Abstract
Aberrant Notch signaling plays a pivotal role in T-cell acute lymphoblastic leukemia (T-ALL) and chronic lymphocytic leukemia (CLL). Amplitude and duration of the Notch response is controlled by ubiquitin-dependent proteasomal degradation of the Notch1 intracellular domain (NICD1), a hallmark of the leukemogenic process. Here, we show that HDAC3 controls NICD1 acetylation levels directly affecting NICD1 protein stability. Either genetic loss-of-function of HDAC3 or nanomolar concentrations of HDAC inhibitor apicidin lead to downregulation of Notch target genes accompanied by a local reduction of histone acetylation. Importantly, an HDAC3-insensitive NICD1 mutant is more stable but biologically less active. Collectively, these data show a new HDAC3- and acetylation-dependent mechanism that may be exploited to treat Notch1-dependent leukemias.
INTRODUCTION
Notch signaling regulates differentiation and tissue homeostasis throughout development and has been particularly well-studied in hematopoiesis (1,2). Dysregulation of the Notch signaling pathway is linked to the development of several cancers (3–6) and mutations within the NOTCH1 gene and the Notch ubiquitin-ligase FBXW7 have been found in leukemias such as T-cell acute lymphoblastic leukemia (T-ALL) and chronic lymphocytic leukemia (CLL) (7–10). While the initial steps of the Notch signaling cascade are fairly well understood, the various events regulating Notch nuclear functions are under intense investigation. Upon ligand–Notch receptor interaction, proteolytic processing results in the release of the Notch Intracellular Domain (NICD), that translocates into the nucleus and activates gene expression by assembling a coactivator complex containing, among others, the transcription factor RBPJ (also known as CSL) and the histone acetyltransferase p300. The signal is terminated by proteasomal degradation of the NICD, followed by transcriptional repression mediated by transcription factor RBPJ and additional co-repressors (11–15). Interestingly, NICD profoundly regulates chromatin marks such as H3K4 methylation and H3K27 acetylation as well as deposition and acetylation of histone variant H2A.Z, which are also tightly controlled by the RBPJ-corepressor complex (15–20). The Notch response is actively terminated by ubiquitin-dependent proteasomal degradation and this in turn is controlled by phosphorylation, methylation and acetylation (9,21–26).
Histone deacetylases (HDACs) regulate gene expression by removing active histone marks (i.e. acetyl groups) from lysine residues and their pharmacological inhibition is currently under investigation for therapeutic purposes (27). Mammalian HDACs are grouped into four different classes based on their enzymatic function, structure and evolutionary conservation (28). HDAC1, 2, 3 and 8 belong to class I HDACs, which are ubiquitously expressed and predominantly localize in the nucleus. While HDAC1 and HDAC2 are part of different multi-subunit complexes, including NuRD, CoREST and Sin3 (29–31), HDAC3 is exclusively found within the NCoR/SMRT complexes (32–37). While the HDAC3-containing NCoR/SMRT complexes have been linked primarily to gene repression, also within the context of the Notch signaling pathway (18,38–41), recent studies also implicate HDAC3 as a positive regulator for gene expression. In fact, HDAC3 is required for the inducible inflammatory gene expression program (42) and as coactivator of IL-1 signaling (43). While the role of HDAC3 in regulating the acetylation of histone proteins has been extensively investigated, evidence for a function in controlling non-histone protein activities remains scarce, with some of the few examples being represented by STAT3 (44) and NF-κB/p65 (43).
Here, we show that HDAC3 loss-of-function results in downregulation of Notch target genes and this is also reflected in a reduction of the transcriptional activation mark H3K27ac. Mechanistically, we reveal that HDAC3-mediated deacetylation of the NICD1 stabilizes the NICD1 protein itself. An HDAC3-independent NICD1 mutant protein shows less ubiquitination, becomes more stable, and possesses reduced biological activity compared to the NICD1 wildtype. Together, we propose that HDAC3 acts as a positive regulator in Notch signaling.
MATERIALS AND METHODS
Cell culture, treatments, transfection and infection
Mouse leukemia preT cells (Beko) were grown in Iscove's Modified Dulbecco Medium (IMDM, Gibco 21980-065) supplemented with 2% fetal bovine serum (Pan Biotech), nonessential amino acids (Gibco), 0.3 mg/ml Primatone, penicillin/streptomycin (Gibco) and 5 mg/l insulin (Sigma-Aldrich). Human CUTLL1 T-ALL cells were kindly provided by Dr F. Radtke (EPFL, Lausanne, Switzerland) and by Dr A. Ferrando (University of Columbia, New York, USA) and they were previously described (45). Human REC-1 cells were commercially acquired (DSMZ, ACC 584). Both CUTLL1 and REC-1 cells were grown in RPMI-1640 (Gibco 61870-010) supplemented with 10% fetal bovine serum (Pan Biotech) and penicillin/streptomycin (Gibco). 293T cells, Phoenix™ packaging cells (Orbigen, Inc., San Diego, CA, USA) and HeLa cells (ATCC: CCL2) were grown in Dulbecco's modified eagle medium (DMEM, Gibco 61965-059) supplemented with 10% fetal bovine serum (Pan Biotech) and penicillin/streptomycin (Gibco). Cells were grown at 37°C under 5% CO2.
Drosophila melanogaster Schneider cells were grown in Schneider's Drosophila medium (Gibco 21720024) supplemented with 10% fetal bovine serum (Gibco 10270-106), penicillin/streptomycin (Gibco) and Glutamine (Gibco 25030-024).
Beko cells were treated with 0.01 μg/ml apicidin (Sigma-Aldrich A8851) for 24 hours, 5 μM MG132 (Calbiochem 474790) for 6 h, 10 μg/ml GSI (DAPT; Alexis ALX-270-416-M025) for 24 h or 50 μg/ml cycloheximide (Applichem A0879.0001) for specific time points as indicated in the respective figure. As control, the respective vehicles were used. In the case of the survival curve, Beko cells were treated for 24 h with: 0.01, 0.02, 0.05 and 0.1 μg/ml apicidin or DMSO as control; 0.05, 0.1, 0.5 and 1 μM SAHA (Selleckchem S1047) or DMSO as control; 1, 10, 20 mM NAM (Sigma-Aldrich 72340-250G) or H2O as control.
In the case of the survival curve, CUTLL1 cells were treated for 24 h with 0.05 μg/ml, 10 μg/ml apicidin or DMSO as control while REC-1 cells were treated for 24 h with 0.05 μg/ml, 1 μg/ml, 10 μg/ml apicidin or DMSO as control.
Phoenix™ packaging cells were transfected using the calcium phosphate method. Briefly, 5 × 106 cells were seeded in a 75 cm2 culture flask in 10 ml medium and incubated at 37°C for 16–24 h. 20 μg of DNA were diluted in 860 μl of sterile H2O. After adding 120 μl of CaCl2, the resulting solution was slowly pipetted dropwise into 1 ml of 2× HBS (50 mM HEPES pH 7.05, 10 mM KCl, 12 mM glucose, 280 mM NaCl, 1.5 mM Na2HPO4) while vortexing. After 20 min of incubation at room temperature, the DNA solution was added to the cells that were pre-incubated for 10 min with chloroquine (Sigma-Aldrich C6628-100G). Six to eight hours post-transfection, the medium was replaced with fresh one.
Retroviral infections of Beko cells were performed as follows: Phoenix™ cells were transfected as described above and 24 h post-transfection the supernatant, containing the retroviral particles, was filtered and supplemented with 2 μg/ml of polybrene (Sigma-Aldrich H9268). The viral suspension was used to infect approximately 5 × 105 cells by centrifugation (1800 rpm, 45 min, 37°C). The infection was repeated four times over a period of 2 days. After infection, cells were analyzed and sorted by FACS or selected with specific antibiotics: puromycin (Serva 33835) and/or histidinol (Sigma-Aldrich H6647).
Apoptosis
Beko cells were treated for 24 h with 0.01 μg/ml apicidin or DMSO as control while CUTLL1 and REC-1 cells were treated for 24 h with 0.05 μg/ml apicidin or DMSO as control. Apoptosis was evaluated using the eBioscience™ Annexin V Apoptosis Detection Kit FITC (Invitrogen 88-8005-72) accordingly to manufacturer′s instructions and data were acquired at a BD FACSCalibur™ flow cytometer. Data were finally analyzed using the FlowJo 8.8.7.
Cell cycle analysis
After washing twice in PBS, cells were fixed in 70% ethanol on dry ice for at least 1 h. For staining, the fixed cells were spun shortly and the pellet was resuspended in 200 μl of staining buffer [50 μg/ml propidium iodide (Merck) in 4 mM sodium citrate, 0.1 mg/ml RNase] for 10 min at 37°C. For subsequent analysis, a FACSCalibur cytometer (BD Biosciences) was used. Cell cycle stages (SubG1/G1/S/G2M) were determined according to the DNA content. The SubG1 cell population was defined as cells with less than 2n DNA content (M4). Cells possessing 2n DNA were assigned to be in G1-phase (M1), those having a DNA content between 2n and 4n were defined as S-phase cells (M2) and those having approximately 4n DNA content were assumed to be in G2M (M3).
Lentiviral shRNA knockdown
HDAC3 knockdown in Beko cells was performed using the pLK0.1 TRC1 shRNA library (Sigma-Aldrich). Briefly, 293T cells were transfected with 3.3 μg of the desired shRNA construct and the packaging vectors psPAX (2.5 μg) and pMD26 (1 μg) using 14 μl of linear PEI (Polyscience 23966). After 48 h of incubation at 37°C, the supernatant of the 293T cells was filtered and used for infections of Beko cells. The selection of the positively infected cells was performed by adding 1 μg/ml puromycin (Serva 33835).
Constructs
All oligonucleotides used for cloning purposes are listed in Supplementary Table S6. PCR products were cloned in the pSC-A-amp/kan (Agilent Technologies 2402055), digested with the desired restriction enzymes (New England Biolabs) and cloned into the destination vectors accordingly to Supplementary Table S7. All plasmids were analyzed by sequencing.
The pcDNA 3.1 Flag2 (Invitrogen) and pGEX6P1 (GE Healthcare) were commercially acquired. The pcDNA3-Flag-mNICD1(wt), pcDNA3.1 Flag-mNICD1 ΔOP, pMT123-HA-8 × –Ubiquitin, pGEX6P1-GST-CT(N1) and pGEX6P1-GST-NT(N1) plasmids were previously described (22,46). The pcDNA3-HA-HDAC3 construct was previously described (47). The pCMV Tag4A Bart1 HDAC1 construct was a generous gift of Dr C. Seiser while the HA-p300 cDNA was kindly provided by Dr M.L. Schmitz (48).
The pMy Bio NCMH oligo pSV40 PURO was generated by insertion of the oligo NCMH (sequence in Supplementary Table S6) into the pMy Bio pSV40 PURO predigested with NotI/HindIII. The pMy Bio NCMXH oligo pSV40 PURO was generated via insertion of the oligo NCMXH (sequence in Supplementary Table S6) into the pMy Bio NCMH oligo pSV40 PURO predigested with NotI/HindIII.
To generate the 8KR (K2050, K2068, K2146, K2147, K2150, K2154, K2161 and K2164) and 6KR (K1764, K1770, K1771, K1772, K1785 and K1935) mutant plasmids, a mouse NICD1 specific 812 bp NotI/HindIII fragment for the 8KR and a mouse NICD1 specific 707 bp Acc65I/AccIII fragment for the 6KR were synthetized at GENEART/Life Technologies and inserted into the corresponding sites of pcDNA3-Flag-mNICD1(wt), pcDNA3-Flag-mNICD1-GFP(wt), pcDNA3-mN1ΔE wt and pCS2-mN1ΔE wt.
The pMy NCMXH BioFlag-NICD1 wt pSV40 PURO and the pMy NCMXH BioFlag-NICD1 8KR pSV40 PURO plasmids were generated by Acc65I/XbaI digestion of the pcDNA3-Flag-mNICD1 wt and the pcDNA3-Flag-mNICD1 8KR, respectively. The digestion products were blunt ended with T4 DNA polymerase (New England Biolabs M0203S) and ligated into the pMy Bio NCMXH oligo pSV40 PURO predigested with MfeI/HindIII.
Protein extracts and Western blotting
Whole cell extracts (WCE) were prepared as follows: cells were washed twice with ice-cold PBS and resuspended in WCE buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% NP-40, 10% glycerol, 0.5 mM Na3VO4, 10 mM NaF, 1 mM PMSF, 1× protease inhibitor cocktail mix). After 15 min incubation on ice, lysates were centrifuged (13 200 rpm, 15 min, 4°C). Protein concentrations were determined by Bradford assay (Sigma-Aldrich) and extracts were analyzed by Western blotting.
To prepare the nuclear extract, Beko cells were washed twice in PBS and lysed in Hypotonic buffer (20 mM HEPES pH 7.9, 20 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.2 mM PMSF). After 20 min incubation on ice, samples were centrifuged (4000 rpm, 10 min, 4°C) and the nuclei were washed twice with ice-cold PBS. Nuclei were resuspended in Hypertonic Buffer (20 mM HEPES pH 7.9, 1 mM MgCl2, 300 mM NaCl, 0.2% NP-40, 25% glycerol, 0.2 mM PMSF, 1× Protease inhibitor mix, 0.3 mM DTT) and incubated 20 min on ice. After centrifugation (14 000 rpm, 5 min, 4°C), the supernatant was collected and boiled in SDS loading buffer.
For Western blotting, proteins were resolved in SDS polyacrylamide gels and transferred to a Hybond-P PVDF membrane (Amersham) by wet blotting. In the case of the p300 western blotting, proteins were transferred over night at 30 V in a transfer buffer lacking of methanol.
Membranes were blocked in 5% milk/TBST (1× TBS, 0.1% Tween 20) before adding the desired antibody diluted in 5% milk/TBST [1:2000 GAPDH (abcam ab8245); 1:5000 FLAG (Sigma-Aldrich F3165); 1:5000 HA (3F10, Roche 1867423); 1:1000 H3 (abcam ab1791); 1:1000 Val1744 cleaved NICD1 (Cell Signaling 2421); 1:1000 NOTCH1 (abcam ab128076); 1:1000 p300 (Cell Signaling 54062); 1:1000 Ac-p300 (Cell Signaling 4771); 1:1000 RBPJ (Cosmo T6709); 1:1000 MAML1 (abcam ab155786); 1:400 HDAC3 (Santa Cruz sc-17795); 1:1000 H3K27ac (Diagenode pAb-174–050); 1:2000 H3K18ac (Cell Signaling 9675); 1:1000 H3K9ac (abcam ab4441); 1:1000 TBP (Santa Cruz sc-273); 1:500 β-ACTIN (Sigma A1978); 1μg/ml VINCULIN (abcam ab130007)]. Membranes were washed in TBST and incubated 1 h at room temperature with secondary antibody diluted 1:5000 in 5% milk / TBST [anti-rat IgG HRP (Jackson ImmunoResearch 112-035-072), anti-mouse IgG HRP (Cell Signaling 7076S), anti-mouse IgG HRP (GE Healthcare NA931V) or anti-rabbit IgG HRP (Cell Signaling 7074S)]. Membranes were finally washed in TBST.
Only in the case of the acetylated-lysine western blotting, the primary acetylated-lysine (Cell Signaling 9441) antibody was diluted 1:300 in 5% BSA/TBST.
In the case of the STREPTAVIDIN blotting, membranes were blocked 1 h at room temperature in 5% BSA/1× TBS. Membranes were incubated with STREPTAVIDIN-HRP (Perkin Elmer NEL750001EA) diluted in 5% BSA/1× TBS for 1 h at room temperature and washed three times, 15 min each, with 1× TBS, 0.5 M NaCl, 0.5% Triton X-100.
All membranes were incubated at room temperature with ECL solution and chemiluminescence was detected with a light sensitive film or with a Vilber Fusion FX7 system.
Ubiquitination assay and acetyltransferase assay
To detect ubiquitination of NICD1, Phoenix™ cells were transfected with pcDNA3-Flag-mNICD1 wt or pcDNA3-Flag-mNICD1 8KR plasmids with or without pMT123-HA-8x–Ubiquitin using the calcium phosphate method as described above. Thirty six hours post-transfection, 20 μM MG132 were added and after 6 h, cells were washed twice with PBS. Cells were lysed in 300 μl of denaturing lysis buffer (20 mM Tris–HCl pH 7.4, 5 mM EDTA, 2% SDS, 10 mM DTT) and, after incubation for 15 min at 99°C, samples were diluted ten times in IP-buffer (50 mM Tris–HCl pH 7.4, 300 mM NaCl, 5 mM EDTA, 1% NP-40, 1 mM PMSF, 1× protease inhibitor cocktail mix). FLAG-tagged proteins were immunoprecipitated with FLAG-M2 affinity gel (Sigma-Aldrich A2220) and beads were washed with IP-buffer and PBS. Immunoprecipitates were analyzed by Western blotting using HA (3F10, Roche 1867423) and FLAG (Sigma-Aldrich F3165) antibodies. Alternatively, Phoenix™ cells were transfected with pcDNA3-Flag-mNICD1 wt or pcDNA3-Flag-mNICD1 8KR plasmids, using the calcium phosphate method. 36 hours post-transfection, 20 μM MG132 were added and after 6 h, cells were washed twice with PBS. Extracts were prepared using WCE buffer and boiled 5 min at 95°C in SDS loading buffer. Samples were analyzed by western blotting.
For the acetyltransferase assays, 2.5 × 106 293T cells were seeded in a 10 cm2 culture dish in 10 ml medium and incubated at 37°C for 16–24 h. 20 μg of DNA were diluted in 325 μl of PBS while 40 μl of linear PEI (Polyscience 23966) were diluted in 309 μl of PBS. The two solutions were mixed together and, after 30 min of incubation at room temperature, the DNA solution was added to the cells. Six to eight hours post-transfection, the medium was replaced with fresh one. Forty eight hours post-transfection, cells were washed with ice-cold PBS, scraped in 500 μl denaturing lysis buffer and boiled for 15 min at 99°C. Lysates were diluted 10 times with IP-Buffer and centrifuged (4000 rpm, 10 min, 4°C). FLAG-tagged proteins were immunoprecipitated with FLAG-M2 affinity gel (Sigma-Aldrich A2220) and beads were washed with WCE buffer. Immunoprecipitates were analyzed by western blotting using HA (3F10, Roche 1867423), FLAG (Sigma-Aldrich F3165) and acetylated-lysine (AcK, Cell Signaling 9441) antibodies.
Cycloheximide chase assay
To perform the cycloheximide (CHX) assay in Beko cells overexpressing the BioNICD1 wt or 8KR mutant, three millions of cells per time point were seeded in a well of a six well plate, treated with 50 μg/ml CHX, collected at the indicated time points and WCE were prepared. In the case of the endogenous NICD1 CHX assay, Beko cells were treated for 24 h with 0.01 μg/ml apicidin or DMSO as control and CHX was added after the first 18 h at a final concentration of 50 μg/ml. Proteins were analyzed by western blotting and quantification was done using ImageJ 1.48v. NICD1 protein abundance was normalized to the loading control GAPDH.
GST protein purification and GST pulldown
GST fusion proteins were expressed in Escherichia coli strain BL21 (Stratagene), purified using standard procedures and stored at −80°C. Proteins were in vitro translated in presence of [35S] methionine (Perkin-Elmer NEG709A500UC) using the reticulocyte lysate-coupled transcription/translation system (Promega L4610) accordingly to manufacturer's instructions. Translation and labeling quality were monitored by SDS-PAGE. GST protein and GST fusion proteins were immobilized on Glutathione Sepharose 4 Fast Flow (GE Healthcare 17-5132-01) and incubated with the in vitro translated proteins in buffer A (40 mM HEPES at pH 7.5, 5 mM MgCl2, 0.2 mM EDTA, 0.5% NP-40, 100 mM KCl) under rotation for 2 h at 4°C. Beads were washed with buffer A, buffer B (40 mM HEPES pH 7.5, 5 mM MgCl2, 0.2 mM EDTA, 0.5% NP-40, 300 mM KCl) and PBS. After washing, beads were boiled in SDS loading buffer and proteins were resolved by SDS-PAGE. Gels were dried and exposed to X-ray films.
Luciferase assay
HeLa cells (2 × 105) were seeded in 48-well plates 24 h prior to transient transfection. Transfection was performed using the Nanofectin transfection reagent (PAA) with 1 μg of reporter plasmid alone or together with NOTCH1 expression plasmids N1ΔE wt or N1ΔE 8KR (amounts are given in the corresponding figure legend). Luciferase activity was determined 24 h after transfection from at least four independent experiments with 20 μl of cleared lysate in an LB 9501 luminometer (Berthold) using the luciferase assay system from Promega.
Immunofluorescence
Imaging was performed by plating HeLa cells at a concentration of 5 × 105 cells/cm2 on chamber coverslips (Nunc). After 16 h, cells were transfected with 250 ng of expression plasmids GFP-NICD1 wt or GFP-NICD1 8KR using the Nanofectin transfection reagent (PAA). Cells were fixed (4% paraformaldehyde) 24 hours after transfection. After DAPI (4′,6-diamidino-2-phenylindole) staining, pictures were acquired using a IX71 fluorescence microscope (Olympus) equipped with a digital camera (C4742, Hamamatsu) and a 100-W mercury lamp (HBO 103W/2, Osram). The following filter sets were used: green channel (ex: HQ470/40, em: HQ525/50), blue channel: (ex: HQ360/40, em: HQ457/50).
Chromatin immunoprecipitation (ChIP), STREPTAVIDIN ChIP, libraries and sequencing
The ChIP-Seq experiments were performed as previously described (49,50) and chromatin from Drosophila melanogaster Schneider cells was used for spike-in purposes (each 25 μg of mouse chromatin, 50 or 25 ng of Drosophila chromatin were used in ChIP versus histone proteins or transcription factors, respectively). 2 μg of anti-His2Av (Active Motif 61686) were added to each immunoprecipitation for spike-in purposes.
In the case of the ChIP-qPCR experiments, Beko cells were washed twice with PBS, fixed for 1 hour at room temperature in 10 mM dimethyladipimate (DMA, Thermo Scientific 20660) dissolved in PBS and, after washing once in PBS, crosslinked in 1% FMA for 30 min at room temperature. The FMA reaction was blocked by adding 1/8 volume of 1 M glycine pH 7.5 and incubating for 5 min at room temperature. Cells were washed twice with PBS and resuspended in 1 ml of SDS Lysis Buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.1). After 10 min of incubation on ice, samples were sonicated using the Covaris System S220 AFA (28 cycles, 30 s ON, 30 s OFF). The chromatin was diluted with ChIP Dilution Buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl pH 8.1, 167 mM NaCl) and pre-cleared with pre-saturated protein-A-Sepharose beads (GE Healthcare 17-5280-02) for 30 min at 4°C. The chromatin was subsequently incubated over night with the proper amount of the desired antibody. Antibodies were immobilized with 40 μl pre-saturated protein-A-Sepharose beads for 1 h at 4°C with shaking. Depending on the antibody, different combinations of the following washing buffers were used: low salt washing buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1, 150 mM NaCl), high salt washing buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1, 500 mM NaCl), LiCl washing buffer (1% NP-40, 1 mM EDTA, 10 mM Tris–HCl pH 8.1, 0.25 M LiCl) and TE buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA). Chromatin was eluted from beads with Elution Buffer (1% SDS, 0.1 M NaHCO3) and crosslinks were reverted at 65°C over night in presence of 180 mM NaCl. After incubation with Proteinase K for 1 h at 45°C, the DNA was purified by phenol/chloroform extraction and precipitated over night at −20°C in presence of yeast tRNA, glycogen and 2-propanol. The DNA pellet was washed with 70% EtOH, dried and dissolved in TE pH 8.0.
The following antibodies were used: HDAC3 (abcam ab7030), H3K27ac (Diagenode pAb-174-050), IgG (Santa Cruz sc-2027; Diagenode kch-504-250) or RBPJ (Cell Signaling 5313).
In the case of the STREPTAVIDIN ChIP, chromatin was diluted and pre-clearing was performed for 1 h at 4°C with IgG magnetic beads M280 (Invitrogen 112.01D) pre-blocked with salmon sperm DNA (Invitrogen) and Fish skin gelatin (Sigma-Aldrich G7041). The chromatin was incubated overnight with M280 STREPTAVIDIN magnetic beads (Invitrogen, 112.06D) pre-blocked with salmon sperm DNA (Invitrogen) and Fish skin gelatin (Sigma-Aldrich G7041). Beads were washed twice with low salt washing buffer, twice with high salt washing buffer and twice with TE buffer. The DNA was eluted, decrosslinked and purified as described above.
ChIP experiments were analyzed on a StepOnePlus™ sequence detector system (Applied Biosystem) using specific oligonucleotides and double-dye probes (Supplementary Table S6).
Libraries were prepared using the Diagenode MicroPlex Library Preparation kit v2 (Diagenode) following manufacturer's instructions with few modifications. Libraries were purified with Agencourt AMPure XP Beads (Beckman Coulter, #A63881), quantified, analyzed on a Tapestaion device (Agilent) and pooled. Finally, sequencing was performed by Centro de Análisis Genómico (CNAG-CRG), Spain.
RNA extraction, reverse transcription, quantitative PCR (qPCR), libraries and sequencing
Total RNA was purified using Trizol reagent (Ambion, 15596018) accordingly to manufacturer's instructions. 1 μg of RNA was retro-transcribed in cDNA using random hexamers and M-MuLV reverse transcriptase (NEB). qPCRs were assembled with Absolute QPCR ROX Mix (Thermo Scientific AB-1139), gene-specific oligonucleotides and double-dye probes (Supplementary Table S6) and analyzed using the StepOnePlus™ sequence detector system (Applied Biosystem). Data were normalized to the housekeeping gene Hypoxanthine Phosphoribosyltransferase 1 (HPRT).
For RNA-Seq purposes, total RNA was purified using the RNeasy Mini Kit (Qiagen #74104), the QIAshredder (Qiagen #79654) and the DNase I (Qiagen #79254) accordingly to manufacturer′s instructions. Libraries were prepared using the TruSeq® Stranded Total RNA LT-Ribo-Zero Gold kit (Illumina RS-122–2301/2) and sequenced on an Illumina HiSeq 1500 with 50 bases single reads.
RNA-Seq and microarray analysis
RNA-Seq reads were quality controlled with FastQC available at http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. After visual inspection of FastQC results, no specific trimming or filtering was applied. Reads were aligned using Tophat 2.0.9 (51) based on Bowtie 2.1.0 (52) to a precompiled index of the mm9 genome downloaded from the Bowtie homepage. Identification of differentially expressed genes was done after import of BAM-alignment files into R using BioConductor packages (53). Therefore, we used the summarize Overlaps function of the Genomic Alignments package (54) in order to extract gene-specific read counts. As gene models, we used mm9 UCSC gene annotations downloaded from Illumina's iGenome site (http://support.illumina.com/sequencing/sequencing_software/igenome.html).
Normalization between experiments and detection of differentially expressed genes was done with the DESeq2 package with default settings (55). Subsequent statistical testing using Wilcoxon-signed rank test and subsequent graphical representation of the analysis were done in R. In order to compare the RNA-Seq data to previously published Affymetrix microarray expression data (22), we downloaded CEL-files from NCBI’s Gene Expression Omnibus (accession no.: GSE62528). We extracted gene expression estimates following the Gene 1.0 ST vignette (http://www.aroma-project.org/vignettes/GeneSTArrayAnalysis/) of the aroma.affymetrix BioConductor package (56). We used RMA background correction and quantile normalization. Gene expression values were log2-transformed and log2-transformed fold change values were calculated by subtracting experimental from the respective control condition. Basic summary statistics for the RNA-Seq experiments are provided in Supplementary Table S2.
ChIP-Seq analysis
Fastq files were controlled for quality issues using fastqc. Read alignment against the mm9 mouse reference genome was done with bowtie2 version 2.2.9 with default settings (52). Duplicate removal was performed using samtools rmdup function. Coverage vectors were generated with deeptools bamCoverage function using FPKM normalization. Visualization of binding profiles was done using the R/BioConductor package Gviz. RBPJ peak calling was done using Macs2 (57) and peakranger (58). For robust detection of peaks, we used the intersection between both methods and both replicates. The resulting set was filtered against blacklisted chromatin regions as detected by ENCODE. For motif identification, we determined the site of maximum coverage within each peak and selected the corresponding DNA sequence from –50 bp to +50 bp around that maximum as input for MEME-ChIP (59). All downstream analysis was done within R (http://www.R-project.org). Quantitative differences in H3K27 acetylation were analyzed using DESeq2 (55). First, Macs2 version 2.1.1 function callpeaks was used for identification of binding peaks using default settings. For each experimental condition (DMSO, GSI, apicidin) the overlap between the two replicates was selected. The three resulting peak sets were merged and reduced using the reduce function of the GenomicRanges package (54). For this unified set of peaks, we extracted the read counts for each experimental sample. DESeq2 was used for normalization and calculation of differential binding events using following settings: betaPrior was set to FALSE and fitType was set to local. We calculated the contrasts apicidin-DMSO and GSI-DMSO. Log2-transformed fold changes between experimental and control treatments were analyzed across all H3K27ac peaks and compared to those H3K27ac peaks overlapping with RBPJ binding sites. A third group of H3K27ac sites was selected based on their overlap with RBPJ binding and the association with a Notch target gene as defined by the approach shown in Figure 1B. Differences were visualized as boxplots. Statistical analysis was done using Wilcoxon rank sum tests. Peak annotation was done using Illumina iGenome annotation for RefSeq genes downloaded as GTF identifying the next transcriptional start site (TSS). The underlying collection of R code and terminal commands are available upon request. Basic summary statistics for the ChIP-Seq experiments are provided in Supplementary Table S3.
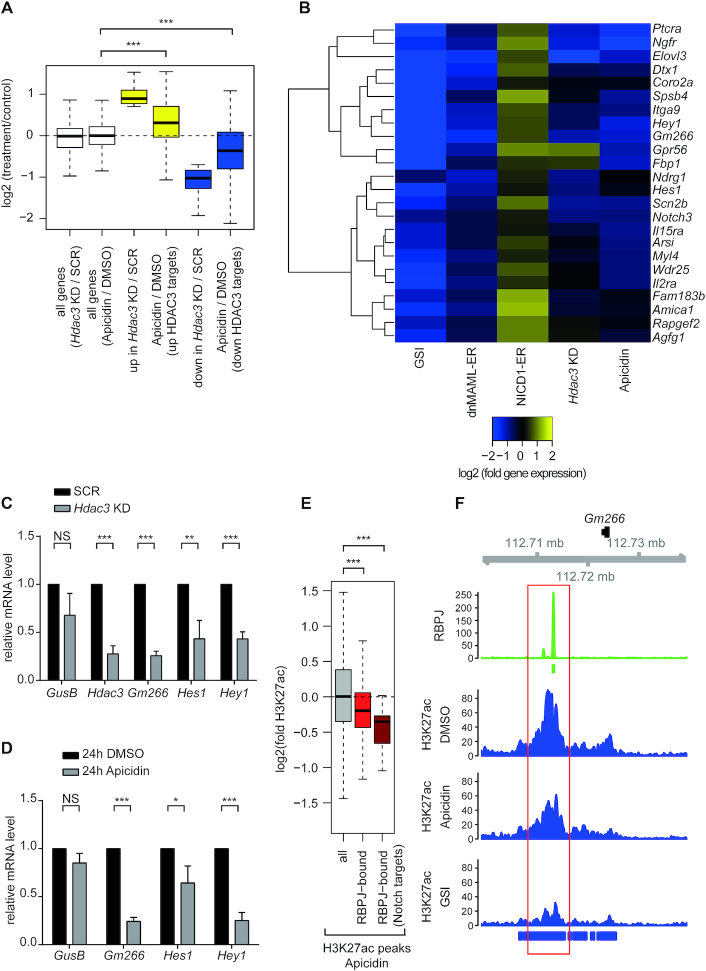
Figure 1.
HDAC3 is a positive regulator of the Notch-dependent gene expression program. (A) Box plot showing the effects of HDAC3 loss-of-function (LoF) by shRNA-mediated knockdown or apidicin treatment on the transcriptome of Beko cells. Beko cells were infected with shRNAs directed against Hdac3 (Hdac3 KD) or scramble (SCR) as control or, alternatively, treated for 24 h with 0.01 μg/ml apicidin or DMSO as control. Upon RNA purification, samples were analyzed by RNA-Seq. Genes up- or downregulated by infection of shRNAs against Hdac3 or apicidin treatment were identified based on log2FC bigger than 0.7 or smaller than –0.7, respectively and adjusted P-value <0.05. Statistical analysis was performed by using the Wilcoxon signed rank test (***P < 0.001). White boxes are representative of all the genes in Hdac3 KD versus scramble (SCR) control or in apicidin versus DMSO control. Yellow and blue boxes indicate up- and downregulated genes, respectively. Up- and downregulated genes upon Hdac3 KD are labeled as ‘up in Hdac3 KD/SCR’ and ‘down in Hdac3 KD/SCR’, respectively. Effects of apicidin treatment are shown for both up- and downregulated HDAC3 targets [‘Apicidin / DMSO (upregulated HDAC3 targets)’ and ‘Apicidin / DMSO (downregulated HDAC3 targets)’]. (B) Heat map showing the effects of HDAC3 LoF at Notch target genes. Notch target genes were identified by analyzing the transcriptome of Beko cells upon inhibition of Notch signaling with γ-secretase inhibitor (GSI) DAPT and analyzing microarray data from Beko cells overexpressing an inducible dominant negative MAML mutant (dnMAML-ER) or an inducible NICD1 (NICD1-ER). Beko cells were treated for 24 h with 10 μg/ml GSI or DMSO as control. (C and D) HDAC3 LoF by (C) shRNA-mediated knockdown or (D) apidicin treatment leads to downregulation of Notch target genes. Beko cells were infected with shRNAs directed against Hdac3 (Hdac3 KD) or scramble (SCR) as control (C) or, alternatively, treated for 24 h with 0.01 μg/ml apicidin or DMSO as control (D). Upon RNA extraction and reverse transcription, cDNAs were analyzed by qPCR using primers specific for GusB, Gm266, Hes1, Hey1 or, in the case of the shRNA-mediated knockdown, Hdac3. Data were normalized to the housekeeping gene Hypoxanthine Guanine Phosphoribosyltransferase (HPRT). Shown is the mean ± SD of three independent experiments (*P < 0.05, **P < 0.01, ***P < 0.001, [NS] not significant, unpaired Student's t-test). (E and F) Apidicin treatment leads to reduced H3K27ac at the RBPJ binding sites in Beko cells. (E) Apicidin treatment leads to reduced H3K27ac at the RBPJ-bound genomic sites. Beko cells were treated for 24 h with 0.01 μg/ml apicidin or DMSO as control and the effects on H3K27ac were investigated by ChIP-Seq comparing all H3K27ac sites with those overlapping RBPJ binding sites and RBPJ-bound Notch targets as shown in the A panel (Wilcoxon rank sum tests P = 9.72e−56 and P = 1.43e−03, respectively, ***P < 0.001). (F) Genome browser snapshot of RPKM-normalized coverage profiles showing the effects of apicidin or GSI treatment on H3K27ac at the RBPJ-bound enhancer of Notch target gene Gm266 in Beko cells. DMSO-treated cells were used as control. Bars mark significantly bound regions as detected by Macs2/peakranger (RBPJ; green) and Macs2 (H3K27ac; blue).
Alignment
Alignment of the NICD1 protein sequences [Homo sapiens (NP_060087.3), Mus musculus (NP_032740.3), Xenopus laevis (NP_001081074.1) and Danio rerio (NP_571377.2)] was done using T-Coffee available at http://tcoffee.vital-it.ch/apps/tcoffee/index.html.
Patient data analysis
Publicly available data were downloaded from NBCI’s Gene Expression Omnibus. The following datasets were analyzed: GSE26713 (60), GSE31048 (61), GSE32018 (62), GSE36907 (63), GSE33469 (64) and GSE33470 (64). Preprocessed table with normalized expression measures were downloaded using the GEOquery package (65) within the statistical programming environment R. In the case of GSE33469 and GSE33470 we combined the corresponding tables into a single data matrix. In all cases data matrices were additionally normalized by quantile normalization. Information about sample annotations were extracted from the ExpressionSet object generated by the GEOquery request. Differences between groups were analyzed for statistical significance by non-parametric Wilcoxon signed-rank test and were visualized as boxplots.
Danio rerio strains and injection procedures
Care and breeding of D. rerio (zebrafish) was conducted as previously described (66). The present study was performed after securing appropriate institutional approvals. It conforms with the Guide for the Care and Use of Laboratory Animals published by the ‘US National Institute of Health’ (NIH Publicaton No. 85-23, revised 1996). For the injection procedures, we used the WIK wildtype strain. Sense capped RNAs encoding N1ΔE wildtype or acetylation-defective N1ΔE 8KR were synthesized using the mMESSAGE mMACHINE system (Ambion). mRNA was diluted (20 ng/μl) in 0.2 M KCl and co-microinjected with 10 ng/μl reporter plasmid [12 × CSLRE-EGFP (22)] into one-cell stage embryos. Siblings from the same pool were injected using the reporter DNA only as control. At 24 h post-fertilization (hpf) pictures were recorded using an Olympus SZX16 Stereomicroscope. Data are presented as mean ± SEM from five independent experiments. Comparisons between experimental groups were performed using nonparametric Mann-Whitney rank sum test. Differences were considered significant if they showed a value of P < 0.05.
RESULTS
HDAC3 is a positive regulator of the Notch signaling response
In higher eukaryotes chromatin configuration at Notch target genes is shaped by the NCoR complex and this process is evolutionary conserved in Drosophila melanogaster (12,18,67). Since the enzyme HDAC3 is an integral part of the NCoR complex, we sought to characterize its role in the Notch signaling pathway by performing shRNA-mediated loss-of-function (LoF) of HDAC3 and apicidin treatment (Figure 1 and Supplementary Figure S1 and Supplementary Tables S1 and S2). These experiments were performed in a preT cell line (Beko), in which NOTCH1 is constitutively active (16,18). The knockdown (KD) of Hdac3 was validated at the mRNA (Figure 1C) and protein level (Supplementary Figure S1A). Subsequently, we investigated in Hdac3 KD cells the effects on gene expression using RNA-Seq analysis (Figures 1A, B, Supplementary Figure S1B, C and Supplementary Tables S1 and S2) and observed 65 up- and 368 downregulated genes (Figure 1A and Supplementary Table S1). Importantly, the effects of either apicidin treatment or Hdac3 KD were similar even if not identical (Figures 1A, Supplementary Figure S1D-E, S1H-I and Supplementary Tables S1 and S2). Any differences observed between apicidin treatment and Hdac3 KD (Supplementary Figure S1H and I) may be due to off-target or non-specific effects or to the residual HDAC3 observed upon KD (Supplementary Figure S1A). The concentration of apicidin used for the RNA-Seq analysis (0.01 μg/ml, 16 nM) did not significantly influence the cell cycle or the viability of Beko cells (Supplementary Figure S1J–L). As next step, we focused on ‘bona fide’ Notch target genes which we previously defined as a) downregulated by γ-secretase inhibitor (GSI), b) downregulated by a dominant negative mutant of the Notch coactivator MASTERMIND [dnMAML (68,69)] and c) upregulated by gain-of-function (GoF) of the NICD1. To put this into a genome-wide context, we performed RNA-Seq analysis upon GSI treatment of Beko cells (Figures 1B, Supplementary Figure S1F-G and Supplementary Table S1 and S2) and compared these data sets to previous loss- and gain-of-function data sets (dnMAML-ER and NICD1-ER, respectively) (22). The resulting short list of ‘bona fide’ Notch target genes is summarized in Figure 1B and Supplementary Table S1. Importantly, when we investigated the effects of Hdac3 KD or apicidin treatment on the expression of these Notch target genes, we found the majority to be downregulated (Figures 1B, Supplementary Figure S1M and Supplementary Table S1). RT-qPCR analysis of Gm266, Hes1 and Hey1 Notch target genes in Beko cells, Hdac3 KD or treated with apicidin, validated the RNA-Seq data (Figures 1C-D), further supporting the positive role of HDAC3 in the regulation of Notch target genes. Importantly, the pan-HDAC inhibitor SAHA (suberoylanilide hydroxamic acid, also known as vorinostat) and the SIRT1 inhibitor NAM (nicotinamide) did not influence the expression of Notch target genes revealing the specificity of the observed changes in gene expression (Supplementary Figure S2).
Given that HDACs are well described as negative regulators of histone acetylation, we investigated the effects of HDAC3 LoF on histone acetylation. As expected, apicidin treatment leads to global increase of H3K27ac, H3K18ac and H3K9ac (Supplementary Figure S3A) and similar effects were observed upon Hdac3 KD (Supplementary Figure S3B). Using Chromatin-IP followed by deep-sequencing (ChIP-Seq), we defined the RBPJ binding profile in Beko cells (Figures 1E, F, Supplementary Figure S3C–H and Supplementary Tables S3–S5). This analysis identified 1486 RBPJ binding sites (Supplementary Figure S3E and Supplementary Table S4) and the RBPJ binding motif was promptly identified (Supplementary Figure S3D), indicating the specificity of the RBPJ ChIP-Seq. In contrast to the global increase in histone acetylation, we observed significantly decreased H3K27ac at the RBPJ binding sites upon apicidin treatment (Figures 1E, F, Supplementary Figure S3F–H and Supplementary Table S5, P = 9.72e–5), as revealed by ChIP-Seq. This decrease was particularly pronounced when focusing on RBPJ binding sites associated with ‘bona fide’ Notch target genes (Figure 1E, F, P = 1.4e–03). It was also observed upon GSI treatment, which served as a positive control (Figures 1F, Supplementary Figure S3C and F–H). Of note, given that the bulk of H3K27ac, H3K18ac and H3K9ac increases upon apicidin treatment (Supplementary Figure S3A) or Hdac3 KD (Supplementary Figure S3B), these data suggest that the Notch pathway is regulated by HDAC3 in a specific manner to positively support gene expression. Altogether, these data define HDAC3 as a positive regulator of Notch signaling.
HDAC3 controls NICD1 protein stability by promoting its deacetylation
We then performed a series of experiments to identify the mechanism(s) by which HDAC3 affects Notch1 function. Since post-translational modifications are known to control NICD protein stability, we investigated the effects of HDAC3 LoF on the endogenous cleaved NICD1 protein. We observed that both shRNA- and apicidin-mediated LoF of HDAC3 lead to a strong decrease in NICD1 protein level in Beko cells (Supplementary Figures S1A and 2A, respectively). In order to investigate NICD1 protein half-life, we blocked protein translation by treating Beko cells with cycloheximide (CHX) and investigated the stability of endogenous cleaved NICD1. We observed that apicidin treatment leads to decreased stability of NICD1 (Figure 2B) and this effect was abrogated by MG132 (Supplementary Figure S4A), suggesting that the HDAC3-mediated NICD1 stabilization involves the blockade of the ubiquitination-dependent proteasomal degradation pathway. Accordingly, ectopic expression of HDAC3 in Beko cells stabilizes the endogenous NICD1 (Supplementary Figure S4B). As a next step, we investigated whether HDAC3 and NICD1 physically interact with each other. We observed a physical interaction between HDAC3 and NICD1 in GST pulldown experiments (Supplementary Figure S5A) and could observe their co-occupancy at the Notch-dependent enhancers of Gm266, Hes1 and Hey1 (Supplementary Figure S5B and C).
Figure 2.
HDAC3 stabilizes NICD1 via deacetylation. (A) Apicidin treatment leads to decreased NICD1 protein level. Beko cells were treated for 24 h with 0.01 μg/ml apicidin or DMSO as control and the whole cell extract (WCE) was analyzed by western blotting versus the endogenous cleaved NICD1 protein or GAPDH as loading control. (B) Apicidin treatment destabilizes the NICD1 protein. Beko cells were treated for 24 h with 0.01 μg/ml apicidin or DMSO as control and, after the first 18 h, protein synthesis was blocked by adding 50 μg/ml cycloheximide (CHX). Samples were collected at the indicated time points. WCE was analyzed by western blotting versus endogenous cleaved NICD1 or GAPDH as loading control. Quantification of the NICD1 levels normalized to GAPDH is shown on the right. The experiment was repeated independently three times. (C) HDAC3 deacetylates NICD1. 293T cells were transfected with plasmids encoding Flag-tagged NICD1 wildtype (Flag-NICD1 wt), HA-tagged HDAC3 (HA-HDAC3) or HA-tagged p300 (HA-p300). WCE were subjected to FLAG immunoprecipitation (FLAG-IP) and the immunoprecipitates were analyzed by western blotting using an acetylated-lysine antibody (AcK) and reblotted (RB) using a Flag antibody.
We thus hypothesized that HDAC3 controls NICD1 directly through regulation of its protein turnover, rather than by altering the NICD1-regulated chromatin environment. Stability of NICD proteins is regulated by their p300-mediated acetylation (24,25). Therefore, we tested the hypothesis whether HDAC3 could increase NICD1 protein stability by deacetylation. We performed acetyltransferase assays by overexpressing combinations of Flag-tagged NICD1 wildtype (wt), HA-tagged HDAC3 and/or HA-tagged p300. After Flag immunoprecipitation (IP) of the NICD1, we analyzed its acetylation by western blotting using a pan-acetyl-lysine (AcK) antibody (Figure 2C). As previously described (24,25), p300 efficiently acetylates the NICD1 protein (Figure 2C, lane 5) and we observed that HDAC3 strongly reduces the p300-mediated acetylation of the NICD1 protein (Figure 2C, lane 6). This effect was specific as HDAC3 did neither affect p300 protein levels nor its acetylation (Supplementary Figure S6). Additionally, when we used an HDAC3 mutant (HDAC3_B), that lacks the catalytic domain, the p300-mediated NICD1 acetylation was not influenced (Supplementary Figure S7A, lane 9) compared to the wildtype HDAC3 (Supplementary Figure S7A, lane 8) or to an HDAC3 mutant that retains its catalytic domain (HDAC3_A) (Supplementary Figure S7A, lane 10). Interestingly, HDAC3_A but not HDAC3_B is able to interact with NICD1 and this interaction does not require amino acids (2294–2531) of NICD1 since a C-terminally truncated NICD1 protein (NICD1 ΔOP) still interacts with HDAC3_A (Supplementary Figure S7B-C). In order to investigate the specificity of HDAC3 in the regulation of the NICD1 acetylation, we performed acetyltransferase assays comparing HDAC3 with another member of the class I family of HDACs, HDAC1 (Supplementary Figure S7D). Unlike HDAC3, HDAC1 did not reduce the p300-mediated NICD1 acetylation (Supplementary Figure S7D, compare lane7 and 8). The HDAC3-mediated regulation of NICD1 was confirmed at the endogenous protein level in Beko cells treated with apicidin (Supplementary Figure S7E): An increased AcK signal is observed upon apicidin treatment compared to the DMSO control (Supplementary Figure S7E). Collectively, these data indicate that HDAC3 stabilizes the NICD1 protein by promoting its deacetylation.
HDAC3 specifically deacetylates specific lysine residues within the NICD1
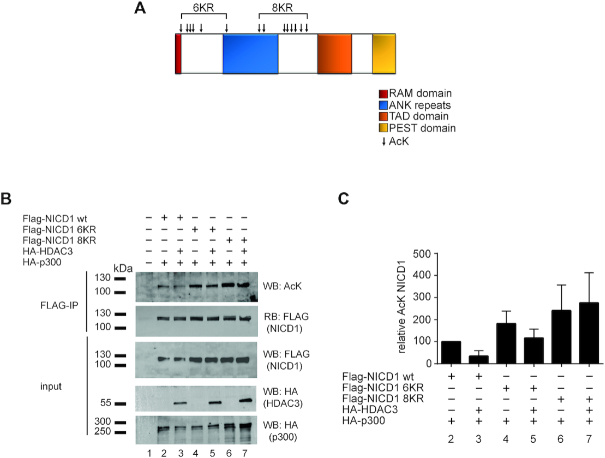
There are 14 different acetyl-lysine residues within the NICD1 that have been identified [(24) and representation in Figure 3A]. In order to dissect the HDAC3-mediated regulation of the NICD1 protein, we generated two different NICD1 batch-mutants where six or eight lysine residues were mutated to arginine residues (NICD1 6KR and NICD1 8KR, respectively; representation in Figure 3A). These mutants, including the NICD1 wt, were used in acetyltransferase assays (Figure 3B and quantification in 3C). We observed that while the 6KR mutant is still sensitive to the deacetylase activity of HDAC3, the 8KR mutant is not deacetylated by HDAC3, suggesting that HDAC3 mainly regulates the acetylation of the eight lysine residues located within the central part of the NICD1 protein (Figure 3B, compare lanes 4 with 5 and lanes 6 with 7 and quantification in 3C). We also observed that the NICD1 8KR mutant is stronger acetylated than its wt counterpart, suggesting that one or more of these eight lysine residues may play an inhibitory role in NICD1 acetylation. These data reveal that HDAC3 regulates the acetylation of the lysine residues close to the ankyrin-repeats in the central part of the NICD1 (Figure 3A).
Figure 3.
HDAC3 deacetylates specific lysine residues within the NICD1. (A) Schematic representation of the lysine residues that are acetylated within the Notch-1 Intracellular Domain (NICD1). Arrows indicate the position of the acetyl-lysines that have been mutated to arginines (R) in the case of the 6KR or 8KR mutants. (B and C) HDAC3 controls the acetylation of eight lysine residues. 293T cells were transfected with plasmids encoding Flag-tagged NICD1 wildtype (Flag-NICD1 wt), Flag-tagged NICD1 6KR mutant (Flag-NICD1 6KR), Flag-tagged NICD1 8KR mutant (Flag-NICD1 8KR), HA-tagged HDAC3 (HA-HDAC3) and/or HA-tagged p300 (HA-p300). WCE were subjected to FLAG immunoprecipitation (FLAG-IP) and the immunoprecipitates were analyzed by Western blotting using an acetylated-lysine antibody (AcK) and reblotted (RB) using a FLAG antibody. Quantification of the AcK levels within the NICD1 proteins is shown in C. Shown is the AcK signal normalized to the total immunoprecipitated NICD1 as percentage relative to the NICD1 wt. The experiment was repeated independently four times.
The NICD1 8KR mutant is more stable compared to the NICD1 wt
Next, we investigated the mechanisms used by HDAC3 to influence the stability of NICD1 protein. Importantly, mutating the HDAC3-regulated eight lysine residues to arginines did not influence the nuclear localization of NICD1 (Supplementary Figure S8A) or its interaction with MAML1 or RBPJ (Supplementary Figure S8B and C). Given that HDAC3 regulates the 8 lysine residues located within the central portion of the NICD1 protein, we hypothesized that the NICD1 8KR mutant could be more stable than its wildtype NICD1 counterpart. To test this, we ectopically expressed Bio-tagged NICD1 wt or NICD1 8KR mutant in Beko cells that express the biotin ligase BirA and performed CHX chase experiments. These experiments revealed that the NICD1 8KR mutant is more stable compared to the NICD1 wt (Figure 4A). In order to investigate whether the differences in NICD1 stability depend on prototypical proteasomal degradation, we expressed FLAG-tagged NICD1 wt or NICD1 8KR mutant in fibroblasts and performed ubiquitination assays in the presence (or absence) of proteasomal inhibitor MG132 (Figures 4B and C). In line with the previous data, we observed that the NICD1 8KR mutant is less ubiquitinated compared to the NICD1 wt (Figures 4B, compare lanes 5 with 6 and 4C, compare lanes 2 with 3). However, the NICD1 8KR mutant is less active in luciferase assays compared to the NICD1 wt (Figure 4D and expression control in Supplementary Figure S8D). Together, HDAC3 enhances the NICD1 protein stability via an ubiquitin-proteasome-dependent mechanism and an HDAC3-independent NICD1 8KR mutant behaves differently in regards to transcriptional output.
Figure 4.
The NICD1 8KR mutant is more stable and less ubiquitinated compared to the NICD1 wt. (A) The NICD1 8KR mutant is more stable compared to the NICD1 wt. Beko cells expressing the biotin ligase BirA, were infected with plasmids encoding BioFlag-NICD1 wildtype (BioNICD1 wt), BioFlag-NICD1 8KR mutant (BioNICD1 8KR) or empty vector as control (Ctrl) and treated with 50 μg/ml of cycloheximide (CHX) for the hours indicated in the figure. WCE was analyzed by STREPTAVIDIN blotting to detect the BioNICD1 wt and 8KR mutant proteins or Western blotting versus GAPDH as loading control. Quantification of the BioNICD1 wt and 8KR mutant levels normalized to GAPDH is shown on the right. The experiment was repeated independently three times. (B, C) The NICD1 8KR mutant is less ubiquitinated compared to the NICD1 wt. (B) Phoenix™ cells were transfected with plasmids encoding Flag-tagged NICD1 wildtype (Flag-NICD1 wt), Flag-tagged NICD1 8KR mutant (Flag-NICD1 8KR) or HA-tagged ubiquitin (HA-Ub) and treated with 20 μM of proteasome inhibitor MG132. WCE were subjected to FLAG immunoprecipitation (FLAG-IP) and the immunoprecipitates were analyzed by Western blotting using an HA antibody and reblotted (RB) with a FLAG antibody. (C) The NICD1 8KR mutant is less ubiquitinated compared to the NICD1 wt. PhoenixTM cells were transfected with plasmids encoding Flag-tagged NICD1 wildtype (Flag-NICD1 wt) or Flag-tagged NICD1 8KR mutant (Flag-NICD1 8KR) and treated with 20 μM of proteasome inhibitor MG132. WCE were analyzed by western blotting using a FLAG antibody or GAPDH as loading control. (D) NICD1 wt is more active compared to NICD1 8KR mutant. Transcriptional activity of N1ΔE wt or N1ΔE 8KR mutant was tested in luciferase assays using the RBPJ-dependent reporter construct pGA891/6. Mean values ± SD (error bars) from six independent experiments are shown ([**] P = 0.003, [***] P < 0.0001, unpaired Student's t-test).
The NICD1 8KR mutant shows a decreased biological activity in vivo
The eight lysine residues within the NICD1 are evolutionary conserved from human to zebrafish (Figure 5A). Hence, in order to investigate the in vivo relevance of our molecular findings, we overexpressed a dominant active NOTCH1, called ΔE (N1ΔE), lacking the extracellular domain which results in constitutive cleavage and release of NICD1. Specifically, we injected mRNAs encoding for N1ΔE wt or the 8KR mutant into one-cell-stage zebrafish (Danio rerio) embryos. Importantly, the eight lysine residues within the NICD1 are evolutionary conserved from human to zebrafish (Figure 5A). To confirm protein expression and activity after N1ΔE mRNA injection, we coinjected a reporter plasmid that expresses the GFP under the control of a Notch-dependent promoter [12× CSLRE-EGFP, (Figure 5B)]. Endogenous zebrafish NOTCH1 was not able to induce GFP expression significantly, whereas both injected N1ΔE mRNAs showed robust GFP activation at 24 hours post-fertilization (24 hpf). Zebrafish embryos injected with N1ΔE wt presented with severely impaired development of eyes and brain structures (Figure 5B and quantification in 5C). A close-up view of the malformations induced by N1ΔE wt compared to N1ΔE 8KR and control-injected embryos is shown in Supplementary Figure S9. In contrast to the N1ΔE wt, embryos injected with N1ΔE 8KR mutant showed a less severe phenotype (Figure 5B, quantification in 5C and Supplementary Figure S9) substantiating in vivo the hypothesis that the 8KR mutant, although more stable, is biologically less active.
Figure 5.

The NICD1 8KR mutant shows decreased biological activity compared to the NICD1 wt in vivo. (A) The eight lysine residues of NICD1, regulated by HDAC3, are evolutionary conserved. T-Coffee alignment of human (H. sapiens, NP_060087.3), mouse (M. musculus, NP_032740.3), frog (X. laevis, NP_001081074.1) and zebrafish (D. rerio, NP_571377.2) NOTCH1 proteins. The grey boxes indicate the eight lysine residues regulated by HDAC3. (B) The NICD1 8KR mutant is less active compared to the NICD1 wt in zebrafish. Zebrafish embryos were injected with cDNA encoding for the membrane bound Notch1ΔE wt (N1ΔE wt) or 8KR (N1ΔE 8KR) mutant. A reporter plasmid where the GFP-encoding gene is under the control of a Notch-dependent promoter was co-injected to monitor the NICD1 activity. (C) Quantification of malformed embryos shown in b. Shown are the means ± SD of the total number of embryos analyzed (n) in five independent experiments (N). **P < 0.001 (nonparametric Mann–Whitney rank sum test).
HDAC3 is highly expressed in human leukemic patient samples and apicidin treatment leads to reduced NICD1 protein levels in human leukemic cells
Our data suggest HDAC3 as an important modulator of NICD1 stability. Given that the NOTCH1 gene is frequently mutated in both T-cell acute lymphoblastic leukemia (T-ALL) (7) and chronic lymphocytic leukemia (CLL) (8), we analyzed the expression of HDAC3 in patient samples. We first observed that HDAC3 expression was significantly higher in T-ALL patients compared to different benign T-cell populations (Figure 6A). Additionally, HDAC3 transcript expression was elevated in pediatric T-ALL patients compared to normal bone marrow (BM) cells (Figure 6B), similarly to DDX5 (Supplementary Figure S10A) that was previously described as a regulator of the Notch pathway and was found to be frequently overexpressed in T-ALL (17,70). Of note, EP300 transcript levels were similar when comparing T-ALL and normal BM samples (Supplementary Figure S10B). Furthermore, we observed increased HDAC3 transcripts in CLL B-cell samples compared to normal B-cells, lymph node tissue and reactive tonsils, but not in comparison to several lymphoma samples with the only exception of mantle cell lymphoma samples (Figure 6C and D). We also note that DDX5 expression was increased (Supplementary Figure S10C) while EP300 expression is not changed (Supplementary Figure S10D) in primary CLL B-cell samples compared to normal B-cells. Of note, HDAC3 expression was comparable in CLL patient samples with mutated or unmutated IgV gene but elevated in comparison to several B-cells subpopulations from peripheral blood (PB, Supplementary Figure S10E-F). Altogether, these data suggest that HDAC3 expression is upregulated in T-ALL and CLL patient samples and thereby supports NOTCH1 stability.
Figure 6.
HDAC3 is highly expressed in human leukemic patient cells and apicidin treatment leads to reduced NICD1 protein levels in human leukemia cell lines. (A) HDAC3 expression is elevated in T-ALL patient samples compared to different sorted normal T-cell populations. Microarray data (GSE33469 and GSE33470) were analyzed to investigate the expression of HDAC3 in T-ALL patient samples and different sorted normal T-cell populations that represent different stages of T-cell development. Statistical analysis was performed with the Wilcoxon signed-rank test (*P < 0.05, **P < 0.01). (B) HDAC3 expression is increased in T-ALL pediatric samples compared to normal bone marrow (BM) cells. Microarray data (GSE26713) were analyzed to investigate the expression of HDAC3 in T-ALL pediatric samples and in normal bone marrow (BM) cells. Statistical analysis was performed with the Wilcoxon signed-rank test (**P < 0.01). (C) HDAC3 expression is increased in CLL B-cells compared to normal B-cells. Microarray data (GSE31048) were analyzed to investigate the expression of HDAC3 in CLL B-cells and normal B-cells. Statistical analysis was performed with the Wilcoxon signed-rank test (**P < 0.01). (D) HDAC3 expression is elevated in CLL compared to other proliferative tissues and mantle cell lymphoma. Microarray data (GSE32018) were analyzed to investigate the expression of HDAC3 in CLL, lymph node, reactive tonsils and other lymphomas. Statistical analysis was performed with the Wilcoxon signed-rank test (**P < 0.01, ***P < 0.001, [NS] not significant, unpaired Student's t-test). (E) Apicidin treatment leads to decreased NICD1 protein level in CUTLL1 cells. CUTLL1 T-ALL cells were treated for 24 h with different concentrations of apicidin (0.05 or 10 μg/ml) or DMSO as control and the whole cell extract (WCE) was analyzed by western blotting versus the endogenous cleaved NICD1 protein or TBP as loading control. (F) Apicidin treatment leads to decreased NICD1 protein level in REC-1 cells. REC-1 mantle cell lymphoma (MCL) cells were treated for 24 h with different concentrations of apicidin (0.05, 1 or 10 μg/ml) or DMSO as control and the whole cell extract (WCE) was analyzed by western blotting versus the endogenous cleaved NICD1 protein or TBP as loading control.
Finally, we validated our findings in human leukemic cell lines: T-ALL CUTLL1 cells and mantle cell lymphoma (MCL) REC-1 cells, both characterized by constitutive Notch activation (45,71). We observed that apicidin treatment leads to strong decrease in NICD1 protein levels in both CUTLL1 (Figures 6E and Supplementary Figure S11) and REC1 (Figures 6F and Supplementary Figure S12) cells.
DISCUSSION
When defining the role of HDAC3 in the regulation of the Notch-response, we discovered that HDAC3 plays an unexpected positive role at Notch target genes. HDAC3 deacetylates the NICD1 itself thereby profoundly influencing its turnover. An HDAC3-independent NICD1 mutant is less ubiquitinylated, more stable but biologically less active.
Regarding NICD deacetylation there are two previous reports (24,25). Palermo et al. (25) showed an acetylation-mediated decrease of NICD3 protein levels that is in line with our results. In the case of NICD3, deacetylation is mediated by HDAC1 and the inhibitor used was trichostatin A (TSA) (25). In our study, the NICD1-deacetylase is HDAC3 and the HDAC inhibitor is apicidin. In another study by Guarani et al. (24), it was shown that SIRT1 deacetylates, in endothelial cells, lysine residues of NICD1 promoting its destabilization. This is in contrast to our observations as HDAC3-mediated NICD1 deacetylation leads to the opposite effect that is an increase in NICD1 protein stability. Potentially, the differences between our results and this study (24) are due to the different target specificity of SIRT1 and HDAC3 but could also be attributed to the differences in Notch activity levels in endothelial cells versus T cells. In preT cells, Notch levels are particularly high while the Notch activity levels could be lower in endothelial cells. In line with the first explanation, we observed that the SIRT1 inhibitor NAM does not influence the expression of Notch target genes in preT cells. Recently, Marcel et al. (72) investigated the role of SIRT1 in Notch signaling further: they found that SIRT1 negatively regulates Notch signaling in regulatory T cells. In contrast, Sirt1 plays a positive role in Drosophila melanogaster by promoting deacetylation of Su(H), the Drosophila homolog of RBPJ (73). Thus, different deacetylases can play selective roles in regulating Notch signaling and this occurs in an even paralog-specific fashion. This scenario is even more complex as recently HDAC4 was described to have a negative function in Notch signaling (74) but whether it influences the NICD1 acetylation is unclear.
Interestingly, there are two recent independent reports analyzing the in vivo role of either Notch1 or Hdac3 in the context of lymphatic valve development in mice: Murtomaki et al. (75) showed that Notch signaling is essential in embryonic endothelial cells mediating lymphatic valve development. Another group analyzed lymphatic valve development in conditional Hdac3 knockout mice using endothelial Cre-mediated deletion (76). They observed that similarly to Notch1, lack of Hdac3 leads to defective lymphatic valve maturation. These studies are in line with our findings that HDAC3 acts in concert with NOTCH1.
HDACs are well-known for their repressive functions acting on chromatin. In agreement with this, HDAC3 loss of function results in increased histone acetylation, implicating HDAC3 as a transcriptional corepressor (77,78). However, in the context of Notch signaling, we show that inhibition of HDAC3 leads to decreased histone acetylation at the Notch-regulated enhancer sites. Our results are explained by the observation that HDAC3 positively influences the stability of the NICD1 protein; this in turn would recruit the histone acetyltransferase p300 at Notch-dependent enhancer sites (17) suggesting an indirect regulation of histone acetylation by HDAC3 in the context of the Notch signaling pathway. Our data are in line with previously published results showing the positive function of HDAC3 in the context of the inflammatory gene expression program in macrophages (42) and in IL-1 signaling (43). Thus, even if these data are in contrast to the current knowledge that suggests HDAC3 as a negative factor, they unveil a new function for HDAC3. This is in line with the emerging view that the vast majority of acetylated proteins are non-histone proteins (79).
Our data support a model where p300-mediated NICD1 acetylation is counteracted by HDAC3 in a wildtype background (Supplementary Figure S13A). As a result of the normal NICD1 turnover, Notch target genes are efficiently transcribed (Supplementary Figure S13A). Loss-of-function (LoF) of HDAC3 results in increased p300-mediated NICD1 acetylation, leading to reduced NICD1 stability and hence to reduced transcription of Notch target genes (Supplementary Figure S13B). In the case of the acetylation-deficient NICD1 mutant, the NICD1 protein is more stable but less active in transcription (Supplementary Figure S13C). This is in line with previous studies demonstrating that reduced turnover of the NICD1 coactivator complex results in reduced transcription of Notch target genes (22,80). Together, our results suggest that amplitude and duration of the Notch response can be significantly altered by HDAC3 inhibition, revealing HDAC3 as a candidate drug target for NOTCH1-mediated diseases such as T-ALL and CLL in which we observed elevated HDAC3 expression.
DATA AVAILABILITY
RNA-Seq and ChIP-Seq data have been deposited at GEO under the accession number GSE106361 and GSE119797, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to P. Käse, T. Schmidt-Wöll, S. Schirmer and R. Rittelmann for excellent technical assistance. We want to thank Dr C. Seiser (Medical University of Vienna, Center for Anatomy and Cell Biology, Division of Cell and Developmental Biology, Austria) and Dr M.L. Schmitz (University of Giessen, Germany) for providing us with plasmids. We want to thank A. Wachtendorf for excellent FACS sorting. We are grateful to Dr F. Radtke (EPFL, Lausanne, Switzerland) and Dr A. Ferrando (University of Columbia, New York, USA) for providing us with cell lines. The authors wish to acknowledge Centro de Análisis Genómico (CNAG-CRG), Spain, for sequencing the ChIP samples.
Authors contributions: F.F., B.D.G., P.K and F.O. performed experiments and analyzed data. M.B, T.Z. and B.D.G. performed the bioinformatic analysis. V.M.-P. and D.M. performed analysis. A.N. and T.S. performed deep-sequencing of the RNA samples. J.M.-S. and M.K. provided reagents. S.J. and P.K. performed the in vivo experiments. F.F., B.D.G. and T.B. designed experiments and wrote the manuscript with contributions from other authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG, German Research Foundation - TRR81/3 – project number A12), Heisenberg program (BO 1639/5-1) and Excellence Cluster for Cardio Pulmonary System (ECCPS) in Giessen to T.B. DFG (SFB 1074/A03, OS 287/4-1) and BMBF (Federal Ministry of Education and Research, research nucleus SyStAR) to F.O. M.K. was supported by grants from the Deutsche Forschungsgemeinschaft [Kr1143/7-3, KR1143/9-1 (KLIFO309), TRR81/2 (B02) and SFB1021 (C02)]. D.M. was supported by the DFG (SFB 1074/B02) and S.J. was supported by the DFG (JU 2859/2-1). B.D.G. is supported by a Research Grant of the University Medical Center Giessen and Marburg (UKGM). Funding for open access charge: DFG collaborative research (TRR81).
Conflict of interest statement. None declared.
REFERENCES
- 1. Radtke F., MacDonald H.R., Tacchini-Cottier F.. Regulation of innate and adaptive immunity by Notch. Nat. Rev. Immunol. 2013; 13:427–437. [DOI] [PubMed] [Google Scholar]
- 2. Rothenberg E.V. Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 2014; 32:283–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koch U., Radtke F.. Notch and cancer: a double-edged sword. Cell Mol. Life Sci. 2007; 64:2746–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aster J.C., Pear W.S., Blacklow S.C.. The varied roles of notch in cancer. Annu. Rev. Pathol. 2017; 12:245–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roy M., Pear W.S., Aster J.C.. The multifaceted role of Notch in cancer. Curr. Opin. Genet. Dev. 2007; 17:52–59. [DOI] [PubMed] [Google Scholar]
- 6. Giaimo B.D., Borggrefe T.. Introduction to molecular mechanisms in notch signal transduction and disease pathogenesis. Adv. Exp. Med. Biol. 2018; 1066:3–30. [DOI] [PubMed] [Google Scholar]
- 7. Weng A.P., Ferrando A.A., Lee W., Morris J.P.t., Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C.. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004; 306:269–271. [DOI] [PubMed] [Google Scholar]
- 8. Puente X.S., Pinyol M., Quesada V., Conde L., Ordonez G.R., Villamor N., Escaramis G., Jares P., Bea S., Gonzalez-Diaz M. et al.. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011; 475:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thompson B.J., Buonamici S., Sulis M.L., Palomero T., Vilimas T., Basso G., Ferrando A., Aifantis I.. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 2007; 204:1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O’Neil J., Grim J., Strack P., Rao S., Tibbitts D., Winter C., Hardwick J., Welcker M., Meijerink J.P., Pieters R. et al.. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 2007; 204:1813–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Borggrefe T., Oswald F.. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol. Life Sci. 2009; 66:1631–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giaimo B.D., Oswald F., Borggrefe T.. Dynamic chromatin regulation at Notch target genes. Transcription. 2017; 8:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bray S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016; 17:722–735. [DOI] [PubMed] [Google Scholar]
- 14. Kovall R.A., Gebelein B., Sprinzak D., Kopan R.. The canonical notch signaling pathway: structural and biochemical insights into shape, Sugar, and Force. Dev. Cell. 2017; 41:228–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan Z., VanderWielen B.D., Giaimo B.D., Pan L., Collins C.E., Turkiewicz A., Hein K., Oswald F., Borggrefe T., Kovall R.A.. Structural and functional studies of the RBPJ-SHARP Complex reveal a conserved corepressor binding site. Cell Rep. 2019; 26:845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liefke R., Oswald F., Alvarado C., Ferres-Marco D., Mittler G., Rodriguez P., Dominguez M., Borggrefe T.. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010; 24:590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jung C., Mittler G., Oswald F., Borggrefe T.. RNA helicase Ddx5 and the noncoding RNA SRA act as coactivators in the Notch signaling pathway. Biochim. Biophys. Acta. 2013; 1833:1180–1189. [DOI] [PubMed] [Google Scholar]
- 18. Oswald F., Rodriguez P., Giaimo B.D., Antonello Z.A., Mira L., Mittler G., Thiel V.N., Collins K.J., Tabaja N., Cizelsky W. et al.. A phospho-dependent mechanism involving NCoR and KMT2D controls a permissive chromatin state at Notch target genes. Nucleic Acids Res. 2016; 44:4703–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu T., Park S.S., Giaimo B.D., Hall D., Ferrante F., Ho D.M., Hori K., Anhezini L., Ertl I., Bartkuhn M. et al.. RBPJ/CBF1 interacts with L3MBTL3/MBT1 to promote repression of Notch signaling via histone demethylase KDM1A/LSD1. EMBO J. 2017; 36:3232–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giaimo B.D., Ferrante F., Vallejo D.M., Hein K., Gutierrez-Perez I., Nist A., Stiewe T., Mittler G., Herold S., Zimmermann T. et al.. Histone variant H2A.Z deposition and acetylation directs the canonical Notch signaling response. Nucleic Acids Res. 2018; 46:8197–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fryer C.J., White J.B., Jones K.A.. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 2004; 16:509–520. [DOI] [PubMed] [Google Scholar]
- 22. Hein K., Mittler G., Cizelsky W., Kuhl M., Ferrante F., Liefke R., Berger I.M., Just S., Strang J.E., Kestler H.A. et al.. Site-specific methylation of Notch1 controls the amplitude and duration of the Notch1 response. Sci Signal. 2015; 8:ra30. [DOI] [PubMed] [Google Scholar]
- 23. Popko-Scibor A.E., Lindberg M.J., Hansson M.L., Holmlund T., Wallberg A.E.. Ubiquitination of Notch1 is regulated by MAML1-mediated p300 acetylation of Notch1. Biochem. Biophys. Res. Commun. 2011; 416:300–306. [DOI] [PubMed] [Google Scholar]
- 24. Guarani V., Deflorian G., Franco C.A., Kruger M., Phng L.K., Bentley K., Toussaint L., Dequiedt F., Mostoslavsky R., Schmidt M.H. et al.. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011; 473:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palermo R., Checquolo S., Giovenco A., Grazioli P., Kumar V., Campese A.F., Giorgi A., Napolitano M., Canettieri G., Ferrara G. et al.. Acetylation controls Notch3 stability and function in T-cell leukemia. Oncogene. 2012; 31:3807–3817. [DOI] [PubMed] [Google Scholar]
- 26. Borggrefe T., Lauth M., Zwijsen A., Huylebroeck D., Oswald F., Giaimo B.D.. The Notch intracellular domain integrates signals from Wnt, Hedgehog, TGFbeta/BMP and hypoxia pathways. Biochim. Biophys. Acta. 2016; 1863:303–313. [DOI] [PubMed] [Google Scholar]
- 27. Li Y., Seto E.. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb. Perspect. Med. 2016; 6:a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haberland M., Montgomery R.L., Olson E.N.. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009; 10:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y., Iratni R., Erdjument-Bromage H., Tempst P., Reinberg D.. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997; 89:357–364. [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D.. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999; 13:1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Humphrey G.W., Wang Y., Russanova V.R., Hirai T., Qin J., Nakatani Y., Howard B.H.. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 2001; 276:6817–6824. [DOI] [PubMed] [Google Scholar]
- 32. Wen Y.D., Perissi V., Staszewski L.M., Yang W.M., Krones A., Glass C.K., Rosenfeld M.G., Seto E.. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:7202–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li J., Wang J., Wang J., Nawaz Z., Liu J.M., Qin J., Wong J.. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000; 19:4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoon H.G., Chan D.W., Huang Z.Q., Li J., Fondell J.D., Qin J., Wong J.. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003; 22:1336–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoon H.G., Chan D.W., Reynolds A.B., Qin J., Wong J.. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell. 2003; 12:723–734. [DOI] [PubMed] [Google Scholar]
- 36. Guenther M.G., Lane W.S., Fischle W., Verdin E., Lazar M.A., Shiekhattar R.. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000; 14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang J., Kalkum M., Chait B.T., Roeder R.G.. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell. 2002; 9:611–623. [DOI] [PubMed] [Google Scholar]
- 38. Kao H.Y., Ordentlich P., Koyano-Nakagawa N., Tang Z., Downes M., Kintner C.R., Evans R.M., Kadesch T.. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998; 12:2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oswald F., Kostezka U., Astrahantseff K., Bourteele S., Dillinger K., Zechner U., Ludwig L., Wilda M., Hameister H., Knochel W. et al.. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 2002; 21:5417–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mikami S., Kanaba T., Ito Y., Mishima M.. NMR assignments of SPOC domain of the human transcriptional corepressor SHARP in complex with a C-terminal SMRT peptide. Biomol. NMR Assign. 2013; 7:267–270. [DOI] [PubMed] [Google Scholar]
- 41. Mikami S., Kanaba T., Takizawa N., Kobayashi A., Maesaki R., Fujiwara T., Ito Y., Mishima M.. Structural insights into the recruitment of SMRT by the corepressor SHARP under phosphorylative regulation. Structure. 2014; 22:35–46. [DOI] [PubMed] [Google Scholar]
- 42. Chen X., Barozzi I., Termanini A., Prosperini E., Recchiuti A., Dalli J., Mietton F., Matteoli G., Hiebert S., Natoli G.. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2865–E2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ziesche E., Kettner-Buhrow D., Weber A., Wittwer T., Jurida L., Soelch J., Muller H., Newel D., Kronich P., Schneider H. et al.. The coactivator role of histone deacetylase 3 in IL-1-signaling involves deacetylation of p65 NF-kappaB. Nucleic Acids Res. 2013; 41:90–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang L., He X., Liu L., Jiang M., Zhao C., Wang H., He D., Zheng T., Zhou X., Hassan A. et al.. Hdac3 interaction with p300 histone acetyltransferase regulates the oligodendrocyte and astrocyte lineage fate switch. Dev. Cell. 2016; 36:316–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Palomero T., Barnes K.C., Real P.J., Glade Bender J.L., Sulis M.L., Murty V.V., Colovai A.I., Balbin M., Ferrando A.A.. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006; 20:1279–1287. [DOI] [PubMed] [Google Scholar]
- 46. Oswald F., Tauber B., Dobner T., Bourteele S., Kostezka U., Adler G., Liptay S., Schmid R.M.. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell Biol. 2001; 21:7761–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang W.M., Tsai S.C., Wen Y.D., Fejer G., Seto E.. Functional domains of histone deacetylase-3. J. Biol. Chem. 2002; 277:9447–9454. [DOI] [PubMed] [Google Scholar]
- 48. de la Vega L., Grishina I., Moreno R., Kruger M., Braun T., Schmitz M.L.. A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol. Cell. 2012; 46:472–483. [DOI] [PubMed] [Google Scholar]
- 49. Giaimo B.D., Ferrante F., Borggrefe T.. Chromatin immunoprecipitation (ChIP) in mouse T-cell lines. J. Vis. Exp. 2017; doi:10.3791/55907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dieguez-Hurtado R., Kato K., Giaimo B.D., Nieminen-Kelha M., Arf H., Ferrante F., Bartkuhn M., Zimmermann T., Bixel M.G., Eilken H.M. et al.. Loss of the transcription factor RBPJ induces disease-promoting properties in brain pericytes. Nat. Commun. 2019; 10:2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics. 2010; doi:10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. et al.. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004; 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J.. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bengtsson H., Simpson K., Bullard J., Hansen K.. aroma.affymetrix: A generic framework in R for analyzingsmall to very large Affymetrix data sets in bounded memory. 2008; https://statistics.berkeley.edu/sites/default/files/tech-reports/745.pdf.
- 57. Feng J., Liu T., Qin B., Zhang Y., Liu X.S.. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012; 7:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Feng X., Grossman R., Stein L.. PeakRanger: a cloud-enabled peak caller for ChIP-seq data. BMC Bioinformatics. 2011; 12:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Homminga I., Pieters R., Langerak A.W., de Rooi J.J., Stubbs A., Verstegen M., Vuerhard M., Buijs-Gladdines J., Kooi C., Klous P. et al.. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011; 19:484–497. [DOI] [PubMed] [Google Scholar]
- 61. Wang L., Shalek A.K., Lawrence M., Ding R., Gaublomme J.T., Pochet N., Stojanov P., Sougnez C., Shukla S.A., Stevenson K.E. et al.. Somatic mutation as a mechanism of Wnt/beta-catenin pathway activation in CLL. Blood. 2014; 124:1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gomez-Abad C., Pisonero H., Blanco-Aparicio C., Roncador G., Gonzalez-Menchen A., Martinez-Climent J.A., Mata E., Rodriguez M.E., Munoz-Gonzalez G., Sanchez-Beato M. et al.. PIM2 inhibition as a rational therapeutic approach in B-cell lymphoma. Blood. 2011; 118:5517–5527. [DOI] [PubMed] [Google Scholar]
- 63. Seifert M., Sellmann L., Bloehdorn J., Wein F., Stilgenbauer S., Durig J., Kuppers R.. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J. Exp. Med. 2012; 209:2183–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Van Vlierberghe P., Ambesi-Impiombato A., Perez-Garcia A., Haydu J.E., Rigo I., Hadler M., Tosello V., Della Gatta G., Paietta E., Racevskis J. et al.. ETV6 mutations in early immature human T cell leukemias. J. Exp. Med. 2011; 208:2571–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Davis S., Meltzer P.S.. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007; 23:1846–1847. [DOI] [PubMed] [Google Scholar]
- 66. Just S., Berger I.M., Meder B., Backs J., Keller A., Marquart S., Frese K., Patzel E., Rauch G.J., Katus H.A. et al.. Protein kinase D2 controls cardiac valve formation in zebrafish by regulating histone deacetylase 5 activity. Circulation. 2011; 124:324–334. [DOI] [PubMed] [Google Scholar]
- 67. Chan S.K.K., Cerda-Moya G., Stojnic R., Millen K., Fischer B., Fexova S., Skalska L., Gomez-Lamarca M., Pillidge Z., Russell S. et al.. Role of co-repressor genomic landscapes in shaping the Notch response. PLos Genet. 2017; 13:e1007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Weng A.P., Nam Y., Wolfe M.S., Pear W.S., Griffin J.D., Blacklow S.C., Aster J.C.. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell Biol. 2003; 23:655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fang T.C., Yashiro-Ohtani Y., Del Bianco C., Knoblock D.M., Blacklow S.C., Pear W.S.. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007; 27:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lin S., Tian L., Shen H., Gu Y., Li J.L., Chen Z., Sun X., You M.J., Wu L.. DDX5 is a positive regulator of oncogenic NOTCH1 signaling in T cell acute lymphoblastic leukemia. Oncogene. 2013; 32:4845–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ryan R.J.H., Petrovic J., Rausch D.M., Zhou Y., Lareau C.A., Kluk M.J., Christie A.L., Lee W.Y., Tarjan D.R., Guo B. et al.. A B cell regulome links notch to downstream oncogenic pathways in small B cell lymphomas. Cell Rep. 2017; 21:784–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Marcel N., Perumalsamy L.R., Shukla S.K., Sarin A.. The lysine deacetylase Sirtuin 1 modulates the localization and function of the Notch1 receptor in regulatory T cells. Sci Signal. 2017; 10:eaah4679. [DOI] [PubMed] [Google Scholar]
- 73. Horvath M., Mihajlovic Z., Slaninova V., Perez-Gomez R., Moshkin Y., Krejci A.. The silent information regulator 1 (Sirt1) is a positive regulator of the Notch pathway in Drosophila. Biochem. J. 2016; 473:4129–4143. [DOI] [PubMed] [Google Scholar]
- 74. Antila C.J.M., Rraklli V., Blomster H.A., Dahlstrom K.M., Salminen T.A., Holmberg J., Sistonen L., Sahlgren C.. Sumoylation of Notch1 represses its target gene expression during cell stress. Cell Death Differ. 2018; 25:600–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Murtomaki A., Uh M.K., Kitajewski C., Zhao J., Nagasaki T., Shawber C.J., Kitajewski J.. Notch signaling functions in lymphatic valve formation. Development. 2014; 141:2446–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Janardhan H.P., Milstone Z.J., Shin M., Lawson N.D., Keaney J.F. Jr, Trivedi C.M.. Hdac3 regulates lymphovenous and lymphatic valve formation. J. Clin. Invest. 2017; 127:4193–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jiao F., Hu H., Yuan C., Jin Z., Guo Z., Wang L., Wang L.. Histone deacetylase 3 promotes pancreatic cancer cell proliferation, invasion and increases drug-resistance through histone modification of P27, P53 and Bax. Int. J. Oncol. 2014; 45:1523–1530. [DOI] [PubMed] [Google Scholar]
- 78. Knutson S.K., Chyla B.J., Amann J.M., Bhaskara S., Huppert S.S., Hiebert S.W.. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008; 27:1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schmitz M.L., de la Vega L.. New insights into the role of histone deacetylases as coactivators of inflammatory gene expression. Antioxid. Redox. Signal. 2015; 23:85–98. [DOI] [PubMed] [Google Scholar]
- 80. Oberg C., Li J., Pauley A., Wolf E., Gurney M., Lendahl U.. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 2001; 276:35847–35853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq and ChIP-Seq data have been deposited at GEO under the accession number GSE106361 and GSE119797, respectively.