Abstract
DNA methyltransferases are primary enzymes for cytosine methylation at CpG sites of epigenetic gene regulation in mammals. De novo methyltransferases DNMT3A and DNMT3B create DNA methylation patterns during development, but how they differentially implement genomic DNA methylation patterns is poorly understood. Here, we report crystal structures of the catalytic domain of human DNMT3B–3L complex, noncovalently bound with and without DNA of different sequences. Human DNMT3B uses two flexible loops to enclose DNA and employs its catalytic loop to flip out the cytosine base. As opposed to DNMT3A, DNMT3B specifically recognizes DNA with CpGpG sites via residues Asn779 and Lys777 in its more stable and well-ordered target recognition domain loop to facilitate processive methylation of tandemly repeated CpG sites. We also identify a proton wire water channel for the final deprotonation step, revealing the complete working mechanism for cytosine methylation by DNMT3B and providing the structural basis for DNMT3B mutation-induced hypomethylation in immunodeficiency, centromere instability and facial anomalies syndrome.
INTRODUCTION
DNA methylation is the key mechanism of epigenetic regulation for various cellular events, including gene expression and genomic imprinting (1,2). In mammals, DNA methylation mainly occurs symmetrically on cytosine residues at the C5 position of CpG dinucleotides on both DNA strands. Mammalian C5-methylation is catalyzed by three DNA methyltransferases (DNMTs)—namely DNMT1, DNMT3A and DNMT3B—all containing a highly conserved methyltransferase catalytic domain (3). DNMT3A and DNMT3B are so-called de novo methyltransferases, as they are responsible for establishing new DNA methylation patterns during embryogenesis and genomic imprints during germ cell development (4,5). The enzymatic activities and multimerization of these two de novo DNMTs are further allosterically stimulated by a catalytically inactive family member, i.e. DNMT3L (6).
Mammalian DNMT3A and DNMT3B share a similar domain organization and high sequence identity (∼85%) in the methyltransferase catalytic domain, but they have distinctive physiological and pathophysiological roles, as well as different enzymatic properties (5,7–9). These two proteins are expressed at different developmental stages and methylate different as well as overlapping targets. DNMT3A is required for the methylation of imprinted genes and dispersed repeated elements, whereas DNMT3B specializes in the methylation of pericentric satellite repeats (2,5,10) and actively transcribed genes within the gene body to prevent spurious transcription initiation (11,12). Homozygous mutations in DNMT3B are associated with immunodeficiency, centromere instability and facial anomalies (ICF) syndrome (13–15), whereas heterozygous mutations in DNMT3A are linked to myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) (16). ICF syndrome-related mutations in DNMT3B result in hypomethylation of long stretches of satellite DNA repeats high in CpG content that are found in pericentromeric regions, leading to chromosome instability, impaired chromosome segregation and perturbed nuclear architecture (17,18).
Biochemically, DNMT3A is a distributive enzyme that methylates DNA by a cooperative mechanism (19,20), whereas DNMT3B performs methylation on DNA in a non-cooperative processive manner, allowing the enzyme to diffuse along DNA and methylate multiple sites before dissociation (21,22). Human DNMT3B also has a distinct strong flanking sequence preference for G at the downstream of CpG sites, unlike that of DNMT3A in vivo (23). Recent emerging evidence has shown that tumor cells generally exhibit genome-wide hypomethylation of repetitive DNA sequences or localized hypermethylation of tumor suppressor gene promoters due to DNMT overexpression, mutations or truncations (24). Therefore, it is important to understand how DNMT3B methylates its DNA targets and the impact of mutations and truncations on its enzymatic activity.
A number of structural investigations have focused on DNMT3A, including crystal structures of the catalytic domain of human DNMT3A–3L complex covalently bound with and without a cytosine-modified DNA. Those studies have revealed how DNMT3A forms a heterotetramer with DNMT3L, how the activity of the catalytic domain is autoinhibited by the ADD domain and how DNMT3A recognizes CpG sites upon binding to the unmethylated histone H3 tail (25–27). However, the molecular mechanism underlying processive DNA methylation by DNMT3B remains elusive. In this study, we purified the catalytic domains of human DNMT3B and DNMT3L, and determined the crystal structure of DNMT3B–3L complex noncovalently bound with and without a DNA duplex bearing two CpGpG sites. These structures faithfully reflect at the atomic level the working mechanism by which DNMT3B recognizes and methylates CpGpG sites. Together with mutational studies, DNA methylation activity assays and crystal structure analyses of mutated DNMT3B as well as DNMT3B–3L bound with a different DNA sequence bearing CpGpT sites, we also provide the structural basis for how DNMT3B processively methylates CpGpG sites with a sequence preference.
MATERIALS AND METHODS
Protein expression and purification
Catalytic domains of human DNMT3B (3B-CD, residues 571–853), human DNMT3L (3L-CD, residues 178–379) and human DNMT3A (residues 627–912) were amplified by using 2× PfuUltra II Hotstart PCR Master Mix (Agilent Technologies) and then cloned into pET28a(+)tev expression vector containing an N-terminal 6xHis-tag and a tobacco etch virus (TEV) cleavage site to generate pET28a(+)tev-3B-CD, pET28a(+)tev-3L-CD and pET28a(+)tev-3A-CD. All point mutations were generated using a QuikChange Kit (Stratagene). The recombinant proteins were expressed in the bacterial strain Rosetta2(DE3)pLysS. Cells were grown at 37°C in LB Broth (Miller) medium and protein expression was induced by addition of 0.4 mM isopropyl-thio-β-d-galactoside (IPTG) at OD600 = 0.6–0.7 for 16–18 h at 18°C. Cells were harvested by centrifugation and stored at −80°C.
To purify human DNMT3B–3L complex, pellets of the bacterial expressed 3B-CD and 3L-CD constructs were mixed, resuspended in 50 mM sodium phosphate (pH 7.4), 500 mM NaCl, 5% glycerol, 5 mM β-mercaptoethanol and ethylenediaminetetraacetic acid (EDTA)-free Protease Inhibitor Cocktail (Roche, Switzerland) and lysed using a microfluidizer (Microfluidics M-110P). After centrifugation at 35 000 × g for 40 min at 4°C, the supernatant was placed in a HisTrap FF column (5 ml, GE Healthcare). After washing with a wash buffer containing 50 mM sodium phosphate (pH 7.4), 500 mM NaCl, 5% glycerol, 5 mM β-mercaptoethanol and 40 mM imidazole, the DNMT3B–3L complex was eluted by means of an imidazole gradient from 40 to 500 mM. The eluted protein was treated with TEV protease and dialyzed overnight at 4°C against a buffer containing 20 mM sodium phosphate (pH 7.4), 225 mM NaCl, 5% glycerol and 5 mM β-mercaptoethanol. The cleaved, untagged DNMT3B–3L complex was further purified by HiTrap Heparin HP (5 ml, GE Healthcare) and HiLoad 16/60 Superdex 200 (GE Healthcare) columns pre-equilibrated in 20 mM Tris–HCl (pH 7.4), 200 mM NaCl, 5% glycerol and 0.5 mM TCEP. All of mutated DNMT3B–3L and DNMT3A–3L complexes were purified using this same protocol.
Protein crystallization and structure determination
Crystals of the human DNMT3B–3L complex were grown under two conditions: (i) the purified DNMT3B–3L complex sample (5.0 mg/ml) was stored in a 1.5-mL Eppendorf tube at 4°C and, after 16 h, crystals of DNMT3B–3L complex appeared in the tube; and (ii) DNMT3B–3L crystals were also grown using the hanging drop vapor diffusion method at 278 K by mixing 1.6 μl of the purified DNMT3B–3L complex sample (5.0 mg/ml) with 0.8 μl of reservoir solution containing 2.0–2.2 M sodium formate. Apart from DNMT3B–3L complex, crystals of the DNMT3B (Q772R)–3L complex were also grown according to condition (ii). To crystallize DNMT3B–3L–DNA complex, DNMT3B–3L complex solution was incubated with S-adenosyl-l-homocysteine (SAH) and a palindromic annealed DNA duplex containing two CpGpG sites (5′-GAATTCGGAAAAATTTTTCCGAATT-3′) or two CpGpT sites (5′-GCATGCGTTCTAATTAGAACGCATG-3′) (Supplementary Table S1) at a 1:3:1.2 molar ratio. Crystals of the DNMT3B–3L–DNA (CpGpG) complex were grown at 278 K by the hanging drop vapor diffusion method by mixing 1.6 μl of the protein–DNA mixture (4.0 mg/ml) with 0.8 μl of reservoir solution containing 50 mM MES (pH 5.7), 160 mM KCl, 10 mM MgSO4 and 14% PEG400. Crystals grew to 0.5–0.6 mm in the longest dimension after 1 week. Crystals of DNMT3B–3L–DNA (CpGpT) complex were also grown by the hanging drop vapor diffusion method using a reservoir solution containing 50 mM MES (pH 5.6), 200 mM KCl, 10 mM MgSO4 and 10% PEG400. Crystals were cryoprotected by their respective reservoir solution plus 25% glycerol before data collection.
The X-ray diffraction datasets for the human DNMT3B–3L–DNA, DNMT3B (Q772R)–3L and DNMT3B–3L complexes were collected at TPS beamline 05A and TLS beamline 15A, National Synchrotron Radiation Research Center, Hsinchu, Taiwan. All diffraction data were indexed, integrated and scaled using HKL2000 (28). The crystal structures were determined by molecular replacement in PHENIX Phaser (29) using the DNMT3A–3L complex structure (PDB entry: 5YX2) as the search model. All initial models were manually rebuilt using the program Coot (30) and then refined in PHENIX. Crystallographic and refinement statistics are listed in Table 1. All structure figures were generated using PyMOL (31) and UCSF ChimeraX (32).
Table 1.
X-ray diffraction and crystal structure refinement statistics for DNMT3B–3L and DNMT3B–3L–DNA complexes
| DNMT3B–3L–DNA (CpGpG) | DNMT3B–3L–DNA (CpGpT) | DNMT3B–3L (i) | DNMT3B–3L (ii) | DNMT3B (Q772R)–3L | |
|---|---|---|---|---|---|
| PDB entry | 6KDA | 6KDB | 6KDL | 6KDP | 6KDT |
| Data collection | |||||
| Space group | P31 | P31 | P31 | C2221 | C2221 |
| Cell dimensions | |||||
| a, b, c (Å) | 193.87, 193.87, 49.74 | 194.82, 194.82, 49.84 | 192.93, 192.93, 50.04 | 72.10, 234.06, 230.95 | 60.35, 240.78, 232.13 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å)a | 30–2.91 | 30–2.85 | 30–3.27 | 30–2.94 | 30–2.85 |
| (3.01–2.91)a | (2.95–2.85)a | (3.39–3.27)a | (3.04–2.94)a | (2.95–2.85)a | |
| R merge | 0.061 (0.745) | 0.047 (0.524) | 0.052 (0.446) | 0.100 (0.538) | 0.127 (0.734) |
| CC1/2 | 0.972 (0.852) | 0.965 (0.807) | 0.957 (0.883) | 0.989 (0.962) | 0.955 (0.824) |
| I/σI | 25.95 (2.05) | 29.05 (1.51) | 17.61 (2.05) | 19.19 (3.15) | 13.24 (2.54) |
| Completeness (%) | 100 (99.8) | 99.2 (92.1) | 98.8 (99.3) | 98.0 (90.3) | 97.1 (97.9) |
| Redundancy | 4.8 (4.2) | 4.5 (3.0) | 2.7 (2.4) | 5.9 (5.0) | 5.2 (5.5) |
| Refinement | |||||
| Resolution (Å) | 29.5–2.91 | 29.61–2.85 | 27.79–3.27 | 29.80–2.94 | 27.37–2.87 |
| No. of reflections | 42 169 | 43 939 | 28 669 | 32 943 | 36 770 |
| R work | 0.1746 (0.2518) | 0.1629 (0.2283) | 0.1790 (0.2339) | 0.2050 (0.2717) | 0.1938 (0.2943) |
| R free | 0.2130 (0.3073) | 0.2005 (0.2984) | 0.2121 (0.3044) | 0.2440 (0.3232) | 0.2422 (0.3620) |
| No. of atoms | |||||
| Macromolecules | 8699 | 8700 | 7460 | 7721 | 7729 |
| Water | 305 | 316 | 190 | 111 | 160 |
| B-factors (Å2) | |||||
| Protein | 75.57 | 82.39 | 89.18 | 56.89 | 56.15 |
| DNA | 167.00 | 219.19 | |||
| Cofactor (SAH) | 27.17 | 38.16 | 67.03 | 39.24 | 57.52 |
| Water | 43.95 | 54.63 | 45.00 | 38.28 | 45.27 |
| RMSDb | |||||
| Bond lengths (Å) | 0.015 | 0.009 | 0.003 | 0.004 | 0.009 |
| Bond angles (°) | 1.34 | 1.11 | 0.64 | 0.64 | 1.00 |
| Ramachandran | |||||
| Favored (%) | 93.69 | 95.22 | 95.83 | 96.53 | 96.22 |
| Allowed (%) | 6.31 | 4.78 | 4.17 | 3.47 | 3.78 |
| Disallowed (%) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
aStatistics for the highest resolution shell are shown in parentheses.
bCategories were defined using PHENIX.
Methylation-coupled restriction enzyme cleavage assay
The 494-bp DNA substrate (sequence is shown in Supplementary Table S1) containing a HpaII recognition site (CCGG) at the 151 bp position was prepared by amplification of the pET28a(+)tev-3B-CD plasmid from 2069 to 2562 bp. The amplified DNA substrate (115 ng) was incubated for 2 h at 37°C with 1 μM wild-type or mutated DNMT3B–3L complex in a reaction buffer of 10 mM Tris–HCl (pH 7.4), 50 mM NaCl, 0.5 mM EDTA, 5% glycerol, 1 mM DTT and 167 μM S-adenosyl-l-methionine (SAM), and then the 494-bp DNA substrate was purified using a QIAquick PCR Purification Kit (Qiagen). The purified 494-bp substrate was treated with HpaII (Catalog R0171S, NEB) overnight and analyzed on a 1.8% agarose gel stained with GelRed (Catalog 41003, Biotium). Band intensities were quantified using ImageJ.
MTase-Glo methyltransferase assay
This experiment was performed using the MTase-Glo Methyltransferase Assay Kit (Promega) in a 384-well white opti-plate system (PerkinElmer). The 24- or 49-bp DNA (2 μM) was incubated with 0.2 μM wild-type, mutated DNMT3B–3L or DNMT3A–3L complex in a reaction buffer containing 20 mM Tris–HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 5% glycerol, 0.1 mg/ml bovine serum albumin (BSA) and 10 μM SAM in a total volume of 4 μl at 37°C for 1 h. DNA sequences are listed in Supplementary Table S1. The reaction was stopped by addition of 1 μl 0.5% trifluoroacetic acid (TFA). After 5 min of incubation with TFA, 1 μl of 6× methyltransferase-Glo reagent was added, mixed well and incubated at room temperature for 30 min. Then, 6 μl of methyltransferase-Glo detection solution was added and incubated at room temperature for another 30 min. The luminescence signals were recorded using an EnSpire Multimode Plate Reader (PerkinElmer).
Fluorescence polarization assay
To determine the DNA-binding affinity (Kd) of DNMT3B–3L and DNMT3A–3L, 10 nM 3′-FAM-labeled CG-3 DNA substrate (49 bp) was incubated with serially diluted DNMT3B–3L or DNMT3B (Q772R)–3L for 5 min in a 30 μl reaction buffer containing 20 mM Tris–HCl (pH 8.0), 50 mM NaCl, 1 mM DTT and 5% glycerol. Fluorescence polarization signal was excited at 485 nm and read at 538 nm using a SpectraMax Paradigm Multi-Mode Microplate Reader. Intensities of the light detected in the parallel (I||) and perpendicular (I⊥) planes with respect to the excitation light were used to calculate fluorescence polarization: mFP = 1000 × (I|| − I⊥)/(I|| + I⊥). Data were fitted to one site-specific binding curve equation using GraphPad Prism software. Dissociation rate constants (koff) were measured by mixing 0.5 μM wild-type or mutated DNMT3B–3L complex with 10 nM 3′-FAM-labeled CG-3, awaiting maximum anisotropy, and then adding an excess of unlabeled CG-3 (1.0 μM, i.e. 100 times of the concentration of 3′-FAM-labeled CG-3). Anisotropy was calculated from the following equation: (I|| − I⊥)/(I|| + 2I⊥). The decrease in anisotropy was recorded over time using a SpectraMax Paradigm Multi-Mode Microplate Reader. Data were fitted to a two-phase exponential decay curve equation using GraphPad Prism software. The DNA-binding dissociation constants (Kd) and dissociation rates (k1 and k2) of DNMT3A–3L and DNMT3A (R831Q)–3L mutants were measured using the same protocol.
RESULTS
Crystal structure of DNMT3B–3L bound with or without DNA
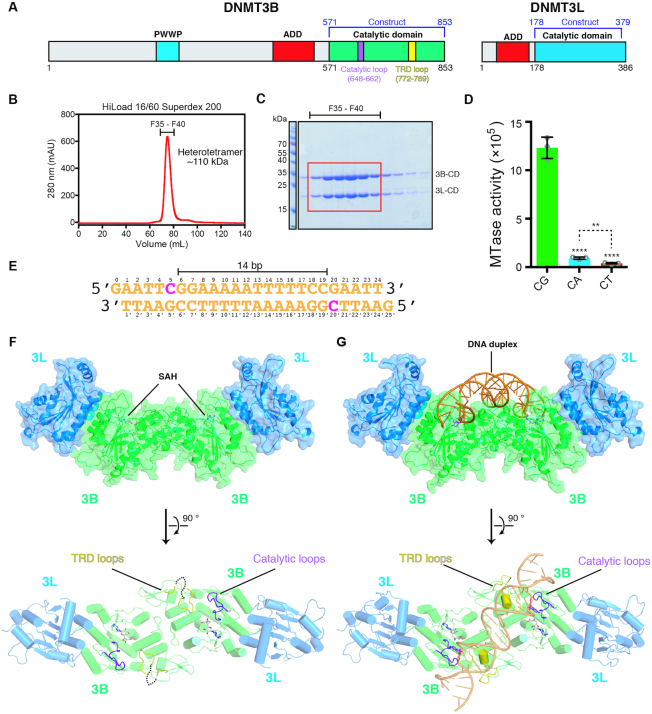
To reveal how DNMT3B methylates DNA, we expressed and co-purified the C-terminal catalytic domains of human DNMT3B (residues 575–853) and DNMT3L (residues 178–379), which was reported to interact with DNMT3B in vitro and in vivo (33–38) (Figure 1A). The two recombinant proteins interacted with each other and formed a stable complex at a one-to-one ratio and with a molecular mass of ∼110 kDa, suggesting that DNMT3B–3L complex is a heterotetramer in solution (Figure 1B and C) (33). The tetrameric DNMT3B–3L complex exhibited a sequence preference for CpG sites as it methylated a 24-bp DNA with 12- or 30-fold higher activity for CpG over CpA or CpT sites, respectively, as shown by our methyltransferase-Glo assay (Figure 1D). The slightly higher in vitro activity for CpA methylation, in comparison to CpT methylation, is consistent with the findings that human DNMT3B is important for CpA methylation in embryonic stem cells guided by DNMT3L (39,40). This DNMT3B–3L complex co-crystallized with the cofactor product, SAH, in the absence and presence of a palindromic 24-bp DNA containing two CpGpG sites separated by 14 bp and a 1-nt 5′ overhang, represents a construct similar to the 14-bp separation of two modified ZpG (Z denoting zebularine) sites of the DNA in the human DNMT3A–3L–DNA complex (25). We designed the DNA sequence based on the flanking sequence preference of human DNMT3B for a G residue downstream of CpG sites (i.e. CpGpG) (Figure 1E) (23). Crystal structures of DNMT3B–3L in DNA-free and DNA-bound forms were determined at 3.2 and 2.9 Å resolutions, respectively (Table 1). Both these crystal structures reveal a heterotetramer presenting a linear assembly of 3L–3B–3B–3L (Figure 1F and G). The 3B–3B and 3B–3L dimeric interface residues of the DNMT3B–3L complex are conserved with respect to those of the DNMT3A–3L complex (Supplementary Figure S1) (26). Two SAH cofactor products are bound in the active sites of DNMT3B, but not in DNMT3L, consistent with the fact that DNMT3L is an inactive enzyme without cofactor-binding ability (41,42).
Figure 1.
Crystal structures of the catalytic domain of DNMT3B–3L complex bound with and without DNA. (A) Domain structures of DNMT3B and DNMT3L. (B) Catalytic domains of human DNMT3B (residues 571–853) and DNMT3L (residues 178–379) were expressed and co-purified as a heterotetramer with a molecular mass of ∼110 kDa. (C) The co-purified recombinant DNMT3B–3L complex exhibited high homogeneity, as revealed by sodium dodecylsulfate–polyacrylamide gel electrophoresis. (D) Activities of recombinant DNMT3B–3L for CpG, CpA and CpT methylation, as measured by methyltransferase-Glo assays (in luminescence, RLU, n = 3; error bars denote standard deviation (SD); **P < 0.01, ****P < 0.0001). Statistical significance (P-values) was determined by two-tailed Student's t-test. (E) The palindromic DNA duplex containing two CpGpG sites was used for co-crystallization with DNMT3B–3L. (F, G) Crystal structures of DNMT3B–3L complex bound without and with DNA revealing two CpGpG sites bound in the active sites of the DNMT3B dimer (in green). The two SAH molecules are represented by a stick model, catalytic loops are shown in purple and target recognition domain (TRD) loops are displayed in yellow.
Two DNA-interacting loops exhibit conformational changes upon DNA binding
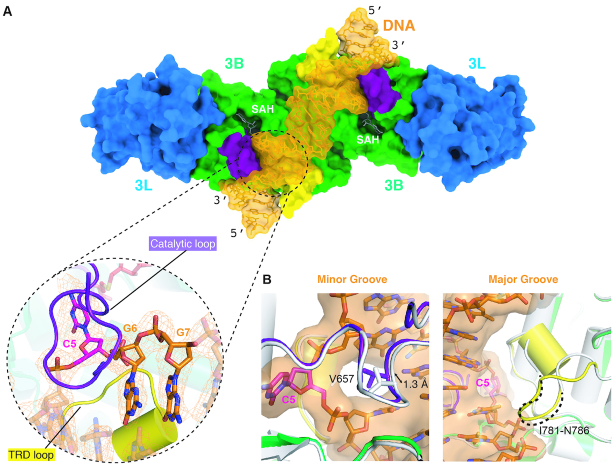
The crystal structure of the DNA-bound form of the human DNMT3B–3L complex [DNMT3B–3L–DNA (CpCpG)] reveals a single DNA duplex with two CpG sites bound in the active sites of the DNMT3B dimer. The bound DNA is snugly enclosed by the catalytic loop and TRD loop of DNMT3B dimer (shown, respectively, in purple and yellow in Figure 2A), but the bound DNA does not contact DNMT3L. The overall structures of the DNA-free and DNA-bound forms of DNMT3B–3L are closely matched, presenting an average RMSD of 0.435 Å for 794 Cα atoms. However, the two DNA-interacting loops exhibit considerable conformational changes upon DNA binding (Figure 2B). Residue V657 in the catalytic loop moves toward the DNA minor groove by ∼1.3 Å upon DNA binding, whereas the disordered region (residues I781–N786) of the TRD loop in the DNA-free form becomes well ordered in the DNA-bound form, as illustrated by the well-defined electron density with an average B-factor of 53.0 Å2 (Supplementary Figure S2), and interacts with DNA at the DNA major groove. Thus, the CpG site is clamped by the catalytic and TRD loops from two different sides of the DNA molecule, with the catalytic loop bound in the minor groove and the TRD loop being bound in the major groove.
Figure 2.
Two DNA-interacting loops in DNMT3B exhibit conformational changes upon DNA binding. (A) DNMT3B–3L complex binds and encloses DNA via the catalytic (purple) and TRD (yellow) loops. The CpG site is clamped by the two loops from two different sides of the DNA molecule, with the catalytic loop bound in the minor groove and the TRD loop being bound in the major groove. The omit electron density maps (Fo − Fc) of DNA (orange) are contoured at the 2σ level. (B) Overlay of the catalytic loop (left-hand panel) and the TRD loop (right-hand panel) of DNA-free (white) and DNA-bound (colored) complexes, which shows how the conformation of the two loops changes upon DNA binding.
Recognition of C in CpG sites
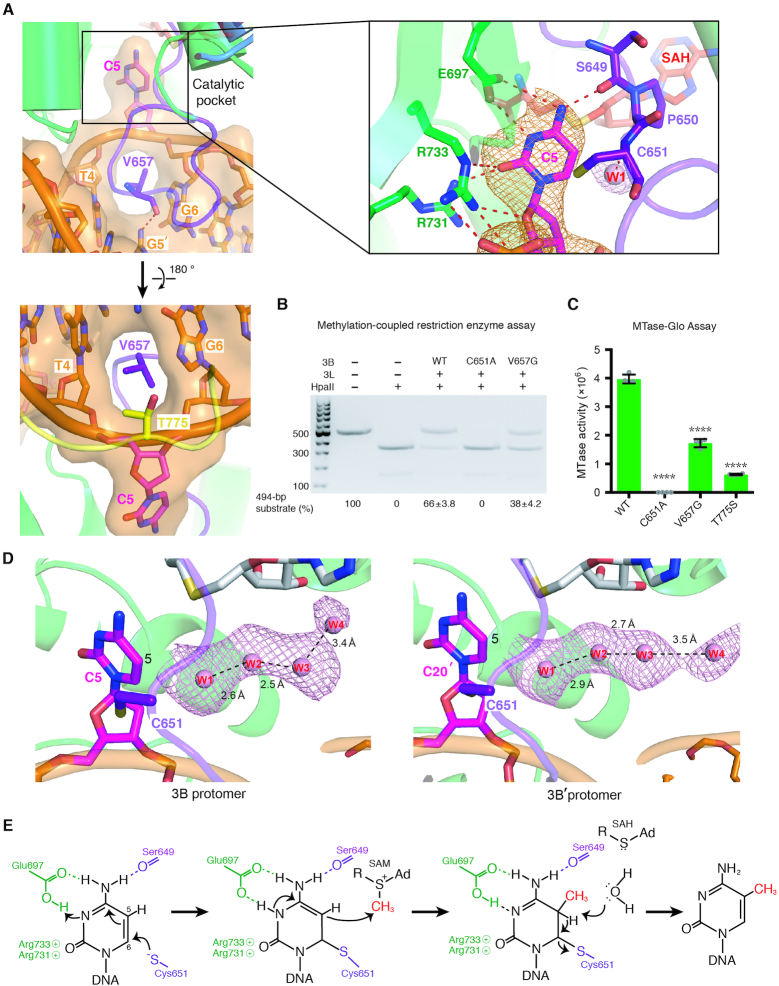
In our DNMT3B–3L–DNA (CpGpG) complex structure, the cytosine of CpG sites is flipped out of the DNA and is inserted deeply into the catalytic pocket of DNMT3B (Figure 3A and Supplementary Figure S3). The catalytic loop residue V657 that is conserved between DNMT3B and DNMT3A inserts into the DNA cavity left by the cytosine (C5) and makes van der Waals interactions with the neighboring DNA bases. V657 also stabilizes the unpaired guanine G5′ in the opposite strand by forming a hydrogen bond between its backbone carbonyl oxygen and the N2 atom of G5′ (Figure 3A, upper left panel). Moreover, the conserved T775 residue in the TRD loop makes hydrophobic interactions with the ribose of the flipped-out cytosine (Figure 3A, lower left panel).
Figure 3.
Specific recognition of the cytosine in CpG sites by DNMT3B. (A) The cytosine (C5) in CpG sites is flipped out of the DNA helix by the catalytic loop. The enlarged panel (upper right) reveals the hydrogen-bond network surrounding the flipped-out cytosine. The omit electron density maps (Fo − Fc) of cytosine (orange) and the nearby water molecule (pink) are contoured at the 4.5σ level. (B) Methyltransferase activities of DNMT3B–3L and the C651A and V657G mutants were estimated by methylation-coupled restriction enzyme HpaII cleavage assays. (C) Methylation activities of wild-type DNMT3B–3L (WT), and the C651A, V657G and T775S mutants were measured by methyltransferase-Glo assays (in luminescence, RLU, n = 4; error bars denote SD; ****P < 0.0001). Statistical significance (P-values) was determined by two-tailed Student's t-test. (D) A water channel located close to the C5 atom of the cytosine of the CpG sites was identified in the two DNMT3B protomers. Cytosines of the CpG sites are shown as magenta sticks, whereas water molecules in the proton wire water channel are shown as pink spheres. The omit electron density maps (Fo − Fc) of water molecules (pink) are contoured at the 2σ level. (E) Proposed working mechanism for methylation by DNMT3B of the cytosine at the C5 position.
In the catalytic pocket, the flipped-out cytosine is located close to C651 and makes hydrogen bonds with S649, E697, R731 and R733 (Figure 3A and Supplementary Figure S3). These particular interactions contribute to cytosine-specific recognition of CpG sites by DNMT3B. C651 presumably represents the catalytic residue mediating nucleophilic attack of the cytosine C6 position. To verify the critical roles of C651 and V657 in DNA methylation, we constructed two respective DNMT3B mutants (C651A and V657G) for methylation-coupled restriction enzyme cleavage assays. The catalytic domain of the DNMT3B–3L complex methylated most of the HpaII cleavage sites of CCGG embedded in our 494-bp DNA substrates, resulting in 66% of the methylated DNA being resistant to cleavage. In contrast, the V657G mutant partially lost its methyltransferase activity, resulting in 38% of DNA remaining uncleaved, whereas the C651A mutant completely lost its enzymatic activity to result in 0% of uncleaved DNA (Figure 3B). The methyltransferase activities of the V657G, C651A and T775S for CpG sites in 24-bp DNA were further assessed by methyltransferase-Glo assays, which revealed a similar trend in that wild-type DNMT3B–3L methylated the CpG sites with ∼2.3- and ∼6.3-fold higher activity than the V657G and T775S mutants, respectively, whereas the C651A mutant had no observable activity (Figure 3C). The low methyltransferase activity of T775S mutant is correlated with the hypomethylation phenotype of ICF syndrome caused by R823G mutation (34), as T775 forms a hydrogen-bond network with R823 in our structure (Figure 6G), indicating that mutation of either T775 or R823 may reduce the stability of the flipped-out cytosine. This structural and biochemical evidence supports that residue C651 is the most critical catalytic residue for DNA methylation (13,22,43), V657 penetrates into the minor groove of DNA and T775 stabilizes the flipped-out cytosine, and that together they contribute to the DNA methylation activity of DNMT3B (25).
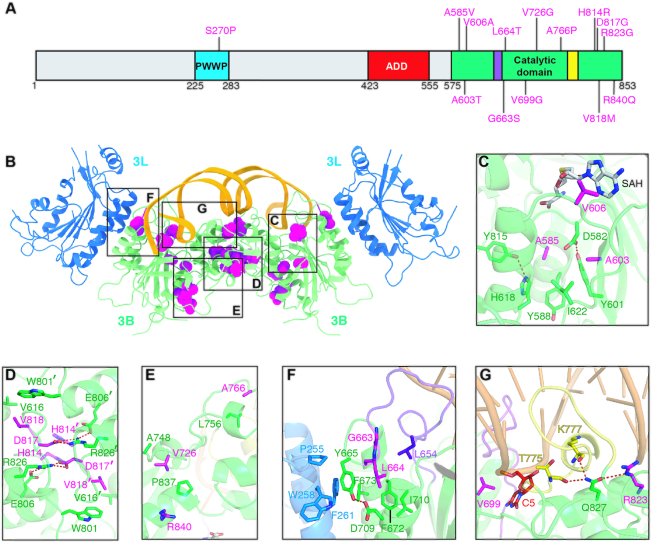
Figure 6.
Structural impact of ICF syndrome-related DNMT3B mutations. (A) Mapping of mutations causing ICF syndrome onto the primary sequence of DNMT3B. (B) Crystal structure of the DNMT3B–3L–DNA complex revealing the locations of ICF syndrome-related mutations. Residues pertaining to ICF syndrome-related mutations are shown as magenta spheres. Zoomed-in views of the boxed areas are displayed in (C)–(G). (C) The mutations—including A585V, A603T and V606A—may disrupt SAM cofactor binding. (D) Mutations in the DNMT3B dimeric interface may impede protein assembly, including H814R, D817G and V818M. (E) Mutations V726G, A766P and R840Q may affect protein stability. (F) Mutations may affect DNMT3B–3L interaction and/or catalytic loop conformation—including G663S and L664T. (G) Mutations may affect cytosine interaction (V699G, R823G) or DNA binding (R823G).
Working mechanism of cytosine methylation by DNMT3B
In the crystal structure of DNMT3B–3L–DNA (CpGpG) complex, DNA is noncovalently bound to DNMT3B, so it represents an enzyme–substrate–cofactor complex before the methyltransferase reaction takes place. Apart from observing all residues and the cofactor in the structure, we also noted that a water molecule (W1) as part of a proton wire water channel—consisting of four well-ordered water molecules linked via hydrogen bonds—is located close to the cytosine C5 position in both protomers of the DNMT3B dimer (Figure 3D). A similar water channel was observed in the active site of bacterial DNMT M.Hhal that was suggested to play a role in the final deprotonation step of methylation (44,45). This water channel was observed for the first time in a mammalian methyltransferase and could be responsible for the deprotonation from C5 during cytosine methylation (44,45). Thus, based on all of these results, we can derive a complete working mechanism at the atomic level for DNA methylation by a mammalian DNMT3B (Figure 3E). The crystal structure of the DNMT3B–3L–DNA complex supports that DNA methylation is initiated by residue C651, which mediates nucleophilic attack on the C6 position, and E697 coordinately facilitates protonation of the N3 of the cytosine ring (3,46). Subsequently, a methyl group is transferred from SAM to C5 of the cytosine ring, followed by deprotonation from C5 by means of an OH− derived from a water molecule that is part of a proton wire water channel to produce 5-methylcytosine.
Recognition of G in CpG sites
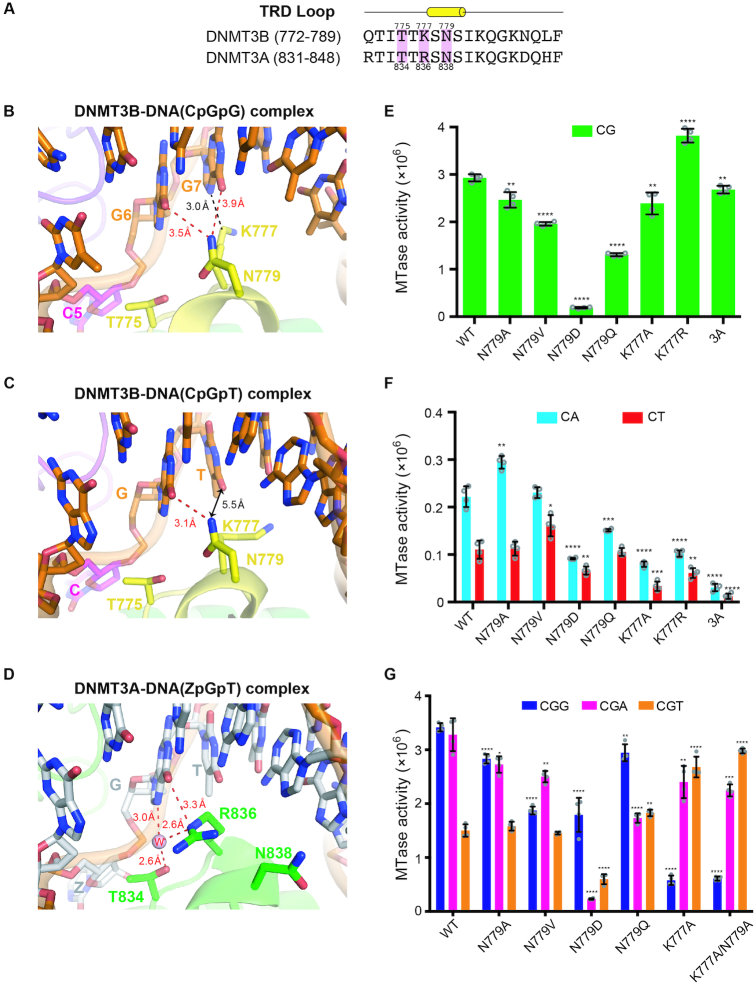
For guanine recognition in CpG sites by human DNMT3B, residue N779 located in the TRD loop specifically interacts with the G6 position of target CpG sites via a weak hydrogen bond (3.5 Å) between the Nδ atom of N779 and the O6 atom of G6 (Figure 4B). N779 is conserved in both DNMT3B and DNMT3A (N838 in human DNMT3A, Figure 4A), but it is residue R836 in DNMT3A, and not N838, that interacts specifically with the guanine in CpG sites (Figure 4D) (25). To establish the role of N779 in CpG-specific methylation, we constructed four site-directed mutants: N779A, N779V, N779D and N779Q. Wild-type DNMT3B–3L exhibited the greatest methylation activity for CpG sites over CpA and CpT sites, whereas the N779D mutant had the lowest activity, demonstrating that N779 is critical for sequence-specific CpG methylation, or a negative-charge side chain (D) is disfavored in the protein–DNA interface (Figure 4E and F). Replacement of N779 with the hydrophobic amino acid A or V, or a larger side-chain residue Q, also reduced methylation of CpG sites and marginally altered methylation of CpA and CpT sites relative to wild-type DNMT3B–3L (Figure 4E and F). Together, these results support that N779 plays a key role in specific recognition of the guanine residue in CpG sites.
Figure 4.
Specific recognition of the guanines in CpGpG sites by DNMT3B. (A) Amino acid sequence alignment of the TRD loop of human DNMT3B and DNMT3A. Key conserved residues are shaded in pink. (B) In the crystal structure of DNMT3B–3L–DNA (CpGpG) complex, residue N779 (Nδ atom) in the TRD loop forms hydrogen bonds with the guanine (G6, O6 atom) of the target CpG site and the flanking guanine (G7, O6 atom) downstream of the CpG site. Residue K777 (Cϵ atom) interacts with the flanking guanine through van der Waals interactions. (C) In the crystal structure of the DNMT3B–3L–DNA (CpGpT) complex, residue N779 (Nδ atom) in the TRD loop forms a hydrogen bond with the guanine (G6, O6 atom) of the target CpG site. Residue K777 does not interact with the flanking thymine beyond a distance of 5.5 Å. (D) Residue R836 of DNMT3A interacts with the guanine of the target ZpG site via a hydrogen bond and water-mediated hydrogen bonds. Methylation activities of DNMT3B, its mutants and DNMT3A for 24-bp DNA containing CpG (E), CpA or CpT (F), as measured by methyltransferase-Glo assays (in luminescence, RLU, n = 4). (G) Methylation activities of DNMT3B and its mutants for 24-bp DNA containing CpGpG, CpGpA or CpGpT, as measured by methyltransferase-Glo assays (in luminescence, RLU, n = 4). For (E)–(G), error bars denote SD; *P < 0.1, **P < 0.01, ***P < 0.001 and ****P < 0.0001. Statistical significance (P-values) was determined by two-tailed Student's t-test.
Recognition of the flanking G downstream CpG sites
Human DNMT3B exhibits a preference for TpCpGpG methylation, with a T residue at the −1 position and a G residue at the +1 position of its target CpG sites (23,47). Residue N779 not only interacts with the guanine (G6) in the CpG site, but also remotely interacts with the flanking guanine at the +1 position with a distance of 3.9 Å between the Nδ and O6 atoms of G7 (Figure 4B). To verify the role of N779 in CpGpG-specific recognition, we further determined the crystal structure of DNMT3B–3L in complex with a DNA bearing CpGpT site [DNMT3B–3L–DNA (CpGpT)], which clearly had a reduced size of electron density for the thymine of the CpGpT site as compared to that of the guanine of DNMT3B–3L–DNA (CpGpG) complex (Table 1 and Supplementary Figure S4). The crystal structure of DNMT3B–3L–DNA (CpGpT) complex reveals that N779 continues to interact with G6 in the CpG site (the O6 atom), but releases its interaction with the flanking T as the distance between the Nδ atom of N779 and the O6 atom of the thymine increases beyond 5.5 Å (Figure 4C). This result confirms that N779 is involved in the recognition of the flanking guanine downstream of CpG sites.
Proximal to N779, residue K777 also makes van der Waals interactions with the flanking guanine of the CpGpG site (Figure 4B), but in human DNMT3A the residue located in the same position is R836, which directly interacts with the guanine of CpG sites. DNMT3A has been shown to have greater CG-specific methyltransferase activity than DNMT3B (8), as also confirmed by our assays (Figure 4E and F). To establish the role of the K777 residue, we constructed two human DNMT3B mutants, i.e. K777A and K777R. The K777A mutant exhibited slightly reduced activity for CpG, CpA and CpT sites compared to wild-type enzyme, whereas the K777R mutant had a significantly enhanced sequence preference for CpG sites (i.e. higher activity for CpG and lower activities for CpA and CpT), suggesting that replacement of K777 with R increases the CpG-specific methylation activity of DNMT3B (Figure 4E and F), making it behave more like human DNMT3A in having a higher CpG specificity.
To analyze the basis for its flanking sequence preference, we further measured the methyltransferase activity of DNMT3B for CpG sites flanked at the +1 position by G, A or T. We found that human DNMT3B–3L catalytic domain exhibited high methyltransferase activity at CpGpG sites (CpGpG ≈ CpGpA > CpGpT), consistent with a recent study showing that mouse DNMT3B preferred to methylate CpGpG and CpGpA sites (48), whereas all of the N779 mutants had slightly reduced or varied specificities for CpGpG site methylation (Figure 4G). In contrast, the K777A mutant and a K777A/N779A double mutant exhibited a flanking sequence preference the reverse of that of wild-type enzyme, exhibiting highest methylation activity for CpGpT sites (CpGpT ≥ CpGpA > CpGpG) (Figure 4G). Taken together, these results suggest that K777 plays a primary role and N779 plays a minor role in recognizing the flanking G residue, resulting in a methylation preference for CpGpG sites.
TRD loop mediates enzymatic processivity of DNMT3B
As DNA binding is mediated by catalytic and TRD loops, it implies that the processivity of DNMT3B may be regulated by these two loops. By comparing the DNA-free form of the catalytic domain of DNMT3A (PDB code: 2QRV) and DNMT3B, we noted that almost the entire TRD loop (residues 832–845) of DNMT3A is disordered, as opposed to the TRD loop of DNMT3B that is mostly visible (only six flexible residues, I781–N786) (Supplementary Figure S5A). However, upon DNA binding, the TRD loop becomes well ordered in both the DNA-bound DNMT3A and DNMT3B structures (Supplementary Figure S5B). This outcome suggests that the less flexible TRD loop of DNMT3B may contribute to stable DNMT3B–DNA complex formation to promote its processive association with DNA during catalytic cycles, a characteristic observed in processive enzymes that generally contain more stable DNA-interacting domains or loops compared to nonprocessive enzymes (49).
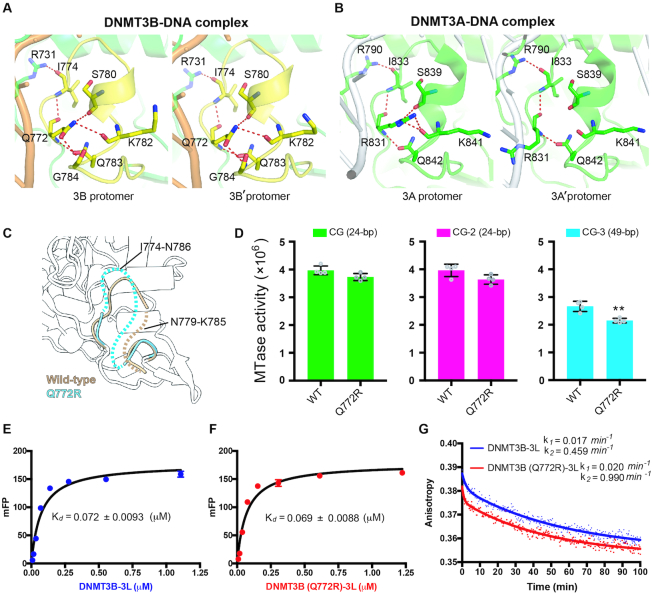
The TRD loop (residues 772–789) in the DNMT3B–3L–DNA complex is stabilized by residue Q772, which forms a hydrogen-bond network with I774, S780, K782, Q783 and G784 in both protomers of the DNMT3B dimer (Figure 5A). However, the TRD loop (residues 831–848) of DNMT3A is partially stabilized by residue R831, which forms a complete hydrogen-bond network with I833, S839, K841 and Q842 of only one of the two protomers (Figure 5B). To verify the role of Q772 in stabilization of the TRD loop, we crystallized wild-type DNMT3B–3L [referred to as DNMT3B–3L (ii)] and a Q772R mutant [DNMT3B (Q772R)–3L] under similar crystallization conditions with an identical C2221 space group (Table 1). We found that the electron density for the TRD loop of the Q772R mutant was missing for residues 774–786, whereas a smaller expanse of the TRD loop of wild-type DNMT3B–3L (ii) was disordered (from residues 779 to 785) (Figure 5C). This structural difference reveals that the Q772R mutant had a more flexible TRD loop than wild-type DNMT3B. We also found that the Q772R mutant presented slightly reduced activity in terms of methylating short 24-bp DNA (CG and CG-2), but significantly lower methylation activity of long 49-bp DNA (CG-3) relative to wild-type DNMT3B–3L (Figure 5D). These crystal structure and biochemical data indicate that the flexible TRD loop of the Q772R mutant significantly decreases the methylation activity of longer DNA stretches by lowering enzyme processivity.
Figure 5.
DNMT3B methylation activity is regulated by TRD loop flexibility. (A) Hydrogen-bond networks mediated by residue Q722 stabilize the TRD loop in both protomers of the DNMT3B dimer in the DNMT3B–3L–DNA complex. (B) Only one complete hydrogen-bond network mediated by residue R831 stabilizes the TRD loop in one of the two protomers of the DNMT3A dimer in the DNMT3A–3L–DNA complex. (C) Superimposition of the crystal structures of wild-type DNMT3B–3L complex (ii) and the DNMT3B (Q772R)–3L complex. Disordered regions in the TRD loop of wild-type DNMT3B–3L (ii) (residues 779–785) and the DNMT3B (Q772R)–3L (residues 774–786) complex are represented by dotted brown or cyan lines, respectively. (D) Methylation activities of DNMT3B–3L and the DNMT3B (Q772R)–3L mutant for DNA of different lengths, as measured by methyltransferase-Glo assays (in luminescence, RLU, n = 4; error bars denote SD; **P < 0.01). Statistical significance (P-values) was determined by two-tailed Student's t-test. (E, F) Dissociation constants (Kd) of DNMT3B–3L and DNMT3B (Q772R)–3L complexes upon binding with the 49-bp DNA (CG-3), as measured by fluorescence polarization (n = 3). (G) Dissociation rate constants (k1 and k2) for DNMT3B–3L and DNMT3B (Q772R)–3L complexes, as determined by measuring the decreasing signal of fluorescence anisotropy of FAM-labeled CG-3 using unlabeled DNA as competitors.
We also measured the DNA-binding affinity and dissociation rates for DNMT3B–3L complexes by fluorescence polarization assays, revealing that DNMT3B–3L dissociated from DNA in two phases with a fast initial rate (k2, likely for nonspecific DNA binding) and a final slow dissociation rate (k1, likely for CpG-specific binding). We found that Q772R mutant dissociated faster from DNA than wild-type DNMT3B–3L (k1 = 0.02 versus 0.017 min−1, k2 = 0.99 versus 0.46 min−1, Figure 5G), even though they shared similar DNA-binding affinities with a dissociation constant (Kd) of 0.069 and 0.072 μM, respectively (Figure 5E and F). A previous study also showed that binding of DNMT3L to DNMT3A lowers the dissociation rate (koff) and enhances kcat of DNMT3A by increasing enzymatic processivity (50). Our result thus supports that Q772 plays a role in regulating the processivity of DNMT3B.
Moreover, we further constructed a DNMT3A (R831Q)–3L mutant to test the role of the TRD loop residue R831 in regulating the enzyme processivity. We found that DNMT3A (R831Q)–3L mutant had highly reduced DNA-binding and methyltransferase activities compared to the wild-type enzyme, as R831 is directly involved in DNA interactions (Supplementary Figure S6) (25). In contrast to the DNMT3B (Q772R)–3L mutant that had increased dissociation rates as compared to wild-type DNMT3B–3L, DNMT3A (R831Q)–3L mutant had slightly decreased DNA dissociation rates as compared to wild-type DNMT3A–3L (k1 = 0.026 versus 0.031 min−1 and k2 = 0.215 versus 0.296 min−1), indicating that this mutant had marginally increased processivity (Supplementary Figure S6C). Taking together all of these results, we conclude that residue Q772 plays a crucial role in stabilizing the TRD loop conformation of DNMT3B and that the TRD loop is involved not only in specific recognition of the guanine residue in CpG sites, but also in regulating DNMT3B processivity.
DISCUSSION
The crystal structures of the human DNMT3B–3L–DNA complex reported in this study reveal not only the critical amino acid residues for CpGpG recognition and methylation, but also a water channel located near the C5 atom of cytosine responsible for the final deprotonation step of DNA methylation, thereby providing a complete working mechanism for DNMT3B methylation. Based on these structures, the impact of DNMT3B mutations in protein folding, enzyme activity, substrate binding, cofactor binding and protein–protein interactions can be assessed. A number of point mutations in DNMT3B have been shown to be responsible for human ICF syndrome (14,51). Most of these mutations have been mapped to the C-terminal catalytic domain of DNMT3B (Figure 6A and B). These mutations abolish or reduce the enzymatic activity of DNMT3B, resulting in DNA hypomethylation (7,13,15,34,52,53). Our DNMT3B–3L–DNA complex crystal structure reveals how these mutations differentially impact enzymatic activity. One group of mutations—including A585V, A603T and V606A—may disrupt SAM cofactor binding, whereas a second group of mutations—H814R, D817G and V818M—may impede protein assembly, as the latter residues form interaction networks in the DNMT3B dimeric interface (Figure 6C and D). Other ICF syndrome-associated mutations may affect protein stability (V726G, A766P, R840Q), DNMT3B–3L interaction, catalytic loop conformation (G663S, L664T), cytosine interaction (V699G, R823G) or DNA binding (R823G) (Figure 6E–G).
Apart from ICF syndrome, overexpression of DNMT3B has been linked to various cancers (54,55), so it is crucial to identify modulators that could specifically inhibit the activity of DNMT3B. A couple of cytidine analogs, azacitidine and decitabine, have been used for the treatment of MDS and AML (56,57), but these drugs do not specifically recognize a single DNMT. Instead, they are incorporated into DNA during DNA synthesis and they inhibit the activities of all DNMTs. Comparing the crystal structures of the DNMT3B–3L–DNA complex with complexes involving DNMT3A reveals in the former a more rigid TRD loop and different residues in the TRD loop for CpGpG-specific interactions, as well as a unique processive methylation mechanism for tandemly repeated CpG. These different structural characteristics and enzymatic properties of DNMT3B may facilitate discovery of DNMT3B modulators for future therapeutic applications.
DATA AVAILABILITY
The coordinates of the structures for human DNMT3B–3L in complex with 24-bp DNA duplex containing CpGpG sites and DNMT3B–3L in complex with 24-bp DNA containing CpGpT sites, DNMT3B–3L complex (i), DNMT3B–3L complex (ii) and DNMT3B (Q772R)–3L complex have been deposited in Protein Data Bank under accession numbers 6KDA, 6KDB, 6KDL, 6KDP and 6KDT, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff members of beamlines BL15A and BL5A in the National Synchrotron Radiation Research Center, Hsinchu, Taiwan, a national user facility supported by the Ministry of Science and Technology of Taiwan. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Core Facility Program for Biotechnology.
Author contributions: C.-C.L. and Y.-P.C. expressed and purified DNMT3B–3L. C.-C.L. and W.-Z.Y. collected the X-ray diffraction data. C.-C.L. determined the crystal structures, designed and performed biochemical experiments, and analyzed data. J.C.K.S. and H.S.Y. provided technical support. C.-C.L. and H.S.Y. wrote the manuscript. H.S.Y. supervised and guided the project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology [108-2311-B-001-009-MY3 to C.-C.L and H.S.Y.; 108-2311-B-001-001 to J.C.K.S. and H.S.Y.]; Academia Sinica [AS-IA-105-L04 to H.S.Y.], Taiwan, R.O.C. Funding for open access charge: Academia Sinica.
Conflict of interest statement. None declared.
REFERENCES
- 1. Law J.A., Jacobsen S.E.. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010; 11:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith Z.D., Meissner A.. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013; 14:204–220. [DOI] [PubMed] [Google Scholar]
- 3. Jurkowska R.Z., Jurkowski T.P., Jeltsch A.. Structure and function of mammalian DNA methyltransferases. ChemBioChem. 2011; 12:206–222. [DOI] [PubMed] [Google Scholar]
- 4. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2017; 19:81–92. [DOI] [PubMed] [Google Scholar]
- 5. Okano M., Bell D.W., Haber D.A., Li E.. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999; 99:247–257. [DOI] [PubMed] [Google Scholar]
- 6. Chedin F., Lieber M.R., Hsieh C.L.. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:16916–16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gowher H., Jeltsch A.. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J. Biol. Chem. 2002; 277:20409–20414. [DOI] [PubMed] [Google Scholar]
- 8. Suetake I., Miyazaki J., Murakami C., Takeshima H., Tajima S.. Distinct enzymatic properties of recombinant mouse DNA methyltransferases Dnmt3a and Dnmt3b. J. Biochem. 2003; 133:737–744. [DOI] [PubMed] [Google Scholar]
- 9. Umehara Y., Hanaoka K., Watanabe D.. Distinct functions of Dnmt3a and Dnmt3b de novo DNA methyltransferases in ES cell proliferation and differentiation. Stem Cell Discov. 2013; 3:127–132. [Google Scholar]
- 10. Li E., Zhang Y.. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014; 6:a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baubec T., Colombo D.F., Wirbelauer C., Schmidt J., Burger L., Krebs A.R., Akalin A., Schubeler D.. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015; 520:243–247. [DOI] [PubMed] [Google Scholar]
- 12. Neri F., Rapelli S., Krepelova A., Incarnato D., Parlato C., Basile G., Maldotti M., Anselmi F., Oliviero S.. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017; 543:72–77. [DOI] [PubMed] [Google Scholar]
- 13. Moarefi A.H., Chedin F.. ICF syndrome mutations cause a broad spectrum of biochemical defects in DNMT3B-mediated de novo DNA methylation. J. Mol. Biol. 2011; 409:758–772. [DOI] [PubMed] [Google Scholar]
- 14. Hansen R.S., Wijmenga C., Luo P., Stanek A.M., Canfield T.K., Weemaes C.M., Gartler S.M.. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jin B., Tao Q., Peng J., Soo H.M., Wu W., Ying J., Fields C.R., Delmas A.L., Liu X., Qiu J. et al.. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum. Mol. Genet. 2008; 17:690–709. [DOI] [PubMed] [Google Scholar]
- 16. Yang L., Rau R., Goodell M.A.. DNMT3A in haematological malignancies. Nat. Rev. Cancer. 2015; 15:152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kondo T., Bobek M.P., Kuick R., Lamb B., Zhu X., Narayan A., Bourc’his D., Viegas-Pequignot E., Ehrlich M., Hanash S.M.. Whole-genome methylation scan in ICF syndrome: hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum. Mol. Genet. 2000; 9:597–604. [DOI] [PubMed] [Google Scholar]
- 18. Walton E.L., Francastel C., Velasco G.. Dnmt3b prefers germ line genes and centromeric regions: lessons from the ICF syndrome and cancer and implications for diseases. Biology. 2014; 3:578–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emperle M., Rajavelu A., Reinhardt R., Jurkowska R.Z., Jeltsch A.. Cooperative DNA binding and protein/DNA fiber formation increases the activity of the Dnmt3a DNA methyltransferase. J. Biol. Chem. 2014; 289:29602–29613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gowher H., Jeltsch A.. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J. Mol. Biol. 2001; 309:1201–1208. [DOI] [PubMed] [Google Scholar]
- 21. Norvil A.B., Petell C.J., Alabdi L., Wu L., Rossie S., Gowher H.. Dnmt3b methylates DNA by a noncooperative mechanism, and its activity is unaffected by manipulations at the predicted dimer interface. Biochemistry. 2018; 57:4312–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Emburgh B.O., Robertson K.D.. Modulation of Dnmt3b function in vitro by interactions with Dnmt3L, Dnmt3a and Dnmt3b splice variants. Nucleic Acids Res. 2011; 39:4984–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wienholz B.L., Kareta M.S., Moarefi A.H., Gordon C.A., Ginno P.A., Chedin F.. DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet. 2010; 6:e1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang W., Xu J.. DNA methyltransferases and their roles in tumorigenesis. Biomark. Res. 2017; 5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Z.M., Lu R., Wang P., Yu Y., Chen D., Gao L., Liu S., Ji D., Rothbart S.B., Wang Y. et al.. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature. 2018; 554:387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jia D., Jurkowska R.Z., Zhang X., Jeltsch A., Cheng X.. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007; 449:248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo X., Wang L., Li J., Ding Z., Xiao J., Yin X., He S., Shi P., Dong L., Li G. et al.. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015; 517:640–644. [DOI] [PubMed] [Google Scholar]
- 28. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 29. Bunkoczi G., Echols N., McCoy A.J., Oeffner R.D., Adams P.D., Read R.J.. Phaser.MRage: automated molecular replacement. Acta Crystallogr. D: Biol. Crystallogr. 2013; 69:2276–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D: Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schrodinger, LLC 2015; The PyMOL Molecular Graphics System, Version 1.8.
- 32. Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E.. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018; 27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gowher H., Liebert K., Hermann A., Xu G., Jeltsch A.. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 2005; 280:13341–13348. [DOI] [PubMed] [Google Scholar]
- 34. Xie Z.H., Huang Y.N., Chen Z.X., Riggs A.D., Ding J.P., Gowher H., Jeltsch A., Sasaki H., Hata K., Xu G.L.. Mutations in DNA methyltransferase DNMT3B in ICF syndrome affect its regulation by DNMT3L. Hum. Mol. Genet. 2006; 15:1375–1385. [DOI] [PubMed] [Google Scholar]
- 35. Pacaud R., Sery Q., Oliver L., Vallette F.M., Tost J., Cartron P.F.. DNMT3L interacts with transcription factors to target DNMT3L/DNMT3B to specific DNA sequences: role of the DNMT3L/DNMT3B/p65-NFkappaB complex in the (de-)methylation of TRAF1. Biochimie. 2014; 104:36–49. [DOI] [PubMed] [Google Scholar]
- 36. Suetake I., Shinozaki F., Miyagawa J., Takeshima H., Tajima S.. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004; 279:27816–27823. [DOI] [PubMed] [Google Scholar]
- 37. Neri F., Krepelova A., Incarnato D., Maldotti M., Parlato C., Galvagni F., Matarese F., Stunnenberg H.G., Oliviero S.. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013; 155:121–134. [DOI] [PubMed] [Google Scholar]
- 38. Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D. et al.. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007; 448:714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arand J., Spieler D., Karius T., Branco M.R., Meilinger D., Meissner A., Jenuwein T., Xu G., Leonhardt H., Wolf V. et al.. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012; 8:e1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ziller M.J., Muller F., Liao J., Zhang Y., Gu H., Bock C., Boyle P., Epstein C.B., Bernstein B.E., Lengauer T. et al.. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011; 7:e1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hata K., Okano M., Lei H., Li E.. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002; 129:1983–1993. [DOI] [PubMed] [Google Scholar]
- 42. Chen Z.X., Mann J.R., Hsieh C.L., Riggs A.D., Chedin F.. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell Biochem. 2005; 95:902–917. [DOI] [PubMed] [Google Scholar]
- 43. Hsieh C.L. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell Biol. 1999; 19:8211–8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J., Lior-Hoffmann L., Wang S., Zhang Y., Broyde S.. DNA cytosine methylation: structural and thermodynamic characterization of the epigenetic marking mechanism. Biochemistry. 2013; 52:2828–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang X., Bruice T.C.. The mechanism of M.HhaI DNA C5 cytosine methyltransferase enzyme: a quantum mechanics/molecular mechanics approach. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:6148–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O’Gara M., Klimasauskas S., Roberts R.J., Cheng X.. Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaL methyltransferase–DNA–AdoHcy complexes. J. Mol. Biol. 1996; 261:634–645. [DOI] [PubMed] [Google Scholar]
- 47. Emperle M., Adam S., Kunert S., Dukatz M., Baude A., Plass C., Rathert P., Bashtrykov P., Jeltsch A.. Mutations of R882 change flanking sequence preferences of the DNA methyltransferase DNMT3A and cellular methylation patterns. Nucleic Acids Res. 2019; 47:11355–11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Norvil A.B., AlAbdi L., Liu B., Forstoffer N. E., Michie A. R., Chen T., Gowher H.. The acute myeloid leukemia variant DNMT3A Arg882His is a DNMT3B-like enzyme. 2019; bioRxiv doi:31 July 2019, preprint: not peer reviewed 10.1101/720557. [DOI] [PMC free article] [PubMed]
- 49. Breyer W.A., Matthews B.W.. A structural basis for processivity. Protein Sci. 2001; 10:1699–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Holz-Schietinger C., Reich N.O.. The inherent processivity of the human de novo methyltransferase 3A (DNMT3A) is enhanced by DNMT3L. J. Biol. Chem. 2010; 285:29091–29100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu G.L., Bestor T.H., Bourc’his D., Hsieh C.L., Tommerup N., Bugge M., Hulten M., Qu X., Russo J.J., Viegas-Pequignot E.. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999; 402:187–191. [DOI] [PubMed] [Google Scholar]
- 52. Ueda Y., Okano M., Williams C., Chen T., Georgopoulos K., Li E.. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development. 2006; 133:1183–1192. [DOI] [PubMed] [Google Scholar]
- 53. Gatto S., Gagliardi M., Franzese M., Leppert S., Papa M., Cammisa M., Grillo G., Velasco G., Francastel C., Toubiana S. et al.. ICF-specific DNMT3B dysfunction interferes with intragenic regulation of mRNA transcription and alternative splicing. Nucleic Acids Res. 2017; 45:5739–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gao J., Wang L., Xu J., Zheng J., Man X., Wu H., Jin J., Wang K., Xiao H., Li S. et al.. Aberrant DNA methyltransferase expression in pancreatic ductal adenocarcinoma development and progression. J. Exp. Clin. Cancer Res. 2013; 32:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang J., Wei X., Wu Q., Xu Z., Gu D., Jin Y., Shen Y., Huang H., Fan H., Chen J.. Clinical significance of the expression of DNA methyltransferase proteins in gastric cancer. Mol. Med. Rep. 2011; 4:1139–1143. [DOI] [PubMed] [Google Scholar]
- 56. Kaminskas E., Farrell A.T., Wang Y.C., Sridhara R., Pazdur R.. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005; 10:176–182. [DOI] [PubMed] [Google Scholar]
- 57. Malik P., Cashen A.F.. Decitabine in the treatment of acute myeloid leukemia in elderly patients. Cancer Manag. Res. 2014; 6:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of the structures for human DNMT3B–3L in complex with 24-bp DNA duplex containing CpGpG sites and DNMT3B–3L in complex with 24-bp DNA containing CpGpT sites, DNMT3B–3L complex (i), DNMT3B–3L complex (ii) and DNMT3B (Q772R)–3L complex have been deposited in Protein Data Bank under accession numbers 6KDA, 6KDB, 6KDL, 6KDP and 6KDT, respectively.