Abstract
Guanine-rich regions of the human genome can adopt non-canonical secondary structures. Their role in regulating gene expression has turned them into promising targets for therapeutic intervention. Ligands based on polyaromatic moieties are especially suitable for targeting G-quadruplexes utilizing their size complementarity to interact with the large exposed surface area of four guanine bases. A predictable way of (de)stabilizing specific G-quadruplex structures through efficient base stacking of polyaromatic functional groups could become a valuable tool in our therapeutic arsenal. We have investigated the effect of pyrene-modified uridine nucleotides incorporated at several positions of the thrombin binding aptamer (TBA) as a model system. Characterization using spectroscopic and biophysical methods provided important insights into modes of interaction between pyrene groups and the G-quadruplex core as well as (de)stabilization by enthalpic and entropic contributions. NMR data demonstrated that incorporation of pyrene group into G-rich oligonucleotide such as TBA may result in significant changes in 3D structure such as formation of novel dimeric topology. Site specific structural changes induced by stacking of the pyrene moiety on nearby nucleobases corelate with distinct thrombin binding affinities and increased resistance against nuclease degradation.
INTRODUCTION
Formation of non-B-DNA four-stranded G-quadruplex structures by guanine-rich oligo- and poly-nucleotides has been known for >50 years (1). Four guanine nucleotides assemble into a plane by means of Hoogsteen hydrogen bonds to form a G-quartet. Subsequently, two or more G-quartets stack to form a G-quadruplex (2–4). G-quartet forming guanine stretches are interconnected by loops, which can be of edge, diagonal or propeller type, creating a tremendous ensemble of possible topologies (5,6). The importance of these polymorphic structures is becoming more evident as new biological roles and involvements in various diseases are discovered (7–9). Recent reports suggest that depending on selection criteria between 65 000 and 180 000 G-quadruplex forming sequences can be found in the human genome (10). As G-quadruplexes have been recognized as promising targets for therapeutic intervention, several approaches have been assessed to selectively bind to various folding topologies and structures as well as affect their thermodynamic stabilities.
Polyaromatic moieties are capable of stabilizing (non-canonical) DNA structures through efficient base stacking, groove binding or intercalation. G-quadruplexes are especially suitable for targeting by planar polyaromatic moieties due to the large exposed surface area of G-quartets (11,12). Polyaromatic groups can stabilize G-quadruplexes as non-covalent ligands, which readily interact and bind to DNA (13–16). Alternatively, they can be covalently linked to DNA at desired position(s) in an oligonucleotide sequence. Fluorescence properties and π–π stacking ability of pyrene makes it a desirable building block in functional oligonucleotides (17–21). Applications of pyrene conjugated oligonucleotides include discrimination of single nucleotide polymorphisms (22–24), stabilization of double-stranded DNA (25–27), detection of RNA (28–30), etc. Changes in fluorescence of pyrene or excimer formation can be used to discriminate between single stranded DNA and G-quadruplex formation (31,32). Pyrene functionalized G-rich oligonucleotides can also serve as K+ ion sensors (33,34). Several studies utilized so called twisted intercalating nucleic acids (TINAs) to achieve stabilization of certain DNA secondary structures through intercalation or stacking (35–37). In short, TINAs are nucleic acid monomers containing a pyrene group, but lacking nucleobase and sugar moieties. TINA monomers can also be used to stabilize G-quadruplex structures (38), which can result in anti-tumour effects (39,40).
The ability to control stability of G-quadruplex structures and thus modulate related biological outcomes offers great potential applications. Stabilization of telomeric G-quadruplexes has been shown to inhibit telomerase activity in cancer cells, while stabilization of promoter G-quadruplexes can potentially be exploited for regulating gene expression (39–44). We have recently shown that a G-quadruplex structure adopted by the vascular endothelial growth factor (VEGF) promoter sequence is disrupted with the introduction of an oxidative lesion. However, the G-quadruplex fold can be recovered by adding a short pyrene conjugated lesion-free G-rich oligonucleotide (45). Achieving the desired effect by covalently attaching polyaromatic groups to oligonucleotides is often highly dependent on the position of modification. There are only a limited number of studies systematically evaluating structure and stability of DNA with incorporated polyaromatic moieties. In the current study we have used the G-quadruplex forming thrombin binding aptamer (TBA) as a well-studied and robust model system to delineate modes of its interaction with polyaromatic moieties as well as (de)stabilizing energetic contributions. TBA exhibits a 15 nucleotide long palindromic DNA sequence, 5′-GGTTGGTGTGGTTGG-3′, that folds into an antiparallel G-quadruplex with a chair-like topology consisting of two G-quartets (Figure 1). The core of the TBA G-quadruplex is flanked by two TT and one TGT loop. T4 and T13 form a non-canonical interloop T-T base pair (46,47). TBA has a high binding affinity towards thrombin protease and acts as its inhibitor and a blood anti-coagulant (48). Due to its potential for biomedical applications, the TBA G-quadruplex has been a subject of many studies exploring effects of different chemical modifications on its stability and affinity towards thrombin (49,50). G-quartet forming guanine residues of TBA can be replaced with different analogues, but these modifications can easily alter G-quadruplex structure. As a result, primary sites of modifications have been loop residues that include changes in loop lengths (51) and sequence (52), LNA (53) and UNA (54) modifications, different internucleotide linkages (55) or inversions in oligonucleotide polarity (56,57) and cyclization (58).
Figure 1.
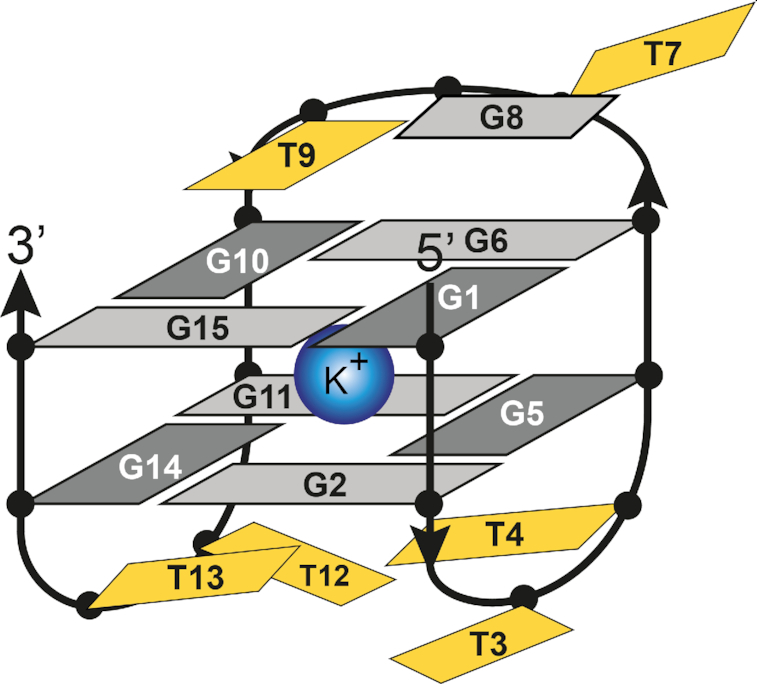
Schematic representation of the TBA G-quadruplex structure. Dark and light grey nucleotides correspond to guanines in syn and anti conformations, respectively. Thymines are shown in yellow. T4 and T13 form uncanonical T–T base pair.
In the current study we have used a commercially available pyrene-modified uridine (Upy) building block (Figure 2) to individually replace all thymine positions in the loops of TBA. We show that only oligonucleotides where the 3′-end loop nucleotide has been substituted (i.e. TUpy or TGUpy) are capable to fold into a single stable G-quadruplex structure. In these cases, an increase in melting temperature (Tm) by 4.8 up to 5.6°C is achieved, which is predominantly the result of favorable stacking interactions between pyrene groups and adjacent G-quartet's guanines. Surprisingly, a substitution in the TGT loop induces formation of two distinct G-quadruplex units in a novel dimeric structure. Position of Upy and respective structural features of TBA analogues are related to their binding to thrombin. Pyrene-modified TBA analogues exhibit increased resistance against nuclease degradation under human serum conditions in comparison to TBA.
Figure 2.
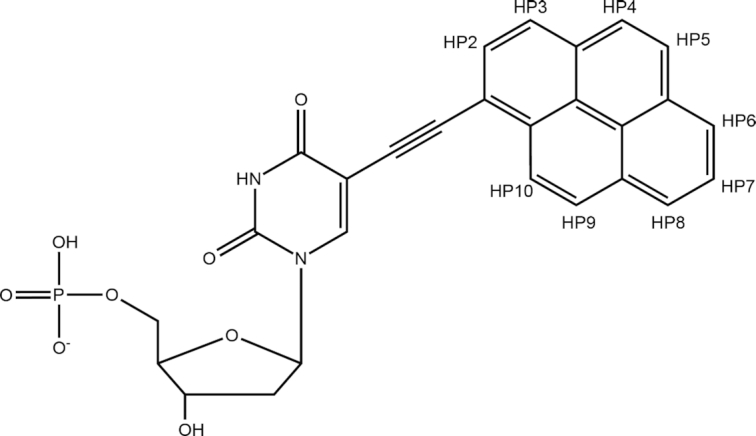
Structural formula of Upy (5-(pyren-1-yl-ethynyl)-dUMP) with the numbering of positions in the pyrene moiety.
MATERIALS AND METHODS
Sample preparation
All DNA oligonucleotides were synthesised by phosphoramidite solid support method on a H-8 DNA synthesizer (K&A Laborgeraete). Pyrene-dU-CE phosphoramidite supplied by Glen Research was used for incorporating thymine nucleotides modified with pyrene group (Upy) into oligonucleotides. Oligonucleotides were removed from solid support with a 2 h incubation in 28% ammonium hydroxide aqueous solution at room temperature. Cyanoethyl protective groups were removed from phosphate groups by a subsequent overnight incubation at 55°C. The solvent was evaporated and samples redissolved in ultrapure water. Oligonucleotide samples were dissolved in MQ water (18.2 MΩ) and purified with FPLC (ÄktaPurifier 10) on a size exclusion HiPrep 26/10 column with MQ water also used for the mobile phase with a flow of 10 ml/min. Samples were injected into the FPLC after initial 6 minutes of column conditioning with the MQ mobile phase, after which the method ran another 16 minutes. FPLC separation was followed by measuring absorbance at 260 nm. Mobile phase was evaporated and oligonucleotides dissolved in NMR solution containing 10% 2H2O and 20, 50 or 100 mM KCl at pH 5.5 and concentrations ranging from 1.0 to 1.5 mM per strand, if not stated otherwise. NMR titration of 100 μM TBA sample in 50 mM KCl solution with sodium salt of 1-pyrenesulfonic acid was performed at 0.00, 0.25, 0.50, 1.00 and 2.00 molar equivalents.
Nuclear magnetic resonance (NMR) spectroscopy
NMR spectra were recorded on Agilent's VNMRS 600 and 800 MHz spectrometers at 25°C. DPFGSE water suppression pulse sequence was used as a part of collection of 1D and 2D NMR spectra. 2D NOESY spectra were acquired with mixing time between 80 and 250 ms and 2D TOCSY spectra with mixing time of 65 ms. 1D 15N- and 13C-edited HSQC spectra were acquired on partially residue specifically 15N and 13C enriched DNA samples (59) (Varian's rna_gNhsqc pulse sequence was used). Raw spectral data was processed with VNMRJ and NMRpipe (60) software packages. Peak assignment was carried out with MestReNova and Sparky (61) programs.
Circular dichroism (CD) spectroscopy
CD spectra were acquired on Chirascan CD spectrometer at 25°C using 0.1 and 2.0 mm light path quartz cuvettes. All samples for CD, except the TBA9D one, were prepared by dilution of the NMR samples to final 50 μM DNA concentration in 50 mM KCl solution. CD spectrum of TBA9D was recorded on a smaller volume of undiluted NMR sample with 100 mM KCl. Spectra were acquired in the wavelength range from 220 to 420 nm.
Thermodynamic analysis
Ultraviolet (UV) absorbance was measured using a Shimadzu UV-1700 and UV-1800 spectrophotometer equipped with a temperature controller. Before measurements, DNA samples were heated to 90°C, cooled to 0°C at a rate of −0.5°C/min and incubated at 0°C for 30 min. Melting curves for TBA, TBA4, TBA9 and TBA13 G-quadruplexes were measured at 295 nm in 50 mM KCl solution at various concentrations ranging from 5 to 500 μM concentrations per strand. Samples were heated and cooled at a rate of 0.5°C/min. ΔH°, TΔS° and ΔG° of TBA, TBA4, TBA9M and TBA13 G-quadruplex structures were calculated from obtained absorbance data at 5, 20 and 40 μM concentrations using the van’t Hoff equation. Melting temperatures of G-quadruplexes (Tm) were obtained with the calculation of the first derivative of the melting curves measured at 20 μM concentration per strand.
PAGE
Before the gel experiments, DNA samples were heated to 80°C, cooled at a rate of –2°C/min and incubated at 4, 25, 37 or 70°C for 10 min. Native gel electrophoresis was carried out on 20% non-denaturing polyacrylamide gels in a buffer containing 80 mM KCl. Loading buffer (40% glycerol and 1% blue dextran) was mixed with 550 μM DNA solutions just before loading on the gel. Gels were run at 120 V for 120–330 min at 4, 25, 37 and 70°C. After, the gels were stained with SYBR® Gold (PerkinElmer Life Sciences) and imaged using a fluorescent imager (FUJIFILM, FLA-5100).
Assays for nuclease resistance
The chemical stability of pyrene-modified TBA was analyzed in the presence of 80% (v/v) human serum type AB (Lonza), which contains various nucleases such as endonucleases and exonucleases. The degradation of 5 μM pyrene-modified TBA by nucleases present in the serum was examined after various incubation times at 37°C. Reactions were quenched at chosen times by the addition of a 5-fold excess volume of stop solution (80 wt% formamide, 10 mM Na2EDTA and 0.1% blue dextran) and incubated at 95°C for 15 min. Before the denaturing PAGE, the precipitate of proteins in the serum was removed by centrifugation. Gels were run at 250 V for 100 min at 25°C and stained with SYBR® Gold (PerkinElmer Life Sciences) and imaged using a fluorescent imager (Fujifilm, FLA-5100).
Binding assay of pyrene-modified TBAs to thrombin by using quartz-crystal microbalance (QCM)
Measurements of the binding of TBAs to thrombin were performed using an AFFINIX Q4 (Initium, Japan) as reported previously with slight modification (45). Human thrombin was purchased from Haematologic Technologies, USA. The solution of thrombin was dialyzed against 50 mM HEPES–NaOH (pH 7.5) containing 10 mM NaCl at 4°C and reacted with EZ-Link™ Sulfo-NHS-Biotin up to 1:10 molar ratio (Thermo Fisher Scientific, USA) for 1 h. The reaction was quenched with the addition of 1/100 volume of 1 M Tris–HCl buffer (pH 7.5), then dialyzed against 50 mM HEPES–NaOH (pH 7.5) containing 10 mM NaCl at 4°C and next purified on a PD-10 column (GE Healthcare, USA). The biotinylated thrombin was immobilized in the NeutrAvidin-coated QCM cell. After immobilization, the solution in the cell was replaced with the 25 mM KCl and 25 mM K phosphate buffer. DNA solutions (100 μM) were annealed in the same buffer and injected into the QCM cell, and frequency changes (ΔF) were recorded at 25°C. Dissociation constant (KD), ΔF values were plotted against the concentration of the injected oligonucleotides. Each series of plots was fitted to the equation of the Langmuir adsorption model –ΔF = [DNA]*(–ΔFmax)/([DNA] + KD), where ΔFmax is the maximal change of frequency at an infinite concentration of DNA oligonucleotide (62).
Fluorescence spectroscopy
Fluorescence properties of 150 μM TBA4, TBA9M and TBA13 G-quadruplexes in a 50 mM KCl solution were measured on QuantaMaster (PTI) spectrofluorometer at 25°C using a quartz cuvette with 1.0 cm path length and excitation wavelength of 330 nm.
RESULTS
Effects of pyrene incorporation are positionally dependent
The use of nucleotides with covalently attached polyaromatic moieties, which stack on the nucleobases of outer G-quartets is a viable strategy for stabilizing G-quadruplex structures. Careful positioning of modified nucleotides is crucial for optimum results. Our study encompasses a series of oligonucleotides based on the TBA sequence, where all thymine residues were individually substituted with Upy (Table 1). Interesting observations were also made with double substituted constructs and TBA0 and TBA16, where Upy nucleotide was added to 5′- and 3′-ends of the parent TBA sequence, respectively.
Table 1.
Names and sequences of oligonucleotides used in this study
| Oligonucleotide | Sequence | ||||||
|---|---|---|---|---|---|---|---|
| TBA | GG | TT | GG | TGT | GG | TT | GG |
| TBA0 | UpyGG | TT | GG | TGT | GG | TT | GG |
| TBA3 | GG | UpyT | GG | TGT | GG | TT | GG |
| TBA4 | GG | TUpy | GG | TGT | GG | TT | GG |
| TBA7 | GG | TT | GG | UpyGT | GG | TT | GG |
| TBA9 | GG | TT | GG | TGUpy | GG | TT | GG |
| TBA12 | GG | TT | GG | TGT | GG | UpyT | GG |
| TBA13 | GG | TT | GG | TGT | GG | TUpy | GG |
| TBA16 | GG | TT | GG | TGT | GG | TT | GGUpy |
| TBA4/9 | GG | TUpy | GG | TGUpy | GG | TT | GG |
| TBA4/13 | GG | TUpy | GG | TGT | GG | TUpy | GG |
| TBA9/13 | GG | TT | GG | TGUpy | GG | TUpy | GG |
Initially TBA and its analogues were dissolved in an H2O/2H2O mixture containing 50 mM KCl, and their ability to fold into a G-quadruplex was assessed with 1D 1H NMR experiments. Spectra of TBA0, TBA3, TBA7, TBA12 and TBA16 contained low-intensity resonances on a broad background, which suggested aggregation and/or multiple structures present in solution (Supplementary Figure S1). On the other hand, NMR spectra of TBA4, TBA9 and TBA13 exhibit favorable spectral properties (Figure 3). 1H NMR spectra of TBA4 and TBA13 contain eight sharp guanine imino resonances in the range from δ 11.5 to 12.5 ppm with chemical shift dispersion comparable to TBA. This is indicative of formation of a single G-quadruplex species with two G-quartet planes. Additionally, spectrum of TBA4 contains lower intensity signals in the range from δ 10.0 to 11.5 ppm, which most likely correspond to thymine imino or guanine amino protons.
Figure 3.
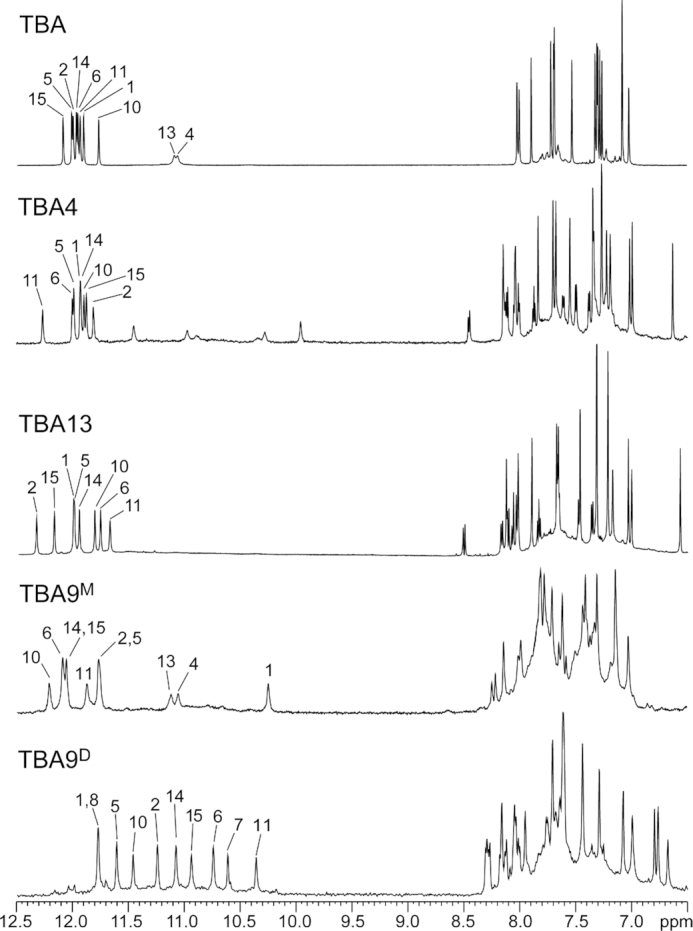
Imino and aromatic regions of 1H NMR spectra of TBA and its analogues acquired at 25°C. Samples of TBA, TBA4 and TBA13 dissolved in 50 mM KCl solution, while samples TBA9M and TBA9D were dissolved in 20 and 100 mM KCl solution, respectively. Signals of imino protons involved in hydrogen bonds are assigned and marked above individual signal.
UV absorbance, fluorescence and CD spectra of TBA, TBA4, TBA9M and TBA13 G-quadruplex samples were acquired also in the wavelength regions that are characteristic for the pyrene moiety (Supplementary Figure S2). TBA did not exhibit any fluorescence properties and pyrene specific absorbance above 320 nm. Minor spectral changes for pyrene-modified TBA analogues suggest different positioning of pyrene moiety in the G-quadruplex structures. Spectral signatures of TBA4 and TBA13 are expectedly identical due to similar environment of their intrinsic pyrene groups. In comparison, TBA9M, exhibits slight blueshift in absorbance and fluorescence spectra as well as higher fluorescence intensity. No distinguishable differences between TBA4, TBA9M and TBA13 are observed in the region of CD spectra corresponding to absorbance of pyrene moieties above 320 nm (Supplementary Figure S2C).
Since TBA forms a stable G-quadruplex structure also in the presence of Na+ cations, we tested the folding ability of Upy analogues in the presence of 50 mM NaCl. However, no imino resonances could be observed in NMR spectra (Supplementary Figure S3), which shows that Na+ cations cannot induce G-quadruplex folding of Upy analogues of TBA. For comparison and control for potential intermolecular interactions of the pyrene moiety with TBA, we performed titration with sodium salt of 1-pyrenesulfonic acid up to 2.0 molar equivalents with respect to TBA. Nine signals of aromatic protons of 1-pyrenesulfonic group can be observed in 1D 1H NMR spectra (Supplementary Figure S4). Surprisingly, there are no observable changes in NMR signals of TBA G-quadruplex confirming the absence of interactions with 1-pyrenesulfonic group.
The pyrene moiety is accommodated at the 3′ ends of TT loops
In order to investigate if substitutions with Upy influence the fold of TBA analogues circular dichroism (CD) spectra were acquired. CD spectra of TBA4 and TBA13 are in agreement with TBA and exhibit maxima at around 245 and 290 nm and a minimum at around 265 nm (Figure 4, Supplementary Figure S10C), which are characteristic for antiparallel G-quadruplex folds (63,64).
Figure 4.
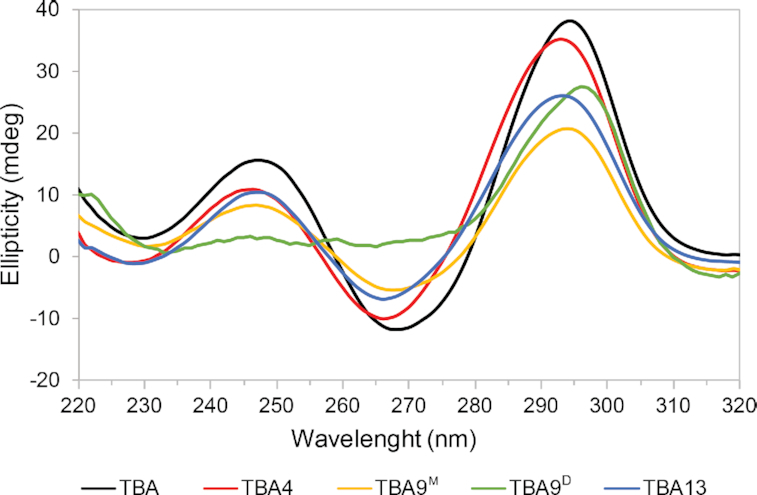
CD spectra characteristic for anti-parallel G-quadruplexes of TBA and its Upy analogues. CD spectra of 50 μM TBA, TBA4, TBA9M and TBA13 oligonucleotides in 50 mM KCl solutions were recorded with a 2.0 mm optical path at 25°C. No signal was observed above 320 nm. CD spectrum of TBA9D was recorded on NMR sample in 100 mM KCl solution with a 0.1 mm optical path at 25°C.
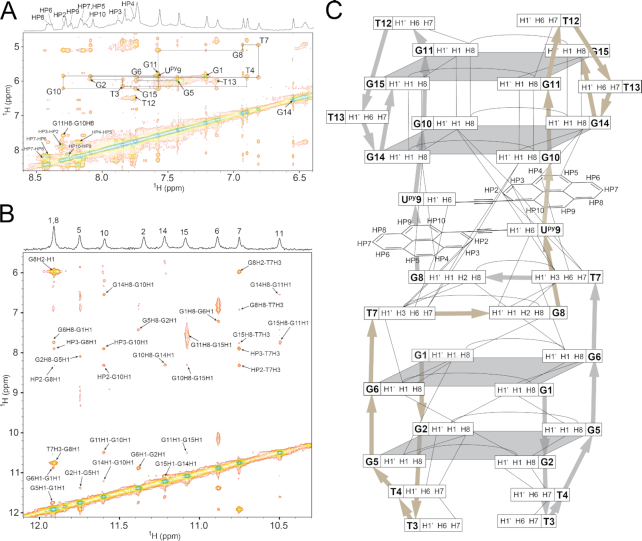
A more detailed structural analysis of TBA4 and TBA13 using 2D NMR revealed structural changes induced by pyrene moieties. Aromatic, anomeric and imino resonances of TBA4 and TBA13 could be assigned by analyzing 2D NOESY spectra. Aromatic/anomeric regions of spectra of both oligonucleotides contain four intense cross-peaks assigned to G1, G5, G10 and G14, which are indicative of their syn conformation (Figure 5). Sequential walk in NOESY spectra of TBA4 is interrupted at G2-T3, T4-G5, T7-G8, T9-G10 and T13-G14 steps. Analogously, in the spectra of TBA13 the sequential walk is interrupted at T4-G5, T7-G8, T9-G10, G11-T12 and T13-G14 steps. Nevertheless, observed NOE connectivities (Figure 6) confirm that the chair-like topology of TBA is retained for TBA4 and TBA13. 1H NMR chemical shifts in the aromatic and anomeric regions of TBA4 and TBA13 do not deviate considerably from TBA. Exceptions are aromatic H8 chemical shifts of G14 and G5 in TBA4 and TBA13, respectively, which exhibit large upfield shifts. G5 H8 in TBA13 and G14 H8 in TBA4 resonate at δ 6.71 and 6.78 ppm, respectively, compared to δ 7.42 and 7.45 ppm in TBA (46).
Figure 5.
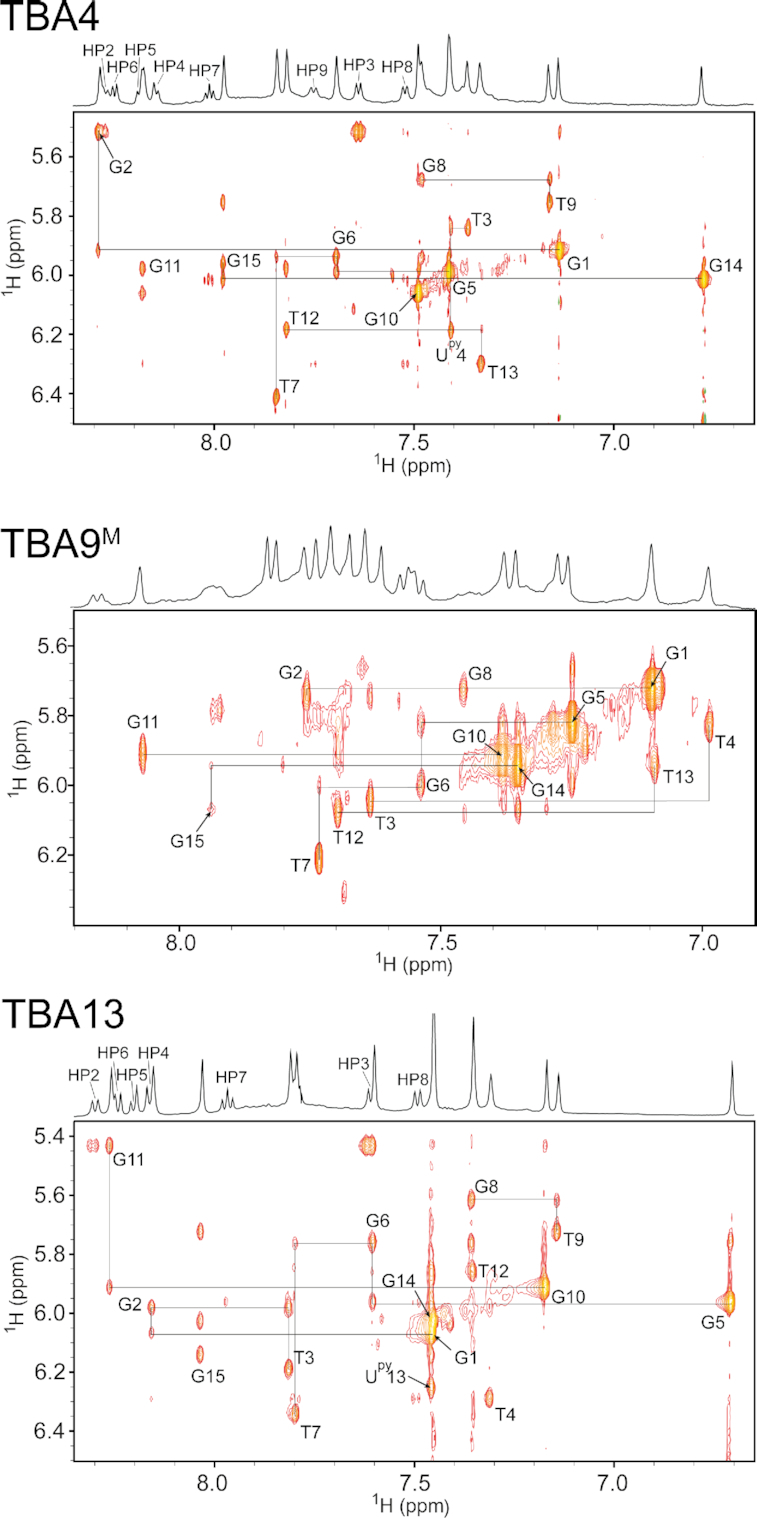
Spectral regions of 2D NOESY spectra (τm = 250 ms) of TBA4, TBA9M and TBA13 showing cross-peaks between aromatic and anomeric protons. Annotated cross-peaks correspond to intranucleotide H8/H6-H1′ NOE contacts. Assignments of pyrene protons are present in 1D traces above NOESY spectra.
Figure 6.
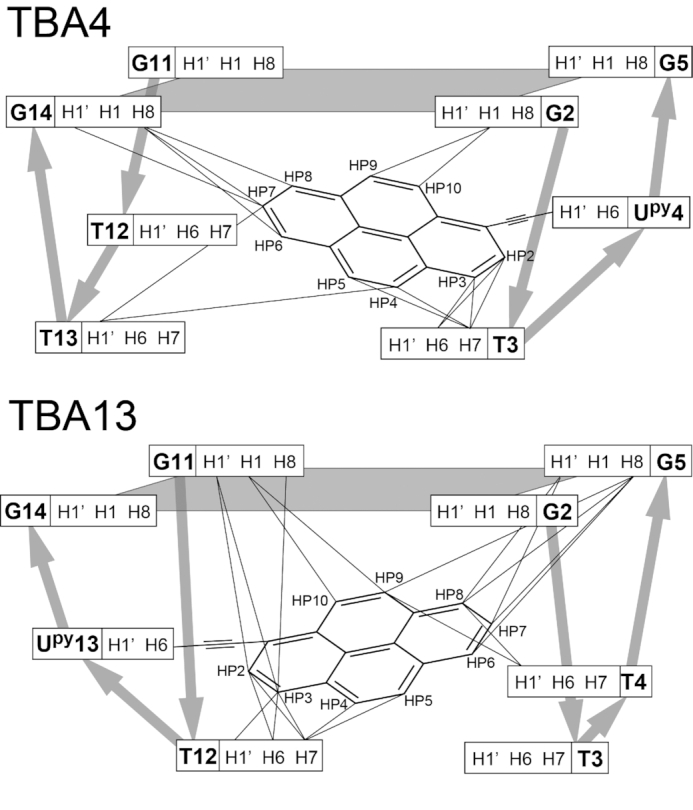
Schematic presentation of all internucleotide NOE correlations with pyrene groups in TBA4 (top) and TBA13 (bottom) G-quadruplexes observed in NOESY spectra (τm = 250 ms). G-quartet planes are shaded. Broad silver arrows show the direction of the phosphodiester backbone.
Pyrene group resonances of Upy analogues of TBA are found in the range from δ 7.5 to 8.6 ppm and overlap with nucleobase aromatic resonances. However, they are easily identifiable due to their multiplet structure, which corresponds to the pyrene spin–spin coupling network. All pyrene proton resonances in TBA4 and TBA13 were assigned with the use of 2D TOCSY spectra.
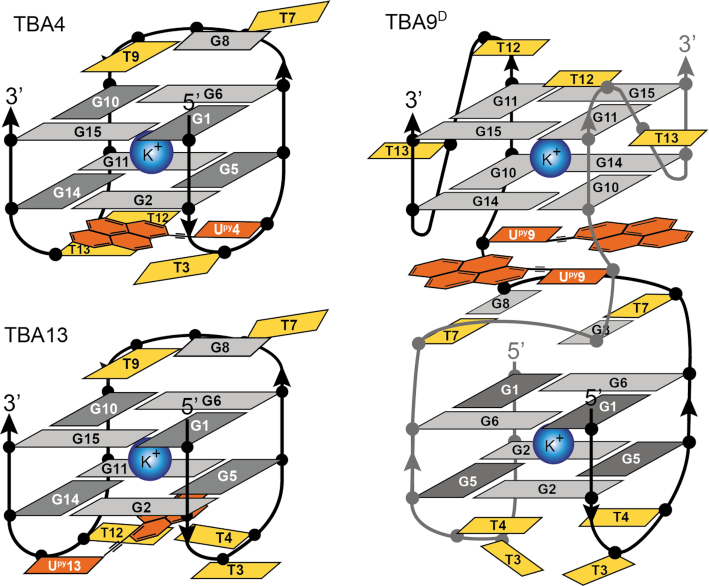
Several NOE contacts between Upy and nearby nucleotides were observed. In TBA4 pyrene protons of Upy4 exhibit NOE cross-peaks with loop nucleotides T3 and T13 as well as G2 and G14 of the adjacent G-quartet (Figure 6). Similarly, in TBA13 cross-peaks were observed between pyrene protons of Upy13 and G5 and G11 involved in G-quartets as well as loop nucleotides T4 and T12 (Figure 6). In total, 14 and 20 inter-nucleotide cross-peaks with pyrene protons were assigned in NOESY spectra of TBA4 and TBA13, respectively. These data suggest that pyrene groups in TBA4 and TBA13 stack on adjacent G-quartets, while loop thymine nucleotides stack on the pyrene groups.
A TBA analogue with double Upy substitutions at positions 4 and 13 (TBA4/13) was also evaluated for G-quadruplex formation. 1D NMR spectrum (Supplementary Figure S5) contains a set of sharp low-intensity imino signals, which suggests that the TBA4/13 oligonucleotide, at least in part, folds into a single G-quadruplex species. However, spectra also exhibit a broad background. We speculate that the part of TBA4/13 is in the form of oligomers facilitated by inter-strand pyrene–pyrene interactions.
Pyrene incorporation into the TGT loop induces dimerization
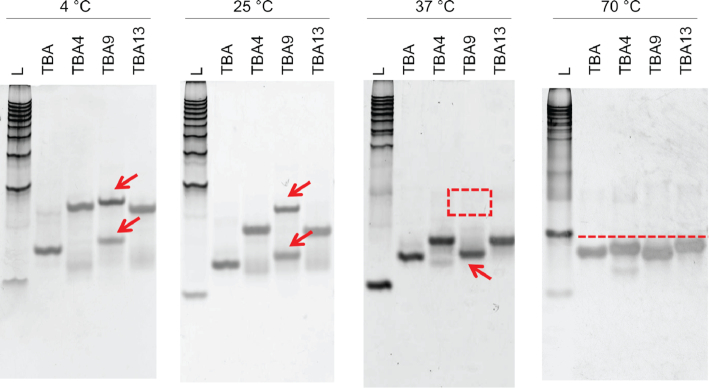
Initial measurements of NMR spectra of TBA9, which contains a Upy substitution in the longer tri-nucleotide loop, were done in a solution containing 50 mM KCl. This resulted in two sets of NMR signals suggesting, that two structures of TBA9 are in equilibrium. To confirm the structure of the TBA9, G-quadruplex structures were separated by native gel electrophoresis (Figure 7). The results showed that, at temperatures of 25°C or lower, two bands of TBA9 were present, while TBA, TBA4 and TBA13 resulted in a single PAGE band. Running a gel at 37°C or higher resulted in single bands for all four tested oligonucleotides.
Figure 7.
Polyacrylamide gel electrophoresis of TBA, TBA4, TBA9 and TBA13. L corresponds to 10–100 base-pair ladder. Gels were run at 4, 25, 37 and 70°C. Red arrows point to TBA9 species. Red square at 37°C indicates the absence of one TBA9 species. Red line at 70°C suggests all oligonucleotides are denatured and migrate at a similar rate.
DOSY NMR experiments revealed that the two structures of TBA9 exhibit different translational diffusion coefficients (Dt). One of them features a Dt of 1.6·10−6 cm2/s, which is in agreement with Dt values for TBA, TBA4 and TBA13. This form corresponds to a monomeric G-quadruplex and was designated as TBA9M. On the other hand, the second structure with a Dt value of 1.2·10−6 cm2/s was assigned a dimeric nature (TBA9D). Different salt concentrations were tested in order to shift the equilibrium towards a single species in solution. TBA9M is the predominant at low K+ ion concentrations and a 20 mM KCl solution was found to give the least of the dimeric form (Figure 3). A higher K+ ion concentration of 100 mM and prolonged incubation were found to shift the equilibrium towards the dimeric TBA9D. After several weeks of incubation at 4°C, NMR spectra contained predominantly TBA9D signals with TBA9M signals reduced to around 5%.
1H NMR spectrum of TBA9M contains one set of imino resonances in the range from δ 10.3 to 12.2 ppm (Figure 3) suggesting a single G-quadruplex species is formed in solution. 2D NOESY spectra were acquired in order to gain further insight into the structure of TBA9M (Figure 5). Cross-peaks were found to exhibit larger linewidths compared to TBA4 and TBA13, which could be attributed to higher dynamics of the Upy and adjacent nucleotides. Nevertheless, we were able to assign imino, aromatic and anomeric 1H resonances of TBA9M with the exception of G8 and Upy9. G1, G5, G10 and G14 were found to be in syn conformation with cross-peaks of G10 being noticeably broader. Chemical shifts of assigned aromatic and anomeric resonances are similar to the values found for TBA. In both cases imino signals of T4 and T13 are observed, thus confirming T4–T13 interloop base pairing in structures of TBA and TBA9M. The most significant change in chemical shift is observed for imino resonance of G1, which is found at δ 10.35 ppm in TBA9M compared to δ 12.05 ppm in TBA (Figure 3). Available NMR data and the CD profile (Figure 4) of TBA9M, which demonstrates characteristic antiparallel signatures of TBA, TBA4 and TBA13, suggest that TBA9M adopts the chair-like TBA fold. Due to broad resonances and signal overlap we were unable to unambiguously assign pyrene resonances of TBA9M and extract NOE connectivities. However, the upfield shift of G1 imino resonance indicates close proximity of the pyrene group, which suggests the pyrene group stacking at the adjacent G1–G6–G10–G15 G-quartet.
The fingerprint imino region of NMR spectra of TBA9D exhibits a distinct resonance pattern (Figure 3). Chemical shift dispersion of imino resonances in the range from δ 10.3 to 11.8 ppm suggests changes in the core G-quartet structure compared to the parent TBA. 2D NMR experiments for TBA9D resulted in good spectral properties. Assignment of correlation peaks in NOESY and TOCSY spectra was confirmed through 15N-edited HSQC spectra on a series of residue-specifically 15N enriched oligonucleotides (Supplementary Figure S6). Imino to aromatic proton NOE contacts were used to establish guanines constituting individual G-quartets (Figure 8). Two distinct G-quadruplex units were identified (Figure 9). Two 5′ segments from G1 to G6 form an antiparallel G-quadruplex unit in a head-to-head arrangement. G-tracts are connected by two edge-type T3–T4 loops and form two G-quartet planes, G1–G6–G1–G6 and G2–G5–G2–G5. Characteristic G(syn)-G(anti) connectivities could be observed in NOESY spectra of TBA9D, with sequential walk interrupted at steps T3–T4 and T4–G5. No imino resonances for T3 or T4 could be observed in NMR spectra suggesting higher levels of dynamics in edge-type loops and subsequently no TT base pairing in comparison to the parent TBA. Two 3′ segments from G10 to G15 form a parallel G-quadruplex unit with two G-quartets, G10–G14–G10–G14 and G11–G15–G11–G15, connected by propeller type T12–T13 loops. All guanines in this unit are in the anti conformation. The sequential walk could be traced from G10 to G15 with a single interruption at the G11–T12 step.
Figure 8.
Spectral regions of a NOESY spectrum (τm = 250 ms) of TBA9D. (A) Aromatic-anomeric region where annotated cross-peaks represent intranucleotide H8/H6-H1′ NOE contacts. (B) NOESY region containing imino proton contacts to pyrene, aromatic and d-ribose protons. (C) Schematic representation of observed internucleotide NOE correlations for the TBA9D G-quadruplex. Broad silver and gold arrows show the direction of the phosphodiester backbone in both TBA9 DNA strands.
Figure 9.
Schematic presentations of structures of TBA4, TBA13 and the dimeric TBA9D G-quadruplexes. Upy residues are in orange. Dark and light grey nucleotides correspond to guanines in syn and anti conformations, respectively. Thymines are shown in yellow.
The two G-quadruplex units of TBA9D are connected by two T7-G8-Upy9 segments. Full sequential walk within the TGUpy segment as well as with adjacent G6 and G10 was observed. Interestingly, the intensity of Upy9 H6-H1′ cross-peak in NOESY spectra is comparable to G1 and G5 indicating that Upy9 adopts a syn conformation. Intense cross-peaks between imino protons of T7 and G8 suggest formation of two inter-strand wobble T7-G8 base pairs. On the basis of NOE cross-peaks, we constructed a model of TBA9D, where two Upy residues form a planar structure and stack with the adjacent G10-G14-G10-G14 quartet of the 3′ G-quadruplex unit and T7-G8 base pairs (Figures 8C and 9). Moreover, the pyrene moieties are sandwiched between T7 and G14 whose aromatic protons exhibit an upfield chemical shift of δ 6.66 and 6.40 ppm, respectively, compared to δ 7.88 and 7.45 ppm in the parent TBA.
In an attempt to stabilize both G-quartets through pyrene stacking we have prepared TBA analogues with double Upy substitutions at positions 4 and 9 (TBA4/9) as well as 9 and 13 (TBA9/13). However, NMR spectra of TBA4/9 and TBA13/9 exhibit only broad resonances and no imino signals (Supplementary Figure S5), which suggests absence of G-quadruplex structures.
Stabilization through enthalpic and entropic contributions
Thermal stability of Upy analogues with respect to parent TBA was assessed by analyzing UV melting curves. Three oligonucleotides containing the pyrene group (TBA4, TBA9M and TBA13) exhibit Tm higher than TBA (Table 2 and Supplementary Figure S7). TBA9M with a TGUpy loop exhibits the highest increase in Tm of 6.4°C, while Tm for TBA4 and TBA13 with TUpy loops increases by 5.6 and 4.8°C, respectively. Thermodynamic analysis shows that in the case of TBA4 and TBA13 higher Tm values are the result of favorable enthalpic contributions related to stacking of pyrene groups on neighboring nucleobases. On the other hand, the increased stability of the TBA9M structure is achieved through entropic contributions of an increased number of equiprobable states. This is in agreement with NMR data where broadening of signals corresponding to Upy9 and adjacent nucleotides was observed.
Table 2.
Thermodynamic parameters for the TBA G-quadruplex and its pyrene-modified analogues
| Oligonucleotide | ΔH° (kcal mol−1) | TΔS° (kcal mol−1) | ΔG°25 (kcal mol−1) | Tm (°C) |
|---|---|---|---|---|
| TBA | −39.2 ± 1.5 | −35.6 ± 1.3 | −3.6 ± 0.2 | 52.0 |
| TBA4 | −44.2 ± 6.8 | −39.7 ± 6.3 | −4.5 ± 0.5 | 57.6 |
| TBA9M | −37.2 ± 4.4 | −33.1 ± 4.1 | −4.1 ± 0.4 | 58.4 |
| TBA13 | −42.2 ± 4.8 | −37.8 ± 4.4 | −4.5 ± 0.4 | 56.8 |
Due to the low DNA concentration used in UV measurements and short incubation time the TBA9 oligonucleotide was present in solution predominantly in the monomeric form (TBA9M). We were unable to perform UV melting experiments at high DNA concentrations, which would facilitate dimerization of TBA9, due to high absorbance of such samples. However, at higher concentrations (400 μM) of TBA9 and additional transition at ∼35°C was be observed (Supplementary Figure S7E), which could be attributed to melting of the dimeric structure.
In order to further assess TBA9D stability we performed an NMR melting experiment with incrementally increasing the temperature from 25 to 55°C (Supplementary Figure S8). As temperature is increased TBA9D gradually transforms into the monomeric TBA9M structure. The 1:1 ratio between TBA9M and TBA9D is achieved at around 45°C above which NMR signals corresponding to both structures decrease in intensity.
Nuclease degradation resistance in human blood serum and interaction of pyrene-modified TBA analogues with thrombin
Pyrene-modified TBA analogues show increased nuclease resistance with respect to the parent TBA as analysed under conditions of human blood serum at 37°C. Degradation of the oligonucleotides was examined after separation using denaturing PAGE (Supplementary Figure S9). After 3 h of incubation, 90% of TBA degraded (Supplementary Figure S9A lane 3 and Supplementary Table S1). In contrast, degradation of pyrene-modified TBAs was markedly inhibited (Supplementary Figure S9B–D, and Supplementary Table S1). In particular, TBA4 and TBA13, which have high thermal stability, had about 4-times as much of full-length oligonucleotides remaining after 3 hours under human serum conditions than TBA. Thus, thermal stabilization of G-quadruplexes with pyrene moiety may be correlated with their higher stability against nuclease degradation.
Binding of pyrene-modified TBAs to the human thrombin was evaluated by means of a quartz-crystal microbalance (QCM) with immobilized thrombin (Supplementary Figure S10A) (65–67). Both parent and modified TBA showed decreases in frequency when bound to thrombin, while the antisense strand of TBA did not show any frequency changes (Supplementary Figure S10B). Analysis of the binding isotherms based on the Langmuir adsorption model indicated that the dissociation constants (KD) of thrombin were 85.1 nM for native TBA, 166 nM for TBA4, 93.6 nM for TBA9M, and 523 nM TBA13 at 25°C (Supplementary Figure S10C and Supplementary Table S2). Therefore, the pyrene-modified TBAs were active for the recognition by thrombin. TBA9M had a similar affinity as the parent TBA. However, TBA4 and TBA13 showed lower affinities than TBA. The pyrene moieties in TBA4 and TBA13 are sterically located in G-quadruplex structures that may face the binding surface of thrombin (68). Therefore, differences in KD values are probably due to the steric hindrance of pyrene moiety for the binding to thrombin.
DISCUSSION
Polyaromatic moieties like pyrene groups seem to be ideal for stacking on G-quartets thus stabilizing G-quadruplex structures. The thrombin protein has two exosites that bind TBA. One binds TBA on the side of the TGT loop and the other on the side of two TT loops (52,69). Therefore, drastic changes on one side of the TBA structure, as in our study with incorporating a relatively bulky pyrene moiety, could still retain TBA’s affinity towards thrombin on the opposite side of the G-quadruplex, while at the same time increase thermodynamic stability. We have used a pyrene conjugated nucleotide (Upy) and incorporated it into loops of the TBA model system. However, individual incorporation of Upy at some loop positions results in destabilization of G-quadruplex structures. On the other hand, replacing any of the thymines at the 3′ ends of loops with Upy increases melting temperatures of G-quadruplex structures chair-type G-quadruplex topology (Figure 9). The Tm increase is moderate with 6.4°C being the maximum, which we believe is due to the low conformational flexibility of Upy residues. The pyrene group is tethered to the uracil nucleobase via a linker. Together these functional groups of Upy form an extended structural unit measuring around 14.5 Å across the diagonal (H6 to HP7). In the case of TUpy loops in TBA4 and TBA13 the pyrene group of the extended structure coupled with a relatively short loop length cannot optimally stack on the adjacent G-quartet. The T7-G8-Upy9 loop in TBA9 is longer and offers more degrees of freedom. In TBA9M more efficient stacking of pyrene groups and adjacent guanines is responsible for slightly higher thermal stability. TBA9M is in equilibrium with dimeric TBA9D structure (Figure 9), whose possibility of formation we also attribute to higher structural reorganization potential of T7–G8–Upy9 loop.
In a study by Mayer et al. higher thermal stability of TBA was achieved with TINA residues, which also consist of pyrene groups but lack deoxy-ribose and nucleobase (38). In TINA the pyrene group is attached to the DNA backbone through functional groups that allow for unrestricted rotation and thus grant a lot of conformational flexibility. Nevertheless, only substitutions with TINAs in the TGT loop of TBA were tolerated to some extent, while substitution of T9 with TINA led to a loss of aptamer activity. CD spectra of the oligonucleotide with TINA at position 9 was not in agreement with the parent TBA and suggested changes in the G-quadruplex conformation. These observations are in agreement with our findings, where Upy substitution in TBA at position 9 can led to dimerization. A different approach by Pedersen et al. used different anthraquinone insertions to replace TT and/or TGT loops (70). Replacing individual loops resulted in structures with lower Tm values. However, when both TT loops were replaced with anthraquinone insertions, the Tm values increased dramatically. Their data suggest this is due to efficient stacking between two anthraquinone groups and not stacking of anthraquinone on the adjacent G-quartet. We believe that our double substituted TBA analogues could suffer from a similar effect due to preferential pyrene-pyrene rather than pyrene-nucleobase stacking. However, due to higher DNA concentrations needed for NMR studies (mM) the pyrene-pyrene interactions could also be inter-strand thus forming oligomers. Stability is not the only factor in designing an efficient aptamer. Recently Montesarchio et al. successfully stabilized TBA by cyclization of the oligonucleotide (58). However, as a result the affinity towards thrombin was decreased due to increased rigidity of the structure. With this consideration in mind position 9 of TBA would be best for a substitution with Upy in TBA due to increased dynamics observed for TBA9.
Although pyrene-modified TBA analogues exhibit weaker interaction with thrombin protease, they may find their use in other applications. TBA4 and TBA13 exhibit weaker binding than the parent TBA, presumably due to steric hindrance imposed by pyrene moiety in the region that binds to thrombin. In the case of monomeric TBA9M binding is comparable to the parent TBA. Lower KD value of modified aptamers for thrombin binding might not be preferred in certain cases. Primary potential at the time of TBA discovery by SELEX method by Bock et al. in 1992 (48) was to act as an anticoagulant agent through thrombin protease inhibition. Recent studies have shown that TBA and its analogues exhibit anti-cancer properties (71), by a not yet understood mechanism, but certainly not through thrombin binding and inhibition, which would in such a case represent unwanted side effect. Therefore, lowering the binding affinity of TBA analogues to thrombin by modifications has potential to increase the TBA activity in another therapeutically important pathway.
Discovery and characterization of new genomic G-quadruplexes in combination with studies that in various ways try to improve their stability and affinity towards other biological molecules are of great importance. These new findings in combination with other types of chemical modifications could lead to the development of very important therapeutic agents for cancer treatments and other disorders where G-quadruplexes play a role.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Slovenian Research Agency [P1-0242, J1-1704]; Japan Society for the Promotion of Science (JSPS); Slovenian Ministry of Education, Science and Sport (MESS) under the Japan-Slovenia Research Cooperative Program; Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT); JSPS; Grant-in-Aid for Scientific Research on Innovative Areas ‘Chemistry for Multimolecular Crowding Biosystems’ [JSPS KAKENHI Grant No. JP17H06351]; MEXT-Supported Program for the Strategic Research Foundation at Private Universities (2014–2019), Japan; Hirao Taro Foundation of Konan Gakuen for Academic Research; Okazaki Kazuo Foundation of Konan Gakuen for Advanced Scientific Research; Chubei Itoh Foundation. The authors acknowledge the CERIC-ERIC Consortium for access to experimental facilities. Funding for open access charge: Slovenian Research Agency [P1-0242, J1-1704].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gellert I., Lipsett M.N., Davies D.R.. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U.S.A. 1962; 48:2013–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sen D., Gilbert W.. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. [DOI] [PubMed] [Google Scholar]
- 3. Neidle S., Balasubramanian S.. Quadruplex Nucleic Acids Royal Society of Chemistry. 2006; Cambridge. [Google Scholar]
- 4. Lane A.N., Chaires J.B., Gray R.D., Trent J.O.. Stability and kinetics of G-quadruplex structures. Nucleic. Acids. Res. 2008; 36:5482–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marušič M., Šket P., Bauer L., Viglasky V., Plavec J.. Solution-state structure of an intramolecular G-quadruplex with propeller, diagonal and edgewise loops. Nucleic. Acids. Res. 2012; 40:6946–6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic. Acids. Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu Y., Brosh R.M.. G-quadruplex nucleic acids and human disease. FEBS J. 2010; 277:3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hänsel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017; 18:279–284. [DOI] [PubMed] [Google Scholar]
- 9. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marsico G., Chambers V.S., Sahakyan A.B., McCauley P., Boutell J.M., Di Antonio M., Balasubramanian S.. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic. Acids. Res. 2019; 47:3862–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arola A., Vilar R.. Stabilisation of G-quadruplex DNA by small molecules. Curr. Top. Med. Chem. 2008; 8:1405–1415. [DOI] [PubMed] [Google Scholar]
- 12. Haider S.M., Neidle S., Parkinson G.N.. A structural analysis of G-quadruplex/ligand interactions. Biochimie. 2011; 93:1239–1251. [DOI] [PubMed] [Google Scholar]
- 13. Monchaud D., Teulade-Fichou M.P.. A hitchhiker's guide to G-quadruplex ligands. Org. Biomol. Chem. 2008; 6:627–636. [DOI] [PubMed] [Google Scholar]
- 14. Kotar A., Wang B., Shivalingam A., Gonzalez-Garcia J., Vilar R., Plavec J.. NMR structure of a triangulenium-based long-lived fluorescence probe bound to a G-quadruplex. Angew. Chem. Int. Ed. 2016; 55:12508–12511. [DOI] [PubMed] [Google Scholar]
- 15. Micheli E., Altieri A., Cianni L., Cingolani C., Iachettini S., Bianco A., Leonetti C., Cacchione S., Biroccio A., Franceschin M. et al.. Perylene and coronene derivatives binding to G-rich promoter oncogene sequences efficiently reduce their expression in cancer cells. Biochimie. 2016; 125:223–231. [DOI] [PubMed] [Google Scholar]
- 16. Zuffo M., Guédin A., Leriche E.D., Doria F., Pirota V., Gabelica V., Mergny J.L., Freccero M.. More is not always better: finding the right trade-off between affinity and selectivity of a G-quadruplex ligand. Nucleic Acids Res. 2018; 46:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winnik F.M. Photophysics of preassociated pyrenes in aqueous polymer solutions and in other organized media. Chem. Rev. 1993; 93:587–614. [Google Scholar]
- 18. Manoharan M., Tivel K.L., Zhao M., Nafisi K., Netzel T.L.. Base-Sequence dependence of emission lifetimes for D141018-30-6NA oligomers and duplexes covalently labeled with pyrene: Relative electron-Transfer quenching efficiencies of A, G, C, and T Nucleosides toward Pyrene. J. Phys. Chem. 1995; 99:17461–17472. [Google Scholar]
- 19. Østergaard M.E., Hrdlicka P.J.. Pyrene-functionalized oligonucleotides and locked nucleic acids (LNAs): Tools for fundamental research, diagnostics, and nanotechnology. Chem. Soc. Rev. 2011; 40:5771–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hrdlicka P.J., Karmakar S.. 25 years and still going strong: 2′-O-(pyren-1-yl)methylribonucleotides - versatile building blocks for applications in molecular biology, diagnostics and materials science. Org. Biomol. Chem. 2017; 15:9760–9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krasheninina O.A., Novopashina D.S., Apartsin E.K., Venyaminova A.G.. Recent advances in nucleic acid targeting probes and supramolecular constructs based on pyrene-modified oligonucleotides. Molecules. 2017; 22:doi:10.3390/molecules22122108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaura M., Hrdlicka P.J.. Locked nucleic acid (LNA) induced effect on the hybridization and fluorescence properties of oligodeoxyribonucleotides modified with nucleobase-functionalized DNA monomers. Org. Biomol. Chem. 2015; 13:7236–7247. [DOI] [PubMed] [Google Scholar]
- 23. Astakhova I.K., Samokhina E., Babu B.R., Wengel J.. Novel (phenylethynyl)pyrene-LNA constructs for fluorescence SNP sensing in polymorphic nucleic acid targets. ChemBioChem. 2012; 13:1509–1519. [DOI] [PubMed] [Google Scholar]
- 24. Astakhova K., Golovin A. V., Prokhorenko I.A., Ustinov A. V., Stepanova I.A., Zatsepin T.S., Korshun V.A.. Design of 2′-phenylethynylpyrene excimer forming DNA/RNA probes for homogeneous SNP detection: the attachment manner matters. Tetrahedron. 2017; 73:3220–3230. [Google Scholar]
- 25. Didion B.A., Karmakar S., Guenther D.C., Sau S.P., Verstegen J.P., Hrdlicka P.J.. Invaders: recognition of double-stranded DNA by using duplexes modified with interstrand zippers of 2′-O-(Pyren-1-yl)methyl-ribonucleotides. ChemBioChem. 2013; 14:1534–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sau S.P., Madsen A.S., Podbevšek P., Andersen N.K., Kumar T.S., Andersen S., Rathje R.L., Anderson B.A., Guenther D.C., Karmakar S. et al.. Identification and characterization of second-generation invader locked nucleic acids (LNAs) for mixed-sequence recognition of double-stranded DNA. J. Org. Chem. 2013; 78:9560–9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guenther D.C., Karmakar S., Hrdlicka P.J.. Bulged invader probes: activated duplexes for mixed-sequence dsDNA recognition with improved thermodynamic and kinetic profiles. Chem. Commun. 2015; 51:15051–15054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamana K., Iwase R., Furutani S., Tsuchida H., Zako H., Yamaoka T., Murakami A.. 5′-Pyrene modified oligonucleotide provides a highly sensitive fluorescent probe of RNA. Nucleic Acids Res. 1999; 27:2387–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamana K., Zako H., Asazuma K., Iwase R., Nakano H., Murakami A.. Fluorescence detection of specific RNA sequences using 2′-pyrene-modified oligoribonucleotides. Angew. Chem. Int. Ed. 2001; 40:1104–1106. [DOI] [PubMed] [Google Scholar]
- 30. Mahara A., Iwase R., Sakamoto T., Yamana K., Yamaoka T., Murakami A.. Bispyrene-conjugated 2′-O-methyloligonucleotide as a highly specific RNA-recognition probe. Angew. Chem. Int. Ed. 2002; 41:3648–3650. [DOI] [PubMed] [Google Scholar]
- 31. Seo Y.J., Lee I.J., Yi J.W., Kim B.H.. Probing the stable G-quadruplex transition using quencher-free end-stacking ethynyl pyrene-adenosine. Chem. Commun. 2007; 27:2817–2819. [DOI] [PubMed] [Google Scholar]
- 32. Park Y., Kim K.T., Kim B.H.. G-Quadruplex formation using fluorescent oligonucleotides as a detection method for discriminating AGG trinucleotide repeats. Chem. Commun. 2016; 52:12757–12760. [DOI] [PubMed] [Google Scholar]
- 33. Nagatoishi S., Nojima T., Juskowiak B., Takenaka S.. A pyrene-labeled G-quadruplex oligonucleotide as a fluorescent probe for potassium ion detection in biological applications. Angew. Chem. Int. Ed. 2005; 44:5067–5070. [DOI] [PubMed] [Google Scholar]
- 34. Ma C., Huang H., Zhao C.. An aptamer-based and pyrene-labeled fluorescent biosensor for homogeneous detection of potassium ions. Anal. Sci. 2010; 26:1261–1264. [DOI] [PubMed] [Google Scholar]
- 35. Filichev V.V., Pedersen E.B.. Stable and selective formation of Hoogsteen-type triplexes and duplexes using twisted intercalating nucleic acids (TINA) prepared via postsynthetic sonogashira solid-phase coupling reactions. J. Am. Chem. Soc. 2005; 127:14849–14858. [DOI] [PubMed] [Google Scholar]
- 36. Gécí I., Filichev V.V., Pedersen E.B.. Synthesis of twisted intercalating nucleic acids possessing acridine derivatives, thermal stability studies. Bioconjug. Chem. 2006; 17:950–957. [DOI] [PubMed] [Google Scholar]
- 37. Le B.T., Filichev V.V., Veedu R.N.. Investigation of twisted intercalating nucleic acid (TINA)-modified antisense oligonucleotides for splice modulation by induced exon-skipping in vitro. RSC Adv. 2016; 6:95169–95172. [Google Scholar]
- 38. Rohrbach F., Fatthalla M.I., Kupper T., Pötzsch B., Müller J., Petersen M., Pedersen E.B., Mayer G.. Chemical maturation of a bivalent aptamer by single domain variation. ChemBioChem. 2012; 13:631–634. [DOI] [PubMed] [Google Scholar]
- 39. Cogoi S., Zorzet S., Rapozzi V., Géci I., Pedersen E.B., Xodo L.E.. MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res. 2013; 41:4049–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cogoi S., Paramasivam M., Filichev V., Géci I., Pedersen E.B., Xodo L.E.. Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells. J. Med. Chem. 2009; 52:564–568. [DOI] [PubMed] [Google Scholar]
- 41. Balasubramanian S., Neidle S.. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009; 13:345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006; 13:1055–1059. [DOI] [PubMed] [Google Scholar]
- 43. Neidle S. Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010; 277:1118–1125. [DOI] [PubMed] [Google Scholar]
- 44. Paramasivam M., Cogoi S., Filichev V.V., Bomholt N., Pedersen E.B., Xodo L.E.. Purine twisted-intercalating nucleic acids: a new class of anti-gene molecules resistant to potassium-induced aggregation. Nucleic Acids Res. 2008; 36:3494–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takahashi S., Kim K.T., Podbevšek P., Plavec J., Kim B.H., Sugimoto N.. Recovery of the formation and function of oxidized G-quadruplexes by a pyrene-modified guanine tract. J. Am. Chem. Soc. 2018; 140:5774–5783. [DOI] [PubMed] [Google Scholar]
- 46. Schultze P., Macaya R.F., Feigon J.. Three-dimensional solution structure of the thrombin-binding DNA Aptamer d(GGTTGGTGTGGTTGG). J. Mol. Biol. 1994; 235:1532–1547. [DOI] [PubMed] [Google Scholar]
- 47. Trajkovski M., Šket P., Plavec J.. Cation localization and movement within DNA thrombin binding aptamer in solution. Org. Biomol. Chem. 2009; 7:4677–4684. [DOI] [PubMed] [Google Scholar]
- 48. Bock L.C., Griffin L.C., Latham J.a., Vermaas E.H., Toole J.J.. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992; 355:564–566. [DOI] [PubMed] [Google Scholar]
- 49. Avino A., Fabrega C., Tintore M., Eritja R.. Thrombin binding aptamer, more than a simple aptamer: chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012; 18:2036–2047. [DOI] [PubMed] [Google Scholar]
- 50. Musumeci D., Montesarchio D.. Polyvalent nucleic acid aptamers and modulation of their activity: a focus on the thrombin binding aptamer. Pharmacol. Ther. 2012; 136:202–215. [DOI] [PubMed] [Google Scholar]
- 51. Smirnov I., Shafer R.H.. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000; 39:1462–1468. [DOI] [PubMed] [Google Scholar]
- 52. Nagatoishi S., Isono N., Tsumoto K., Sugimoto N.. Loop residues of thrombin-binding DNA aptamer impact G-quadruplex stability and thrombin binding. Biochimie. 2011; 93:1231–1238. [DOI] [PubMed] [Google Scholar]
- 53. Virno A., Randazzo A., Giancola C., Bucci M., Cirino G., Mayol L.. A novel thrombin binding aptamer containing a G-LNA residue. Bioorganic Med. Chem. 2007; 15:5710–5718. [DOI] [PubMed] [Google Scholar]
- 54. Pasternak A., Hernandez F.J., Rasmussen L.M., Vester B., Wengel J.. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 2011; 39:1155–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saccà B., Lacroix L., Mergny J.L.. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005; 33:1182–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martino L., Virno A., Randazzo A., Virgilio A., Esposito V., Giancola C., Bucci M., Cirino G., Mayol L.. A new modified thrombin binding aptamer containing a 5′-5′ inversion of polarity site. Nucleic Acids Res. 2006; 34:6653–6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pagano B., Martino L., Randazzo A., Giancola C.. Stability and binding properties of a modified thrombin binding aptamer. Biophys. J. 2008; 94:562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Riccardi C., Meyer A., Vasseur J.J., Russo Krauss I., Paduano L., Oliva R., Petraccone L., Morvan F., Montesarchio D.. Stability is not everything: The case of the cyclisation of a thrombin-binding aptamer. ChemBioChem. 2019; 20:1789–1794. [DOI] [PubMed] [Google Scholar]
- 59. Phan A.T., Patel D.J.. A site-specific low-enrichment 15N, 13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002; 124:1160–1161. [DOI] [PubMed] [Google Scholar]
- 60. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 61. Lee W., Tonelli M., Markley J.L.. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015; 31:1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Takahashi S., Matsuno H., Furusawa H., Okahata Y.. Direct monitoring of allosteric recognition of type IIE restriction endonuclease EcoRII. J. Biol. Chem. 2008; 283:15023–15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Karsisiotis A.I., Hessari N.M.A., Novellino E., Spada G.P., Randazzo A., Webba Da Silva M.. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed. 2011; 50:10645–10648. [DOI] [PubMed] [Google Scholar]
- 64. Nagatoishi S., Tanaka Y., Tsumoto K.. Circular dichroism spectra demonstrate formation of the thrombin-binding DNA aptamer G-quadruplex under stabilizing-cation-deficient conditions. Biochem. Biophys. Res. Commun. 2007; 352:812–817. [DOI] [PubMed] [Google Scholar]
- 65. Takahashi S., Iida M., Furusawa H., Shimizu Y., Ueda T., Okahata Y.. Real-time monitoring of cell-free translation on a quartz-crystal microbalance. J. Am. Chem. Soc. 2009; 131:9326–9332. [DOI] [PubMed] [Google Scholar]
- 66. Takahashi S., Tsuji K., Ueda T., Okahata Y.. Traveling time of a translating ribosome along messenger RNA monitored directly on a quartz crystal microbalance. J. Am. Chem. Soc. 2012; 134:6793–6800. [DOI] [PubMed] [Google Scholar]
- 67. Takahashi S., Furusawa H., Ueda T., Okahata Y.. Translation enhancer improves the ribosome liberation from translation initiation. J. Am. Chem. Soc. 2013; 135:13096–13106. [DOI] [PubMed] [Google Scholar]
- 68. Russo Krauss I., Merlino A., Randazzo A., Novellino E., Mazzarella L., Sica F.. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012; 40:8119–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pica A., Russo Krauss I., Merlino A., Nagatoishi S., Sugimoto N., Sica F.. Dissecting the contribution of thrombin exosite i in the recognition of thrombin binding aptamer. FEBS J. 2013; 280:6581–6588. [DOI] [PubMed] [Google Scholar]
- 70. Gouda A.S., Amine M.S., Pedersen E.B.. Synthesis and molecular modeling of thermally stable DNA G-quadruplexes with anthraquinone insertions. Eur. J. Org. Chem. 2017; 2017:3092–3100. [Google Scholar]
- 71. Esposito V., Russo A., Vellecco V., Bucci M., Russo G., Mayol L., Virgilio A., Galeone A.. Thrombin binding aptamer analogues containing inversion of polarity sites endowed with antiproliferative and anti-motility properties against Calu-6 cells. Biochim. Biophys. Acta - Gen. Subj. 2018; 1862:2645–2650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.