Abstract
Objective
To evaluate a reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and compare it with RT-PCR.
Methods
We designed primers specific to the orf1ab and S genes of SARS-CoV-2. Total viral RNA was extracted using the QIAamp Viral RNA Mini Kit. We optimized the RT-LAMP assay, and evaluated it for its sensitivity and specificity of detection using real-time turbidity monitoring and visual observation.
Results
The primer sets orf1ab-4 and S-123 amplified the genes in the shortest times, the mean (±SD) times were 18 ± 1.32 min and 20 ± 1.80 min, respectively, and 63°C was the optimum reaction temperature. The sensitivities were 2 × 101 copies and 2 × 102 copies per reaction with primer sets orf1ab-4 and S-123, respectively. This assay showed no cross-reactivity with 60 other respiratory pathogens. To describe the availability of this method in clinical diagnosis, we collected 130 specimens from patients with clinically suspected SARS-CoV-2 infection. Among them, 58 were confirmed to be positive and 72 were negative by RT-LAMP. The sensitivity was 100% (95% CI 92.3%–100%), specificity 100% (95% CI 93.7%–100%). This assay detected SARS-CoV-2 in a mean (±SD) time of 26.28 ± 4.48 min and the results can be identified with visual observation.
Conclusion
These results demonstrate that we developed a rapid, simple, specific and sensitive RT-LAMP assay for SARS-CoV-2 detection among clinical samples. It will be a powerful tool for SARS-CoV-2 identification, and for monitoring suspected patients, close contacts and high-risk groups.
Keywords: SARS-CoV-2, RT-LAMP, COVID-19, Detection, Visual
Introduction
The outbreak of a cluster of respiratory infections designated as coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), had a significant impact on both the health and economy of China [[1], [2], [3], [4]]. To date (1 April 2020), up to 754 948 SARS-CoV-2 cases have been confirmed, including more than 36 571 deaths (https://www.who.int/home). Human-to-human transmission was confirmed through a case of five patients in a family cluster [5]. The virus transmitted rapidly via aerial droplets, contact and fomites [6].
SARS-CoV-2 has been classified as a β-coronavirus of group 2B with higher similarity, over 96% identical at the whole-genome level, to two bat-derived coronavirus strains than to other known human coronaviruses [7,8]. The genome of the novel virus comprises a 5ʹ untranslated region, replicase complex (orf1ab), Spike gene (S gene), E gene, M gene, N gene, 3ʹ untranslated region, and several unidentified non-structural open reading frames [9].
Since the outbreak of SARS-CoV-2 infection, real-time RT-PCR assays have played an important role in clinical diagnosis and the investigation of suspected cases. However, such methods are laborious, time-consuming and require specialized instruments, and are therefore not able to satisfy the current rapid growth and demands of testing the large number of suspected patients, asymptomatic patients and close contacts. A rapid, simple, and sensitive assay is urgently needed [10].
Loop-mediated isothermal amplification (LAMP) was developed by Notomi et al. in 2000 [11]. It is a rapid, sensitive and effective visual nucleic acid amplification method. Recently, this method has been widely applied for the detection of influenza virus, Middle East respiratory syndrome-CoV, West Nile virus, Ebola virus, Zika virus, yellow fever virus and a variety of other pathogens [[12], [13], [14], [15], [16], [17]]. The test relies on auto-cycling strand displacement DNA synthesis using four to six specific primers to recognize six to eight sequences of the target gene in only 1 hour.
In this study, we developed a reverse transcription LAMP (RT-LAMP) assay to detect SARS-CoV-2 in individuals with COVID-19. We designed five sets of primers that target the orf1ab gene and the spike gene for optimization of the assay. We also evaluated the sensitivity and specificity of this method. Finally, 130 clinical samples collected from patients were analysed for clinical diagnosis.
Materials and methods
Viruses and bacterial strains
A total of 60 respiratory pathogens including human coronavirus HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1, SARS-CoV (pseudo-virus) and Middle East respiratory syndrome-CoV (pseudo-virus); influenza A-H3N2, H1N1, influenza B, parainfluenza viruses types 1/2/3/4, adenoviruses types 1/2/3/4/5/6/7, respiratory syncytial virus types A/B, human metapneumovirus, human bocavirus, rhinovirus types A/B/C, as well as respiratory pathogens such as Mycoplasma pneumoniae strains M129/FH, Haemophilus influenzae ATCC 49247, Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae and Pseudomonas aeruginosa were used in this study.
RNA extraction
Total viral RNA was extracted from the specimens using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The extracted RNAs were stored at –80°C.
Preparation of the artificial SARS-CoV-2 gene
The full-length 3822-bp spike gene (21 563–25 384) and a 500-bp sequence (13 791–14 290) of the orf1ab gene of SARS-CoV-2 (GenBank Accession No. MN908947.3) were synthesized (Sangon Biotec Co., Ltd., Shanghai, China) and cloned into vector pUC57. Pseudo-viruses of 2019-nCov-S and 2019-nCov-ORF1ab (Xiamen Zeesan Biotech Co., Ltd, Xiamen, China) were also used. The copy number of the pseudo-viruses was calculated using the following formula: copies/μL = 6.02 × 1023 × 10−9 × concentration (ng/μL)/(fragment length × 340). Then, ten-fold serial dilutions of the pseudo-viruses ranging from 1 × 108 copies/μL to 1 copy/μL were prepared.
Primer design
We aligned 103 complete genomes of SARS-CoV-2 obtained from four databases, GenBank, GISAID, GWH and NMDC (https://bigd.big.ac.cn/ncov), to confirm the conservation of the selected sequence. Homology analysis was conducted using BLAST (National Centre for Biotechnology Information). Five sets of primers specific to the orf1ab and S genes were designed using the software PrimerExplorer V5 at the website: http://primerexplorer.jp/e/. The primers included an outer forward primer (F3), an outer backward primer (B3), a forward inner primer (FIP) and a backward inner primer (BIP). A loop forward primer (LF) and/or a loop backward primer (LB) were also designed to accelerate the reaction. We also designed primers to amplify the two regions by conventional PCR assay (orf1ab-F: 5ʹ-CAGACCTCGTCTATGCTTTAAGGC-3ʹ; orf1ab-R: 5ʹ-CCCTGGTCAAGGTTAATATAGGCA-3ʹ; S–F: 5ʹ-CTTCCCTCAGTCAGCACCTC-3ʹ; S-R: 5ʹ-AACCAGTGTGTGCCATTTGA-3ʹ). The primers were synthesized by Shanghai Invitrogen Biological Engineering, Co., Ltd (Shanghai, China) and purified by high-performance liquid chromatography.
RT-LAMP assay
A Loopamp RNA amplification kit (Eiken Chemical Co., Ltd., Tokyo, Japan) was used to perform the RT-LAMP reaction. The reaction volume of 25 μL contained 2 × Reaction Mix 12.5 μL, Enzyme Mix (a mix of Bst DNA polymerase and AMV reverse transcriptase) 1.0 μL, primers FIP and BIP (40 pmol), primers F3 and B3 (5 pmol), primers LF and/or LB (20 pmol), fluorescent detection reagent 1 μL (when needed), and template RNA 2 μL. The mixture was incubated at 63°C for 60 min. The process was monitored using a Loopamp Real-time Turbidimeter (LA-230; Eiken Chemical Co., Ltd., Tochigi, Japan). Turbidity readings at an optical density at 650 nm were obtained every 6 seconds, and the reaction was considered positive when the turbidity values were >0.1. For visual observation, 1 μL of fluorescent calcein was added to the mixture, and a colour change from orange to green was observed by the naked eye for a positive reaction.
Real-time RT-PCR assay
A commercial real-time RT-PCR kit (BGI PathoGenesis Pharmaceutical Technology, Shenzhen, China) was used as the reference standard for the RT-LAMP. The total reaction volume was 30 μL and the reaction was set up according to the manufacturer's protocol. The reaction procedure was 50°C for 20 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. A threshold cycle (Ct value) < 38 was determined to indicate a positive sample.
Clinical specimens
One hundred and thirty clinical specimens were collected from individuals with pneumonia and suspected SARS-CoV-2 infection, including swabs and bronchoalveolar lavage fluid. Initial processing of all specimens was handled in a validated biological safety cabinet, and performed by trained staff wearing appropriate personal protective equipment. The extracted RNA was divided into two parts for amplification. The RT-LAMP assays and real-time RT-PCR were performed simultaneously in a biosafety level 2 laboratory, as detailed in the WHO Laboratory biosafety manual, third edition. The amplified products of the RT-LAMP were detected by turbidity monitoring as well as visual observation. To assess turbidity, the time of the positivity detection was counted. Mean and standard deviation were used to describe the data and 95% CI were presented where relevant. This work was approved by the research board of the Ethics Committee of the Capital Institute of Paediatrics, Beijing, China. All patients provided informed consent in accordance with the Declaration of Helsinki.
Results
Optimizing the RT-LAMP assay
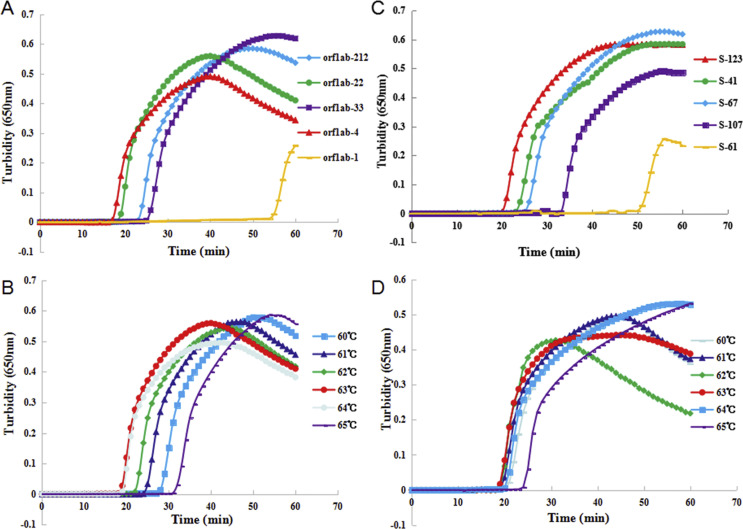
The conserved regions of the orf1ab gene and S gene of SARS-CoV-2 with low mutation frequencies were candidate target sequences (see Supplementary material, Fig. S1). The pseudo-viruses were used as the template (concentration 1 ng/μL) to optimize the RT-LAMP reaction conditions. Five sets of primers were designed to respectively detect the orf1ab gene (orf1ab-212, orf1ab-4, orf1ab-33, orf1ab-22 and orf1ab-1) and the spike gene (S-107, S-123, S-67, S-41 and S-61) of SARS-CoV-2 using a real-time LAMP turbidimeter. As shown in Fig. 1 , the primer sets orf1ab-4 and S-123 amplified the genes in the shortest times, the mean (±SD) times were 18 ± 1.32 min and 20 ± 1.80 min, respectively, so these two primer sets were chosen as the optimal primers for RT-LAMP detection. All of the primer sets used in this study are listed in the Supplementary material (Table S1).
Fig. 1.
The most appropriate primers and reaction temperature for the reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay. (A) Most suitable primer set for the RT-LAMP assay to amplify the orf1ab gene. (B) Optimal reaction temperature for the RT-LAMP assay with primer orf1ab-4. (C) Most suitable primer set for the RT-LAMP assay to amplify the S gene. (D) Optimal reaction temperature for the RT-LAMP assay with primer S-123. The reaction volume was 25 μL contained 2 μL RNA template, and the template concentration was 1 ng/μL.
To screen for the optimum temperature, the reaction was incubated at six different temperatures (60°C–65°C) for 60 min. As presented in Fig. 1, the highest amplification efficiency occurred at 63°C. Therefore, 63°C was confirmed as the optimum reaction temperature for this assay.
Sensitivity test for the RT-LAMP assay
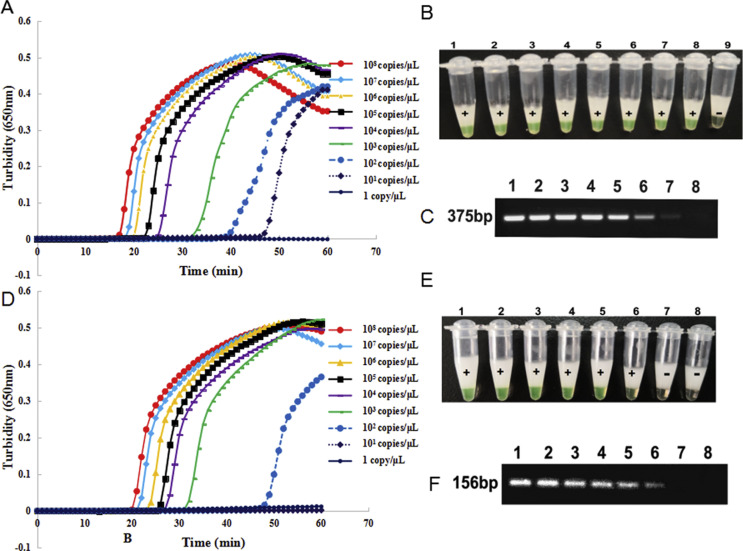
The sensitivity of the RT-LAMP assay using primer sets orf1ab-4 and S-123 was evaluated using turbidity monitoring and visual observation. Ten-fold serial dilutions of the pseudo-viruses, ranging from 1 × 108 copies/μL to 1 copy/μL (concentration of template input) were detected by the RT-LAMP assays. As illustrated in Fig. 2 , the time taken for positive detection ranged from 18 min at 1 × 108 copies/μL to 48 min at 1 × 101 copies/μL using primer set orf1ab-4, whereas for primer set S-123, the time taken for positive detection ranged from 21 min at 1 × 108 copies/μL to 50 min at 1 × 102 copies/μL. Hence, sensitivity of the assays was 2 × 101 copies and 2 × 102 copies per reaction at 63°C within 60 min with primer sets orf1ab-4 and S-123, respectively. The detection limit was the same with visual observation, with all positive reactions changing to green and negative reactions remaining orange as observed by the naked eye. But sensitivity of the conventional PCR was 2 × 102 copies and 2 × 103 copies per reaction with primer sets targeting orf1ab gene and S gene, respectively.
Fig. 2.
Sensitivity of the reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay and conventional PCR for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection. (A,B) Sensitivity of the RT-LAMP assay using primer set orf1ab-4 to target the orf1ab gene for SARS-CoV-2 detection. (D,E) Sensitivity of the RT-LAMP assay using primer set S-123 to target the S gene for SARS-CoV-2 detection. (C,F) Sensitivity of the conventional PCR assay targeting the orf1ab and S genes for SARS-CoV-2 detection. The detection was monitored by turbidity using a Loopamp real-time turbidimeter, and was judged by the naked eye depending on a colour change from orange to green. (B), Lanes 1–9: 108, 107, 106, 105, 104, 103, 102, 101 and 100 copies/μL; (E) lanes 1–8: 107, 106, 105, 104, 103, 102, 101 and 100 copies/μL; (C) and (F) lanes 1–8: 108, 107, 106, 105, 104, 103, 102 and 101 copies/μL. In the sensitivity test, 60 min can be used as the cut-off for the visual detection.
Specificity test for RT-LAMP assay
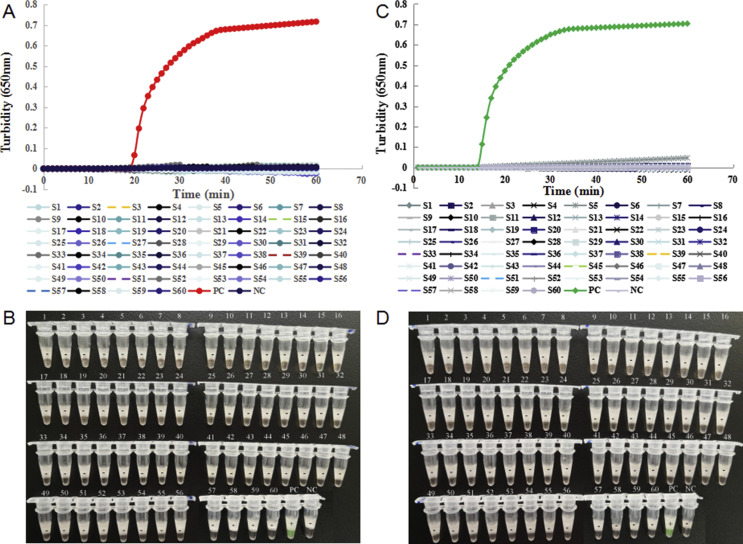
The specificity of the RT-LAMP assays was evaluated using 60 strains of human respiratory pathogens. Fig. 3 reveals that only the pseudo-viruses tested positive, the nucleic acids of other respiratory pathogens and the blank control all tested negative by the RT-LAMP assay with primer sets orf1ab-4 and S-123, using turbidity monitoring and visual observation. Therefore, the RT-LAMP assay showed no cross-reactivity with other respiratory pathogens.
Fig. 3.
Specificity of the reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection. (A,B) Specificity of the RT-LAMP assay with primer set orf1ab-4 targeted the orf1ab gene for SARS-CoV-2 detection. (C,D) Specificity of the RT-LAMP assay with primer set S-123 targeted the S gene for SARS-CoV-2 detection. The detection was monitored by turbidity using a Loopamp real-time turbidimeter (A, C), and was judged by the naked eye depending on a colour change from orange to green (B, D). S1: human coronavirus (HCoV) -229E-1; S2: HCoV-229E-2; S3: HCoV-NL63-1; S4: HCoV-NL63-2; S5: HCoV-NL63-3; S6: HCoV-OC43-1; S7: HCoV-OC43-2; S8: HCoV-HKU1-1; S9: HCoV-HKU1-2; S10: H3N2-1; S11: H3N2-2; S12: H3N2-3; S13: H3N2-4; S14: H1N1-1; S15: H1N1-2; S16: influenza B-1; S17: influenza B-2; S18: influenza B-3; S19: parainfluenza (PIV) -1-1; S20: PIV-1-2; S21: PIV-2-1; S22: PIV-2-2; S23: PIV-3-1; S24: PIV-3-2; S25: PIV-4-1; S26: PIV-4-2; S27: adenovirus (ADV) -1-1; S28: ADV-1-2; S29: ADV-2-1; S30: ADV-2-2; S31: ADV-3-1; S32: ADV-3-2; S33: ADV-4; S34: ADV-5-1; S35: ADV-5-2; S36: ADV-6; S37: ADV-7-1; S38: ADV-7-2; S39: respiratory syncytial virus (RSV) A-1; S40: RSV A-2; S41: RSV B-1; S42: RSV B-2; S43: human metapneumovirus HMPV-1; S44: HMPV-2; S45: human bocavirus (BoV) -1; S46: BoV-2; S47: rhinovirus (Rh) A-1; S48: Rh A-2; S49: Rh B-1; S50: Rh B-2; S51: Rh C; S52: Mycoplasma pneumoniae (MP) -FH; S53: MP-M129; S54: Haemophilus influenzae; S55: Staphylococcus aureus; S56: Klebsiella pneumoniae; S57: Streptococcus pneumoniae; S58: Pseudomonas aeruginosa; S59: SARS-CoV; S60: Middle East respiratory syndrome-CoV; PC, positive control (pseudo-virus); NC, negative control (distilled water).
Clinical sample detection
The 130 swabs and bronchoalveolar lavage fluid samples obtained from individuals with pneumonia and suspected SARS-CoV-2 infection were simultaneously investigated by RT-LAMP with the two primer sets (orf1ab-4 and S-123) and by real-time RT-PCR. Among them, 58 individuals were confirmed to be infected with SARS-CoV-2, while 72 tested negative (Table 1 ). The sensitivity was 100% (95% CI 92.3%–100%) and specificity was 100% (95% CI 93.7%–100%)
Table 1.
Comparison between the reverse transcription loop-mediated isothermal amplification assay and RT-PCR for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection from clinical samples
| Results | RT-LAMP |
Real-time RT-PCR | |
|---|---|---|---|
| orf1ab gene | S gene | ||
| Positive samples | 58 | 58 | 58 (Ct < 38) |
| Mean ± SD (min) | 25.36 ± 5.09 | 27.21 ± 5.74 | |
| Rangea (min) | 16–35 | 18–40 | |
| Negative samples | 72 (no change in colour) | 72 (no change in colour) | 72 (no peak) |
RT-LAMP, reverse transcription loop-mediated isothermal amplification; RT-PCR, reverse transcription-polymerase chain reaction; SD, standard deviation.
Range: the shortest detection time – the longest detection time.
Discussion
This study established a rapid, sensitive and specific assay for SARS-CoV-2 detection by RT-LAMP. To improve the sensitivity, we chose two target genes, orf1ab and S, to detect viral RNA in clinical samples. To ensure the stability of the detection assay, we aligned 103 complete virus genomes, then selected conversed regions without gene mutations in the orf1ab (245 bp) and S (216 bp) genes. To reduce the reaction time, we designed loop primers. Briefly, six primers targeting eight regions generated a self-priming dumbbell-shaped template upon isothermal incubation with strand-displacing polymerase, resulting in the rapid production of large quantities of complex amplicon [18].
Since the release of the first SARS-CoV-2 genome sequence via the community online resource, virological.org, on 10 January (Wuhan-Hu-1, GenBank Accession no.: MN908947), more than 2500 coronavirus sequences have been uploaded to date. These data make it possible for researchers to develop assays to detect viral RNA, to quarantine the source of infection, to explore the origin and native host(s), and to reveal the likely receptor-binding properties of this novel coronavirus.
The novel RT-LAMP assay presented positive results in the mean (±SD) time of 26.28 ± 4.48 min; whereas the RT-PCR assay requires 1–2 h after viral RNA preparation [19]. The sensitivity of this assay was 20 copies per reaction, similar to that reported for the RT-PCR assay in a previous study [20]. The RT-LAMP assay showed no cross-reactivity with other respiratory pathogens, so the diagnostic specificity of this method was higher than that reported for the serology test [21].
To assess the applicability of the assay for the clinical diagnosis of SARS-CoV-2 infection, an evaluation of 130 clinical samples was conducted. Sensitivity and specificity were both 100%. In fact, compared with the RT-PCR method, the RT-LAMP is easy to handle, and does not require skilled personnel or specialized instruments. The visual RT-LAMP assay can complete detection within 60 min, which was faster than RT-PCR assay, which required more than 80 min. In addition, the positive results were directly visible and so easier to observe.
Strengths of this study include that the RT-LAMP assay is an important point-of-care testing detection method. The results are easy to read. Positive results could be judged by the naked eye according to a colour change from orange to green, while negative results remained orange. Moreover, the designed RT-LAMP primers targeted five/six independent regions. In addition, the new method can detect the virus in a single step at a constant temperature of 63°C, so the assay can be performed using a small instrument at the bedside or in the field.
The main limitation of the current study is that we aligned only 103 complete genomes of SARS-CoV-2 obtained from four databases when designing the primers. With the spread of this virus, the accuracy of this RT-LAMP assay will be affected by mutations occurring in the primer sequence region of the target gene. So, it is necessary to monitor the mutant sites of the virus genome by whole-genome sequencing.
The established RT-LAMP assay has important implications for clinical practice. COVID-19 is widely distributed throughout 203 countries, areas or territories, and the numbers of confirmed cases and deaths continue to rise [22,23]. Therefore, under the current public health emergency, early diagnosis is essential to control the outbreak and to select appropriate treatment. At present, RT-PCR is widely applied for clinical diagnosis [19,20,24,25]. However, there are various drawbacks to this methodology, such as low sensitivity, the occurrence of false negatives, and virus loading in oral swabs [26]. As a result of time constraints and clinical needs, the quality of the commercial kits has not been further verified and evaluated using a large number of clinical samples. The RT-PCR assay is also laborious to perform, time-consuming and requires specialized equipment. To overcome these problems, a more rapid, convenient and less complex assay is needed.
In conclusion, we developed an RT-LAMP assay for SARS-CoV-2 detection that demonstrated high diagnostic sensitivity and specificity among clinical samples. The RT-LAMP assay is a powerful tool for SARS-CoV-2 identification not only in experimental laboratories, but also in hospitals as it does not require complex equipment. It may prove useful in monitoring suspected patients, close contacts and high-risk groups.
Transparency declaration
All authors disclose no conflicts of interest.
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant number 31670035), Mega-projects of Science and Technology Research of China (grant numbers 2017ZX100102, 2018ZX10101-003-002) to JY, and CAMS innovation Fund for Medical Sciences (CIFMS) (grant number 2016-I2M-1-008), Beijing Talents Fund (2018000021469G280), and Public service development and reform pilot project of Beijing Medical Research Institute (BMR2019-11).
Contribution of authors
JY and DL designed the experiments and edited the manuscript. CY, JC, BD, GX, SL and WZ performed and developed the RT-LAMP. HY, LH, LC, LZ, YS, NL, HZ, YF, SL and QZ performed the experiments. CY and JC wrote the manuscript.
Acknowledgements
We are grateful to Weijun Chen of Beijing macroµ-test Bio-Tech Co., Ltd. and to Yiming Li of the Chinese PLA General Hospital for providing and detecting the clinical samples from COVID-2019 patients and controls. We thank Kate Fox, DPhil, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Editor: F. Allerberger
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.04.001.
Contributor Information
D. Liu, Email: liud@wh.iov.cn.
J. Yuan, Email: yuanjing6216@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
Sequence comparison and gene analysis of the orf1b and S genes.
References
- 1.Giovanetti M., Benvenuto D., Angeletti S., Ciccozzi M. The first two cases of 2019-nCoV in Italy: where they come from? J Med Virol. 2020;92:518–521. doi: 10.1002/jmv.25699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H. Washington State 2019-nCoV case investigation team. first case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard Stoecklin S., Rolland P., Silue Y., Mailles A., Campese C., Simondon A. First cases of coronavirus disease 2019 (COVID-19) in France: surveillance, investigations and control measures, January 2020. Euro Surveill. 2020;25:2000094. doi: 10.2807/1560-7917.ES.2020.25.6.2000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu P., Zhu J., Zhang Z., Han Y., Huang L. A familial cluster of infection associated with the 2019 novel coronavirus indicating potential person-to-person transmission during the incubation period. J Infect Dis. 2020:jiaa077. doi: 10.1093/infdis/jiaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization . WHO; Geneva: 2020. Novel coronavirus (2019-nCoV) situation report-7: 27 January 2020. [Google Scholar]
- 7.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benvenuto D., Giovanetti M., Salemi M., Prosperi M., De Flora C., Junior Alcantara L.C. The global spread of 2019-nCoV: a molecular evolutionary analysis. Pathog Glob Health. 2020;114:64–67. doi: 10.1080/20477724.2020.1725339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reusken C.B.E.M., Broberg E.K., Haagmans B., Meijer A., Corman V.M., Papa A. Laboratory readiness and response for novel coronavirus (2019-nCoV) in expert laboratories in 30 EU/EEA countries, January 2020. Euro Surveill. 2020;25:2000082. doi: 10.2807/1560-7917.ES.2020.25.6.2000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge Y., Wu B., Qi X., Zhao K., Guo X., Zhu Y. Rapid and sensitive detection of novel avian-origin influenza A (H7N9) virus by reverse transcription loop-mediated isothermal amplification combined with a lateral-flow device. PLoS One. 2013;8:e69941. doi: 10.1371/journal.pone.0069941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang P., Wang H., Cao Z., Jin H., Chi H., Zhao J. A Rapid and specific assay for the detection of MERS-CoV. Front Microbiol. 2018;9:1101. doi: 10.3389/fmicb.2018.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Z., Wang H., Wang L., Li L., Jin H., Xu C. Visual detection of West Nile virus using reverse transcription loop-mediated isothermal amplification combined with a vertical flow visualization strip. Front Microbiol. 2016;7:554. doi: 10.3389/fmicb.2016.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chotiwan N., Brewster C.D., Magalhaes T., Weger-Lucarelli J., Duggal N.K., Ruckert C. Rapid and specific detection of Asian- and African-lineage Zika viruses. Sci Transl Med. 2017;9:eaag0538. doi: 10.1126/scitranslmed.aag0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Wang X., Liu W., Wei X., Lin W., Li E. Survey and visual detection of Zaire ebolavirus in clinical samples targeting the nucleoprotein gene in Sierra Leone. Front Microbiol. 2015;6:1332. doi: 10.3389/fmicb.2015.01332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwallah A., Inoue S., Muigai A.W., Kubo T., Sang R., Morita K. A real-time reverse transcription loop-mediated isothermal amplification assay for the rapid detection of yellow fever virus. J Virol Methods. 2013;193:23–27. doi: 10.1016/j.jviromet.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Notomi T., Mori Y., Tomita N., Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol. 2015;53:1–5. doi: 10.1007/s12275-015-4656-9. [DOI] [PubMed] [Google Scholar]
- 19.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbe Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. China novel coronavirus investigating and research team. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn J Infect Dis. 2020 doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 25.Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbe Infect. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.