Abstract
Measuring minimal residual disease in cancer has applications for prognosis, monitoring treatment and detection of recurrence. Simple sequence-based methods to detect nucleotide substitution variants have error rates (about 10−3) that limit sensitive detection. We developed and characterized the performance of MASQ (multiplex accurate sensitive quantitation), a method with an error rate below 10−6. MASQ counts variant templates accurately in the presence of millions of host genomes by using tags to identify each template and demanding consensus over multiple reads. Since the MASQ protocol multiplexes 50 target loci, we can both integrate signal from multiple variants and capture subclonal response to treatment. Compared to existing methods for variant detection, MASQ achieves an excellent combination of sensitivity, specificity and yield. We tested MASQ in a pilot study in acute myeloid leukemia (AML) patients who entered complete remission. We detect leukemic variants in the blood and bone marrow samples of all five patients, after induction therapy, at levels ranging from 10−2 to nearly 10−6. We observe evidence of sub-clonal structure and find higher target variant frequencies in patients who go on to relapse, demonstrating the potential for MASQ to quantify residual disease in AML.
INTRODUCTION
Accurate counting of nucleic acid templates is often critical for assessing biological phenomena, in particular when measuring the amount of a non-host genome. When the non-host genome is a pathogen whose genomic sequence is vastly different from the host genome, many methods are available, almost all based on polymerase chain reaction (PCR). However, when measuring minimal residual disease (MRD) for malignancies, the typical cancer genome differs from the host germline genome in only a few positions. This similarity presents formidable problems for detection and quantitation of variants. Because of sequence errors and amplification biases, PCR alone is insufficient for accurately detecting or quantifying rare variants. However, performance can be improved by coupling PCR to an additional ‘protocol’ such as limiting dilution, the counting of PCR cycles, or counting the number of different tags added to initial templates (1–3).
Current approaches for detecting and measuring MRD include multi-parametric flow cytometry (4–6), FISH (7), PCR detection of fusion transcripts (8,9) and targeted sequencing of common mutations (10–16). Each of these methods have their utility, but many are limited in their sensitivity, specificity and/or applicability to all patients. Improving the measurement of MRD in cancer should lead to better informed treatment decisions. In this paper, we present and demonstrate a protocol and analysis pipeline for multiplex accurate sensitive quantitation (MASQ), a method that can accurately count single nucleotide variants (SNVs) at many positions against a background of millions of host genomes.
We developed the MASQ protocol to satisfy six important properties. First, the method is quantitative, achieved by adding a unique sequence tag to the initial templates, which results in an accurate count not distorted by amplification. Second, it has a low error rate, below 10−6, achieved by demanding multiple read consensus for each template tag, thus removing or correcting amplification and sequencing-platform error. Utilization of a proofreading polymerase also reduces error generated during PCR. Third, MASQ enriches for target loci, which facilitates the use of a large amount of input material. This is necessary to examine millions of templates per locus and achieve high sensitivity. Fourth, it can assay many loci simultaneously, on the same starting material, which increases both the sensitivity and specificity in detecting low levels of residual disease. This approach also makes maximal use of valuable patient samples. Fifth, the method achieves a high yield, uniform across all loci, by performing many rounds of linear amplification prior to exponential amplification. Sixth, it enables an empirical error model, based on the error counts at non-target control positions, thus improving the accuracy of target variant frequency estimates by correcting for remaining error, and reducing false positives by providing accurate detection thresholds.
The last few years have seen an influx of new protocols and informatics that use next generation sequencing to detect rare variants (17). These methods satisfy some of these desirable qualities but not others (Supplementary Table S1). Duplex sequencing (18) and Illumina TruSight kits use tagged primers to achieve accurate quantitation and low error rates. However, both methods use capture hybridization to enrich for target loci, which imposes a restrictive limit on the amount of input DNA and also suffers from poor yield. Other methods modify the standard PCR protocol and/or informatics to obtain error rates of ∼10−4 (19–23), however these methods are quantitatively imprecise at variant allele frequencies below 1:5000. In contrast, the MASQ protocol and informatics satisfy the full range of desirable qualities. To establish this, we performed a set of tests to demonstrate the performance characteristics of the MASQ protocol while varying a range of conditions such as template concentration, DNA volume and number of loci queried.
We also demonstrate a use case for MASQ in five patients with acute myeloid leukemia (AML), measuring the proportion of leukemic cells present in the blood and bone marrow at presentation, during clinical remission and at relapse. We use whole genome sequencing (WGS) to identify hundreds of patient-specific variants, selecting ∼30 per patient to target with MASQ. Having hundreds of variants to choose from allows us to select for those with advantageous error profiles while broadly sampling clonal tumor heterogeneity. Our results support previous observations that although a patient is in cytological remission, with no detectable leukemic blasts, the patient still has measurable MRD, and that levels of such may be a predictor of long term response (13–14,24–29). We observe clustering of variant frequencies following treatment, which may prove critical in understanding and predicting relapse (30,31).
MATERIALS AND METHODS
The MASQ method depends on the interaction of bench protocols and informatics. We first apply a set of computational algorithms to identify patient-specific SNV target variants where the tumor genome differs from the normal genome. We select from those SNVs a set of target loci that satisfy constraints about fragment length, mutation context, sequence uniqueness and proximity to a restriction enzyme cut-site. A final algorithm identifies a set of compatible restriction enzymes and designs patient-specific primer sequences. The bench protocol uses those restriction enzymes and sequence primers to generate a sequence library where most sequence reads cover the targeted loci and where each target read has a sequence tag that uniquely identifies its originating molecule. A second informatics pipeline analyses the read data, collecting reads from the same locus, identifying reads from the same initial molecule and correcting sequencing errors by consensus where possible. Counting the SNVs observed after error correction provides an accurate quantitative measure of the tumor genome while counting variants observed at neighboring positions in the locus inform our models for sequence error. These error profiles feed back into the locus selection procedure and are important in the statistical interpretation of counts at the target positions.
Identifying target variants
To identify tumor-specific SNVs, we compare whole genome sequence data from the tumor with that of a paired normal DNA sample. For the five AML patients, we used the remission blood sample as the normal tissue. To identify tumor-specific sequence variants, we used a custom software pipeline detailed in the Supplementary Methods. Based on our analyses (see ‘Results’ section), not all variant sites are equally good candidates for error correction. Moreover, in order to multiplex, a batch of target variants must share a compatible set of restriction endonucleases (REs) and primer sites (see Figure 1). We developed an algorithm that, given a list of candidate variants from the tumor genome, finds a large set of target loci that satisfy protocol and error optimization requirements (Supplementary Methods and Figure S1). Several hundred candidate target variants were available for each of five AML samples in our study. From these hundreds of candidate variants, 25–30 compatible target variants per patient were chosen for MASQ. To demonstrate the efficiency of primer design, we simulated target variant selection given different numbers of input SNVs (Supplementary Figure S2).
Figure 1.
MASQ Protocol. (A) In step 1, genomic input DNA (gray and blue lines) is cleaved with a set of REs near the target variants (vertical red hash). The cleavage sites serve in steps 2–3 as the entry point to ‘guided elongation’, which is the method by which a ‘varietal tag’ sequence (VT) and a universal primer sequence (UP) is added to a specific strand (blue) of the target locus using the target-specific primer A (green) with a blocked 3′ end that cannot be extended. These, and all the following steps, are performed in multiplex mode for a compatible batch of targets and REs. In steps 4–5, multiple copies of the elongated target locus are generated by linear amplification using a biotinylated primer. The biotin is used to enrich linear copies of the targets (step 6). Exponential PCR uses the universal primer and a target specific primer for each locus (step 7). Sequencing libraries are prepared from PCR products using standard methods (step 8). (B) Target variants chosen from all candidates must have the allowed distance to the enzyme cut site and specified range for amplicon length, as specified in the figure.
MASQ bench protocol
The bench procedure is illustrated in Figure 1. The protocol steps are performed in multiplex mode for a compatible batch of target loci and REs. In step 1, input DNA is cleaved with a set of REs near the target variants (vertical red hash). The cleavage sites serve in steps 2–3 as the entry point to ‘guided elongation’, which is the method utilized to add a ‘varietal tag’ sequence (VT) and a universal primer sequence (UP) to each of the original target templates. A varietal tag is one from a diverse set of random sequences that when added to its target sequence renders an effectively unique combination of nucleotides (also known as unique molecular identifier, UMI) (1). Guided elongation consists of hybridizing a ‘guide’ oligonucleotide to a specific template so it can be elongated at the cut site. In steps 4–5, multiple copies of the elongated target locus are generated by linear amplification using a biotinylated primer. The biotin permits the enrichment of linear copies of the targets by capture with streptavidin beads (step 6). Exponential PCR is carried out using the universal primer and a target-specific primer for each locus (step 7). Sequencing libraries are prepared from PCR products (step 8). Further details are found in the Supplementary Methods and primer sequences are listed in Supplementary Table S2.
MASQ informatics protocol
Sequencing reads are processed through the following computational pipeline as illustrated in Supplementary Figure S3: Reads from a single assay are each assigned to one of the multiplexed target loci. Replicate reads with the same template varietal tag are aggregated. The number of reads per template (RPT) is tabulated. A consensus rule is applied to call a base at each position of the template, both at target and control positions. The consensus base is either the expected host base or one of the three possible variant bases. The consensus rule applies when the RPT > 1. In practice, we restrict our attention to templates with RPT ≥ 2. A consensus base is called if 80% of the replicate reads agree at that template position, otherwise we make no base call, thus resulting in both the removal and correction of errors. At each position, the number of consensus bases corresponding to each of the four possible bases is counted. At control positions, we assume that variant base observations are the result or error and so we calculate empirical error rates for each of the variant bases. This error rate is the number of consensus variant bases at that position divided by the total number of templates with consensus calls at that position. Template positions are further grouped by their 64 trinucleotide sequence contexts. Target surrogates are defined as the control positions in the templates that match the trinucleotide context of the target variant. The relevant surrogate error rates are used to adjust the counts of each target variant, considering that a portion of the count may be derived from error. Surrogate error rates are also used to assess whether the aggregate count at the target variant exceeds that possible by error.
Modeling noise and estimating frequency by surrogate sampling
Statistical methods are required to evaluate whether a cancer genome is present in a sample, and if so, to estimate its proportion relative to the host germline genome. We perform these tasks for a single target variant by sampling ‘error’ from all its surrogate positions. Additionally, we extend this method to aggregate signal over multiple target positions.
Despite low error rates on the order of 10−6, when sequencing hundreds of thousands of templates per locus, variant bases will still be observed, albeit infrequently, at control positions. These originate either from low-level pre-existing somatic mutation, or from events arising during any of the steps leading to library preparation and the final sequence acquisition. Regardless of origin, we call these ‘background error’ or just ‘error’. We infer the error rate at the target positions from the error rate at surrogate positions, i.e. control positions with the same trinucleotide context as target positions. Typically, any given assay has sufficient surrogate positions to determine the background error rate for each context. Because error rates for surrogate positions match well from assay to assay (Supplementary Table S3), there is an option to derive this error estimation from other assays. However, in this report, we only utilize surrogates from within the same assay.
For each target variant, over many iterations, we randomly sample from the background error rates of the surrogate positions. This allows us to estimate what proportion of the target variant count likely derived from error. By subtracting the estimated error count from the observed target count, we obtain an adjusted target count. We assume the adjusted count is drawn from a binomial distribution, and use that to estimate the frequency distribution. We perform multiple independent samplings and average the resulting distributions. The mean and 90% confidence interval are obtained from this distribution (see Supplementary Methods for details).
RESULTS
Not all mutations are equally easy to measure since some sequence contexts will generate a higher rate of background error than others. Using information from control positions, we determine that the major factors influencing background error are the sequence context (flanking nucleotides) and the specific base change. Determining background error rates as a function of context is important for selecting variants with desirable error profiles. We then explore the key performance characteristics of the MASQ protocol: proportion of reads on target, uniformity of coverage and accuracy of measurement. We test these parameters while varying the number of loci tested, the amount of input DNA and the proportion of variant genome. We demonstrate the performative advantage of MASQ over standard methods, comparing to a standard PCR library and to the MASQ data without tag counts and error correction. In the last section, we apply these methods to a small illustrative study in AML.
Mutational context
To determine the error rate characteristics of MASQ, we performed eight separate assays that differed in target loci, DNA source and depth of coverage. Both RPT and trinucleotide sequence context contribute to variation in error rate. Figure 2A shows the error rate as a function of RPT from each of these eight assays. The curve in red represents average error rate across all assays and all positions. Templates with only one read have an overall error rate of 10−3. By demanding consistency in the sequence of multiple reads from the same template, much lower error rates can be observed. In principle, with multiple first round linear copies, followed by highly redundant sequencing of each linear copy, all errors could be eliminated except those arising from template damage or from consistent machine error. Error rates drop as RPT increases, averaging to below 10−5 for positions in templates with at least two reads (Figure 2A, right side). Considerably better error rates are achieved at positions with preferred nucleotide contexts, as next discussed.
Figure 2.
Sources of error. (A) Error rate versus RPT. Templates are grouped into bins based on the number of RPT (X-axis). The error rate for templates in a bin is the average error rate at each control positions and each possible variant base. They are determined by the consensus rules and plotted on the Y-axis. Each line represents one of eight assays (set A and set C, as described in ‘Performance’ section). The red line indicates the error rates when all assays are combined into one dataset. (B) Error rate versus RPT is broken down by all possible 64 trinucleotide sequence contexts and each of their three possible variant bases (192 in total). Error rates are shown from the combined dataset. Lines are colored by the central base substitution. (C) Summary table for all 64 trinucleotide sequence contexts and each of their three possible variant bases of their mean error rate in the combined dataset. Red indicates higher error rates and blue indicates lower error rates. Black boxes surround the high error sequence contexts. (D) Error rate versus RPT for each of eight assays and the combined dataset, separated into high (blue) and low (green) error sequence contexts.
Analyzing 64 different trinucleotide contexts with three central base substitutions results in 192 distinct error rates; all are plotted in Figure 2B and summarized over eight assays in Figure 2C. The 192 individual rates from each assay, with a 2 RPT cut-off, are found in Supplementary Table S3. Error rates vary over three orders of magnitude, and more than half of the 192 variant possibilities have error rates <10−6. The primary determinant of the rate is the central nucleotide substitution, which is color-coded in Figure 2B. For example, G to T variants, in any trinucleotide context, have high error rates. This may reflect spontaneous depurination of G in the template (32). Many DNA polymerases insert an A when confronted with an apurinic site, resulting in a G to T conversion. Some surrounding contexts matter, as exemplified by a high error rate (10−4) for CG to TG, whereas CA to TA has a lower error rate (10−5). This may reflect the spontaneous deamination of 5-OH-methyl-C to T occurring in vitro (33). The strand-specific nature of the CpG errors was confirmed by targeting each strand independently (Supplementary Figure S4). This error rate analysis by trinucleotide context enables one to choose more reliable variants to assess (Figure 2D), and also improves statistical modeling.
Analytic sensitivity by serial dilution and surrogate sampling
We assess the analytical sensitivity of MASQ and illustrate the surrogate sampling method (see ‘Materials and Methods’ section) using a set of 10-fold serial dilution assays of one sample spiked into another (1 in 102 to 1 in 105). Twenty heterozygous variants, present in the spiked-in genome (LCL1) and absent in the host genome (SKN1), are the target variants in these assays. These estimates are shown for each variant in Figure 3A, and tabulated in Supplementary Table S4. In the lowest dilution assay, single variant estimates range from 2.4 × 10−6 to 2.5 × 10−5, which in all but one instance exceeds the maximum error rate from the surrogates. In 8 out of 20 variants the minimum of the confidence interval is below the maximum error rate from surrogates.
Figure 3.
Estimation of target variant frequencies. (A) Target variant estimations for 10-fold serial dilution experiments. Colored dots indicate the single variant estimates, with vertical black bars indicating the 90% confidence interval for the estimate. Horizontal colored lines indicate the aggregate estimate for that assay using all target variants. Points and lines are colored by the dilution level of the sample. The yellow triangles represent the maximum observed error frequency of any surrogate matching the same sequence context as the variant from the 1:10^5 dilution assay. Target loci are sorted identically for all dilution samples, by the difference between the variant estimate and maximum surrogate for the 1:105 dilution sample. (B) Illustration of the method of aggregate estimation for the 1:105 dilution sample. (Main Panel) Histogram of aggregate error counts derived from 10 000 simulations in which a matching surrogate for each of the target variants is selected, and their error counts are summed. A zoomed view of the aggregate error count distribution is in the left inset panel. The right inset panel indicates the posterior distribution for the aggregate frequency estimate. The pink line shows the range of aggregate scores equivalent to the frequency estimate range when multiplied by the total number of templates observed. The black star indicates the target variant aggregate score.
By taking an aggregate measure of signal over all variant positions we obtain much greater power. To do this we use the same framework as described above for single variants (see Figure 3B). In each iteration, we randomly choose a surrogate for each of the target variants, and sum their sampled error counts into an aggregate score. The distribution of these aggregate counts for the 1:105 dilution assay is shown in Figure 3B and the left inset panel. We then compare the aggregate count of the target variants to the distribution of aggregate scores from the simulation. If the target count is contained within the simulated values, we calculate its P-value as the proportion of the distribution greater than the target score. If the target score is outside the range, we report the P-value limit and calculate the number of standard deviations, z, beyond the mean of the distribution. For the final dilution, we obtain a P-value < 10−4 and z = 52. Lastly, the aggregate estimate of the likely proportion is calculated as for the single variants. This distribution is shown in Figure 3B, right inset panel, and the mean estimate is indicated by the colored lines in Figure 3A.
Performance
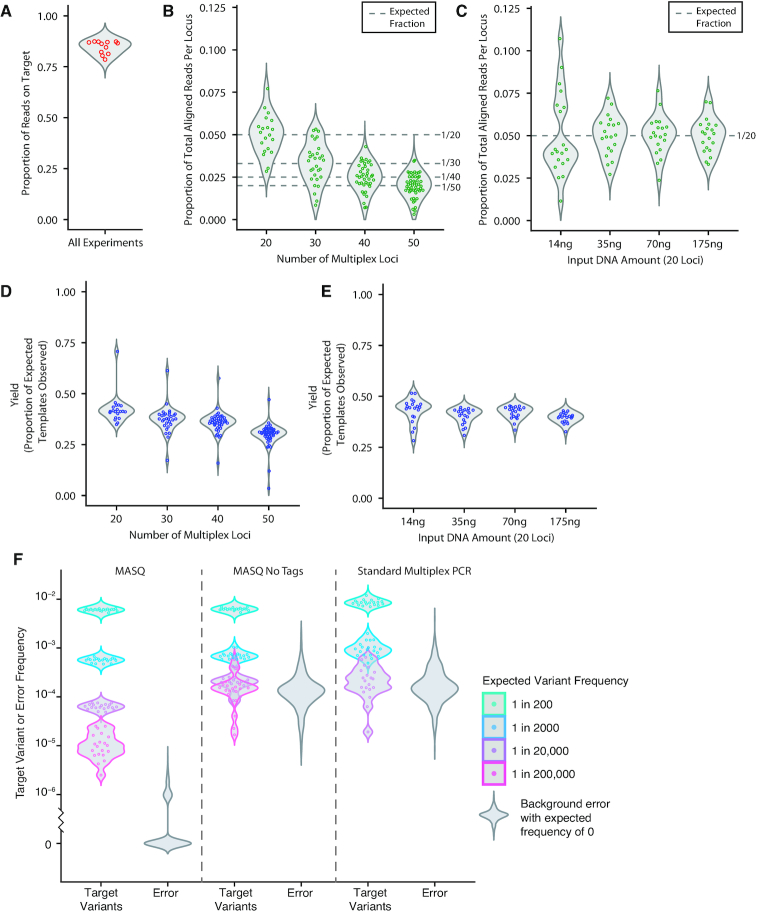
The performance of MASQ was evaluated on assays using various amounts of input DNA and varying numbers of targets. The performance criteria include: the yield of templates observed, the evenness of reads per locus and the efficiency of sequencing, namely, the proportion of all reads from a library that match a template. Details on these performance characteristics for each assay are summarized in Supplementary Table S5. Figure 4 shows data from 12 assays. In four assays (set A) the number of multiplexed loci varied from 20 to 50 using nested subsets of the 50 loci with 1.5 μg input. In four assays (set B) the amount of input DNA varied from 14 to 175 ng for 20 fixed loci. Four assays (set C) derive from the dilution assays previously discussed (1.4 to 14 μg input, 20 loci).
Figure 4.
Performance metrics for MASQ. (A–E) Performance metrics from 12 different assays are shown. (A) Proportion of reads on-target. Reads pairs that are on-target have both the expected sequence structure and align to one of the target loci. (B) Proportion of total aligned reads that map to each single target locus are plotted for each locus from four assays. The number of loci vary from 20 to 50. A total of 1.5 μg input DNA was used for all assays. (C) Proportion of total aligned reads that map to each single target locus are plotted for each locus from four assays. These assays have a constant 20 target loci but vary the input DNA amount from 14 to 175 ng. For a comparison to 1.5 μg input DNA at 20 loci see Panel B. (D) The yield, or proportion of expected template molecules recovered, is shown for set A, which varies in number of loci. (E) The yield, or proportion of template molecules recovered, is shown for set B, which varies in input DNA amount. For both D and E, the number of expected template molecules is calculated using the amount of DNA input in nanograms and the approximation of 3.59 picograms per haploid human genome. (F) Dilution assay target variant frequencies and error distributions for MASQ, MASQ without varietal tags for error correction or counting, and standard multiplex PCR. Violin plots are colored by dilution level. Background error distributions are in gray.
Figure 4A shows that, in each of the 12 assays, nearly all (80–90%) reads have the expected sequence structure and map to one of the target loci. These reads map relatively evenly to each of the target loci regardless of the number of loci (Figure 4B, set A) or the amount of input DNA (Figure 4C, set B). The exception is some unevenness at the lowest amount of DNA input, more as number of reads than as the number of uniquely tagged templates. The uniquely tagged template counts are even more tightly distributed than read counts, regardless of number of loci (Figure 4D, set A) or input DNA (Figure 4E, set B). The proportion of template molecules recovered is nearly 50% when only 20 loci are examined at any input level (Figure 4E), dropping to half that value when 50 loci are examined (Figure 4D).
Additionally, the distributions of RPT were analyzed as a function of overall sequencing coverage. Supplementary Figure S5 shows for each of the three sets of MASQ assays, the proportion of tags with at least 2 reads as a function of read depth (expressed as average RPT). Down-sampling of existing datasets shows that with an average of 2 RPT, 53% of the templates have at least 2 RPT and hence can be error corrected. This proportion increases to 84% at an average of 5 RPT and 93% at an average of 10 RPT.
Comparison to alternative methods
To assess the advantages of MASQ over other approaches, we compared our results to standard multiplex PCR using the same dilution assay approach. We performed PCR at the three concentrations where the amount of input DNA needed to observe the variant was within the acceptable parameters for PCR. We also re-analyzed the MASQ sequence data ignoring varietal tag information. This foregoes error correction and template count and the resulting ‘no-tag MASQ’ results closely mirror those of standard PCR.
We first note that multiplex PCR generates a similar proportion of reads on target as MASQ (90–95%). However, multiplex PCR lacks the first-strand target enrichment used in MASQ and therefore the target reads are less uniformly distributed by locus than in MASQ. For PCR, the coefficient of variation in coverage is 0.8 (compared to 0.2 for MASQ) and the locus with the least coverage is at 3% of the mean coverage (compared to 33% for MASQ, see Supplementary Figure S6). Importantly, when measuring the frequency of an allele, we find a dramatic reduction in accuracy for a dilution in the range of 1 in 10 000 (Figure 4F), consistent with literature on PCR (19–23). This results from the background error rate increasing to an average of 10−4 per position. When we examine the MASQ data but ignore tag information, we obtain a degradation of performance similar to what we see with PCR, namely the loss in quantitative accuracy and high background error rates (see Figure 4F; Supplementary Figures S7–9). In both the standard PCR and no-tag MASQ datasets, target variants start becoming difficult to distinguish from background error when they are present below 1 in 1000 (Supplementary Figure S8); and at frequencies of 1 in 10 000 and below, sensitivity and specificity of standard multiplex PCR and no-tag MASQ are dramatically worse than MASQ, which maintains a perfect sensitivity at 98% specificity for target variants present at a frequency of 1 in 200 000 (Supplementary Figure S9).
Application: Minimal residual disease in AML
We have applied MASQ to the clinically relevant question of measuring MRD in AML. In this pilot study, DNA from five AML patients was assayed at three disease stages: presentation, remission and relapse (where applicable). The remission sample was taken immediately after induction therapy, but prior to consolidation therapy (34). All five patients had a complete cytological response to induction therapy, with 0–1% blast cells detected by cytological exam. Two patients relapsed <1 year after presentation (pt27 and pt17), and expired shortly after relapse. One patient (pt49) relapsed 2 years after presentation, followed by a second remission and second relapse. The final two patients (pt12 and pt57) remain in long term remission more than 8 years after initial diagnosis. A summary of these clinical landmarks is shown in Figure 5A, and additional clinical information is listed in Supplementary Table S6. DNA from presentation and relapse was isolated from peripheral blood or bone marrow, depending on availability. DNA isolated from bone marrow and from blood were both assayed for the remission time point (Figure 5).
Figure 5.
Target variant frequencies in five AML patients. (A) Violin plots show the distribution of target variant frequency estimates for each patient and time point. Clinical histories are listed below as a table. Each section represents one patient, with presentation, remission and relapse (when applicable and available) samples shown from left to right. At the remission time point, results from both blood and bone marrow are shown. The points plotted each represent one of 24–30 variants assayed per sample. The black horizontal dashed lines indicate the aggregate estimate when all variants are taken together. (B) Target variant frequency estimates are again plotted, but on a customized linear scale and sorted in the order of their frequency at remission in blood samples. Black vertical bars indicate the 90% confidence interval for each variant estimate. The black horizontal dashed lines indicate the aggregate estimate when all variants are taken together.
To measure MRD in each patient, target somatic variants specific to each individual's leukemic cells were identified. Variants were selected by comparing WGS from presentation samples and ‘normal’ samples obtained at remission. Leukemic variants were chosen from variants detectable via WGS at presentation that were dramatically reduced in frequency, or not detectable, in the remission WGS data (Supplementary Table S7). The WGS analysis resulted in 272 to 1118 candidate variants per patient, of which 4 to 14 were in exonic regions, consistent with previous WGS studies of AML (35,36). For the relapse cases, 92% (pt27) and 88% (pt17) of variants detected at presentation in the WGS data are present at relapse. Further selection of target variants was based on rates of error for each trinucleotide substitution, as well as for compatibility with MASQ. Between 27 and 30 variants per individual were selected to multiplex in one MASQ assay. These variant sites were first verified by non-quantitative PCR on DNA from presentation and remission. Quality control on MASQ data based on performance metrics was used to eliminate a minority of poorly performing loci from further analysis (24–30 loci per patient in final dataset). Overall performance (Supplementary Table S5) and trinucleotide error rate profiles for the AML remission datasets (Supplementary Figure S10) are highly similar to those shown in Figures 2 and 4.
Frequency estimates of the leukemic target variants in each AML sample are presented in Figure 5A (and in Supplementary Table S8). Each dot represents a single target variant, and the violin plot shows the overall density of assayed variants for each sample. Two patients (pt27 and pt17) were assayed at presentation, at remission and at relapse. Three patients (pt49, pt12 and pt57) were assayed at presentation and at remission. We also tested two MASQ variant sets (from pt12 and pt57) on a negative control sample (see ‘Materials and Methods’ section). As expected, the frequencies reported in a negative control precisely reflect the expectation from surrogate positions (Supplementary Figure S11).
At presentation and relapse, the target variant frequencies are highest, and all but one of the frequencies fall into the range of 0.20 to 0.48. In contrast, as expected after a complete response to induction therapy, target variant frequencies drop significantly and vary from 3 × 10−6 to 0.05. Of the 288 variants assayed in remission, 4 have 90% confidence intervals including zero. Furthermore, two patients (pt12 and pt57) who achieved long-term remission (>8 years) have much lower overall variant frequencies at remission than those patients who relapsed, consistent with the belief that the extent of responsiveness to induction therapy correlates positively with patient outcome. Aggregate frequency estimates for each sample are also shown.
Overall variant concentrations in blood and bone marrow, from the same remission time point, are highly similar, with bone marrow concentrations slightly higher in four of five cases. The rank order of the variants across source material, as shown in Figure 5B, are nearly identical. These results attest to the near equivalence of blood and bone marrow and to the robustness of quantitation achieved in these assays.
Not all variants are present at equal frequencies within one sample. Figure 5B further examines the relationship between variant frequencies at different disease stages by sorting each patient's variants by decreasing frequency in blood during remission, maintaining the order across all time points. As observed most clearly in remission, the within-sample frequencies appear in all cases to cluster into discrete groups rather than a continuous spread. In three patients, this clustering is evident upon presentation or relapse.
Typically, there is a major cluster in remission, represented by the largest number of variants at one frequency. We present the aggregate estimates for these major clusters in Supplementary Figure S12 and Table S8. In all remission samples, there are variants more abundant than the major cluster. We hypothesize that these patterns are explained by residual phylogenetic lineages distinguished by nested sets of variants on the path to leukemia that may respond differently to the treatment regimen.
Importantly, in the relapse cases, all the variants assayed with MASQ increased from remission to relapse, approaching levels observed at presentation, suggesting the therapy did not completely eradicate the leukemic blasts.
To further demonstrate the importance of the varietal tags in MASQ for error correction and accurate quantitation, we re-analyzed the MASQ AML data while ignoring the tag information. The resulting variant frequencies, and background error distributions, are shown in Supplementary Figures S13 and S14. Error rates rise from below 10−6 in MASQ to above 10−4 in the ‘no-tag MASQ’ data. For patients 12 and 57, whose remission variant frequencies lie largely at 10−4 and below, variants that are easily distinguished from background error in MASQ become nearly indistinguishable from error in the absence of varietal tag correction. Furthermore, the tight clustering of variant frequencies in remission in patients 17, 12 and 57 is obscured in the no-tag MASQ data.
DISCUSSION
The MASQ protocol is designed to assay multiple genomic variants with great accuracy and at high depth of coverage against a background of normal genomes. To detect a variant genome at very low frequency requires both (i) a low error rate and (ii) observing a sufficient number of distinct molecules. These constraints were the driving factors in the development of the MASQ protocol. To obtain a low error rate, we choose loci that have advantageous error properties, use varietal tags to correct sequencing error by tagging the original template molecule and apply a rigorous error model to derive accurate quantitation. To obtain high yield over the target loci, we use linear amplification, enrich with biotinylated primers and simultaneously assay multiple loci. These properties make MASQ a useful assay for a variety of possible applications. In addition to quantifying residual disease in AML, MASQ has applications in counting circulating tumor cells in solid cancers, measuring tumor fraction in cell-free DNA (cfDNA) from plasma and quantifying tumor load in surgical margins. Outside of cancer applications, MASQ could be applied to counting fetal cells in maternal tissues or measuring low levels of mosaicism.
Measuring tumor load and heterogeneity
Our primary application for MASQ is the highly accurate and sensitive measurement of tumor load in the cancer patient. For solid cancers, tumor load is primarily assessed by imaging and for leukemia, by cytometry. As leukemia is a cancer of the blood, and solid cancer DNA is at least sometimes found in blood (37–41,34), we chose to detect sequence variants from the neoplasia against a large background of host germline DNA in blood. If successful, such a method could better inform treatment decisions. For leukemia patients, if residual disease persists, additional consolidation therapy including a bone marrow transplant may be warranted to eradicate all disease.
To increase the robustness and sensitivity of our assays, we chose to target many cancer variants. Despite the relatively low rate of sequence variation in AML (35,36,42–44), by opting for WGS we identified hundreds of variants from which to choose targets. Testing multiple loci increases the effective depth of the assay, increases robustness and enables subclonal resolution. A single variant can generate false positives because of spurious somatic variation or false negatives if its loss is the result of clonal drift or subclonal drug response. For MASQ, we chose to detect SNVs, by far the most abundant variants in solid cancers and leukemias. The selected SNVs were identified by WGS rather than targeted gene panels or whole exome sequencing, where not enough variants can be found (in our cases: 0–3 variants in recurrently mutated genes, 4–14 variants in exonic regions and 272–1118 variants per patient in the WGS data.) We did not focus here on common ‘pathogenic’ AML variants, although these could be included among the target variants in future studies using MASQ. By selecting multiple target variants from across the entire genome, we can integrate signal from multiple variants and at the same time capture the diversity of subclonal response to treatment.
Protocol properties
To achieve reduced error sequencing, one strand of the target template is elongated with random tags prior to any amplification and sequencing. Identical tags distinguish which reads derive from the same original template. After linear and then exponential amplification many reads per template are generated, and a template sequence is inferred only when those identically tagged reads are in agreement over a position. Following a thorough analysis of the error rates of all variants and tri-nucleotide contexts, certain nucleotide variant contexts are excluded, largely avoiding C or G changing to T. Under these conditions, we achieve error rates of 10−6 and below.
The MASQ method has several attractive properties: each locus can be quantified accurately. The yield is robust; about one third of the expected number of all input templates are observed. Multiplexing works well; both the number of reads per locus and the number of RPT are relatively uniform. At least 50 loci can be measured simultaneously, over a large range of input DNA amounts. Sequencing costs constrain the number of multiplexed loci, although we have not yet encountered an upper limit in terms of performance. Robust performance permits statistical modeling over aggregated data. Primer design is a bit complex, but we provide algorithms and code that yield suitable primers for the great majority of candidate target variants.
Comparison to other methods
Previously, SNVs have been of limited utility for measuring MRD because the error rates of PCR and sequencing are too high, on the order 10−3. Recent approaches to read filtering and variant selection reduce the error rates to the range of 10−4 to 10−5 but suffer from imprecision when the allele frequency falls below 1 part in 5000 (19). Duplex sequencing is a molecular tagging method that uses double-stranded asymmetric primers to reach error rates as low as 10−9. The utility of this method in a targeted capture framework, however, is constrained by low yield (<1%) and limited input (250 ng or ∼50 000 genomes) enabling a query of ∼500 genomes per reaction (18). In contrast, MASQ has error rates commensurate with its depth of coverage, capable of querying 180 000 genomes in a single reaction (2 μg or ∼600 000 genomes with 25–30% yield.) Combined with the power of simultaneously assaying multiple loci, MASQ provides a sensitivity and specificity that surpasses other error-correcting genomic methods (19–23,45–47).
Limitations of MASQ
While the MASQ error rate of 10−6 is sufficient for sensitive detection down to one part in a million, were we to sample on the order of 108 template molecules, such as are present in a 10 ml blood sample, lower error rates would be desirable. We attribute our sensitivity limit to two primary factors: template damage and early-round error while copying templates.
We already avoid template damage by preferentially selecting for target variants in particular sequence contexts. For example, we avoid target variants such as C or G changing to a T, as these loci frequently encounter deamination or depuration, leading to consistent errors upon copying. Sensitivity might be improved by first destroying damaged templates enzymatically (48,49). Duplex sequencing, discussed above, controls for template damage by copying from both strands of the same original molecule (50). Unfortunately, this approach is not easily incorporated into our method of targeted amplification.
Early-round amplification error is another factor that limits sensitivity. The clear evidence for this is that error is reduced as we sequence more copies of the original template (Figure 3A). Ideally, consensus rules should be applied only to first round copies. MASQ uses multiple first rounds of copying, but we cannot know whether identical tags for a read derive from one or many first copies of the original template. One approach that might address this issue is to add a second varietal tag such that each first round copy receives its own unique tag in addition to the common template tag. This could improve consensus base calling and reduce error rates.
Another approach for reducing error rates is to select insertion-deletion or ‘indel’ variants. Analysis of indel error in MASQ data suggest that consensus error for small deletions are <10−8 (Supplementary Table S9). Although indels are about a tenth as abundant as SNVs, even in AML, a neoplasia with a low mutation rate, we find hundreds of candidate indels (see Supplementary Table S10).
There have been reports that cancer-specific mutations from solid tumors are observable in the plasma component of blood as cfDNA (37–41). Existing evidence suggests that the fragment sizes for cfDNA are in the range of 120–200 bp (51). While the MASQ fragments reported here range from 100–300 bp, the same MASQ protocol can be applied to amplify variants in the shorter size range common to cfDNA. This could broaden the utility of MASQ to measuring disease burden in solid cancers as well.
Application to AML
Detection of MRD has proven clinical utility for AML, as a endpoint in chemotherapy trials, as a surrogate marker for response and for detecting and predicting relapse (4,13–14,24–27). We tested MASQ in a small pilot study, measuring MRD in five patients who entered remission following induction therapy. We detected leukemic variants in all five patients while in remission, at levels ranging from 10−2 to nearly 10−6. In the two patients that relapsed, virtually all of the variants that we assayed increased in frequency upon relapse. Nothing of statistical significance can be expected in such a small study. Moreover, interpreting MRD is more complicated than simply measuring the aggregate concentration of leukemic variants (30–31,52–54), as we and others observe evidence of subclonal variation in therapeutic response. Nevertheless, we observe a trend correlating levels of MRD and future relapse: for the aggregate signal, for the main cluster of variant frequencies and for the minimum variant frequencies.
The MASQ protocol performs as well on our patient samples as it does in our controlled laboratory experiments, with similar yields, alignment rates and proportion of reads on-target (Supplementary Table S5). The trinucleotide context error profiles are very similar (Supplementary Figure S10). Quantitation from blood very closely matches bone marrow, indicating that assaying variants in blood is likely to suffice for detection of MRD (5,55). Although the latter is slightly higher, the rank order of variant frequencies are nearly identical. The quantitation is such that we observe similar clustering of variant frequencies in blood and bone. In fact, the clustering of frequencies is an interesting and potentially useful feature of patient response.
Monitoring multiple leukemia-associated variants in an MRD assay enables robustness to technical and biological variation, increased sensitivity and accurate quantitation of leukemic and pre-leukemic (56–58) subclones across the course of treatment. With MASQ as a tool for MRD monitoring in AML, future studies can explore the clinical utility of this highly sensitive and quantitative assay to monitor patients and improve patient outcome.
DATA AVAILABILITY
Sequencing data has been deposited at the European Genome-phenome Archive (EGA), under accession number EGAS00001003732. Code is available at https://github.com/amoffitt/MASQ.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Breast Cancer Research Foundation [18-174 to M.W.]; Simons Foundation, Life Sciences Founders Directed Giving Research [519054 to M.W.]; Cold Spring Harbor Laboratory and Northwell Health Affiliation (to M.W.); Cold Spring Harbor Laboratory Cancer Gene Discovery and Cancer Biology Postdoctoral Training Program [National Cancer Institute NRSA] (to A.M.); Cold Spring Harbor Laboratory Cancer Center Support Grant [5P30CA045508]; Simons Center for Quantitative Biology at Cold Spring Harbor Laboratory (to A.K and D.L). Funding for open access charge: Simons Foundation, Life Sciences Founders Directed Giving Research [519054].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hicks J., Navin N., Troge J., Wang Z., Wigler M.. Varietal counting of nucleic acids for obtaining genomic copy number information. 2016; US Patent US20140065609A.
- 2. Wang A.M., Doyle M.V., Mark D.F.. Quantitation of mRNA by the polymerase chain reaction. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:9717–9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ramakers C., Ruijter J.M., Deprez R.H.L., Moorman A.F.. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003; 339:62–66. [DOI] [PubMed] [Google Scholar]
- 4. Kern W., Voskova D., Schoch C., Hiddemann W., Schnittger S., Haferlach T.. Determination of relapse risk based on assessment of minimal residual disease during complete remission by multiparameter flow cytometry in unselected patients with acute myeloid leukemia. Blood. 2004; 104:3078–3085. [DOI] [PubMed] [Google Scholar]
- 5. Maurillo L., Buccisano F., Spagnoli A., Del Poeta G., Panetta P., Neri B., Del Principe M.I., Mazzone C., Consalvo M.I., Tamburini A. et al.. Monitoring of minimal residual disease in adult acute myeloid leukemia using peripheral blood as an alternative source to bone marrow. Haematologica. 2007; 92:605–611. [DOI] [PubMed] [Google Scholar]
- 6. Miyazaki T., Fujita H., Fujimaki K., Hosoyama T., Watanabe R., Tachibana T., Fujita A., Matsumoto K., Tanaka M., Koharazawa H. et al.. Clinical significance of minimal residual disease detected by multidimensional flow cytometry: serial monitoring after allogeneic stem cell transplantation for acute leukemia. Leuk. Res. 2012; 36:998–1003. [DOI] [PubMed] [Google Scholar]
- 7. Gallo J., Robson L., Watson N., Sharma P., Smith A.. Comparison of metaphase and interphase FISH monitoring of minimal residual disease with MLL gene probe: case study of AML with t (9; 11). Ann. Genet. 1999; 42:109–112. [PubMed] [Google Scholar]
- 8. Ommen H.B., Schnittger S., Jovanovic J.V., Ommen I.B., Hasle H., Østergaard M., Grimwade D., Hokland P.. Strikingly different molecular relapse kinetics in NPM1c, PML-RARA, RUNX1-RUNX1T1, and CBFB-MYH11 acute myeloid leukemias. Blood. 2010; 115:198–205. [DOI] [PubMed] [Google Scholar]
- 9. Schnittger S., Weisser M., Schoch C., Hiddemann W., Haferlach T., Kern W.. New score predicting for prognosis in PML-RARA+, AML1-ETO+, or CBFBMYH11+ acute myeloid leukemia based on quantification of fusion transcripts. Blood. 2003; 102:2746–2755. [DOI] [PubMed] [Google Scholar]
- 10. Cloos J., Goemans B.F., Hess C.J., van Oostveen J.W., Waisfisz Q., Corthals S., de Lange D., Boeckx N., Hahlen K., Reinhardt D. et al.. Stability and prognostic influence of FLT3 mutations in paired initial and relapsed AML samples. Leukemia. 2006; 20:1217–1220. [DOI] [PubMed] [Google Scholar]
- 11. Kronke J., Schlenk R.F., Jensen K.O., Tschurtz F., Corbacioglu A., Gaidzik V.I., Paschka P., Onken S., Eiwen K., Habdank M. et al.. Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: a study from the German-Austrian acute myeloid leukemia study group. J. Clin. Oncol. 2011; 29:2709–2716. [DOI] [PubMed] [Google Scholar]
- 12. Salipante S.J., Fromm J.R., Shendure J., Wood B.L., Wu D.. Detection of minimal residual disease in NPM1-mutated acute myeloid leukemia by next-generation sequencing. Mod. Pathol. 2014; 27:1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schnittger S., Kern W., Tschulik C., Weiss T., Dicker F., Falini B., Haferlach C., Haferlach T.. Minimal residual disease levels assessed by NPM1 mutation–specific RQ-PCR provide important prognostic information in AML. Blood. 2009; 114:2220–2231. [DOI] [PubMed] [Google Scholar]
- 14. Shayegi N., Kramer M., Bornhäuser M., Schaich M., Schetelig J., Platzbecker U., Röllig C., Heiderich C., Landt O., Ehninger G. et al.. The level of residual disease based on mutant NPM1 is an independent prognostic factor for relapse and survival in AML. Blood. 2013; 122:83–92. [DOI] [PubMed] [Google Scholar]
- 15. Kohlmann A., Nadarajah N., Alpermann T., Grossmann V., Schindela S., Dicker F., Roller A., Kern W., Haferlach C., Schnittger S.. Monitoring of residual disease by next-generation deep-sequencing of RUNX1 mutations can identify acute myeloid leukemia patients with resistant disease. Leukemia. 2014; 28:129–137. [DOI] [PubMed] [Google Scholar]
- 16. Zuffa E., Franchini E., Papayannidis C., Baldazzi C., Simonetti G., Testoni N., Abbenante M.C., Paolini S., Sartor C., Parisi S.. Revealing very small FLT3 ITD mutated clones by ultra-deep sequencing analysis has important clinical implications in AML patients. Oncotarget. 2015; 6:31284–31294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salk J.J., Schmitt M.W., Loeb L.A.. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 2018; 19:269–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmitt M.W., Fox E.J., Prindle M.J., Reid-Bayliss K.S., True L.D., Radich J.P., Loeb L.A.. Sequencing small genomic targets with high efficiency and extreme accuracy. Nat. Methods. 2015; 12:423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma X., Shao Y., Tian L., Flasch D.A., Mulder H.L., Edmonson M.N., Liu Y., Chen X., Newman S., Nakitandwe J.. Analysis of error profiles in deep next-generation sequencing data. Genome Biol. 2019; 20:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stasik S., Schuster C., Ortlepp C., Platzbecker U., Bornhäuser M., Schetelig J., Ehninger G., Folprecht G., Thiede C.. An optimized targeted next-generation sequencing approach for sensitive detection of single nucleotide variants. Biomol. Detect. Quant. 2018; 15:6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerstung M., Beisel C., Rechsteiner M., Wild P., Schraml P., Moch H., Beerenwinkel N.. Reliable detection of subclonal single-nucleotide variants in tumour cell populations. Nat. Commun. 2012; 3:e811. [DOI] [PubMed] [Google Scholar]
- 22. Wei Z., Wang W., Hu P., Lyon G.J., Hakonarson H.. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011; 39:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilm A., Aw P.P.K., Bertrand D., Yeo G.H.T., Ong S.H., Wong C.H., Khor C.C., Petric R., Hibberd M.L., Nagarajan N.. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012; 40:11189–11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Corbacioglu A., Scholl C., Schlenk R.F., Eiwen K., Du J., Bullinger L., Frohling S., Reimer P., Rummel M., Derigs H.G. et al.. Prognostic impact of minimal residual disease in CBFB-MYH11-positive acute myeloid leukemia. J. Clin. Oncol. 2010; 28:3724–3729. [DOI] [PubMed] [Google Scholar]
- 25. Pastore F., Levine R.L.. Next-generation sequencing and detection of minimal residual disease in acute myeloid leukemia: ready for clinical practice. JAMA. 2015; 314:778–780. [DOI] [PubMed] [Google Scholar]
- 26. Walter R.B., Buckley S.A., Pagel J.M., Wood B.L., Storer B.E., Sandmaier B.M., Fang M., Gyurkocza B., Delaney C., Radich J.P. et al.. Significance of minimal residual disease before myeloablative allogeneic hematopoietic cell transplantation for AML in first and second complete remission. Blood. 2013; 122:1813–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walter R.B., Gooley T.A., Wood B.L., Milano F., Fang M., Sorror M.L., Estey E.H., Salter A.I., Lansverk E., Chien J.W. et al.. Impact of pretransplantation minimal residual disease, as detected by multiparametric flow cytometry, on outcome of myeloablative hematopoietic cell transplantation for acute myeloid leukemia. J. Clin. Oncol. 2011; 29:1190–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klco J.M., Miller C.A., Griffith M., Petti A., Spencer D.H., Ketkar-Kulkarni S., Wartman L.D., Christopher M., Lamprecht T.L., Helton N.M. et al.. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA. 2015; 314:811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Young A.L., Wong T.N., Hughes A.E., Heath S.E., Ley T.J., Link D.C., Druley T.E.. Quantifying ultra-rare pre-leukemic clones via targeted error-corrected sequencing. Leukemia. 2015; 29:1608–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ding L., Ley T.J., Larson D.E., Miller C.A., Koboldt D.C., Welch J.S., Ritchey J.K., Young M.A., Lamprecht T., McLellan M.D. et al.. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012; 481:506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Uy G.L., Duncavage E.J., Chang G.S., Jacoby M.A., Miller C.A., Shao J., Heath S., Elliott K., Reineck T., Fulton R.S. et al.. Dynamic changes in the clonal structure of MDS and AML in response to epigenetic therapy. Leukemia. 2017; 31:872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kunkel T.A. Mutational specificity of depurination. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:1494–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen J.-C., Rideout W.M. III, Jones P.A.. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 1994; 22:972–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Kouchkovsky I., Abdul-Hay M.. Acute myeloid leukemia: a comprehensive review and 2016 update. Blood Cancer J. 2016; 6:e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013; 368:2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tyner J.W., Tognon C.E., Bottomly D., Wilmot B., Kurtz S.E., Savage S.L., Long N., Schultz A.R., Traer E., Abel M. et al.. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018; 562:526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abbosh C., Birkbak N.J., Wilson G.A., Jamal-Hanjani M., Constantin T., Salari R., Le Quesne J., Moore D.A., Veeriah S., Rosenthal R. et al.. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017; 545:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coombes R.C., Page K., Salari R., Hastings R.K., Armstrong A.C., Ahmed S., Ali S., Cleator S.J., Kenny L.M., Stebbing J. et al.. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin. Cancer Res. 2019; 25:4255–4263. [DOI] [PubMed] [Google Scholar]
- 39. Garcia-Murillas I., Schiavon G., Weigelt B., Ng C., Hrebien S., Cutts R.J., Cheang M., Osin P., Nerurkar A., Kozarewa I. et al.. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015; 7:302ra133. [DOI] [PubMed] [Google Scholar]
- 40. Tie J., Wang Y., Tomasetti C., Li L., Springer S., Kinde I., Silliman N., Tacey M., Wong H.-L., Christie M. et al.. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016; 8:346ra392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Phallen J., Sausen M., Adleff V., Leal A., Hruban C., White J., Anagnostou V., Fiksel J., Cristiano S., Papp E.. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017; 9:eaan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brewin J., Horne G., Chevassut T.. Genomic landscapes and clonality of de novo AML. N. Engl. J. Med. 2013; 369:1472–1473. [DOI] [PubMed] [Google Scholar]
- 43. Ley T.J., Mardis E.R., Ding L., Fulton B., McLellan M.D., Chen K., Dooling D., Dunford-Shore B.H., McGrath S., Hickenbotham M. et al.. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008; 456:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mardis E.R., Ding L., Dooling D.J., Larson D.E., McLellan M.D., Chen K., Koboldt D.C., Fulton R.S., Delehaunty K.D., McGrath S.D.. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009; 361:1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Waalkes A., Penewit K., Wood B.L., Wu D., Salipante S.J.. Ultrasensitive detection of acute myeloid leukemia minimal residual disease using single molecule molecular inversion probes. Haematologica. 2017; 102:1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang K., Ma Q., Jiang L., Lai S., Lu X., Hou Y., Wu C.-I., Ruan J.. Ultra-precise detection of mutations by droplet-based amplification of circularized DNA. BMC Genomics. 2016; 17:e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ståhlberg A., Krzyzanowski P.M., Jackson J.B., Egyud M., Stein L., Godfrey T.E.. Simple, multiplexed, PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing. Nucleic Acids Res. 2016; 44:e105–e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B.. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 1977; 252:3286–3294. [PubMed] [Google Scholar]
- 49. Liu Y., Prasad R., Beard W.A., Kedar P.S., Hou E.W., Shock D.D., Wilson S.H.. Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase β. J. Biol. Chem. 2007; 282:13532–13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A.. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cristiano S., Leal A., Phallen J., Fiksel J., Adleff V., Bruhm D.C., Jensen S.Ø., Medina J.E., Hruban C., White J.R.. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019; 570:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hughes A.E., Magrini V., Demeter R., Miller C.A., Fulton R., Fulton L.L., Eades W.C., Elliott K., Heath S., Westervelt P.. Clonal architecture of secondary acute myeloid leukemia defined by single-cell sequencing. PLos Genet. 2014; 10:e1004462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lindsley R.C., Mar B.G., Mazzola E., Grauman P.V., Shareef S., Allen S.L., Pigneux A., Wetzler M., Stuart R.K., Erba H.P. et al.. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015; 125:1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Walter M.J., Shen D., Ding L., Shao J., Koboldt D.C., Chen K., Larson D.E., McLellan M.D., Dooling D., Abbott R. et al.. Clonal architecture of secondary acute myeloid leukemia. N. Engl. J. Med. 2012; 366:1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Leroy H., de Botton S., Grardel-Duflos N., Darre S., Leleu X., Roumier C., Morschhauser F., Lai J.L., Bauters F., Fenaux P. et al.. Prognostic value of real-time quantitative PCR (RQ-PCR) in AML with t(8;21). Leukemia. 2005; 19:367–372. [DOI] [PubMed] [Google Scholar]
- 56. Terwijn M., Zeijlemaker W., Kelder A., Rutten A.P., Snel A.N., Scholten W.J., Pabst T., Verhoef G., Lowenberg B., Zweegman S. et al.. Leukemic stem cell frequency: a strong biomarker for clinical outcome in acute myeloid leukemia. PLoS One. 2014; 9:e107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jan M., Snyder T.M., Corces-Zimmerman M.R., Vyas P., Weissman I.L., Quake S.R., Majeti R.. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 2012; 4:149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Corces-Zimmerman M.R., Hong W.-J., Weissman I.L., Medeiros B.C., Majeti R.. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:2548–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data has been deposited at the European Genome-phenome Archive (EGA), under accession number EGAS00001003732. Code is available at https://github.com/amoffitt/MASQ.