Abstract
The coronavirus disease 2019 (COVID-19) pandemic now has >2,000,000 confirmed cases worldwide. COVID-19 is currently diagnosed using quantitative RT-PCR methods, but the capacity of quantitative RT-PCR methods is limited by their requirement of high-level facilities and instruments. We developed and evaluated reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays to detect genomic RNA of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative virus of COVID-19. RT-LAMP assays reported in this study can detect as low as 100 copies of SARS-CoV-2 RNA. Cross-reactivity of RT-LAMP assays to other human coronaviruses was not observed. A colorimetric detection method was adapted for this RT-LAMP assay to enable higher throughput.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative viral pathogen responsible for coronavirus disease 2019 (COVID-19), of which the outbreaks resulted in 2,245,872 confirmed cases involving 85,506 deaths worldwide as of April 20, 2020 (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). As the name suggests, SARS-CoV-2 is closely related to a group of severe acute respiratory syndrome–related coronaviruses, namely subgenus Sarbecovirus, showing 96% identity to a bat coronavirus.1 , 2
COVID-19 can be diagnosed through computed tomographic scan of suspicious patients, and a confirmatory laboratory test is performed using published quantitative RT-PCR (RT-qPCR) methods3, 4, 5, 6 and recommendations from The World Health Organization (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance, last accessed April 2, 2020). Although RT-qPCR methods are used as the gold standard for detection of pathogens because of their high sensitivity and specificity, there are still some caveats. Briefly, laboratory-level facilities, with reliable supply of electricity, expensive instruments, and trained personnel, are required to properly perform RT-qPCR tests. These restrictions hinder use of RT-qPCR methods for various point-of-care situations, where pathogen detection might be required.7
To overcome such cost restriction of RT-qPCR and still detect nucleic acids from the pathogen, isothermal amplification methods have been developed.8 Among such methods, the loop-mediated isothermal amplification (LAMP) method has some advantages to be applied for point-of-care testing.9 Well-optimized LAMP assay shows sensitivity comparable to that of PCR, <10 copies per reaction.10 Intercalating fluorescent dyes are compatible with LAMP reaction so that amplification can be observed in real-time.11 Because the amplification efficiency of LAMP reaction is high, changes in reaction mixture components make it possible to detect the result with colorimetric detection methods.12, 13, 14 Moreover, unpurified sample can be directly used for LAMP.15, 16, 17 This indicates that high-throughput test is possible when use of unpurified specimen is combined with noninstrumental (eg, colorimetric) detection.
In this study, one-step reverse transcription LAMP (RT-LAMP) methods to detect SARS-CoV-2 were designed and evaluated. A pair of LAMP primer sets specific to SARS-CoV-2 and accompanying optimized reaction conditions was provided. The leuco crystal violet method was applied to achieve colorimetric detection of LAMP reaction.13
Materials and Methods
Viral RNA Preparation
SARS-CoV-2 viral RNA was prepared, as previously described.18 Human coronavirus (hCoV)–229E and hCoV-OC43 viral RNA were isolated from culture media of infected MRC-5 cells (ATCC, Manassas, VA; CCL-171). Middle East respiratory syndrome coronavirus (MERS-CoV) RNA was isolated from cell pellet lysate of infected Vero cells (ATCC; CCL-81).
Viral RNA Titration
In vitro transcribed standard RNA for SARS-CoV-2 was prepared, as previously described.18 To prepare standard RNA for hCoV-229E, first amplicon of PCR (forward primer, 5′-GCTAGTGGATGATCATGCTTTG-3′; reverse primer, 5′-TGGGGCCATAAACTGTTCTATTAC-3′) was cloned to pBluescript II KS (+) plasmid with BamHI and XhoI. Then, in vitro transcription template was prepared by restriction enzyme cut with BglI and XhoI and purification after agarose gel electrophoresis. For hCoV-OC43 and MERS-CoV, amplicons of PCR (OC43 forward primer, 5′-AGCAACCAGGCTGATGTCAATACC-3′; OC43 reverse primer, 5′-AGCAGACCTTCCTGAGCCTTCAAT-3′; MERS-CoV, UpE region19) were synthesized and cloned to pBIC-A plasmid (Bioneer, Daejeon, Republic of Korea). In vitro transcription template for hCoV-OC43 and MERS-CoV was prepared by restriction enzyme cut with BamHI-XhoI or SspI-XhoI, respectively. In vitro transcriptions were performed with EZ T7 High Yield In Vitro Transcription kit (Enzynomics, Daejeon, Republic of Korea), per manufacturer's instructions. RNA products were then purified using Agencourt RNAClean XP (Beckman Coulter, Brea, CA). Standard RNA copy numbers were calculated from concentration measured by NanoDrop Lite (Thermo Scientific, Waltham, MA). All restriction enzymes were purchased from Enzynomics.
To evaluate genomic copy number of viral RNAs, dilutions of standard RNAs and viral RNAs in TE buffer (10 mmol/L Tris-Cl, pH 8.0, and 1 mmol/L EDTA) are subjected to one-step RT-qPCR. RT-qPCRs were performed using LightCycler 96 instrument (Roche, Basel, Switzerland) and following reagents: Luna Universal One-Step RT-qPCR Kit [New England Biolabs (NEB), Ipswich, MA] for hCoV-229E and hCoV-OC43, THUNDERBIRD Probe qPCR Master Mix (Toyobo, Osaka, Japan) for MERS-CoV, and Luna Universal Probe One-Step RT-qPCR Kit (NEB) for SARS-CoV-2.
Reverse Transcription
cDNA of SARS-CoV-2 was made using SuperScript IV Reverse Transcriptase (Invitrogen, Waltham, MA) following manufacturer's instructions with modifications. Briefly, 10 pmol of random hexamer was used as reverse transcription primer, and reaction was performed as follows: 20 minutes at 25°C, 30 minutes at 55°C, and 10 minutes at 80°C.
LAMP and RT-LAMP Reaction
LAMP reaction was performed with reaction mixture containing following components: 1.6 μmol/L forward inner primer/backward inner primer, 0.2 μmol/L F3/B3 primers, 0.4 μmol/L loop forward/loop backward primers, 1× Isothermal Amplification Buffer II [NEB; 20 mmol/L Tris-HCl, pH 8.8, 10 mmol/L (NH4)2SO4, 150 mmol/L KCl, 2 mmol/L MgSO4, and 0.1% Tween 20], 6 mmol/L MgSO4 (NEB; final, 8 mmol/L Mg2+), 1.4 mmol/L each dNTP (Enzynomics), 0.4 μmol/L SYTO-9 (Invitrogen), and 6 U Bst 3.0 DNA polymerase (NEB) in total 15-μL reaction volume. For RT-LAMP, 10 U of SuperScript IV Reverse Transcriptase (Invitrogen) was added. For end-point colorimetric detection of LAMP reaction, tweaked version of 5× stock leuco crystal violet solution13 containing 0.5 mmol/L crystal violet (Sigma, St. Louis, MO; C0775), 60 mmol/L sodium sulfite (Sigma; S0505), and 5 mmol/L β-cyclodextrin (Sigma; C4767) was directly added to LAMP reaction mixture to 1× concentration. When using WarmStart Colorimetric LAMP 2X Master Mix (NEB), same concentrations of primers and 0.4 μmol/L SYTO-9 were added. Isothermal incubation and fluorescence signal measurement were performed using LightCycler 96 instrument at 69°C with additional heat inactivation (5 minutes at 95°C) and melting curve analysis steps. Fluorescence signals were measured for every minute during 60 (screening and optimization) or 30 [limit of detection (LoD) and cross-reactivity] minutes of incubation. Any changed conditions are specified for each experiment.
Results
LAMP Primer Design
Five SARS-CoV-2 sequences (MN908947, MN938384, MN988713, MN985325, and MN975262) and seven SARS-CoV sequences (NC_004718, AY613947, AY313906, AY559094, AY502924, AY278491, and AY502927) were aligned by MEGA software version 7.20 SARS-CoV-2–specific regions for LAMP primer design are manually selected: two regions from Nsp3 (3055 to 3591 and 6172 to 7273), two regions from Spike (21540 to 22549 and 22890 to 23779), and one region from Orf8 (27824 to 28396). Whole Nucleocapsid gene region is also included as Nucleocapsid is usual target of molecular diagnosis because of the abundance of its mRNA.21 Two to five basic LAMP primer sets are designed and selected with PrimerExplorer V5 (http://primerexplorer.jp/lampv5e/index.html, last accessed February 22, 2020) for each target region. Loop primers are designed by PrimerExplorer V5 or manually selected. Sixteen LAMP primer sets, with loop forward primer and loop backward primer showing proper melting temperature, were selected and subjected to further screening.
LAMP Primer Screening
First round of screening was performed using WarmStart Colorimetric LAMP 2X Master Mix with cDNA, of which corresponding RNA concentration is 8.3 × 104 copies/reaction. Of 16 primer sets, nine were selected by threshold time from the results of 40 minutes’ incubation at 65°C and subjected to further screening.
Second round of screening was performed using Bst 3.0. The same amount of cDNA as the first round of screening is used. Two primer sets that showed relatively early non-specific amplification are discarded, and the remaining seven primer sets are subjected to further screening.
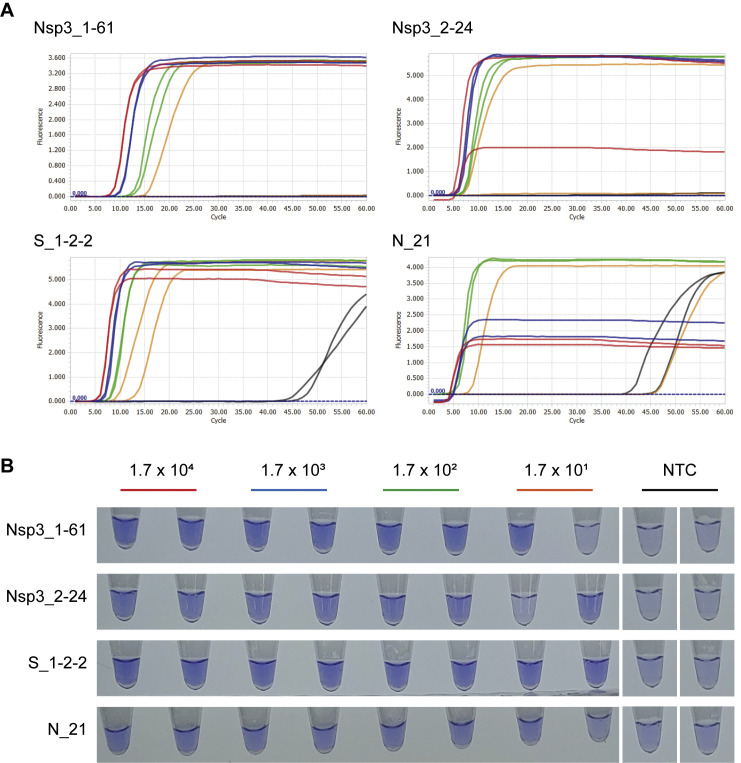
Third and fourth rounds of screenings were performed by checking sensitivity to dilutions of cDNA and RNA, respectively. Five of seven primer sets showed specific amplification for at least one replicate of duplicate, with cDNA concentration corresponding to 1.7 × 101 copies of input RNA (Supplemental Figure S1). Next, sensitivity of the five primer sets to RNA dilutions was evaluated in RT-LAMP using Bst 3.0 and SuperScript IV Reverse Transcriptase. Two primer sets, both targeting Nsp3, showed best sensitivity that showed specific amplification at 10−6 dilution of RNA (Figure 1 ). Two additional primer sets, one targeting Spike and the other targeting Nucleocapsid, were added for reaction optimization experiments as they showed fast threshold time for cDNA and to keep ranges of target genes. Primer sequences are represented in Table 1 .
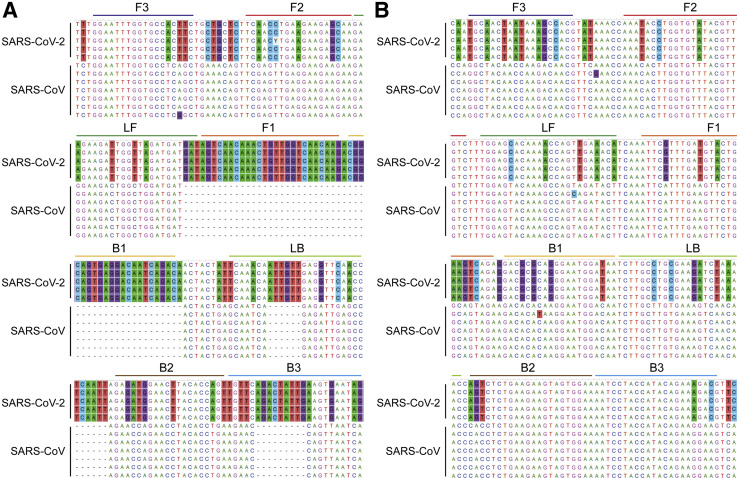
Figure 1.
Loop-mediated isothermal amplification (LAMP) primer positions on aligned sequences of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and SARS-CoV. Primer binding sites of Nsp3_1-61 (A) and Nsp3_2-24 (B) LAMP primer sets are depicted on aligned sequences of five SARS-CoV-2 (from the top, MN908947, MN938384, MN988713, MN985325, and MN975262) and seven SARS-CoV (from the top, NC_004718, AY613947, AY313906, AY559094, AY502924, AY278491, and AY502927). Conserved sites are toggled at 50% level by MEGA software version 7 so that SARS-CoV-2 specific residues versus SARS-CoV are background colored. Different color bars are used as follows to distinguish binding sites: F3, blue; F2, red; F1, orange; loop forward (LF), green; loop backward (LB), light green; B1, light orange; B2, brown; and B3, light blue.
Table 1.
LAMP Primers of Which Assays Were Optimized and LoD Evaluated
| Set name | Primer | Sequence |
|---|---|---|
| Nsp3_1-61 | F3 | 5′-GGAATTTGGTGCCACTTC-3′ |
| B3 | 5′-CTATTCACTTCAATAGTCTGAACA-3′ | |
| FIP | 5′-CTTGTTGACCAACAGTTTGTTGACTTCAACCTGAAGAAGAGCAA-3′ | |
| BIP | 5′-CGGCAGTGAGGACAATCAGACACTGGTGTAAGTTCCATCTC-3′ | |
| LF | 5′-ATCATCATCTAACCAATCTTCTTC-3′ | |
| LB | 5′-TCAAACAATTGTTGAGGTTCAACC-3′ | |
| Nsp3_2-24 | F3 | 5′-TGCAACTAATAAAGCCACG-3′ |
| B3 | 5′-CGTCTTTCTGTATGGTAGGATT-3′ | |
| FIP | 5′-TCTGACTTCAGTACATCAAACGAATAAATACCTGGTGTATACGTTGTC-3′ | |
| BIP | 5′-GACGCGCAGGGAATGGATAATTCCACTACTTCTTCAGAGACT-3′ | |
| LF | 5′-TGTTTCAACTGGTTTTGTGCTCCA-3′ | |
| LB | 5′-TCTTGCCTGCGAAGATCTAAAAC-3′ | |
| S_1-2-2 | F3 | 5′-CTGACAAAGTTTTCAGATCCTCAG-3′ |
| B3 | 5′-AGTACCAAAAATCCAGCCTCTT-3′ | |
| FIP | 5′-TCCCAGAGACATGTATAGCATGGAATCAACTCAGGACTTGTTCTTACC-3′ | |
| BIP | 5′-TGGTACTAAGAGGTTTGATAACCCTGTTAGACTTCTCAGTGGAAGCA-3′ | |
| LF | 5′-CCAAGTAACATTGGAAAAGAAA-3′ | |
| LB | 5′-GTCCTACCATTTAATGATGGTGTTT-3′ | |
| N_21 | F3 | 5′-GCCAAAAGGCTTCTACGCA-3′ |
| B3 | 5′-TTGCTCTCAAGCTGGTTCAA-3′ | |
| FIP | 5′-TCCCCTACTGCTGCCTGGAGGCAGTCAAGCCTCTTCTCG-3′ | |
| BIP | 5′-TCTCCTGCTAGAATGGCTGGCATCTGTCAAGCAGCAGCAAAG-3′ | |
| LF | 5′-TGTTGCGACTACGTGATGAGGA-3′ | |
| LB | 5′-ATGGCGGTGATGCTGCTCT-3′ |
BIP, backward inner primer; FIP, forward inner primer; LB, loop backward; LF, loop forward; LAMP, loop-mediated isothermal amplification; LoD, limit of detection.
LAMP Reaction Optimization
To optimize RT-LAMP reaction with Bst 3.0, first, optimal concentrations of dNTP and Mg2+ were evaluated for each primer set. Three dNTP- Mg2+ concentration combinations are tested at 69°C: dNTP (1.4 mmol/L) and Mg2+ (8 mmol/L), dNTP (1.4 mmol/L) and Mg2+ (6 mmol/L), and dNTP (1 mmol/L) and Mg2+ (6 mmol/L). The copy number of RNA template was 1000 copies per reaction. Best concentration combination was selected for each primer set by whether both of duplicates show specific amplification and by threshold time. Average threshold time (Tt_av) and corresponding dNTP and Mg2+ concentrations are as follows: dNTP (1 mmol/L) and Mg2+ (6 mmol/L) for Nsp3_1-61 (Tt_av = 11.93 minutes), dNTP (1.4 mmol/L) and Mg2+ (8 mmol/L) for Nsp3_2-24 (Tt_av = 7.74 minutes), dNTP (1.4 mmol/L) and Mg2+ (6 mmol/L) for S_1-2-2 and N_21 (Tt_av = 11.08 minutes and 5.50 minutes, respectively).
Next, whether amplification is improved at lower temperature (65°C) was evaluated, because optimal reaction temperature of SuperScript IV Reverse Transcriptase is 50°C to 55°C and the manufacturer recommends 65°C to 72°C for optimal performance of Bst 3.0. Notably, Nsp3_1-61 and S_1-2-2 primer sets show improved threshold time (Tt_av = 8.92 minutes and 8.49 minutes, respectively).
Assessing LoD and Cross-Reactivity
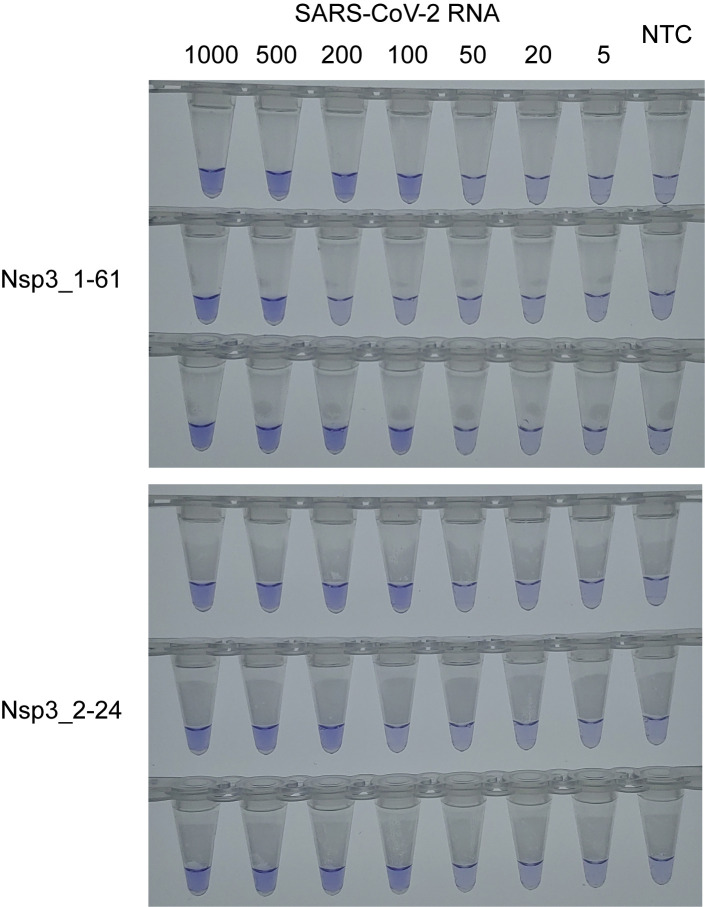
The LoD of optimized RT-LAMP assays was assessed through 5 to 1000 RNA copies/reaction in triplicate. Among four primer sets subjected to reaction optimization, S_1-2-2 and N_21 sets showed relatively poor sensitivity. For Nsp3_1-61 and Nsp3_2-24 primer sets, LoD was additionally evaluated with twofold SuperScript IV Reverse Transcriptase (20 U/reaction). LoD and Tt_av were improved by increasing reverse transcriptase amount (Table 2 and Supplemental Figure S2). As a result, both Nsp3_1-61 and Nsp3_2-24 RT-LAMP assays could detect as low RNA concentration as 100 copies per reaction.
Table 2.
LoD of RT-LAMP Assays Targeting SARS-CoV-2
| RNA copies | SSIV, 10 U/rxn |
SSIV, 20 U/rxn |
||||||
|---|---|---|---|---|---|---|---|---|
| Nsp3_1-61 |
Nsp3_2-24 |
Nsp3_1-61 |
Nsp3_2-24 |
|||||
| Tt_av | RPR | Tt_av | RPR | Tt_av | RPR | Tt_av | RPR | |
| 1000 | 13.11 | 3:3 | 9.31 | 3:3 | 10.7 | 3:3 | 7.53 | 3:3 |
| 500 | 13.47 | 3:3 | 8.16 | 2:3 | 10.63 | 3:3 | 7.59 | 3:3 |
| 200 | 13.65 | 2:3 | 11.19 | 2:3 | 11.34 | 2:3 | 7.78 | 3:3 |
| 100 | 14.62 | 3:3 | — | — | 12.26 | 2:3 | 8.47 | 2:3 |
| 50 | — | — | — | — | — | — | — | — |
| 20 | 13.88 | 1:3 | — | — | — | — | — | — |
| 5 | — | — | — | — | — | — | — | — |
| NTC | — | — | — | — | — | — | — | — |
—, no amplification observed; LoD, limit of detection; RPR, ratio of positive replicates; RT-LAMP, reverse transcription loop-mediated isothermal amplification; rxn, reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SSIV, SuperScript IV Reverse Transcriptase; Tt_av, average threshold time in minutes; NTC, no template control.
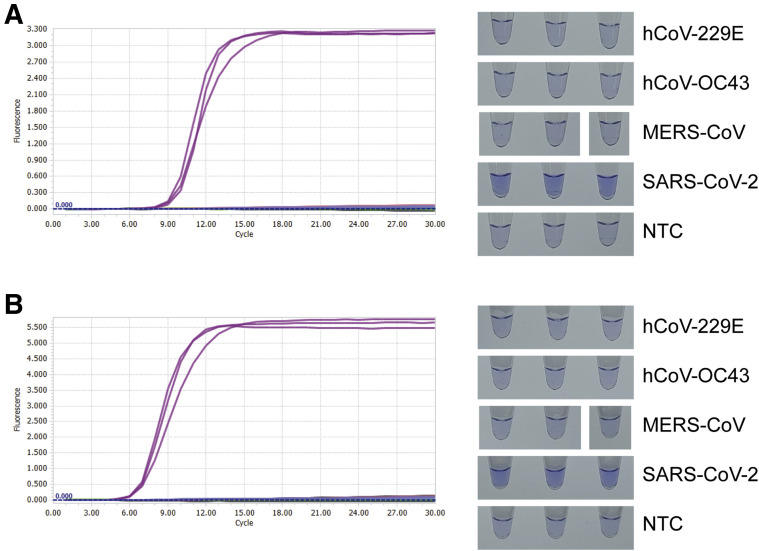
The cross-reactivity of two SARS-CoV-2 RT-LAMP assays targeting Nsp3 to other human coronaviruses was not found for hCoV-229E, hCoV-OC43, and MERS-CoV (Figure 2 ).
Figure 2.
Cross-reactivity to other coronaviruses tested for reverse transcription loop-mediated isothermal amplification (LAMP) assay targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Real-time amplification fluorescence signal and end-point leuco crystal violet colorimetric detection results of cross-reactivity test for Nsp3_1-61 (A) and Nsp3_2-24 (B) LAMP primer sets. Reactions are performed with 20 U/reaction of reverse transcriptase and optimized temperature, dNTP concentration, and Mg2+ concentration for each primer set. RNA copy number of each viral RNA is as follows: human coronavirus (hCoV)–229E, 1.6 × 106; hCoV-OC43, 1.6 × 106; Middle East respiratory syndrome coronavirus (MERS-CoV), 4.5 × 106; and SARS-CoV-2, 2.5 × 103. NTC, no template control.
Discussion
In this study, LAMP primer sets targeting SARS-CoV-2 were designed and screened. Reaction was also optimized for selected primer sets. In summary, a pair of RT-LAMP assays for detection of SARS-CoV-2 with LoD of 100 copies per reaction was designed and evaluated. These RT-LAMP assays showed specificity to SARS-CoV-2 versus alphacoronavirus (hCoV-229E), betacoronavirus (hCoV-OC43), and MERS-CoV. Although specificity to SARS-CoV could not be tested because a proper sample was not in hand, specificity of the RT-LAMP assays is easily expectable from the mismatching bases in primer binding sites (Figure 1). Especially, both of F1/B1 sites of Nsp3_1-61 primer set are in SARS-CoV-2 specific region, of which aligning SARS-CoV sequence does not exist.
About the sensitivity of LAMP assays, note that average threshold time is not well correlated with LoD. In fact, average threshold times of S_1-2-2 and N_21 primer sets for 1000 copies were faster than that of Nsp3_1-61: 7.33 (ratio of positive replicates, 2:3) and 5.06 (ratio of positive replicates, 1:3) minutes, respectively. This disrelation is previously reported.22 One peculiar observation is that both S_1-2-2 and N_21 showed good sensitivity to cDNA template (Supplemental Figure S1), unlike to RNA. Observed difference of sensitivity to RNA and cDNA seems significant even when accounting for slight amplification that might happen during reverse transcription. The reason may include stochastic nature of forming proper LAMP amplification intermediate-dumbbell structure,22 higher stability of DNA/RNA double strand than DNA/DNA double strand, and more.
The LoD of RT-LAMP assays suggested in this study, 100 copies per reaction, may not be enough for sensitive screening of suspicious patients. This relatively high LoD would be from the target sequences used for primer design that are selected by specificity versus SARS-CoV. Indeed, target GC percentage and melting temperature of primers had to be adjusted to get enough number of starting sets for primer screening during LAMP primer design to give less-optimal primer sets. Therefore, there might be better target sequences for LAMP assay of SARS-CoV-2 in the viewpoint of sensitivity. However, expected high specificity of RT-LAMP assays suggested in this study would be a good feature for a confirmatory test. In addition, considering high viral load of SARS-CoV-2 at early stage after symptom onset23 suggested RT-LAMP assays may still be useable for screening tests.
In conclusion, highly specific RT-LAMP assays for detection of SARS-CoV-2 were developed and evaluated. The results of these RT-LAMP assays can be detected within 30 minutes after amplification reaction begins. In addition, optimized reaction conditions were provided, to which leuco crystal violet colorimetric detection method is applied that can be used for point-of-care tests.
Acknowledgment
We thank Korea Centers for Disease Control and Prevention for kind and rapid sharing of isolated strain of severe acute respiratory syndrome coronavirus 2.
Footnotes
Supported by National Research Council of Science and Technology grant CRC-16-01-KRICT by the Ministry of Science and ICT of the Republic of Korea.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2020.03.006.
Supplemental Data
Supplemental Figure S1.
Sensitivity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) loop-mediated isothermal amplification (LAMP) assays to cDNA dilutions. Real-time amplification fluorescence signal (A) and end-point leuco crystal violet (LCV) colorimetric detection results (B) of four SARS-CoV-2 LAMP primer sets that were subjected to reaction optimization. Color of amplification signal curve and LCV detection label for each cDNA dilution are matched. Designated copy number is corresponding reverse transcription input RNA copy number. Some low fluorescence of amplification signal plateaus is due to baseline correction process of LightCycler 96 software. NTC, no template control.
Supplemental Figure S2.
Colorimetric detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reverse transcription loop-mediated isothermal amplification limit of detection (LoD) tests for Nsp3_1-61 and Nsp3_2-24. Leuco crystal violet colorimetric detection results of LoD tests for Nsp3_1-16 and Nsp3_2-24. A 20 U/reaction of reverse transcriptase was used. NTC, no template control.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nayak S., Blumenfeld N.R., Laksanasopin T., Sia S.K. Point-of-care diagnostics: recent developments in a connected age. Anal Chem. 2017;89:102–123. doi: 10.1021/acs.analchem.6b04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niemz A., Ferguson T.M., Boyle D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011;29:240–250. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shirato K., Yano T., Senba S., Akachi S., Kobayashi T., Nishinaka T., Notomi T., Matsuyama S. Detection of middle east respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol J. 2014;11:139. doi: 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oscorbin I.P., Belousova E.A., Zakabunin A.I., Boyarskikh U.A., Filipenko M.L. Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification (qLAMP) Biotechniques. 2016;61:20–25. doi: 10.2144/000114432. [DOI] [PubMed] [Google Scholar]
- 12.Goto M., Honda E., Ogura A., Nomoto A., Hanaki K. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 13.Miyamoto S., Sano S., Takahashi K., Jikihara T. Method for colorimetric detection of double-stranded nucleic acid using leuco triphenylmethane dyes. Anal Biochem. 2015;473:28–33. doi: 10.1016/j.ab.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Tanner N.A., Zhang Y., Evans T.C., Jr. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques. 2015;58:59–68. doi: 10.2144/000114253. [DOI] [PubMed] [Google Scholar]
- 15.Nie K., Qi S.X., Zhang Y., Luo L., Xie Y., Yang M.J., Zhang Y., Li J., Shen H., Li Q., Ma X.J. Evaluation of a direct reverse transcription loop-mediated isothermal amplification method without RNA extraction for the detection of human enterovirus 71 subgenotype C4 in nasopharyngeal swab specimens. PLoS One. 2012;7:e52486. doi: 10.1371/journal.pone.0052486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nyan D.C., Ulitzky L.E., Cehan N., Williamson P., Winkelman V., Rios M., Taylor D.R. Rapid detection of hepatitis B virus in blood plasma by a specific and sensitive loop-mediated isothermal amplification assay. Clin Infect Dis. 2014;59:16–23. doi: 10.1093/cid/ciu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee D., Shin Y., Chung S., Hwang K.S., Yoon D.S., Lee J.H. Simple and highly sensitive molecular diagnosis of Zika virus by lateral flow assays. Anal Chem. 2016;88:12272–12278. doi: 10.1021/acs.analchem.6b03460. [DOI] [PubMed] [Google Scholar]
- 18.Jung Y.J., Park G.-S., Moon J.H., Ku K., Beak S.-H., Kim S., Park E.C., Park D., Lee J.-H., Byeon C.W., Lee J.J., Maeng J.-S., Kim S.J., Kim S.I., Kim B.-T., Lee M.J., Kim H.G. Comparative analysis of primer-probe sets for the laboratory confirmation of SARS-CoV-2. BioRxiv. 2020 doi: 10.1021/acsinfecdis.0c00464. [DOI] [PubMed] [Google Scholar]
- 19.Corman V.M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M., van Boheemen S., Gopal R., Ballhause M., Bestebroer T.M., Muth D., Muller M.A., Drexler J.F., Zambon M., Osterhaus A.D., Fouchier R.M., Drosten C. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiscox J.A., Cavanagh D., Britton P. Quantification of individual subgenomic mRNA species during replication of the coronavirus transmissible gastroenteritis virus. Virus Res. 1995;36:119–130. doi: 10.1016/0168-1702(94)00108-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khorosheva E.M., Karymov M.A., Selck D.A., Ismagilov R.F. Lack of correlation between reaction speed and analytical sensitivity in isothermal amplification reveals the value of digital methods for optimization: validation using digital real-time RT-LAMP. Nucleic Acids Res. 2016;44:e10. doi: 10.1093/nar/gkv877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]