Abstract
The recent emergence of a novel coronavirus (SARS-CoV-2) in China has caused significant public health concerns. Recently, ACE2 was reported as an entry receptor for SARS-CoV-2. In this study, we present the crystal structure of the C-terminal domain of SARS-CoV-2 (SARS-CoV-2-CTD) spike (S) protein in complex with human ACE2 (hACE2), which reveals a hACE2-binding mode similar overall to that observed for SARS-CoV. However, atomic details at the binding interface demonstrate that key residue substitutions in SARS-CoV-2-CTD slightly strengthen the interaction and lead to higher affinity for receptor binding than SARS-RBD. Additionally, a panel of murine monoclonal antibodies (mAbs) and polyclonal antibodies (pAbs) against SARS-CoV-S1/receptor-binding domain (RBD) were unable to interact with the SARS-CoV-2 S protein, indicating notable differences in antigenicity between SARS-CoV and SARS-CoV-2. These findings shed light on the viral pathogenesis and provide important structural information regarding development of therapeutic countermeasures against the emerging virus.
Keywords: SARS-CoV-2, CTD, receptor binding domain, receptor, ACE2, crystal structure, immunogenicity
Graphical Abstract
The crystal structure of the C-terminal domain of the SARS-CoV-2 spike protein in complex with human ACE2 reveals insights into the mechanisms of binding of this virus and its differences from SARS.
Introduction
Emerging and re-emerging viruses are a significant threat to global public health (Gao, 2018). Since the end of 2019, Chinese authorities have reported a cluster of human pneumonia cases in Wuhan City, China (Wang et al., 2020), and the disease was designated coronavirus disease 2019 (COVID-19). These cases showed symptoms such as fever and dyspnea and were diagnosed as viral pneumonia (Tan et al., 2020, Zhu et al., 2020). Whole-genome sequencing results showed that the causative agent was a novel coronavirus that was initially named 2019-nCoV by the World Health Organization (WHO) (Wu et al., 2020, Zhou et al., 2020, Zhu et al., 2020). Later, the International Committee on Taxonomy of Viruses (ICTV) officially designated the virus SARS-CoV-2 (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020), although many virologists argue that HCoV-19 is more appropriate (Jiang et al., 2020). As of February 24, 2020, 79,331 laboratory-confirmed cases have been reported to the WHO globally, with 77,262 cases in China, including 2,595 deaths (https://www.who.int/). In addition, 29 other countries have confirmed imported cases of SARS-CoV-2 infection (https://www.who.int/), raising great public health concerns worldwide.
SARS-CoV-2 is the seventh coronavirus that is known to cause human disease. Coronaviruses (CoVs) are a group of large and enveloped viruses with positive-sense, single-stranded RNA genomes (Lai et al., 2007, Lu and Liu, 2012). The viruses can be classified into four genera: alpha, beta, gamma, and deltaCoVs (Woo et al., 2009; https://talk.ictvonline.org/). Previously identified human CoVs that cause human disease include the alphaCoVs hCoV-NL63 and hCoV-229E and the betaCoVs HCoV-OC43, HKU1, severe acute respiratory syndrome CoV (SARS-CoV), and Middle East respiratory syndrome CoV (MERS-CoV) (Lu et al., 2015, Wevers and van der Hoek, 2009). Both alphaCoVs and the betaCoVs HCoV-OC43 and HKU1 cause self-limiting common cold-like illnesses (Chiu et al., 2005, Gorse et al., 2009, Jean et al., 2013, Jevšnik et al., 2012). However, SARS-CoV and MERS-CoV infection can result in life-threatening disease and have pandemic potential. During 2002–2003, SARS-CoV initially emerged in China and swiftly spread to other parts of the world, causing more than 8,000 infections and approximately 800 related deaths worldwide (WHO, 2004). In 2012, MERS-CoV was first identified in the Middle East and then spread to other countries (Ksiazek et al., 2003, Zaki et al., 2012). As of November 2019, a total of 2,494 MERS cases with 858 related deaths have been recorded in 27 countries globally (https://www.who.int/emergencies/mers-cov/en/). Notably, new cases of MERS-CoV infecting humans are still being reported (https://www.who.int/csr/don/archive/disease/coronavirus_infections/en/). SARS-CoV and MERS-CoV are zoonotic pathogens originating from animals. Detailed investigations indicate that SARS-CoV is transmitted from civet cats to humans and MERS-CoV from dromedary camels to humans (Azhar et al., 2014, Ge et al., 2013, Guan et al., 2003). The source of SARS-CoV-2, however, is still under investigation but linked to a wet animal market (Zhu et al., 2020, The 2019-nCoV Outbreak Joint Field Epidemiology Investigation Team and Li, 2020).
Virus infections initiate with binding of viral particles to host surface cellular receptors. Receptor recognition is therefore an important determinant of the cell and tissue tropism of a virus. In addition, the gain of function of a virus to bind to the receptor counterparts in other species is also a prerequisite for inter-species transmission (Lu et al., 2015). Interestingly, with the exception of HCoV-OC43 and HKU1, both of which have been shown to engage sugars for cell attachment (Li et al., 2005), the other human CoVs recognize proteinaceous peptidases as receptors. HCoV-229E binds to human aminopeptidase N (hAPN) (Li et al., 2019), and MERS-CoV interacts with human dipeptidyl peptidase 4 (hDPP4 or hCD26) (Lu et al., 2013, Raj et al., 2013). Although they belong to different genera, SARS-CoV and hCoV-NL63 interact with human angiotensin-converting enzyme 2 (hACE2) for virus entry (Hofmann et al., 2005, Li et al., 2003, Wu et al., 2009). After the outbreak of COVID-19, Chinese scientists promptly determined that SARS-CoV-2 also utilizes hACE2 for cell entry (Zhou et al., 2020).
In CoVs, the entry process is mediated by the envelope-embedded surface-located spike (S) glycoprotein (Lu et al., 2015). This S protein would, in most cases, be cleaved by host proteases into the S1 and S2 subunits, which are responsible for receptor recognition and membrane fusion, respectively (Lai et al., 2007). S1 can be further divided into an N-terminal domain (NTD) and a C-terminal domain (CTD), both of which can function as a receptor-binding entity (e.g., SARS-CoV and MERS-CoV utilize the S1 CTD to recognize the receptor (also called receptor binding domain [RBD]) (Li et al., 2005, Lu et al., 2013), whereas mouse hepatitis CoV engages the receptor with its S1 NTD (Taguchi and Hirai-Yuki, 2012)). The region in SARS-CoV-2 S protein that is responsible for hACE2 interaction remains unknown.
In this study, by utilizing immunostaining and flow cytometry assays, we first identify the S1 CTD (SARS-CoV-2-CTD) as the key region in SARS-CoV-2 that interacts with the hACE2 receptor. We subsequently solved a 2.5-Å crystal structure of SARS-CoV-2-CTD in complex with hACE2, which reveals a receptor-binding mode similar overall to that observed for the SARS-CoV RBD (SARS-RBD). However, SARS-CoV-2-CTD forms more atomic interactions with hACE2 than SARS-RBD, which correlates with data showing higher affinity for receptor binding. Notably, a panel of monoclonal antibodies (mAbs) as well as murine polyclonal antisera against SARS-S1/RBD were unable to bind to the SARS-CoV-2 S protein, indicating notable differences in antigenicity between SARS-CoV and SARS-CoV-2, suggesting that the previously developed SARS-RBD-based vaccine candidates are unlikely to be of any clinical benefit for SARS-CoV-2 prophylaxis. Taken together, these data shed light on viral entry and pathogenesis and will hopefully inspire new targeted treatments for this emerging pathogen.
Results
SARS-CoV-2 Applies the CTD to Interact with hACE2
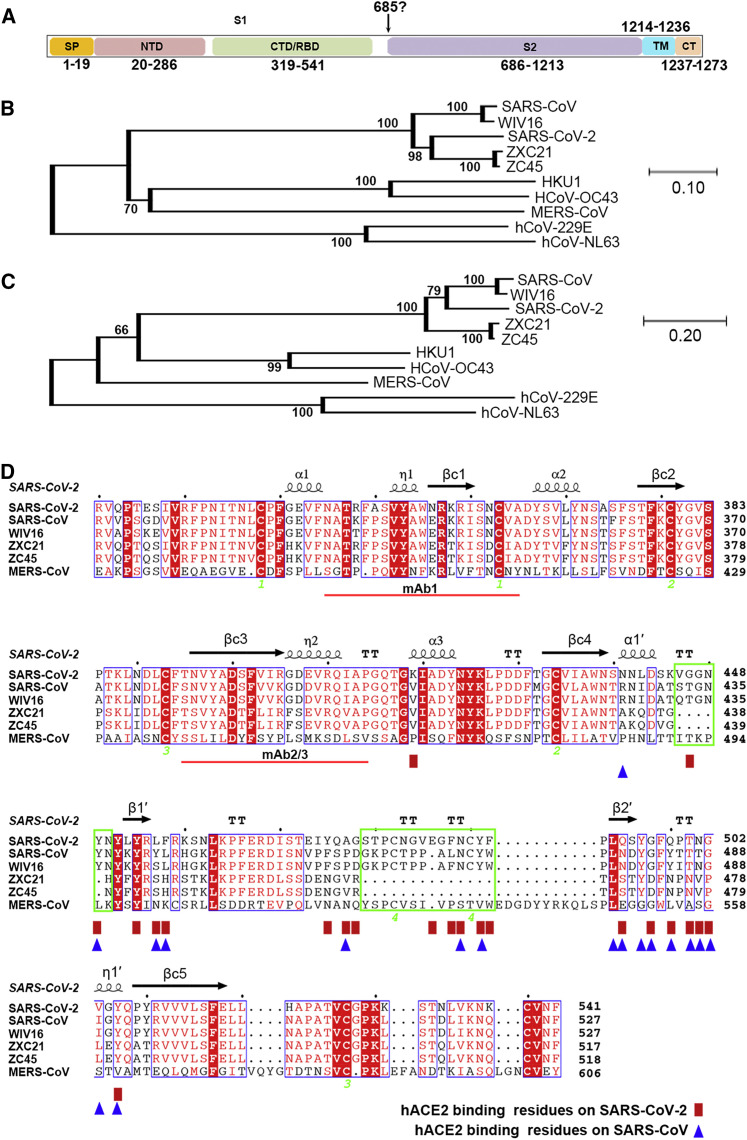
Through bioinformatics analysis, the SARS-CoV-2 S protein has been shown to display characteristic CoV S features, including a S1 region containing the NTD and CTD, S2, a transmembrane region, and a short cytoplasmic domain (Figure S1 A). Phylogenetic studies reveal that SARS-CoV-2 belongs to a group containing SARS-CoV as well as two bat-derived SARS-like viruses, ZC45 and ZCX21 (Figures S1B–S1D). Recently, hACE2 was reported to be the receptor of SARS-CoV-2 (Zhou et al., 2020). Because SARS-CoV utilizes its S1 CTD, otherwise known as the RBD, to recognize the same receptor, we decided to test whether the CTD in SARS-CoV-2 is also the key region for interaction with its receptor hACE2.
Figure S1.
Phylogenetic Analysis of SARS-CoV-2 and Sequence Alignments at the CTD Region, Related to Figures 2 and 3
(A) Schematic representation of the SARS-CoV-2 S protein based on the SARS-CoV S protein.
(B) Phylogenetic tree generated using MEGA (Tamura et al., 2013) with the S protein sequences.
(C) Phylogenetic tree generated using MEGA (Tamura et al., 2013) with the CTD region.
(D) Structure-based sequence alignment. The secondary structure elements were defined based on an ESPript (Robert and Gouet, 2014) algorithm and are labeled based on the SARS-CoV-2-CTD structure reported in this study. Spiral lines indicate α or 310 helices, and arrows represent β strands. The Arabic numerals 1-4 indicate cysteine residues that pair to form disulfide bonds. The red rectangles and blue triangles indicate the residues in the SARS-CoV-2-CTD and the SARS-RBD that interact with hACE2, respectively. Two deletions present in the ZXC21 and ZC45 external subdomains were highlighted with green boxes. The red lines indicate the epitopes recognized by mAb1 or mAb2/3.
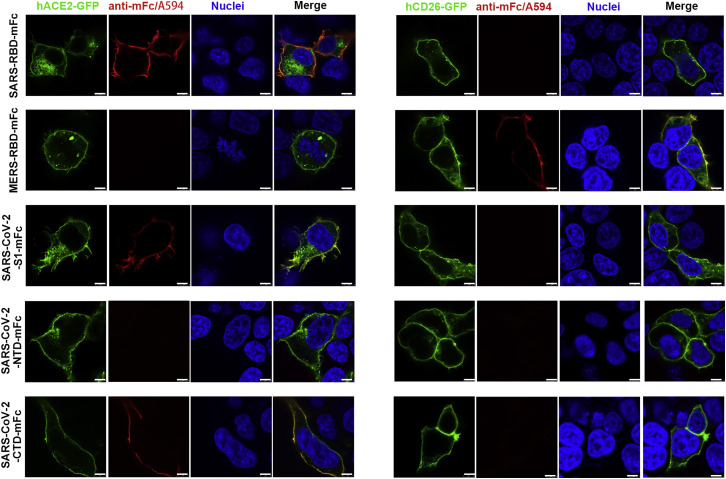
We prepared a series of mouse Fc (mFc)-fused SARS-CoV-2 S protein preparations, including S1 (SARS-CoV-2-S1), the NTD (SARS-CoV-2-NTD), and the CTD, and subsequently visualized their binding to GFP-tagged hACE2 expressed on the cell surface via confocal fluorescence microscopy. As a control, we also prepared the Fc fusion proteins for SARS-RBD and MERS-RBD and tested these in parallel with the SARS-CoV-2 proteins. As expected, SARS-RBD showed co-localization with hACE2 and MERS-RBD with hCD26. For the novel CoV proteins, SARS-CoV-2-S1 and SARS-CoV-2-CTD co-localized with hACE2 on the cell surface. The SARS-CoV-2-NTD protein, however, was incapable of binding to hACE2. In addition, none of the SARS-CoV-2 proteins interacted with hCD26 (Figure 1 ).
Figure 1.
SARS-CoV-2-S1 and SARS-CoV-2-CTD Co-localize with hACE2
HEK293T cells were transfected with pEGFP-N1-hACE2 (left panels, hACE2-GFP) or pEGFP-C1-hCD26 (right panels, hCD26-GFP). Twenty-four hours later, the cells were incubated with supernatant containing mFc-tagged SARS-CoV-2-S1 (SARS-CoV-2-S1-mFc), SARS-CoV-2-NTD (SARS-CoV-2-NTD-mFc), SARS-CoV-2-CTD (SARS-CoV-2-CTD-mFc), MERS-RBD (MERS-RBD-mFc), or SARS-RBD (SARS-RBD-mFc) proteins and subsequently incubated with anti-mouse IgG (mIgG) antibody conjugated with A594 (anti-mIgG/A594). Nuclei were stained with DAPI. All images were obtained by confocal microscopy using a Leica SP8 (×100 oil immersion objective lens). The scale bar in each panel indicates 8 μm. The data shown are representative of two independent experiments.
See also Figure S2.
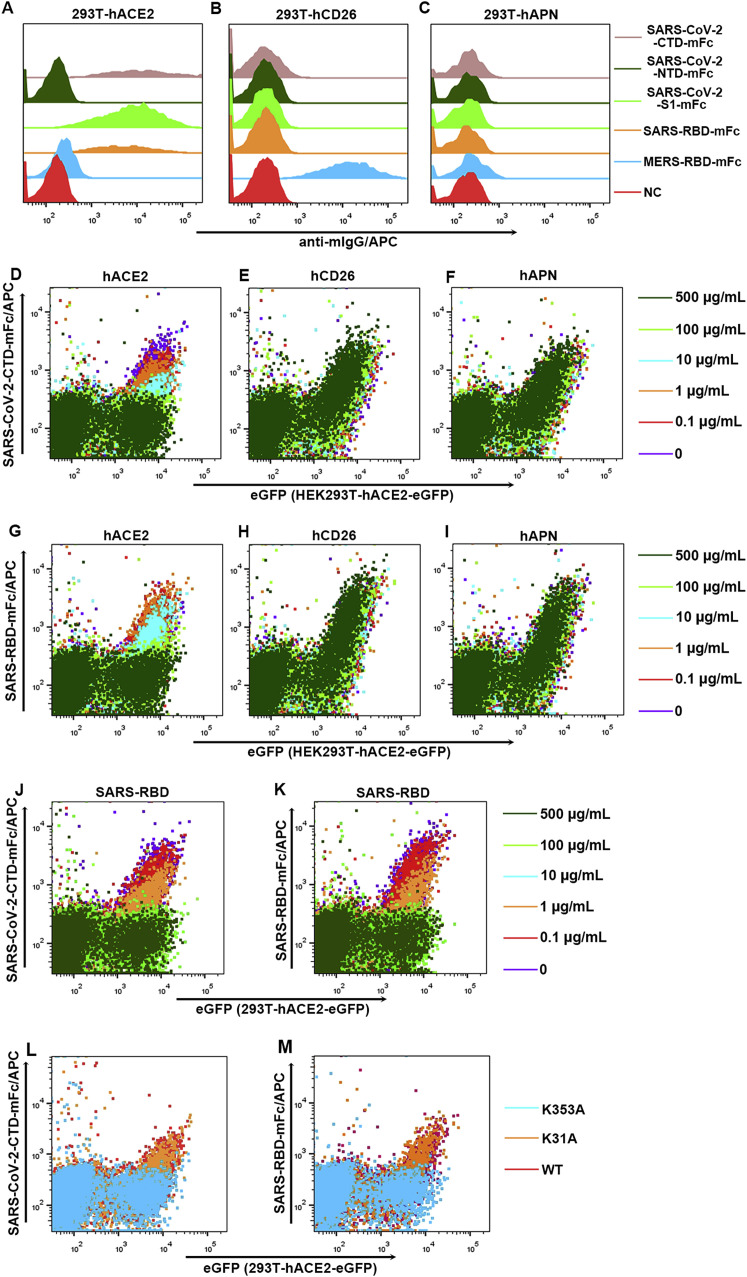
We further tested binding of the viral proteins to cell-surface hACE2 via flow cytometry. Consistently, SARS-CoV-2-S1 and SARS-CoV-2-CTD, but not SARS-CoV-2-NTD, showed strong affinity for hACE2 (Figure S2 A). None of the novel CoV proteins interacted with hCD26 or hAPN (Figures S2B and S2C). In addition, soluble hACE2, but not hCD26 or hAPN, was shown to inhibit the interaction between viral proteins, with cells expressing hACE2 in a dose-dependent manner (Figures S2D–S2I). Taken together, these results clearly demonstrate that SARS-CoV-2 is capable of binding, via the viral CTD, to hACE2.
Figure S2.
Characterization of Binding between SARS-CoV-2 and hACE2 by Flow Cytometry, Related to Figures 1 and 2
(A-C) Supernatant containing the indicated mFc-fusion proteins were incubated with HEK293T cells transiently expressing eGFP-tagged hACE2 (A), hCD26 (B) or hAPN (C), respectively. Anti-mIgG/APC was used to detect the mFc-fusion protein binding to the cells. Culture supernatant of HEK239T cells was used as negative control and marked as NC. For each sample, eGFP positive cells were first gated and then used to analyze fluorescence intensity of APC.
(D-F) Supernatant containing SARS-CoV-2-CTD-mFc proteins were pre-incubated with soluble hACE2 (D), hCD26 (E) or hAPC (F) at the indicated concentrations before addition to HEK293T cells transfected with pEGFP-N1-hACE2. mFc-fusion protein binding to HEK293T cells were detected by anti-mIgG/APC.
(G-I) Supernatant containing SARS-RBD-mFc proteins were pre-incubated with soluble hACE2 (G), hCD26 (H) or hAPC (I) at the indicated concentrations before addition to HEK293T cells transfected with pEGFP-N1-hACE2. mFc-fusion protein binding to HEK293T cells were detected by anti-mIgG/APC.
(J-K) HEK293T cells transfected with pEGFP-N1-hACE2 were pre-incubated with soluble SARS-RBD at the indicated concentration, before the addition of supernatant containing either SARS-CoV-2-CTD-mFc (J) or SARS-RBD-mFc (K). mFc-fusion protein binding to HEK293T cells were detected by anti-mIgG/APC.
(L-M) HEK293T cells transfected with pEGFP-N1-hACE2 (WT), or the mutants containing K353A (K353A) or K31A (K31A) were incubated with supernatant containing either SARS-CoV-2-CTD-mFc (L) or SARS-RBD-mFc (M). mFc-fusion protein binding to HEK293T cells were detected by anti-mIgG/APC.
All data shown are representative of two independent experiments.
The fluorescence signals were monitored by BD FACSCanto and the results were analyzed using FlowJo V10 (https://www.flowjo.com/solutions/flowjo/downloads).
Complex Structure between SARS-CoV-2 and hACE2
We then attempted to study the structural basis of the virus-receptor interaction. We prepared the SARS-CoV-2-CTD/hACE2 complex by in vitro mixture of the two proteins and isolated complexes via size exclusion chromatography. The complex structure was solved to 2.5-Å resolution (Table 1 ), with one SARS-CoV-2-CTD binding to a single hACE2 molecule in the asymmetric unit. For hACE2, clear electron densities could be traced for 596 residues from S19 to A614 of the N-terminal peptidase domain as well as glycans N-linked to residues 53, 90, and 322 (Figure 2 A).
Table 1.
Data Collection and Refinement Statistics
| SARS-CoV-2-CTD and hACE2 | |
|---|---|
| Data Collection | |
| Space group | P41212 |
| Cell Dimensions | |
| a, b, c (Å) | 104.45, 104.45, 229.79 |
| a, b, c (°) | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50.00–2.50 (2.59–2.50) |
| Unique reflections | 44,981 (43,84) |
| Completeness (%) | 100.0 (100.0) |
| Rmerge | 0.129 (1.147) |
| I/σI | 26.7 (3.3) |
| CC1/2 (%) | 0.999 (0.867) |
| Redundancy | 21.6 (22.3) |
| Refinement | |
| Resolution (Å) | 34.50-2.50 |
| Number of reflections | 44861 |
| Rwork/Rfree | 0.1846/0.2142 |
| Number of Atoms | |
| Protein | 6,461 |
| Ligand/ion | 1 |
| Water | 322 |
| B-factors | |
| Protein | 44.1 |
| Ligand/ion | 38.3 |
| Water | 40.4 |
| RMSDs | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.799 |
| Ramachandran Statistics (%) | |
| Favored | 98.60 |
| Allowed | 1.02 |
| Disallowed | 0.38 |
Values in parentheses are for the highest-resolution shell.
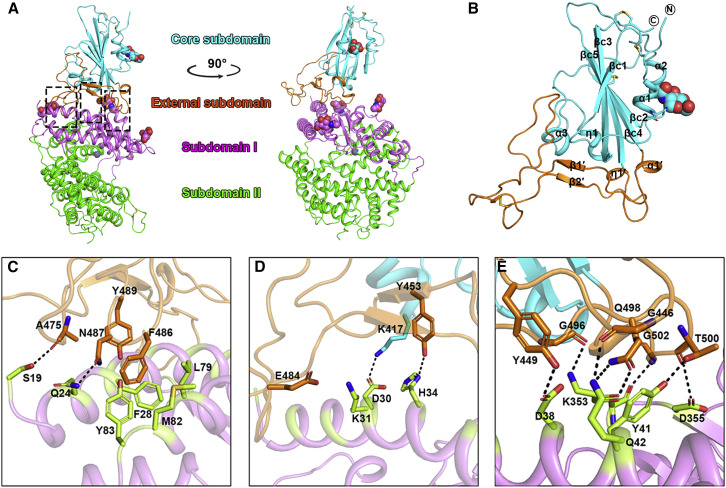
Figure 2.
The Complex Structure of SARS-CoV-2-CTD Bound to hACE2
(A) A cartoon representation of the complex structure. The core subdomain and external subdomain in SARS-CoV-2-CTD are colored cyan and orange, respectively. hACE2 subdomain I and II are colored violet and green, respectively. The right panel was obtained by anticlockwise rotation of the left panel along a longitudinal axis. The contacting sites are further delineated in (C)–(E) for the amino acid interaction details.
(B) A carton representation of the SARS-CoV-2-CTD structure. The secondary structural elements are labeled according to their occurrence in sequence and location in the subdomains. Specifically, the β strands constituting the core subdomain are labeled with an extra c, whereas the elements in the external subdomain are labeled with an extra prime symbol. The disulfide bonds and N-glycan linked to N343 are shown as sticks and spheres, respectively.
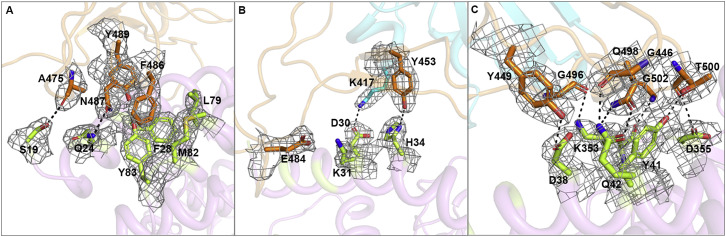
(C–E) Key contact sites are marked with the left, middle and right box in (A) and further delineated for interaction details, respectively. The residues involved are shown and labeled.
See also Figures S1, S2, and S3 and Table S1.
In the complex structure, the SARS-CoV-2-CTD contains 195 consecutive density-traceable residues spanning T333 to P527 together with N-linked glycosylation at N343. Similar to other reported betaCoV CTD structures, this protein also exhibits two structural domains (Han et al., 2017). One is the conserved core subdomain with five antiparallel β strands and a conserved disulfide bond between βc2 and βc4 (Figures 2B and S1D). The other is the external subdomain, which is dominated by a disulfide bond-stabilized flexible loop that connects two small β strands. The complex structure data show that SARS-CoV-2-CTD utilizes its external subdomain to recognize subdomain I in the hACE2 NTD (Figure 2A; Towler et al., 2004).
Further analysis was performed to identify key residues involved in complex formation. Amino acids located within the van der Waals (vdw) contact distance (4.5-Å-resolution cutoff) between the viral ligand and receptor were selected (Table 2 ), and a series of hydrophilic residues located along the interface were found to form a solid network of H-bond and salt bridge interactions (Figure 2). These strong polar contacts include the SARS-CoV-2-CTD residue A475 interacting with hACE2 residue S19, N487 with Q24 (Figures 2C and S3 A), E484 with K31, and Y453 with H34 (Figures 2D and S3B). Residue K417, located in helix α3 of the CTD core subdomain, was shown to contribute ionic interactions with hACE2 D30 (Figures 2D and S3B). Notably, the bulged loops in SARS-CoV-2-CTD, the α1’/β1’ loop and β2′/η1’ loop, properly position several residues (G446, Y449, G496, Q498, T500, and G502) in close proximity with hACE2 amino acids D38, Y41, Q42, K353, and D355, forming a concentration of H-bonds (Figures 2E and S3C). Further virus-receptor contacts include SARS-CoV-2-CTD Y489 and F486 packing against hACE2 residues F28, L79, M82, and Y83, forming a small patch of hydrophobic interactions at the interface (Figures 2C and S3A). Overall, the virus-receptor engagement is dominated by polar contacts mediated by the hydrophilic residues. In support of this hypothesis, a single K353A mutation was sufficient to abolish these interactions (Figure S2L).
Table 2.
Comparison of hACE2 Binding to SARS-CoV-2-CTD and SARS-RBD
| hACE2 | SARS-CoV-2-CTD | SARS-RBD |
|---|---|---|
| S19 (7/1) | A475 (3,1), G476 (4) | P462 (1) |
| Q24 (24/6) | A475 (4), G476 (5), N487 (15, 1) | N473 (6, 1) |
| T27 (15/8) | F456 (5), Y473 (1), A475 (2), Y489 (7) | L443 (3), Y475 (5) |
| F28 (7/7) | Y489 (7) | Y475 (7) |
| D30 (10/2) | K417 (4, 1), L455 (2), F456 (4) | Y442 (2) |
| K31 (19/12) | L455 (2), F456 (5), E484 (1), Y489 (6), F490 (2), Q493 (3) | Y442 (6), Y475 (6) |
| H34 (20/10) | Y453 (5, 1), L455 (9), Q493 (6) | Y440 (5, 1), Y442 (1), N479 (4) |
| E35 (8/0) | Q493 (8) | – |
| E37 (7/4) | Y505 (7) | Y491 (4) |
| D38 (15/11) | Y449 (9, 1), G496 (5), Q498 (1) | Y436 (9, 2), G482 (1), Y484 (1) |
| Y41 (23/25) | Q498 (8), T500 (7, 1), N501 (8, 1) | Y484 (9), T486 (8, 1), T487 (8) |
| Q42 (16/9) | G446 (4, 1), Y449 (4, 1), Q498 (8, 3) | Y436 (5, 1), Y484 (4) |
| L45 (4/3) | Q498 (3), T500 (1) | Y484 (2), T486 (1) |
| L79 (2/2) | F486 (2) | L472 (2) |
| M82 (9/4) | F486 (9) | L472 (4) |
| Y83 (20/10) | F486 (11), N487 (8, 1), Y489 (1) | N473 (8, 2), Y475 (2) |
| Q325 (0/4) | – | R426 (2), I489 (2) |
| E329 (0/6) | – | R426 (6, 1) |
| N330 (8/11) | T500 (8) | T486 (11, 1) |
| K353 (50/48) | G496 (7, 1), N501 (11), G502 (4, 1), Y505 (28) | Y481 (1), G482 (3),Y484 (2), T487 (11), G488 (6, 1), Y491 (25) |
| G354 (11/10) | Y502 (7), Y505 (4) | G488 (7), Y491 (3) |
| D355 (9/15) | T500 (8, 1), G502 (1) | T486 (8), T487 (3), G488 (4) |
| R357 (3/4) | T500 (3) | T486 (4) |
| R393 (1/1) | Y505 (1) | Y491 (1) |
| Total | 288 (16) | 213 (11) |
The numbers in parentheses of hACE2 residues represent the number of vdw contacts between the indicated residue with SARS-CoV-2-CTD (the former) and SARS-RBD (the latter). The numbers in parentheses of either ligand residues represent the numbers of wdw contacts the indicated residues conferred. The numbers with underline suggest numbers of potential H-bonds between the pairs of residues. wdw contact was analyzed at a cutoff of 4.5 Å and H-bonds at a cutoff of 3.5 Å. See also Table S1.
Figure S3.
Representative Electron Density Maps at the Binding Interface, Related to Figure 2
The electron densities of residues at the interaction interface between SARS-CoV-2-CTD and hACE2. The density maps are drawn in gray mesh contoured at 1 sigma. The core and external subdomains are colored cyan and orange, respectively. hACE2 is marked in violet. Residues in hACE2 that interact with the SARS-CoV-2-CTD are highlighted in lemon.
Comparison of the Binding Interfaces between hACE2/SARS-CoV-2-CTD and hACE2/SARS-RBD
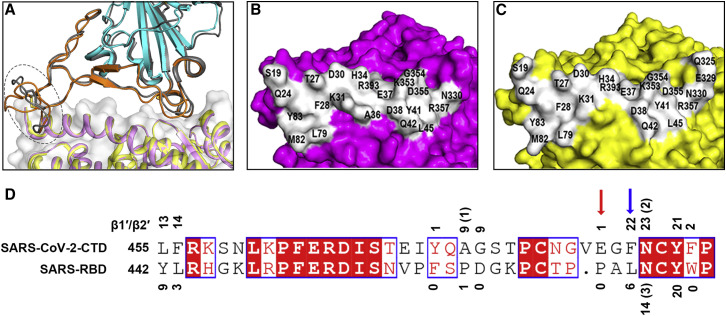
SARS-CoV-2-CTD exhibits significant structural homology to its SARS-CoV homolog, in agreement with high sequence identity between the two molecules (~73.9%) (Figure S1C). Superimposition of the SARS-CoV-2-CTD structure onto a previously reported SARS-RBD structure (PDB: 2GHV) revealed a root-mean-square deviation (RMSD) of 0.475 Å for 128 equivalent Cα atoms (Figure 3 A). In comparison with the SARS-RBD, the majority of the secondary structure elements are well superimposed in SARS-CoV-2-CTD, with the exception of the β1′/β2′ loop, which showed the most sequence variation between the two ligands (Figures 3A and S1D).
Figure 3.
Comparison of the SARS-CoV-2-CTD/hACE2 and SARS-RBD/hACE2 Binding Sites
(A) Overall similar receptor binding modes were observed for SARS-CoV-2-CTD and SARS-RBD. Superimposition of the structure of SARS-CoV-2-CTD (external subdomain in orange and core subdomain in cyan) bound to hACE2 (violet) and a complex structure of SARS-RBD (in gray) with hACE2 (yellow) are shown. The loop exhibiting variant conformations is highlighted by a dashed oval.
(B) hACE2 displayed in surface view. Residues that interact with the SARS-CoV-2-CTD are marked.
(C) hACE2 displayed in surface view. Residues that interact with the SARS-RBD are marked.
(D) Residues substitutions in SARS-CoV-2-CTD slightly strengthen the interaction with the receptor compared to the SARS-RBD. The amino acid sequences of the loop specified in (A) were aligned between the SARS-CoV-2-CTD and the SARS-RBD. The numbers show the vdw contacts between the receptor with the indicated SARS-CoV-2-CTD residues (above the sequence) or SARS-RBD residues (below the sequence). Numbers in parentheses indicate the number of potential H-bonds conferred by the indicated residues. The red and blue arrows represent the amino acids that form ionic and aromatic-aromatic interactions with the receptor, respectively.
The overall structure of the SARS-CoV-2-CTD/hACE2 complex is very similar to the previously reported structure of SARS-RBD bound to the same receptor with an RMSD of 0.431 Å for 669 equivalent Cα atoms (Li et al., 2005; Figures 3A–3C). Consistent with this high degree of similarity, the soluble SARS-RBD blocks the interaction between the SARS-CoV-2 ligand with hACE2 in a concentration-dependent manner (Figures S2J and S2K). Further detailed comparison of the receptor binding interface between the two viruses reveals that, among the 24 residues in hACE2 that make vdw contacts with either CTD, 15 amino acids display more contacts with the SARS-CoV-2-CTD (Table 2). The SARS-CoV-2-CTD binding interface also has more residues than SARS-RBD (21 versus 17) that directly interact with hACE2, forming more vdw contacts (288 versus 213) as well as H-bonds (16 versus 11) (Tables 2 and S1). Consistently, SARS-CoV-2-CTD in complex with hACE2 buries larger surface areas than SARS-RBD (1773 Å2 versus 1686 Å2).
Notably, the most variable loop (β1’/β2′ loop) contributes substantially more vdw contacts in SARS-CoV-2-CTD than for the SARS-RBD (115 versus 53) (Figure 3D; Table S1). Specifically, F486 in SARS-CoV-2, instead of I472 in SARS-RBD, forms strong aromatic-aromatic interactions with hACE2 Y83, and E484 in the SARS-CoV-2-CTD, instead of P470 in the SARS-RBD, forms ionic interactions with K31 (Figure 3D).
The Interaction between SARS-CoV-2-CTD and hACE2 Is Specific and Displays 4-Fold Stronger Affinity Compared with the SARS-RBD
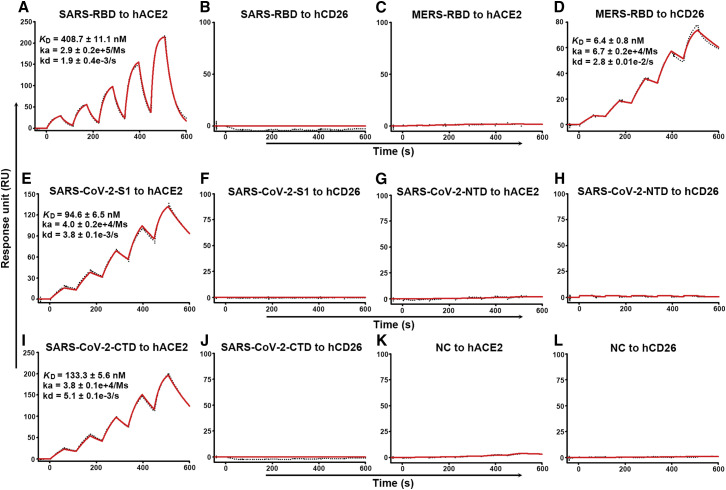
In light of the increased atomic interactions between hACE2 with the SARS-CoV-2-CTD compared with the SARS-RBD, we speculated that the former should bind to the receptor with stronger affinity than the latter. To test this hypothesis, we performed real-time surface plasmon resonance (SPR) assays. The mFc-tagged S-domain proteins were captured by anti-mouse IgG (mIgG) antibodies that were immobilized on the chip and tested for binding with gradient concentrations of the soluble ectodomain proteins of hACE2 and hCD26. As assay controls, SARS-RBD and MERS-RBD were found to readily interact with their respective canonical receptors (Figures 4A and 4D). SARS-CoV-2-S1 and SARS-CoV-2-CTD bound to hACE2 but not to hCD26 (Figures 4E, 4F, 4I, and 4J). The recorded binding profiles revealed typical slow-on/slow-off kinetics, as observed with the SARS-CoV and MERS-CoV proteins. The equilibrium dissociation constants (KD) of SARS-CoV-2-S1 and SARS-CoV-2-CTD binding to hACE2 were calculated to be 94.6 ± 6.5 nM and 133.3 ± 5.6 nM, respectively. These values represent ~4-fold higher binding affinities than that observed for the SARS-RBD engaging the same receptor, which was determined to be 408.7 ± 11.1 nM (Figure 4). Taken together, the increased atomic interactions between the hACE2 and SARS-CoV-2-CTD binding region leads to ~4-fold higher binding affinity compared with the SARS-RBD.
Figure 4.
Specific Interactions between SARS-CoV-2-S1 and SARS-CoV-2-CTD with hACE2, Characterized by SPR
The indicated mFc-tagged proteins in the supernatant were captured by anti-mIgG antibodies that were immobilized on the chip and subsequently tested for binding with gradient concentrations of hACE2 or hCD26, with the following binding profiles shown.
(A) SARS-RBD binding to hACE2.
(B) SARS-RBD binding to hCD26.
(C) MERS-RBD binding to hACE2.
(D) MERS-RBD binding to hCD26.
(E) SARS-CoV-2-S1 binding to hACE2.
(F) SARS-CoV-2-S1 binding to hCD26.
(G) SARS-CoV-2-NTD binding to hACE2.
(H) SARS-CoV-2-NTD binding to hCD26.
(I) SARS-CoV-2-CTD binding to hACE2.
(J) SARS-CoV-2-CTD binding to hCD26.
(K) Culture supernatant of HEK293T cells without transfection (NC) binding to hACE2.
(L) Culture supernatant of HEK293T cells without transfection (NC) binding to hCD26.
The values shown are the mean ± SD of three independent experiments.
SARS-CoV-2 Exhibits Distinct Epitope Features in the RBD from SARS-CoV
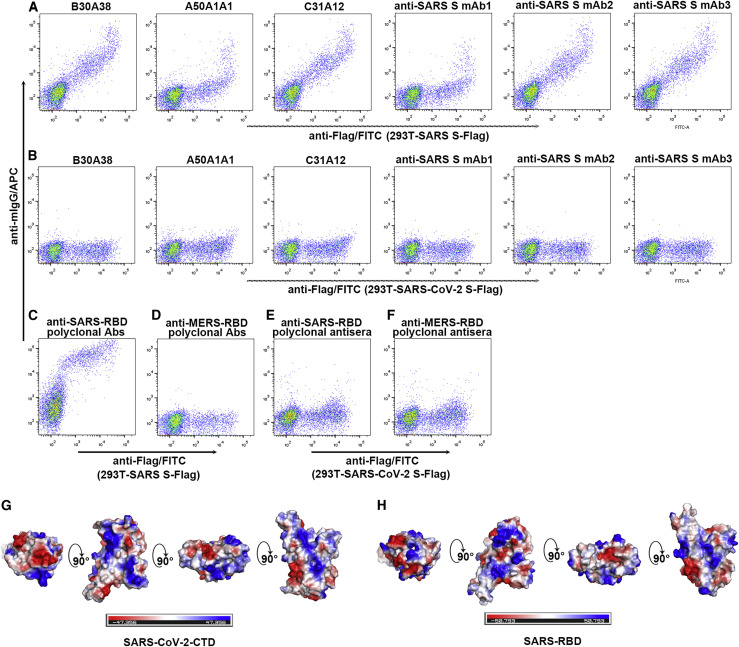
To conclude, we set out to investigate the epitope features of SARS-CoV-2 S by using a panel of murine mAbs directed against SARS-CoV S, including the B30A38, A50A1A1, and C31A12 antibodies, which recognize SARS-CoV S1, and mAbs 1–3, which recognize the SARS-RBD (Figure S1D; Wen et al., 2004, Zhang et al., 2009). Using flow cytometry, all six mAbs were observed to effectively bind to cells expressing SARS-CoV S. None of the mAbs, however, interacted with SARS-CoV-2 S (Figures 5A and 5B).
Figure 5.
Different Antigenicity between the SARS-CoV-2 S and SARS-CoV S Proteins
(A and B) HEK293T cells were transfected with pCAGGS plasmids containing Flag-tagged SARS-CoV S (A) or SARS-CoV-2 S (B). The indicated purified murine mAbs were subsequently added to the transfected cells before they were fixed, permeabilized, and stained with anti-Flag/fluorescein isothiocyanate (FITC).
(C and D) HEK293T cells were transfected with pCAGGS plasmids expressing Flag-tagged SARS-CoV S. The murine polyclonal sera against SARS-RBD (C) or MERS-RBD (D) were subsequently added to the transfected cells before they were fixed, permeabilized, and stained with anti-Flag/FITC.
(E and F) HEK293T cells were transfected with pCAGGS plasmids expressing Flag-tagged SARS-CoV-2 S. The murine polyclonal sera against SARS-RBD (E) or MERS-RBD (F) were subsequently added to the transfected cells before they were fixed, permeabilized, and stained with anti-Flag/FITC.
(G) Electrostatic surface view of SARS-CoV-2-CTD. The first panel represents the top view. The others are yielded by rotation of the former panel along a horizontal axis.
(H) Electrostatic surface view of SARS-RBD. The first panel represents the top view. The others are yielded by rotation of the former panel along a horizontal axis.
In comparison with a limited number of mAbs, polyclonal antibodies provide a more comprehensive view on potential epitope differences. In light of the determinant role of SARS-RBD and MERS-RBD in receptor recognition (Li et al., 2003, Li et al., 2005, Lu et al., 2013, Raj et al., 2013), the majority of neutralizing antibodies have been shown to target the RBD, exerting neutralization activity by disrupting virus/receptor engagement (Du et al., 2009, Wang et al., 2016). We therefore further prepared murine polyclonal antibodies against SARS-RBD and MERS-RBD. These two viral RBDs share very limited sequence identity and exhibit distinct structural characteristics in the RBD external subdomain that mediates receptor binding (Li et al., 2005, Lu et al., 2013). In the positive control, anti-SARS-RBD antibodies, but not anti-MERS-RBD antibodies, potently bound to cells expressing SARS-CoV S, as expected (Figures 5C and 5D). Nonetheless, neither of the antibody preparations bound to SARS-CoV-2 S (Figures 5E and 5F). In agreement with this observation, although SARS-CoV-2-CTD is structurally similar to the SARS-RBD structures (Figure 3), the electrostatic surface potential maps of these proteins were different (Figure 5G and 5H), which might explain the differing immunogenicity of the two ligands. Therefore, the results highlight distinct epitope features between SARS-RBD and SARS-CoV-2-CTD, although both can engage hACE2.
Discussion
The recent emergence of SARS-CoV-2 infection in China has led to major public health concerns. ACE2 has been reported to be the receptor for this novel CoV (Hoffmann et al., 2020, Zhou et al., 2020). In this study, we determined the key region in SARS-CoV-2 that is responsible for the interaction with the receptor and solved the crystal structure of SARS-CoV-2-CTD in complex with hACE2.
Considering the newly identified SARS-CoV-2, a total of seven human CoVs have been reported so far. Of these viruses, three (hCoV-NL63, SARS-CoV, and SARS-CoV-2) have been shown to utilize the hACE2 receptor for cell entry. The complex structures of hCoV-NL63 CTD and SARS-RBD bound to hACE2 have been reported previously (Li et al., 2005, Wu et al., 2009). Although hCoV-NL63 CTD and SARS-RBD are structurally distinct, the two viral ligands recognize and engage sterically overlapping sites in the receptor (Li, 2015). The complex structure of SARS-CoV-2-CTD together with hACE2 reveals that the majority of binding sites of SARS-CoV-2 in hACE2 also overlap the SARS-CoV binding site. The observations favor a scenario where these CoVs have evolved to recognize a “hotspot” region in hACE2 for receptor binding.
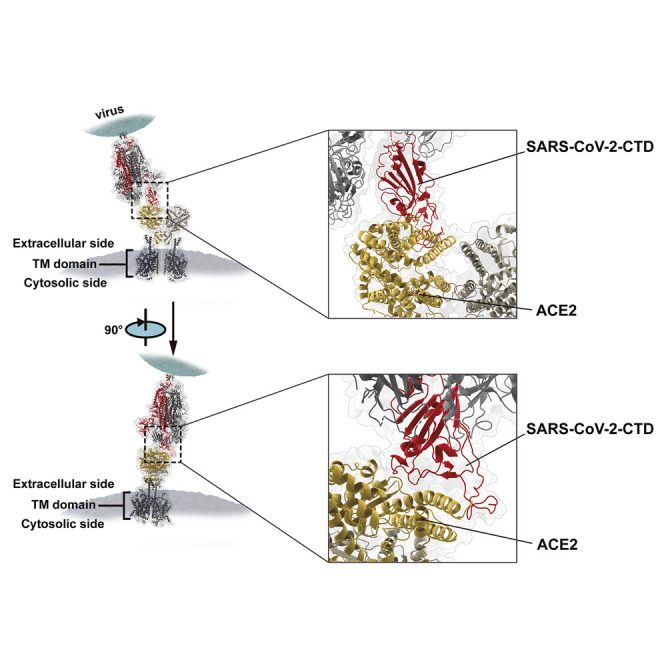
During the revision of our manuscript, the full-length hACE2 structure was reported to form a dimer in the presence of B0AT1 (an amino acid transporter), as revealed by cryoelectron microscopy (cryo-EM) analysis (Yan et al., 2020). They also reported the cryo-EM structure of dimeric hACE2-B0AT1 bound to two SARS-CoV-2-CTDs, with each molecule bound to an hACE2 monomer, with a local resolution of 3.5 Å at the interface. Our crystal structure of SARS-CoV-2-CTD/hACE2 is well superimposed with the cryo-EM structure, with an RMSD of 1.019 Å over 722 pairs of Cα atoms. Notably, two cryo-EM structures of trimeric SARS-CoV-2 S proteins were also published recently, with the receptor binding region buried or exposed (Walls et al., 2020, Wrapp et al., 2020), which is consistent with the structural features of MERS-CoV and SARS-CoV S proteins (Yuan et al., 2017). Further structure alignments show that the crystal structure of SARS-CoV-2-CTD in the complex also fits well with its counterparts in the cryo-EM structures, with RMSDs of 0.724 Å (exposed state) and 0.742 Å (buried states) related to PDB: 6VSB and 0.632 Å (exposed state) and 0.622 Å (buried state) related to PDB: 6VYB, respectively. These results indicate that the crystal structure of the complex is consistent with the respective cryo-EM structures and provide more detailed binding information.
Considering the high sequence identity between SARS-CoV-2-CTD and SARS-RBD, atomic comparisons of the two viral ligands binding the same receptor were performed. Atomic details reveal more interactions in SARS-CoV-2-CTD/hACE2 than in SARS-RBD/hACE2, including more engaged residues, more vdw contacts, more H-bonds, as well as larger buried surface areas. Interestingly, the β1’/β2′ loop, which is the most variable region between SARS-CoV-2-CTD and SARS-RBD, confers more interactions to SARS-CoV-2-CTD/hACE2, including strong interactions, such as aromatic-aromatic interactions and ionic interactions, in contrast to the SARS-RBD β1’/β2′ loop. A recently published paper also indicates that the SARS-CoV-2 S protein binds hACE2 with higher affinity than the SARS-CoV S protein (Wrapp et al., 2020), which was shown in this report as well.
Proteolysis of the S protein into S1 and S2 is another prerequisite for CoVs infection. MERS-Uganda and the bat CoV HKU4 can readily interact with hCD26, but they both require protease activation for cell entry (Kam et al., 2009, Matsuyama and Taguchi, 2009, Menachery et al., 2015, Menachery et al., 2020, Wang et al., 2014). A recent study shows that, in contrast with SARS-CoV S, which does not contain furin recognition sites between S1 and S2, SARS-CoV-2 S contains one potential cleavage site and could be efficiently processed into S1 and S2 (Hoffmann et al., 2020). The serine protease TMPRSS2 has been reported to contribute to priming of the SARS-CoV-2 S protein, and a TMPRSS2 inhibitor approved for clinical use was able to block entry. The authors postulated that the TMPRSS2 inhibitor might be a treatment option (Hoffmann et al., 2020).
Although SARS-CoV and SARS-CoV-2 share more than 70% sequence identity in the S protein, and both engage hACE2 via the CTD, we find that the two viruses CTDs are antigenically distinct. When using a panel of mAbs targeting SARS-CoV S1/CTD, none of the antibodies were able to recognize SARS-CoV-2 S. mAb1 and mAb2/mAb3 used in the above assay have been determined to bind to SARS-CoV S protein 330–350 and 380–399, respectively (Zhang et al., 2009). However, the binding sites for the other three mAbs (B30A38, A50A1A1, and C31A12), which were generated using SARS-CoV S1 as the immunogen, remain elusive. Consistently, a recently published paper also reported similar results showing that three SARS-RBD-directed mAbs, S230, m396, and 80R, were unable to bind to SARS-CoV-2 (Wrapp et al., 2020). Furthermore, we also demonstrate that polyclonal antisera directed against SARS-RBD do not recognize the S protein of SARS-CoV-2. A comparison of the two viral ligands shows that they display divergent electrostatic potential, which likely results in differing immunogenicity despite both ligands showing a similar protein fold.
Considering the key role of the CTD in receptor binding, this receptor engagement entity is an ideal immunogen for vaccine development. For instance, SARS-RBD and MERS-RBD proteins have been shown to efficiently induce production of neutralizing antibodies (Du et al., 2009, Wang et al., 2016). However, because of the observed differences in antigenicity and electrostatic distribution between SARS-CoV and SARS-CoV-2, it is unclear whether previously developed SARS-RBD-based vaccine candidates, such as subunit vaccines, will confer effective SARS-CoV-2 prophylaxis. During the revision of our manuscript, other studies have reported that SARS-CoV S-elicited polyclonal antibodies in mice and patients potently neutralized SARS-CoV-2 S-mediated entry into cells (Hoffmann et al., 2020, Walls et al., 2020). Notably, the S2 regions between SARS-CoV and SARS-CoV-2 exhibit higher sequence identity (~90%) and also contain neutralizing epitopes (Duan et al., 2005, Wang et al., 2015). Thus, the efficacy of SARS-CoV vaccines targeting S proteins for SARS-CoV-2 prophylaxis requires further evaluation and study.
In conclusion, CoVs are zoonotic pathogens and infect humans via inter-species transmission. SARS-CoV and MERS-CoV are two notorious examples of CoVs crossing the species barrier and resulting in human infection. Previous studies have shown that the two viruses first jumped from their natural hosts (bats) to an intermediate adaptive animal (e.g., dromedary camels for MERS-CoV) before infecting humans (Azhar et al., 2014, Wang et al., 2014). Delineating this cross-species transmission route could be highly instructive for disease control. Nevertheless, the natural host and the intermediate adaptive animal, if any, for SARS-CoV-2 remains unknown. The structural information for SARS-CoV-2-CTD and hACE2 shown in this study should shed light on the viral inter-species transmission route by characterizing the interactions between S and hACE2 of different species in the future.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC Goat Anti-mIgG | BioLegend | Cat# 405308; RRID: AB_315011 |
| Anti-His/APC | Miltenyi Biotec | Cat# 130-119-820; RRID: AB_2751870 |
| Anti-Flag/FITC | Miltenyi Biotec | Cat# 130-119-683; RRID: AB_2811325 |
| Goat anti-mIgG/Alexa Fluor 594 | ZSGB-BIO | ZF-0513 |
| DAPI | Beyotime | Cat# C1002 |
| Murine anti-Flag antibody | Sigma-Aldrich | F1804; RRID: AB_262044 |
| Fixation/Permeabilization Solution Kit | BD Biosciences | Cat# 554714 |
| Bacterial and Virus Strains | ||
| Escherichia coli (E. coli) strain DH5α | TIANGEN | Cat# CB101-02 |
| MAX Efficiency DH10Bac Competent E. coli | Invitrogen | Cat# 10361-012 |
| Chemicals, Peptides, and Recombinant proteins | ||
| PEI | Alfa | A04043896-1g |
| Recombinant SARS-CoV-2-S1 protein fused with mFc, spike residues 20-685, accession number: EPI_ISL_402119 | This paper | N/A |
| Recombinant SARS-CoV-2-NTD protein fused with mFc, spike residues 20-286, accession number: EPI_ISL_402119 | This paper | N/A |
| Recombinant SARS-CoV-2-CTD protein fused with mFc, spike residues 319-541, accession number: EPI_ISL_402119 | This paper | N/A |
| Recombinant MERS-RBD protein fused with mFc, spike residues 367-606, accession number: JX869050 | This paper | N/A |
| Recombinant SARS-RBD protein fused with mFc, spike residues 306-527, accession number: NC_004718 | This paper | N/A |
| Recombinant hCD26 protein, residues 39−766, accession number: NP_001926 | This paper | N/A |
| Recombinant hACE2 protein, residues 19−615, accession number: BAJ21180 | This paper | N/A |
| Recombinant hAPN protein, residues 66−967, accession number: NP_001141 | This paper | N/A |
| Critical Commercial Assays | ||
| HisTrap HP 5 mL column | GE Healthcare | Cat# 17524802 |
| HiLoad 16/600 Superdex 200 pg | GE Healthcare | Cat# 28989335 |
| Series S Sensor Chip CM5 | GE Healthcare | Cat# 29149603 |
| HiTrap Protein G HP | GE Healthcare | Cat# 17040503 |
| Mouse Antibody Capture Kit | GE Healthcare | Cat# BR-1008-38 |
| Deposited Data | ||
| Crystal structure of SARS-CoV-2-CTD/hACE2 | This paper | PDB: 6LZG; NMDC: NMDCN0000001 |
| Experimental Models: Cell Lines | ||
| Sf9 Cells, SFM Adapted | Invitrogen | Cat# 11496015 |
| Hi5 cells | Invitrogen | Cat# B85502 |
| Huh7 cells | Institute of Basic Medical Sciences CAMS | 3111C0001CCC000679 |
| HEK293T cells | ATCC | ATCC CRL-3216 |
| Recombinant DNA | ||
| pEGFP-N1 | MiaoLingPlasmid | Cat# P0133 |
| pEGFP-N1-hACE2, accession number: BAJ21180 | This paper | N/A |
| pEGFP-C1 | MiaoLingPlasmid | Cat# P0134 |
| pEGFP-C1-hCD26, accession number: NP_001926 | This paper | N/A |
| pEGFP-C1-hAPN, accession number: NP_001141 | This paper | N/A |
| pFastbac1 | Invitrogen | 10360014 |
| pFastbac-hCD26-His, residues 39−766, accession number: NP_001926 | This paper | N/A |
| pFastbac-hACE2-His, residues 19−615, accession number: BAJ21180 | This paper | N/A |
| pFastbac-hAPN-His, residues 66−967, accession number: NP_001141 | This paper | N/A |
| pCAGGS | MiaoLingPlasmid | Cat# P0165 |
| pCAGGS-SARS-CoV-2 S-Flag, accession number: EPI_ISL_402119 | This paper | N/A |
| pCAGGS-MERS-CoV-S-Flag, accession number: JX869050 | This paper | N/A |
| pCAGGS-SARS-CoV-S-Flag, accession number: NC_004718 | This paper | N/A |
| pCAGGS-SARS-CoV-2-S1-mFc, residues 20-685, accession number: EPI_ISL_402119 | This paper | N/A |
| pCAGGS-SARS-CoV-2-NTD-mFc, residues 20-286, accession number: EPI_ISL_402119 | This paper | N/A |
| pCAGGS-SARS-CoV-2-CTD-mFc, residues 319-541, accession number: EPI_ISL_402119 | This paper | N/A |
| pCAGGS-MERS-RBD-mFc, residues 367-606, accession number: JX869050 | This paper | N/A |
| pCAGGS-SARS-RBD-mFc, residues 306-527, accession number: NC_004718 | This paper | N/A |
| Software and Algorithms | ||
| PyMOL software | Molecular Graphics System, Version 1.8 Schrödinger | https://pymol.org/2/ |
| MEGA version X | Tamura et al., 2013 | https://www.megasoftware.net/ |
| BIAcore® 8K Evaluation software | GE Healthcare | N/A |
| FlowJo V10 | FLOWJO | https://www.flowjo.com/solutions/flowjo/downloads |
| ESPript 3 | Robert and Gouet, 2014 | http://espript.ibcp.fr/ESPript/ESPript/ |
| Graphpad Prism 6 | GraphPad Software | https://www.graphpad.com/ |
| HKL2000 | Otwinowski and Minor, 1997 | N/A |
| Phaser | Read, 2001 | N/A |
| COOT | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/peemsley/coot/ |
| Phenix | Adams et al., 2010 | http://www.phenix-online.org/ |
| MolProbity | Williams et al., 2018 | N/A |
| SigmaPlot | Systat Software, Inc | https://systatsoftware.com/products/sigmaplot/ |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jianxun Qi (jxqi@im.ac.cn).
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
The number of replicates carried out for each experiment is described in the figure/table legends.
Experimental Model and Subject Details
Cells
HEK293T cells (ATCC CRL-3216) and Huh7 cells (Institute of Basic Medical Sciences CAMS 3111C0001CCC000679) were cultured at 37°C in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
Method Details
Gene cloning
The plasmids used for protein expression and purification were separately constructed by insertion of the coding sequences of hCD26 (residues 39−766, accession number NP_001926), hACE2 (residues 19−615, accession number: BAJ21180) and hAPN (residues 66−967, accession number: NP_001141) into the baculovirus transfer vector pFastbac1 (Invitrogen) using the EcoRI and XhoI restriction sites. All proteins contained an N-terminal gp67 signal peptide and a C-terminal 6 × His tag.
The pEGFP-C1-hCD26 and pEGFP-C1-hAPN plasmids were constructed by cloning the coding region of hCD26 or hAPN into pEGFP-C1 using restriction enzymes XhoI and SmaI, respectively. Similarly, the hACE2 protein was fused to eGFP by cloning the coding region into pEGFP-N1.
Recombinant proteins SARS-CoV-2-S1-mFc, SARS-CoV-2-NTD-mFc, SARS-CoV-2-CTD-mFc, MERS-RBD-mFc and SARS-RBD-mFc were used in assays of flow cytometry (FACS), immunostaining and surface plasmon resonance (SPR). The coding sequences of SARS-CoV-2-S1 (residues 1-685, GISAID: EPI_ISL_402119), SARS-CoV-2-NTD (residues 1−286, GISAID: EPI_ISL_402119), SARS-CoV-2-CTD (residues 319−541, GISAID: EPI_ISL_402119), MERS-RBD (residues 367-606, GenBank: JX869050) and SARS-RBD (residues 306-527, GenBank: NC_004718) tagged with the mFc domain of mouse IgG were individually cloned into the pCAGGS expression vector using the EcoRI and XhoI restriction sites. For the secretion of SARS-CoV-2-CTD, SARS-RBD and MERS-RBD, signal peptides from the parental virus were used.
The full-length coding region of SARS-CoV-2 S, SARS-CoV S and MERS-CoV S protein with a C-terminal Flag tag was cloned into the pCAGGS vector using the EcoRI and SmaI restriction sites (pCAGGS-SARS-CoV-2 S-Flag, pCAGGS-SARS S-Flag and pCAGGS-MERS S-Flag).
Protein expression and purification
The Bac-to-Bac baculovirus expression system (Invitrogen) was used to express the proteins for FACS and SPR analysis. The constructed pFastbac1 vectors were transformed into DH10Bac competent cells to generate recombinant bacmids (Zhang et al., 2010). Transfection of bacmids and virus amplification were conducted in Sf9 cells, while Hi5 cells were used for protein expression. The supernatants of Hi5 cells were collected 48 h post-infection, and soluble proteins were purified by metal affinity chromatography using a HisTrap HP 5 mL column (GE Healthcare). The samples were then pooled and further purified via size exclusion chromatography with a Superdex 200 column (GE Healthcare) in a buffer composed of 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl.
The mFc recombinant proteins were expressed in HEK293T cells. pCAGGS plasmid containing MERS-RBD coding sequences were transiently transfected into cells. After 4 d expression, supernatants were collected, centrifuged, and mixed with the same volume of binding buffer containing 20 mM Na3PO4 (pH 7.0). The mixtures were then filtered through 0.22-μm filters and passed through a HiTrap rProtein A FF (GE Healthcare) affinity chromatography column at a maximum flow rate of 1 mL/min. The bound protein was eluted with 0.1 M glycine-HCl (pH 3.0) and collected into tubes containing 200 μL 1 M Tris-HCl (pH 9.0). mFc fusion proteins were further purified by gel filtration in PBS and concentrated and stored at −80°C. To prepare the mouse Fc-fusion proteins of SARS-CoV-2-S1, SARS-CoV-2-NTD, SARS-CoV-2-CTD and, SARS-RBD, HEK293T cells were transfected with pCAGGS plasmid containing the coding sequence for the indicated protein. 24 h later, the supernatant containing the indicated protein were collected, concentrated and then used for FACS, immunostaining and SPR assays.
Flow cytometry
For the binding test, plasmids containing hCD26, hACE2 or hAPN that were fused with eGFP were transfected into HEK293T cells using PEI (Alfa) according to the manufacturer’s instructions. 2 × 105 cells were collected 24 h after transfection, suspended in PBS (with 0.5% FBS) and incubated with the individual mFc-fusion proteins-containing supernatant at 37°C for 30 min, followed by washing with PBS twice and further incubation with anti-mIgG/APC antibodies (1:500, Miltenyi Biotec). After washing, the cells were analyzed using a BD FACSCalibur. The cells incubated with only the secondary antibody were used as negative controls. For the binding-blocking assay, the supernatant containing the indicated mFc-fusion proteins were preincubated with hACE2, hCD26 or hAPN at concentration of 100, 10, 1, or 0.1 μg/mL at 37°C for 1 h before the addition to the cells.
When SARS-RBD and MERS-RBD were used for binding competition assay, 2 × 105 HEK293T cells with transient expression of hACE2 were incubated with SARS-RBD or MERS-RBD at concentrations of 100, 10, 1 and 0.1 μg/mL at 37°C for 1 h. The supernatant containing the indicated proteins were subsequently added. After washing with PBS (with 0.5% FBS), anti-mIgG/APC (1:500, Miltenyi Biotec) antibodies were used to detect the binding.
To test whether anti-SARS-CoV S antibodies bound to SARS-CoV-2 S protein, HEK293T cells were first transfected with pCAGGS containing Flag-tagged SARS-CoV-2 S protein. HEK293T cells expressing SARS-CoV S protein were used as positive control. The purified murine Abs were then used at final concentration of 10 μg/mL to stain 3 × 105 cells. After washing with PBS with 0.5% FBS, anti-mIgG/APC (1:500, Miltenyi Biotec) was added. After washing, the cells were fixed and permeabilized with Fixation/Permeabilization solution (BD Biosciences), and stained with anti-Flag/FITC (1:100, Miltenyi Biotec) at 37°C for 30 min. After washing, the cells were subjected to BD FACSCanto for fluorescent detection. The data was subsequently analyzed using FlowJo V10.
Previously immunized murine polyclonal antibodies were also applied to test binding to SARS-CoV-2. 2 × 105 HEK293T cells expressing Flag-tagged SARS-CoV-2 S proteins were incubated with murine MERS-RBD-immunized sera (1:10 dilution) or SARS-RBD-immunized sera (1:10 dilution) at 37°C for 30 min. The cells were sequentially washed twice, incubated with anti-mIgG/APC (1:500, Miltenyi Biotec), washed again, and fixed and permeabilized with Fixation/Permeabilization solution (BD Biosciences). Then anti-Flag/FITC (1:100 dilution, Miltenyi Biotec) was added to the cells and stained for another 30 min, before washing and fluorescence analysis by BD FACSCanto.
SPR analysis
The interaction between indicated mFc-fusion protein with hACE2 or hCD26 was monitored by SPR using a BIAcore 8K (GE Healthcare) carried out at 25°C in single-cycle mode. The CM5 biosensor chip (GE Healthcare) was first immobilized with anti-mIgG antibody for flow cells (Fc) 1 and 2, according to manufacturer’s amine-coupling chemistry protocol (GE Healthcare). The indicated mFc-tagged protein was then injected and captured on Fc 2. Fc 1 was used as the negative control. Both hACE2 and hCD26 used for this assay were in buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, and 0.005% (v/v) Tween 20. Concentrated supernatant containing SARS-CoV-2-S1-mFc, SARS-CoV-2-NTD-mFc, SARS-CoV-2-CTD-mFc and SARS-RBD-mFc and purified MERS-RBD-mFc were individually captured by the antibody immobilized on the CM5 chip at approximately 200-500 response units. Various concentrations of hCD26s and hACE2s were then flowed through the chip and the real-time response was recorded. The concentrations of hCD26 were 6.25, 12.5, 25, 50 and 100 nM when testing interactions with MERS-RBD. To test the interaction with SARS-CoV-2-S1-mFc, SARS-CoV-2-NTD-mFc, SARS-CoV-2-CTD-mFc and SARS-RBD-mFc, 50, 100, 200, 400 and 800 nM of hCD26 were used. The concentrations of hACE2 were 50, 100, 200, 400 and 800 nM. After each reaction, the chip was re-generated using pH 1.7 glycine. The equilibrium dissociation constants (binding affinity, K D) for each pair of interaction were calculated using BIAcore® 8K evaluation software (GE Healthcare). The K D values were calculated using the model of 1:1 (Langmuir) binding mode. The graphics were prepared using SigmaPlot 10.0.
Indirect immunofluorescence analysis and confocal microscopy
For indirect immunofluorescence analysis, HEK293T cells were pre-seeded in a 15 mm culture dish and transfected with plasmids containing either eGFP-tagged hACE2 or hCD26. 24 h later, the cells were washed three times with PBS, fixed with 4% paraformaldehyde in PBS for 10 min, washed three times with PBS, and then blocked in PBS containing 1% bovine serum albumin for 1 h. The cells were then incubated with concentrated supernatant containing indicated proteins or purified MERS-RBD-mFc proteins (10 μg/mL). Cells were then washed three times with PBS and incubated with goat anti-mIgG conjugated with Alexa Fluor 594 (1:200, ZSGB-BIO) at room temperature for 1 h. Nuclei were stained with DAPI (5 μg/mL, Beyotime). The cells were then visualized on a Leica SP8 confocal microscope.
Immunization of mice
Both MERS-RBD and SARS-RBD were expressed and purified as previously reported (Li et al., 2005, Raj et al., 2013). Five BALB/c mice were immunized intra-muscularly with 10 μg MERS-RBD or SARS-RBD resuspended in PBS solution (pH 7.4) in the presence of MF59. Three weeks later, mice were boosted. The antisera were collected 2 weeks after the boost and kept at −20°C before use. The antisera mixture from five mice were used to evaluate the binding of S protein in flow cytometry assay.
Crystallization
Crystallization trials were performed by sitting-drop method with 0.8 μL protein mixing with 0.8 μL reservoir solution at 18°C. The initial crystallization screenings were carried out using the commercially available kits. Diffractable crystals of the SARS-CoV-2-CTD/hACE2 complex was finally obtained in a solution consisting of 0.1 M MES pH 6.5, 10% w/v PEG 5000 MME and 12% v/v 1-propanol with a protein concentration of 15 mg/ml.
Data collection and structure determination
Diffraction data was collected at Shanghai Synchrotron Radiation Facility (SSRF) BL17U (wavelength, 0.97919 Å). For data collection, the crystals were cryo-protected by briefly soaking in reservoir solution supplemented with 20% (v/v) glycerol before flash-cooling in liquid nitrogen. The dataset was processed with HKL2000 software (Otwinowski and Minor, 1997). The complex structure of SARS-CoV-2-CTD with hACE2 was determined by the molecular replacement method using Phaser (Read, 2001) with previously reported SARS-RBD complex structure (PDB: 2AJF). The atomic models were completed with Coot (Emsley and Cowtan, 2004) and refined with phenix.refine in Phenix (Adams et al., 2010), and the stereochemical qualities of the final models were assessed with MolProbity (Williams et al., 2018). Data collection, processing, and refinement statistics are summarized in Table 1. All structural figures were generated using Pymol software (https://pymol.org/2/).
Sequences used in the alignments
The GenBank accession numbers of the sequences used for analyzing the conservation among betaCoVs are the following: MERS-CoV, GenBank: JX869050; SARS-CoV, GenBank: NC_004718; ZXC21, GenBank: AVP78042.1; ZC45, GenBank: AVP78031.1; WIV16, GenBank: ALK02457.1; hCoV-NL63, GenBank: Q6Q1S2; hCoV-229E, GenBank: P15423; HKU1, GenBank: Q0ZME7; HCoV-OC43, GenBank: U3M6B4; SARS-CoV-2, GISAID: EPI_ISL_402119.
Quantification and Statistical Analysis
Binding studies
K D values for SPR experiments were obtained with BIAcore® 8K Evaluation Software (GE Healthcare), using a 1:1 binding model. The values shown are the mean ± SD of three independent experiments.
Flow cytometry analysis
All of the experiments were performed twice; one representative of each experiment was shown in Figures 1, 5, and S2.
Data and Code Availability
The accession number for the atomic coordinates and diffraction data reported in this study is PDB: 6LZG. The data have also been deposited in the China Natinal Microbiology Data Center (NMDC) at (http://www.nmdc.cn/) with accession number NMDCN0000001.
Acknowledgments
We are grateful to W.N.D. Gao for revising the manuscript. We thank Y. Chen and Z. Yang (Institute of Biophysics, Chinese Academy of Sciences [CAS]) for technical support for the SPR analysis. We thank X. Zhang (Institute of Microbiology, CAS) for help with performing confocal imaging. We are grateful to L. Dai (Beijing Institute of Life Sciences, CAS) and K. Wen (Southern Medical University) for kindly providing the mAbs/pAbs against SARS-CoV. We thank L.Wang for help with the phylogenetics analysis. This work was supported by the Strategic Priority Research Program of CAS (XDB29010202 and XDB29040302), the National Key Research and Development Program of China (2016YFD0500305), the National Natural Science Foundation of China (81922044, 81673358 and 81973228), and the Zhejiang University Special Scientific Research Fund for COVID-19 Prevention and Control (2020XGZX031). Q.W. is supported by Youth Innovation Promotion Association CAS grant 2018119. G.L. is supported by the special research fund on COVID-19 of Sichuan Province (2020YFS0010) and the special research fund on COVID-19 of West China Hospital Sichuan University (HX-2019-nCoV-004). J.Y is supported by Foundation of the NSFC Innovative Research Group grant 81621091.
Author Contributions
Q.W., J.Y., and J.Q. initiated and coordinated the project. Q.W. designed the experiments. Q.W., Y.Z., L.W., S.N., and C.S. conducted the experiments with assistance from Z.Z., C.Q., and Y.H.. Y.Z. and S.N. grew the complex crystals. J.Q. collected the diffraction data and determined the complex structure with help from Q.S.W. and H.Z. L.W. performed the immunostaining. C.S. performed the flow cytometry assays. Y.Z. and Q.W. conducted the SPR analysis. K.-Y.Y. provided the purified mAbs against the SARS-CoV S protein. Q.W. and G.L. analyzed the data and wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: April 9, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.03.045.
Supplemental Information
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- Chiu S.S., Chan K.H., Chu K.W., Kwan S.W., Guan Y., Poon L.L., Peiris J.S. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin. Infect. Dis. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J., Yan X., Guo X., Cao W., Han W., Qi C., Feng J., Yang D., Gao G., Jin G. A human SARS-CoV neutralizing antibody against epitope on S2 protein. Biochem. Biophys. Res. Commun. 2005;333:186–193. doi: 10.1016/j.bbrc.2005.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gao G.F. From “A”IV to “Z”IKV: Attacks from emerging and re-emerging pathogens. Cell. 2018;172:1157–1159. doi: 10.1016/j.cell.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.-Y., Li J.-L., Yang X.-L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorse G.J., O’Connor T.Z., Hall S.L., Vitale J.N., Nichol K.L. Human coronavirus and acute respiratory illness in older adults with chronic obstructive pulmonary disease. J. Infect. Dis. 2009;199:847–857. doi: 10.1086/597122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Han X., Qi J., Song H., Wang Q., Zhang Y., Wu Y., Lu G., Yuen K.Y., Shi Y., Gao G.F. Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein. Virology. 2017;507:101–109. doi: 10.1016/j.virol.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. S0092-8674(20)30229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean A., Quach C., Yung A., Semret M. Severity and outcome associated with human coronavirus OC43 infections among children. Pediatr. Infect. Dis. J. 2013;32:325–329. doi: 10.1097/INF.0b013e3182812787. [DOI] [PubMed] [Google Scholar]
- Jevšnik M., Uršič T., Zigon N., Lusa L., Krivec U., Petrovec M. Coronavirus infections in hospitalized pediatric patients with acute respiratory tract disease. BMC Infect. Dis. 2012;12:365. doi: 10.1186/1471-2334-12-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Shi Z., Shu Y., Song J., Gao G.F., Tan W., Guo D. A distinct name is needed for the new coronavirus. Lancet. 2020;395:949. doi: 10.1016/S0140-6736(20)30419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.W., Okumura Y., Kido H., Ng L.F., Bruzzone R., Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS ONE. 2009;4:e7870. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lai M.M.C., Perlman S., Anderson L.J. Coronaviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott Williams & Wilkins; 2007. pp. 1305–1335. [Google Scholar]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li Z., Tomlinson A.C., Wong A.H., Zhou D., Desforges M., Talbot P.J., Benlekbir S., Rubinstein J.L., Rini J.M. The human coronavirus HCoV-229E S-protein structure and receptor binding. eLife. 2019;8:e51230. doi: 10.7554/eLife.51230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Liu D. SARS-like virus in the Middle East: a truly bat-related coronavirus causing human diseases. Protein Cell. 2012;3:803–805. doi: 10.1007/s13238-012-2811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Taguchi F. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J. Virol. 2009;83:11133–11141. doi: 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Dinnon K.H., 3rd, Yount B.L., Jr., McAnarney E.T., Gralinski L.E., Hale A., Graham R.L., Scobey T., Anthony S.J., Wang L. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J. Virol. 2020;94 doi: 10.1128/JVI.01774-19. e01774–e01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read R.J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku316. W320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Hirai-Yuki A. Mouse hepatitis virus receptor as a determinant of the mouse susceptibility to MHV infection. Front. Microbiol. 2012;3:68. doi: 10.3389/fmicb.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Zhao X., Ma X., Wang W., Niu P., Xu W., Gao G.F., Wu G. Notes from the field: A novel coronavirus genome identified in a cluster of pneumonia cases - Wuhan, China 2019-2020. China CDC Weekly. 2020;2:61–62. [PMC free article] [PubMed] [Google Scholar]
- The 2019-nCoV Outbreak Joint Field Epidemiology Investigation Team. Li Q. Notes from the Field: An Outbreak of NCIP (2019-nCoV) Infection in China - Wuhan, Hubei Province, 2019–2020. China CDC Weekly. 2020;2:79–80. [PMC free article] [PubMed] [Google Scholar]
- Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. S0092-8674(20)30262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y., Ji W., Li Y., Wu Y., Wang J. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi W., Joyce M.G., Modjarrad K., Zhang Y., Leung K., Lees C.R., Zhou T., Yassine H.M., Kanekiyo M. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015;6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wong G., Lu G., Yan J., Gao G.F. MERS-CoV spike protein: Targets for vaccines and therapeutics. Antiviral Res. 2016;133:165–177. doi: 10.1016/j.antiviral.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen K., Mei Y.B., Qiu L.W., Liao Z.Y., Yuen K.Y., Che X.Y. Preparation and characterization of monoclonal antibodies against S1 domain at N-terminal residues 249 to 667 of SARS-associated coronavirus S1 protein. J. First Mil. Med. Univ. 2004;24:1–6. [PubMed] [Google Scholar]
- Wevers B.A., van der Hoek L. Recently discovered human coronaviruses. Clin. Lab. Med. 2009;29:715–724. doi: 10.1016/j.cll.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Cumulative number of reported probable cases of severe acute respiratory syndrome (SARS) 2004. https://www.who.int/csr/sars/country/en/
- Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lai K.K., Huang Y., Lee P., Luk G.S., Dyrting K.C., Chan K.H., Yuen K.Y. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 2009;83:908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Li W., Peng G., Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. USA. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020 doi: 10.1126/science.abb2762. Published online March 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang X., Wang J., Wen K., Mou Z., Zou L., Che X., Ni B., Wu Y. Antibody binding site mapping of SARS-CoV spike protein receptor-binding domain by a combination of yeast surface display and phage peptide library screening. Viral Immunol. 2009;22:407–415. doi: 10.1089/vim.2009.0046. [DOI] [PubMed] [Google Scholar]
- Zhang W., Qi J., Shi Y., Li Q., Gao F., Sun Y., Lu X., Lu Q., Vavricka C.J., Liu D. Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus. Protein Cell. 2010;1:459–467. doi: 10.1007/s13238-010-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the atomic coordinates and diffraction data reported in this study is PDB: 6LZG. The data have also been deposited in the China Natinal Microbiology Data Center (NMDC) at (http://www.nmdc.cn/) with accession number NMDCN0000001.