Abstract
By interacting with proteins and nucleic acids, the vast family of mammalian circRNAs is proposed to influence many biological processes. Here, RNA sequencing analysis of circRNAs differentially expressed during myogenesis revealed that circSamd4 expression increased robustly in mouse C2C12 myoblasts differentiating into myotubes. Moreover, silencing circSamd4, which is conserved between human and mouse, delayed myogenesis and lowered the expression of myogenic markers in cultured myoblasts from both species. Affinity pulldown followed by mass spectrometry revealed that circSamd4 associated with PURA and PURB, two repressors of myogenesis that inhibit transcription of the myosin heavy chain (MHC) protein family. Supporting the hypothesis that circSamd4 might complex with PUR proteins and thereby prevent their interaction with DNA, silencing circSamd4 enhanced the association of PUR proteins with the Mhc promoter, while overexpressing circSamd4 interfered with the binding of PUR proteins to the Mhc promoter. These effects were abrogated when using a mutant circSamd4 lacking the PUR binding site. Our results indicate that the association of PUR proteins with circSamd4 enhances myogenesis by contributing to the derepression of MHC transcription.
INTRODUCTION
Even though >90% of the mammalian genome is transcribed, only ∼2% of transcripts encode protein (1). Recent progress in high-throughput RNA-sequencing and bioinformatic analysis has found that many of the noncoding transcripts form covalently closed circular (circ)RNAs. Tens of thousands of circRNAs have been identified in different cell types, and many display tissue-specific expression patterns and evolutionary conservation (2,3). CircRNAs have been shown to influence cellular processes such as proliferation, differentiation, senescence, as well as maintenance of cell pluripotency (4–6). Accordingly, several circRNAs have been implicated in physiology and disease (7–11). Despite this progress, the functions of circRNAs in most biological processes are unknown.
The skeletal muscle comprises nearly 40% of adult tissue, initially forming during embryonic development and continually regenerating throughout life via myogenesis (12). This process involves the specification of mesodermal precursor cells (satellite cells) into myoblasts, followed by subsequent differentiation and fusion of these cells into multinucleated myotubes. Early myogenesis is governed transcriptionally by the myogenic regulatory factors (MRFs) MYOD and MYF5, which mediate the initial specification of skeletal myoblasts, and by myogenin (MYOG), MRF4, and myocyte-specific enhancer factors (MEF2A and MEF2C), which induce differentiation of these specified cells (13). Later myogenic stages involve the fusion of the mononucleated myoblasts into multinucleated myotubes, the reorganization of cytoskeletal proteins, and the transcriptional expression of myosin heavy chain (MHC) proteins, which function in muscle contraction. In addition to the myogenic transcription factors, myogenesis is tightly regulated at the post-transcriptional level through the actions of RNA-binding proteins (RBPs) and noncoding RNAs (ncRNA) such as miRNAs, lncRNAs, and circRNAs (14–16).
Numerous chronic age-associated muscular disorders, including sarcopenia, osteoarthritis, chronic heart failure, and chronic obstructive pulmonary disease (17–20), have been associated with aberrant myogenesis. We previously identified age-associated changes in circRNAs expressed in monkey skeletal muscle with advancing age and proposed that some of these circRNAs might influence muscle function during aging (21). Here, we employed RNA-sequencing analysis to identify circRNAs differentially expressed during muscle differentiation and conserved in human muscle cells. We found that depletion of the highly expressed circSamd4 delayed myogenic progression and influenced late stages of muscle differentiation. Affinity pulldown followed by proteomic analysis identified circSamd4-interacting proteins including purine-rich binding proteins (PUR) alpha and beta (PURA, PURB), two proteins that bind to and repress myosin heavy chain (Mhc) promoter sequences. We propose that the progressive elevation in circSamd4 during myogenesis contributes to sequestering PUR proteins, in turn facilitating a time-dependent derepression of the Mhc promoter and the increase in MHC production.
MATERIALS AND METHODS
Cell culture, differentiation, modulation of circSamd4 levels
Proliferating C2C12 myoblasts were cultured in growth medium (GM), DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Gibco), and antibiotics penicillin and streptomycin (Life Technologies). Proliferating, subconfluent C2C12 myoblasts were induced to differentiate by culture to high density and switch to differentiation medium [(DM), DMEM with 2% horse serum] for 3–6 days. Human myoblasts (22) were cultured in human skeletal muscle GM (Promocell) consisting of equal parts [DMEM with 10% FBS, 2 mM l-glutamine, 25 ng/ml FGFβ, 1 ng/ml EGF, and penicillin and streptomycin] and [Ham's F10 media containing 20% FBS] and antibiotics penicillin and streptomycin, and differentiated as described above with DM media. For silencing, 50 nM of control (Ctrl) or circSamd4 siRNAs (IDT) were transfected using Lipofectamine® 2000 (Thermo Fisher Scientific) into proliferating myoblasts 12 h before inducing differentiation. For overexpression, 1 μg of empty vector pcDNA3, pcDNA3-circSamD4 or deletion mutant pcDNA3-circSamD4Δ3 was transfected by electroporation (Amaxa Biosystems™) before inducing differentiation.
Identification of circRNA from RNA-seq analysis
The RNA from GM and DM cells was purified using the miRNeasy Mini kit (Qiagen) following the manufacturer's instructions. The RNA quality and quantity were assessed with the Agilent 2100-Bioanalyzer followed by removal of rRNA with the rRNA Depletion Nano kit (Qiagen). The cDNA library was prepared with the Ovation RNA-Seq System V2 (NuGEN) kit, and sequencing was performed on an Illumina HiSeq 2500 instrument (23); data are deposited in GSE92632 and GSE136004. Sequencing reads were mapped to the mouse genome (mm9) using TopHat2 (version 2.0.14) followed by identification of non-linear fusion junction reads using TopHat2. CircRNAs expressed in GM and DM samples were identified with the CIRCexplorer program (v1.10) as described previously (23).
RNA isolation and reverse transcription followed by quantitative PCR (RT-qPCR) analysis
RNA was isolated using TRIzol (Thermo Fisher Scientific) and subjected to reverse transcription (RT) and real-time quantitative (q)PCR analysis. After sequencing, divergent primers were designed to measure the relative expression levels of circRNAs and identify the junction sequence. Total RNA (2 μg) isolated from proliferating or differentiated cells was either left untreated or treated with RNase R (20 units, Epicentre) for 30 min at 37°C; RNA samples were then assayed by RT-qPCR analysis. Copy number was calculated by comparing Ct values of circSamd4 with the concentration of transcripts of known abundance in cells, assuming 1500 copies of Gapdh mRNA per cell; these estimates were comparable to those described (24,25). Forward and reverse primers (Supplementary Table S1) were used to sequence PCR products and identify the junction sequence.
Expression of circRNAs in old and young human skeletal muscle
Quadriceps skeletal muscle was collected from lean elderly individuals (age 79 years ± 2; n = 7) intraoperatively during total knee arthroplasty surgery, and from lean young adults (age 25 years ± 4; n = 7) using a Bergström needle (UK Research Ethics Committee 14/ES/1044). The muscle tissue was snap-frozen in liquid nitrogen and ∼100 mg ground into a fine powder using a pestle and mortar under liquid nitrogen. Powdered tissue was immediately transferred to 1 ml of Trizol® reagent (Thermo Fisher Scientific) and homogenized using a Qiagen Tissue Ruptor. Total RNA was then extracted as described in the manufacturer's protocol (Trizol, Thermo Fisher Scientific) and resuspended in 30 μl of RNase-free water (Thermo Fisher Scientific). RNA concentration was quantified using a Nanodrop 2000 (Life Technologies).
For reverse transcription (RT), cDNA was synthesized from 60 ng of template RNA using a Tetro cDNA Synthesis Kit (Bioline) with random hexamers, as per the manufacturer's instructions. Relative RNA abundance was determined by two-step RT-qPCR analysis normalized to GAPDH mRNA levels; qPCR analysis was performed using a SensiFAST™ SYBR® No-ROX Kit (Bioline) following the manufacturer's protocol, using the primers listed in the Supplementary Figure S1; for GAPDH mRNA, the primers (sequence not disclosed) were from PrimerDesign Ltd. All reactions were performed using a Bio-Rad cycler (Bio-Rad) with a total reaction volume of 5 μl. Samples were assayed in triplicate and a non-template control was included for each gene of interest.
Plasmids
Briefly, plasmids to overexpress the circSamd4 (pcDNA3-circSamd4) and deletion mutant circSamd4Δ3 (pcDNA3-circSamd4Δ3) were constructed by cloning the circSamd4 full and truncated sequences in the circRNA mini vector ZKSCAN1 (24) (Addgene #60649). Genomic DNA was isolated using DNeasy Blood & Tissue Kit (69504, Qiagen) and 25 ng of genomic DNA was used to amplify ∼519-bp genomic sequences upstream and downstream of circSamd4 genomic coordinates using forward and reverse primers actgGGATCCTCGTGCTTGCGCCGGTTCCAA and actgaagcttGAATCATTAACCAATGGCAAC, respectively. The PCR products were cloned into the pcDNA3.0 circRNA mini vector ZKSCAN1. Constructs were verified by sequencing.
Antisense oligomer (ASO) pulldown and mass spectrometry analysis
For ASO pulldown, whole-cell lysates were incubated with 3 μg of a biotinylated control ASO (5′Biot-TTGGAACCGGCGCAAGCACGAGAATCATTAACCAATGGCA), or an ASO complementary to the circSamd4 junction (Biot-TGGAACCGGCGCAAGCACGAGAATCATTAACCAATGGCAA) in 1× TENT buffer (10 mM Tris–HCl at pH 8.0, 1 mM EDTA at pH 8.0, 250 mM NaCl, 0.5% [v/v] Triton X-100) containing fresh protease and RNase inhibitors for 1 h at 25°C with rotation. The biotin-ASO complexes were then pulled down following addition of Streptavidin-coupled Dynabeads (50 μl, Invitrogen) and incubation for 30 min at 25°C with rotation. After extensively washing the Dynabeads, the samples were divided such that RNA and protein were isolated using TriPure (Sigma-Aldrich) and 2× SDS elution dye, respectively, added directly to the washed Dynabead-biotin-ASO complexes. The enrichment of RNAs in the circSamD4-ASO pulldown was assessed by RT-qPCR analysis and the circSamD4-ASO-associated proteins were subjected to mass spectrometry analysis. The presence of these enriched proteins was confirmed by Western blot analysis with and without digestion with RNase A, RNase R or both RNases.
Mass spectrometry raw data files were collected from analysis of each of four pulldown samples from human myoblasts and from each of four pulldown samples from mouse myoblasts. Proteins were searched against the Human UniProtKB protein sequence database (20 608 entries, 2016) and the mouse UniProtKB mouse protein sequences database (16 685 entries, 2016), respectively, obtained from NCBI website using the Proteome Discoverer 1.4 software (Thermo Scientific), using the SEQUEST and percolator algorithms. The searches were performed with the following parameters: Carbamidomethylation (+57.021 Da) of cysteines was fixed modification, and Oxidation and Deamidation Q/N-deamidated (+0.98402 Da) were set as dynamic modifications; the minimum peptide length was specified to be five amino acids; the precursor mass tolerance was set to 15 ppm, and fragment mass tolerance was set to 0.05 Da. The maximum false peptide discovery rate was specified as 0.01. The resulting Proteome Discoverer Report and assembled proteins with peptides sequences and matched spectrum counts are summarized in Supplementary Table S2.
Western blot analysis
Whole-cell lysates prepared in RIPA buffer were separated by SDS-PAGE and transferred onto nitrocellulose membranes (Invitrogen iBlot Stack). Incubations with primary antibodies recognizing MHC, MYOG, HSP90, LMNB2 (Lamin B) (Santa Cruz Biotech), MEF2C (Millipore), PURA, PURB, YB1, YBOX3 (Abcam) and HNRNPM (Invitrogen) were followed by incubations with the appropriate secondary antibodies conjugated with horseradish peroxidase (GE Healthcare). Signals were developed using Enhanced Chemiluminescence (ECL), and digitized images were captured using Kwik Quant Imager (Kindle Biosciences).
Immunofluorescence and single-molecule in situ hybridization microscopy
Cells were plated in 35-mm imaging dishes with glass bottoms (Idibi) in 100 μl PBS, fixed with 100 μl of 4% paraformaldehyde for 15 min at room temperature, washed twice with 100 μl of PBS, permeabilized and blocked with 0.2% Triton X-100 and 1% goat serum, incubated with 50 μl of anti-myosin antibody (1:500 Santa Cruz Biotech) and washed twice in PBS, incubated with 50 μl anti-mouse Cy3/AlexaFluor488 secondary antibody (Thermo Scientific), diluted 1:300 in PBS, and finally washed twice in PBS. Nuclei were stained using ProLong® Gold Antifade Mountant with DAPI (Thermo Scientific) and signals were visualized using a confocal microscope (ZEISS 710 LSM or 880 LSM) using a 65× objective for image acquisition.
For single-molecule in situ hybridization analysis, target probe sequences (BaseScope), preamplifier, amplifier, wash buffer, and target retrieval buffers are proprietary (Advanced Cell Diagnostics). Undifferentiated C2C12 myoblasts were harvested by standard methods, counted, and seeded in glass-bottom dishes (Cellvis, 55 mm, #0) at 300 000 cells per dish and cultured at 37°C/5% CO2. Twenty-four hours after seeding, undifferentiated cells were fixed (undifferentiated controls) or media was changed to DMEM with 2% horse serum and Penicillin/Streptomycin for differentiation. Differentiation proceeded for 4 days before fixation. Samples were fixed in 10% neutral buffered formalin for 30 min at 25°C, washed three times with PBS, and dehydrated in 50%, then 70%, and then 100% ethanol. Fixed and dehydrated samples were stored in 100% ethanol at –20°C until staining. Samples were rehydrated in 70% ethanol, followed by 50% ethanol and finally in PBS. Samples were incubated with hydrogen peroxide for 10 min at 25°C; after rinsing twice in PBS, samples were hybridized and stained following the manufacturer's recommendations. Briefly, samples were treated with Protease III (ACD Bio) at 1:15 in PBS for 10 min at 25°C followed by rinsing in twice in PBS. Hybridization cassette (ACD Bio) and HybEZ oven (ACD Bio) were used for subsequent hybridization, preamplification and amplification. Single ZZ probes (ACD Bio, BaseScope) targeting linear Samd4 mRNA (Ch1 – Green) or circular circSamd4 (Ch2 - Red) were designed to target outside the circularized sequence or to the backsplice junction, respectively. Samd4 mRNA or positive control [Ppib (Ch1) or Polr2a (Ch2)] probes were hybridized in the HybEZ oven set at 40°C for 2 h, followed by preamplification and amplification steps based on the ACD protocol. Chromogenic detection was performed using Duplex Green/Fast Red (ACD). Samples were then counterstained with hematoxylin 7211 (Richard Allen) for 10 s, rinsed briefly in water, then in 0.02% ammonia for 30 s, followed by another rinse with water. Coverslips were added with mounting medium (Vectamount) and cured for 24 h before imaging. Brightfield images were taken at 20× or 40× with a Nikon upright microscope equipped with a digital RGB camera.
Ribonucleoprotein immunoprecipitation (RIP) analysis
The association of circSamd4 with endogenous PURA and PURB in cells was analyzed by ribonucleoprotein (RNP) immunoprecipitation (RIP) as described (27). Briefly, cells were lysed in polysome extraction buffer (PEB; 20 mM Tris–HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2 and 0.5% NP-40) supplemented with RNase and protease inhibitors. The cytoplasmic lysates were incubated with PURA (Abcam) and PURB (Abcam) or control IgG (Santa Cruz Biotech) antibodies at 4°C followed by incubation with protein-A-sepharose for 2 h at 4°C with rotation. After washes with ice-cold NT2 buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40], the RNA in the RNP complexes was isolated using TriPure and used for RT-qPCR analysis as described above.
Biotin pulldown analysis
C2C12 whole-cell lysates (200 μg) were incubated with 250 ng of non-overlapping biotinylated RNA fragments (F1 to F8) custom-made from IDT to cover the entire sequence of circSamd4 for 30 min at 25°C with rotation. Complexes were isolated with Streptavidin-coupled Dynabeads (Invitrogen) and the pulldown material was analyzed by western blot analysis using PURA and PURB antibodies.
Cell fractionation
Fractionation of nuclear and cytoplasmic components of C2C12 was performed according to the manufacturer's instructions (NE-PER kit, Thermo Fisher ScientificNE-PER kit, Thermo Fisher ScientificNE-PER kit, Thermo Fisher ScientificNE-PER kit, Thermo Fisher Scientific). The cytoplasmic and nuclear fractions were subjected to analysis of protein, RNA, and ribonucleoprotein complexes.
Luciferase assays
The dual MHC promoter reporter construct was obtained from GeneCopoeia and contains the MHC promoter driving expression of Gaussia luciferase with an SV40 promoter driving expression of Renilla luciferase. The reporter plasmid (pMH2-LUC-Renilla) (MPRM31645-PL01) (500 ng) along with 1 μg of either empty vector (pcDNA3), deletion mutant (pcDNA3-circSamd4Δ3) or wild-type (pcDNA3-circSamd4) plasmids, were transfected using Lipofectamine 2000® in 0.3 × 106 pre-seeded cells in a six-well plate; cells were harvested 48 h later in PLB lysis buffer (Promega) and RLuc and GLuc activities were measured by Dual Glo luciferase assay (Promega) following the manufacturer's protocol.
ChIP-qPCR analysis
ChIP assays were performed using a CHIP-IT High sensitivity kit (Active Motif, Inc.) as instructed by the manufacturer. Briefly, differentiated C2C12 cells were crosslinked with 1% formaldehyde for 10 min at 25°C, washed, and treated with glycine Stop-Fix solution. After resuspending in cell lysis buffer and incubating on ice for 10 min, pellets were sonicated to generate genomic DNA fragments. The chromatin was incubated for 16 h at 4°C with anti-PURA, anti-PURB (Abcam), or control IgG (Santa Cruz Biotechnology) antibodies. Protein G beads were then added to the antibody-chromatin complex and incubated for 3 h at 4°C. After extensive washes, the immunoprecipitated DNA complexes were eluted from the beads. DNA was purified by proteinase K digestion and the presence of Mhc II promoter (−270 to −450) regions as well as negative control Syn2 region (TTS +200) (primers listed in Supplementary Table S1) were measured by qPCR analysis following the percentage input method as described (28,29).
Statistical analysis
All experiments were repeated at least 3 times unless otherwise stated. Quantitative data are represented as the mean ± SEM, and compared statistically by unpaired student's t test, using Prism GraphPad (7.0). Statistical significance is indicated in the figures as * (P < 0.05) and ** (P < 0.005). Dedicated statistics were applied to RNA-seq data analysis as specified in the legends.
RESULTS
The levels of conserved circRNA circSamd4 increase in differentiated myoblasts
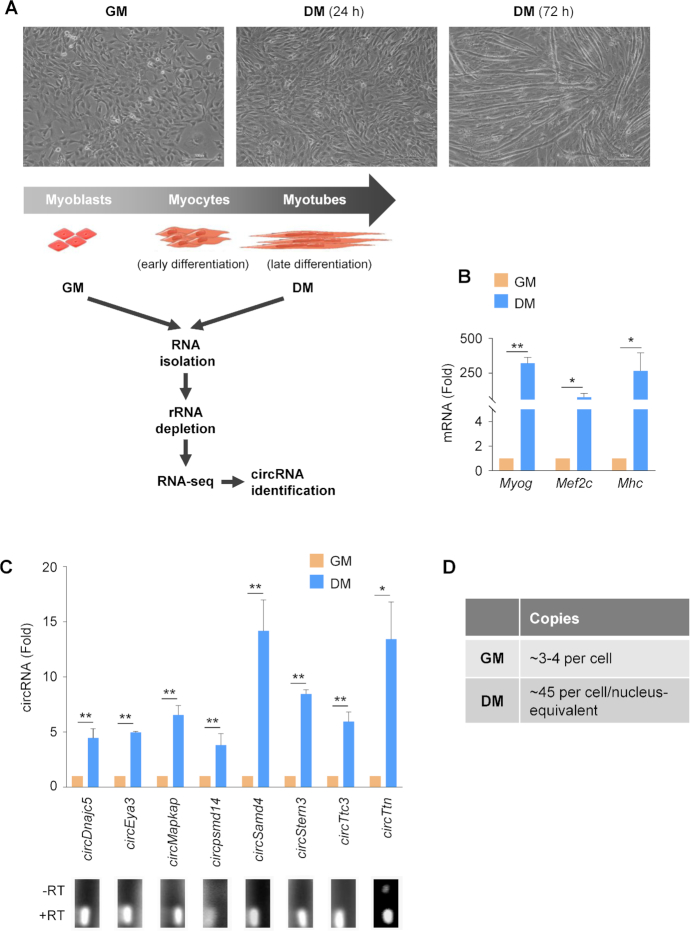
We used the mouse C2C12 myoblast model of myogenesis to investigate circRNAs implicated in skeletal muscle differentiation. C2C12 cells proliferate rapidly in normal growth media (GM), but upon reaching confluency and exposure to differentiation media (DM) in low serum, they stop dividing and progressively fuse, becoming multinucleated myotubes (myofibers) with a contractile phenotype (Figure 1A, top). To identify circRNAs differentially expressed during myogenesis, total RNA was isolated from C2C12 GM and DM (3 days) cultures and the circRNAs were identified by RNA-Seq as described (Materials and Methods) (Figure 1A, schematic; deposited in GSE92632 and GSE136004). The circRNAs were identified by the CIRCexplorer algorithm.
Figure 1.
circRNAs expressed differentially during myogenesis. (A) Myoblast differentiation program; phase-contrast micrographs depicting cultures of C2C12 mouse myoblasts in proliferating conditions [growth medium (GM)], in early myogenesis after 24 h in differentiation medium (DM), and in late myogenesis, including myotubes, after 72 h in DM. Schematic, total RNA was prepared from proliferating C2C12 cells (GM) and from C2C12 cultures differentiating for 72 h (DM); after digestion of linear RNA, RNA pools enriched in circRNAs were sequenced (GSE92632 and GSE136004), and the circRNAs most highly expressed and most differentially abundant between GM and DM were validated by reverse transcription (RT) followed by real-time quantitative (q)PCR analysis. (B) Expression levels of myogenic markers Myog, Mef2c, Mhc mRNAs in C2C12 cultures, as assessed by RT-qPCR analysis. mRNA levels were normalized to the levels of housekeeping transcript Gapdh mRNA. (C) Expression levels of the indicated circRNAs, chosen among the most differentially abundant in DM relative to GM and most highly expressed overall. RT-qPCR analysis was performed by using divergent primers (Materials and Methods) in proliferating (GM) and differentiated (DM, 72 h) cultures. mRNA levels were normalized to the levels of housekeeping transcript Gapdh mRNA. Bottom, representative PCR products on 2% agarose gels; product specificity was assessed from reactions carried out in the presence or absence of reverse transcriptase (RT). (D) Quantification of copy numbers of circSamd4 in C2C12 cultures in GM (per cell) and DM (per cell/nucleus-equivalent) by RT-qPCR analysis. Data in (B, C) are the means ± SEM from three independent experiments. *P < 0.05, **P < 0.005.
To monitor this myogenesis model, we studied the expression levels of myogenic markers MEF2C and MYOG, which regulate transcriptionally the myogenic protein expression program. The expression levels of these myogenic markers, as well as the myogenic protein myosin heavy chain (MHC), were monitored by reverse transcription (RT) followed by real-time quantitative (qPCR) analysis. As shown, the corresponding mRNAs were drastically higher in C2C12 cultures by 3 days in DM relative to proliferating C2C12 (GM) cultures (Figure 1B, graph).
Among the circRNAs upregulated in differentiated C2C12 cells, we validated circRNAs that were abundant in this paradigm and those that were highly enriched in DM relative to GM. The levels of a subset of the validated circRNAs is shown (Figure 1C, including representative PCR products). We focused further efforts on circSamd4 (circID-mmu_circ_0000529, hsa_circ_0004846), a circRNA derived from exon 3 of the mouse gene Samd4 (NM_001037221.2; exon 2 in other transcript variants from the same mouse gene) that encodes the RBP SAMD4, for a number of reasons. There is 93% sequence similarity between human circSAMD4 (circSAMD4A) and mouse circSamd4 by BLAT analysis using circBase, indicating that it is conserved between both species (Supplementary Figure S1A). Moreover, in the human myoblast line KM155 (22), expression of circSAMD4 increased similarly with myogenesis (DM relative to GM conditions; Supplementary Figure S1B) and followed similar kinetics (Supplementary Figure S1C). In addition, circSAMD4 expression levels increased with age in human skeletal muscle, while SAMD4 mRNA levels only changed moderately with age (Supplementary Figure S1D). Other connections to myogenesis include: (i) SAMD4 protein suppressed hallmarks of myotonic dystrophy (30); (ii) mutation in the mouse Samd4 gene caused myopathy and uncoupled mitochondrial respiration (31); (iii) loss of SAMD4 in mice resulted in multiple developmental defects, including delayed bone development and decreased osteogenesis (32) and (iv) circSAMD4 was recently implicated in the enhanced proliferation and stemness traits of osteosarcoma cells (33). The number of circSamd4 copies in C2C12 myoblasts as well as the nucleus-equivalent copies in C2C12 myotubes was estimated as explained in Materials and Methods and revealed ∼3–4 copies of circSamd4 per cell in GM populations, and ∼45 copies of circSamd4 per cell/nucleus-equivalent in DM conditions (Figure 1D).
circSamd4
circSamd4 is generated by circularization of exon 3 of the Samd4 gene located on chromosome 14 (mm9), mmu_circ_0000529. It is 519 nucleotides long and arises from head-to-tail splicing of exon 3 from NM_001037221.2, chr14 (hg19), region 47635951–47636470 (Figure 2A). In humans, circSAMD4 arises from backsplicing of exon 3 of SAMD4A pre-mRNA (hsa_circ_0004846, NM_015589.6); it is also located on chromosome 14, 55168779–55169298 and is 519 nucleotides long. Of note, the same exon is exon 2 in other transcript variants from the same mouse and human genes. The circSamd4 PCR product was sequenced to verify the presence of the backspliced junction (Figure 2A). Digestion using RNase R to degrade the linear RNA depleted almost all Samd4 mRNA, but not circSamd4 (Figure 2B), serving to confirm the circular nature of circSamd4. Despite the observed ∼14-fold increase in circSamd4 levels (Figures 1C and 2C) during myogenesis, Samd4 mRNA levels increased only 3-fold, suggesting that circSamd4 expression was selectively upregulated during myogenesis (Figure 2C). The relative distribution of circSamd4 in the nucleus and cytoplasm of C2C12 myoblasts and differentiated C2C12 myotubes was first assessed by RT-qPCR analysis, and included measurement of control transcripts Actb mRNA and lncRNA 7sk to monitor the preparation of cytoplasmic and nuclear RNAs, respectively. As shown (Figure 2D), circSamd4 levels increased markedly overall with differentiation, and it was found in both compartments, but was predominantly cytoplasmic.
Figure 2.
Characterization of circSamd4. (A) Schematic of exon 3 of Samd4 mRNA, located on mouse chromosome 14 and giving rise to a 519-nucleotide long circSamd4, and detail of the sequence junction. (B) RT-qPCR results showing the abundance of circSamd4 and Samd4 mRNA in total RNA from C2C12 cells (GM) that were either left untreated or treated with RNase R. RNA levels were normalized to the levels of Gapdh mRNA. (C) Relative levels of Samd4 mRNAs as measured by RT-qPCR analysis in GM and DM (72 h) C2C12 cultures. RNA levels were normalized to the levels of Gapdh mRNA in the same samples. (D) Relative levels of circSamd4 in the nuclear (Nuc) and cytoplasmic/sarcoplasmic (Cyto) compartments of C2C12 GM and DM (72 h) cultures; the quality of the fractionation was assessed by monitoring the levels of a predominantly cytoplasmic transcript (Actb mRNA) and a predominantly nuclear lncRNA (7sk). (E) Brightfield micrographs to visualize circSamd4 (red) and Samd4 mRNA (blue) in proliferating (GM) and differentiated (DM) C2C12 cultures using the BaseScope single-molecule detection system (Advanced Cell Diagnostics, CA) (Materials and Methods). Nuclei are counterstained with hematoxylin. Data in (B, C) are the means ±SEM from three or more independent experiments. **P < 0.005.
This general pattern of distribution was verified by visualizing circSamd4 as well as Samd4 mRNA in proliferating and differentiated C2C12 cultures using the BaseScope single-molecule in situ hybridization (Materials and Methods). As shown in Figure 2E, Samd4 mRNA signals were more abundant than circSamd4 signals in proliferating myoblasts. By contrast, circSamd4 signals increased overall during differentiation, and were found in both the nuclei and the cytoplasm of myotubes, but were more abundant in the cytoplasm.
circSamd4 promotes myogenesis
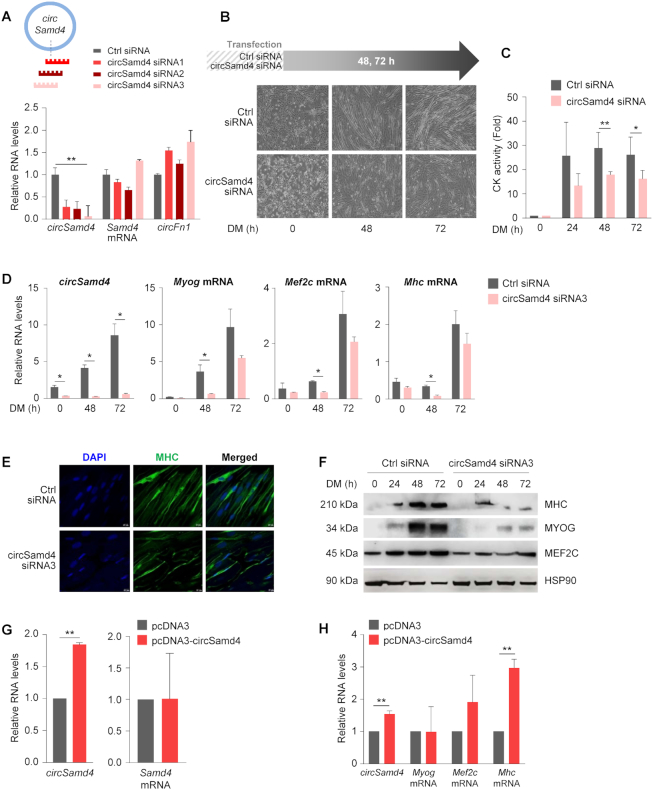
To investigate the biological function of circSamd4 in myoblast differentiation, we designed several small interfering (si)RNAs targeting circSamd4, each spanning the backsplice junction (Figure 3A). Transfection of each of the three siRNAs reduced circSamd4 levels significantly, but siRNA3 displayed the most potent reduction and did not suppress the levels of linear counterpart Samd4 mRNA or the circRNA circFn1 (Figure 3A and Supplementary Figure S2A). Given that circSamd4-directed siRNA1 and siRNA2 reduced Samd4 mRNA levels moderately in C2C12 cells (Figure 3A, Supplementary Figure S2A–C), circSamd4 siRNA3 was chosen for subsequent studies.
Figure 3.
Silencing circSamd4 reduces the levels of myogenic markers. (A) Schematic representation of 3 siRNAs designed to target the circSamd4 junction. RT-qPCR analysis of the indicated circular and linear RNAs; C2C12 in GM were transfected with siRNAs, 8 h later the medium was switched to DM and 48 h after that, cells were collected for analysis. RNA levels were normalized to the levels of Gapdh mRNA. (B–D) Effect of silencing circSamd4 on C2C12 differentiation. Ctrl and circSamd4 siRNAs were transfected into C2C12 (GM) cells and differentiation was initiated 24 h later (time 0 h); 48 and 72 h into DM, phase-contrast micrographs were taken to monitor myotube formation (B), the levels of creatine kinase (CK) were measured at 0, 24, 48 and 72 h (C), and RNA was extracted to measure by RT-qPCR analysis the levels of circSamd4 as well as myogenic markers Myog, Mef2c, and Mhc mRNAs, normalized to Gapdh mRNA levels (D). (E, F) The effect of silencing circSAMD4 in human KM155 myoblasts (Supplementary Figure S1) on myogenesis was measured by immunofluorescence analysis of the myotube marker MHC in DM (72 h); nuclei were labeled with DAPI (E), and the levels of the myogenic proteins MHC, MYOG, and MEF2C were measured by western blot analysis (F). (G, H) Forty-eight hours after transfecting pcDNA3 (empty control) or pcDNA3-circSamd4 into C2C12 cells, RT-qPCR analysis was used to assess the expression levels of circSamd4 and Samd4 mRNA (G) as well as those of myogenic markers Myog, Mef2c, and Mhc mRNAs, which were normalized to the levels of Gapdh mRNA (H). Data in (A, C, D, G, H) are the means ± SEM from three independent experiments. *P < 0.05, **P < 0.005.
After reducing circSamd4 levels by transfection with circSamd4 siRNA followed by exposure to DM conditions, microscopic analysis revealed that silencing circSamd4 delayed myotube formation (Figure 3B). The activity levels of the myogenesis marker creatine kinase (CK) (Figure 3C), as well as the abundance of the myogenic markers Myog, Mef2c and Mhc mRNAs (Figure 3D) were also decreased when circSamd4 was silenced. These results suggest that circSamd4 contributes to the progression through myogenesis. In human KM155 myoblasts, knockdown of circSAMD4 by siRNA3 similarly suppressed myogenesis, as determined by monitoring the levels of SAMD4, MYOG, and MHC mRNAs as well as CK activity (Supplementary Figure S2D–G), suggesting that circSAMD4 also promoted human myogenesis. Immunofluorescence analysis provided additional evidence that myogenesis, as assessed by the myogenic marker MHC, was impaired in human myoblasts following circSAMD4 silencing (Figure 3E). Western blot analysis further revealed that MHC and other myogenic markers (MYOG, MEF2C) were also reduced in human myoblasts in DM conditions after circSAMD4 siRNA transfections (Figure 3F). We then assessed whether ectopic overexpression of circSamd4 in C2C12 influenced myogenesis by employing an expression vector designed to specifically express circRNAs (Materials and Methods). Although the magnitude of circSamd4 overexpression achieved in C2C12 cells was modest (Figure 3G, left), it enhanced Mhc mRNA levels (Figure 3H), while it did not raise endogenous Samd4 mRNA levels or ectopic linear exon 3 levels (Figure 3G, right; Supplementary Figure S3). Collectively, these data further support the notion that circSamd4 contributes to myogenesis.
circSamd4 associates with PURA and PURB, transcriptional repressors of MHC production
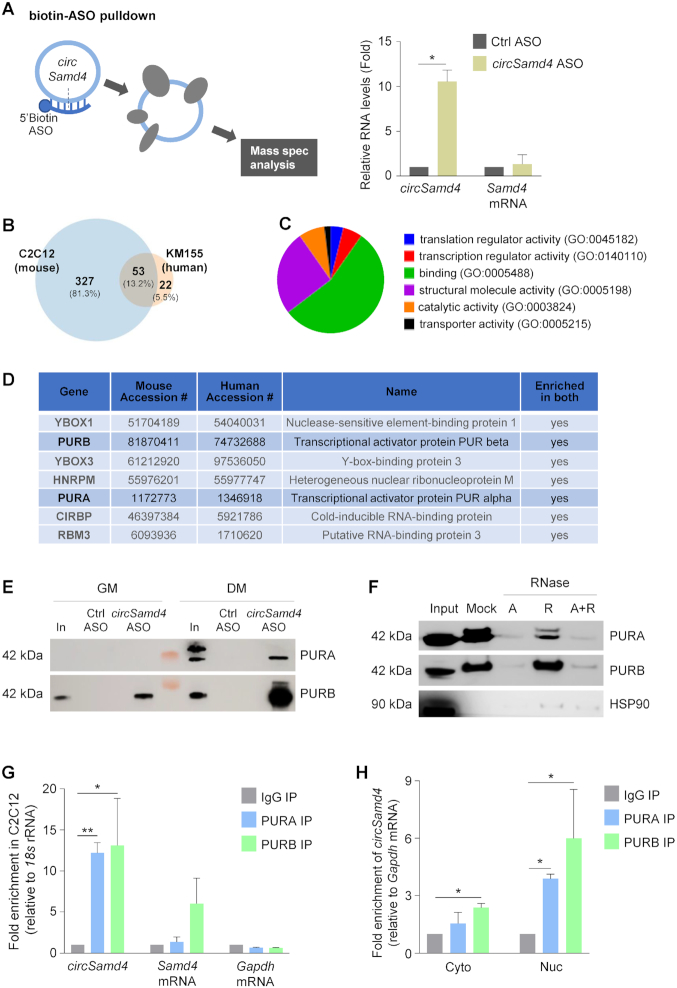
To investigate how circSamd4 modulates myogenesis, we sought to identify interacting partners, specifically circSamd4-associated proteins. We designed an antisense oligomer (ASO) complementary to the region spanning the circSamd4 junction region in order to pull down the endogenous circSamd4 from C2C12 myoblasts (Figure 4A, left) as well as circSAMD4 from human KM155 myoblasts (Supplementary Figure S4). Following pulldown and confirmation by RT-qPCR analysis that circSamd4 was selectively enriched in the pulldown material (Figure 4A, right), while Samd4 mRNA was not enriched, we identified the associated proteins by mass spectrometry analysis (Figure 4B). Comparison of the ∼380 putative interacting proteins in mouse myoblasts and the ∼75 putative interacting proteins in human myoblasts (Supplementary Figures S4A, B and Supplementary Table S2) revealed a total of 53 proteins enriched in both the circSamd4 (mouse myoblasts) and circSAMD4 (human myoblasts) pulldowns preferentially in DM relative to GM (Figure 4B and Supplementary Table S2). Gene Ontology (GO) analysis revealed that some of the shared 53 proteins were capable of binding DNA and/or RNA (Figure 4C). Phenotype cluster analysis further showed that the proteins enriched in both groups play key roles in RNA processing (Supplementary Figure S4C).
Figure 4.
circSamd4 interacts with transcription factors PURA and PURB. (A) Left, biotinylated antisense oligomers (ASOs) complementary to the junction of circSamd4 and control (Ctrl) were incubated with C2C12 in GM as well as in DM. After affinity pulldown using streptavidin beads, circSamd4-interacting proteins were identified by mass spectroscopy. Right, levels of circSamd4 enrichment in ASO pulldown samples, as assessed by RT-qPCR analysis of circSamd4 (relative to the enrichment of Gapdh mRNA, a transcript that does not bind the ASOs and encodes a housekeeping protein) in the pulldown samples. Enrichment in Samd4 mRNA was monitored in parallel. (B) Venn diagram representation of the proteins significantly enriched in the mass spec datasets identified after pulldown of circSamd4 ASO vs Ctrl ASOs in mouse C2C12 myoblasts (left) and in human KM155 myoblasts (right) (Supplementary Figure S3; Materials and Methods). 53 proteins were shared in both mass spec datasets (intersection). (C) Pie chart represents GO functional categories of the 53 proteins at the intersection of both datasets. (D) Top 7 proteins shared in the mass spec datasets from mouse and human myoblast pulldowns (Supplementary Table S2 and Supplementary Figure S3A and S3B). (E) Western blot analysis of the interaction of circSamd4 with PURA and PURB present in DM and GM lysates, as assessed after biotin-ASO pulldown followed by western blot analysis of PURA and PURB levels in the pulldown material. Input (In), 5 μg of GM or DM lysates (C2C12). (F) The interaction of PURA and PURB with circSamd4 was assessed as explained in panel (E), except that lysates were treated with RNase A and/or RNase R before pulldown followed by western blot analysis. (G) For RIP (ribonucleoprotein immunoprecipitation) analysis, IP was performed using DM C2C12 lysates and either IgG (control) antibodies or antibodies recognizing PURA or PURB; the presence of circSamd4 and Samd4 mRNA (as well as control Gapdh mRNA) in the IP material was measured by RT-qPCR analysis. Enrichments were normalized to the levels of 18s rRNA. (H) After fractionating nuclei and cytoplasm, RIP analysis was carried out as explained in (G) to measure the interaction of PURA and PURB with circSamd4 in the nucleus and the cytoplasm. Data in (A,G,H) are the means ±SEM from three independent experiments. *P < 0.05, **P < 0.005.
Among the shared proteins, those displaying maximum peptide counts and playing potential roles in myogenesis or skeletal muscle metabolism included two candidates in the PUR family, PURA (Purα) and PURB (Purβ) (Figure 4D). These proteins were particularly interesting, as previous reports had suggested that PURA and PURB, which bind both RNA and DNA elements, were capable of binding the proximal promoters of the myosin heavy chain (Mhc) promoter in C2C12 cells and negatively regulated MHC production (34). Additionally, PUR proteins regulated MHC production transcriptionally in cardiac myocytes and were proposed to mediate a cardiac-restricted MHC production by preventing its expression in non-muscle cells (35).
We confirmed the interactions of PUR proteins with circSamd4 using two different assays. First, we carried out pulldown analysis using ASOs, streptavidin-coated beads, and C2C12 lysates (GM and DM), followed by PURA and PURB western blot analysis. As shown in Figure 4E, circSamd4-directed ASO but not Ctrl ASO pulled down PURA and PURB, and this binding was increased in DM conditions. Other nucleic acid-binding proteins identified in the screen, including YBOX1 and YBOX3 were not enriched after testing of individual proteins in the circSamd4 ASO pulldown samples (Supplementary Figure S4D), perhaps because YBOX proteins might interact with PUR proteins instead, as shown previously (36). Additionally, low levels of HNRNPM were recovered in both ASO fractions (Supplementary Figure S4D). We also confirmed that PUR RNP complexes contained circSamd4 and not Samd4 mRNA by digesting the samples with RNase A (which degrades all RNAs) and/or RNase R (which degrades linear RNAs preferentially). As shown in Figure 4F, PURA and PURB were no longer present if the complex was digested with RNase A, but they remained in the complex after digestion with RNase R, supporting the notion that they associated with the circular RNA.
Second, we studied these interactions by performing ribonucleoprotein (RNP) immunoprecipitation (RIP) analysis using antibodies that recognized PURA or PURB (Supplementary Figure S4E) and lysates from differentiated C2C12 cultures under IP conditions that preserved native complexes. We found a 12- to 14-fold enrichment in the levels of circSamd4 RNA in PURA and PURB immunoprecipitated (IP) samples relative to the levels of circSamd4 in the IgG IP samples (Figure 4G); importantly, Samd4 mRNA was moderately enriched in PURB RIP samples (although not in PURA RIP samples) (Figure 4G). The abundant transcript Gapdh mRNA, encoding the housekeeping protein GAPDH, was not enriched in the IP materials, supporting the specificity of the association of circSamd4 with PURA and PURB (Figure 4G). After fractionating nuclear and cytoplasmic compartments, RIP analysis revealed that PURA and PURB were capable of interacting with circSamd4 in both the nucleus and the cytoplasm (Figure 4H). As shown in Supplementary Figure S4F, the levels of PURA and PURB increased with differentiation, and in DM they were detected in both compartments.
In control experiments, additional circRNAs that were initially identified as being upregulated during DM (Figure 1C) were not significantly enriched in PURA or PURB RIP samples (Supplementary Figure S5A), and YBOX3 RIP analysis did not show significant binding to circSamd4 or Samd4 mRNA (Supplementary Figure S5B). Finally, the levels of PURA and PURB, as well as the corresponding mRNAs, increased slightly with differentiation (Supplementary Figure S5C,D). Taken together, these data indicate that PURA and PURB preferentially associate with circSamd4 during myogenesis.
A specific region in circSamd4 promotes efficient binding of PUR proteins
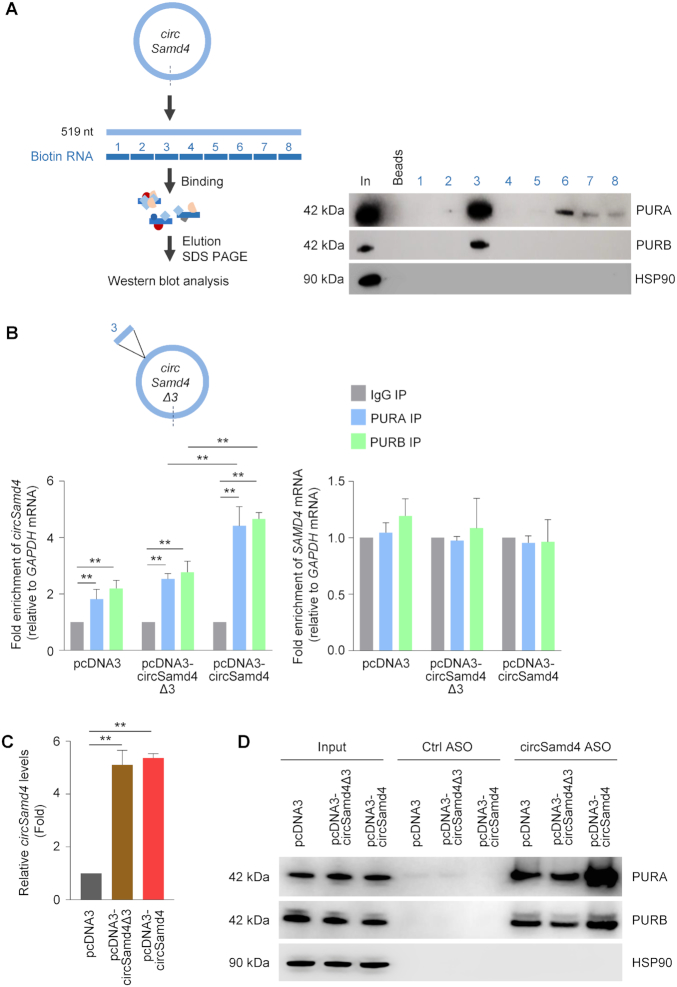
In order to map the region(s) of interaction between circSamd4 and PUR proteins, we prepared nonoverlapping biotinylated RNA fragments spanning the body of the circRNA (Materials and Methods). After incubation of each biotinylated RNA with whole-cell lysates prepared from differentiated C2C12 cultures, pulldown analysis using streptavidin beads followed by PURA and PURB western blot analysis revealed that both proteins associated most avidly with segment 3 (Figure 5A); we also observed weak binding of PURA to segments 6–8 (Figure 5A). The overall RNA-binding propensity and the prediction of potential RNA-binding regions of these PUR proteins were assessed using catRAPID analysis (37) (Supplementary Figure S6), yielding an overall interaction score of approximately 0.6 and supporting the observed RNA-binding properties of these proteins. Interestingly, the sequence of circSamd4 segment 3 contains some of the trinucleotides (GGA, AAG, CGG, CAG) identified by SELEX (38) as being preferentially bound by PURA, although they were not found as triplet repeats within circSamd4.
Figure 5.
PUR proteins predominantly associate with a specific region of circSamd4. (A) Schematic of biotin pulldown of PURA and PURB using nonoverlapping biotinylated RNAs (each ∼65 nt long) tiling the length of circSamd4. Following incubation of the biotinylated RNAs with C2C12 DM lysates, bound proteins were pulled down using streptavidin beads and the presence of PURA and PURB was assessed by western blot analysis. Input (In) was 2 μg of DM lysates; Beads only (Beads), no biotinylated RNA. (B, C) Schematic, circSamd4Δ3 bearing a deletion of segment 3. By 24 h after transfecting HeLa cells with vectors (2 μg) to overexpress circSamd4 and circSamd4(Δ3), RIP analysis was used to measure circSamd4 (left graph) and SAMD4 mRNA (right graph) bound to PURA and PURB relative to the levels of GAPDH mRNA (B). The levels of circSamd4Δ3 and circSamd4 expressed in HeLa cells 24 h after transfection of plasmids pcDNA3-circSamd4Δ3 and pcDNA3-circSamd4, respectively, were measured by RT-qPCR analysis and normalized to GAPDH mRNA levels (C). (D) Following overexpression of circSamd4 and circSamd4Δ3 as explained in panel (B), biotin-ASO pulldown was carried out to confirm the interaction of PURA and PURB with circSamd4 in C2C12 lysates as shown by Western blot analysis. The levels of the housekeeping protein HSP90 were assessed as a background control protein. Input was 2 μg of DM lysates. Data in (B,C) are the means ±SEM from three independent experiments. **P < 0.005.
Further evidence that endogenous PUR proteins bound segment 3 was obtained by employing constructs that overexpressed full-length circSamd4 (pcDNA3-circSamd4) or a truncated circRNA lacking fragment 3, circSamd4Δ3 (expressed from pcDNA-circSamd4Δ3). To test if fragment 3 was the region of interaction, we did not use C2C12 cells, as modulating circSamd4 alone had confounding effects on myogenesis. Instead, we transfected the constructs into human cervical carcinoma HeLa cells and 24 h later performed RIP analysis using anti-PURA and anti-PURB antibodies; as shown (Figure 5B, left), only the full-length circSamd4 was enriched in the PURA and PURB IP samples, as binding to the truncated RNA circSamd4Δ3 was equivalent to the binding observed with the empty vector. The interaction of PUR proteins with linear Samd4 mRNA was unchanged regardless of whether the full-length or the truncated circRNA was overexpressed (Figure 5B, right) further supporting the notion that PURA and PURB associated with segment 3 of circSamd4. Both circSamd4Δ3 and circSamd4 were expressed at approximately the same levels (Figure 5C). Additional analysis of the consequences of PURA and PURB interacting with circSamd4 was pursued by overexpressing these plasmids, as well as empty vector pcDNA3 (Figure 5C) in HeLa cells followed by ASO pulldown. In keeping with earlier data, both PURA and PURB were more enriched in pulldowns from cultures expressing full-length circSamd4 than in cultures expressing circSamd4Δ3, indicating that this segment was important for protein binding (Figure 5D). Together, these data suggest that region 3 in circSamd4 is important for efficient binding of PURA and PURB.
PURA and PURB repress the transcription of Mhc mRNA, circSamd4 alleviates this repression
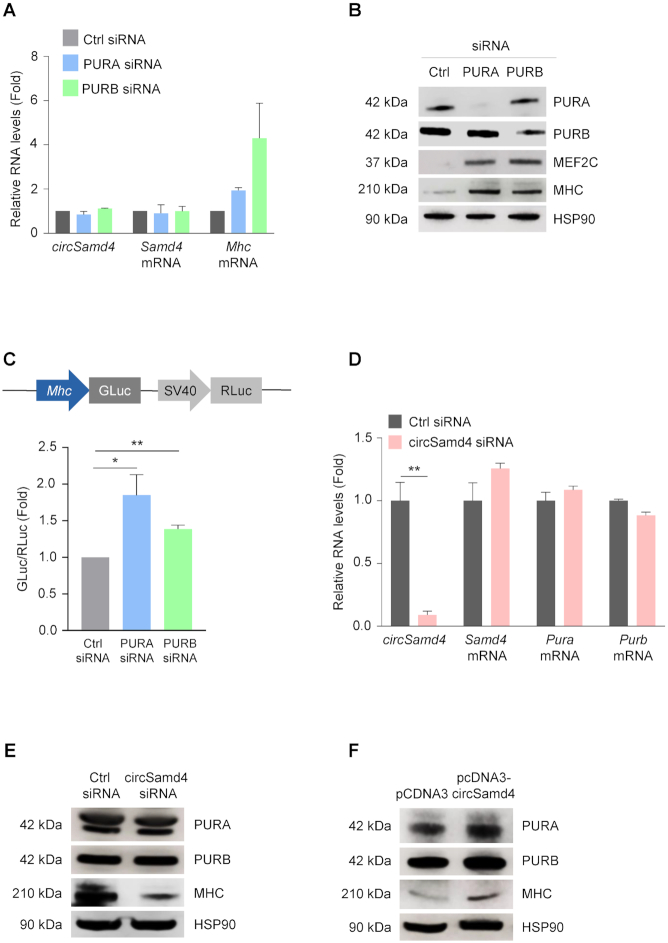
While PUR proteins can promote transcription of target genes, they can also suppress transcription, as shown for Mhc (34). Accordingly, silencing PURA or PURB in C2C12 myoblasts led to increased expression of both Mhc RNA (Figure 6A) and MHC protein (Figure 6B), supporting earlier reports that PURA and PURB suppress transcription of the Mhc gene (34); under these conditions, circSamd4 and Samd4 mRNA levels remained unaltered. Further evidence of this regulatory paradigm came from analysis of a heterologous luciferase reporter in which the Mhc promoter drove Gaussia luciferase (GLuc) activity; Renilla luciferase (RLuc) expressed from the same vector, served as the transfection control. As shown in Figure 6C, silencing of either PURA or PURB significantly elevated Mhc-driven expression of luciferase 1.8- and 1.3-fold, respectively, supporting the notion that PURA and PURB repress MHC expression. Importantly, the converse did not occur, as circSamd4 silencing in C2C12 cells did not influence the production of Pura or Purb mRNAs (Figure 6D) or PURA or PURB proteins (Figure 6E). Likewise, overexpression of circSamd4 in C2C12 cells did not influence the levels of PURA or PURB (Figure 6F). These data indicate that PURA and PURB repress Mhc transcription and hence MHC expression levels.
Figure 6.
PUR proteins suppress MHC production transcriptionally. (A, B) C2C12 myoblasts were transfected with siRNAs to silence PURA and PURB; 48 h later, the levels of circSamd4, Samd4 mRNA, and Mhc mRNA were assessed by RT-qPCR analysis and normalized to Gapdh mRNA (A). Western blot analysis was used to evaluate the levels of PURA, PURB, MEF2C, MHC, and loading control HSP90 (B). (C) Schematic of the Mhc promoter reporter construct (pmh2-GLuc-RLuc) (GeneCopoeia Inc.) used to evaluate MHC transcription. GM C2C12 cells were transfected with 1 μg of plasmid pmh2-GLuc-RLuc in cells expressing normal levels or silenced PURA and PURB as explained in panel (A); 8 hours later, cells were switched to DM media, and 24 h after that, promoter activity was measured. GLuc levels were normalized to RLuc levels in each group; data were then plotted as the difference in GLuc/RLuc ratios in the Ctrl siRNA transfection group compared with PURA or PURB silencing groups. (D) C2C12 cells were transfected with control (Ctrl) siRNA or circSamd4 siRNA; 48 h later, the expression levels of Pura and Purb mRNAs, normalized to the levels of Gapdh RNA, were measured by RT-qPCR analysis and plotted. (E, F) Effect of silencing circSamd4 on the expression of PURA, PURB, and MHC; 48 h after transfecting GM C2C12 cells with control (Ctrl) siRNA or circSamd4 siRNA, the levels of circSamd4, and Samd4, Pura, and Purb mRNAs were assessed by RT-qPCR analysis (E) and the levels of proteins PURA, PURB, MHC, and loading control HSP90 by western blot analysis (F). Data in (A,C,D) are the means ±SEM from three or more independent experiments. *P < 0.05, **P < 0.005.
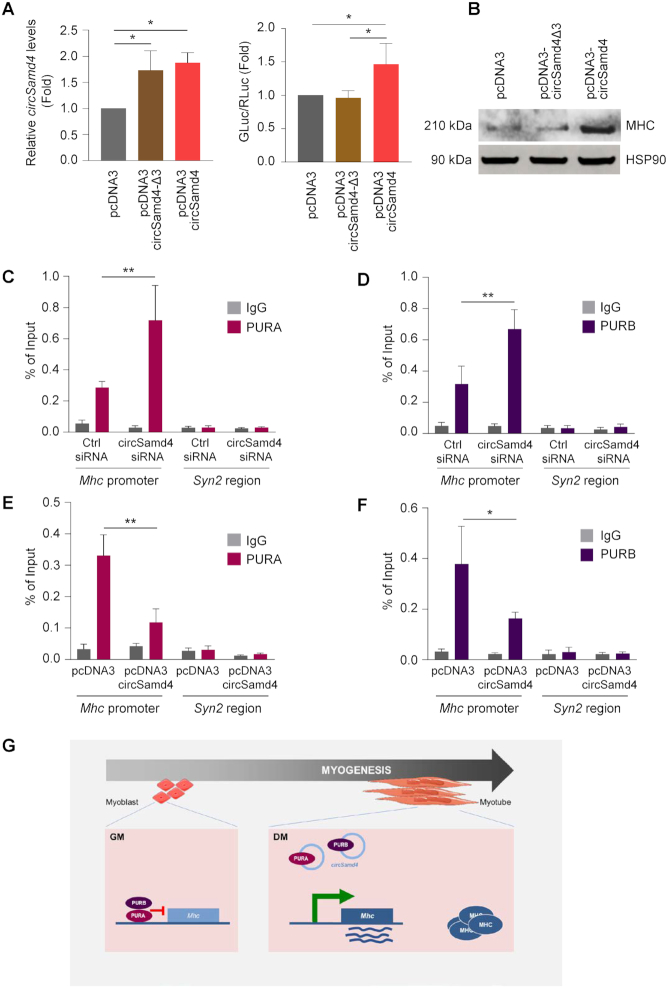
We then hypothesized that binding of circSamd4 might impact upon PURA and PURB function. To test this possibility, we used the luciferase reporter construct in the presence of either full-length circSamd4 or circSamd4Δ3. As shown, only expression of full-length circSamd4 was able to promote reporter gene expression in differentiated C2C12 cultures, while expression of the deletion mutant circSamd4Δ3 or the empty vector control failed to do so (Figure 7A). Furthermore, expression of circSamd4 increased MHC expression in C2C12 cells, while expression of circSamd4Δ3 did not (Figure 7B). These data suggest that circSamd4 might be sequestering PURA and PURB proteins, thereby enabling MHC production during myogenesis, while deletion mutant circSamd4Δ3, by being unable to bind PURA and PURB, failed to derepress MHC production.
Figure 7.
circSamd4 binds PUR proteins, derepressing Mhc gene transcription. (A) Activity of the Mhc promoter, as determined by monitoring luciferase activity after transfecting 500 ng of reporter pMH2-LUC-Renilla (Figure 6C) into C2C12 cells that had been transfected 24 h earlier with parent vector pcDNA3 or with vectors expressing circSamd4 or circSamd4-Δ3 (left graph). Eight h after transfection of the reporter construct, the cells were switched to DM media for 24 h and promoter activity was measured; as above (Figure 6C), GLuc was normalized to RLuc and these ratios were plotted relative to the ratios measured in the empty vector group (right graph). (B) The levels of MHC in the different transfection groups shown in panel (A) were assessed by western blot analysis and HSP90 levels were shown as the loading control. (C–F) ChIP-qPCR analysis of the association of PURA and PURB with the Mhc proximal promoter region (−270/−450). Following crosslinking (Materials and Methods), antibodies recognizing PURA (C, E) or PURB (D, F) or control nonspecific rabbit IgG antibodies were used to immunoprecipitate sheared chromatin from C2C12 myoblasts in which circSamd4 was silenced (C, D) or overexpressed (E, F). Specific oligonucleotide primers were used to amplify the region of interaction with PURA and PURB. ChIP-qPCR data were calculated using percentage of input; the TTS region of the neuron-specific gene Syn2 (region +200 TTS), which encodes the protein SYN2 (Synapsin), was assayed as a non-target negative control. (G) Summary model. In myoblasts (GM), virtually undetectable levels of circSamd4 permit the repression of the Mhc promoter by PURA and PURB. As myogenesis progresses (DM), the rise in circSamd4 leads to binding of a fraction of PURA and PURB away from the Mhc promoter, thereby helping to derepress Mhc transcription and enabling the accumulation of Mhc mRNA and the production of MHC, a key protein in mature myotubes. Data in (A, C–F) are the means ±SEM from three or more independent experiments. *P < 0.05, **P < 0.005.
To test these possibilities directly, we assessed if circSamd4 influenced the binding of PUR proteins to the Mhc promoter. Chromatin immunoprecipitation (ChIP) was performed followed by qPCR analysis (Materials and Methods) of the conserved Mhc proximal promoter region containing the dNRE-S elements, a major binding site of PURA and PURB in C2C12 myoblasts (34). This analysis revealed that binding of PURA and PURB to the Mhc promoter increased significantly upon depletion of circSamd4 in C2C12 cells under DM conditions (Figures 7C and D), supporting the view that circSamd4 can relieve the transcriptional inhibition of Mhc transcription by PURA and PURB. Conversely, overexpression of circSamd4 in C2C12 cells had the opposite effect, as ChIP analysis revealed that PURA and PURB binding to the Mhc promoter decreased after circSamd4 overexpression (Figures 7E and F). Collectively, these data support a model whereby in proliferating myoblasts (GM) PURA and PURB suppress the Mhc promoter and MHC production. During myogenesis, circSamd4 levels rise and circSamd4 associates with a fraction of nuclear and/or cytoplasmic PURA and PURB, two transcriptional repressors of the myogenic protein MHC, which is required for myotube formation. By contributing to the derepression of MHC production, circSamd4 helps to establish later stages of myogenesis (Figure 7G).
DISCUSSION
CircRNAs are increasingly recognized as being implicated in gene regulatory networks influencing development and disease. Here, we investigated circRNAs associated with skeletal myogenesis, a process whereby myoblasts differentiate into myotubes. We found that circSamd4 increased robustly during skeletal muscle differentiation and associated with PURA and PURB, two proteins that bind and transcriptionally repress the Mhc promoter and thereby suppress myogenic differentiation. We propose a model (Figure 7G) in which circSamd4, accumulating during myogenesis, associates with PURA and PURB, thereby contributing at least in part to the derepression of Mhc transcription, the rise in MHC levels, and the promotion of myogenesis. To support this model, we present evidence that silencing circSamd4 delayed myogenesis, reduced the expression of myogenic factors (e.g. Myog, Mef2c and Mhc mRNAs, as well as the encoded proteins) (Figure 3) and helped to preserve the binding of PURA and PURB to the Mhc promoter (Figure 7C and D), keeping MHC levels low; conversely, overexpression of circSamd4 lowered PURA and PURB binding to the Mhc promoter and increased MHC production (Figure 7B, E and F). A mutant circSamd4 lacking the site of interaction with PURA/B (circSamd4Δ3) was unable to rescue the effects of circSamd4 on myogenesis.
The mechanism whereby circSamd4 promotes myogenesis remains to be studied in molecular detail. For example, how specifically the production of circSamd4 from Samd4 pre-mRNA rises during differentiation while Samd4 mRNA levels rise comparatively less (Figure 2). The possible involvement of myogenic splicing factors in the production of circSamd4 is not known at present. Whether some of the additional circSamd4-binding proteins that have splicing activity, such as HNRNPM (Figure 4D), TRA2B, SRSF6 or others (Supplementary Figure S4A, B; Supplementary Table S2) are implicated in the generation of circSamd4 also warrants future investigation.
The lower abundance of circSamd4 in the nucleus (estimated at ∼2 molecules) suggests that nuclear circSamd4 cannot alone suppress the binding of all of the nuclear PURA and PURB to the Mhc promoter. Instead, circSamd4 likely cooperates with other factors to prevent the transcriptional repression of Mhc by PURA and PURB, even though these cooperating factors were not identified in this study. Alternatively, given that cytoplasmic circSamd4 is more abundant (∼20-fold than the nuclear circSamd4; Figure 2D and E), it is possible that circSamd4 binds to and sequesters PURA/B in the cytoplasm, and thus it prevents PURA/B from repressing Mhc transcription in the nucleus, or perhaps circSamd4 helps export PUR proteins to the cytoplasm. Supporting these possibilities, PURA and PURB were also found to reside in both compartments (supplementary Figure S4F) and circSamd4 was found to interact with PUR proteins in both the nucleus and the cytosol (Figure 4H). Additional related questions that warrant study in biochemical detail include how the affinity of PURA and PURB for circSamd4 compares to its affinity for the Mhc promoter during myogenesis. In a recent study, circSAMD4 was found to promote osteosarcoma by associating with microRNA miR-1244 (33). Whether PURA and PURB binding influences the binding of myogenic microRNAs to circSAMD4 or circSamd4 also deserves future study.
PURA and PURB are highly conserved proteins, from bacteria to mammals, and are found in virtually every tissue (www.genecards.org), although PURA appears to be more abundant. The PUR protein family also includes PURG, although PURG was not found to bind circSamd4 in our screen, and it seems less abundant in tissues overall. PURA and PURB interact with both single-stranded DNA and RNA and influence molecular processes such as DNA replication, gene transcription, cytoplasmic RNA transport, and compartmentalized mRNA translation (39,40). Besides mRNAs, PURA was found to interact with lncRNA BC1 in rodents and the cognate lncRNA BC200 in humans (41,42), as well as with other proteins (43). It will be interesting to study whether circSamd4 mobilizes PURA and PURB to the cytoplasm of myoblasts and enables cytoplasmic functions for PURA and PURB in mRNA transport or translation, particularly during later stages of myogenesis when MHC is strongly expressed.
Previous work indicated that PUR proteins repressed the transcription of α-MHC mRNA and the translation of α-MHC in cardiac myocytes and may thus prevent its expression in non-muscle cells (35). In this regard, it will also be important to establish the possible impact of PURA and PURB, and by extension of circSamd4, on the production of the different sarcomeric MHC gene variants (34) and to find out if other promoters inhibited by PUR proteins are also derepressed by an increase in circSamd4 levels. In addition, endogenous PURA and PURB were necessary to fully repress full-length smooth muscle ACTA (α-Actin) promoter in cultured fibroblasts but to a lesser extent in vascular smooth muscle cells (44). Indeed, the effects of PUR-circSamd4-mediated gene regulation may be broadly relevant to other tissue types such as cardiomyocytes, fibroblasts, neurons, myeloid cells, and vascular smooth muscle cells in which PUR proteins are expressed and are functional (35,40,44,45).
Future studies analyzing comprehensively circSamd4-interacting proteins and RNAs may uncover its role in skeletal muscle formation and in muscle disorders such as myotonic dystrophy (DM) and facioscapulohumeral muscular dystrophy (FSHD). As several chronic skeletal muscle dysfunctions are associated with changes in MHC production, interventions to modulate circSamd4 function may be therapeutically valuable in disease conditions. Finally, work is underway to understand the mechanisms driving the age-dependent decline in MHC levels, given that circSAMD4 abundance increases with age (Supplementary Figure S1D). A comprehensive understanding of circSamd4 biogenesis, interaction partners, and impact on myogenic gene expression programs will guide novel circRNA-centered interventions in age- and disease-associated loss of muscle function.
DATA AVAILABILITY
RNA-seq data has been deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/) with accession numbers GSE92632 and GSE136004.
Supplementary Material
ACKNOWLEDGEMENTS
The authors appreciate the assistance provided by Dr Anup Mahurkar from the University of Maryland and Dr Dhiraj Kumar from MD Anderson Cancer Institute.
Author contributions: P.R.P. and A.C.P. conceived the study; P.R.P., K.A. and M.G. designed experiments and analyzed data; P.R.P., J.H.Y. A.C.P., R.M., D.T., T.N., D.H., D.A. and X.Y performed experiments; A.C.P., J.H.Y., J.H.N., K.M.K., J.L.M. and M.W.C. provided technical support; S.D. performed informatics analysis; P.S., K.A., S.W.J. and E.H., contributed intellectually; and P.R.P. and M.G. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIA-IRP, NIH; S.W.J. received funding from Versus Arthritis [21530, 21812]; D.H., D.A. and E.H. were supported by NIH/NCI [P30CA016087, S10 OD021747NIH, R01CA2022027]; Leveraged Finance Fights Melanoma-MRA Team Science Award. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Atianand M.K., Fitzgerald K.A.. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol. Med. 2014; 20:623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338. [DOI] [PubMed] [Google Scholar]
- 3. Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O.. Cell-type specific features of circular RNA expression. PLoS Genet. 2013; 9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bachmayr-Heyda A., Reiner A.T., Auer K., Sukhbaatar N., Aust S., Bachleitner-Hofmann T., Mesteri I., Grunt T.W., Zeillinger R., Pils D.. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 2015; 5:8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gu X., Li M., Jin Y., Liu D., Wei F.. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017; 18:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu C.Y., Li T.C., Wu Y.Y., Yeh C.H., Chiang W., Chuang C.Y., Kuo H.C.. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017; 8:1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang D., Yang K., Yang M.. Circular RNA in aging and age-related diseases. Adv. Exp. Med. Biol. 2018; 1086:17–35. [DOI] [PubMed] [Google Scholar]
- 8. Zhao Z.J., Shen J.. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017; 14:514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lukiw W.J. Circular RNA (circRNA) in Alzheimer's disease (AD). Front. Genet. 2013; 4:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Floris G., Zhang L., Follesa P., Sun T.. Regulatory role of circular RNAs and neurological disorders. Mol. Neurobiol. 2017; 54:5156–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A. et al.. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016; 7:12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frontera W.R., Ochala J.. Skeletal muscle: a brief review of structure and function. Calcif. Tissue Int. 2015; 96:183–195. [DOI] [PubMed] [Google Scholar]
- 13. Molkentin J.D., Olson E.N.. Defining the regulatory networks for muscle development. Curr. Opin. Genet. Dev. 1996; 6:445–453. [DOI] [PubMed] [Google Scholar]
- 14. Das A., Das A., Das D., Abdelmohsen K., Panda A.C.. Circular RNAs in myogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 2019; doi:10.1016/j.bbagrm.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Y., Chen X., Sun H., Wang H.. Long non-coding RNAs in the regulation of skeletal myogenesis and muscle diseases. Cancer Lett. 2018; 417:58–64. [DOI] [PubMed] [Google Scholar]
- 16. Xu M., Chen X., Chen D., Yu B., Li M., He J., Huang Z.. Regulation of skeletal myogenesis by microRNAs. J. Cell Physiol. 2020; 235:87–104. [DOI] [PubMed] [Google Scholar]
- 17. Carvalho R.F., Cicogna A.C., Campos G.E., De Assis J.M., Padovani C.R., Okoshi M.P., Pai-Silva M.D.. Myosin heavy chain expression and atrophy in rat skeletal muscle during transition from cardiac hypertrophy to heart failure. Int. J. Exp. Pathol. 2003; 84:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fink B., Egl M., Singer J., Fuerst M., Bubenheim M., Neuen-Jacob E.. Morphologic changes in the vastus medialis muscle in patients with osteoarthritis of the knee. Arthritis Rheum. 2007; 56:3626–3633. [DOI] [PubMed] [Google Scholar]
- 19. Ottenheijm C.A., Heunks L.M., Dekhuijzen P.N.. Diaphragm muscle fiber dysfunction in chronic obstructive pulmonary disease: toward a pathophysiological concept. Am. J. Respir. Crit Care Med. 2007; 175:1233–1240. [DOI] [PubMed] [Google Scholar]
- 20. Pette D., Staron R.S.. Transitions of muscle fiber phenotypic profiles. Histochem. Cell Biol. 2001; 115:359–372. [DOI] [PubMed] [Google Scholar]
- 21. Abdelmohsen K., Panda A.C., De S., Grammatikakis I., Kim J., Ding J., Noh J.H., Kim K.M., Mattison J.A., de Cabo R., Gorospe M.. Circular RNAs in monkey muscle: age-dependent changes. Aging. 2015; 7:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Munk R., Martindale J.L., Yang X., Yang J.H., Grammatikakis I., Di Germanio C., Mitchell S.J., de Cabo R., Lehrmann E., Zhang Y. et al.. Loss of miR-451a enhances SPARC production during myogenesis. PLoS One. 2019; 14:e0214301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Panda A.C., De S., Grammatikakis I., Munk R., Yang X., Piao Y., Dudekula D.B., Abdelmohsen K., Gorospe M.. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017; 45:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanno J., Aisaki K., Igarashi K., Nakatsu N., Ono A., Kodama Y., Nagao T.. ‘Per cell’ normalization method for mRNA measurement by quantitative PCR and microarrays. BMC Genomics. 2006; 7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kramer M.C., Liang D., Tatomer D.C., Gold B., March Z.M., Cherry S., Wilusz J.E.. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015; 29:2168–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pandey P.R., Rout P.K., Das A., Gorospe M., Panda A.C.. RPAD (RNase R treatment, polyadenylation, and poly(A)+ RNA depletion) method to isolate highly pure circular RNA. Methods. 2019; 155:41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen Z., Viboolsittiseri S.S., O’Connor B.P., Wang J.H.. Target DNA sequence directly regulates the frequency of activation-induced deaminase-dependent mutations. J. Immunol. 2012; 189:3970–3982. [DOI] [PubMed] [Google Scholar]
- 29. Chen P., Zhao J., Wang Y., Wang M., Long H., Liang D., Huang L., Wen Z., Li W., Li X. et al.. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013; 27:2109–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Haro H.M., Al-Ramahi I., Jones K.R., Holth J.K., Timchenko L.T., Botas J.. Smaug/SAMD4A restores translational activity of CUGBP1 and suppresses CUG-induced myopathy. PLoS. Genet. 2013; 9:e1003445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen Z., Holland W., Shelton J.M., Ali A., Zhan X., Won S., Tomisato W., Liu C., Li X., Moresco E.M., Beutler B.. Mutation of mouse Samd4 causes leanness, myopathy, uncoupled mitochondrial respiration, and dysregulated mTORC1 signaling. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:7367–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Niu N., Xiang J.F., Yang Q., Wang L., Wei Z., Chen L.L., Yang L., Zou W.. RNA-binding protein SAMD4 regulates skeleton development through translational inhibition of Mig6 expression. Cell Discov. 2017; 3:16050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yanbin Z., Jing Z.. CircSAMD4A accelerates cell proliferation of osteosarcoma by sponging miR-1244 and regulating MDM2 mRNA expression. Biochem. Biophys. Res. Commun. 2019; 516:102–111. [DOI] [PubMed] [Google Scholar]
- 34. Ji J., Tsika G.L., Rindt H., Schreiber K.L., McCarthy J.J., Kelm R.J. Jr, Tsika R.. Puralpha and Purbeta collaborate with Sp3 to negatively regulate beta-myosin heavy chain gene expression during skeletal muscle inactivity. Mol. Cell Biol. 2007; 27:1531–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gupta M., Sueblinvong V., Raman J., Jeevanandam V., Gupta M.P.. Single-stranded DNA-binding proteins PURalpha and PURbeta bind to a purine-rich negative regulatory element of the alpha-myosin heavy chain gene and control transcriptional and translational regulation of the gene expression. J. Biol. Chem. 2003; 278:44935–44948. [DOI] [PubMed] [Google Scholar]
- 36. Lasham A., Lindridge E., Rudert F., Onrust R., Watson J.. Regulation of the human fas promoter by YB-1, Puralpha and AP-1 transcription factors. Gene. 2000; 252:1–13. [DOI] [PubMed] [Google Scholar]
- 37. Livi C.M., Klus P., Delli P.R., Tartaglia G.G.. catRAPID signature: identification of ribonucleoproteins and RNA-binding regions. Bioinformatics. 2016; 32:773–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aumiller V., Graebsch A., Kremmer E., Niessing D., Forstemann K.. Drosophila Pur-alpha binds to trinucleotide-repeat containing cellular RNAs and translocates to the early oocyte. RNA Biol. 2012; 9:633–643. [DOI] [PubMed] [Google Scholar]
- 39. Daniel D.C., Johnson E.M.. PURA, the gene encoding Pur-alpha, member of an ancient nucleic acid-binding protein family with mammalian neurological functions. Gene. 2018; 643:133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hariharan S., Kelm R.J. Jr, Strauch A.R.. The Puralpha/Purbeta single-strand DNA-binding proteins attenuate smooth-muscle actin gene transactivation in myofibroblasts. J. Cell Physiol. 2014; 229:1256–1271. [DOI] [PubMed] [Google Scholar]
- 41. Ohashi S., Kobayashi S., Omori A., Ohara S., Omae A., Muramatsu T., Li Y., Anzai K.. The single-stranded DNA- and RNA-binding proteins pur alpha and pur beta link BC1 RNA to microtubules through binding to the dendrite-targeting RNA motifs. J. Neurochem. 2000; 75:1781–1790. [DOI] [PubMed] [Google Scholar]
- 42. Johnson E.M., Kinoshita Y., Weinreb D.B., Wortman M.J., Simon R., Khalili K., Winckler B., Gordon J.. Role of Pur alpha in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J. Neurosci. Res. 2006; 83:929–943. [DOI] [PubMed] [Google Scholar]
- 43. Gallia G.L., Darbinian N., Tretiakova A., Ansari S.A., Rappaport J., Brady J., Wortman M.J., Johnson E.M., Khalili K.. Association of HIV-1 Tat with the cellular protein, Puralpha, is mediated by RNA. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:11572–11577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Knapp A.M., Ramsey J.E., Wang S.X., Godburn K.E., Strauch A.R., Kelm R.J. Jr. Nucleoprotein interactions governing cell type-dependent repression of the mouse smooth muscle alpha-actin promoter by single-stranded DNA-binding proteins Pur alpha and Pur beta. J. Biol. Chem. 2006; 281:7907–7918. [DOI] [PubMed] [Google Scholar]
- 45. Johnson E.M., Daniel D.C., Gordon J.. The pur protein family: genetic and structural features in development and disease. J. Cell Physiol. 2013; 228:930–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data has been deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/) with accession numbers GSE92632 and GSE136004.