Abstract
Emerging evidence indicates that epigenetic regulators are critically required for the maintenance of tissue-specific stem cells and that the epigenetic marks are altered in stem cells during physiological aging. Intriguingly, aging-associated stem cell functional decline can be reversed by manipulating epigenetic factors that become dysregulated during aging. These observations lend support to the stem cell theory of aging, which postulates that aging is the result of the inability of tissue-specific stem cells to replenish the tissues with functional differentiated cells that maintain the function of a tissue, and open a new era of research on the epigenetics of stem cell aging that may represent therapeutic potential. Recent advances in single cell technologies are revolutionizing our mechanistic understanding of rare populations of cells, such as stem cells, and offer an unprecedented opportunity to address this challenge.
Introduction
The past two decades have witnessed a significant revision of a traditional view that aging is a random and passive process. Mounting evidence suggests that the aging process is controlled by evolutionarily conserved genetic pathways [1,2]. The intense effort to identify genetic regulators of organismal aging has led to the revelation that the rate of aging is controlled, in part, by several epigenetic modifiers and metabolic factors [3]. Given that epigenetic regulators require various metabolites as cofactors for chromatin modifications [4], these findings raise the possibility that, during the aging process, epigenetic changes result in the dysregulation of gene expression and the subsequent loss of cellular function. This model is attractive, because it provides a potential explanation for the well-documented lifespan- and healthspan-extending effects of calorie restriction [5].
Every cell changes with age. However, tissue-specific stem cells receive much attention in aging research due to their unique properties of self-renewal and differentiation to give rise to the progeny that support tissue structures and functions. The fact that stem cells persist throughout the lifespan, to repair tissues and maintain homeostasis, argues that they may have an essential role in the aging process. In this regard, the aging-associated accumulation of damage in stem cells can be passed along to their progeny and account for tissue degeneration and dysfunction.
The property of maintaining tissue structures and functions has earned stem cells a prominent position in the pantheon of aging research [6]. However, recent human studies suggest that the impact of stem cell aging has been underestimated. For example, the peripheral blood of young individuals is generated from a diverse pool of active hematopoietic stem cells (HSCs), whereas aged individuals lose this diversity as clones arise and take over the population, a phenomenon termed ‘clonal hematopoiesis of intermediate potential’ (CHIP) [7]. Surprisingly, individuals with CHIP are not only at a higher risk for developing aging-associated hematologic malignancies, but also display early mortality, likely due to an increased risk of cardiovascular diseases. Thus, the effect of HSC aging may go beyond the blood system, and impact distant tissues and overall organismal aging. Given the implication of epigenetic alteration as a hallmark of aging, it is natural to inquire whether epigenetic drift (see Glossary) is a driver of stem cell exhaustion during aging. Here, we present emerging evidence supporting the epigenetics of stem cell aging and clarify the trends and outstanding questions in this burgeoning field.
Opportunities
Early evidence supporting the epigenetic regulation of stem cell aging has begun to emerge, and several epigenetic regulators have been shown to be critically required for stem cell maintenance during aging. The sirtuin family members are linked to metabolism and epigenetic regulation through their unique deacetylase activity that is dependent on NAD+. The expression of SIRT2, SIRT3, and SIRT7 is repressed in aged HSCs [8–10]. Genetic studies show that Sirt2- and Sirt3-deficient mice have age-dependent defects in HSC maintenance, while Sirt6- and Sirt7-deficient mice exhibit HSC defects that resemble aspects of HSC aging [8–11]. The expression of Tet2, a regulator of DNA methylation, is reduced in aged neural stem cells (NSCs) and such a reduction can be reversed by parabiosis, a surgical procedure that has been shown to reverse aging-associated degeneration [12]. Tet2 ablation in mice causes compromised neurogenesis and cognition. In Drosophila, the heterochromatin regulator Piwi limits aging-related changes in intestinal stem cells (ISCs) [13]. Consistent with the genetic evidence, biochemical studies from stem cells of various tissue origins have demonstrated aging-associated alterations in DNA methylation, as well as histone acetylation and methylation marks [12,14–17]. Together, these genetic and biochemical studies support the notion that epigenetic regulation is likely to be a mechanism contributing to stem cell aging that is conserved across tissues and species.
The study of stem cell biology has been challenging, because stem cells represent rare cell populations and many conventional biochemical assays are not applicable. Furthermore, the requirement of the niche to maintain stem cell identity renders further barriers to address this problem through in vitro expansion. The realization of stem cell population heterogeneity casts another layer of complexity. Until recently, stem cell studies relied primarily on genetic approaches and biochemical studies that can provide direct mechanistic insights have been limited. However, recent advances in genomic and single cell technologies offer an opportunity to obtain direct evidence for the epigenetic regulation of stem cell aging with unprecedented sensitivity and precision. This technical advancement, together with the early genetic and biochemical evidence, will usher in a new era of research on the epigenetics of stem cell aging.
Gaps and Challenges
What Are the Best Approaches and Stem Cell Models to Measure Epigenetic Changes due to Aging?
A comprehensive understanding of the epigenetics of stem cell aging requires the identification of the pattern of epigenetic marks that change in stem cells during aging, the demonstration of the kinetics of such changes throughout lifespan, and the elucidation of their impact on chromatin packaging and 3D organization. Refined technologies using smaller cell numbers will improve the resolution of the epigenetic atlas of stem cell aging. Coupling the kinetics of epigenetic changes and functional deterioration of stem cell aging will substantiate or dispute the theory of epigenetics of stem cell aging. The comparison of aging-associated epigenetic drift in stem cells across tissues or species will likely enrich our understanding of stem cell aging through the identification of shared and unique regulatory mechanisms, ultimately informing the most effective approaches for rejuvenation. Sex as a biological variable should be considered, because sexual divergence in aging-related epigenetic changes has been noted [15]. Lessons can also be obtained from studies of long-lived species, such as naked mole rats, where the epigenome is more stable, leading to maintained genome organization and the prevention of malignant transformation [18]. Another valuable model is induced pluripotent stem cells (iPSCs) derived from patients with progeria, where reprogramming erases the epigenetic defects and resets the epigenetic landscape to a revitalized pluripotent state [19].
What Is the Relationship between Epigenetic Perturbations and Stem Cell Aging?
Studies of known epigenetic regulators have provided proof-of-principle for the concept that epigenetic alterations contribute to stem cell aging, and they offer an entry point to more deeply dissect the regulatory mechanisms involved [8,9,11–13]. However, the epigenetic regulation of stem cell aging is likely to be controlled by a multifactorial network. Thus, the identification of the bona fide regulators of this process is a prerequisite to understand these biological processes at a deep molecular level. The bona fide epigenetic regulators of stem cell aging must fulfill the following criteria: (i) their activities must be changed in stem cells during aging and the epigenetic marks governed by these factors should be altered; and (ii) their gain- or loss-of-function should result in altered stem cell fate and function that mirror some aspects of stem cell aging. Expanding our knowledge of how the epigenomic landscape changes with age through the survey of additional epigenetic modifications in stem cells should aid the identification of candidate regulators.
Given that metabolism is intimately linked to epigenetic regulation [4], it is equally important to determine the metabolic alterations that occur upon stem cell aging. Analysis of aging-associated metabolic alterations may provide insights into how epigenetic factors become dysregulated in stem cells, and also begs the question as to what causes the metabolic alterations. The role of the mitochondria as a metabolic hub and their ability to produce free radicals that initiate stress make the mitochondria likely suspects, but this remains to be tested.
It is increasingly appreciated that genetic mosaicism increases with age [7]. In clonal hematopoiesis, several epigenetic regulatory genes (DNMT3A, TET2, and ASXL1) frequently acquire somatic mutations. Thus, DNA damage could be a source of the epigenetic drift that occurs in aged stem cells, and targeted error-corrected gene-sequencing approaches can detect mutant clones in a heterogenous population of cells. In addition to mutations in epigenetic factors, DNA damage may elicit cellular responses to alter the epigenetic landscape [20]. Transposable elements can also disturb the genomic landscape and their role in epigenetic drift warrants investigation.
The drivers of epigenetic drift in stem cells may also come from the extrinsic signals. As an example, the robustness of circadian rhythms is reduced with age, contributing to aging-associated physiological decline. Aging rewires the circadian gene expression in stem cells [21]. How distal tissues communicate with stem cells to rewire the clock, and whether the circadian rewiring is related to epigenetic drift in stem cells, are topics that remain to be explored.
In addition to the sources of epigenetic drift in aged stem cells, it is equally important to understand how epigenetic alterations lead to the functional decline of aged stem cells. Given that mitochondrial stress [8], loss of proteostasis [9], and DNA damage [22] have been put forth as causes of stem cell aging, the investigation of how epigenetic drift results in the accumulation of cellular damage is a priority. Potential clues may come from dysregulation in gene expression, including cryptic genes [23], and the altered downstream cellular pathways, such as the developmental pathways [17]. Given that cellular damage can drive epigenetic drift in aged stem cells and some epigenetic regulators of stem cell aging have been shown to prevent cellular damages [8,9], a likely scenario is a feed-forward loop that perpetuates the epigenetic drift and cellular damage (Figure 1, Key Figure). This line of investigations will likely have profound implications for understanding the extent to which stem cell aging can be reversed.
Figure 1.
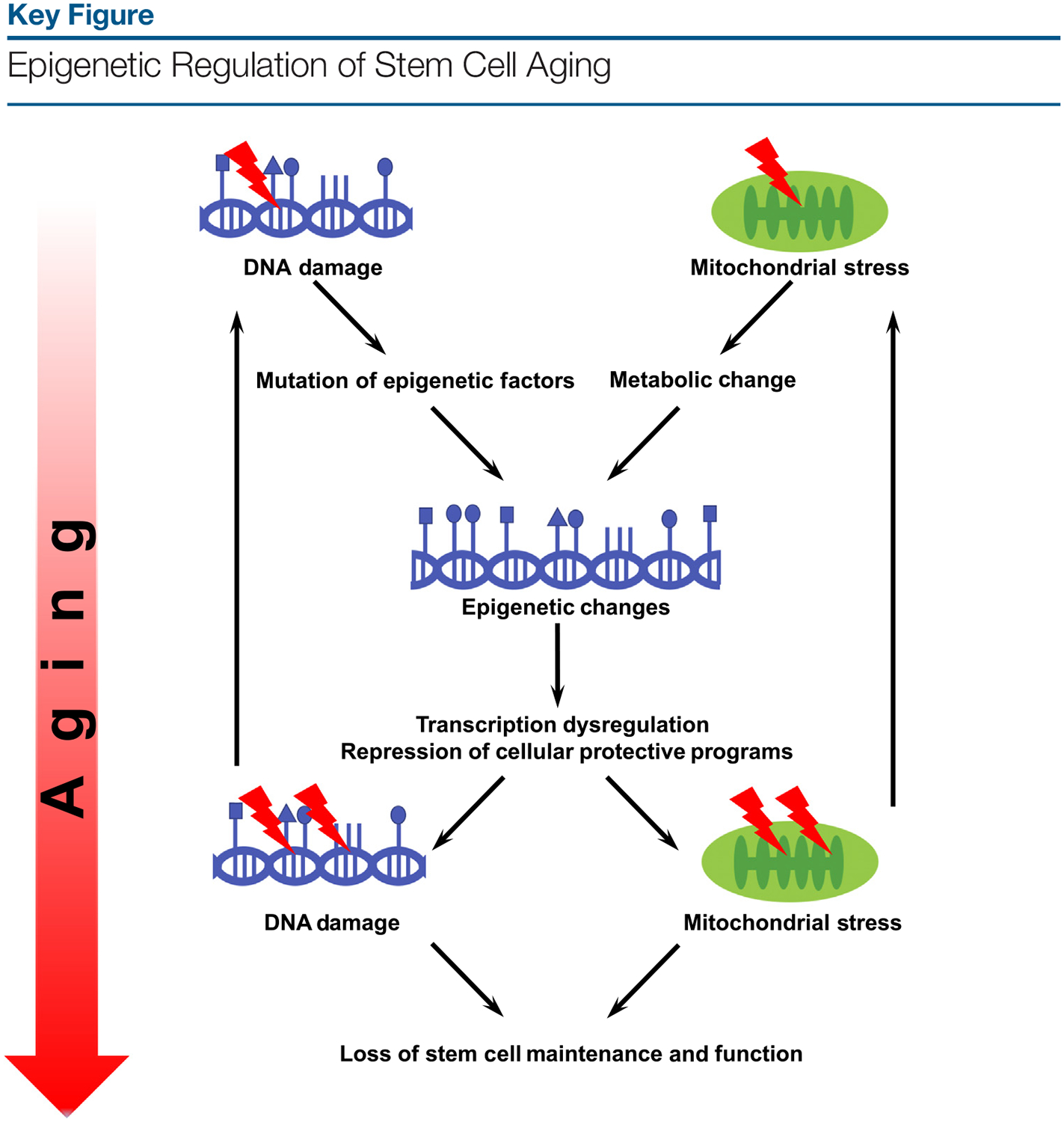
During the aging process, mitochondrial stress causes metabolic changes, while DNA damage causes mutations in epigenetic factors, resulting in epigenetic changes, transcriptional dysregulation, and repression of cellular protective programs that safeguard mitochondrial and genomic integrity. This forms a feed-forward loop that perpetuates the epigenetic changes and cellular damage that ultimately lead to loss of stem cell maintenance and function.
What Are Potential Interventions to Reverse or Overcome Stem Cell Aging?
Boosting mitochondrial protective programs that become dysregulated in aged stem cells and repressing mitochondrial stresses can reverse the functional decline of stem cell aging [8,9]. Thus, stem cell aging is likely due to the repression of cellular protective programs and can be reversed by targeting these cellular protective programs. Reverting the alteration in epigenetic stress responses in aged stem cells by targeting the epigenetic modifying enzymes can improve the function of aged stem cell [17]. The identification of bona fide epigenetic regulators of stem cell aging may lead to novel interventions to reverse stem cell aging. In addition to insights from physiological stem cell aging, potential interventions impacting the epigenome may also be derived from reverse engineering of known antiaging approaches, such as calorie restriction and exercise. Rejuvenation strategies that broadly target stem cell aging in multiple tissue types could have pleiotropic beneficial effects. However, although the successful reversal of stem cell aging relies on the efficacy and precision of the epigenetic changes, one should also be wary of potential untoward effects, such as oncogenic transformation.
An important consideration is the proper assessment of stem cell aging and rejuvenation. The multiplexed nature of epigenetic regulation of stem cell aging posits that manipulation of a single mechanism is unlikely to fully recapitulate or revert physiological stem cell aging. A complete understanding of key mechanisms of epigenetic regulation of stem cell aging is accomplished when physiological stem cell aging is faithfully reconstituted or reverted with the manipulation of these combined mechanisms. Stem cell aging and rejuvenation are ultimately assessed by their functional capacity to self-renew, differentiate, and regenerate the tissues that are changed by natural aging. The functional assessment can be complemented by the measurement of an array of molecular markers of stem cell aging to demonstrate the rate-limiting step of stem cell aging. The recent success in the development of biomarkers of aging based on DNA methylation is instrumental, because such epigenetic clocks enable accurate estimate of biological age across tissues [24]. Such technologies may be applied to stem cells. Alternatively, stem cell-specific epigenetic clocks could be developed for assessing stem cell aging and rejuvenation.
What Are the Gaps in Our Knowledge for Age-Related Changes in Epigenetic Status of Stem Cells and Their Relationship to Certain Diseases or Afflictions?
Animal models where bona fide epigenetic regulators of stem cell aging are specifically ablated in stem cells are valuable tools to assess the impact of epigenetics of stem cell aging on distal tissues and organismal aging. For example, clonal expansion has been recapitulated in a mouse model reconstituted with HSCs deficient for Tet2, which is frequently mutated in the blood cells of individuals with clonal hematopoiesis, and this mouse model develops accelerated atherosclerosis and heart failure due to aberrant activation of the NLRP3 inflammasome in macrophages [25,26]. The impact of age-related changes in epigenetics of stem cells on the development of other age-related diseases and the underlying mechanisms remain to be explored. The observation that the NLRP3 inflammasome becomes aberrantly activated in aged HSCs raises the possibility that epigenetic drift ultimately manifests as dysregulated inflammatory signaling in aged HSCs, which give rise to mature blood cells that are prone to inflammation and negatively affect distant tissues [10]. This is just the tip of the iceberg in understanding how stem cell aging impacts distant tissues and overall healthspan. The significance of this line of research is that it provides the rationale for stem cell-based therapies to extend healthspan. While current epidemiological studies have demonstrated that individuals with clonal hematopoiesis have shortened lifespan, studies in centenarians may provide unique insights into the relationship between epigenetics of stem cell aging and longevity.
Concluding Remarks
Given the importance of stem cell aging in the degeneration and dysfunction of aging tissues and the reversible nature of epigenetic regulation, a comprehensive understanding of the epigenetics of stem cell aging is central to the basic biology of aging (see Outstanding Questions). In addition, this understanding is pivotal to the realization of the extent to which aging-associated conditions are reversible and, ultimately, to the therapeutic potential of the biology of aging.
Outstanding Questions.
What are the best approaches and stem cell models to measure epigenetic changes due to aging?
What is the relationship between epigenetic perturbations and stem cell aging?
What are potential interventions to reverse or overcome stem cell aging?
What are the gaps in our knowledge for age-related changes in epigenetic status of stem cells and their relationship to certain diseases or afflictions?
Highlights.
Epigenetic marks are altered in stem cells during physiological aging.
Epigenetic regulators are critically required for the maintenance of tissue-specific stem cells.
Aging-associated stem cell functional decline can be reversed by altering the epigenetic landscape.
Epigenetics of stem cell aging can impact distant tissues and organismal aging.
Acknowledgments
We thank Isabel E. Beerman, Weiwei Dang, Willard Freeman, Vera Gorbunova, R. David Hawkins, Heinrich Jasper, Philipp Oberdoerffer, K. Lenhard Rudolph, Paolo Sassone-Corsi, William L Stanford, Saul A Villeda, and Kenneth Walsh for their input to clarify the trends and outstanding questions in the field at the NIA Workshop on Epigenetics of Stem Cell Aging. Supported by NIH Grants R01 DK101885 (D.C.), R01 DK117481 (D.C.), AG063404 (D.C.), and National Institute of Food and Agriculture (D.C.).
Glossary
- DNA damage
alteration in the chemical structure of DNA, such as a break in a DNA strand, a base missing from the DNA backbone, or a chemically changed base
- Epigenetic drift
aging is associated with global alterations of epigenetic markers and this altered epigenomic state is referred to as epigenetic drift
- Mitochondrial stress
challenges associated with mitochondrial physiology, including imbalance between reactive oxygen species (ROS) generation and scavenging, and mitochondrial protein folding stress
- Proteostasis
the concept that integrated cellular pathways control protein biogenesis, folding, trafficking, and degradation to maintain cellular protein homeostasis
References
- 1.Kenyon C (2005) The plasticity of aging: insights from long-lived mutants. Cell 120, 449–460 [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Otin C et al. (2013) The hallmarks of aging. Cell 153, 1194–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guarente L and Kenyon C (2000) Genetic pathways that regulate ageing in model organisms. Nature 408, 255–262 [DOI] [PubMed] [Google Scholar]
- 4.Kaelin WG Jr. and McKnight SL (2013) Influence of metabolism on epigenetics and disease. Cell 153, 56–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weindruch R and Walford R (1988) The Retardation of Aging and Disease by Dietary Restriction, Charles C. Thomas Publishing [Google Scholar]
- 6.Goodell MA and Rando TA (2015) Stem cells and healthy aging. Science 350, 1199–1204 [DOI] [PubMed] [Google Scholar]
- 7.Jan M et al. (2017) Clonal hematopoiesis. Semin. Hematol 5, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown K et al. (2013) SIRT3 reverses aging-associated degeneration. Cell Rep. 3, 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohrin M et al. (2015) Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347, 1374–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo H et al. (2019) Mitochondrial stress-initiated aberrant activation of the NLRP3 inflammasome regulates the functional deterioration of hematopoietic stem cell aging. Cell Rep. 26, 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H et al. (2016) SIRT6 controls hematopoietic stem cell homeostasis through epigenetic regulation of Wnt signaling. Cell Stem Cell 18, 495–507 [DOI] [PubMed] [Google Scholar]
- 12.Gontier G et al. (2018) Tet2 rescues age-related regenerative decline and enhances cognitive function in the adult mouse brain. Cell Rep. 22, 1974–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sousa-Victor P et al. (2017) Piwi is required to limit exhaustion of aging somatic stem cells. Cell Rep. 20, 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beerman I et al. (2013) Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell 1, 413–425 [DOI] [PubMed] [Google Scholar]
- 15.Masser DR et al. (2017) Sexually divergent DNA methylation patterns with hippocampal aging. Aging Cell 16, 1342–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasumarthy KK et al. (2017) Methylome analysis of human bone marrow MSCs reveals extensive age- and culture-induced changes at distal regulatory elements. Stem Cell Rep. 9, 999–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schworer S et al. (2016) Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature 540, 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan L et al. (2017) Naked mole rat cells have a stable epigenome that resists iPSC reprogramming. Stem Cell Rep. 9, 1721–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z et al. (2017) Reprogramming progeria fibroblasts reestablishes a normal epigenetic landscape. Aging Cell 16, 870–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J et al. (2018) Replication stress shapes a protective chromatin environment across fragile genomic regions. Mol. Cell 69, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solanas G et al. (2017) Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell 170, 678–692 [DOI] [PubMed] [Google Scholar]
- 22.Rossi DJ et al. (2007) Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 [DOI] [PubMed] [Google Scholar]
- 23.Sen P et al. (2015) H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 29, 1362–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath S and Raj K (2018) DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet 19, 371–384 [DOI] [PubMed] [Google Scholar]
- 25.Sano S et al. (2018) Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1beta/NLRP3 inflammasome. J. Am. Coll. Cardiol 71, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuster JJ et al. (2017) Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]


