Abstract
N 6-Methyladenosine (m6A) is the most abundant RNA modification in mammal mRNAs and increasing evidence suggests the key roles of m6A in human tumorigenesis. However, whether m6A, especially its ‘reader’ YTHDF1, targets a gene involving in protein translation and thus affects overall protein production in cancer cells is largely unexplored. Here, using multi-omics analysis for ovarian cancer, we identified a novel mechanism involving EIF3C, a subunit of the protein translation initiation factor EIF3, as the direct target of the YTHDF1. YTHDF1 augments the translation of EIF3C in an m6A-dependent manner by binding to m6A-modified EIF3C mRNA and concomitantly promotes the overall translational output, thereby facilitating tumorigenesis and metastasis of ovarian cancer. YTHDF1 is frequently amplified in ovarian cancer and up-regulation of YTHDF1 is associated with the adverse prognosis of ovarian cancer patients. Furthermore, the protein but not the RNA abundance of EIF3C is increased in ovarian cancer and positively correlates with the protein expression of YTHDF1 in ovarian cancer patients, suggesting modification of EIF3C mRNA is more relevant to its role in cancer. Collectively, we identify the novel YTHDF1-EIF3C axis critical for ovarian cancer progression which can serve as a target to develop therapeutics for cancer treatment.
INTRODUCTION
N 6-Methyladenosine (m6A) is the most prevalent internal modification for eukaryotic mRNA and influences nearly every stage of RNA metabolism, including splicing, decay, export and translation (1–5). m6A RNA methylation is catalyzed by a multicomponent protein complex consisting of the ‘writers’ METTL3, METTL14 and WTAP, among which METTL3 methyltransferase is the key catalytic subunit (6). On the other hand, two demethylases FTO and ALKBH5 act as ‘erasers’ which are responsible for the removal of m6A modification on mRNA. The interaction between ‘writers’ and ‘erasers’ determines the dynamic and reversible feature of m6A modification (7,8). In addition, YT521-B homology (YTH) domain-containing proteins including YTHDF1-3, YTHDC1 and YTHDC2 have recently been recognized as ‘readers’ to specifically identify the m6A modification and are responsible for the outcomes of m6A-modified mRNAs (9). In various cellular contexts, individual reader binds to distinct subset of m6A-modified mRNAs and affects gene expression (9,10). Increasing studies uncover that m6A modification is implicated in diverse biological processes including tissue development, embryonic stem cell self-renewal, and fate determination (7,11,12). Importantly, dysregulation of m6A modification is closely related with initiation and progression of several cancers, such as lung cancer, hepatocellular carcinoma and acute myeloid leukemia (13–15). Notably, the m6A modification pathway exerts either oncogenic or tumor suppressive effect in different scenarios (14,16,17).
Ovarian cancer is the fifth leading cause of cancer-related death in women worldwide and has the highest mortality rate among gynecological cancers (18). Ovarian cancer patients are usually diagnosed at the late stage with poor prognosis and high relapse rate. Thus, extraordinary studies are required to identify and validate specific biomarkers and therapeutic targets for ovarian cancer management. Here, we focused on ovarian cancer to investigate how m6A acted on protein translation and subsequently regulate cancer progression.
We first observed that YTHDF1 expression was aberrantly up-regulated in ovarian cancer and YTHDF1 was essential for ovarian cancer cell growth and metastasis. The multi-omics analysis identified the translation initiation factor EIF3C as the direct target of YTHDF1 in ovarian cancer cells. YTHDF1 regulated EIF3C’s translation in an m6A-dependent manner and affected the overall protein translation in ovarian cancer cells. Thus, our data demonstrated the critical oncogenic roles of m6A reader YTHDF1 in cancer development.
MATERIALS AND METHODS
Tumor samples
Formalin-fixed paraffin-embedded (FFPE) samples of serous epithelial ovarian cancer and normal ovarian tissues, and frozen specimens of serous ovarian cancer were obtained from Daping Hospital. Fresh ovarian surface epithelium brushings were obtained from the normal ovaries of donors during surgery for other benign gynecological diseases at Daping Hospital. All of these samples were examined by experienced pathologists who confirmed the diagnosis of disease samples. Patients in this study provided written informed consent. This study of patient specimens was approved by the Institutional Review Board of Daping Hospital at Army Medical University. The comprehensive clinical and pathological information of ovarian cancer patients were shown in Supplementary Table S1.
Gene expression and survival analysis in ovarian cancer datasets
The CSIOVDB, a microarray gene expression database containing 3431 human ovarian cancer tissues with clinicopathological characteristics and follow-up information, was used to validate the expression of genes and their correlation with pathological differentiation degree, and clinical outcome of ovarian cancer patients (19). The cBioPortal (cBio Cancer Genomics Portal) TCGA dataset was used to examine gene expression and mutations with three cohorts of high-grade serous ovarian cancer datasets: Nature 2011 (20), PanCancer Altas (21) and Provisional.
Cell culture
HEK293T, A2780 and SKOV3 cells were purchased from National Infrastructure of Cell Line Resource (Beijing, China). HEK293T and SKOV3 cells were cultured in DMEM (GIBCO, USA) and A2780 cells were cultured in RPMI 1640 (GIBCO, USA) medium. The medium was supplemented with 10% fetal bovine serum (FBS; GIBCO, USA), penicillin (100 U/ml; GIBCO, USA) and streptomycin (200 μg/ml; GIBCO, USA). All the cells were maintained at 37°C in 5% CO2 cell culture incubator.
Plasmids
For shRNA plasmids used in lentivirus-mediated interference, complementary sense and antisense oligonucleotides encoding shRNAs targeting YTHDF1 or EIF3C were synthesized, annealed and cloned into pLKO.1 vector (#10878, Addgene). The related sequences of shRNAs were shown in Table S2 of the Additional file 6. EIF3C expression plasmid was constructed via cloning EIF3C gene into pCDH-CMV-MCS-EF1-copGFP vector (#CD511B-1, SBI). YTHDF1-wt (YTHDF1-FLAG) and YTHDF1-mut (K395A, Y397A) expression plasmids were cloned into pCMV6 vector (OriGene, USA). pCMV6-EIF3A, pCMV6-EIF3B and pCMV6-EIF3D were purchased from OriGene company. EIF3C-HA and mutation expression plasmids were obtained via cloning EIF3C with HA tag into pcDNA3.1 vector (IDOBIO, China).
Cell transfection and lentiviral infection
For transient transfection, cells were transfected with expression vectors by using Lipofectamine 3000 (Invitrogen, USA). For lentivirus production, lentiviral vectors were co-transfected into HEK293T cells with packaging vectors psPAX2 (#12260, Addgene) and pMD2.G (#12259, Addgene) using lipofectamine LTX (Invitrogen, USA). Infectious lentivirus particles were harvested at 48 h after transfection, filtered through 0.45 μm PVDF filters and transduced into cells.
RNA isolation and RT-qPCR
Total RNA was extracted from cells and tissues using Trizol (Invitrogen, USA) according to the manufacturer's instruction. For RT-qPCR, RNA was reverse transcribed to cDNA by using a Reverse Transcription Kit (Takara, Dalian, China). The levels of RNA transcripts were analyzed by the Bio-rad CFX96 real-time PCR system (Bio-rad, USA). All samples were normalized to GAPDH or 18s rRNA. Primers used in RT-qPCR were listed in Supplementary Table S2.
Western blot and antibodies
A2780 or SKOV3 cells were washed twice with cold PBS and pelleted. The pellet was resuspended in lysis buffer, incubated on ice with frequent vortex for 10 min, and the lysate was obtained by centrifugation at 12 000 g for 10 min. Proteins were fractionated by SDS-PAGE, transferred onto PVDF membranes, blocked in 5% nonfat milk or BSA in TBS/Tween-20, and then blotted with specific antibodies. Antibodies used were as follows: anti-YTHDF1 (1:500, Proteintech), anti-GAPDH (1:5000, Proteintech), anti-EIF3C (1:500, Proteintech), anti-EIF3A (1:500, Proteintech), anti-EIF3B (1:200, Proteintech), anti-EIF3D (1:500, Proteintech), anti-EIF3E (1:500, Proteintech), anti-EIF3G (1:500, Proteintech), anti-EIF3H (1:500, Proteintech), anti-EIF3I (1:200, Proteintech), anti-HA-tag (1:1000, Affinity) and anti-FLAG-tag (1:1000, Sigma-Aldrich).
Immunohistochemistry, scoring and HE staining
FFPE samples were used to create three paraffin-embedded tissue microarrays. Sections were also stained with hematoxylin and eosin (HE). Arrays were dewaxed and then doused with endogenous peroxidase 3% hydrogen peroxide. The epitope retrieval was conducted with 10 mM sodium citrate (pH 6) followed by blocking using PBS supplemented with 5% BSA. The specific antibody was incubated at 4°C overnight, and the secondary antibody was incubated at room temperature for 30 min. Cells were washed with PBS, and then incubated with the appropriate secondary antibody for 30 min at 37°C. Peroxidase activity was revealed by 3,3-diaminobenzidine and cells were counter-stained with hematoxylin. Cells were viewed and photographed with a Zeiss UV LSM 510 confocal microscope. A composite score was determined as the previous study (22). Antibodies used were as follows: anti-YTHDF1 (1:500, Proteintech), anti-EIF3C (1:500, Proteintech), Ki-67 (9449T, Cell Signaling Technology), Caspase-3 (9664T, Cell Signaling Technology).
Cell growth and proliferation assays
Cell viability was detected by adding 10% CCK8 (DOJINDO, Japan) into the infected cells plated in 96-well plates and incubation at 37°C for 2 h at 0, 24, 48, 72 and 96 h. The absorbance of each well was measured by a microplate reader set at 450 and 630 nm. All experiments were performed in triplicate.
For colony formation assay, 1 × 103 A2780 or 3 × 103 SKOV3 infected cells were maintained in each well of six-well plates and the medium was refreshed every 3 days. Colonies were fixed with methanol after 10 days for A2780 cells or 12 days for SKOV3 cells, then stained with 0.1% crystal violet (Sigma-Aldrich, USA) for 30 min and washed with PBS. The numbers of colonies containing more than 50 cells were counted.
EdU assays were performed using the EdU Cell Proliferation Assay Kit (Cat.C10310-1, Ruibo, China) according to the manufacturer's instructions.
Transwell migration and invasion assays
Migration assays were performed using a 24-well Transwell chamber system (Corning, USA). 5 × 104 cells were seeded in the upper chamber of an insert with 0.4 ml serum-free culture media in 24-well plates. 0.6 ml culture media with 20% FBS were added to the lower chamber. After incubation for 24 h, cells were fixed in methanol for 15 min and then stained with 0.1% crystal violet (Sigma-Aldrich, USA) for 30 min. After rinsing with water, the membranes of the chambers were mounted, covered on slides. For invasion assays, the 24-well Transwell chamber system (Corning, USA) and the upper chamber of an insert was coated with Matrigel (Sigma-Aldrich, USA) before plating cells. Migrated or invaded cells were imaged and counted under a 20× microscope.
Apoptosis assay
Cells were stained by using Annexin V/PI Cell Apoptosis Kit (DOJINDO, Japan) according to the manufacturer's recommendations and apoptosis was analyzed by BD Accuri C6 Flow cytometer (BD Biosciences, USA).
Nascent protein synthesis assay
Nascent protein synthesis was detected by using Click-iT HPG Alexa Fluor Protein Synthesis Assay Kits (Invitrogen, USA) according to the manufacturer's recommendations. Briefly, cells were cultured in l-methionine-free medium with l-homopropargylglycine (HPG) at 37°C for 30 min. Cells were washed once with PBS and fixed by 3.7% formaldehyde incubation at room temperature for 15 min. After washed twice with 3% BSA, cells were incubated with 0.5% Triton X-100 for 20 min. Subsequently, Click-iT reaction cocktail mix was added with incubation for 30 min protected from light. Nucleus was stained for another 30 min and proceeded to imaging and analysis.
In vivo tumorigenesis and metastasis assays
All animal studies were performed with an approved protocol from the Guide for the Care and Use of Laboratory Animals (NIH publications Nos. 80-23, revised 1996) and according to the institutional ethical guidelines for animal experiments. For tumorigenesis assay, A2780 cells were infected with shRNAs or empty vector, collected and resuspended in PBS. 5 × 106 cells in PBS were injected subcutaneously into one side of the posterior flanks of Balb/C nude mice at 6–8 weeks old. Tumor growth and volume were detected every 3 days. After 4 weeks, mice were sacrificed and weight of xenografts was examined. For metastasis assays, 1 × 106 cells in PBS were injected into the abdominal cavity of three groups of Balb /c mice. After 4 weeks, mice were sacrificed and the numbers of metastatic nodules were counted.
Gene-specific m6A qPCR and m6A sequencing (m6A-seq)
Total RNAs were extracted and purified by using PolyTtract mRNA Isolation System (Promega, Hong Kong). After fragmentation, RNA was incubated with m6A antibody for immunoprecipitation according to the standard protocol of the Magna methylated RNA immunoprecipitation m6A Kit (Merck Millipore, Germany). Enrichment of m6A containing mRNA was then analyzed either through RT-qPCR or high-throughput sequencing. Primers to m6A negative region of EEF1A was used as the negative control and primers to m6A postive region of EEF1A was used as the positive control according to the standard protocol of the Magna methylated RNA immunoprecipitation m6A Kit (Merck Millipore, Germany). For high-throughput sequencing, purified RNA fragments were used for library construction with the NEBNext Ultra RNA library Prep kit for Illumina (New England BioLabs) and were sequenced with Illumina HiSeq X Ten platform. Library preparation and high-throughput sequencing were performed by Novogene (Beijing, China).
RNA sequencing (RNA-seq)
RNA-seq was processed according to the instructions of NEBNext Ultra RNA Library Prep Kit for Illumina (New England BioLabs). Briefly, total RNAs were isolated from YTHDF1-depleted or control A2780 cells using Trizol reagent. Poly(A) RNA was subsequently purified by using PolyTtract mRNA Isolation System and used to generate cDNA libraries. All samples were sequenced on Illumina HiSeq X Ten platform. Sequence reads were mapped to the human genome version hg38 by using Illumina sequence analysis pipeline. The average gene expression values of three independent studies were used for following analysis.
RNA immunoprecipitation and high-throughput sequencing (RIP-seq)
Cells were washed twice with PBS, collected and then the pellet was resuspended in IP lysis buffer (150 mM KCl, 25 mM Tris (pH 7.4), 5 mM EDTA, 0.5 mM DTT, 0.5% NP40, 1× protease inhibitor, 1 U/μl RNase inhibitor). The lysate was harvested by centrifugation at 12 000 g for 10 min after incubation for 30 min. Antibodies and 40 μl of protein G beads (Invitrogen, USA) were added into the lysate followed by incubation overnight at 4°C. After washed three times with wash buffer (150 mM KCl, 25 mM Tris (pH 7.4), 5 mM EDTA, 0.5 mM DTT, 0.5% NP40), co-precipitated RNAs were extracted by Trizol reagent, ethanol-precipitated with glycogen (Invitrogen, USA). The enrichment of RNAs was normalized to IgG. For sequencing, rRNAs was depleted by using the NEBNext rRNA depletion kit (New England BioLabs). cDNA libraries were produced by employing NEBNext Ultra RNA Library Prep Kit for Illumina (New England BioLabs) and sequenced on Illumina HiSeq X Ten platform. Each group was sequenced in duplicate.
Enhanced UV crosslinking, immunoprecipitation and high-throughput sequencing (eCLIP-seq)
eCLIP was performed as described previously with minor modifications (23). Briefly, cells were UV-crosslinked at 150 mJ and 254 nm wavelength in 10 cm plates with 10 ml of cold PBS. Then cells were pelleted, flash frozen in liquid nitrogen, and stored at −80°C. The pellet was lysed with lysis buffer followed by further RNase I, and Turbo DNase treatment as described. The lysate was incubated with specific antibody overnight at 4°C for immunoprecipitation. 40 μl of protein G beads (Invitrogen, USA) was added and incubated for 2 h followed by washes as described. Following end repair and 3′ adaptor ligation, size selection was conducted using Nupage 4–12% Bis–Tris protein gels followed by transfer to nitrocellulose membranes. RNAs on nitrocellulose were harvested and reverse transcribed using SuperScript III (Thermo Fisher, USA). cDNA libraries were then prepared as described and sequenced by using Illumina Hiseq 1000 with pair end 150 bp read length.
Polysome profiling
Cells were incubated with 100 μg/ml cycloheximide (CHX, Merck Millipore, Germany) at 37°C for 15 min. Cells were pelleted and lysed on ice with lysis buffer. The lysate was collected, loaded onto a 10/50% (w/v) sucrose gradient solution prepared in lysis buffer, and then centrifuged at 4°C for 4 h at 27 500 rpm (Beckman, rotor SW28). The sample was then fractioned and analyzed by Gradient Station (BioCamp) equipped with an ECONO UV monitor (BioRad) as well as fraction collector (FC203B, Gilson). RNA was purified by Trizol from each fraction and subjected to RT-qPCR analysis.
Protein stability
To evaluate protein stability, ovarian cancer cells were treated with 100 μg/ml CHX during indicated times and harvested. Then protein expression of EIF3C or MYC was determined by western blot analysis.
Statistics
Unless otherwise noted, the data are expressed as mean ± SD. The significance of difference was evaluated using Student's t test or unpaired two sided t test. The Kaplan–Meier method and the log-rank test were applied to estimate overall survival, progression-free survival, and their differences involved. Quantifications of histological experiments were analyzed by means of a generalized linear mixed effects model with logit link by testing for non-zero regression coefficients and by using a Chi-square test. All of the analyses were performed by SPSS (Statistical Package for the Social Sciences) version 18.0 (Chicago, IL, USA). When the P value is <0.05, the results were considered to be statistically significant.
RESULTS
YTHDF1 is highly expressed in ovarian cancer
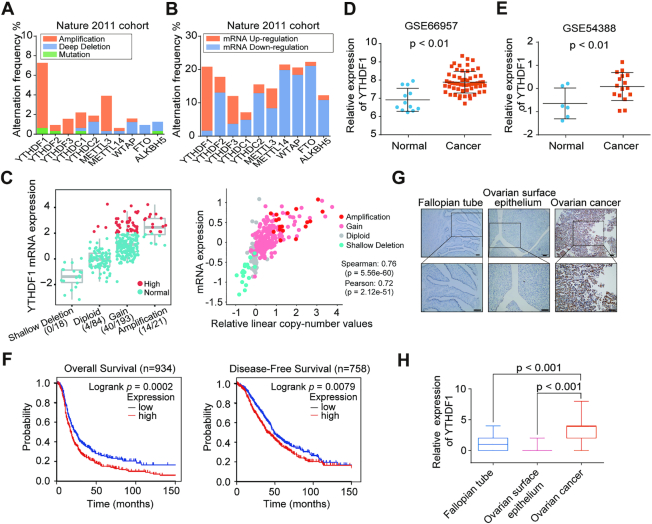
To investigate the roles of the m6A-associated genes in ovarian cancer development, we first analyzed the genetic alterations and expression levels of these genes by using the cBioPortal (cBio Cancer Genomics Portal) TCGA dataset which included three cohorts of high-grade serous ovarian cancer datasets (Nature 2011, PanCancer Altas and Provisional). Interestingly, these genes displayed different mutations and expression patterns, among which YTHDF1 gene was most frequently amplified and its expression was significantly elevated, and the copy number status of YTHDF1 was positively correlated with its mRNA expression in all three cohorts (Figure 1A–C, and Supplementary Figure S1A–D). In TCGA pan-cancer database, the amplification of YTHDF1 increased YTHDF1 expression and were frequently observed in various cancers (Supplementary Figure S1E and F). Then we analyzed expression of YTHDF1 in ovarian cancer according to two GEO datasets (GSE66957 and GSE54388) and found that YTHDF1 was up-regulated in ovarian cancer compared to normal ovarian epithelium cells (Figure 1D and E). Furthermore, we performed the Kaplan–Meier survival analysis by using CSIOVDB and found that ovarian cancer patients with high YTHDF1 expression exhibited the poorer overall survival and disease-free survival (Figure 1F). Since high-grade serous ovarian cancer accounts for the most common histological type of epithelial ovarian cancer, we particularly examined YTHDF1 expression in the high-grade serous ovarian cancers at different stages. Our data indicated that compared to FIGO stage I or II ovarian cancer patients, patients at FIGO stage III and IV displayed the increased expression of YTHDF1 (Supplementary Figure S2A). Thus, we analyzed the association between YTHDF1 expression and prognosis within these high-grade serous ovarian cancer patients at stage III and IV, and found that patients with higher YTHDF1 expression tended to have a shorter overall and progression-free survival (Supplementary Figure S2B). Consistently, the public ovarian cancer cohorts (kmplot.com) profiled on Affymetrix microarray for ovarian cancer revealed that YTHDF1 expression was negatively correlated with overall and progression-free survival of ovarian cancer patients (Supplementary Figure S2C). Again, the high-grade serous ovarian cancer patients at FIGO stage III and IV with higher YTHDF1 expression have the poor overall and progression-free survival (Supplementary Figure S2D).
Figure 1.
Increased YTHDF1 expression is associated with poor prognosis of ovarian cancer patients. (A) Gene mutation rates of m6A-associated genes in ovarian cancer according to cBioPortal datasets. (B) Gene expression of m6A-associated genes in ovarian cancer according to cBioPortal dataset (TCGA Nature 2011). (C) Correlation analysis between gene expression and the copy number status in TCGA Nature 2011 ovarian cancer dataset. (D and E) Relative RNA levels of YTHDF1 in ovarian cancer and normal ovarian surface epithelium in GEO datasets. (F) Kaplan–Meier analysis of ovarian cancer patients in CSIOVDB for the correlations between YTHDF1 expression and disease-free survival as well as overall survival. (G) Representative immunohistochemical images of YTHDF1 expression in primary ovarian cancer tissues, fallopian tube, and normal ovarian surface epithelium. Scale bar, 100 μm. (H) Relative YTHDF1 protein expression in fallopian tube, ovarian surface epithelium, and ovarian cancer specimens assessed by immunohistochemistry. Data are shown as means ± S.D.
We then compared the expression of YTHDF1 mRNA in 35 fresh frozen ovarian cancer specimens and 12 normal ovarian surface epithelium specimens collected from Daping Hospital by RT-qPCR. In line with previous findings from the GEO datasets, we found markedly increased expression of YTHDF1 in ovarian cancers compared with normal ovarian tissues (Supplementary Figure S2E). To examine the expression of YTHDF1 in ovarian cancer more precisely, we performed tissue microarray including ovarian surface epithelial tissues, fallopian tube specimens, and 134 ovarian cancer specimens, although the majority of cases are high-grade serous ovarian cancers. The result revealed that the expression of YTHDF1 protein was highly enhanced in ovarian cancer samples (Figure 1G and H). Together, these results suggest that m6A reader YTHDF1 is overexpressed in ovarian cancer and correlated with the poor prognosis of ovarian cancer patients.
YTHDF1 regulates proliferation, migration and invasion of ovarian cancer cells
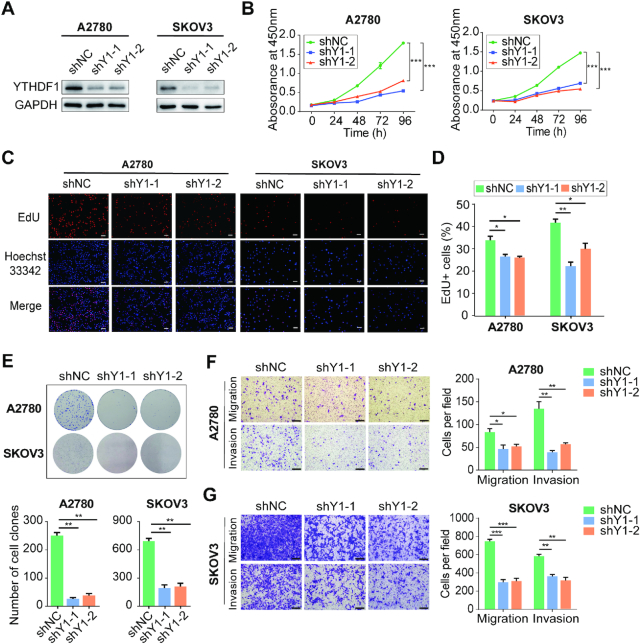
To explore the function of YTHDF1 in ovarian cancer, we sought to characterize the altered cellular phenotypes in ovarian cancer cells depleted of YTHDF1. YTHDF1 was efficiently knocked down by two shRNAs (shY1-1 and shY1-2) in both A2780 and SKOV3 ovarian cancer cells (Figure 2A). The knockdown of YTHDF1 significantly impaired cell growth of A2780 and SKOV3 cells as determined by CCK8 assays (Figure 2B). Furthermore, the EdU staining indicated ovarian cancer cells’ proliferation was remarkably decreased upon YTHDF1 silencing (Figure 2C and D). Similarly, the colony-formation assays revealed that clonogenic capacity was inhibited after knockdown of YTHDF1 (Figure 2E), and depletion of YTHDF1 could induce apoptosis in A2780 and SKOV3 cells (Supplementary Figure S3A and B). In addition, cell migration and invasion assays demonstrated YTHDF1 deficiency impaired the migration and invasion abilities of A2780 and SKOV3 cells (Figure 2F and G). These results indicate that YTHDF1 plays important roles in ovarian cancer cells’ proliferation, migration and invasion.
Figure 2.
Inhibition of YTHDF1 suppresses cell growth and migration in vitro. (A) Western blot analysis for YTHDF1 expression in A2780 and SKOV3 cells infected with two independent shRNAs targeting YTHDF1 or a control shRNA. (B) CCK8 assays were performed to determine cell growth after YTHDF1 was knocked down in A2780 and SKOV3 cells. (C) EdU assays of A2780 and SKOV3 cells described in (A). Scale bar, 100 μm. (D) Quantification of EdU positive cells shown in (C). (E) Colony formation assays of A2780 and SKOV3 cells described in (A). (F) Knockdown of YTHDF1 decreased the abilities of migration and invasion of A2780 cells. Scale bar, 200 μm. (G) Knockdown of YTHDF1 decreased the abilities of migration and invasion of SKOV3 cells. Data are shown as means ± S.D. *P < 0.05, **P < 0.01, ***P < 0.001.
YTHDF1 deficiency inhibits tumorigenesis and metastasis of ovarian cancer cells in vivo
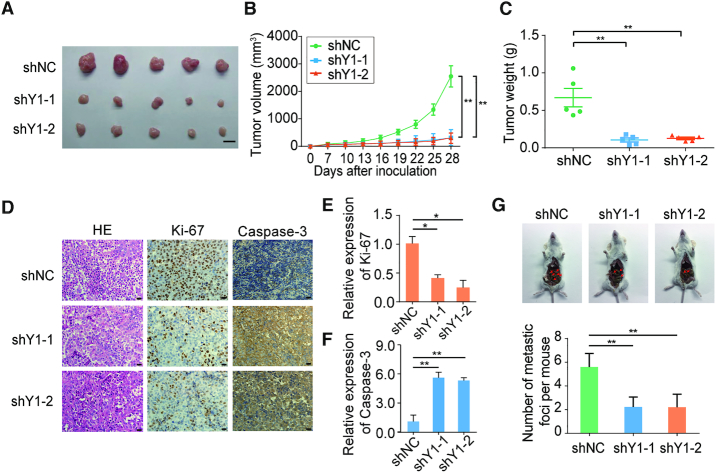
To evaluate the oncogenic role of YTHDF1 in ovarian cancer in vivo, we applied both subcutaneous tumorigenesis model and peritoneal metastatic xenograft model. First, YTHDF1 deficient A2780 ovarian cancer cells and control cells were injected subcutaneously in nude mice, respectively. After 4 weeks, the mice were sacrificed and the tumors were isolated (Supplementary Figure S3C). The tumors derived from the YTHDF1-deficient groups were substantially smaller than those from the control group (Figure 3A). Meanwhile, the average tumor volume and weight at sacrifice were markedly decreased in mice with YTHDF1 knockdown compared with the control mice (Figure 3B and C). We also evaluated the cell proliferation and apoptosis index in these solid tumors. The YTHDF1-silenced tumors exhibited decreased Ki-67 signal, but increased caspase-3 activity as compared to the control cells (Figure 3D–F). Next, we utilized an orthotopic model to examine peritoneal metastasis of ovarian cells after 4 weeks post-inoculation (Supplementary Figure S3D). Injection of YTHDF1-silenced cells substantially abrogated the capacity of the cells to form secondary tumors in abdominal cavity (Figure 3G). Collectively, these results demonstrate the oncogenic role of YTHDF1 in ovarian cancer by regulating cell proliferation and metastasis.
Figure 3.
YTHDF1 regulates tumorigenesis and metastasis of ovarian cancer cells in mice. (A–C) Inhibition of YTHDF1 impaired the growth of xenografted tumors. A2780 cells with empty vector or YTHDF1 knockdown vectors were subcutaneously injected into nude mice. (B) Tumor size was measured every 4 days and growth curves were plotted. Tumors were dissected from the nude mice of each group and photographed at 28 days after transplantation and the weight of tumors was measured (C). Scale bar, 10 mm. (D–F) Representative immunohistochemistry results and quantification of Ki-67 and Caspase-3 positive staining in xenografted tumors. Scale bar, 20 μm. (G) Representative images and quantification of abdominal cavity metastatic tumors derived from YTHDF1 knockdown or control A2780 cells. Data are shown as means ± S.D. *P < 0.05, **P < 0.01, ***P < 0.001.
Identification of the YTHDF1 targets in ovarian cancer
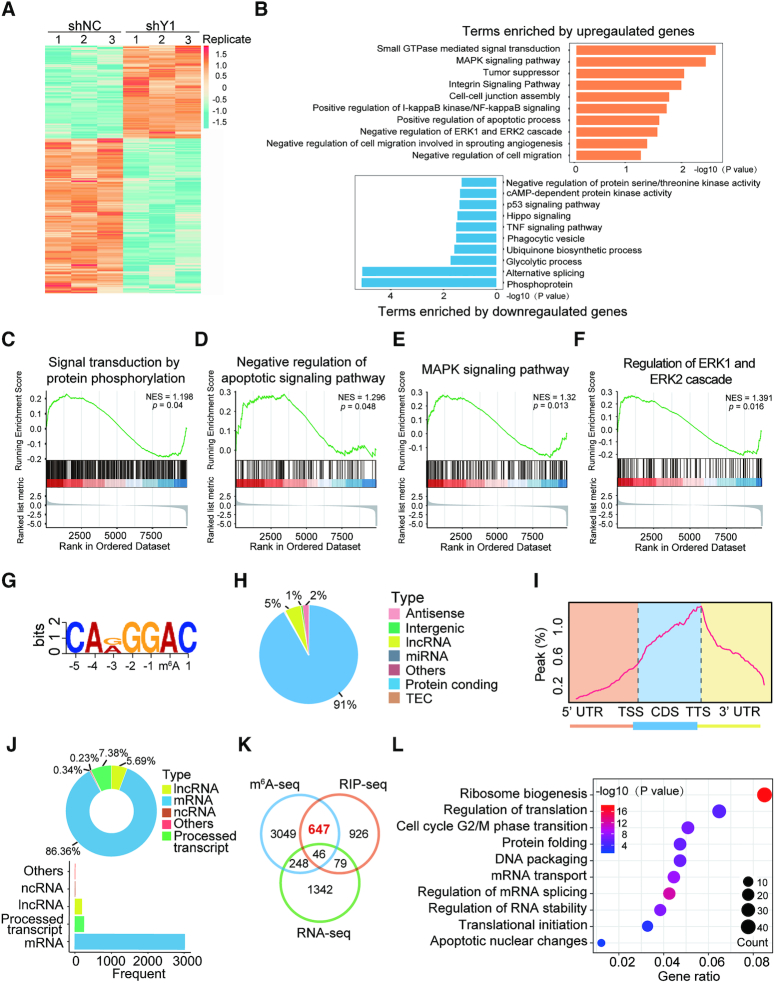
To explore the underlying mechanisms of YTHDF1 in ovarian cancer development, we first performed RNA-seq analysis with YTHDF1 knockdown and control A2780 cells. YTHDF1 depletion resulted in 1715 genes altered globally, including 633 up-regulated genes and 1082 down-regulated genes (Figure 4A, Supplementary Figure S4A and Supplementary Table S3). Gene ontology (GO) analysis showed that several enriched pathways such as MAPK signaling pathway, tumor suppressing, and regulation of cell migration (Figure 4B), and gene set enrichment analysis (GSEA) also revealed that genes altered by YTHDF1 were associated with signal transduction of protein phosphorylation, apoptotic signaling pathway, MAPK pathway, ERK cascade and cell activation (Figure 4C-F and Supplementary Figure S4B–F), supporting the regulatory roles of YTHDF1 in tumorigenesis of ovarian cancer.
Figure 4.
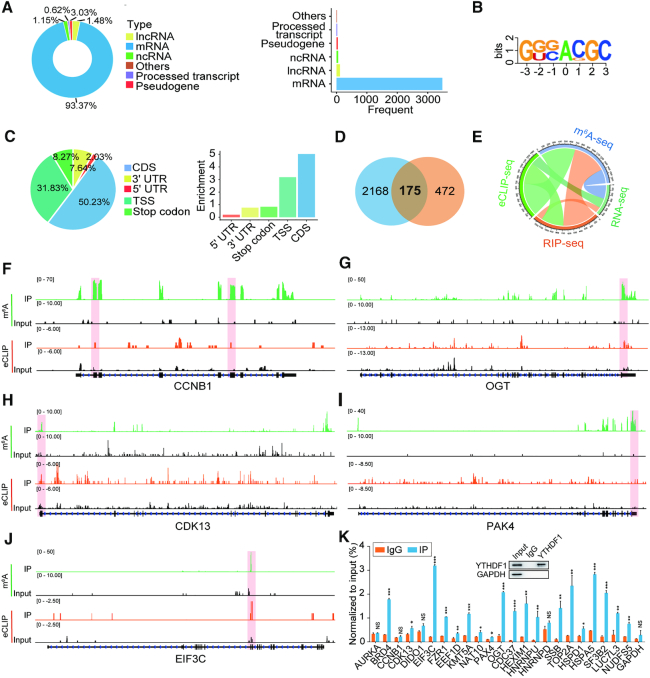
Identification of the YTHDF1 targets in ovarian cancer cells. (A) Heatmap of differentially expressed genes (DEGs) identified by RNA-seq. (B) GO enrichment analysis of DEGs. (C–F) GSEA plots showing the pathways of DEGs altered by YTHDF1 were involved in ovarian cancer cells. (G) The m6A motif detected by the MEME motif analysis with m6A-seq data. (H) Percentages of various RNA species by m6A modifications. (I) Metagene profiles of m6A enrichment across mRNA transcriptome in ovarian cancer cells. (J) Distribution of YTHDF1 targeted transcripts as identified by RIP-seq. (K) Overlapping analysis of genes identified by m6A-seq, RIP-seq, and RNA-seq. (L) GO analysis of genes described in (K).
It is well-recognized that YTHDF1 acts as an m6A reader, YTHDF1 functions via binding and affecting m6A methylated transcripts (5,10,24,25). Thus we applied m6A-seq in A2780 cells and RIP-seq with the antibody specific to FLAG in a stable FLAG-tagged YTHDF1-expressing A2780 cells. By m6A-seq analysis, we identified 8104 m6A peaks out of 3990 genes (Supplementary Table S4). MEME algorithm analysis identified the m6A consensus motif (GGAC), indicating the successful enrichment of m6A-modified mRNA (Figure 4G). These m6A modifications were predominately located in protein-coding transcripts (91%) and enriched near the stop codons (Figure 4H and I). In line with other m6A-seq results, the m6A peaks we identified were abundant in mRNA open reading frames (ORFs) and around the stop codons (Supplementary Figure S4G). Functional annotation of these mRNAs revealed several distinct gene clusters including RNA splicing and translation initiation (Supplementary Figure S4H). RIP-seq revealed 1,698 potential candidate targets of YTHDF1, among which 93% were mRNAs and these genes were enriched in different pathways (Figure 4J, Supplementary Figure S4I, J, and Supplementary Table S5). Interestingly, overlapping the genes from RNA-seq, m6A-seq, and RIP-seq revealed that 693 genes bound by YTHDF1 were tagged with m6A, among which 647 (93.4%) genes were not altered upon YTHDF1 knockdown as shown by RNA-seq (Figure 4K). These results suggested that YTHDF1 did not affect the RNA abundance of its targets, consistent with the finding that YTHDF1 regulated protein synthesis by interacting with m6A modified mRNA as reported in other studies (5,24,25). In addition, functional annotation showed that these 647 genes were involved in multiple RNA metabolic processes including translation, mRNA splicing, and RNA homeostasis (Figure 4L).
EIF3C is the m6A modification target of YTHDF1
In order to explore the direct interactions between YTHDF1 and its targeted transcripts comprehensively, we performed eCLIP-seq by using the antibody specific to FLAG in the stable FLAG-tagged YTHDF1-expressing A2780 cells and identified 2,343 targeted transcripts, and the majority of these transcripts were mRNAs (Figure 5A, Supplementary Figure S5A and Supplementary Table S6). Comprehensive analysis of YTHDF1 targets from RIP-seq and eCLIP-seq revealed that 438 identified targets from RIP-seq were confirmed by eCLIP-seq, suggesting there was the indirect or non-specific interaction between YTHDF1 and the transcripts (Supplementary Figure S5B). Nevertheless, KEGG pathway analysis showed that these mRNA were mainly mapped to apoptosis and mTOR signaling pathway, which was similar to the RIP-seq results (Supplementary Figure S5C). The m6A consensus motif GGAC and YTHDF1 binding preference were also identified from eCLIP-seq (Figure 5B, C and Supplementary Figure S5D). Of note, 175 genes were identified as the direct targets of YTHDF1 by overlapping the results from eCLIP-seq and 647 genes identified above (Figure 5D). The Circos plot showed that YTHDF1-targeted genes from RIP-seq or from eCLIP-seq were m6A modified, but their transcription was not changed upon YTHDF1 knockdown (Figure 5E), indicating that YTHDF1 might regulate translation rather than RNA abundance in ovarian cancer cells.
Figure 5.
EIF3C is the target of YTHDF1 in ovarian cancer cells. (A) Distribution of YTHDF1 targeted transcripts as identified by eCLIP-seq. (B) The consensus sequences of YTHDF1-binding sites detected by HOMER motif analysis with eCLIP-seq data. (C) The distribution (left) and enrichment (right) of YTHDF1-binding sites within different gene regions. Enrichment was measured by the proportion of m6A peaks normalized to the length of the region. (D) Venn diagram illustrated overlapping of the targets (647) of YTHDF1 identified by RIP-seq, m6A-seq, and RNA-seq analysis and the targets (2343) identified by eCLIP-seq. (E) Circos plot showing the targets of YTHDF1 from RIP-seq, eCLIP-seq, m6A-seq, and RNA-seq data. (F–J) Distribution of m6A peaks and YTHDF1-binding peaks across transcripts. (K) CLIP-qPCR detected the interaction between YTHDF1 and the target genes. Data are shown as means ± S.D. *P < 0.05, **P < 0.01, ***P < 0.001, NS, not significant.
Next, we analyzed the distribution of m6A peaks and eCLIP peaks across 175 YTHDF1-binding mRNAs, and some of them were closely associated with tumorigenesis. We found that most of YTHDF1 binding sites in these transcripts including EIF3C fitted well with the m6A sites (Figure 5F-J and Supplementary Figure S5E). GO analysis revealed that YTHDF1-binding transcripts were most closely associated with regulation of translation (Figure 4L and Supplementary Figure S5F). EIF3C is one of the subunits of eukaryotic initiation factor 3 (EIF3) orchestrating initiation factor and ribosome interactions for translation, and its translational control contributes to tumorigenesis. Moreover, EIF3C was associated with malignant behaviors in multiple cancers including hepatocellular carcinoma, colon cancer, and breast cancer (26–28). CLIP-qPCR also showed that EIF3C mRNA was enriched most prominently by YTHDF1 (Figure 5K). These data indicate that EIF3C is a direct target of YTHDF1 in ovarian cancer cells.
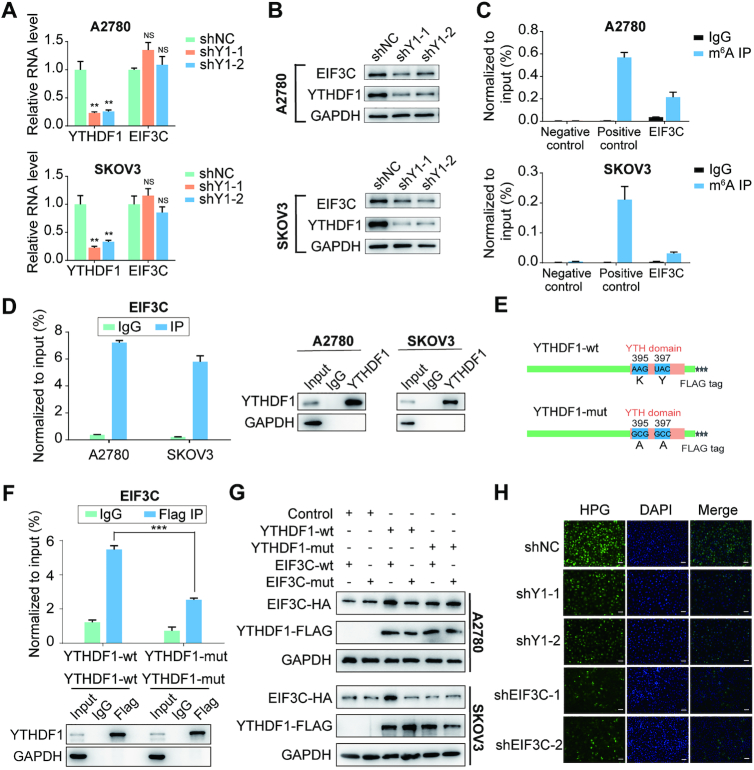
YTHDF1 regulates EIF3C expression and the overall translational output in ovarian cancer cells
To confirm YTHDF1 regulating EIF3C expression in ovarian cancer cells, we initially examined the transcription and translation of EIF3C upon YTHDF1 loss. As expected, YTHDF1 silencing by shRNAs decreased the protein abundance of EIF3C without affecting its RNA level in both A2780 and SKOV3 ovarian cancer cells (Figure 6A and B). Then we assessed the m6A modification status of EIF3C mRNA by the gene-specific m6A assay and a significant enrichment of EIF3C mRNA was observed (Figure 6C). Moreover, RIP-qPCR confirmed the interaction between YTHDF1 and EIF3C mRNA in both A2780 and SKOV3 ovarian cancer cells (Figure 6D). Since the protein level but not the mRNA level of EIF3C declined upon YTHDF1 deficiency, we speculated that YTHDF1 might regulate either protein stability or translation efficiency of EIF3C. To test this, control or YTHDF1-depleted ovarian cells were treated with protein translation inhibitor cycloheximide (CHX). Western blot analysis revealed that knockdown of YTHDF1 had no effect on the stability of EIF3C protein in both A2780 and SKOV3 cells, excluding the possibility that YTHDF1 regulates EIF3C protein stability (Supplementary Figure S6A). However, polysome profiling showed depletion of YTHDF1 resulted in a moderate shift of EIF3C mRNA to non-polysome fractions with reduced EIF3C mRNA in translation fractions, supporting that YTHDF1 regulates protein synthesis of EIF3C (Supplementary Figure S6B).
Figure 6.
YTHDF1 regulates EIF3C translation in an m6A-dependent manner. (A) Relative RNA level of EIF3C in A2780 and SKOV3 upon YTHDF1 knockdown. (B) Western blot detected the protein level of EIF3C in A2780 and SKOV3 cells upon YTHDF1 knockdown. (C) Gene-specific m6A qPCR validation of m6A levels in A2780 and SKOV3 cells. Primers to m6A negative region of EEF1A as the negative control and primers to m6A postive region of EEF1A as the positive control. (D) YTHDF1 RIP followed by RT-qPCR confirmed the interaction between YTHDF1 and EIF3C mRNA. (E) Schematic representation of wild-type (YTHDF1-wt) and mutant (YTHDF1-mut) YTHDF1 constructs. (F) RIP-derived RNA and protein in A2780 cells were measured by RT-qPCR and western blot, respectively. GAPDH was used as the negative control in western blot assays. (G) Western blot confirmed HA-tagged EIF3C expression in A2780 or SKOV3 cells co-transfected with empty vector, wild-type or mutant Flag-tagged YTHDF1 and wild-type or mutant HA-tagged EIF3C. (H) Nascent protein synthesis was detected by HPG incorporation upon YTHDF1 knockdown or EIF3C knockdown in A2780 cells. Scale bar, 100 μm. Data are shown as means ± S.D. *P < 0.05, **P < 0.01, ***P < 0.001, NS, not significant.
As EIF3C mRNA was modified by m6A, we next questioned whether YTHDF1 regulating EIF3C expression was m6A dependent. YTHDF1 is known to bind m6A sites through its m6A-binding pockets in YTH domain, mutations in K395 and Y397 could abrogate the binding capacity of YTHDF1 with mRNA (29). By introducing two point mutations K395A and Y397A in YTH domain of YTHDF1 with FLAG tag (YTHDF1-mut), ovarian cancer cells were transfected with the YTHDF1 wide-type (YTHDF1-wt) or YTHDF1-mut constructs, respectively (Figure 6E). Subsequently, RIP by using the antibody against FLAG followed by qPCR revealed EIF3C mRNA was immunoprecipitated effectively in cells transfected with YTHDF1-wt, but the interaction between YTHDF1 mutants and EIF3C mRNA were significantly decreased (Figure 6F and Supplementary Figure S6C), suggesting the m6A-binding pocket were crucial for YTHDF1 to bind EIF3C mRNA. Moreover, we found that YTHDF1-wt but not YTHDF1-mut could increase the protein expression of EIF3C (Supplementary Figure S6D). Additionally, we constructed HA-tagged EIF3C expression vector (EIF3C-wt) and its mutant with m6A sites mutation (EIF3C-mut). Western blot analysis revealed that YTHDF1-wt but not YTHDF1-mut could enhance expression of EIF3C-wt, while YTHDF1-wt had mild effect on expression of EIF3C-mut (Figure 6G and Supplementary Figure S6E).
Since EIF3C is the key subunit of EIF3 complex which is essential for protein synthesis, we next explore whether YTHDF1 could affect the overall translational output in ovarian cancer cells. To test this, we analyzed new protein synthesis by using HPG incorporation assay. Compared to control cells, HPG incorporated into synthesizing proteins was declined upon EIF3C knockdown (Figure 6H and Supplementary Figure S6F). Similarly, YTHDF1-deficient cells displayed a declined HPG signal compared with control cells (Figure 6H), indicating YTHDF1 could affect overall protein synthesis. Collectively, these data suggest that YTHDF1 promotes overall protein synthesis via regulating EIF3C protein expression in ovarian cancer cells.
EIF3C plays an oncogenic role in ovarian cancer cells
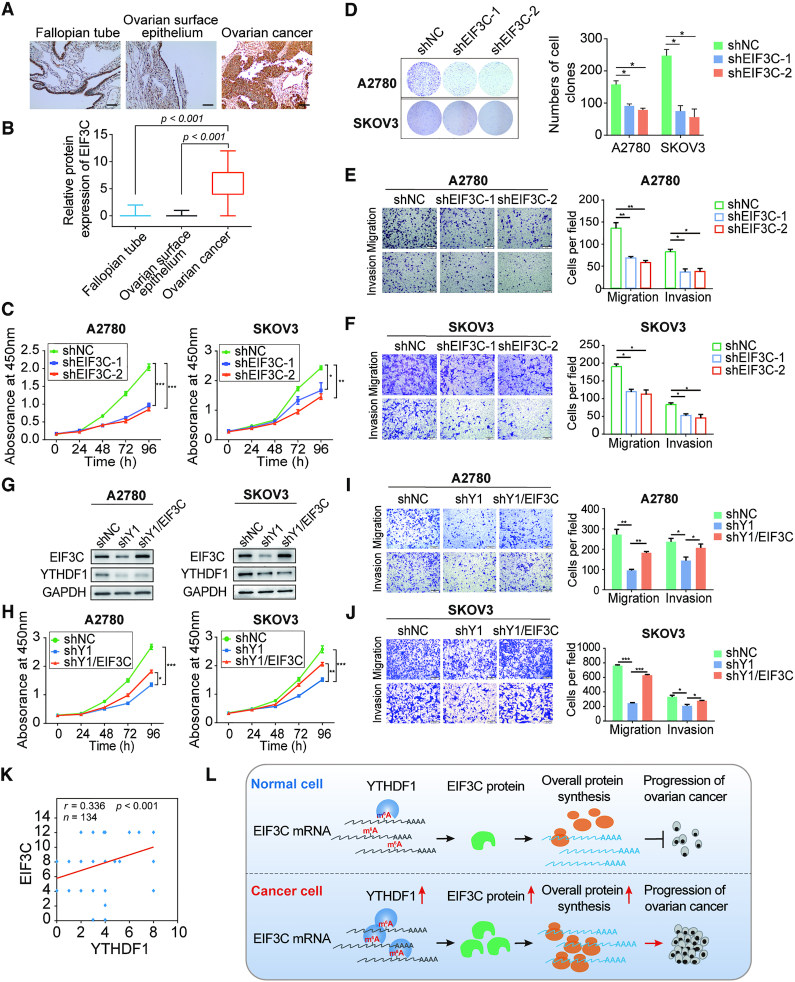
Since the functions of EIF3C in ovarian cancer cells remain unclear, we first examined the RNA level of EIF3C in ovarian cancer samples compared with normal ovarian surface epithelium samples by applying GEO database and CSIOVDB. Intriguingly, the RNA level of EIF3C was comparable in GSE54388 dataset but showed an increase in GSE66957 dataset in ovarian cancer samples (Supplementary Figure S7A). Similar results were found in CSIOVDB as the transcription level of EIF3C was not highly correlated with clinicopathologic features (Supplementary Figure S7B–E). Then we examined the protein expression of EIF3C in ovarian cancer tissues by using tissue microarray. Surprisingly, the protein expression of EIF3C in ovarian cancer was significantly elevated as compared to the fallopian tube (Figure 7A and B).
Figure 7.
YTHDF1 promotes cell growth and migration of ovarian cancer cells dependent on EIF3C. (A) Representative immunohistochemical images of EIF3C expression in primary ovarian cancer tissues, fallopian tube, and normal ovarian surface epithelium. Scale bar, 100 μm. (B) Relative EIF3C protein expression in fallopian tube, ovarian surface epithelium, and ovarian cancer specimens assessed by immunohistochemistry. (C) CCK8 assays were performed upon EIF3C knockdown in A2780 and SKOV3 cells. (D) Colony formation assays of A2780 and SKOV3 cells upon EIF3C knockdown. (E) EIF3C knockdown decreased the abilities of migration and invasion of A2780 cells. Scale bar, 200 μm. (F) EIF3C knockdown decreased the abilities of migration and invasion of SKOV3 cells. Scale bar, 200 μm. (G) Western blot detected the protein levels of YTHDF1 and EIF3C in YTHDF1-deficient A2780 and SKOV3 cells upon overexpression of EIF3C. (H) Cell growth was measured by CCK8 assays in A2780 and SKOV3 cells described in (g). (I) Transwell assays were used to measure abilities of migration and invasion of A2780 cells described in (G). Scale bar, 200 μm. (J) Transwell assays were used to measure abilities of migration and invasion of SKOV3 cells described in (G). (K) Spearman's rank correlation between YTHDF1 and EIF3C protein in ovarian cancer patients (n = 134) by use of tissue microarray. (L) Proposed model underlying the roles of YTHDF1-mediated EIF3C translation in ovarian cancer. Data are shown as means ± S.D. *P < 0.05, **P < 0.01, ***P < 0.001.
To further examine the role of EIF3C in ovarian cancer, we analyzed the cellular phenotypes including cell growth, colony formation ability, cell migration and invasion upon EIF3C knockdown in both A2780 and SKOV3 cells (Supplementary Figure S6F and S7F). Notably, cell growth and colony formation capacities were markedly suppressed upon EIF3C knockdown in both ovarian cancer cell lines (Figure 7C and D). Furthermore, EIF3C deficiency also reduced cell migration and invasion of ovarian cancer cells (Figure 7E and F). These results indicate that EIF3C promotes oncogenesis in ovarian cancer cells.
Ectopic expression of EIF3C ameliorates the tumor suppressive effect of YTHDF1 deficiency in ovarian cancer cells
Next, we overexpressed EIF3C in YTHDF1-deficient A2780 and SKOV3 cells, and EIF3C expression was restored in both cell lines (Figure 7G). YTHDF1 knockdown impaired cell growth and colony formation, whereas over-expression of EIF3C could reverse such effect (Figure 7H and Supplementary Figure S7G). Consistently, both cell migration and invasion suppressed by YTHDF1 deficiency were re-established after EIF3C overexpression (Figure 7I and J). These results suggest that EIF3C is a critical downstream target of YTHDF1 to facilitate ovarian cancer progression.
To test whether the interaction between YTHDF1 and EIF3C is specific in ovarian cancer progression, we evaluated the expression of several other subunits of EIF3 complex upon YTHDF1 knockdown and found that none of these subunits was affected (Supplementary Figure S7H). However, overexpression of EIF3A, 3B, or 3D alone in YTHDF1-deficient cells indeed promoted cell growth and migration although the effect of EIF3B on cell migration and invasion was quite weak. These results were consistent with these subunits’ roles of promoting tumorigenesis in previous studies, but also indicated that these subunits were not directly involved in YTHDF1-mediated tumorigenesis in ovarian cancer (Supplementary Figure S7I–L). Finally, we analyzed the correlation between EIF3C protein expression and YTHDF1 protein expression in ovarian cancer tissues. As expected, the protein abundance of YTHDF1 was positively correlated to the expression of EIF3C (Figure 7K and Supplementary Figure S7M). Taken together, our data confirm that EIF3C is critical for YTHDF1 to relay the oncogenic signal in ovarian cancer. Thus, we propose that increased YTHDF1 expression enhances overall protein synthesis by regulating EIF3C translation and promotes tumorigenesis in ovarian cancer cells (Figure 7L).
DISCUSSION
Dysregulation of m6A methylation closely correlates with various cancers’ development. And some studies also reported that the m6A methyltransferases (writers) or demethylases (erasers) either enhanced or suppressed tumorigenesis in different cancers (13,14,16,30–32). For example, the m6A demethylase FTO promotes leukemic oncogene-mediated cell transformation and leukemogenesis. FTO inhibitors suppress proliferation and induce apoptosis of human acute myeloid leukemia (AML) cells, thus exhibits a potential therapeutic for AML (13,33).
The biological function of m6A RNA methylation is highly dependent on the cellular context. A previous study demonstrated the METTL3 promotes ovarian carcinoma growth and invasion, whereas it was independent of its methyltransferase activity (34). In our study, we found that YTHDF1 gene was frequently amplified and up-regulated in high-grade serous ovarian cancer in TCGA cohorts. Moreover, YTHDF1 was also amplified and its expression increased accordingly in many cancer types, suggesting that YTHDF1 might be an important oncogene and which is selected during cancer evolution. Our finding is consistent with the role of YTHDF1 in non-small cell lung cancer (35) and oncogene expression driven by gene amplification might be a key evemt during cancer evolution (36). By analyzing public ovarian cancer databases, we found that YTHDF1 expression correlated with tumor grades, FIGO stages, and overall survival. Although the YTHDF1 expression was also reported to be up-regulated in both colorectal cancer and hepatocellular carcinoma (37,38), its detail functions and mechanisms in cancers were unclear. Our study demonstrated that YTHDF1 was crucial for proliferation and metastasis of ovarian cancer cells A2780 and SKOV3 both in vitro and in vivo. Albeit A2780 and SKOV3 are most frequently used for ovarian cancer studies, particularly for high-grade serous ovarian cancer, they are probably originated from endometrioid or clear cell ovarian cancer (39). These models might have some limitations to dissect the tumorigenesis of ovarian cancer precisely, but the functions of m6A regulators in these ovarian cancer cell lines are informative to our understanding of how m6A regulates ovarian cancer progression. Our results suggest that targeting YTHDF1 might represent a promising strategy for ovarian cancer treatment. Nevertheless, selection of the cell line models for the distinct cancer subtypes needs to be considered to increase the robustness of preclinical studies in future investigations.
To dissect the mechanisms of YTHDF1 in ovarian cancer, we applied a multi-omics screening strategy by combining m6A-seq, RIP-seq and eCLIP-seq as well as RNA-seq. m6A methylome in ovarian cancer cells was analyzed and the targets of YTHDF1 were identified. Results from eCLIP-seq showed that YTHDF1-binding sites were also distributed in the coding region and untranslated regions, as supported by the findings that the YTHDF1-binding sites in dendritic cells are enriched in the coding region as well as untranslated regions, and YTHDF1 preferentially binds the sites enriched near the stop codon and 3′ UTR in the mouse hippocampus (24,40). Moreover, m6A marks appear in the start codon regions and coding regions were also observed in the other tissues, such as cerebral cerebellum and muscles (41–43). YTHDF1 is known to regulate mRNA translation by recruiting the initiation factor EIF3 (5). However, YTHDF1 was reported to bind to the m6A site in CDS region of Snail mRNA and enhance its translation by recruiting the translation elongation factor eEF-2 (44), indicating that YTHDF1 could function via regulating translation elongation as well as translation initiation. Intriguingly, about 40% of targets from RIP overlapped with m6A-containing RNAs, but this could not prove that YTHDF1 is prone to binding unmethylated mRNAs because we did not exclude the indirect or non-specific targets from RIP results. Moreover, our YTHDF1 CLIP results collected the motif of m6A, supporting the interaction between YTHDF1 and m6A-marked transcripts. Nearly 30% peaks of YTHDF1 CLIP results were overlapped with m6A peaks in the mouse hippocampus (40), suggesting that YTHDF1 might regulate gene expression via an m6A-independent manner. Interestingly, the m6A reader YTHDF3 serves as an RNA binding protein bound to FOXO3 mRNA and promoted its translation independent of m6A modification (45). Thus, other potential mechanisms of YTHDF1 require further investigations. The YTH domain family proteins including YTHDF1 and YTHDF2 directly bind and recognize m6A methylation on RNA. In the cytoplasm, YTHDF1 promotes its targets’ translation by recruiting initiation factors and facilitating ribosome loading, but YTHDF2 is prone to induce degradation of its targets by localizing these targets to processing bodies. In Hela cells, YTHDF1 shared only 46% of the targets with YTHDF2 as reported in previous studies. Though YTHDF1 and YTHDF2 share the YTH domain which is responsible for interacting m6A sites on RNA, they have other different functions for their different domains. For example, YTHDF2 promotes its targets’ degradation by recruiting CCR4–NOT deadenylase complex, and it is YTHDF2 N region but not the YTHDF2 C region (the YTH domain) that interacts with CNOT1 directly, demonstrating the critical role of YTHDF2 N region for its function. Whether the different domains affect the localization of the YTH domain family proteins and their recognition of different m6A-containing RNAs needs to be further investigated. In different cellular contexts, m6A-modified RNAs might be recognized by the different YTH domain proteins and other factors might be also involved in the recognition. In our study, we found that knockdown of YTHDF2 could not change the mRNA level of EIF3C (data not shown). These data support that EIF3C is the direct target of YTHDF1 in ovarian cancer cells.
In our study, transcriptom analysis upon YTHDF1 knockdown in ovarian cancer cells revealed that the level of most YTHDF1-binding transcripts were unchanged, suggesting that YTHDF1 might control mRNA translation in ovarian cancer cells. Our data suggested that YTHDF1 could regulate the translation of EIF3C mRNA and overall translational output in ovarian cancer cells. m6A methylome analysis also identified that multiple translation initiation factors were modified by m6A, indicating m6A modification could directly regulate the expression of translation associated factors. Global translation rates are generally enhanced in cancer cells and altered translational control has been a fundamental response to oncogenic stimulation (46). Various factors involved in translation control have been linked to cancer progression (47–49). For example, EIF3A was shown to be overexpressed in in breast, cervix, lung and stomach cancers and the other subunits including 3B, 3C, and 3H were also up-regulated in specific cancers (47,50–53). Here, we showed that EIF3C expression was modified by m6A and regulated by YTHDF1 in ovarian cancer cells. Moreover, EIF3C was essential for growth and migration of ovarian cancer cells. EIF3C was also involved in metastasis of cervical cancer (54), overexpression of EIF3C partially rescued the phenotypes of YTHDF1-deficient cells in vitro, demonstrating the importance of EIF3C in ovarian cancer development. However, our results also showed that EIF3C could not completely rescue the effect of YTHDF1 knockdown on ovarian cancer cell metastasis and cell growth, suggesting that other factors might be responsible for YTHDF1 regulation. Indeed, we found many other candidates as the targets of YTHDF1 and these candidates also involved in metastasis of cancers. YTHDF1 might regulate multiple genes’ expression and the action of YTHDF1 on ovarian cancer might be a global effect of various targets, which needs to be further validated.
In summary, our study identifies the translation initiation factor EIF3C is the direct target of YTHDF1 in ovarian cancer cells. YTHDF1 controls EIF3C’s translation in an m6A-dependent manner and affect the overall protein translation as an oncogenic regulator. Thus, targeting YTHDF1 might be a promising candidate for ovarian cancer therapy.
DATA AVAILABILITY
All datasets generated in this work have been deposited to the Sequenced Read Archive (SRA) under accession number PRJNA543980, PRJNA543997, PRJNA544021, and PRJNA544187.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Drs.Ma Y.N., Wang X.S. and Gao Y.F. (CAMS & PUMC, Beijing) to support this study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2018YFC1313400 to P.Y.]. Funding for open access charge: National Key R&D Program of China: [2018YFC1313400 to P.Y.]
Conflict of interest statement. None declared.
REFERENCES
- 1. Roundtree I.A., Evans M.E., Pan T., He C.. Dynamic RNA modifications in gene expression regulation. Cell. 2017; 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lin S., Choe J., Du P., Triboulet R., Gregory R.I.. The m(6)a methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016; 62:335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lesbirel S., Viphakone N., Parker M., Parker J., Heath C., Sudbery I., Wilson S.A.. The m(6)A-methylase complex recruits TREX and regulates mRNA export. Sci. Rep. 2018; 8:13827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. et al.. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C.. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. et al.. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H. et al.. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G. et al.. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liao S., Sun H., Xu C.. YTH domain: a family of N(6)-methyladenosine (m(6)A) readers. Genomics Proteomics Bioinformatics. 2018; 16:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C.. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017; 27:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vu L.P., Pickering B.F., Cheng Y., Zaccara S., Nguyen D., Minuesa G., Chou T., Chow A., Saletore Y., MacKay M. et al.. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017; 23:1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Batista Pedro J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley Donna M., Lujan E., Haddad B., Daneshvar K. et al.. m6A RNA modification controls cell fate transition in mammalian embryonic stem Cells. Cell Stem Cell. 2014; 15:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao B.S., Wang X., Beadell A.V., Lu Z., Shi H., Kuuspalu A., Ho R.K., He C.. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017; 542:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma J.-z., Yang F., Zhou C.-c., Liu F., Yuan J.-h., Wang F., Wang T.-t., Xu Q.-g., Zhou W.-p., Sun S.-h.. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulatingN6-methyladenosine-dependent primary microRNA processing. Hepatology. 2017; 65:529–543. [DOI] [PubMed] [Google Scholar]
- 15. Choe J., Lin S., Zhang W., Liu Q., Wang L., Ramirez-Moya J., Du P., Kim W., Tang S., Sliz P. et al.. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018; 561:556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weng H., Huang H., Wu H., Qin X., Zhao B.S., Dong L., Shi H., Skibbe J., Shen C., Hu C. et al.. METTL14 inhibits hematopoietic Stem/Progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018; 22:191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu J., Eckert M.A., Harada B.T., Liu S.M., Lu Z., Yu K., Tienda S.M., Chryplewicz A., Zhu A.C., Yang Y. et al.. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018; 20:1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bowtell D.D., Bohm S., Ahmed A.A., Aspuria P.J., Bast R.C. Jr, Beral V., Berek J.S., Birrer M.J., Blagden S., Bookman M.A. et al.. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer. 2015; 15:668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tan T.Z., Yang H., Ye J., Low J., Choolani M., Tan D.S., Thiery J.P., Huang R.Y.. CSIOVDB: a microarray gene expression database of epithelial ovarian cancer subtype. Oncotarget. 2015; 6:43843–43852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011; 474:609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cancer Genome Atlas Research, N. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M.. The cancer genome atlas Pan-Cancer analysis project. Nat. Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li L., Luo Q., Xie Z., Li G., Mao C., Liu Y., Wen X., Yin N., Cao J., Wang J. et al.. Characterization of the expression of the RNA binding protein eIF4G1 and its clinicopathological correlation with serous ovarian cancer. PLoS One. 2016; 11:e0163447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K. et al.. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han D., Liu J., Chen C., Dong L., Liu Y., Chang R., Huang X., Liu Y., Wang J., Dougherty U. et al.. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019; 566:270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhuang M., Li X., Zhu J., Zhang J., Niu F., Liang F., Chen M., Li D., Han P., Ji S.J.. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 2019; 47:4765–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emmanuel R., Weinstein S., Landesman-Milo D., Peer D.. eIF3c: a potential therapeutic target for cancer. Cancer Lett. 2013; 336:158–166. [DOI] [PubMed] [Google Scholar]
- 27. Lee H., Chen C., Ho C., Lee S., Chang C., Chen K., Jou Y.. EIF3C-enhanced exosome secretion promotes angiogenesis and tumorigenesis of human hepatocellular carcinoma. Oncotarget. 2018; 9:13193–13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li T., Li S., Chen D., Chen B., Yu T., Zhao F., Wang Q., Yao M., Huang S., Chen Z. et al.. Transcriptomic analyses of RNA-binding proteins reveal eIF3c promotes cell proliferation in hepatocellular carcinoma. Cancer Sci. 2017; 108:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Min J.. Structural basis for the discriminative recognition ofN6-Methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 2015; 290:24902–24913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheng M., Sheng L., Gao Q., Xiong Q., Zhang H., Wu M., Liang Y., Zhu F., Zhang Y., Zhang X. et al.. The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-kappaB/MYC signaling network. Oncogene. 2019; 38:3667–3680. [DOI] [PubMed] [Google Scholar]
- 31. Song H., Feng X., Zhang H., Luo Y., Huang J., Lin M., Jin J., Ding X., Wu S., Huang H. et al.. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019; 15:1419–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang C., Samanta D., Lu H., Bullen J.W., Zhang H., Chen I., He X., Semenza G.L.. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E2047–E2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang Y., Su R., Sheng Y., Dong L., Dong Z., Xu H., Ni T., Zhang Z.S., Zhang T., Li C. et al.. Small-Molecule targeting of oncogenic FTO Demethylase in acute myeloid leukemia. Cancer Cell. 2019; 35:677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hua W., Zhao Y., Jin X., Yu D., He J., Xie D., Duan P.. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol. Oncol. 2018; 151:356–365. [DOI] [PubMed] [Google Scholar]
- 35. Shi Y., Fan S., Wu M., Zuo Z., Li X., Jiang L., Shen Q., Xu P., Zeng L., Zhou Y. et al.. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat. Commun. 2019; 10:4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chowdhry S., Zanca C., Rajkumar U., Koga T., Diao Y., Raviram R., Liu F., Turner K., Yang H., Brunk E. et al.. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature. 2019; 569:570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nishizawa Y., Konno M., Asai A., Koseki J., Kawamoto K., Miyoshi N., Takahashi H., Nishida N., Haraguchi N., Sakai D. et al.. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018; 9:7476–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao X., Chen Y., Mao Q., Jiang X., Jiang W., Chen J., Xu W., Zhong L., Sun X.. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018; 21:859–868. [DOI] [PubMed] [Google Scholar]
- 39. Beaufort C.M., Helmijr J.C., Piskorz A.M., Hoogstraat M., Ruigrok-Ritstier K., Besselink N., Murtaza M., van I.W.F., Heine A.A., Smid M. et al.. Ovarian cancer cell line panel (OCCP): clinical importance of in vitro morphological subtypes. PLoS One. 2014; 9:e103988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shi H., Zhang X., Weng Y.L., Lu Z., Liu Y., Lu Z., Li J., Hao P., Zhang Y., Zhang F. et al.. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018; 563:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 42. Ma C., Chang M., Lv H., Zhang Z.-W., Zhang W., He X., Wu G., Zhao S., Zhang Y., Wang D. et al.. RNA m6A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018; 19:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tao X., Chen J., Jiang Y., Wei Y., Chen Y., Xu H., Zhu L., Tang G., Li M., Jiang A. et al.. Transcriptome-wide N (6) -methyladenosine methylome profiling of porcine muscle and adipose tissues reveals a potential mechanism for transcriptional regulation and differential methylation pattern. BMC Genomics. 2017; 18:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin X., Chai G., Wu Y., Li J., Chen F., Liu J., Luo G., Tauler J., Du J., Lin S. et al.. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019; 10:2065. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45. Zhang Y., Wang X., Zhang X., Wang J., Ma Y., Zhang L., Cao X.. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Truitt M.L., Ruggero D.. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer. 2016; 16:288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hershey J.W. The role of eIF3 and its individual subunits in cancer. Biochim. Biophys. Acta. 2015; 1849:792–800. [DOI] [PubMed] [Google Scholar]
- 48. Spilka R., Ernst C., Mehta A.K., Haybaeck J.. Eukaryotic translation initiation factors in cancer development and progression. Cancer Lett. 2013; 340:9–21. [DOI] [PubMed] [Google Scholar]
- 49. Liu S., Hausmann S., Carlson S.M., Fuentes M.E., Francis J.W., Pillai R., Lofgren S.M., Hulea L., Tandoc K., Lu J. et al.. METTL13 Methylation of eEF1A increases translational output to promote tumorigenesis. Cell. 2019; 76:491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yin J.Y., Zhang J.T., Zhang W., Zhou H.H., Liu Z.Q.. eIF3a: A new anticancer drug target in the eIF family. Cancer Lett. 2018; 412:81–87. [DOI] [PubMed] [Google Scholar]
- 51. Liu R.Y., Dong Z., Liu J., Yin J.Y., Zhou L., Wu X., Yang Y., Mo W., Huang W., Khoo S.K. et al.. Role of eIF3a in regulating cisplatin sensitivity and in translational control of nucleotide excision repair of nasopharyngeal carcinoma. Oncogene. 2011; 30:4814–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yin J.Y., Shen J., Dong Z.Z., Huang Q., Zhong M.Z., Feng D.Y., Zhou H.H., Zhang J.T., Liu Z.Q.. Effect of eIF3a on response of lung cancer patients to platinum-based chemotherapy by regulating DNA repair. Clin. Cancer Res. 2011; 17:4600–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang X., Wang H., Zhao S., Sun P., Wen D., Liu T., Liu H., Yang Z., Ma Z.. Eukaryotic translation initiation factor EIF3H potentiates gastric carcinoma cell proliferation. Tissue Cell. 2018; 53:23–29. [DOI] [PubMed] [Google Scholar]
- 54. Hu C., Wang Y., Li A., Zhang J., Xue F., Zhu L.. Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR-545/EIF3C axis. J. Cell. Physiol. 2019; 234:9225–9232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated in this work have been deposited to the Sequenced Read Archive (SRA) under accession number PRJNA543980, PRJNA543997, PRJNA544021, and PRJNA544187.