Abstract
Aims:
One-third of DCM patients experience ventricular tachycardia (VT), but a clear biological basis for this has not been established. The purpose of this study was to identify transcriptome signatures and enriched pathways in the hearts of dilated cardiomyopathy (DCM) patients with VT.
Methods and Results:
We used RNA-sequencing in explanted heart tissue from 49 samples: 19 DCM patients with VT, 16 DCM patients without VT, and 14 non-failing controls. We compared each DCM cohort to the controls and identified the genes that were differentially expressed in DCM patients with VT but not without VT. Differentially expressed genes were evaluated using pathway analysis, and pathways of interest were investigated by qRT-PCR validation, Western blot, and microscopy. There were 590 genes differentially expressed in DCM patients with VT that are not differentially expressed in patients without VT. These genes were enriched for genes in the TGFß1 and TP53 signaling pathways. Increased fibrosis and activated TP53 signaling was demonstrated in heart tissue of DCM patients with VT.
Conclusions:
Our study supports that distinct biological mechanisms distinguish ventricular arrhythmia in DCM patients.
Keywords: heart failure, RNA-seq, arrhythmia, ventricular tachycardia, dilated cardiomyopathy
Introduction
Cardiac arrhythmias frequently complicate dilated cardiomyopathy (DCM), causing physical symptoms, a need for medications and/or device therapy, and sudden cardiac death[1]. In DCM, life-threatening arrhythmias can present early in disease,[2] and approximately one-third of patients experience frequent ventricular arrhythmias, which may occur independent of left ventricular (LV) dysfunction[3]. Although arrhythmia-prone patients can be identified and stratified clinically once arrhythmias develop, there is a significant knowledge gap in understanding the biological mechanisms that contribute to ventricular arrhythmias in arrhythmogenic DCM patients.
Molecular and histological studies of human heart tissue from DCM patients with and without history of ventricular arrhythmias showed that patients with a positive history had increased hypertrophy, evidence of oxidative stress, and fibrosis. Microarray analysis showed differences in gene expression that included increased expression of select genes encoding extracellular matrix proteins (FGF18, COL4A2, COL12A1) and ion channels (KCNN2, TRPM7); a formal analysis for enrichment of differentially expressed genes in key biological pathways has not been performed and prior results were not compared against non-failing (NF) controls.[4]
We and others have previously showed that although there is a common heart failure gene expression signature for heart failure, there are also distinct gene expression signatures that distinguish DCM from ischemic cardiomyopathy that suggest distinct disease mechanisms[5–9]. Here, we hypothesized that within DCM, sub-phenotypic gene expression signatures exist that distinguish the arrhythmogenic DCM (aDCM) from the non-arrhythmogenic DCM (naDCM) state. To test this hypothesis, we performed RNA-sequencing (RNA-seq) on 49 explanted human hearts and used a multi-analytic approach to discern aDCM- and naDCM-specific expression profiles. Using pathway analysis, our results support that distinct disease mechanisms exist within DCM that separate arrhythmogenic and non-arrhythmogenic biological sub-phenotypes of DCM, including activation of TGFβ1 and TP53 signaling.
2. Methods
2.1. Tissue collection
Heart tissues were collected as described previously[9]. Briefly, the Colorado Multicenter Institutional Review Board approved the protocol for the collection, storage, and analysis of human tissue at the University of Colorado Hospital as part of the Division of Cardiology Cardiac Tissue Biobank (COMIRB, protocol 01–568). Transplant-listed patients signed written consent for use of their explanted hearts for research purposes. Explanted tissue, absent of scarring or infarcted segments, was collected according to tissue section and flash frozen in the operating room. NF donor hearts were similarly collected and frozen from hearts that were not utilized for transplantation for non-cardiac reasons. In this case, the heart was harvested from a recently deceased individual by the local organ procurement agency. Family members of NF organ donors signed written consent for research use of explanted cardiac tissue. Tissue samples were stored at −80 C until the time of analysis. In all samples described here, LV free wall tissue was utilized. Corresponding clinical information was stored in a secure, deidentified database.
2.2. Patient cohorts
General clinical etiology of DCM in patients transplanted for heart failure with reduced ejection fraction was determined by medical history. Arrhythmogenic patients had history of VT requiring ICD placement. Each patient had at least one appropriate shock within one year prior to transplant. The non-arrhythmogenic had no report of VT at any point throughout their medical history. NF organ donor hearts were defined by no major cardiac history and a LV echocardiography-based shortening fraction of ≥ 25%. Statistical differences between cohort clinical characteristics were calculated by either Mann-Whitney U Test or Fisher’s Exact Test, where appropriate, using a 0.05 significance level and a two-sided p-value.
2.3. RNA extraction
RNA was extracted from heart tissue as reported previously[9]. Briefly, approximately 20mg of tissue, macroscopically free of fat, fibrosis, and blood, was mechanically homogenized in TRIzol reagent (Thermo Fisher Scientific, Waltham, MA). Total RNA was extracted, samples were DNase treated, and RNA was quantified at 260nm. RNA integrity (RIN) was measured and all samples were required to demonstrate RIN ≥ 7.0, with a range from 7.0–9.3.
2.4. RNA sequencing
RNA sequencing was performed as reported previously[10], and a subset of the data deposited in the GEO database under GSE116250 were used in this study. Briefly, PolyA transcripts were isolated from 1μg total RNA using oligo-dT beads and cDNA libraries were constructed using the TruSeq Stranded mRNA Library Prep Kit and protocol from Illumina (Illumina Inc., San Diego, CA). Libraries were sequenced single-read with an Illumina HiSeq 2500 for 50 cycles. Reads were filtered for quality and aligned to the GRchr37/hg19 version of the reference human genome using gSNAP[11]. Expression in terms of RPKM (reads per kilobase of transcript per million reads mapped) was derived using Cufflinks[12] and Ensembl’s GRch37.82 GTF. Due to the high proportion of cardiac mRNA reads known to map to the mitochondria,[13] mitochondrial genes were removed from the GTF file for a final set of 57,974 annotations.
2.5. Statistical analysis
Statistical analyses were performed in R. Expressed genes were defined as genes with mean RPKM ≥ 5 in all groups. Differential expression was analyzed using Linear Model ANOVA. Differentially expressed genes (DEGs) were defined as genes with a difference in RPKM between groups ≥ 5 and a p-value adjusted for Benjamini-Hochberg false discovery rate (FDR) ≤ 0.05. A multiple linear regression model was used to adjust for age and sex covariates of gene expression in R. Data was transformed log2(RPKM+1). Disease status and sex were categorical variables and age was a continuous variable. The Mann-Whitney test was used in RNAscope quantification. For all other experiments, Student’s t-test (qRT-PCR) or one-way ANOVA with Tukey Method were used. Error bars represent means ± one standard deviation, and p ≤ 0.05 was considered significant. Sample sizes are indicated in figure legends. Genes were clustered using Spearman rank correlation and average linkage using the “hclust” function with the heatmap.2 package in R. Principal component analysis was calculated using the prcomp() function in R.
2.6. Pathway analysis
DEGs were interpreted using Ingenuity Pathway Analysis (IPA; Qiagen, Redwood City, CA). A dataset of Ensembl gene identifiers and fold changes was uploaded for Core Analysis. Association between upstream regulators was assessed and visualized in STRING v10[14] with the exception of the addition of the interaction between TGFβ1 and mir-193[15]. The Cardiac Conduction gene list (GO:0061337) was downloaded from the Gene Ontology AmiGO2 database v2.5.12[16–18].
2.7. qRT-PCR
cDNA was synthesized from total RNA using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). The mRNA levels of selected targets were quantified by qRT-PCR using SYBR Select Master Mix (Thermo Fisher Scientific) and normalized to GAPDH and 18s.
2.8. Second harmonic generation (SHG) and two-photon excitation fluorescent (TPEF) microscopy
Heart tissues were fixed in 10% formalin (Fisher) and embedded in paraffin. 5 μm thick sections were scanned in 20 random regions of interest. Images were acquired at 20x using a Zeiss 780 microscope (Carl Zeiss, Jena, Germany) equipped with a titanium:sapphire Chameleon Ultra II laser (Coherent, Santa Clara, CA). The average laser power of 7% at 800 nm (tuned for SHG) with 140 fs pulse duration and 80 MHz repetition rate was used. SHG signal was detected on a non-descanned detector (NDD) following transmission through a filter cube containing a narrow band 390 – 410 nm emission filter (hq400/20m-2p, Chroma Technology, Bellows Falls, VT). Quantification of collagen was done using ImageJ (NIH, http://imagej.nih.gov/ij/). The green (autofluorescence) and red (fibrillar collagens) channels were separated and a threshold was set for the collagen. The mean intensity and percent area were quantified using the threshold value.
2.9. Western blotting
Tissues were homogenized in RIPA buffer (150mM sodium chloride, 1% NP-40, 0.5% sodium dyoxycholate, 0.1% SDS, 50mM Tris pH 7.6) with a 1:100 protease inhibitor cocktail (Sigma P8340). Tissues were mechanically homogenized using a IKA T25 Ultra-Turaxx homogenizer and lysed with gentle agitation at 4°C for one hour. Samples were denatured in 1x Laemmli buffer. A total of 35mg (for anti-TP53) or 100mg (for anti-MDM2) of protein per sample was resolved by SDS-polyacrylamine gel electrophoresis (10% gel) and transferred to a PVDF membrane. Membranes were blocked for one hour with 5% milk in TBST. They were incubated overnight at 4˚C in anti-TP53 (Santa Cruz
Biotechnology sc-6243, 1:200) and anti-MDM2 (Santa Cruz Biotechnology sc-56154, 1:500) primary antibodies or with anti-GAPDH (Invitrogen AM4300, 1:8000) primary antibody at room temperature for one hour. These were followed by a one-hour incubation with secondary antibodies anti-mouse IgG peroxidase (Sigma-Aldrich A2554, 1:5000) or anti-rabbit IgG peroxidase (Sigma-Aldrich A0545, 1:5000). Protein bands were visualized using a FluorChem 8900 (Alpha Innotec). Densiometric quantification of blots was performed using ImageLab (Bio Rad Laboratories).
2.10. RNAScope
RNAScope detection was performed according the manufacturer’s protocol (Advanced Cell Diagnostics, Hayward, CA). Briefly, formalin-fixed paraffin embedded human hearts were cut into 5 μm thick tissue sections. Slides were deparaffinized in xylene, followed by rehydration in absolute ethanol washes. Following citrate buffer (Advanced Cell Diagnostics) antigen retrieval, slides were rinsed in deionized water and immediately treated with protease plus (Advanced Cell Diagnostics) at 40° C for 30 min in a HybEZ hybridization oven (Advanced Cell Diagnostics). Probes directed against POSTN and SERPINE2 mRNA and control probes were applied at 40°C in the following order: target probes, preamplifier, amplifier, and label probe for 10 minutes. After each hybridization step, slides were washed two times at room temperature. Chromogenic detection was performed followed by counterstaining with hematoxylin (American MasterTech Scientific, Lodi, CA). Staining was visualized using an Aperio CS2 slide scanner using a 40x lens (Leica Biosystems, Buffalo Grove, IL). For all samples (naDCM n = 4, aDCM n = 4), 30 fields of view were quantified.
3. Results
3.1. Clinical characteristics of patients
Forty-nine human hearts were investigated from: 19 aDCM patients, 16 naDCM patients, and 14 NF (Additional File 1). Clinical criteria for aDCM and naDCM patients are described in the Methods. Table 1 summarizes clinical characteristics between the patient groups. As expected, the aDCM cohort had a significantly greater proportion of patients taking amiodarone (p=0.04), having an implantable cardioverter defibrillator (ICD; p=0.01), and experiencing ventricular tachycardia (p<0.00001).
Table 1.
Clinical Characteristics of aDCM and naDCM cohorts
| Characteristics | naDCM (n=16) | aDCM (n=19) | p-value |
|---|---|---|---|
| Male sex, n (%) | 12 (75) | 16 (84) | 0.68 |
| Age at transplant | 47 ± 14 | 51 ± 12 | 0.29 |
| Race | |||
| Caucasian, n (%) | 13 (81) | 16 (84) | 1.00 |
| Black/African American, n (%) | 2 (13) | 1 (5) | 0.58 |
| Unknown, n (%) | 1 (6) | 2 (11) | 1.00 |
| Ethnicity | |||
| Not Hispanic or Latino, n (%) | 13 (81) | 11 (59) | 0.17 |
| Hispanic or Latino, n (%) | 1 (6) | 4 (21) | 0.35 |
| Unknown, n (%) | 2 (13) | 4 (21) | 0.67 |
| NYHA* | 3.2 ± 0.7 | 3.3 ± 0.6 | 0.79 |
| LVEF (%)* | 19 ± 9 | 17 ± 6 | 0.45 |
| Medications | |||
| Antiarrhythmic therapy, n (%) | 13 (81) | 17 (89) | 0.64 |
| Amiodarone, n (%) | 2 (13) | 9 (64) | 0.04 |
| Beta Blockers, n (%) | 10 (63) | 8 (42) | 0.22 |
| ACE inhibitor, n (%) | 8 (67) | 9 (64) | 1.00 |
| Device Therapy | |||
| ICD, n (%) | 11 (69) | 19 (100) | 0.01 |
| LVAD/BiVAD, n (%) | 8 (50) | 8 (42) | 0.74 |
| Comorbidities | |||
| History of smoking, n (%) | 8 (50) | 8 (47)* | 1.00 |
| Diabetes, n (%) | 1 (6) | 5 (26) | 0.19 |
| *BMI ≥ 25, n (%) | 6 (40) | 10 (67) | 0.5 |
| LVIDs (cm)* | 5.4 ± 1.3 | 5.8 ± 1.4 | 0.76 |
| LVIDd (cm)* | 6.1 ± 1.5 | 6.5 ± 1.4 | 0.74 |
| QRS duration (ms) | 144 ± 37 | 137 ± 49 | 0.68 |
| QT interval (corrected) | 505 ± 76 | 498 ± 57 | 0.90 |
| Arrhythmia† | |||
| Ventricular tachycardia, n (%) | 0 (0) | 19 (100) | < 0.00001 |
| Ventricular fibrillation, n (%) | 0 (0) | 4 (21) | 0.11 |
| Atrial fibrillation, n (%) | 4 (25) | 8 (42) | 0.48 |
unknown for some patients.
within 1 year of explant (aDCM) or never in medical record (naDCM).
Plus-minus values are means ± one SD. BMI=body mass index; ICD=implantable cardioverter defibrillator; LVAD/BiVAD=left/biventricular assist device; LVEF=left ventricular ejection fraction; NYHA=New York Heart Association
3.2. Principal component analysis of the cohorts
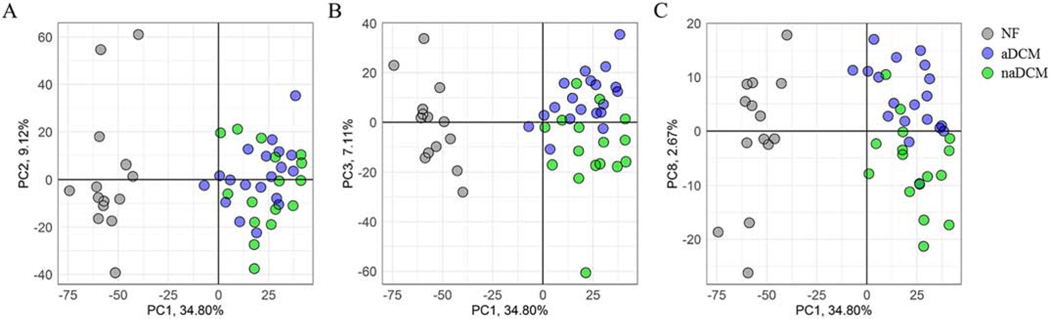
To investigate gene expression differences between arrhythmogenic and non-arrhythmogenic sub-phenotypes in DCM, we performed poly-A RNA-seq on explanted LV tissue samples (Figure 1, Additional File 2). To broadly understand gene expression relationships between cohorts and visualize global sample clustering for all expressed genes, we investigated the principal components for all 3,956 DEGs relative to NF. The first principal component explains 34.8% of the variance and separates the samples according to NF and DCM (Figure 2). The second component, which explains 9.1% of variance, does not separate the sub-phenotypes. However, the third and eighth principal components, which explain 7.1% (Figure 2a) and 2.7% (Figure 2b) respectively, cluster the DCM samples into two semi-distinct sub-phenotype groups. The data suggest that globally, differences in NF and DCM gene expression on the first component drive the most variance within the data, but that more subtle differences in gene expression separate the sub-phenotypes on the third and eighth components and in total explain nearly 10% of the variance in gene expression. All 49 principal components are included in Additional File 3. We used IPA to investigate genes with the highest factor loadings (±0.03) in PC3 (n = 209) and PC8 (n = 240) to determine if they were enriched for biological signatures. Only 8 genes are shared between these two datasets. PC3 genes were highly enriched for Oxidative Phosphorylation (p=3.16−40) and Mitochondrial Dysfunction (p=1.00−36) while PC8 genes were enriched for TGFβ1 (p=1.93−9) and TP53 (p=9.44−5).
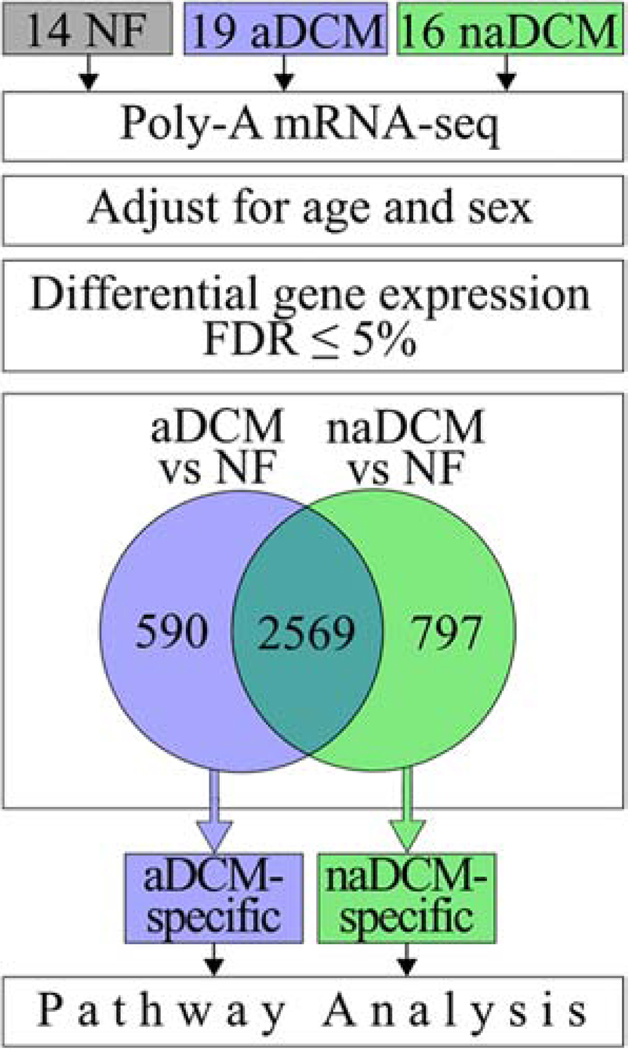
Figure 1. Schematic of RNA-seq analysis [COLOR, 1 column].
mRNA from 49 human hearts was extracted, sequenced, and adjusted for covariates. 14 NF (grey), 19 aDCM (blue) and 16 naDCM (green) explanted LVs were analyzed. DEGs at a 5% FDR were compared between each sub-phenotype and NF controls. Sub-phenotypic genes were identified and used in pathway analysis.
Figure 2. Principal component analysis of 3,956 DEGs in aDCM and naDCM compared to NF [COLOR, 2 column].
A) Principal components 1 and 2 show delineation of NF and DCM on component 1. B) Principal components 1 and 3 show delineation of aDCM and naDCM on component 3. C) Principal components 1 and 8 show delineation of aDCM and naDCM on component 8. NF samples are grey, aDCM are blue and naDCM are green. Each axis is labeled by the principal component and the proportion of variance explained by that co mponent.
3.3. Phenotype-specific expression signatures
3.3.1. Identifying phenotype-specific DEGs
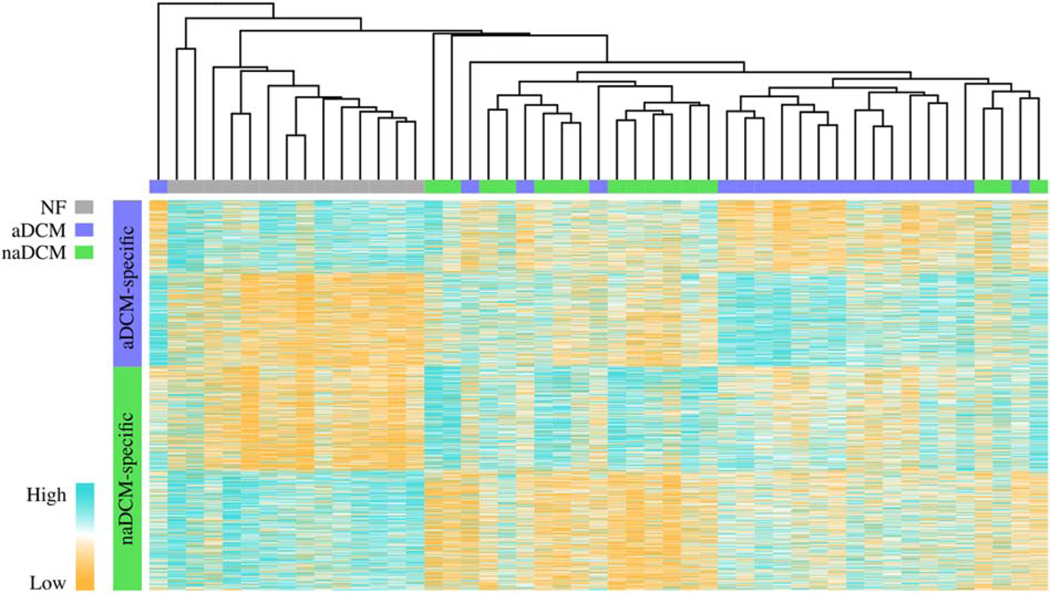
Our main hypothesis was that aDCM and naDCM patients would exhibit distinct transcriptome signatures. When aDCM and naDCM are compared directly, there are only 4 significant DEGs (5% FDR): UCHL1 and PAK1 are upregulated in aDCM, and JPH2 and NDUFS7 are upregulated in naDCM. However, our principal component analysis demonstrated that there are subtler changes in gene expression, accounting for a small but significant proportion of variance, that cluster the samples by sub-phenotype and are biologically relevant. To determine if these signatures represent a distinction between aDCM and naDCM or simply natural disease progression, we removed genes common to failing hearts, by excluding DEGs shared between each sub-phenotype and the control. These 2,569 genes were all in the same fold change direction in each sub-phenotype relative to NF. The aDCM vs NF analysis yielded 590 aDCM-specific DEGs and the naDCM vs NF comparison yielded 797 naDCM-specific DEGs (Figure 1, Additional File 4). Of the 4 DEGs identified by comparing aDCM and naDCM directly, PAK1 is aDCM-specific, JPH2 and NDUFS27 are naDCM-specific, and UCHL1 is not significant. To validate the disease specificity of these gene profiles, we performed unsupervised hierarchical clustering of the combined genes for all samples to visualize gene expression clustering. The samples segregate mostly into three large distinct clusters by NF, aDCM, and naDCM (Figure 3) where 15/19 aDCM samples group within the same large cluster. This shows that although the unshared DEGs are not statistically significant between the direct aDCM vs naDCM comparison, the trends of their gene expression across the samples are sufficient to cluster by sub-phenotypes.
Figure 3. Unsupervised hierarchical clustering of aDCM- and naDCM-specific DEGs compared to NF [COLOR, 2 column].
Unsupervised hierarchical clustering by 590 aDCM-specific and 797 naDCM-specific genes demonstrates sub-phenotype clustering. NF samples are grey, aDCM are blue and naDCM are green. Orange indicates relatively high expression while blue indicates relatively low expression.
3.3.2. Canonical pathway analysis for phenotype-specific DEGs
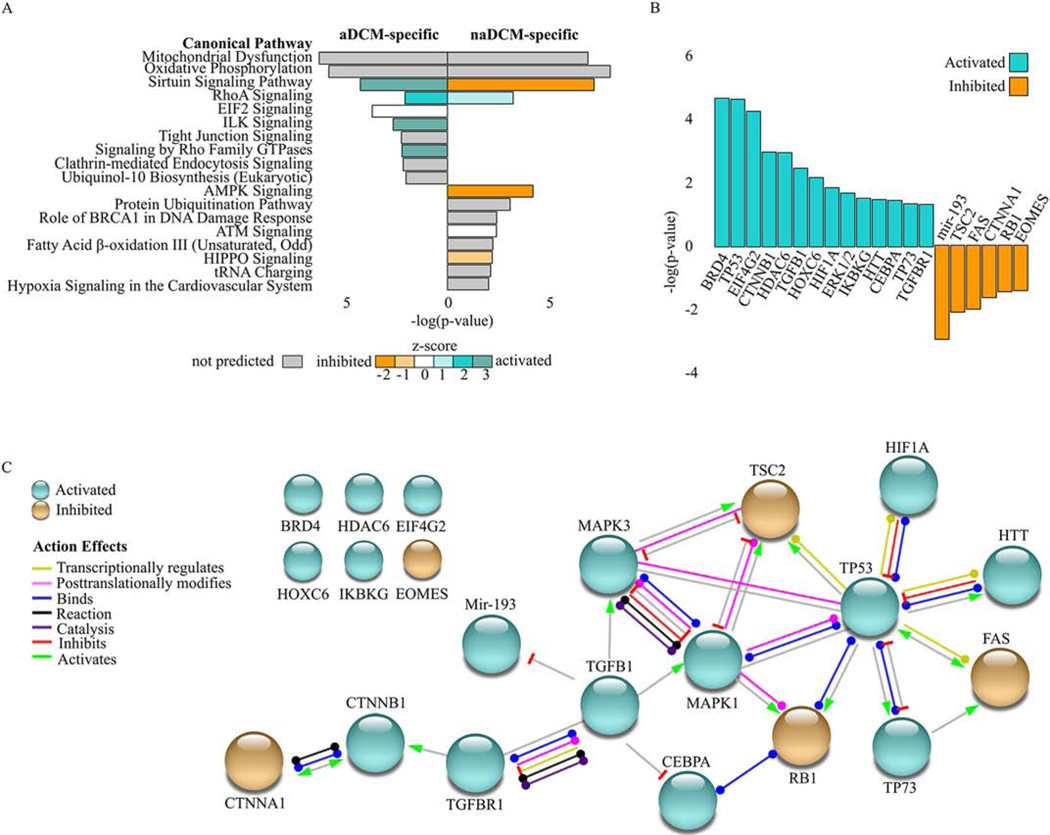
We used IPA to investigate if the 590 aDCM-specific and 797 naDCM-specific DEGs were enriched for biologically relevant pathways. At a stringency of p≤0.05, 30 pathways were predicted enriched in aDCM and 35 in naDCM (Additional File 5). The most significant pathways with p≤0.01 are displayed in Figure 4A. The three most significantly enriched pathways in each analysis are significant in both sub-phenotypes: Mitochondrial Dysfunction, Oxidative Phosphorylation, and Sirtuin Signaling. (Figure 4A). These three pathways all relate to dysregulated energy metabolism in the mitochondria, which we have shown previously is a characteristic common to distinct heart failure etiologies[9].
Figure 4. Pathway analysis in sub-phenotype-specific DEGs [COLOR, 2 column].
A) Enriched IPA pathways (p≤0.05) with aDCM-specific (left) and naDCM-specific (right) analyses. Bars are filled according to z-score: teal indicates higher (activated), orange indicates lower (inhibited). Pathways without a z-score are grey, pathways with a z-score of zero are white. B) Enriched upstream regulators in aDCM. C) Network of enriched upstream regulators adapted from STRING. Nodes (circles) represent regulators and edges (lines) represent biological actions or effects that connect each node.
3.3.3. Upstream regulator analysis for phenotype-specific DEGs
Because mitochondrial dysregulation could be a downstream effect in heart failure rather than a primary causative DCM mechanism, we considered upstream regulators that might explain the gene expression changes within the sub-phenotypes. Using IPA, we investigated whether our gene lists were enriched for biologically relevant upstream regulators. Using a cutoff of p≤0.05 and an absolute activation z-score ≥2, there are 11 significant regulators in naDCM (3 activated, 8 inhibited; Additional File 6) and 20 in aDCM (14 activated, 6 inhibited; Figure 4B). We used these significant regulators in aDCM to generate a connectivity network, where TP53 and TGFβ1 directly interact with a majority of the other regulators (Figure 4C).
3.3.3. Targeted gene analysis for aDCM-specific DEGs
Due to the previously established involvement of electrophysiological changes in aDCM, we used a targeted approach to identify genes related to cardiac electrophysiology. We compared the Cardiac Conduction gene list from Gene Ontology AmiGO2, which consists of 157 human genes, to our aDCM-specific genes and found that six genes overlapped: ATP2B4, CORIN, GJA5, KCNJ11, SLC9A1, TMEM65 (fold changes=1.28, −2.52, 1.84, −1.62, 1.44, −1.36 respectively).
3.4. Increased expression of TGFβ1 targets and collagen accumulation in aDCM
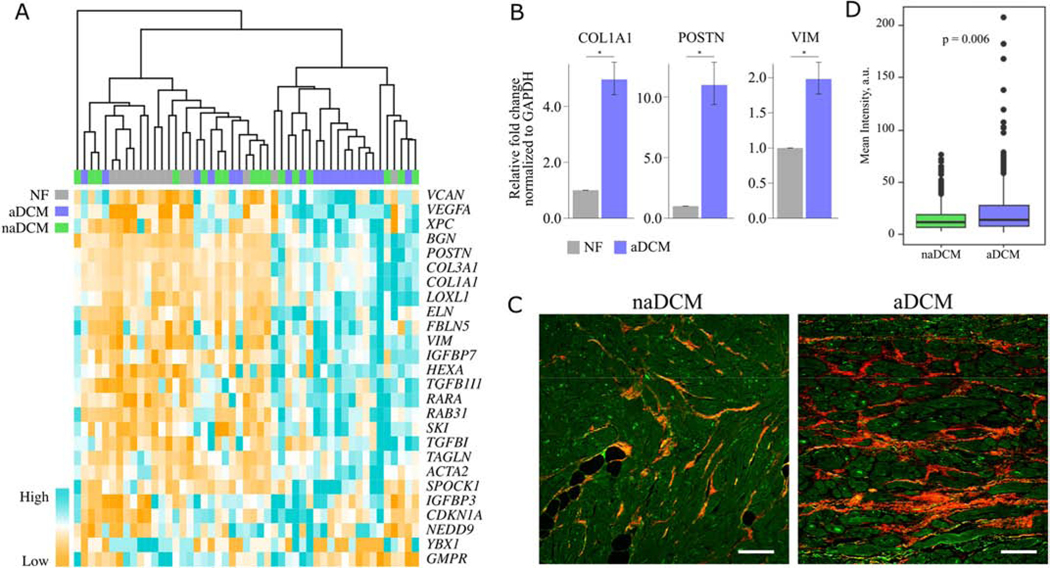
Because TGFβ1 is a known regulator of cardiac fibrosis and increased cardiac fibrosis has previously been reported in DCM patients with ventricular tachycardia (VT)[4], we sought to validate its role as a distinguishing mechanism between aDCM and naDCM. Unsupervised hierarchical clustering using to TGFβ1 target gene expression from IPA demonstrates that the samples cluster into two large groups with 13/19 aDCM samples clustering together (Figure 5A). To validate upregulation of TGFβ1 targets, we confirmed that POSTN, COL1A1, and VIM, which are all cardiac fibroblast markers[19], had increased expression in aDCM relative to NF hearts (Figure 5B). To determine whether increased cardiac fibrosis also distinguished our aDCM and naDCM cohorts, we used second harmonic generation imaging to quantitate the collagen in LV explanted heart tissue (Figure 5C, Figure 5D)[20, 21]. aDCM patients showed significantly higher levels of collagen (p=0.006; Figure 5D).
Figure 5. Increased expression of TGFβ1 targets and collagen accumulation in aDCM [COLOR, 2 column].
A) Unsupervised hierarchical clustering of TGFβ1 targets from IPA clusters most aDCM together. B) qRT-PCR validation of TGFβ1 targets COL1A1, POSTN, and VIM in mRNA from NF (n=4) and aDCM (n=3) LV. C) Representative images from second generation harmonic imaging of collagen in LV tissue from naDCM (n=10) and aDCM (n=13) demonstrating increased collagen signal. Scale bar represents 100μm, and the collagen signal is quantified in D. *p < 0.05
3.5. TP53 signaling is activated in aDCM
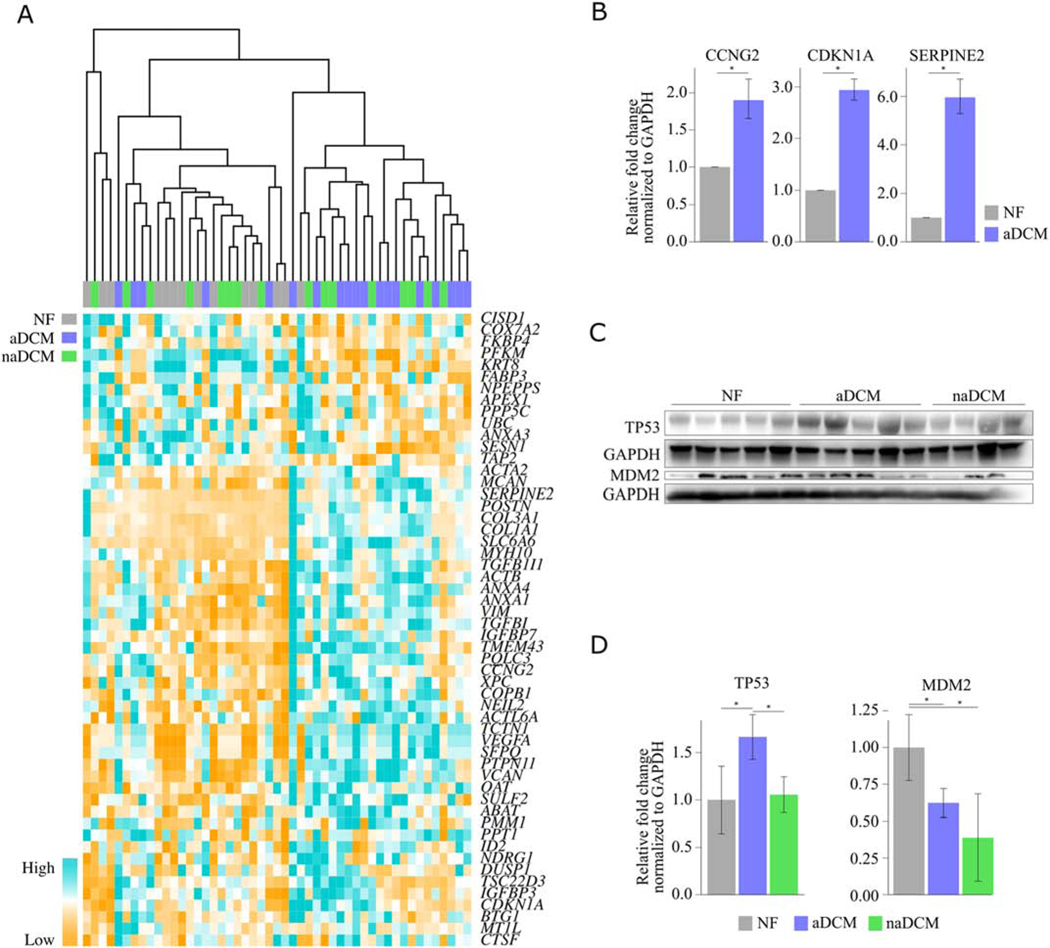
TP53 is a tumor suppressor that responds to cellular stresses by regulating gene expression to control the cell cycle, apoptosis, and DNA repair. Activated TP53 has previously been noted in DCM, specifically associated with mutations in LMNA, which can cause aDCM[22, 23] and was an enriched pathway in an RNA-seq from biopsy tissue in patients with atrial fibrillation compared to normal sinus rhythm[24]. Unsupervised hierarchical clustering of TP53 target gene expression demonstrates that the samples cluster into two large groups with 14/19 aDCM samples clustering together (Figure 6A). Interestingly, five of the six samples that do not cluster with the majority of aDCM are the same as those that also do not cluster for the TGFβ1 heatmap (Figure 5a). Clustering of all TP53 and TGFβ1 genes together demonstrated similar results with 14/19 aDCM samples clustering together (Additional File 7). We further validated these findings by confirming upregulation of targets in aDCM relative to NF: CDKN1A (p21), a cyclin-dependent kinase inhibitor and primary mediator of p53-induced cell cycle response; CCNG2, involved in cell cycle progression; XPC, involved in nucleotide excision in response to DNA damage; and SERPINE2, involved in proliferation (Figure 6B). TP53 signaling is tightly regulated in normal cells, where TP53 protein is typically present at low levels because MDM2 targets it for ubiquitination and destruction. However, stressors like DNA damage trigger rapid stabilization and activation of TP53, resulting in changes in gene expression. Stabilized TP53 induces expression of MDM2 in a feedback loop so that the pathway remains tightly regulated. Although neither TP53 nor MDM2 mRNA was differentially expressed in the RNA-seq, TP53 protein was significantly more expressed in aDCM compared to both naDCM and NF (p=0.02 and 0.007, respectively; Figure 6C, D), and MDM2 protein was significantly less expressed in aDCM compared to NF (p=0.04; Figure 6C, D) but was also significantly less expressed in naDCM compared to NF (p=0.003; Figure 6C, D). In aDCM, these results together suggest that MDM2 may be inhibited, allowing TP53 levels to remain higher than normal.
Figure 6. TP53 signaling is activated in aDCM [COLOR, 2 column].
A) Unsupervised hierarchical clustering of TP53 targets from IPA clusters most aDCM together B) qRT-PCR validation of TP53 targets CCNG2, CDKN1A, SERPEINE2, and XPC in mRNA from NF (n=4) and aDCM (n=3–5) LV. C) Western blot of TP53 and MDM2 in NF, aDCM, and naDCM LV protein extracts. Blots normalized to GAPDH and quantified in D. *p < 0.05
3.6. Increased TGFβ1 and TP53 target expression in heart tissue
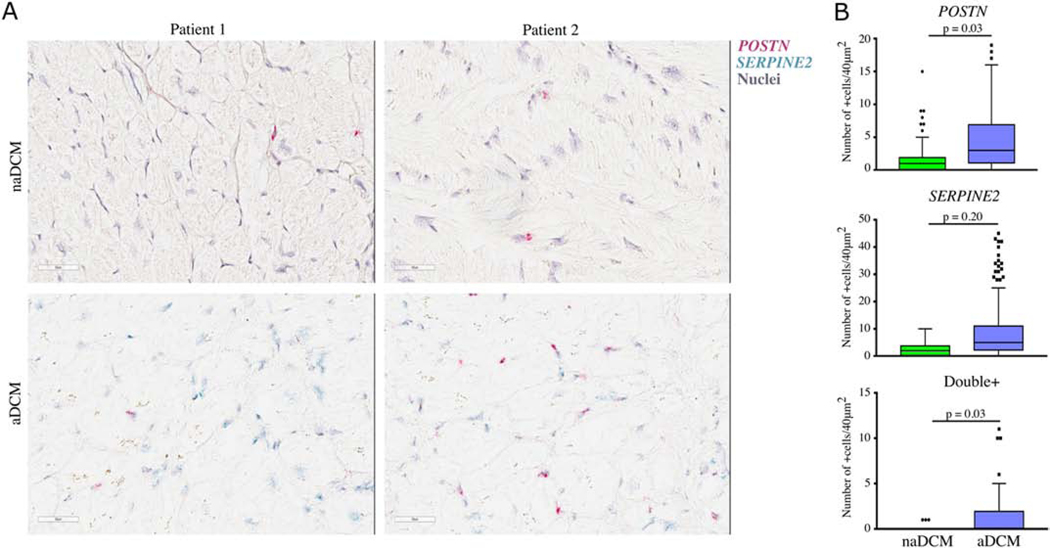
To confirm our RNA-Seq results we selected two key mRNAs, POSTN from TGFß1 and SERPINE2 from TP53 to validate using RNAscope in explanted LV tissue. We detected a significant increase in the number of cells expressing POSTN (p=0.03) and co-expressing POSTN and SERPINE2 (p=0.03), as well as an increasing trend expressing SERPINE2 (p=0.20) in aDCM patients (Figure 7).
Figure 7. Localization of TGFß1 and TP53 targets in LV tissue [COLOR, 2 column].
mRNA for POSTN (red) and SERPINE2 (blue) was A) detected and B) quantified in naDCM (n=4) and aDCM (n=4) LV heart tissue. Double+ cells contained both POSTN and SERPINE2 mRNA. Scale bar represents 50μm.
4. Discussion
4.1. aDCM and naDCM have distinct gene expression signatures
Prior studies have identified gene expression changes that occur in the failing human heart and are distinct from the healthy state, and that failing hearts share common gene expression signatures. However, differences in gene expression exist within heart failure that are able to accurately classify hearts by underlying disease etiologies, such as ischemic or non-ischemic disease[9]. Here, we showed that once the common, overlapping expression shared by all DCM patients is removed, we can identify biologically relevant signatures within DCM that reflect underlying arrhythmogenic (590 genes) and non-arrhythmogenic states (797 genes). It is interesting to note that five of the six aDCM samples that do not cluster together in the TGFß1 and TP53 gene expression heatmaps (Figures 5a and 6a) are the same in both analyses. Perhaps there are sub-groups that exist within our sub-phenotypes that have distinct mechanisms, or perhaps this reflects an intermediate or transition phase from naDCM to aDCM. These data are encouraging that transcriptome analyses can reveal biologically distinct subgroups within the overall clinical heart failure presentation and potential targets for novel therapies.
4.2. Genes involved in cardiac conduction are significantly differentially expressed in aDCM
Comparison of our gene list with genes involved in cardiac conduction revealed six DEGs. ATP2B4 encodes a calcium-transporting ATPase involved in intracellular calcium homeostasis. CORIN encodes a protein that converts pro-atrial natriuretic peptide to its active form, and serum corin levels have been associated with major adverse cardiac events including rehospitalization due to worsening heart failure and death[25]. CORIN had a −2.52 fold change in aDCM compared to NF and was additionally significantly negatively associated with age. GJA5 is also known as connexin40, a gap junction protein, that has been associated with progressive familial heart block type 1[26]. KCNJ11 encodes a potassium voltage-gated channel subunit. SLC9A1 encodes a sodium-hydrogen exchanger. TMEM65 encodes an inner mitochondrial membrane protein. Recessive mutations in TMEM65 cause mitochondrial myopathy[27]. Additionally, PAK1 is the only DEG identified in both the aDCMnaDCM direct comparison as well as the aDCM-specific genes. It is upregulated in aDCM relative to naDCM (fold change = 1.87) and NF (fold change = 1.69). PAK1 encodes p21(RAC1) activated kinase 1, which is required for ventricular calcium homeostasis and normal SERCA2a levels and function[28]. Pak1−/− mice and mice with cardiomyocyte-specific Pak1 deletions experience increased atrial and ventricular arrhythmias[28–30]. Similarly, increased PAK1 may cause dysregulated calcium flux and promote ventricular arrhythmia.
4.3. Age and sex have a significant contribution to aDCM-specific gene expression
Of the 590 aDCM-specific genes, 44 are significantly associated with age and 52 with sex (p ≤ 0.05). Within the context of TP53 signaling, PFKM, VCAN, and PTPN11, are significantly associated with age and VEGFA, SFPQ, TCTN1, KRT8, and FABP3 are significantly associated with sex. Within TGFβ1 signaling, YBX1 is significantly associated with sex and VCAN and VEGFA are also components of this signaling pathway. Even within the aDCM and naDCM sub-phenotypes, there are differences in gene expression based on age and sex. Future studies with increased sample sizes will be better equipped to statistically refine differences in how age and sex may contribute to differing mechanisms within these pathways.
4.4. Upstream regulators may play a role in gene expression changes that distinguish aDCM from naDCM mechanisms
Several enriched upstream regulators were predicted as activated or inhibited in aDCM relative to naDCM (Figure 4B). Mir-193 levels have been negatively correlated with fibrosis, and TGFβ stimulates downregulation of mir-193[15, 31]. BRD4 is an epigenetic regulator that binds acetylated chromatin to promote transcription. In cardiomyocytes, BRD4 promotes expression of genes associated with pathological hypertrophy and TGFβ-mediated fibrosis in response to stress[32–34]. CTNNB1 encodes β-catenin, a protein involved in signal transduction. It duel-functions at the intercalated disc in cell-cell adhesion and in the nucleus as an effector of gene transcription in Wnt signaling. Although inhibition of Wnt/β-catenin has been demonstrated in arrhythmogenic right ventricular cardiomyopathy (ARVC),[35] activation has been associated with cardiac fibrosis[36, 37]. HDAC6 is a histone deacetylase that controls acetylation of α-tubulins, which are structural cytoskeletal polymers. Increased HDAC6 activity models causes α-tubulin deacetylation, depolymerization, and degradation, which were confirmed in human tissue from patients with atrial fibrillation. Further, in vivo HDAC6 inhibition prevents electrical remodeling and contractile dysfunction[38]. MAPK3 and MAPK1 are also known as ERK1/2 and are important regulators of cell cycle progression, proliferation, and survival. Activation of ERK1/2 has previously been reported in LMNA-related cardiomyopathy models,[39, 40] and pharmacological inhibition improves cardiac function[41–43]. Activation of ERK1/2 also crosstalks with RB1, which is the tumor suppressor retinoblastoma 1 predicted to be inhibited. Interestingly, A-type lamins and RB1 directly interact and this is required for proper localization and stability of RB1[44, 45]. This association can be disrupted by ERK1/2, which can inactivate RB1 and induce cell cycle entry and proliferation[46].
4.5. TGFβ1 signaling is activated in aDCM
TGFβ1 is a master regulator of fibrosis in many tissues[47, 48] through induction of myofibroblast differentiation[49]. In the heart, TGFβ1 can alter the electrophysiology of cardiac myofibroblasts by promoting pro-arrhythmic intercellular interactions between myofibroblasts and cardiomyocytes[50]. A transgenic mouse model of TGFβ1 overexpression results in cardiac hypertrophy and interstitial fibrosis,[51] and a heterozygous knockout protects against age-associated cardiac fibrosis[52]. TGFβ1 was predicted as activated in our aDCM cohort, and we showed that aDCM patients have higher levels of collagen on average than naDCM (Figure 5C, Figure 5D) and that aDCM patients trend towards higher levels of POSTN mRNA (Figure 7). Additionally, TGFβ1 is known to interact with a number of the other predicted upstream regulators, including activating the non-canonical ERK1/2 (MAPK3 and MAPK1) map kinase pathway (Figure 4C)[47, 53, 54]. ERK1/2 activation has previously been reported in LMNA-related cardiomyopathy[39, 40]. Whether or not the fibrotic signaling we detected precedes fibrotic changes or is merely a manifestation of existent fibrosis remains to be determined.
4.6. TP53 signaling is activated in aDCM
TP53 is a transcriptional regulator and a prominent tumor suppressor. Increased protein expression of TP53, decreased protein expression of its negative regulator MDM2, enriched downstream expression of TP53 target genes, and specific increased LV expression of SERPINE2 in aDCM patients suggest that TP53 is activated in aDCM. Its role in the heart, in either a healthy state or a pathogenic one, has not been well characterized. In endothelial cells and macrophages, activation of the sympathetic nervous system via β2-adrenergic signaling increased expression of p53, resulting in cardiac inflammation[55]. P53 accumulation has been described in failing human hearts from DCM patients, but clinical characteristics including arrhythmia were not described[56]. A transgenic mouse model of an ACTC1 missense mutation demonstrated accumulation of TP53 and irregular calcium handling. Arrhythmia was suggested as a potential cause of death.[57] In accordance with our findings, in a zebrafish LMNA knockdown model, TP53 and CDKN1A are both upregulated, supporting TP53 activation.[58] Recently, cardiomyocyte-specific conditional knockout of TP53 in mice led to hypertrophy and decreased contractile function[59]. This was accompanied by repression of extracellular matrix and fibrosisrelated genes (including TGFβ1, TGFβ2, TGFβ3, POSTN, and numerous collagens, including COL1A1) and increased expression of genes related to mitochondrial biogenesis and oxidative phosphorylation[59]. Similarly, our results showed that activation of TP53 is accompanied by corresponding changes in TGFβ1, fibrosis, and mitochondrial dysfunction. The results of this study are summarized in Additional File 8.
4.7. Limitations
The limitations concerning RNA-seq studies in end-stage, explanted human heart tissue have been described in detail and also apply to the current study[9]. In particular, it is important to note that adult heart tissue is comprised of many different cell types, including an estimated 20–40% cardiomyocytes[60, 61], with 65% endothelial cells, 15% fibroblasts, and 10% leukocytes comprising the noncardiomyocytes[62]. The RNA-seq is this study represents the transcriptome of whole tissue, but each cell type likely has a distinct profile. We demonstrated significant increased co-localization of two important signaling molecules in the work, but further investigation of cell-specific effects is a logical next. Additionally, this work demonstrates distinct RNA-seq profiles between NF and DCM and within DCM, but further expression validation and will be necessary to establish aDCM and naDCM as distinct biological sub-phenotypes. There are also several clinical limitations. Significantly more aDCM patients were receiving amiodarone than naDCM (Table 1; n=9 and n=2, p=0.04) and had ICDs (n=19 and n=11, p=0.01), which could potentially impact gene expression between groups. Amiodarone has been reported to decrease expression of Na+/K+ ATPase subunits α2 and β2 (ATP1A2 and ATP1B2)[63], sodium channels (SCN4A, SCN5A, SCN1B), connexin43 (GJA1), the calcium channel CACNA1C, various potassium channels (KCNA5, KCNB1, KCND2) and increase expression of other potassium channels (KCNA4, KCNK1, KCNAB1, KCNE3,[64] KCNJ2[65]). Our aDCM-specific DEGs do not include any of these genes. Moreover, in our unsupervised hierarchical clustering analyses with both TGFβ1 and TP53 target genes, we do not see any distinct clustering differences between patients with ICDs or taking amiodarone. Additionally, a study of the effect of amiodarone on non-CMs demonstrated that amiodarone treatment does not affect expression of TP53[66], and long-term amiodarone therapy does not affect LV remodeling or interstitial fibrosis[67]. Lastly, our clinical cohorts were selected based on patients that had never experienced VT or that had experienced VT within a year of transplant, but does not negate naDCM patients who may have been at risk of developing VT.
4.6. Conclusions
We used RNA-seq, pathway analysis, histology, and molecular studies to demonstrate that clinical distinctions in DCM arrhythmogenic sub-phenotypes result in biologically relevant differences. In our study, aDCM and naDCM were revealed to have distinct gene expression signatures, including significant differences in the TGFβ1 and TP53 pathways, which were validated by dysregulated protein expression and histological evidence of differing degrees of fibrosis. Notably, explanted LV tissue demonstrates significant increased expression and colocalization of TGFβ1 and TP53 targets, suggesting a direct link between these signaling pathways and ventricular arrhythmia. The underlying biology of end-stage heart failure differs between DCM patients with and without an arrhythmogenic phenotype, supporting distinct heart failure disease mechanisms.
Supplementary Material
Highlights.
About 1:3 of dilated cardiomyopathy patients experience ventricular tachycardia
Our understanding of the biological basis of these arrhythmias is incomplete
RNA-seq demonstrates discrete arrhythmia and non-arrhythmia expression profiles
These expression signatures are enriched for distinct biological pathways
TP53 and TGFβ1 pathways distinguish arrhythmia and non-arrhythmia cohorts
6. Acknowledgements
The authors would like to thank Dr. Peter Buttrick and the University of Colorado’s Division of Cardiology for ongoing maintenance of the human cardiac tissue biobank as well as the patients and organ donors who provided the heart tissues used in this study.
5. Funding
This work was supported by the National Institutes of Health [UL1 TR002535, R01 HL69071, R01 116906 (LM), R01 HL109209 (MRGT)] and the Fondation Leducq [14-CVD 03]. Contents are the authors’ sole responsibility and do not necessarily represent official NIH views.
Abbreviations
- DCM
dilated cardiomyopathy
- LV
left ventricular
- RNA-seq
RNA-sequencing
- NF
non-failing
- aDCM
arrhythmogenic DCM
- naDCM
non-arrhythmogenic DCM
- ICD
Implantable cardioverter-defibrillator
- DEG
differentially expressed genes
Footnotes
Disclosures
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
8. References
- 1.Stecker EC, et al. , Public health burden of sudden cardiac death in the United States. Circ Arrhythm Electrophysiol, 2014. 7(2): p. 212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Losurdo P, Stolfo D, Merlo M, Barbati G, Gobbo M, Gigli M, Ramani F, Pinamonti B, Zecchin M, Finocchiaro G, Mestroni L, Sinagra G, Early arrhythmic events in idiopathic dilated cardiomyopathy. JACC Clin EP, 2016. 2(5): p. 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spezzacatene A, et al. , Arrhythmogenic Phenotype in Dilated Cardiomyopathy: Natural History and Predictors of Life-Threatening Arrhythmias. J Am Heart Assoc, 2015. 4(10): p. e002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parajuli N, et al. , Determinants of ventricular arrhythmias in human explanted hearts with dilated cardiomyopathy. Eur J Clin Invest, 2015. 45(12): p. 1286–96. [DOI] [PubMed] [Google Scholar]
- 5.Kittleson MM, et al. , Gene expression analysis of ischemic and nonischemic cardiomyopathy: shared and distinct genes in the development of heart failure. Physiol Genomics, 2005. 21(3): p. 299–307. [DOI] [PubMed] [Google Scholar]
- 6.Kuner R, et al. , Genomic analysis reveals poor separation of human cardiomyopathies of ischemic and nonischemic etiologies. Physiol Genomics, 2008. 34(1): p. 88–94. [DOI] [PubMed] [Google Scholar]
- 7.Blaxall BC, et al. , Differential gene expression and genomic patient stratification following left ventricular assist device support. J Am Coll Cardiol, 2003. 41(7): p. 1096–106. [DOI] [PubMed] [Google Scholar]
- 8.Kittleson MM, et al. , Identification of a gene expression profile that differentiates between ischemic and nonischemic cardiomyopathy. Circulation, 2004. 110(22): p. 3444–51. [DOI] [PubMed] [Google Scholar]
- 9.Sweet ME, et al. , Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure. BMC Genomics, 2018. 19(1): p. 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Begay RL, et al. , Filamin C Truncation Mutations Are Associated With Arrhythmogenic Dilated Cardiomyopathy and Changes in the Cell–Cell Adhesion Structures. JACC Clin EP, 2018. 4(4): p. 504–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu TD and Nacu S, Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics, 2010. 26(7): p. 873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trapnell C, et al. , Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol, 2010. 28(5): p. 511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang KC, et al. , Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation, 2014. 129(9): p. 1009–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szklarczyk D, et al. , STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res, 2015. 43(Database issue): p. D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy S, et al. , miR-30c and miR-193 are a part of the TGF-beta-dependent regulatory network controlling extracellular matrix genes in liver fibrosis. J Dig Dis, 2015. 16(9): p. 513–24. [DOI] [PubMed] [Google Scholar]
- 16.Carbon S, et al. , AmiGO: online access to ontology and annotation data. Bioinformatics, 2009. 25(2): p. 288–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mi H, et al. , PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res, 2019. 47(D1): p. D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Gene Ontology C, The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res, 2019. 47(D1): p. D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doppler SA, et al. , Cardiac fibroblasts: more than mechanical support. J Thorac Dis, 2017. 9(Suppl 1): p. S36–S51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman JE, et al. , Pyrroloquinoline quinone prevents developmental programming of microbial dysbiosis and macrophage polarization to attenuate liver fibrosis in offspring of obese mice. Hepatol Commun, 2018. 2(3): p. 313–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranjit S, et al. , Label-free fluorescence lifetime and second harmonic generation imaging microscopy improves quantification of experimental renal fibrosis. Kidney Int, 2016. 90(5): p. 1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moiseeva O, et al. , Mutant lamin A links prophase to a p53 independent senescence program. Cell Cycle, 2015. 14(15): p. 2408–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen SN, et al. , DNA Damage Response/TP53 Pathway Is Activated and Contributes to the Pathogenesis of Dilated Cardiomyopathy Associated With LMNA (Lamin A/C) Mutations. Circ Res, 2019. 124(6): p. 856–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang DY, et al. , Identification of microRNA-mRNA dysregulations in paroxysmal atrial fibrillation. Int J Cardiol, 2015. 184: p. 190–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, et al. , Plasma Corin as a Predictor of Cardiovascular Events in Patients With Chronic Heart Failure. JACC Heart Fail, 2016. 4(8): p. 664–9. [DOI] [PubMed] [Google Scholar]
- 26.Makita N, et al. , A connexin40 mutation associated with a malignant variant of progressive familial heart block type I. Circ Arrhythm Electrophysiol, 2012. 5(1): p. 163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nazli A, et al. , A mutation in the TMEM65 gene results in mitochondrial myopathy with severe neurological manifestations. Eur J Hum Genet, 2017. 25(6): p. 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, et al. , Pak1 is required to maintain ventricular Ca(2)(+) homeostasis and electrophysiological stability through SERCA2a regulation in mice. Circ Arrhythm Electrophysiol, 2014. 7(5): p. 938–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeSantiago J, et al. , Loss of p21-activated kinase 1 (Pak1) promotes atrial arrhythmic activity. Heart Rhythm, 2018. 15(8): p. 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeSantiago J, et al. , p21-Activated kinase1 (Pak1) is a negative regulator of NADPH-oxidase 2 in ventricular myocytes. J Mol Cell Cardiol, 2014. 67: p. 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang L, et al. , Circulating microRNAs as biomarkers for diffuse myocardial fibrosis in patients with hypertrophic cardiomyopathy. J Transl Med, 2015. 13: p. 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiltoir JI, et al. , BET acetyl-lysine binding proteins control pathological cardiac hypertrophy. J Mol Cell Cardiol, 2013. 63: p. 175–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stratton MS, et al. , Signal-Dependent Recruitment of BRD4 to Cardiomyocyte Super-Enhancers Is Suppressed by a MicroRNA. Cell Rep, 2016. 16(5): p. 1366–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stratton MS, Haldar SM, and McKinsey TA, BRD4 inhibition for the treatment of pathological organ fibrosis. F1000Res, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Gras E, et al. , Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest, 2006. 116(7): p. 2012–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizutani M, Wu JC, and Nusse R, Fibrosis of the Neonatal Mouse Heart After Cryoinjury Is Accompanied by Wnt Signaling Activation and Epicardial-to-Mesenchymal Transition. J Am Heart Assoc, 2016. 5(3): p. e002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang FL, Fang M, and Yutzey KE, Loss of beta-catenin in resident cardiac fibroblasts attenuates fibrosis induced by pressure overload in mice. Nat Commun, 2017. 8(1): p. 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D, et al. , Activation of histone deacetylase-6 induces contractile dysfunction through derailment of alpha-tubulin proteostasis in experimental and human atrial fibrillation. Circulation, 2014. 129(3): p. 346–58. [DOI] [PubMed] [Google Scholar]
- 39.Muchir A, et al. , Activation of MAPK pathways links LMNA mutations to cardiomyopathy in EmeryDreifuss muscular dystrophy. J Clin Invest, 2007. 117(5): p. 1282–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu W, et al. , Mitogen-activated protein kinase inhibitors improve heart function and prevent fibrosis in cardiomyopathy caused by mutation in lamin A/C gene. Circulation, 2011. 123(1): p. 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi JC, et al. , Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci Transl Med, 2012. 4(144): p. 144ra102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muchir A, et al. , Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum Mol Genet, 2009. 18(2): p. 241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muchir A, et al. , Treatment with selumetinib preserves cardiac function and improves survival in cardiomyopathy caused by mutation in the lamin A/C gene. Cardiovasc Res, 2012. 93(2): p. 311–9. [DOI] [PubMed] [Google Scholar]
- 44.Markiewicz E, et al. , Lamin A/C binding protein LAP2alpha is required for nuclear anchorage of retinoblastoma protein. Mol Biol Cell, 2002. 13(12): p. 4401–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson BR, et al. , A-type lamins regulate retinoblastoma protein function by promoting subnuclear localization and preventing proteasomal degradation. Proc Natl Acad Sci U S A, 2004. 101(26): p. 967–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez J, et al. , ERK1/2 MAP kinases promote cell cycle entry by rapid, kinase-independent disruption of retinoblastoma-lamin A complexes. J Cell Biol, 2010. 191(5): p. 967–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng XM, Nikolic-Paterson DJ, and Lan HY, TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol, 2016. 12(6): p. 325–38. [DOI] [PubMed] [Google Scholar]
- 48.Border WA and Noble NA, Transforming growth factor beta in tissue fibrosis. N Engl J Med, 1994. 331(19): p. 1286–92. [DOI] [PubMed] [Google Scholar]
- 49.Desmouliere A, et al. , Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol, 1993. 122(1): p. 103–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salvarani N, et al. , TGF-beta1 (Transforming Growth Factor-beta1) Plays a Pivotal Role in Cardiac Myofibroblast Arrhythmogenicity. Circ Arrhythm Electrophysiol, 2017. 10(5): p. e004567. [DOI] [PubMed] [Google Scholar]
- 51.Rosenkranz S, et al. , Alterations of beta-adrenergic signaling and cardiac hypertrophy in transgenic mice overexpressing TGF-beta(1). Am J Physiol Heart Circ Physiol, 2002. 283(3): p. H1253–62. [DOI] [PubMed] [Google Scholar]
- 52.Brooks WW and Conrad CH, Myocardial fibrosis in transforming growth factor beta(1)heterozygous mice. J Mol Cell Cardiol, 2000. 32(2): p. 187–95. [DOI] [PubMed] [Google Scholar]
- 53.Hyman KM, et al. , Transforming growth factor-beta1 induces apoptosis in vascular endothelial cells by activation of mitogen-activated protein kinase. Surgery, 2002. 132(2): p. 173–9. [DOI] [PubMed] [Google Scholar]
- 54.Yu L, Hebert MC, and Zhang YE, TGF-beta receptor-activated p38 MAP kinase mediates Smad-independent TGF-beta responses. EMBO J, 2002. 21(14): p. 3749–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida Y, et al. , p53-Induced inflammation exacerbates cardiac dysfunction during pressure overload. J Mol Cell Cardiol, 2015. 85: p. 183–98. [DOI] [PubMed] [Google Scholar]
- 56.Song H, et al. , Increased p53 protein expression in human failing myocardium. J Heart Lung Transplant, 1999. 18(8): p. 744–9. [DOI] [PubMed] [Google Scholar]
- 57.Toko H, et al. , Ca2+/calmodulin-dependent kinase IIdelta causes heart failure by accumulation of p53 in dilated cardiomyopathy. Circulation, 2010. 122(9): p. 891–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koshimizu E, et al. , Embryonic senescence and laminopathies in a progeroid zebrafish model. PLoS One, 2011. 6(3): p. e17688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mak TW, et al. , p53 regulates the cardiac transcriptome. Proc Natl Acad Sci U S A, 2017. 114(9): p. 2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou P and Pu WT, Recounting Cardiac Cellular Composition. Circ Res, 2016. 118(3): p. 368–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bergmann O, et al. , Dynamics of Cell Generation and Turnover in the Human Heart. Cell, 2015. 161(7): p. 1566–75. [DOI] [PubMed] [Google Scholar]
- 62.Pinto AR, et al. , Revisiting Cardiac Cellular Composition. Circ Res, 2016. 118(3): p. 400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hensley CB, et al. , Amiodarone decreases Na,K-ATPase alpha 2 and beta 2 expression specifically in cardiac ventricle. J Mol Cell Cardiol, 1994. 26(4): p. 417–24. [DOI] [PubMed] [Google Scholar]
- 64.Le Bouter S, et al. , Long-term amiodarone administration remodels expression of ion channel transcripts in the mouse heart. Circulation, 2004. 110(19): p. 3028–35. [DOI] [PubMed] [Google Scholar]
- 65.Ji Y, et al. , Class III antiarrhythmic drugs amiodarone and dronedarone impair KIR 2.1 backward trafficking. J Cell Mol Med, 2017. 21(10): p. 2514–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di Matola T, et al. , Amiodarone induces cytochrome c release and apoptosis through an iodine-independent mechanism. J Clin Endocrinol Metab, 2000. 85(11): p. 4323–30. [DOI] [PubMed] [Google Scholar]
- 67.Zagorianou A, et al. , The effect of long-term amiodarone administration on myocardial fibrosis and evolution of left ventricular remodeling in a porcine model of ischemic cardiomyopathy. Springerplus, 2016. 5(1): p. 1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.