., Wang T., Chen J., Wang K. Next-generation sequencing library preparation method for identification of RNA viruses on the Ion Torrent Sequencing Platform. Virus Genes. 2018;54(4):536–542. doi: 10.1007/s11262-018-1568-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Harris H., Borucki M.K., Torres C., Slezak T.R., Allen J.E. Ultra-deep mutant spectrum profiling: improving sequencing accuracy using overlapping read pairs. BMC Genomics. 2013;14:96. doi: 10.1186/1471-2164-14-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Budhauliya R., Das B., Chatterjee S., Vanlalhmuaka, Veer V. Next-generation sequencing in clinical virology: discovery of new viruses. World J. Virol. 2015;4(3):265–276. doi: 10.5501/wjv.v4.i3.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinis J.M., Florek N.W., Fatola O.O., Moncla L.H., Mutschler J.P., Charlier O.K., Meece J.K., Belongia E.A., Friedrich T.C. Deep sequencing reveals potential antigenic variants at low frequencies in influenza a virus-infected humans. J. Virol. 2016;90(7):3355–3365. doi: 10.1128/JVI.03248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolled-Filhart M.P., Lee M., Jr, Ou-Yang C.W., Haraksingh R.R., Lin J.C. Computational and bioinformatics frameworks for next-generation whole exome and genome sequencing. Sci. World J. 2013;2013 doi: 10.1155/2013/730210. 730210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Sheldon J., Perales C. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 2012;76(2):159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editorial The long view on sequencing. Nat. Biotechnol. 2018;36(4):287. doi: 10.1038/nbt.4125. [DOI] [PubMed] [Google Scholar]
- Fernandez-Cassi X., Rusinol M., Martinez-Puchol S. Viral concentration and amplification from human serum samples prior to application of next-generation sequencing analysis. Methods Mol. Biol. 2018;1838:173–188. doi: 10.1007/978-1-4939-8682-8_13. [DOI] [PubMed] [Google Scholar]
- Forth L.F., Hoper D. Highly efficient library preparation for Ion Torrent sequencing using Y-adapters. Biotechniques. 2019;67(5):229–237. doi: 10.2144/btn-2019-0035. [DOI] [PubMed] [Google Scholar]
- Fox E.J., Reid-Bayliss K.S., Emond M.J., Loeb L.A. Accuracy of next generation sequencing platforms. Next Gener. Seq. Appl. 2014:1. doi: 10.4172/jngsa.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet R., Fabre F., Michalakis Y., Blanc S. The number of target molecules of the amplification step limits accuracy and sensitivity in ultra deep sequencing viral population studies. J. Virol. 2017 doi: 10.1128/JVI.00561-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardy J.L., Loman N.J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018;19(1):9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller R., Estada U., Peris J.B., Andreu I., Bou J.V., Garijo R., Cuevas J.M., Sabariegos R., Mas A., Sanjuan R. Highly heterogeneous mutation rates in the hepatitis C virus genome. Nat. Microbiol. 2016;1(7):16045. doi: 10.1038/nmicrobiol.2016.45. [DOI] [PubMed] [Google Scholar]
- Gerstung M., Beisel C., Rechsteiner M., Wild P., Schraml P., Moch H., Beerenwinkel N. Reliable detection of subclonal single-nucleotide variants in tumour cell populations. Nat. Commun. 2012;3:811. doi: 10.1038/ncomms1814. [DOI] [PubMed] [Google Scholar]
- Godoy C., Tabernero D., Sopena S., Gregori J., Cortese M.F., Gonzalez C., Casillas R., Yll M., Rando A., Lopez-Martinez R., Quer J., Gonzalez-Aseguinolaza G., Esteban R., Riveiro-Barciela M., Buti M., Rodriguez-Frias F. Characterization of hepatitis B virus X gene quasispecies complexity in mono-infection and hepatitis delta virus superinfection. World J. Gastroenterol. 2019;25(13):1566–1579. doi: 10.3748/wjg.v25.i13.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17(6):333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M., Miller C.A., Griffith O.L., Krysiak K., Skidmore Z.L., Ramu A., Walker J.R., Dang H.X., Trani L., Larson D.E., Demeter R.T., Wendl M.C., McMichael J.F., Austin R.E., Magrini V., McGrath S.D., Ly A., Kulkarni S., Cordes M.G., Fronick C.C., Fulton R.S., Maher C.A., Ding L., Klco J.M., Mardis E.R., Ley T.J., Wilson R.K. Optimizing cancer genome sequencing and analysis. Cell Syst. 2015;1(3):210–223. doi: 10.1016/j.cels.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck N.C., Kirpach J., Kiefer C., Farinelle S., Maucourant S., Morris S.A., Rosenberg W., He F.Q., Muller C.P., Lu I.N. Applying unique molecular identifiers in next generation sequencing reveals a constrained viral quasispecies evolution under cross-reactive antibody pressure targeting long alpha helix of hemagglutinin. Viruses. 2018;10(4) doi: 10.3390/v10040148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Wu J., Dressman D.C., Iacobuzio-Donahue C., Markowitz S.D., Velculescu V.E., Diaz L.A., Jr, Kinzler K.W., Vogelstein B., Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464(7288):610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head S.R., Komori H.K., LaMere S.A., Whisenant T., Van Nieuwerburgh F., Salomon D.R., Ordoukhanian P. Library construction for next-generation sequencing: overviews and challenges. Biotechniques. 2014;56(2) doi: 10.2144/000114133. 61-64, 66, 68, passim. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt J.B., Patwardhan R.P., Turner E.H., Lee C., Shendure J. Parallel, tag-directed assembly of locally derived short sequence reads. Nat. Methods. 2010;7(2):119–122. doi: 10.1038/nmeth.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houldcroft C.J., Beale M.A., Breuer J. Clinical and biological insights from viral genome sequencing. Nat. Rev. Microbiol. 2017;15(3):183–192. doi: 10.1038/nrmicro.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M., Metzner K.J., Geissberger F.D., Shah C., Leemann C., Klimkait T., Boni J., Trkola A., Zagordi O. MinVar: a rapid and versatile tool for HIV-1 drug resistance genotyping by deep sequencing. J. Virol. Methods. 2017;240:7–13. doi: 10.1016/j.jviromet.2016.11.008. [DOI] [PubMed] [Google Scholar]
- Hwang S., Kim E., Lee I., Marcotte E.M. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci. Rep. 2015;5:17875. doi: 10.1038/srep17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth C.J.R., Roy S., Beale M.A., Tutill H., Williams R., Breuer J. On the effective depth of viral sequence data. Virus Evol. 2017;3(2) doi: 10.1093/ve/vex030. vex030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura J., Kurosaki M., Higuchi M., Takada H., Nakakuki N., Itakura Y., Tamaki N., Yasui Y., Suzuki S., Tsuchiya K., Nakanishi H., Takahashi Y., Maekawa S., Enomoto N., Izumi N. Resistance-associated NS5A variants of hepatitis C virus are susceptible to interferon-based therapy. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0138060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabara C.B., Jones C.D., Roach J., Anderson J.A., Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc. Natl. Acad. Sci. 2011;108(50):20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James K.L., de Silva T.I., Brown K., Whittle H., Taylor S., McVean G., Esbjornsson J., Rowland-Jones S.L. Low-bias RNA sequencing of the HIV-2 genome from blood plasma. J. Virol. 2019;93(1) doi: 10.1128/JVI.00677-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S.R., Schmitt M.W., Fox E.J., Kohrn B.F., Salk J.J., Ahn E.H., Prindle M.J., Kuong K.J., Shen J.C., Risques R.A., Loeb L.A. Detecting ultralow-frequency mutations by duplex sequencing. Nat. Protoc. 2014;9(11):2586–2606. doi: 10.1038/nprot.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 2011;108(23):9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou R., Lam H., Duan H., Ye L., Jongkam N., Chen W., Zhang S., Li S. Benefits and challenges with applying unique molecular identifiers in next generation sequencing to detect low frequency mutations. PLoS One. 2016;11(1) doi: 10.1371/journal.pone.0146638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Katano H., Nakajima N., Tobiume M., Ainai A., Sekizuka T., Hasegawa H., Tashiro M., Sasaki Y., Arakawa Y., Hata S., Watanabe M., Sata T. Characterization of quasispecies of pandemic 2009 influenza A virus (A/H1N1/2009) by de novo sequencing using a next-generation DNA sequencer. PLoS One. 2010;5(4) doi: 10.1371/journal.pone.0010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyeyune F., Gibson R.M., Nankya I., Venner C., Metha S., Akao J., Ndashimye E., Kityo C.M., Salata R.A., Mugyenyi P., Arts E.J., Quinones-Mateu M.E. Low-frequency drug resistance in HIV-Infected ugandans on antiretroviral treatment is associated with regimen failure. Antimicrob. Agents Chemother. 2016;60(6):3380–3397. doi: 10.1128/AAC.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Waterman M.S. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2(3):231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- Langmead B., Nellore A. Cloud computing for genomic data analysis and collaboration. Nat. Rev. Genet. 2018;19(5):325. doi: 10.1038/nrg.2018.8. [DOI] [PubMed] [Google Scholar]
- Lavezzo E., Barzon L., Toppo S., Palu G. Third generation sequencing technologies applied to diagnostic microbiology: benefits and challenges in applications and data analysis. Expert Rev. Mol. Diagn. 2016;16(9):1011–1023. doi: 10.1080/14737159.2016.1217158. [DOI] [PubMed] [Google Scholar]
- Lee E.R., Parkin N., Jennings C., Brumme C.J., Enns E., Casadella M., Howison M., Coetzer M., Avila-Rios S., Capina R., Marinier E., Van Domselaar G., Noguera-Julian M., Kirkby D., Knaggs J., Harrigan R., Quinones-Mateu M., Paredes R., Kantor R., Sandstrom P., Ji H. Performance comparison of next generation sequencing analysis pipelines for HIV-1 drug resistance testing. Sci. Rep. 2020;10(1):1634. doi: 10.1038/s41598-020-58544-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung R.K., Dong Z.Q., Sa F., Chong C.M., Lei S.W., Tsui S.K., Lee S.M. Quick, sensitive and specific detection and evaluation of quantification of minor variants by high-throughput sequencing. Mol. Biosyst. 2014;10(2):206–214. doi: 10.1039/c3mb70334g. [DOI] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Chng K.R., Boey E.J., Ng A.H., Wilm A., Nagarajan N. INC-Seq: accurate single molecule reads using nanopore sequencing. Gigascience. 2016;5(1):34. doi: 10.1186/s13742-016-0140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Si H.R., Zhu Y., Yang X.L., Anderson D.E., Shi Z.L., Wang L.F., Zhou P. Discovery of bat coronaviruses through surveillance and probe capture-based next-generation sequencing. mSphere. 2020;5(1) doi: 10.1128/mSphere.00807-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou D.I., Hussmann J.A., McBee R.M., Acevedo A., Andino R., Press W.H., Sawyer S.L. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc. Natl. Acad. Sci. U.S.A. 2013;110(49):19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinier E., Enns E., Tran C., Fogel M., Peters C., Kidwai A., Ji H., Van Domselaar G. Quasitools: a collection of tools for viral quasispecies analysis. bioRxiv. 2019 [Google Scholar]
- Marston D.A., McElhinney L.M., Ellis R.J., Horton D.L., Wise E.L., Leech S.L., David D., de Lamballerie X., Fooks A.R. Next generation sequencing of viral RNA genomes. BMC Genomics. 2013;14:444. doi: 10.1186/1471-2164-14-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrone J.T., Lauring A.S. Measurements of intrahost viral diversity are extremely sensitive to systematic errors in variant calling. J. Virol. 2016;90(15):6884–6895. doi: 10.1128/JVI.00667-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy K., Zagordi O., Bull R., Luciani F., Beerenwinkel N. Accurate single nucleotide variant detection in viral populations by combining probabilistic clustering with a statistical test of strand bias. BMC Genomics. 2013;14:501. doi: 10.1186/1471-2164-14-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheyev A.S., Tin M.M. A first look at the Oxford Nanopore MinION sequencer. Mol. Ecol. Resour. 2014;14(6):1097–1102. doi: 10.1111/1755-0998.12324. [DOI] [PubMed] [Google Scholar]
- Minoche A.E., Dohm J.C., Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol. 2011;12(11):R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwenifumbo J.C., Marra M.A. Cancer genome-sequencing study design. Nat. Rev. Genet. 2013;14(5):321–332. doi: 10.1038/nrg3445. [DOI] [PubMed] [Google Scholar]
- Ng S.H., Braxton C., Eloit M., Feng S.F., Fragnoud R., Mallet L., Mee E.T., Sathiamoorthy S., Vandeputte O., Khan A.S. Current perspectives on high-throughput sequencing (HTS) for adventitious virus detection: upstream sample processing and library preparation. Viruses. 2018;10(10) doi: 10.3390/v10100566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabinger S., Dander A., Fischer M., Snajder R., Sperk M., Efremova M., Krabichler B., Speicher M.R., Zschocke J., Trajanoski Z. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform. 2014;15(2):256–278. doi: 10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J., Chen J. Application of next generation sequencing for the detection of human viral pathogens in clinical specimens. J. Clin. Virol. 2017;86:20–26. doi: 10.1016/j.jcv.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlotsky J.M. Molecular diagnosis of viral hepatitis. Gastroenterology. 2002;122(6):1554–1568. doi: 10.1053/gast.2002.33428. [DOI] [PubMed] [Google Scholar]
- Peng Q., Vijaya Satya R., Lewis M., Randad P., Wang Y. Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genomics. 2015;16:589. doi: 10.1186/s12864-015-1806-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R., Oliveira J., Sousa M. Bioinformatics and computational tools for next-generation sequencing analysis in clinical genetics. J. Clin. Med. 2020;9(1) doi: 10.3390/jcm9010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada-Cespedes S., Seifert D., Beerenwinkel N. Recent advances in inferring viral diversity from high-throughput sequencing data. Virus Res. 2017;239:17–32. doi: 10.1016/j.virusres.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Prachayangprecha S., Schapendonk C.M., Koopmans M.P., Osterhaus A.D., Schurch A.C., Pas S.D., van der Eijk A.A., Poovorawan Y., Haagmans B.L., Smits S.L. Exploring the potential of next-generation sequencing in detection of respiratory viruses. J. Clin. Microbiol. 2014;52(10):3722–3730. doi: 10.1128/JCM.01641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson J.M.O., Gohl D.M., Landman S.R., Roth M.E., Meissner M.E., Peterson T.S., Hodges J.S., Beckman K.B., Mansky L.M. Single-strand consensus sequencing reveals that HIV type but not subtype significantly impacts viral mutation frequencies and spectra. J. Mol. Biol. 2017;429(15):2290–2307. doi: 10.1016/j.jmb.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert K., Langmead B., Weese D., Evers D.J. Alignment of next-generation sequencing reads. Annu. Rev. Genomics Hum. Genet. 2015;16:133–151. doi: 10.1146/annurev-genom-090413-025358. [DOI] [PubMed] [Google Scholar]
- Sahoo M.K., Lefterova M.I., Yamamoto F., Waggoner J.J., Chou S., Holmes S.P., Anderson M.W., Pinsky B.A. Detection of cytomegalovirus drug resistance mutations by next-generation sequencing. J. Clin. Microbiol. 2013;51(11):3700–3710. doi: 10.1128/JCM.01605-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi F., Baronio R., Idrogo-Lam R., Vu H., Hall L.V., Kaiser P., Lathrop R.H. CHOPER filters enable rare mutation detection in complex mutagenesis populations by next-generation sequencing. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0116877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salk J.J., Schmitt M.W., Loeb L.A. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 2018 doi: 10.1038/nrg.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathiamoorthy S., Malott R.J., Gisonni-Lex L., Ng S.H.S. Selection and evaluation of an efficient method for the recovery of viral nucleic acids from complex biologicals [corrected] Npj Vaccines. 2018;3:31. doi: 10.1038/s41541-018-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 2012;109(36):14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert D., Di Giallonardo F., Topfer A., Singer J., Schmutz S., Gunthard H.F., Beerenwinkel N., Metzner K.J. A comprehensive analysis of primer IDs to study heterogeneous HIV-1 populations. J. Mol. Biol. 2016;428(1):238–250. doi: 10.1016/j.jmb.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Shendure J., Balasubramanian S., Church G.M., Gilbert W., Rogers J., Schloss J.A., Waterston R.H. DNA sequencing at 40: past, present and future. Nature. 2017;550(7676):345–353. doi: 10.1038/nature24286. [DOI] [PubMed] [Google Scholar]
- Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014;15(2):121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- Singanallur N.B., Anderson D.E., Sessions O.M., Kamaraj U.S., Bowden T.R., Horsington J., Cowled C., Wang L.F., Vosloo W. Probe capture enrichment next-generation sequencing of complete foot-and-mouth disease virus genomes in clinical samples. J. Virol. Methods. 2019;272 doi: 10.1016/j.jviromet.2019.113703. [DOI] [PubMed] [Google Scholar]
- Song L., Huang W., Kang J., Huang Y., Ren H., Ding K. Comparison of error correction algorithms for Ion Torrent PGM data: application to hepatitis B virus. Sci. Rep. 2017;7(1):8106. doi: 10.1038/s41598-017-08139-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopena S., Godoy C., Tabernero D., Homs M., Gregori J., Riveiro-Barciela M., Ruiz A., Esteban R., Buti M., Rodriguez-Frias F. Quantitative characterization of hepatitis delta virus genome edition by next-generation sequencing. Virus Res. 2018;243:52–59. doi: 10.1016/j.virusres.2017.10.003. [DOI] [PubMed] [Google Scholar]
- Sutar J., Padwal V., Sonawani A., Nagar V., Patil P., Kulkarni B., Hingankar N., Deshpande S., Idicula-Thomas S., Jagtap D., Bhattacharya J., Bandivdekar A., Patel V. Effect of diversity in gp41 membrane proximal external region of primary HIV-1 Indian subtype C sequences on interaction with broadly neutralizing antibodies 4E10 and 10E8. Virus Res. 2019;273 doi: 10.1016/j.virusres.2019.197763. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Kawada J.I., Okuno Y., Hayano S., Horiba K., Torii Y., Takahashi Y., Umetsu S., Sogo T., Inui A., Ito Y. Comprehensive detection of viruses in pediatric patients with acute liver failure using next-generation sequencing. J. Clin. Virol. 2017;96:67–72. doi: 10.1016/j.jcv.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Treangen T.J., Salzberg S.L. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2011;13(1):36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbist B.M., Thys K., Reumers J., Wetzels Y., Van der Borght K., Talloen W., Aerssens J., Clement L., Thas O. VirVarSeq: a low-frequency virus variant detection pipeline for Illumina sequencing using adaptive base-calling accuracy filtering. Bioinformatics. 2015;31(1):94–101. doi: 10.1093/bioinformatics/btu587. [DOI] [PubMed] [Google Scholar]
- Verhoeven J.T.P., Canuti M., Munro H.J., Dufour S.C., Lang A.S. ViDiT-CACTUS: an inexpensive and versatile library preparation and sequence analysis method for virus discovery and other microbiology applications. Can. J. Microbiol. 2018;64(10):761–773. doi: 10.1139/cjm-2018-0097. [DOI] [PubMed] [Google Scholar]
- Wei Z., Wang W., Hu P., Lyon G.J., Hakonarson H. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011;39(19):e132. doi: 10.1093/nar/gkr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield Z.J., Andino R. Characterization of viral populations by using circular sequencing. J. Virol. 2016;90(20):8950–8953. doi: 10.1128/JVI.00804-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm A., Aw P.P., Bertrand D., Yeo G.H., Ong S.H., Wong C.H., Khor C.C., Petric R., Hibberd M.L., Nagarajan N. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40(22):11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.J., Reifman J. A quantitative quasispecies theory-based model of virus escape mutation under immune selection. Proc. Natl. Acad. Sci. U.S.A. 2012;109(32):12980–12985. doi: 10.1073/pnas.1117201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Leonard S.R., Mammel M.K., Elkins C.A., Kulka M. Towards next-generation sequencing analytics for foodborne RNA viruses: examining the effect of RNA input quantity and viral RNA purity. J. Virol. Methods. 2016;236:221–230. doi: 10.1016/j.jviromet.2016.07.013. [DOI] [PubMed] [Google Scholar]
- Yeo Z.X., Chan M., Yap Y.S., Ang P., Rozen S., Lee A.S. Improving indel detection specificity of the Ion Torrent PGM benchtop sequencer. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0045798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagordi O., Klein R., Daumer M., Beerenwinkel N. Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies. Nucleic Acids Res. 2010;38(21):7400–7409. doi: 10.1093/nar/gkq655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagordi O., Bhattacharya A., Eriksson N., Beerenwinkel N. ShoRAH: estimating the genetic diversity of a mixed sample from next-generation sequencing data. BMC Bioinf. 2011;12:119. doi: 10.1186/1471-2105-12-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanini F., Brodin J., Albert J., Neher R.A. Error rates, PCR recombination, and sampling depth in HIV-1 whole genome deep sequencing. Virus Res. 2017;239:106–114. doi: 10.1016/j.virusres.2016.12.009. [DOI] [PubMed] [Google Scholar]
- Zaraket H., Saito R., Suzuki Y., Baranovich T., Dapat C., Caperig-Dapat I., Suzuki H. Genetic makeup of amantadine-resistant and oseltamivir-resistant human influenza A/H1N1 viruses. J. Clin. Microbiol. 2010;48(4):1085–1092. doi: 10.1128/JCM.01532-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Jones C., Mieczkowski P., Swanstrom R. Primer ID validates template sampling depth and greatly reduces the error rate of next-generation sequencing of HIV-1 genomic RNA populations. J. Virol. 2015;89(16):8540–8555. doi: 10.1128/JVI.00522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus, I, Research, T A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.