Abstract
RNAs are post-transcriptionally modified by dedicated writer or eraser enzymes that add or remove specific modifications, respectively. Mass spectrometry (MS) of RNA is a useful tool to study the modification state of an oligonucleotide (ON) in a sensitive manner. Here, we developed an ion-pairing reagent free chromatography for positive ion detection of ONs by low- and high-resolution MS, which does not interfere with other types of small compound analyses done on the same instrument. We apply ON-MS to determine the ONs from an RNase T1 digest of in vitro transcribed tRNA, which are purified after ribozyme-fusion transcription by automated size exclusion chromatography. The thus produced tRNAValAAC is substrate of the human tRNA ADAT2/3 enzyme and we confirm the deamination of adenosine to inosine and the formation of tRNAValIACin vitro by ON-MS. Furthermore, low resolution ON-MS is used to monitor the demethylation of ONs containing 1-methyladenosine by bacterial AlkB in vitro. The power of high-resolution ON-MS is demonstrated by the detection and mapping of modified ONs from native total tRNA digested with RNase T1. Overall, we present an oligonucleotide MS method which is broadly applicable to monitor in vitro RNA (de-)modification processes and native RNA.
INTRODUCTION
Ribonucleic acids (RNA) contain a vast variety of chemical modifications, which derive from the four canonical nucleosides adenosine, guanosine, uridine and cytidine. Modifications are introduced by dedicated enzymes, sometimes referred to as RNA writers. In analogy, RNA erasers exist, which demethylate adenosine in messenger RNA (mRNA) (1,2) or transfer RNA (tRNA) (3,4). Many RNA modifying enzymes add methyl groups to either the nucleobase or the ribose. For example, tRNA methyltransferase 1 (TRMT1) is responsible for the dimethylation of guanosine to 2,2-dimethylguanosine in tRNA (m22G) (5) and methyltransferase like proteins 3/14 (METTL3/METTL14) methylate position 6 of adenosine and the epitranscriptomic mark m6A forms in mRNA. In addition to RNA modification by methylation, the conversion of adenosine to inosine by deaminases such as the adenosine deaminase tRNA specific enzyme 2/3 (ADAT2/3) has been reported (6). Interestingly, many neurological diseases are connected with mutations in RNA modifying enzymes such as TRMT1 (5) and ADAT2/3 (7). In addition to the active decoration of RNA with modifications, the removal by active demethylation is also possible. Demethylation of m6A and its ribose methylated variant m6Am has been reported in human mRNA (1,2). Bacterial RNAs, including tRNA and rRNA, are also methylated, however there are no reports of active demethylation of enzymatically methylated sites. In many bacteria, the methylation of adenosine at position N1 (1-methyladenosine, m1A) is not found in DNA or tRNA. However, alkylation stress can lead to direct m1A formation in DNA and RNA. Due to methylating agents, which chemically methylate nucleic acids, bacteria have the ability for active demethylation. This m1A is removed by the alpha-ketoglutarate-dependent dioxygenase AlkB (8). In human tRNAs, m1A is found in 42% of all tRNAs at position 58. m1A58 has been reported to be substrate to the human homologues of AlkB, namely ALKBH1 (3) and ALKBH3 (9).
The detection of modified moieties in RNA is possible by chemical means (10), by sequencing (11) and mass spectrometry (MS) (12). Even with the ever-rising number of sequencing techniques, which detect modified nucleosides in whole transcriptomes, MS remains the key technique for characterization of modified nucleosides. RNA MS analytics can be subdivided into three major principles. The first relies on complete enzymatic digestion of the RNA into the nucleoside building block and is highly sensitive with lower limits of detection (LLOD) in the fmol and amol range (nucleoside-MS). This technique is commonly used for detection (12), quantification (13) or discovery (14,15) of modified nucleosides. The second uses enzymes, which only partially digest the RNA and smaller oligonucleotides (ON) emerge (in this manuscript referred to as ON-MS). In the case of ON-MS, some of the sequence context surrounding a modified nucleoside is preserved and the technique is used to place modified nucleosides in known and unknown RNA sequences. Here, the pioneering work of the McCloskey (16,17), Limbach (18) and Suzuki (19) lab have largely contributed to the establishment of ON-MS. Disadvantages of this bottom-up approach are the loss of (most) sequence information, congestion of peaks in a small mass range, which mess up spectra, as well as the detection loss of low-abundant-modified RNAs by domination of their unmodified counterparts (20–22). The third principle of MS-based RNA analytics is the analysis of full-length RNAs (top-down-MS) at sophisticated mass spectrometers which has been pioneered by the Breuker lab (23–26). Top-down-MS of RNA is suited for many types of modification and it reveals the sample heterogeneity. It provides a high sequence coverage and information without a time-consuming digestion step. The disadvantage of top-down approaches is the need for RNA shorter ∼60 nts and the available dissociation steps in the MS field are low-efficiency processes. Another challenge is the underdevelopment of back-end bioinformatic tools. Nevertheless, quantitation of modified nucleobases has been also shown by top-down-MS (27). Both bottom-up- and top-down-MS approaches utilize MS/MS analysis and the underlying RNA dissociation reactions have been reviewed (28). From the perspective of instrumentation, high-resolution mass spectrometers are ideally suited for the MS and MS/MS analysis of RNA and its fragments. Instruments, such as time-of-flight, iontrap or orbitrap MS, deliver the necessary resolution to determine the charge state of the ONs and they allow sequence prediction based on their accuracy. Low resolution instruments, such as triple quadrupole MS, are not commonly applied for oligonucleotide MS analysis as they lack the resolution to distinguish similar ONs and to clearly determine the charge state of an ionized ON.
In contrast to top-down-MS, bottom-up-MS relies on the chromatographic separation of the ONs to solve the problem of congested MS spectra. The liquid chromatography is achieved by using an RP-18-based column and an ion-pairing reagent such as triethyl-ammonium-acetate (TEAA), which separates the oligonucleotides by their length. The eluting oligonucleotides are then analyzed in negative ionization mode by a high-resolution mass spectrometer and sequenced according to their fragmentation pattern (29,30) or modification footprints (31). With TEAA dependent flow chromatography (32), it is now possible to analyze 2–5 ng of purified tRNA isoacceptors (∼80–200 fmol pure tRNA) and determine the sequence and modification status of these tRNAs (33). Due to the common use of organic bases as ion-pairing reagents for ON separation and thus negative ionization, oligonucleotide MS is mostly used in labs with liquid chromatography coupled mass spectrometry (LC-MS) instruments dedicated for ON analysis. For other labs, the limitation of using ion-pairing reagents is the difficulty of their removal from the instrument. Residual ion-pairing reagents stay on the LC system and interfere with other types of chromatography and in addition they reduce the sensitivity of the mass spectrometer. To overcome this problem ion-pairing reagent free chromatography can be used on a reverse phase (RP) column (34). Here, the retention behavior of oligonucleotides is unexplored. Another alternative is chromatography on a hydrophilic interaction liquid chromatography (HILIC) column which will allow separation in a similar fashion to ion-pairing chromatography (35). Both methods are reported in combination with negative ionization mode MS detection and are thus not applicable for labs with mass spectrometers that preferably operate in positive ionization mode. Recently, a method was presented which used positive ionization detection of oligonucleotides after ion-pairing chromatography (36). Thus, laboratories have currently the option of doing classical ON-MS (ion-pairing reagent chromatography in negative ionization mode), ion-pairing reagent free ON-MS in negative mode or ion-pairing chromatography in positive mode. Thus, there is currently no method available which overcomes both limitations for a broad application of ON-MS.
A general challenge for MS-based modification analysis is its non-quantitative nature. For quantification, the signal intensity of an analyte must correlate with its concentration or amount. In MS, the signal intensity depends of course on the amount of analyte, but in addition on a multitude of other parameters such as salt load, ionization properties of the analyte, instrument parameters and so on. These detection fluctuations make quantification by MS a challenging task, which can only be done by using stable isotope labeled internal standards (SILIS) of the analyte of interest. For nucleoside-MS, this problem has been overcome by synthetic (37) or biosynthetic (13,38) preparation of stable isotope labeled nucleosides. For ON-MS, biosynthetic approaches have been reported (39,40). Another elegant way to solve the problem was presented by the Limbach lab (41). They performed the enzymatic digest of the unknown RNA in the presence of H218O, which results in oxygen-18 incorporation into the oligonucleotide 3′-phosphate. As a third option, in vitro transcribed RNA is prepared in the presence of stable isotope-labeled nucleoside triphosphates (NTP) that is then used as an internal standard (42,43).
Although ON-MS has become a powerful tool for analysis of RNA modifications within their sequence context, it is not commonly applied. Due to the benefits of ON-MS, we developed a TEAA free chromatography, which separates the ONs not by length, but by chemical composition of the sequence. In this manuscript, we describe the development and separation principle of the method using synthetic ONs. Detection is achieved by low-resolution and high-resolution MS in positive ionization mode. We present MS/MS data for the analyzed ONs and determine the LLOD in various detection modes. We describe the automated purification of unlabeled and stable isotope labeled in vitro transcripts of tRNAValAAC and tRNASerUGA by ribozyme fusion transcription. These in vitro transcripts and native tRNA from HEK cells are analyzed by our ON-MS method. To verify correct folding of the produced tRNAs, we use the adenosine-to-inosine deaminating enzyme ADAT2/3 on tRNAValAAC. Inosine formation is observed by both nucleoside-MS and ON-MS. Importantly, these experiments are done on the same day using the same instrument which highlights the compatibility of our ON-MS method with sensitive small compound analysis. Furthermore, we use the developed ON-MS method to monitor the demethylation of short oligonucleotides containing 1-methyladenosine by bacterial AlkB in vitro.
Overall, we provide a method for automated purification of RNA transcripts and a broadly applicable ON-MS method for instruments commonly used for other types of small compound analysis, especially nucleoside analysis.
MATERIALS AND METHODS
Salts, reagents and nucleosides
All salts, solvents and reagents were obtained from Sigma Aldrich (Munich, Germany) at molecular biology grade unless stated otherwise. All solutions and buffers were made with water from a Millipore device (Milli-Q, Merck, Darmstadt, Germany). Nucleosides: adenosine (A), cytidine (C), guanosine (G) and uridine (U) were purchased from Sigma Aldrich. 1-methyladenosine (m1A) and inosine (I) were purchased from Carbosynth (Newbury, UK).
Oligonucleotides
All oligonucleotides were delivered in a stock concentration of 100 μM in water and are listed in Supplementary Table S1.
AlkB in vitro assay
An aliquot of bacterial AlkB protein (Peak Proteins, Cheshire, UK) was thawed on ice. Every assay was performed in a volume of 50 μl with a final concentration of 50 mM TRIS HCl pH 7.5, 15 mM KCl, 2 mM L-ascorbate, 300 μM α-ketoglutarate, 300 μM Fe(II) (NH4)2(SO4)2 × 6H2O. L-ascorbate, α-ketoglutarate and diammonium iron (II) sulfate hexahydrate stock solutions were made afresh. A total of 10 μM of the synthetic RNA oligonucleotide was incubated with 1 μM AlkB enzyme. All assays were incubated at 37°C for 1 h and immediately stopped afterward by filtering through a molecular weight cut-off filter (VWR, Partnumber: 516-0229) for oligonucleotides or RNA precipitation for tRNA in vitro transcripts, respectively.
tRNA digestion for nucleoside mass spectrometry
Up to 1 μg RNA in 30 μl aqueous digestion mix were digested to single nucleosides by using 0.2 u Alkaline Phosphatase, 0.02 u Phosphodiesterase I (VWR, Radnor, PA, USA) and 0.2 u Benzonase in 5 mM TRIS (pH 8.0) and 1 mM MgCl2. Furthermore, 0.5 μg tetrahydrouridine (Merck, Darmstadt, Germany), 1 μM butylated hydroxytoluene and 0.1 μg pentostatine were added. The mixture was incubated with the RNA for 2 h at 37°C and filtered through 96-well filterplates (AcroPrep™ Advance 350 10 K Omega™, PALL Corporation, New York, USA) at 4°C for 30 min at 3000 × g, or through single tubes (VWR, Partnumber: 516-0229) at room temperature for 7 min at 5000 × g. The filtrate was mixed with 1/10 Vol. of 10× yeast SILIS (stable isotope labeled internal standard) (38) for absolute quantification.
Nucleoside mass spectrometry
For nucleoside-MS measurements, a liquid chromatography unit (1290 Infinity II, Agilent Technologies, Waldbronn, Germany) equipped with a diode-array detector (DAD, Agilent Technologies) was used that was interfaced with a triple quadrupole mass spectrometer (G6470A, Agilent Technologies) via an electrospray ionization (ESI) source (Jet Stream, Agilent Technologies). For separation of nucleosides, a Synergi Fusion-RP column (Phenomenex®, Torrance, California, USA; Synergi® 2.5 μm Fusion-RP 100 Å, 150 × 2.0 mm) at 35°C and a flow rate of 0.35 ml/min were used. The eluents were 5 mM NH4OAc, brought to pH 5.3 with glacial acetic acid (buffer A), and pure acetonitrile (buffer B). The gradient started at 100% A for 1 min, followed by an increase of solvent B to 10% over 5 min. From 5 to 7 min, solvent B was increased to 40% and was maintained for 1 min before returning to 100% solvent A in 0.5 min and a 2.5 min re-equilibration period. The QQQ mass spectrometer was operated in dynamic multiple reaction monitoring (dMRM) mode between 1.1 min and 9 min with a cell accelerator voltage of 5 eV. Operating parameters: positive-ion mode, skimmer voltage of 15 V, cell accelerator voltage of 5 V, N2 gas temperature of 230°C and N2 gas flow of 6 l/min, sheath gas (N2) temperature of 400°C with a flow of 12 l/min, capillary voltage of 2500 V, nozzle voltage of 0 V and nebulizer at 40 psi. The detailed mass spectrometric parameters for each nucleoside are given in Supplementary Table S2.
Calibration for nucleoside mass spctrometry
For calibration, synthetic nucleosides Cytidine, Uridine, Guanosine, Adenosine, 1-methyladenosine (m1A) and Inosine (I) were weighed and dissolved in water to a stock concentration of 1–10 mM. Calibration solutions ranging from 0.15 to 500 pmol for each canonical nucleoside and from 0.15 to 500 fmol for each modified nucleoside were prepared by serial dilution (1:10). The calibration solutions were mixed with 1/10 Vol. of 10× yeast SILIS and analyzed by nucleoside-MS. Data were analyzed using Agilent's Quantitative or Qualitative Software. The absolute amount determined for m1A and I was normalized to the amount of injected RNA as determined by the absolute abundance of all four canonical nucleosides (38).
Mammalian cells
HEK 293T and HeLa ACC 57 cells (DSMZ, Braunschweig, Germany) were cultured in Dulbecco’s Modified Eagle Medium (DMEM). DMEM medium was prepared by dissolving 8.4 g DMEM powder D5030 in 1 l pure water. Before sterile filtration, carbonate and phenol red were added to a final concentration of 3.7 g/l NaHCO3 and 0.0159 g/l phenol red. Stocks of glucose (225 g/l) and L-glutamine (15 g/l) were prepared and sterile filtered. These solutions were added to the DMEM medium before usage to a final concentration of 4.5 g/l glucose, 0.584 g/l L-glutamine and 10% fetal calf serum (FCS). The methionine concentration was 0.15 g/l in the final media. For splitting, the cells were treated with TrypLE Express (Gibco, Carlsbad, CA, USA). The cells were incubated and cultivated at 10% CO2 atmosphere.
RNA isolation
HEK and HeLa cells were harvested directly in cell culture flasks using 1 ml TRI-Reagent® per 25 cm2. Isolation was performed according to the manufacturer's protocol with chloroform (Roth, Karlsruhe, Germany). The RNA was finally dissolved in 30 μl water.
PCR
All polymerase chain reactions (PCR) were performed in a total volume of 50 μl with a final concentration of 1-fold Phusion Buffer HF (New England Biolabs, Ipswich, MA, USA) and 0.8 μM forward and reverse primer. The sequence of templates and primers are given in Supplementary Table S1. Additionally, 1 μl dNTPs, 0.5 μl Phusion polymerase and 100 ng of the desired DNA template were added. All samples were amplified with the same PCR program: 95°C for 2 min, 95°C for 30 s for 20 amplification cycles, 57°C for 30 s for 20 times and 68°C for 1 min for 20 times. At the end of the program, the PCR reaction was incubated at 68°C for 1 min and was cooled down to 4°C. Every PCR reaction was performed twice and pooled afterward for the T7 in vitro transcription.
T7 invitro transcription
The total volume of the T7 in vitro transcription was 200 μl. A total of 100 μl PCR product were added to T7 buffer mix and T7 enzyme (TranscriptAid T7 High Yield Transcription Kit, Thermo Fisher Scientific, Waltham, MA, USA) and 1.6 μl of each rNTP (14N-rNTPs were provided by the kit, 15N-rNTPs were purchased by Silantes, Munich, Germany, Partnumber.: 121306100). The mixture was incubated for 2 h at 37°C and 600 rpm. After 2 h incubation, the sample was treated with 2 μl T7 enzyme mix and 5 μl 50 mM MgCl2 and incubated for additional 2 h. After another 2 h incubation, the sample was treated again with 2 μl T7 enzyme mix and 5 μl 50 mM MgCl2 and incubated for additional 2 h to improve the yield of the transcription. In total, the transcription was finished after 6 h. DNA template was removed by addition of 4 μl DNase 1, which is provided in the kit, 1 h at 37°C. In the next step, MgCl2 was added with a final concentration of 5 mM and the sample was incubated at 60°C for 1 h to auto-catalytically cleave the precursor in vitro transcript into its target tRNA. Prior to RNA precipitation, the sample was centrifuged at 5000 × g for 5 min at room temperature to remove the insoluble pyrophosphate of the transcription reaction. The supernatant was precipitated by addition of 0.1 Vol. of 5 M ammonium acetate and 2.5 Vol. of ice-cold ethanol (100%) followed by overnight incubation at −20°C. The RNA was pelleted by centrifugation (12 000 × g, 40 min, 4°C), washed with 70% ethanol and resuspended in 30 μl water.
Size exclusion chromatography (SEC)
For purification of tRNA from total RNA or purification of tRNA in vitro transcripts from precursor products, size exclusion chromatography (SEC) was used on an Agilent 1100 HPLC system (Degasser, G1279A; Quad Pump, G1311A; ALS, G1313A; COLCOM, G1316A; VWD, G1314A; Analyt FC, G1364C). For method development, three columns with 7.8 × 300 mm, 2.7 μm particle size from Agilent (Waldbronn, Germany) were tested. The SEC-3 (Partnumber: 5190-2511 reported by Dedon lab (44)), the AdvanceBio 300 Å (Partnumber: 1180-5301) and the AdvanceBio 130 Å (Partnumber: 1180-5350).
The column temperature was set to 40°C for native tRNA purification using the Advance Bio 300 Å and 60°C for in vitro transcript purification using the AdvanceBio 130 Å. For elution, a 1 ml/min isocratic flow of 0.1 M ammonium acetate was used. Eluting RNA was detected at 254 nm with a diode array detector. The eluted RNA was collected by a fraction collector and the eluent was evaporated (GeneVac, EZ-2 PLUS, Ipswich, UK) to a volume of ∼50 μl before ethanol precipitation. The purified RNA was dissolved in 30 μl water for further enzymatic assays or MS analysis.
Handling guide for SEC columns
For prolongation of the column lifetime, it is essential to avoid sudden pressure changes. Thus we recommend to run a conditioning method, which slowly increases the flow rate from 0 ml/min to 1 ml/min within 20 min. For tRNA purification from total RNA, a column temperature of 40°C is sufficient. For purification of in vitro transcripts, 60°C yielded better separation results. Note: Column lifetime is shortened at 60°C and thus long exposures to high temperatures should be avoided. After use, the column was stored in 0.05% NaN3.
RNA concentration and quality measurements
The RNA concentration was determined with an Implen Nanophotometer NP 80 (Munich, Germany). For quality control of SEC purified in vitro transcripts, the Agilent 2100 Bioanalyzer (Small RNA analysis chip, Partnumber: 5067-1548, Agilent, Waldbronn, Germany) was used.
Isoacceptor purification
The procedure was adapted from Hauenschild et al. (45). For tRNA isoacceptor purification, pre-purified total tRNA was used. The sequence of the biotinylated 2′-deoxyoligonucleotide probes is listed in Supplementary Table S1.
Purification of ADAT2/3
Open Reading Frame sequences encoding for human ADAT2 and ADAT3 were cloned into the petDuet plasmid. ADAT2 was cloned into Multiple Cloning Site (MCS) 1 which allows fusion with a HIS tag. ADAT3 was cloned into MCS2. BL21DE3 RIPL Escherichia coli cells (Agilent) were transformed with the above plasmid. Starter culture was grown to an O.D of 0.1–0.5, and inoculated into a larger culture to an O.D of 0.05 and cells were induced with Isopropyl-β-D-thiogalactoside (IPTG) at an O.D. of 0.6–0.8. After induction, E. coli were grown at 20°C for 15 h. Cells were harvested and lysed by sonication in a buffer containing 20 mM TrisHCl pH 7.6, 5% glycerol, 0.1% Triton, 1 mM Dithiothreitol (DTT), 0.1 mM (Phenylmethylsulfonylfluorid) PMSF, 500 mM NaCl, 25 mM imidazole. Cell lysates were oscillated with HisPur Colbalt Resin (Thermo Scientific # 89964) at 4°C for 2 h. Beads were washed 3× with buffer containing 20 mM TrisHCl pH 7.6, 5% Glycerol, 0.1% Triton, 1 mM DTT, 0.1 mM PMSF, 300 mM NaCl and 25 mM imidazole. Elution of protein was carried out with above buffer containing 300 mM imidazole. Elution buffer was exchanged to buffer containing 20 mM TRIS, 5% Glycerol, 0.1% Triton, 150 mM NaCl and 1 mM DTT. Expression and purification was verified by sodium dodecyl sulphate-PAGE (polyacrylamide gel electrophoresis) followed by western blotting. Quantification of purified protein was carried out by Coomassie with BSA size standards. Final concentration of ADAT2/3 was ∼8 ng/μl.
ADAT2/3 assay
An aliquot of ADAT2/3 protein was thawed on ice. A total of 6 μl tRNA substrate (115 ng/μl) were incubated in 6 μl water and 6 μl melting mix. The melting mix buffer was prepared with 30 mM TRIS pH 7.5 and 1 mM ethylenediaminetetraacetic acid. To denature the tRNA, the mix was heated to 95°C for 2 min and immediately placed on ice for 3 min for tRNA folding. In the next step, 3 μl folding mix was added and incubated for 20 min at 37°C. The folding mix buffer was prepared with final concentrations of 333 mM HEPES pH 7.5, 20 mM MgCl2 and 333 mM NaCl. A total of 9 μl of ADAT2/3 was added to obtain a total reaction volume of 30 μl and incubated for 1 h at 37°C and immediately stopped afterward by RNA precipitation with 300 μl LiClO4 in acetone (2%). After incubation at room temperature for 5 min, the sample was centrifuged at 5000 × g for 5 min. The supernatant was discarded and the tRNA pellet was solved in 20 μl milliQ water for further analysis.
tRNA digestion for oligonucleotide mass spectrometry
RNase T1 was diluted to a 10 U/μl solution by mixing 2 μl of the RNase T1 stock (186 U/μl, Sigma-Aldrich, Munich, Germany) with 35.2 μl TRIS pH 7.5 (25 mM). The diluted RNase T1 should be stored at 4°C. Up to 1 μg RNA was digested with RNase T1 at 37°C for 1 h in a total volume of 50 μl with final concentrations of 25 mM TRIS pH 7.5, 100 mM NaCl and 1 U/μl RNase T1 and 0.2 u/μl Alkaline Phosphatase. The digested samples were filtered through a molecular weight 10 kDa cut-off filter (VWR, Dreieich, Germany, Partnumber: 516-0229) and analyzed by MS.
Oligonucleotide mass spectrometry
For oligonucleotide MS measurements, the instrument described in the nucleoside-MS section was used. For separation of oligonucleotides, a Synergi Fusion-RP column (Phenomenex®, Torrance, CA, USA; Synergi® 2.5 μm Fusion-RP 100 Å, 150 × 2.0 mm) at 35°C and a flow rate of 0.35 ml/min were used. The eluents were 10 mM NH4OAc (pH 7.0) (buffer A) and pure acetonitrile (buffer B) (Carl-Roth, Karlsruhe, Germany; LC-MS grade, purity ≥95.95). The gradient started at 100% buffer A, followed by an increase of B to 5% over 10 min. From 10 to 12 min, buffer B was increased to 50% and was maintained for 1 min before returning to 100% buffer A and a 4 min re-equilibration period. For source optimization experiments a shorter gradient was used starting at 100% buffer A, increased to 2% buffer A over 1 min, increased to 10% A by 4 min, then to 50% A by 5 min, held at 50% A for 0.5 min then returned to 100% A. The same re-equilibration period as the previous method was used. The QQQ mass spectrometer was operated in full scan (MS2Scan), between 500–1000 m/z with a fragmentor voltage of 100 V and a cell accelerator voltage of 5 V in positive ionization mode. For determination of CID spectra, collision energies of 5–40 eV were used and the instrument operated in Product Ion Scan mode. Mass transition of ONs were used in a targeted MRM method. Data were analyzed using Agilent's Qualitative Analysis Software.
High-resolution mass spectra of oligonucleotide ions were recorded by a Thermo Finnigan LTQ Orbitrap XL with a heated electrospray ionization (HESI) source was operated in positive ionization mode with a capillary voltage of −10 V and temperature of 310°C. The spray voltage to 3.3 kV, and the atmospheric pressure chemical ionization (APCI) temperature was set to 135°C. Sheath, auxiliary and sweep gases were set to the following respectively 5, 35 and 7 arbitrary units. MS1 specta were collected from 200 to 1000 m/z and data-dependent acquisition (DDA) set to acquire MS2 spectra of the top three most abundant ions. Data acquisition and analysis was completed on the Thermo Xcalibur software platform.
RESULTS
Exploring the applicability of a TEAA-free chromatography for oligonucleotide mass spectrometry
In a first step, we wanted to find a TEAA (triethylammoniumacetate) and other ion-pairing reagents free chromatography for separation of oligonucleotides which might be compatible with commonly used mass spectrometers and small compound analysis. A literature search, to avoid ion-pairing reagents, revealed a method from 1994, where an RP-18 column was used for separation of tRNA isoacceptors (46). Here, the elution was achieved by a gradient of simple ammonium acetate and acetonitrile, which is comparable to the separation procedure of nucleosides (38). Inspired by this early work, we tested the separation of synthetic oligonucleotides (ON) on our nucleoside column (Phenomenex, Fusion-RP) using a 10 mM ammonium acetate buffer and acetonitrile for elution. Due to the negatively charged phosphate backbone of RNA, we did not expect good retention on a hydrophobic RP-18 column. However, due to the special column material of the tested column, we observed excellent retention for the tested ONs. As the UV chromatogram (λ260 nm) in Figure 1A shows, the tested 8-mer ONs elute first, followed by 9-mer ONs and 5-mer ONs. To understand which properties of the ONs influence the retention behavior, we used ONs, which only differ in one nucleobase in their sequence. As shown in Figure 1A, the cytidine containing 8-mer elutes first, while the exchange to a uridine leads to better retention. The permutated 8-mer with adenosine is retained most on the column. In the 5-mer, we observe a better retention of the cytidine containing ON compared to its U containing permutation. In case of the 9-mer ONs, the deamination of one adenosine into inosine leads to reduced retention. In these sequence contexts, we clearly see a dependence of the retention behavior on the chemistry of the present nucleobases and not on the length of the ON. From our data, we conclude that the retention behavior of ONs in our TEAA-free system depends on two factors: the distribution of nucleobases and the sequence. We observe no rules which allow prediction of ON elution.
Figure 1.
Separation and detection of synthetic oligonucleotides (ON) by MS. (A) UV chromatogram (λ 260 nm) of various ONs (20 pmol injection) separated by our ion-pairing reagent free chromatography. (B) Detection of the ONs from (A) by MS using positive ionization. (C) Detection of the ONs from (A) by MS using negative ionization. (D) Mass spectrum of the first eluting ON from (A) in positive ion mode (black) and negative ion mode (gray) on a low-resolution triple quadrupole instrument. The charge state is determined by the pattern of sodium charges in the spectrum. (E) Mass spectrum of the first eluting ON from (A) in positive ion mode (black) on a high resolution Orbitrap instrument.
In a next step, we connected our developed chromatographic system with our low resolution triple quadrupole (QQQ) mass spectrometer. We tuned the instrument in both negative and positive ionization mode using the tune mix supplied by the manufacturer and determined the optimal source parameters for sensitive detection of ON 13 (CCUAACACG) (Supplementary Figure S1). With these parameters, we scanned for the eluting ONs in a mass range of m/z 500–1000 in positive and negative ionization mode. We received MS signals of all tested ONs below 10 nt in both positive and negative ionization mode (Figure 1B/C). Our instrument is commonly operated in positive ionization mode, and thus it was no surprise to find a substantially higher sensitivity in positive ionization mode. We attribute this observation to the fact that the instrument is only operated in positive ion mode with the respective buffers for ideal protonation in the ESI source. Thus, the ionization efficiency in negative ion mode is reduced. On other instruments, the sensitivity is higher in negative mode compared to positive mode (47). We recommend assessment of the most sensitive ionization mode on every mass spectrometer by running a mixture of short synthetic ONs. The addition of the modifier 1,1,1,3,3,3-Hexafluor-2-propanol (HFIP) did not improve the detection in positive or negative ion mode. Due to the common use of our instrument in positive ion mode and the higher sensitivity, we decided to use the positive ionization mode for further analyses. We used the mongo-oligo mass calculator tool (https://mods.rna.albany.edu/masspec/Mongo-Oligo) to compare our experimental signals with the predicted signals. For the tested ONs, we observed the predicted m/z signals, commonly of the +2 charge state for 5 mers, +3 charge for 8 mers and +4 charge for 10 mers (Supplementary Figure S2). The charge state of an ON is commonly calculated with the natural abundance of carbon-13 on high-resolution instruments. The difference between the 12C-m/z value and the 13C-m/z value is ∼1 in a +1 charge state, ∼0.5 in a +2 charge state, ∼0.3 in a +3 charge state, etc. On a low-resolution instrument, such as the used QQQ MS, the natural isotope abundance is not resolved and cannot be used to predict the charge state. As an alternative, the difference between the (multi-) protonated ion and the sodium charged ion can be used. In a +1 charge state, the difference is 22 units, in a +2 charge state it is 11 units (charged by one protonation and one sodium cation), in a +3 charge state it is ∼7 units (two protons and one sodium cation), etc. Our mass spectrum for the C-permutated 8-mer ONs revealed a main signal at m/z 808.1 (protonated species) and a m/z 816 for the Na+-charge and thus we conclude a +3 charge state of this ON (see Figure 1D). For the 8-mer ONs, other charge states are of minor abundance (see full mass spectra in Supplementary Figure S3).
In addition to the analyses of ONs by low resolution MS, we wanted to test the compatibility of our method with a high resolution mass spectrometer. The instrument was optimized with the 9-mer ON 13 and the optimal parameters were chosen according to the results given in Supplementary Figure S1. As shown in Figure 1E and Supplementary Figure S2, all tested ONs were detectable by high-resolution MS in positive ionization mode. On this instrument Na+ adducts were of minor abundance and protonated species dominated the MS spectra.
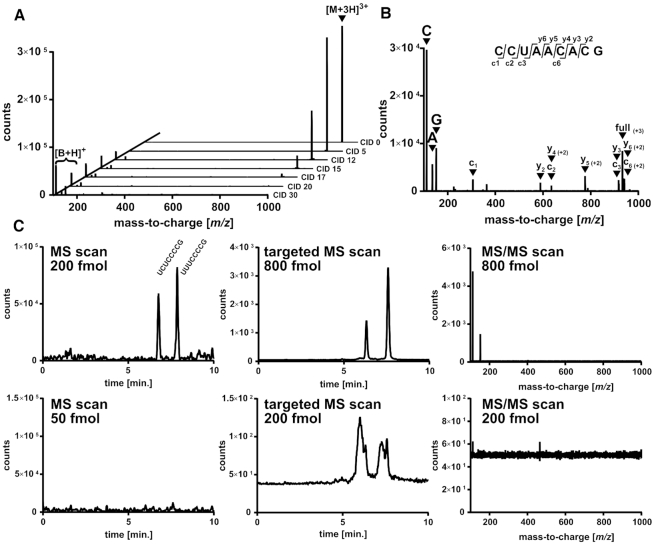
MS of oligonucleotides is commonly used to determine the sequence and location of modified nucleosides (26,48). For this purpose, fragmentation of the ONs by collision-induced-dissociation (CID) is a useful tool and the behavior of ONs in positive and negative ionization mode is well described (47). On the low resolution QQQ instrument, we analyzed ON 13 and ramped CID energies from 0 to 30 eV. The recorded product ion scans are displayed in Figure 2A. With increasing collision energy, the signal of the precursor ion decreases, while multiple new signals appear in the spectra. At higher collision energies, the most prominent product ions in positive ionization mode are protonated nucleobase fragment ions. At energies around 15–20 eV, c and y ions become apparent (Figure 2B). Due to the low resolution of a QQQ instrument, the charge state of these c- and y-ions cannot be determined and thus QQQ MS is not suitable for de novo sequencing. The signal to fragment matching in Figure 2B and for other ONs in Supplementary Figure S4 was only possible due to the known sequence of the ON and the use of the Mongo oligonucleotide tool (https://mods.rna.albany.edu/masspec/Mongo-Oligo). CID analysis on the high resolution MS produced more c- and y- fragment ions of known charge states and thus de novo sequencing is theoretically possible (Supplementary Figure S5).
Figure 2.
Collision-induced dissociation (CID) and sensitivity of oligonucleotides on a low resolution triple quadrupole mass spectrometer (30 pmol injection). (A) MS/MS spectra of the 8-mer CCUAACACG at various collision energies (CID 0–30 eV). The emergence of base fragments is indicated with [B+H]+. (B) MS/MS spectrum of CCUAACACG at a CID energy of 15 eV. The resulting fragments are assigned to the known sequence by using (https://mods.rna.albany.edu/masspec/Mongo-Oligo). Base fragments are abbreviated with C—cytosine, A—adenine and G—guanine. (C) LLOD for the ONs U(U/C)UCCCCG in MS scan mode (left), targeted MS mode (middle) of the mass transition 808.1→112 and 808.5→112 and MS/MS scan mode using the precursor m/z 808.1 and 808.5. The upper graphs are above the LLOD, the lower graphs are just below the LLOD.
Low resolution instruments such as our used QQQ mass spectrometer are commonly used for the sensitive detection and quantification of small compounds. In our hands, the LLOD and lower limits of quantification (LLOQ) of modified nucleosides is in the single digit fmol range or even amol range (38). For ON analysis, we injected dilutions of ON 13 and ON 14 and found LLODs/LLOQs of 200 fmol in MS2Scan mode, 800 fmol in MS/MS mode and 800 fmol in Product Ion (PI) Scan mode for sequence determination purposes (Figure 2C). For isolated tRNA isoacceptors, the lowest sensitivity was achieved with a TEAA dependent nano-LC setup (33). With nano-LC, 2–5 ng tRNA (∼80–200 fmol) are sufficient for analysis which is comparable to our results.
Although the sensitivity towards ONs is lower compared to small molecule analysis, it is sufficient to analyze ONs derived from synthesis, in vitro experiments or potentially even native RNAs.
Preparation of in vitro transcribed tRNA by SEC
To expand the field of application, we wanted to use our method on partial RNA digests of full-length in vitro transcripts and also native RNAs. Thus, we decided to produce two human tRNAs by ribozyme-fusion in vitro transcription and analyze the transcripts after RNase T1 digestion by ON-MS. After transcription, the cleaved transcript is commonly purified by PAGE and the ribozyme and the full-length transcript are removed (49). However, PAGE purified RNA always contains small amounts of polyacrylamide, which potentially interacts with later MS. Thus, we adapted a previously described method based upon size exclusion chromatography (SEC, SEC-3 column) that is rapid, automatable and produces clean RNA (44). We have found the AdvanceBio SEC column to be more robust and thus recommend this column for tRNA purification from total RNA. We supply a detailed handling guide to prolong column lifetime to several hundred injections in the materials and methods section. A comparative profile of an RNA ladder (1000 nts to 50 nts) loaded onto these two 300 Å columns is shown in Figure 3A and B. Especially at the used column temperature of 60°C, both columns nicely separate the 50 and 80 nts marker from the longer RNAs >150 nts. As expected, native tRNA elutes at the same time as the 80 nts marker, which indicates that secondary structures play no role in the separation of RNAs. Thus, these columns are ideal for separation of large RNAs such as ribosomal RNA (rRNA) and mRNA from the smaller tRNAs. The AdvanceBio column is also available with a 130 Å pore size. We tested the separation efficiency of this column with the ladder and as it is to be expected, the large RNA markers can no longer be separated due to reduced interaction with the smaller pores (Figure 3C). The 130 Å column is less suitable for the separation of total RNA. However, it has a potential for the separation of RNAs smaller 80 nts, e.g. tRNA and tRNA-derived fragments (tRF or tiRNA) (Supplementary Figure S6).
Figure 3.
SEC of RNA for automated purification (A)-(C) Separation of an RNA ladder (black) of a: 1000 nts, b: 500 nts, c: 300nts, d: 150 nts, e: 80 nts and f: 50 nts. For comparison, the elution of total tRNA is shown in green. (A) SEC-3, 300 Å (B) AdvanceBio SEC 300 Å (C) AdvanceBio SEC 130 Å (D) Purification of T7 in vitro transcribed tRNAValAAC (blue) and tRNASerUGA (red) on a 130 Å column at 60°C. Peak 1: tRNA-ribozyme fusion product, Peak 2: tRNA, Peak 3: ribozyme; (E) Quality control of the T7 transcription mix before purification (pre) and after SEC separation (post) for tRNAValAAC and tRNASerUGA on an automated gel-electrophoresis system. 1: tRNA-ribozyme fusion product, 2: tRNA, 3: ribozyme
For the sake of column preservation, we tested the chromatographic resolution at lower temperatures. We find sufficient separation of the small 80 nts from the larger RNAs (>150 nts) at 40°C (Supplementary Figure S7). We have applied the AdvanceBio 300 Å SEC column at 40°C in several studies for the purification of tRNA from total RNA (5,50–51). The purification was always reliable and thus we recommend this column with the developed parameters for tRNA purification.
For the purification of tRNA from ribozyme-fusion in vitro transcription, both 300 Å and 130 Å pore size columns have been tested. tRNASerUGA is an 85 nts long tRNA and fused with the ribozyme fusion transcript is 135 nts in length. The fusion transcript auto-catalytically cleaves itself into the 85 nts long tRNA and the 40 nts long ribozyme. As shown in Figure 3D, the tRNA peak is separated from the full-length transcript, while the cleaved ribozyme partly co-elutes with the tRNA (130 Å column at 60°C). We collected the tRNA peak and analyzed the fraction by high-resolution automated electrophoresis (Bioanalyzer). Here, we find that some of the ribozyme (40 nts) is still detectable in the tRNA fraction but the full-length transcript (135 nts) is completely removed (Figure 3E). If necessary, the collection of the tRNA peak is possible in a smaller time window or second purification round and thereby the ribozyme can be completely removed (Supplementary Figure S8). After successful purification of the comparably long tRNASer, we were curious whether purification of a tRNA with short variable loop is possible by our automated system. For this purpose, we prepared a fusion-transcript of tRNAValAAC. The full-length transcript is 124 nts long, the tRNA itself 76 nts and the ribozyme again 40 nts. For this tRNA, the size difference between tRNA and the ribozyme is only 36 nts. As a rule of thumb, SEC separation of biomolecules is possible as long as they differ in size by a factor of two. This pre-requisite is not met in case of tRNAVal and as expected, the separation of the produced tRNA and the ribozyme is not possible (Figure 3D, blue chromatogram). The ribozyme co-elutes as a shoulder and subsequently, more ribozyme can be found in the purified tRNA transcript (Figure 3E). Similar to the presented solution for tRNASer, a reduced collection time window can help to remove the remaining ribozyme if complete removal of the ribozyme is necessary for subsequent experiments.
Overall, SEC purification of ribozyme-fusion transcribed tRNAs is an alternative to purification by PAGE. The complete transcription mix can be loaded onto the SEC column without pre-purification and within 20 min the pure tRNA fraction is received in a volume of 500–1000 μl in 0.1 M ammonium acetate. Concentration of the tRNA product is possible by subsequent solvent evaporation and/or ethanol precipitation.
Analysis of RNase T1 treated tRNA by oligonucleotide mass spectrometry
With the tRNA transcripts in hand, we were ready to test our LC-MS method by injecting a partial digest of the produced tRNAs. For this purpose, 1 μg of each transcript was incubated with RNase T1. The enzyme was removed by molecular weight cut-off filtration and 200 ng (∼8 pmol) of the resulting ON mixture were injected onto the LC-QQQ set-up in scanning mode. As expected from our experiments with synthetic ONs, we observe separation of the RNase T1 derived ON mixture as seen for the total ion chromatogram shown in Figure 4A (tRNAValAAC) and 4b (tRNASerUGA). The color-coded tRNA sequences are shown in Supplementary Figure S9. For tRNAValAAC, we prepared transcripts containing stable isotope labeled nucleotides 15N3-cytidine or 15N5-guanosine. The fragments of the unlabeled and labeled transcripts elute at the same retention time. The low resolution mass spectra of the eluting peaks allowed the assignment of the ON sequences, which are derived from various sequences of the tRNA. As expected, we observe differences in the m/z value due to the stable isotopes in the 15N containing fragments compared to the unlabeled fragments (Figure 4A). Thus, our method is capable of separating RNase T1 derived tRNA digests. In this context, we observed a low MS abundance of the 9 mer fragment (#4) compared to the 8 mer fragment (#6) from tRNAVal. This is explained by the different amount of acetonitrile (ACN) at these retention times. While the 9-mer elutes at around 4.5% ACN, the 8-mer elutes later and with ∼ 25% ACN. With higher amounts of ACN, the ionization efficiency is increased and thus later eluting compounds have a higher signal intensity. When the gradient is eliminated by direct injection of ONs or elution at isocratic ACN conditions (Supplementary Figure S2), the abundance of all injected synthetic ONs is comparable and thus we conclude that the ionization efficiency is not majorly impacted by the composition of canonical nucleosides.
Figure 4.
MS chromatograms of in vitro transcribed (IVT) tRNAs after RNase T1 digestion. (A) Extracted ion chromatograms of tRNAValAAC IVT without stable isotopes (light ivt), with 15N3-Cytidine or 15N5-Guanosine. The sequences of the numbered peaks were determined by low resolution MS. The retention time (Rt), charge state and m/z values for the unlabeled and stable isotope labeled sequences are given in the table. (B) Total ion chromatogram of tRNASerUGA IVT. The sequences of the numbered peaks were determined by low-resolution MS and are listed below the chromatogram.
RNA and especially tRNA are heavily post-transcriptionally modified. To study the applicability of our method to native RNA, we treated total tRNA from HEK cell culture with RNase T1, separated the resulting fragments with our chromatographic method and analyzed the effluent by high-resolution MS. Here, the mass resolving power of the Orbitrap instrument is necessary to confirm the sequence and modification status of the observed tRNA derived ONs. With this approach, we could identify several modified ONs from total tRNA. The corresponding chromatograms and MS/MS spectra are shown in Figure 5.
Figure 5.
Extracted ion chromatograms and MS/MS spectra of modified RNA fragments from HEK 293 cell total tRNA digested with RNase T1. Spectra were recorded on a high-resolution Orbitrap instrument at CID energies of 35 eV. Fragments were assigned using (https://mods.rna.albany.edu/masspec/Mongo-Oligo) calculations from known modified tRNA sequences (acquired from modomics database).
ADAT2/3 deaminates tRNAValAAC at position 34 in vitro
Since tRNA modifying enzymes depend on a correctly folded tRNA substrate for their activity, we tested whether the SEC purified IVTs fold into the expected tRNA shape and are thus usable in in vitro modification assays. For these assays, we used human ADAT2/3 enzyme, which has been shown to catalyze the deamination of wobble adenosine at position 34 of in vitro transcribed tRNAValAAC to inosine (52). We incubated tRNAValAAC with purified ADAT2/3 expecting deamination of A34 to I34. The presumably deaminated tRNA was digested by RNase T1 and we screened for the tRNA sequence covering position 34, namely CCUAACACG and CCUIACACG (see Figure 6A). Surprisingly, we only found the unmodified sequence CCUAACACG and not the inosine containing sequence (Figure 6B). Due to this unexpected result, we digested an aliquot of the ADAT2/3 treated tRNA to nucleosides and switched to quantitative nucleoside analysis. While no inosine was detected in the untreated tRNA by nucleoside MS, we could detect an 8.0% conversion of adenosine to inosine per tRNA molecule in the ADAT2/3 modified transcript (Figure 6F). From this finding we conclude that our SEC purified tRNA folds correctly and is thus recognized as a substrate by the tRNA modifying enzyme ADAT2/3.
Figure 6.
In vitro modification of tRNAValAAC by ADAT2/3. (A) 3D structure of tRNAValAAC in green. In red, the RNase T1 fragment of the ADAT2/3 target site is shown. (B) RNase T1 digest of untreated (left) and ADAT2/3 treated (right) tRNAValAAC. In black, the signal for the resulting A-containing ON is shown and in gray, the signal of the I-containing ON. (C) Calibration curve of the CCUI fragment for external calibration quantification by low-resolution oligonucleotide MS. (D) Extracted ion chromatograms of RNase T1 digested tRNAValAAC before (black) and after (red) ADAT2/3 treatment. In gray the synthetic CCUI oligo is shown as a retention time control. (E) Extracted ion chromatograms of RNase T1 digested 15N3-cytidine-labeled tRNAValAAC before (black) and after (red) ADAT2/3 treatment. In gray the signal of the spiked in synthetic CCUI oligo is shown. (F) Quantification of ADAT2/3 treated tRNA reveals the abundance of inosine per tRNA (%I / tRNA). Quantification was achieved by isotope dilution MS of a complete nucleoside digest (Nucs-MS), by external calibration and by isotope standard spike-in for oligonucleotide MS.
Intrigued by the absence of the inosine modified ON in the Oligo-MS analysis and with the goal to assign the location of the A-to-I editing in tRNAValAAC, we utilized the synthetic 9-mer ONs presented in Figure 1A. These ONs represent the sequences expected from an RNase T1 digestion of tRNAValAAC (unmodified: CCUAACACG and modified: CCUIACACG). RNase T1 is a commonly used endoribonuclease, which specifically cleaves RNA after guanosine moieties. Guanosine and adenosine differ at position 6 and position 2 of their purine structures. Inosine and guanosine are identical at position 6 but differ at position 2. Due to the high chemical similarity of inosine and guanosine, we wondered whether RNase T1 is capable of inosine recognition and subsequent cleavage. A literature search revealed that the detection of inosine by sequencing is commonly done utilizing RNase T1 (53,54). In MS-based RNA modification studies, this knowledge is not yet widespread. To test the impact of RNase T1 on I containing RNA in MS, we incubated the A and I containing ONs with RNase T1 and analyzed the resulting mixture. For CCUAACACG, we observe one prominent peak which corresponds to the full-length ON and a second peak (less than 30%), corresponding to the 3′cleavage product ACACG (Supplementary Figure S11). In contrast, full-length CCUIACACG, is barely detectable after RNase T1 treatment. Instead, two new peaks are found (Supplementary Figures S10 and 11). One corresponds to the 3′ fragment ACACG and the other to the inosine containing 5′ cleavage product CCUI (Supplementary Figure S10). From our observation it is now clear, that RNase T1 cleaves both guanosine and inosine containing RNA sequences which impacts bottom-up oligonucleotide mass sepctrometry. Based upon our results, the substrate preference of RNase T1 can now be summarized as G > I > A.
In case of tRNAValAAC, RNase T1 will then result in an additional cleavage after I34 (Supplementary Figure S12). We re-analyzed ADAT2/3 treated and untreated tRNA by oligonucleotide MS and screened for the newly identified ONs. We detected the CCUI fragment in unlabeled and 15N-Cytidine-labeled tRNA digests after ADAT2/3 treatment, which indicates the A-to-I conversion of position 34 of the tRNA. Due to the similar mass of the CCUI and the CCUA fragment (1 Da), the result must be confirmed in the +1 charge state on a low resolution instrument or by high resolution MS. Analysis of the sample on the Orbitrap instrument revealed indeed formation of CCUI and thus confirms our findings from low resolution MS (Supplementary Figure S13). For quantification of the deamination reaction, we used the synthetic ON CCUIACACG and prepared an external calibration with its RNase T1 digest and re-analayzed the samples on the QQQ MS. We detected 7.6% inosine formation using this external calibration method (Figure 6C and D). Mass spectrometric quantification is ideally done using stable isotope labeled internal standards. Here, we used our 15N3-cytidine labeled transcript of tRNAVal as the substrate and the unlabeled synthetic ON as the internal standard. With this method, we find a 6.2% conversion of A34 to I34 (Figure 6E). This result is 1.3-fold lower compared to our accurate nucleoside-MS method (Figure 6F), which is acceptable for most potential applications of our method. Our data suggests that our method is capable of absolute quantification, but further studies using internal standards as suggested by (39–43) must be performed to determine the accuracy and precision for a given biological context.
The activity of AlkB is sequence dependent
Another context which benefits from ON-MS on a low resolution MS, is in vitro modification/demodification experiments. Here, we applied our method to the analysis of 1-methyladenosine (m1A)-modified ONs to study the demethylation by AlkB (8). With the goal to study the substrate specificity of AlkB by ON-MS, we designed three m1A-containing ONs. In human tRNA, m1A is commonly found at position 58. Sequence overlay of 13 random human tRNAs revealed the weight matrix of the nucleobases surrounding m1A58 in human tRNAs (Supplementary Figure S14). The first ON (UUCG(m1A)UUC), was designed to match this sequence surrounding m1A58 from human tRNAs and is thus referred to as ON-human. The second ON reflects a mutated version of the ON-human, where three preserved bases are exchanged (ON-mutated, UAGG(m1A)UUG). The last ON, ON-ribo, reflects the sequence surrounding the only possible m1A site of E. coli 16S rRNA; (CGUC(m1A)CAC). This site is found to be methylated in bacteria resistant to certain antibiotics (55). With this experiment, we wanted to evaluate the activity of AlkB toward these three ONs and whether a sequence dependence is observable.
All three ONs were incubated with 10 μM AlkB. The subsequent ON-MS analysis revealed the formation of demethylated ONs as shown in Figure 7A–C. The demethylated ONs elute at a later retention time compared to the respective m1A modified ONs on our TEAA-free chromatography due to the lost positive charge of m1A. For ON-human and ON-ribo, we observe only trace formation of the demethylated ON (Figure 7A/C). In contrast, we observed more demethylated ON for ON-mutated. At first glance, our observation indicates that the m1A site of E. coli rRNA, and the ON with the sequence of eukaryotic tRNA m1A58 are less suitable substrates to AlkB compared to the completely unnatural substrate, ON-mutated. However, without the use of stable isotope labeled internal standards, MS is non-quantitative and the data may not be interpreted in such a way.
Figure 7.

in vitro AlkB assay with different m1A containing oligonucleotides (ON) (A–C) left: ON-MS of m1A containing ONs after incubation with 10 μM AlkB and their assigned m/z values, charge states and sequences. (A–C) right: amount of m1A per ON determined by isotope dilution MS of fully digested ONs with and without 1 μM AlkB treatment; bars represent the mean, error bars the standard deviation, n = 3 biological replicates, a two-tailed, unpaired students t-test was performed (ON-human: P = 0.0067; ON-mutated: P < 0.0001; ON-ribo: P < 0.0001).
To provide a quantitative method, we digested the AlkB treated and untreated ONs to nucleosides and performed nucleoside-MS with our established stable isotope dilution MS (38). For this purpose, we equilibrated the system for 1 h by flushing the instrument with our aqueous nucleoside-MS buffer and performed our quantitative nucleoside experiments. Before AlkB treatment, we find 0.23 m1A per ON-human, 0.12 m1A per ON-mutated and 0.19 m1A per ON-ribo. At a low AlkB concentration (1 μM), we observe 9.8% demethylation for the ON-human (P = 0.0067). The ON-mutated shows 12.9% demethylation (P < 0.0001) and the ON-ribo 18.6% demethylation (P < 0.0001). The absolute quantification data is in stark contrast to the non-quantitative results observed by the oligonucleotide MS. Intriguingly, AlkB seems to prefer the only potential m1A site found in bacteria, namely m1A1408 of the 16 S rRNA. Our observation is confirmed by incubation of the same ONs with higher amounts of AlkB (10 μM, Supplementary Figure S15).
DISCUSSION
In this manuscript we present a method for the separation of oligonucleotides and subsequent detection by oligonucleotide MS (ON-MS). While methods for ON-MS have been available for several years, key hurdles toward their broader application were the use of ion-pairing reagents such as TEAA and negative ionization. We have overcome these hurdles by successfully using a modified RP-18 column with a simple ammonium acetate buffer in combination with acetonitrile. These are common solvents for LC-MS instruments, which do not contaminate the system or interfere with subsequent analysis of other compounds. The broad applicability is demonstrated by our analyses of synthetic ONs, RNase T1 digestion derived RNA fragments of in vitro transcripts and native tRNA on both low and high-resolution mass spectrometers. A higher resolving power is required to distinguish mass differences of oligonucleotides, their sequence and modification content. A key challenge is that the nucleotide bases uridine and cytidine (and adenosine and inosine) have a mass difference of only 1 Da, which is already the resolution limit of the QQQ MS. For the analysis of unknown sequences, the mass accuracy and resolving power of the mass spectrometer must therefore be sufficient to resolve this difference (56). A second benefit of high resolution MS, is the determination of charge states even in MS/MS experiments which allows sequence determination and localization of modified nucleosides as demonstrated in Figure 5.
Nevertheless, low resolution MS instruments are valuable for the analysis of oligonucleotides of known sequence which have been prepared for e.g. in vitro analysis of RNA modifying (Figure 6) or demethylating enzymes (Figure 7).
A single sample run is <15 min and ONs below 10 nt are separated according to their chemistry, but the elution order is not predictable. Thus 2D analysis of sequences as shown by the Szostak lab is not possible with our method (57). In our hands, switching from ON-MS to sensitive nucleoside-MS is possible within 60 min to allow system and column equilibration. A comparison of our TEAA free and the common ON-MS method is given in Table 1.
Table 1.
Advantages and Limitations of TEAA-free and other commonly used ON-MS methods
| TEAA-based ON-MS | TEAA free low-resolution MS | TEAA free high-resolution MS | |
|---|---|---|---|
| Analysis of native RNA | |||
| • Known sequence | yes | yes | yes |
| • Unknown sequence | yes | no | yes |
| Analysis of synthetic ONs | yes | yes | yes |
| Analysis of ONs longer 10 nts | no | no | no |
| Compatibility with small compound analyses | no | yes | yes |
| LC and MS contamination | yes | no | no |
| 2D analysis (57) | yes | no | no |
| Sensitivity | Lower compared to nucleoside MS | Lower compared to nucleoside MS | Lower compared to nucleoside MS |
| Quantitative accuracy and precision | (39–43) | Not fully determined | Not fully determined |
For the analysis of RNA modifying enzymes, the preparation and use of in vitro transcribed RNA is highly useful. In this manuscript, we use ribozyme-fusion transcription of a 76 nts long tRNA and an 85 nts long tRNA. Instead of laborious PAGE purification, we use automated SEC. With nucleoside-MS (Figure 6F), we show that the thus purified tRNA is recognized and converted by ADAT2/3, which demonstrates the correct folding of the tRNA. With the goal to determine the position of A-I conversion, the produced tRNAs are digested by RNase T1 and the resulting fragments are separated by our TEAA-free chromatography. It is noteworthy that our chromatography allows the separation of ONs where only one oxygen is replaced with an amino group. The RNase T1 derived fragments are assigned by the detected m/z values in positive ionization mode by low resolution MS (Figures 4 and 6). For such in vitro experiments, there is no immediate need for high-resolution MS as the resulting fragments and MS/MS products can be predicted and resolved by low resolution MS. In case of the studied A-I conversion, the target fragment differs in only 1 Da, which can be resolved by low-resolution MS in the +1 charge state, but not in higher charge states. In such cases, the use of high-resolution instruments might become necessary for confirmation (Supplementary Figure S13).
We observe efficient cleavage after inosine by RNase T1 in the A-I converted tRNA and synthetic ONs as expected from approaches to sequence inosine containing RNA (53,54). This substrate diversity of RNase T1 is not commonly described in the bottom-up-MS literature but due to the high abundance of inosine in tRNA (6) and mRNA (58) it should be kept in mind when using RNase T1.
Altogether, we present various MS-based approaches for the bottom-up analysis of RNA which are easily transferrable to laboratories which avoided ON-MS in the past due to the problems introduced by ion-pairing reagents and negative ion mode. We demonstrate that switching from ON-MS to small compound analysis and back is possible after a brief equilibration procedure (Figures 6 and 7). With this, ON-MS can be routinely used on any LC-MS system to study RNA modifications in their sequence context.
DATA AVAILABILITY
https://mods.rna.albany.edu/masspec/Mongo-Oligo, http://modomics.genesilico.pl/.
Supplementary Material
ACKNOWLEDGEMENTS
S.K. is grateful to Agilent Technologies (Waldbronn, Germany) for providing test columns during method development. F.H. and S.K. thank Thomas Carell and his group for instrument time and scientific discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [CIPSMwoman, KE1943/3-1, KE1943/4-1 to S.K.]; National Science Foundation CAREER Award [1552126 to J.R, D.F.]. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G. et al.. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H. et al.. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu F., Clark W., Luo G., Wang X., Fu Y., Wei J., Wang X., Hao Z., Dai Q., Zheng G. et al.. ALKBH1-Mediated tRNA demethylation regulates translation. Cell. 2016; 167:816–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wei J., Liu F., Lu Z., Fei Q., Ai Y., He P.C., Shi H., Cui X., Su R., Klungland A. et al.. Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell. 2018; 71:973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dewe J.M., Fuller B.L., Lentini J.M., Kellner S.M., Fu D.. TRMT1-Catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol. Cell Biol. 2017; 37:e00214-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gerber A.P., Keller W.. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999; 286:1146–1149. [DOI] [PubMed] [Google Scholar]
- 7. Ramos J., Han L., Li Y., Hagelskamp F., Kellner S.M., Alkuraya F.S., Phizicky E.M., Fu D.. Formation of tRNA wobble inosine in humans is disrupted by a millennia-old mutation causing intellectual disability. Mol. Cell Biol. 2019; 39:e00203–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ougland R., Zhang C.M., Liiv A., Johansen R.F., Seeberg E., Hou Y.M., Remme J., Falnes P.O.. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell. 2004; 16:107–116. [DOI] [PubMed] [Google Scholar]
- 9. Ueda Y., Ooshio I., Fusamae Y., Kitae K., Kawaguchi M., Jingushi K., Hase H., Harada K., Hirata K., Tsujikawa K.. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 2017; 7:doi:10.1038/srep42271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heiss M., Kellner S.. Detection of nucleic acid modifications by chemical reagents. RNA Biol. 2017; 14:1166–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Motorin Y., Helm M.. Methods for RNA modification mapping using deep sequencing: Established and new emerging technologies. Genes (Basel). 2019; 10:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takeda N., Pomerantz S.C., McCloskey J.A.. Detection of ribose-methylated nucleotides in enzymatic hydrolysates of RNA by thermospray liquid chromatography-mass spectrometry. J. Chromatogr. 1991; 562:225–235. [DOI] [PubMed] [Google Scholar]
- 13. Kellner S., Ochel A., Thuring K., Spenkuch F., Neumann J., Sharma S., Entian K.D., Schneider D., Helm M.. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res. 2014; 42:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kellner S., Neumann J., Rosenkranz D., Lebedeva S., Ketting R.F., Zischler H., Schneider D., Helm M.. Profiling of RNA modifications by multiplexed stable isotope labelling. Chem. Commun. (Camb.). 2014; 50:3516–3518. [DOI] [PubMed] [Google Scholar]
- 15. Dal Magro C., Keller P., Kotter A., Werner S., Duarte V., Marchand V., Ignarski M., Freiwald A., Muller R.U., Dieterich C. et al.. A vastly increased chemical variety of RNA modifications containing a thioacetal structure. Angew. Chem. Int. Ed. Engl. 2018; 57:7893–7897. [DOI] [PubMed] [Google Scholar]
- 16. Limbach P.A., Crain P.F., McCloskey J.A.. Molecular mass measurement of intact ribonucleic acids via electrospray ionization quadrupole mass spectrometry. J. Am. Soc. Mass Spectrom. 1995; 6:27–39. [DOI] [PubMed] [Google Scholar]
- 17. Guymon R., Pomerantz S.C., Crain P.F., McCloskey J.A.. Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: the complete modification map of 16S rRNA of Thermus thermophilus. Biochemistry. 2006; 45:4888–4899. [DOI] [PubMed] [Google Scholar]
- 18. Hossain M., Limbach P.A.. Multiple endonucleases improve MALDI-MS signature digestion product detection of bacterial transfer RNAs. Anal. Bioanal. Chem. 2009; 394:1125–1135. [DOI] [PubMed] [Google Scholar]
- 19. Ikeuchi Y., Kimura S., Numata T., Nakamura D., Yokogawa T., Ogata T., Wada T., Suzuki T., Suzuki T.. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 2010; 6:277–282. [DOI] [PubMed] [Google Scholar]
- 20. Meng Z., Limbach P.A.. Shotgun sequencing small oligonucleotides by nozzle-skimmer dissociation and electrospray ionization mass spectrometry. Eur. J. Mass Spectrom. (Chichester). 2005; 11:221–229. [DOI] [PubMed] [Google Scholar]
- 21. Hossain M., Limbach P.A.. Mass spectrometry-based detection of transfer RNAs by their signature endonuclease digestion products. RNA. 2007; 13:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taucher M., Ganisl B., Breuker K.. Identification, localization, and relative quantitation of pseudouridine in RNA by tandem mass spectrometry of hydrolysis products. Int. J. Mass Spectrom. 2011; 304:91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taucher M., Rieder U., Breuker K.. Minimizing base loss and internal fragmentation in collisionally activated dissociation of multiply deprotonated RNA. J. Am. Soc. Mass Spectrom. 2010; 21:278–285. [DOI] [PubMed] [Google Scholar]
- 24. Huang T.Y., Liu J., McLuckey S.A.. Top-down tandem mass spectrometry of tRNA via ion trap collision-induced dissociation. J. Am. Soc. Mass Spectrom. 2010; 21:890–898. [DOI] [PubMed] [Google Scholar]
- 25. Taucher M., Breuker K.. Top-down mass spectrometry for sequencing of larger (up to 61 nt) RNA by CAD and EDD. J. Am. Soc. Mass Spectrom. 2010; 21:918–929. [DOI] [PubMed] [Google Scholar]
- 26. Taucher M., Breuker K.. Characterization of modified RNA by top-down mass spectrometry. Angew. Chem. Int. Ed. Engl. 2012; 51:11289–11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Glasner H., Riml C., Micura R., Breuker K.. Label-free, direct localization and relative quantitation of the RNA nucleobase methylations m6A, m5C, m3U, and m5U by top-down mass spectrometry. Nucleic Acids Res. 2017; 45:8014–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schurch S. Characterization of nucleic acids by tandem mass spectrometry—the second decade (2004-2013): From DNA to RNA and modified sequences. Mass Spectrom. Rev. 2016; 35:483–523. [DOI] [PubMed] [Google Scholar]
- 29. Juhasz P., Roskey M.T., Smirnov I.P., Haff L.A., Vestal M.L., Martin S.A.. Applications of delayed extraction matrix-assisted laser desorption ionization time-of-flight mass spectrometry to oligonucleotide analysis. Anal. Chem. 1996; 68:941–946. [DOI] [PubMed] [Google Scholar]
- 30. Chou C.W., Limbach P.A., Cole R.B.. Fragmentation pathway studies of oligonucleotides in matrix-assisted laser desorption/ionization mass spectrometry by charge tagging and H/D exchange. J. Am. Soc. Mass Spectrom. 2002; 13:1407–1417. [DOI] [PubMed] [Google Scholar]
- 31. Farand J., Beverly M.. Sequence confirmation of modified oligonucleotides using chemical degradation, electrospray ionization, time-of-flight, and tandem mass spectrometry. Anal. Chem. 2008; 80:7414–7421. [DOI] [PubMed] [Google Scholar]
- 32. Suzuki T., Ikeuchi Y., Noma A., Suzuki T., Sakaguchi Y.. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007; 425:211–229. [DOI] [PubMed] [Google Scholar]
- 33. Suzuki T., Suzuki T.. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014; 42:7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Popova A.M., Williamson J.R.. Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry. J. Am. Chem. Soc. 2014; 136:2058–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lobue P.A., Jora M., Addepalli B., Limbach P.A.. Oligonucleotide analysis by hydrophilic interaction liquid chromatography-mass spectrometry in the absence of ion-pair reagents. J. Chromatogr. A. 2019; 1595:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weng G., Sun B., Liu Z., Wang F., Pan Y.. Analysis of oligonucleotides by ion-pair reversed-phase liquid chromatography coupled with positive mode electrospray ionization mass spectrometry. Anal. Bioanal. Chem. 2019; 411:4167–4173. [DOI] [PubMed] [Google Scholar]
- 37. Bruckl T., Globisch D., Wagner M., Muller M., Carell T.. Parallel isotope-based quantification of modified tRNA nucleosides. Angew. Chem. Int. Ed. Engl. 2009; 48:7932–7934. [DOI] [PubMed] [Google Scholar]
- 38. Borland K., Diesend J., Ito-Kureha T., Heissmeyer V., Hammann C., Buck A.H., Michalakis S., Kellner S.. Production and application of stable Isotope-Labeled internal standards for RNA modification analysis. Genes (Basel). 2019; 10:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Waghmare S.P., Dickman M.J.. Characterization and quantification of RNA post-transcriptional modifications using stable isotope labeling of RNA in conjunction with mass spectrometry analysis. Anal. Chem. 2011; 83:4894–4901. [DOI] [PubMed] [Google Scholar]
- 40. Kung A.W., Kilby P.M., Portwood D.E., Dickman M.J.. Quantification of dsRNA using stable isotope labeling dilution liquid chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 2018; 32:590–596. [DOI] [PubMed] [Google Scholar]
- 41. Li S., Limbach P.A.. Method for comparative analysis of ribonucleic acids using isotope labeling and mass spectrometry. Anal. Chem. 2012; 84:8607–8613. [DOI] [PubMed] [Google Scholar]
- 42. Paulines M.J., Limbach P.A.. Stable isotope labeling for improved comparative analysis of RNA digests by mass spectrometry. J. Am. Soc. Mass Spectrom. 2017; 28:551–561. [DOI] [PubMed] [Google Scholar]
- 43. Taoka M., Nobe Y., Hori M., Takeuchi A., Masaki S., Yamauchi Y., Nakayama H., Takahashi N., Isobe T.. A mass spectrometry-based method for comprehensive quantitative determination of post-transcriptional RNA modifications: the complete chemical structure of Schizosaccharomyces pombe ribosomal RNAs. Nucleic Acids Res. 2015; 43:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chionh Y.H., Ho C.H., Pruksakorn D., Ramesh Babu I., Ng C.S., Hia F., McBee M.E., Su D., Pang Y.L., Gu C. et al.. A multidimensional platform for the purification of non-coding RNA species. Nucleic Acids Res. 2013; 41:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hauenschild R., Tserovski L., Schmid K., Thuring K., Winz M.L., Sharma S., Entian K.D., Wacheul L., Lafontaine D.L., Anderson J. et al.. The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res. 2015; 43:9950–9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kanduc D. Fractionation of rat liver tRNA by reversed-phase high performance liquid chromatography: isolation of Iso-tRNAs(Pro). Prep. Biochem. 1994; 24:167–174. [DOI] [PubMed] [Google Scholar]
- 47. Riml C., Glasner H., Rodgers M.T., Micura R., Breuker K.. On the mechanism of RNA phosphodiester backbone cleavage in the absence of solvent. Nucleic Acids Res. 2015; 43:5171–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gaston K.W., Limbach P.A.. The identification and characterization of non-coding and coding RNAs and their modified nucleosides by mass spectrometry. RNA Biol. 2014; 11:1568–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wyatt J.R., Chastain M., Puglisi J.D.. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991; 11:764–769. [PubMed] [Google Scholar]
- 50. Heiss M., Reichle V.F., Kellner S.. Observing the fate of tRNA and its modifications by nucleic acid isotope labeling mass spectrometry: NAIL-MS. RNA Biol. 2017; 14:1260–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reichle V.F., Weber V., Kellner S.. NAIL-MS in E. coli determines the source and fate of methylation in tRNA. Chembiochem. 2018; 19:2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Torres A.G., Pineyro D., Rodriguez-Escriba M., Camacho N., Reina O., Saint-Leger A., Filonava L., Batlle E., Ribas de Pouplana L.. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015; 43:5145–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cattenoz P.B., Taft R.J., Westhof E., Mattick J.S.. Transcriptome-wide identification of A >I RNA editing sites by inosine specific cleavage. RNA. 2013; 19:257–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Morse D.P., Bass B.L.. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry. 1997; 36:8429–8434. [DOI] [PubMed] [Google Scholar]
- 55. Macmaster R., Zelinskaya N., Savic M., Rankin C.R., Conn G.L.. Structural insights into the function of aminoglycoside-resistance A1408 16S rRNA methyltransferases from antibiotic-producing and human pathogenic bacteria. Nucleic Acids Res. 2010; 38:7791–7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thomas B., Akoulitchev A.V.. Mass spectrometry of RNA. Trends Biochem. Sci. 2006; 31:173–181. [DOI] [PubMed] [Google Scholar]
- 57. Bjorkbom A., Lelyveld V.S., Zhang S., Zhang W., Tam C.P., Blain J.C., Szostak J.W.. Bidirectional direct sequencing of noncanonical RNA by Two-Dimensional analysis of mass chromatograms. J. Am. Chem. Soc. 2015; 137:14430–14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Paul M.S., Bass B.L.. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998; 17:1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
https://mods.rna.albany.edu/masspec/Mongo-Oligo, http://modomics.genesilico.pl/.