Summary
The outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in China and rapidly spread worldwide. To prevent SARS-CoV-2 dissemination, understanding the in vivo characteristics of SARS-CoV-2 is a high priority. We report a ferret model of SARS-CoV-2 infection and transmission that recapitulates aspects of human disease. SARS-CoV-2-infected ferrets exhibit elevated body temperatures and virus replication. Although fatalities were not observed, SARS-CoV-2-infected ferrets shed virus in nasal washes, saliva, urine, and feces up to 8 days post-infection. At 2 days post-contact, SARS-CoV-2 was detected in all naive direct contact ferrets. Furthermore, a few naive indirect contact ferrets were positive for viral RNA, suggesting airborne transmission. Viral antigens were detected in nasal turbinate, trachea, lungs, and intestine with acute bronchiolitis present in infected lungs. Thus, ferrets represent an infection and transmission animal model of COVID-19 that may facilitate development of SARS-CoV-2 therapeutics and vaccines.
Keywords: 2019-novel coronavirus, 2019-nCoV, severe acute respiratory syndrome coronavirus 2, SARS-CoV-2, novel coronavirus disease, COVID-19, virus shedding, transmission, ferrets
Graphical Abstract
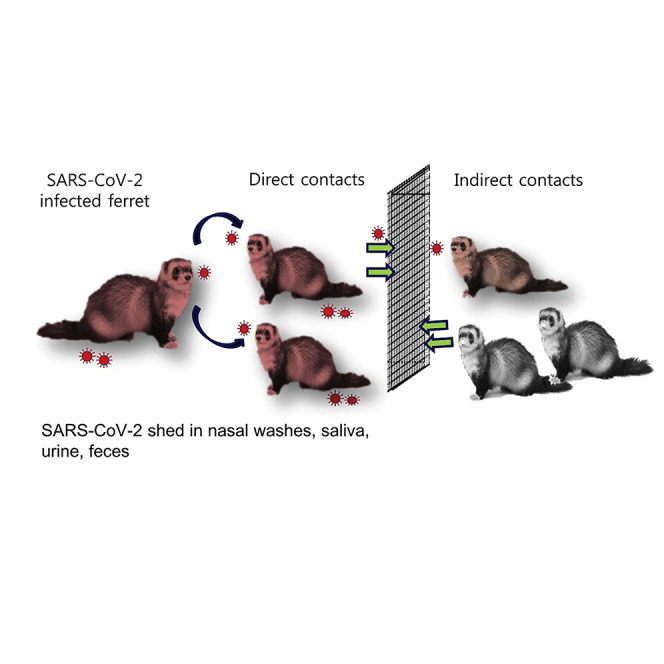
Highlights
-
•
SARS-CoV-2-infected ferrets exhibit elevated body temperature and virus replication
-
•
SARS-CoV-2 is shed in nasal washes, saliva, urine and feces
-
•
SARS-CoV-2 is effectively transmitted to naive ferrets by direct contact
-
•
SARS-CoV-2 infection leads acute bronchiolitis in infected ferrets
The outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rapidly spreads, leading to a pandemic infection. Kim et al. show that ferrets are highly susceptible to SARS-CoV-2 infection and effectively transmit the virus by direct or indirect contact, recapitulating human infection and transmission.
Main Text
Coronaviruses (CoVs) are a large family of viruses that cause respiratory and intestinal infections in animals and humans (Masters and Perlman, 2013). Of the four genera—alphacoronavirus, betacoronavirus, gammacoronavirus, and deltacoronavirus—alphacoronavirus and betacoronavirus are commonly associated with respiratory illness in humans and gastroenteritis in animals (Cui et al., 2019). CoVs were not typically considered to be highly pathogenic in humans until the outbreaks of Severe Acute Respiratory Syndrome CoV (SARS-CoV) (Zhong et al., 2003), Middle East Respiratory Syndrome CoV (MERS-CoV) (Zaki et al., 2012), and more recently, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
In late December of 2019, a novel coronavirus disease (COVID-19) was identified in Wuhan City, Hubei Province, China from patients with severe pneumonia (Zhu et al., 2020). Deep sequencing analysis of lower respiratory tract samples revealed the identity of the causative agent as a newly emerged strain of betacoronavirus, temporarily named 2019 novel coronavirus (2019-nCoV) and later renamed as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses (ICTV) (ICTV, 2020). As of March 23, there have been approximately 81,601 confirmed cases of COVID-19 in China with over 3,276 deaths (WHO, 2020b). The SARS-CoV-2 has been found to have high human-to-human transmission through close contact with infected patients, leading to rapid global spread by infected travelers from China. As of March 23, 2020, SARS-CoV-2 cases have been confirmed in at least 171 countries with a steady increase in the number of laboratory confirmed cases (251,329 cases) outside of China suggesting that non-pharmaceutical intervention strategies have not ultimately been successful in limiting spread. Therefore, an animal model that recapitulates the COVID-19 clinical symptoms in human infection is urgently needed in order to decipher the transmission routes and pathobiology of this virus and to allow testing of pharmaceutical interventions.
Given that SARS-CoV-2 shares higher sequence homology with SARS-CoV (79% homology) than with MERS-CoV (50% homology), the entry receptor for SARS-CoV, human Angiotensin-converting enzyme 2 (hACE2), was considered as a receptor candidate for SARS-CoV-2 (Lu et al., 2020). Correspondingly, Bao et al. (2020) reported weight loss and virus replication in lungs of hACE2 transgenic mice following SARS-CoV-2 infection; however, no other clinical symptoms such as cough or fever were observed. In order to understand the rapid spreading characteristics of SARS-CoV-2, additional animal models that mimic high human-to-human transmission of SARS-CoV-2 infections are warranted. Given that ferret ACE2 has been shown to contain critical SARS-CoV binding residues (Wan et al., 2020), we performed infection and direct and indirect contact transmission studies using a ferret model previously developed for influenza virus infections (Park et al., 2018; Bouvier, 2015).
To demonstrate ferret-to-ferret transmission in an experimental setting, ferrets (n = 2) were inoculated via the intranasal (IN) route with 105.5 TCID50 of NMC-nCoV02, a strain that was isolated from a COVID-19-confirmed patient in South Korea in February of 2020. To evaluate the transmission mode of the virus, naive ferrets (n = 2/group) were placed in direct contact (DC) (co-housed) or indirect contact (IC) (housed in cages with a permeable partition separating them from infected ferrets) with infected ferrets two days after the primary infection. Clinical features of SARS-CoV-2 infections were recorded. This study was repeated in three independent trials (total n = 24; direct infection [n = 6], DC [n = 6], IC [n = 6], and PBS control [n = 6] ferrets).
NMC-nCoV02-infected ferrets had elevated body temperatures, from 38.1°C to 40.3°C, between 2 and 8 dpi; these returned to normal by 8 dpi (Figure 1 A). While reduced activity was observed in NMC-nCoV02-infected ferrets between 2 and 6 dpi with occasional coughs, there was no detectable body weight loss, nor were there any fatalities during the experimental period. Interestingly, all six DC ferrets showed increased body temperatures (∼39°C) with reduced activity between 4 and 6 days post contact (dpc) and no detectable body weight loss (Figures S1A and S1B). However, none of the IC ferrets showed increased body temperature or weight loss over the 12 days of the studies (Figures S1C and S1D). These data indicate that the efficient establishment of COVID-19 clinical features in ferrets exposed to infected animals requires direct contact, recapitulating human-to-human transmission.
Figure 1.
Temperature Changes, Weight Loss, Survival, Viral Shedding, and Immunohistochemistry of Tissues of NMC-nCoV02-Infected Ferrets
(A–C) Six ferrets were inoculated intranasally with 105.5 TCID50 of virus. (A) Temperature changes, (B) number of viral RNA copies, and (C) infectious virus titers were measured in tissues of NMC-nCoV02-infected ferrets (n = 6/group). Each tissue (n = 3 per group) was collected at 4, 8, and 12 dpi. Viral loads in nasal turbinate, trachea, lung, kidney, and intestine were titered using quantitative real-time PCR and TCID50. Data are presented as mean ± SEM.
(D) Serum neutralizing (SN) antibody titers (GMT) against NMC-nCoV02 (100 TCID50) were measured onto Vero cells after 12 days of experiment (n = 6 per group). Data are presented as geometric mean ± SD. Tissues were harvested on day 4 after inoculation and immunohistochemistry was performed with a mouse polyclonal antibody.
(E–H) Tissues of PBS control ferrets; (E) nasal turbinate, (F) trachea, (G) lung, and (H) intestine.
(I–L) Tissues of NMC-nCoV02 infected ferrets: (I) Nasal turbinate, (J) Trachea, (K) lung, and (L) Intestine.
The presence of NMC-nCoV02 antigen was determined by IHC with mouse polyclonal antibody. Magnification ×400. Asterisks indicate statistical significance compared with PBS control group by the two-way ANOVA with Sidaks multiple comparisons test (A), the two way ANOVA with Dunnett’s multiple comparisons test (B and C), or one-way ANOVA Dunnett’s multiple comparisons test (∗ indicates p < 0.05, ∗∗ indicates p < 0.001, and ∗∗∗ indicates p < 0.0001).
To investigate SARS-CoV-2 replication and shedding in each group of ferrets, we collected blood, nasal washes, saliva, urine, and fecal specimens every other day for 12 days. Collected ferret secretions were resuspended in cold phosphate-buffered saline (PBS) containing antibiotics (5% penicillin/streptomycin; GIBCO). For virus titration, total RNA was extracted from the collected samples using the RNeasy Mini kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions (QIAGEN, 2012) and cDNA was synthesized with a cDNA synthesis kit (Omniscript Reverse Transcriptase; QIAGEN, Hilden, Germany). To quantitate viral RNA copy number, quantitative real-time RT-PCR (qRT-PCR) was performed targeting the spike (Table 1 ) and ORF1a (Table S1) genes as previously described (Zhu et al., 2020) using the SYBR Green kit (iQ™ SYBR Green supermix kit, Bio-Rad, Hercules, CA, USA). The number of viral RNA copies was calculated by comparison to the number of copies of a standard control. In the NMC-nCoV02 infected group, viral spike RNA was detected in all specimens at 2 dpi. The highest amount of viral RNA was detected in nasal washes and peaked at 4 dpi (3.83 log10 copies/mL), persisting until 8 dpi before dropping below detection limits at 10 dpi (Table 1). The virus was also detected in saliva specimens from 2 dpi (1.73 log10 copies/mL) through 8 dpi. Although viral spike RNA was detected in sera of infected ferrets, the viral copy number was low (peaked titer 0.35 log10 copies/mL) and dropped below detection limits earlier than in nasal wash and saliva specimens. To evaluate the infectious virus titer in each specimen, collected nasal washes and saliva specimens were inoculated onto Vero cells for virus isolation. In IN infected ferret group, NMC-nCoV02 was isolated from both saliva and nasal washes specimens as early as 2 dpi and persisted until 4 and 6 dpi, respectively (Table 1). Nasal washes specimens showed higher virus titers (1.83–2.88 log10 TCID50/mL) than saliva specimens (0.82–0.92 log10 TCID50/mL). In DC ferret group, virus was isolated from the nasal washes at 4 dpc (2.4 log10 TCID50/mL) and 6 dpc (1.0 log10 TCID50/mL) but not in saliva specimens (Table 1). Because gastrointestinal involvement is a characteristic of coronavirus infections of animals and humans (Leung et al., 2003), we also collected fecal and urine specimens. Viral RNA was detected in a majority of collected specimens in both IN-infected and DC groups as early as 2 dpc (Table 1). Similarly to the IN infected group, the DC group had the highest virus copy numbers (3.27 log10 copies/mL) in nasal washes, with RNA detected through 8 dpc. In addition, viral RNA was detected in saliva and fecal specimens of the DC group for 8 days, whereas the urine specimens contained detectable viral RNA until 4 dpc. For the IC group, 2 out of 6 ferrets were positive for viral RNA in nasal washes and fecal specimens at 4 dpc, although viral RNA copy numbers were lower (0.53 and 0.52 log10 copies/mL, respectively) than in DC ferrets. Due to the cytotoxicity of urine and fecal specimens of ferrets, we could not assess virus isolation and titer in Vero cells. To evaluate the presence of infectious NMC-nCoV02 in urine and fecal specimens, urine or fecal specimens (at 4 dpi) of IN-infected DC or IC ferrets were centrifuged to remove the debris, and the supernatants were inoculated into naive ferrets (n = 3) per each specimen. Nasal washes from specimen-inoculated ferrets were collected at 2, 4, and 6 dpi and infected onto Vero cells for virus isolation. Noticeably, NMC-nCoV02 was isolated from the nasal wash specimens of 2 out of 3 urine-specimen-treated or fecal-specimen-treated ferrets (Table 1). However, we failed to re-isolate virus from the ferrets infected with the fecal specimens of IC ferrets. These results indicate that ferret is highly susceptible for the infection of SARS-CoV-2 derived from body fluids, and infectious SARS-CoV-2 sheds through urine and fecal specimens of infected ferrets.
Table 1.
Quantitation of Viral RNA in Specimens (Serum, Feces, Nasal Wash, Saliva, and Urine) from Each Group of Ferrets
| Route | Ferret groups | Days post treatment; log10 copies/mL (log10 TCID50/mL)a |
|||||
|---|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | 12 | ||
| Serum | Infected | 0.35 ± 0.08 | 0.35 ± 0.08 | - | - | - | - |
| DC | - | - | - | - | - | - | |
| IC | - | - | - | - | - | - | |
| Naive | - | - | - | - | - | - | |
| Nasal washes | Infected | 2.67 ± 1.01∗∗(2.17 ± 0.94∗) | 3.83 ± 0.94∗∗∗(2.88 ± 0.84∗∗∗) | 2.67 ± 0.63∗∗(1.83 ± 0.63∗) | 1.40 ± 1.06 | - | - |
| DC | 0.67 ± 0.34∗ | 3.27 ± 1.31(2.40 ± 1.17) | 1.48 ± 0.23(1.00 ± 0.25) | 1.38 ± 1.00 | - | - | |
| IC | - | 0.53 ± 0.36 | 0.39 ± 0.17∗ | 0.38 ± 0.16 | - | - | |
| Naive | - | - | - | - | - | - | |
| Saliva | Infected | 1.73 ± 0.54∗∗(0.92 ± 0.38) | 1.67 ± 0.94∗(0.82 ± 0.62) | 0.60 ± 0.47 | 0.50 ± 0.49 | - | - |
| DC | 0.52 ± 0.33 | 0.85 ± 0.48∗ | 0.53 ± 0.21 | 0.38 ± 0.2 | - | - | |
| IC | - | - | - | - | - | - | |
| Naive | - | - | - | - | - | - | |
| Urine | Infected | 0.81 ± 0.56 | 0.87 ± 0.53 (2/3)b | 0.52 ± 0.40 | 0.35 ± 0.12 | - | - |
| DC | 0.72 ± 0.42 | 1.08 ± 0.81 (2/3) | - | - | - | - | |
| IC | - | - | - | - | - | - | |
| Naive | - | - | - | - | - | - | |
| Fecal | Infected | 1.37 ± 0.38∗ | 1.51 ± 0.52∗∗ (2/3) | 0.77 ± 0.73 | 0.53 ± 0.38 | - | - |
| DC | 0.42 ± 0.10 | 1.40 ± 0.51∗ (2/3) | 0.92 ± 1.04 | 0.80 ± 0.80 | - | - | |
| IC | - | 0.52 ± 0.44 (0/3) | 1.08 ± 0.73∗ | - | - | - | |
| Naive | - | - | - | - | - | - | |
Infected: NMC-nCoV02 infected group; DC, directly contacted group; IC, indirectly infected group. Asterisks indicate statistical significance compared with naive sample by the Ordinary one-way ANOVA with Dunnett’s multiple comparisons test (∗ indicates p < 0.05, ∗∗ indicates p < 0.001, and ∗∗∗ indicates p < 0.0001).
Virus spike RNA gene detection limit and viral titer limit were 0.3 log10 copies/mL and 0.8 log10 TCID50/mL, respectively.
Isolated viruses from nasal wash samples inoculated in ferrets.
To assess the replication of SARS-CoV-2 in ferret organs, an additional 12 ferrets were infected with NMC-nCoV02 or PBS via the IN route and 3 ferrets were sacrificed at 4, 8, and 12 dpi. Nasal turbinate, trachea, lung, kidney, and intestine tissues were collected using individual scissors to avoid cross contamination. The highest viral RNA levels were detected in nasal turbinate (4.2 log10 copies/g) and lung tissue (1.53 log10 copies/g) at 4 dpi. Viral RNA was also detected in intestine (0.93 log10 copies/g) and kidney (0.87 log10 copies/g) at 4 dpi. At 8 dpi, viral RNA was still detected in nasal turbinate, trachea, lungs, kidney, and intestine (Figure 1B). In correlation with viral RNA copy numbers (Figure 1B), the highest infectious virus titer was detected in nasal turbinate (3.23 log10 TCID50/g) and lung tissue (1.4 log10 TCID50/g) at 4 dpi, whereas infectious virus recovery failed from trachea, kidney, and intestine tissues, which carried less than 1.13 log10 viral RNA copies/g (Figure 1C). Finally, infectious NMC-nCov02 was isolated from nasal turbinate (2.07 log10 TCID50/g) and trachea (1.07 log10 TCID50/g) at 8 dpi but not from other tissues at 8 dpi (Figure 1C). However, both viral RNA detection and virus recovery failed in all tested tissues at 12 dpi. These results suggest that virus isolation from infected tissues is closely related to viral RNA copy number.
To further confirm viral replication in infected ferrets, immunohistochemistry (IHC) and histopathological examinations were conducted (Figure 1 and Figure S2). Briefly, tissue samples were collected from NMC-nCoV02 infected or PBS-treated ferrets at 4 dpi and incubated in 10% neutral-buffered formalin for virus inactivation and tissue fixation before they were embedded in paraffin. The embedded tissues were sectioned and dried for 3 days at room temperature. To detect the viral antigens by IHC, mouse polyclonal antibody raised by the immunization of mice with inactivated NMC-nCoV02 virions was used as a primary antibody. Slides were viewed using the Olympus BX53 (Olympus, Tokyo, Japan) microscope with DP controller software to capture images. IHC analyses showed that a number of cells in the nasal turbinate, trachea, lung, and intestine sections of NMC-nCoV02-infected ferrets (Figures 1I–1L), but not PBS-treated control ferrets (Figures 1E–1H), were positive for SARS-CoV-2 antigen. Further, the lung histopathology showed that, compared with PBS-treated ferrets, NMC-nCoV02-infected ferrets at 4 dpi showed increased immune infiltration and cell debris in the alveolar wall, bronchial epithelium, and bronchial lumen (Figure S2), evidencing acute bronchiolitis by NMC-nCoV02 infection.
After 12 days of infection, all remaining ferrets, including IN infection (n = 6), DC (n = 6), and IC (n = 6), had returned to normal ranges of body temperature and body weight, and all specimens were negative for viral RNA. To evaluate the seroconversion rate of each group, sera were collected from all remaining ferrets and a serum-neutralizing (SN) antibody assay against NMC-nCoV02 (100 TCID50) was conducted on Vero cells. Although IN infection group showed the highest mean SN titers compared the other groups, the SN titers of both IN infection and DC groups ranged between 32 and128 (Figure 1D). On the other hand, only 1 of 6 IC ferrets showed a positive SN titer of 16. Taken together, this demonstrates the presence of SARS-CoV-2 in multiple sources from infected ferrets, potentially explaining the rapid transmission to naive hosts in close contact with the infected hosts.
Given the rapid geographical spread of COVID-19, the WHO declared the SARS-CoV-2 outbreak a public health emergency of international concern (PHEIC) on the 30th of January, 2020 (WHO, 2020a) and labeled the COVID-19 outbreak a pandemic by the 12th of March, 2020 (WHO, 2020). Most confirmed COVID-19 patients at this time reported close epidemiological association (direct or indirect) with other COVID-19 patients. Interestingly, a growing number of individuals with no travel history to China and no direct contact with infected patients have become infected (Lim et al., 2020). To understand how this virus rapidly spreads within a community, and to inform infection control messaging, it is essential to develop an experimental animal model that can support the active infection, shedding, and transmission of SARS-CoV-2 to sentinel animals. In this study, we established an infection and transmission ferret animal model for COVID-19. The SARS-CoV-2 was found to efficiently infect ferrets and induce moderate increases in body temperature (∼38.5-40.3°C). Moreover, we were able to detect viral RNA in blood (for 4 dpi), nasal washes (for 8 dpi), urine (for 8 dpi), and fecal (for 8 dpi) specimens. Findings suggest that SARS-CoV-2 can be shed through multiple routes of body discharge specimens, with these potentially serving as sources for viral transmission to those in close contact with infected individuals.
Interestingly, ferrets in direct contact with SARS-CoV-2-infected ferrets were positive for SARS-CoV-2 infection as early as 2 dpc, suggesting that rapid transmission occurred even prior to infected ferrets reaching their highest viral RNA copy numbers in nasal washes at 4 dpi. Transmission also occurred prior to peak body temperature and body weight loss in infected animals, which is consistent with the infectiousness of individuals during asymptomatic periods. With regard to potential airborne transmission of SARS-CoV-2, viral RNA was detected in nasal washes and fecal specimens in IC ferrets and persisted for 4 days after indirect contact; only one of the two positive animals seroconverted. These data show that airborne transmission is likely but is considerably less robust than direct contact transmission.
Following the fortuitous discovery of the natural susceptibility of ferrets to human influenza viruses, ferret models were found to highly reproduce the human disease manifestation of several respiratory viruses, including respiratory syncytial virus, parainfluenzaviruses, and SARS-CoV-1 (Capraro et al., 2008, Chan et al., 2018, Enkirch and von Messling, 2015, Park et al., 2018). In addition to the presence of the respective viral receptors, the anatomic proportions of the ferret upper and lower respiratory tracts, the density of submucosal glands in the bronchial wall, and the number of generations of terminal bronchioles all reproduce the condition in the human respiratory tract (Enkirch and von Messling, 2015). This further supports the significance of ferrets as animal model for human respiratory viral infection. We demonstrated that SARS-CoV-2-infected ferrets showed high virus titers in upper respiratory tracts (nasal washes) and consequently transmitted to naive ferrets by direct contact at high efficiency, suggesting that SARS-CoV-2 ferret model recapitulates aspects of human infection and transmission. Further, as suspected in recent COVID-19 patients (Kim et al., 2020, Xu et al., 2020), we detected the infectious viruses in urine and fecal specimens of virus-infected ferrets. However, there are also limitations in the SARS-CoV-2 ferret model, as SARS-CoV-2 infected ferrets showed only mild clinical symptoms and relatively lower virus titers in lungs of infected animals than SARS-CoV-1-infected or MERS-CoV-infected hACE2 or hDPP4 transgenic mice (Glass et al., 2004 and Li et al., 2017). On the other hand, it is also possible that SARS-CoV-2 replicates weaker but persists longer in vivo than SARS-CoV-1, ultimately leading an asymptomatic carrier with a persistent infection to effectively spread the virus. Therefore, given the rapid spreading characteristics of SASRS-CoV-2 in humans, ferret model would be a useful tool to evaluate the efficacy of prophylactic anti-virals and preventive vaccines.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| In-house mouse polyclonal antibody | This study | N/A |
| Bacterial and Virus Strains | ||
| SARS-CoV-2; NMC-nCoV02 | This study | N/A |
| Biological Samples | ||
| Ferret nasal wash samples | This study | See Table 1 |
| Ferret blood samples | This study | See Table 1 |
| Ferret saliva samples | This study | See Table 1 |
| Ferret urine samples | This study | See Table 1 |
| Ferret fecal samples | This study | See Table 1 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trypsin | Thermo Fisher Scientific | Cat#15090-046 |
| Carbo-free blocking Solution | VECTOR | Cat#SP-5040 |
| iQ SYBR green supermix | Biorad | Cat#1708882 |
| Penicillin-Streptomycin | GIBCO | Cat#15140-122 |
| Critical Commercial Assays | ||
| Omniscript RT kit | QIAGEN | Cat#205113 |
| RNeasy mini kit | QIAGEN | Cat#74106 |
| Vecstain ABC kit | VECTOR | Cat#PK-6102 |
| DAB substrate kit, peroxidase | VECTOR | Cat#SK-4100 |
| Experimental Models: Cell Lines | ||
| African green monkey: Vero cells | ATCC | Cat#ATCC CCL-81; RRID: CVCL_0059 |
| Experimental Models: Organisms/Strains | ||
| Ferret (Mustela putorius furo) | ID BIO | N/A |
| Oligonucleotides | ||
| SARS-CoV-2 S F: attcaagactcactttcttccaca | This study | See Table 1 |
| SARS-CoV-2 S R: tgtttaaagcttgtgcattttggttgacc |
This study | See Table 1 |
| SARS-CoV-2 ORF1a F: ccctgtgggttttacacttaa |
This study | See Table S1 |
| SARS-CoV-2 S ORF1a R: tcagctgatgcacaatcgt |
This study | See Table S1 |
| Software and Algorithms | ||
| GraphPad Prism 8.3.1 | N/A | https://www.graphpad.com/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Young Ki Choi (choiki55@chungbuk.ac.kr).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data Code and Availability
This study did not generate any unique datasets or code.
Experimental Model and Subject Details
Experimental Animals
Male and female ferrets, 12- to 20- month old and sero-negative for influenza A viruses, MERS-CoV, and SARS-CoV (ID Bio Corporation) were maintained in the isolator (woori IB Corporation) in BSL3 of Chungbuk National University. All ferrets were group hosed with a 12 h light/dark cycle and allowed access to diet and water. All animal studies were carried out in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) in Chungbuk National University.
Growth and Isolation of Virus
Virus was isolated from an isolate of SARS-CoV-2 from a COVID-19 confirmed patient in Korea. To infect the animal, viruses were propagated on the Vero cells in the DMEM medium (GIBCO) supplemented with 1%penecillin/streptomycin (GIBCO) and TPCK trypsin (0.5ug/mL; Worthington Biochemical) at 37°C for 72 h. Propagated viruses were stored at −80°C freezer for future usage.
Method Details
Study Design for Animal-to-Animal Transmission
12 −24 month old male and female ferrets, which were confirmed as Influenza A (H1N1, H3N1), MERS-CoV, and SARS-CoV antibody free ferrets by the standard enzyme-linked immunosorbent assay (ELISA) previously described elsewhere (El-Duah et al., 2019, Park et al., 2014, Woo et al., 2005), were infected through intranasal (IN) route with NMC2019-nCoV02 virus, an isolate of SARS-CoV-2 from a COVID-19 confirmed patient in Korea, 2020 February, at a dose of 105.5 TCID50 per ferrets (n = 2). At one-day post-infection, one naive direct contact (DC) and indirect contact (IC) ferrets were introduced into the cage, while IC ferrets were separated from inoculated animals with a partition, which allowed air to move, and without direct contact between animals. This study was conducted with three independent trials. Blood, fecal, nasal wash, saliva, and urine specimens were collected every other day for 12 days from each group of ferrets to detect SARS-CoV-2. Further, to investigate whether each collected specimen contained infectious live virus, we inoculated it onto Vero cells.
To access the replication of the virus in ferrets following SARS-CoV-2 infection in various organs, additional 9 ferrets were infected with SARS-CoV-2 by IN route. Three ferrets were sacrificed at 4, 8 and 12 dpi were and their lung, liver, spleen, kidney, and intestinal tissues were collected with individual scissors to avoid cross contamination.
Quantitative Real-Time RT-PCR (qRT-PCR) to Detect SARS-CoV-2 RNA
Collected ferret secretions were resuspended with cold phosphate-buffered saline (PBS) containing antibiotics (5% penicillin/streptomycin; GIBCO). For virus titration, total RNA was extracted from the collected samples using the RNeasy Mini® kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. A cDNA synthesis kit (Omniscript Reverse Transcriptase; QIAGEN, Hilden, Germany) was used to synthesize single strand cDNA using total viral RNA. To quantify viral RNA and viral copy number, quantitative real-time RT-PCR (qRT-PCR) was performed for the partial Spike gene (Table 1) and ORF1a (Table S1) with the SYBR Green kit (iQ™ SYBR Green supermix kit, Bio-Rad, Hercules, CA, USA), and the number of viral RNA copies was calculated and compared to the number of copies of the standard control.
Immunohistochemistry (IHC)
Tissue samples were collected from PBS control and NMC-nCoV02 infected ferrets and incubated in 10% neutral-buffered formalin for fixation before they were embedded in paraffin based to standard procedures. The embedded tissues were sectioned and dried for 3 days at room temperature. To detect the viral antigen by immunohistochemistry, mouse polyclonal antibody developed by inactivated NMC-nCoV02 was used as the primary antibody. Antigen was visualized using the biotin-avidin system (Vector Labs). Slides were viewed using the Olympus IX 71 (Olympus, Tokyo, Japan) microscope with DP controller software to capture images.
Quantification and Statistical Analysis
Statistical Analysis
The statistical significance of infected and contact samples compared with naive sample was assessed by two-way ANOVA with Sidaks multiple comparisons test and one way ANOVA Dunnett’s multiple comparisons test. While for the comparison of the significance of viral copy number or titer among samples, we use the two-way ANOVA with Dunnett’s multiple comparisons test.
Data plotting, interpolation and statistical analysis were performed using GraphPad Prism 8.2 (GraphPad Software, La Jolla, CA). Statistical details of experiments are described in the figure legends. A p value less than 0.05 is considered statistically significant.
Acknowledgement
All animal experiments were approved by the Medical Research Institute, a member of the Laboratory Animal Research Center of Chungbuk National University (LARC) (approval number CBNUA-1352-20-02), and were conducted in strict accordance and adherence to relevant policies regarding animal handling as mandated under the Guidelines for Animal Use and Care of the Korea Center for Disease Control (K-CDC). Viruses were handled in an enhanced biosafety level 3 (BSL3) containment laboratory as approved by the Korean Centers for Disease Control and Prevention (KCDC-14-3-07).
This work was supported by National Research Foundation of Korea (NRF-2018M3A9H4056536, 2020R1A2C 3008339), the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program (KGM9942011), the National Institute of Health (AI140705, AI140718, AI152190, and AI116585), and the National Institute of Allergy and Infectious Diseases (Centers of Excellence for Influenza Research and Surveillance [CEIRS] contract number HHSN272201400006C).
Author Contributions
Conceptualization: Y.I. Kim, S.J. Park, R.J. Webby, J.U. Jung, and Y.K. Choi; Investigation: Y.I. Kim, S.G. Kim, S.M. Kim, E.H. Kim, S.J. Park, K.M. Yu, J.H. Chang, E.J. Kim, M.A.B. Casel, V.D. Lai, S.H. Lee, J. Um, Y. Kim, B.S. Chin, J.S. Park, H.W. Jeong, S.S. Foo, H. Poo, I.P. Mo, O.J. Lee, M.S. Song, and Y.K. Choi; Writing: Y.I. Kim, S.J. Park, R.J. Webby, J.U. Jung, Y.K. Choi.
Declaration of Interests
Jae U. Jung is a scientific advisor of the Vaccine Stabilization Institute, a California corporation.
Published: April 6, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2020.03.023.
Contributor Information
Jae U. Jung, Email: jaeujung@med.usc.edu.
Young Ki Choi, Email: choiki55@chungbuk.ac.kr.
Supplemental Information
References
- Bao L., Deng W., Huang B., Gao H., Ren L., Wei Q., Yu P., Xu Y., Liu J., Qi F. The Pathogenicity of 2019 Novel Coronavirus in hACE2 Transgenic Mice. bioRxiv. 2020 [Google Scholar]
- Bouvier N.M. Animal models for influenza virus transmission studies: a historical perspective. Curr. Opin. Virol. 2015;13:101–108. doi: 10.1016/j.coviro.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capraro G.A., Johnson J.B., Kock N.D., Parks G.D. Virus growth and antibody responses following respiratory tract infection of ferrets and mice with WT and P/V mutants of the paramyxovirus Simian Virus 5. Virology. 2008;376:416–428. doi: 10.1016/j.virol.2008.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.F., Carolan L.A., Korenkov D., Druce J., McCaw J., Reading P.C., Barr I.G., Laurie K.L. Investigating viral interference between influenza A virus and human respiratory syncytial virus in a ferret model of infection. J. Infect. Dis. 2018;218:406–417. doi: 10.1093/infdis/jiy184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Duah P., Meyer B., Sylverken A., Owusu M., Gottula L.T., Yeboah R., Lamptey J., Frimpong Y.O., Burimuah V., Folitse R. Development of a whole-virus ELISA for serological evaluation of domestic livestock as possible hosts of human coronavirus NL63. Viruses. 2019;11:43. doi: 10.3390/v11010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkirch T., von Messling V. Ferret models of viral pathogenesis. Virology. 2015;479-480:259–270. doi: 10.1016/j.virol.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass W.G., Subbarao K., Murphy B., Murphy P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 2004;173:4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- ICTV . 2020. Naming the 2019 Coronavirus. [Google Scholar]
- Kim J.Y., Ko J.-H., Kim Y., Kim Y.-J., Kim J.-M., Chung Y.-S., Kim H.M., Han M.-G., Kim S.Y., Chin B.S. Viral Load Kinetics of SARS-CoV-2 Infection in First Two Patients in Korea. J. Korean Med. Sci. 2020;35:e86. doi: 10.3346/jkms.2020.35.e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.K., To K.F., Chan P.K., Chan H.L., Wu A.K., Lee N., Yuen K.Y., Sung J.J. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Wohlford-Lenane C.L., Channappanavar R., Park J.-E., Earnest J.T., Bair T.B., Bates A.M., Brogden K.A., Flaherty H.A., Gallagher T. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc. Natl. Acad. Sci. USA. 2017;114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Jeon S., Shin H.-Y., Kim M.J., Seong Y.M., Lee W.J., Choe K.-W., Kang Y.M., Lee B., Park S.-J. Case of the Index Patient Who Caused Tertiary Transmission of COVID-19 Infection in Korea: the Application of Lopinavir/Ritonavir for the Treatment of COVID-19 Infected Pneumonia Monitored by Quantitative RT-PCR. J. Korean Med. Sci. 2020;35:e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Perlman S. Fields Virology. 2013. Coronaviridae; pp. 825–858. [Google Scholar]
- Park S.-J., Kim E.-H., Pascua P.N.Q., Kwon H.-I., Lim G.-J., Decano A., Kim S.M., Song M.K., Shin E.-C., Choi Y.-K. Evaluation of heterosubtypic cross-protection against highly pathogenic H5N1 by active infection with human seasonal influenza A virus or trivalent inactivated vaccine immunization in ferret models. J. Gen. Virol. 2014;95:793–798. doi: 10.1099/vir.0.058636-0. [DOI] [PubMed] [Google Scholar]
- Park S.-J., Kim E.-H., Kwon H.-I., Song M.-S., Kim S.M., Kim Y.-I., Si Y.-J., Lee I.-W., Nguyen H.D., Shin O.S. Altered virulence of Highly Pathogenic Avian Influenza (HPAI) H5N8 reassortant viruses in mammalian models. Virulence. 2018;9:133–148. doi: 10.1080/21505594.2017.1366408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QIAGEN . 2012. RNeasy Mini Handbook. [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS Coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. WHO Director-General’s opening remarks at the media briefing on COVID-19. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020. [Google Scholar]
- WHO 2019-nCoV outbreak is an emergency of international concern. 2020. http://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/01/2019-ncov-outbreak-is-an-emergency-of-international-concern [DOI] [PMC free article] [PubMed]
- WHO Coronavirus disease 2019 (COVID-19) Situation Report – 63. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200323-sitrep-63-covid-19.pdf?sfvrsn=d97cb6dd_2
- Woo P.C., Lau S.K., Wong B.H., Tsoi H.W., Fung A.M., Kao R.Y., Chan K.H., Peiris J.S., Yuen K.Y. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 2005;43:3054–3058. doi: 10.1128/JCM.43.7.3054-3058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020 doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon L., Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



