Abstract
CRISPR/Cas9 has become a powerful tool for genome editing in zebrafish that permits the rapid generation of loss of function mutations and the knock-in of specific alleles using DNA templates and homology directed repair (HDR). We examined the efficiency of synthetic, chemically modified gRNAs and demonstrate induction of indels and large genomic deletions in combination with recombinant Cas9 protein. We developed an in vivo genetic assay to measure HDR efficiency and we utilized this assay to test the effect of altering template design on HDR. Utilizing synthetic gRNAs and linear dsDNA templates, we successfully performed knock-in of fluorophores at multiple genomic loci and demonstrate transmission through the germline at high efficiency. We demonstrate that synthetic HDR templates can be used to knock-in bacterial nitroreductase (ntr) to facilitate lineage ablation of specific cell types. Collectively, our data demonstrate the utility of combining synthetic gRNAs and dsDNA templates to perform homology directed repair and genome editing in vivo.
INTRODUCTION
CRISPR/Cas9 has been used for a wide range of experimental applications, and zebrafish has been a key model organism to test and validate strategies for genome editing (1,2). Repair of CRISPR-generated double-stranded breaks (DSBs) by non-homologous end joining (NHEJ) leads to insertions and deletions (indels) which may result in loss of function of the targeted gene product. By supplying an exogenous DNA template, DSBs can be repaired through homology directed repair (HDR), allowing for precision genome editing, including base pair changes and insertion of protein tags. Prior studies reporting genome editing in zebrafish have used single-stranded donor oligonucleotides (ssODN) to knock-in short DNA sequences (3–6) or plasmid-based donor vectors to knock-in fluorophores (7–10). The efficiency of transmitting fluorophore knock-in through the germline has varied widely. Reports have described an increased efficiency of CRISPR/Cas9 targeting in human cells using chemically modified synthetic gRNAs (11,12), and increased HDR efficiency in medaka using chemically modified dsDNA templates (13). We evaluated synthetic gRNAs and found that they are highly effective in leading to indels after microinjection in zebrafish embryos, consistent with a recent report (14). We used synthetic gRNAs in combination with synthetic DNA templates and we developed an assay for HDR in zebrafish utilizing an mitfa mutant, b692. Using this assay, we quantitatively compared multiple templates for HDR and correlate phenotypic and molecular efficiency. Unexpectedly, we find that minor differences in HDR template design are associated with significant changes in genome editing efficiency. We designed templates and performed precise genomic editing to generate in-frame gene fusions with fluorophores using synthetic gRNAs, and linear dsDNA templates. Knock-in alleles were transmitted through the germline at efficiencies of 14–25%. Finally, we used HDR to target bacterial nitroreductase (ntr) to the liver-specific gene fabp10a. We transmitted a genome edited fabp10a-mScarlet-ntr allele through the germline and demonstrate that treatment of embryos with metronidazole leads to lineage ablation of hepatocytes. These results demonstrate that the combination of synthetic gRNAs and dsDNA templates result in efficient genome editing in zebrafish.
MATERIALS AND METHODS
gRNA and HDR template sequence selection
Gene-specific gRNAs sequences were selected using a combination of prediction tools including gRNA Scorer 1.0 and 2.0 (15,16), GuideScan (17), or CRISPRz (18). We selected gRNA sequences with zero predicted off targets with one base pair mismatch. Design principles for HDR templates included a mutated gRNA recognition sequence, incorporation of barcoded nucleotides, homology arms, and incorporation of heterologous DNA sequence encoding EGFP or mScarlet. In some cases, repetitive sequence in non-coding regions of the genome constrained the length of homology arms, as templates could not be synthesized by the manufacturer. Annotated HDR templates have been deposited in Genbank (Accessions MN832870-MN832873). HDR templates were ordered as gBlock Gene Fragments (Integrated DNA Technologies [IDT]). For single stranded DNA templates Ultramer DNA Oligonucleotides (IDT) were used. In HDR templates containing two gRNA sites (b692 assay), the b692 recognition site was appended to the template end. Similarly the ISce site was appended to the end of the template noted to contain this sequence. gBlocks were resuspended in nuclease-free water to a concentration of 50 ηg/μl and stored at −20°C. gRNA sequences are listed in Supplementary Table S1.
Preparation and characterization of gRNAs
Gene-specific crRNAs were synthesized by IDT as Alt-R® CRISPR-Cas9 crRNAs. A bipartite synthetic gRNA was heteroduplexed using gene-specific crRNAs and a tracrRNA according to manufacturer recommendations. For simplicity, the bipartite synthetic gRNA is referred to as a gRNA throughout the manuscript. Individual gRNAs were evaluated to ensure efficient induction of indels by microinjection (as below) and a clonal analysis from individual embryos. All gRNAs used induced indels in a minimum of 8/10 clones analyzed from a pool of 10 embryos.
Microinjection
For global editing with a single gRNA, 250 ρg gRNA and 500 ρg recombinant Cas9 protein (rCas9, PNA Bio CP01) were injected into the yolk of one-cell stage embryos. Cas9 nickase (D10A Cas9 nickase protein with NLS, PNABio CN01) was purchased from PNA Bio. To generate deletions, 250 ρg gRNA 1, 250 ρg gRNA 2 and 500 ρg rCas9 were injected into the yolk of one-cell stage embryos. For HDR injections, 250 ρg gRNA, 500 ρg rCas9 and 37.5 ρg HDR template were injected into one-cell stage embryos. Throughout the manuscript rCas9 refers to recombinant Cas9 protein.
Assessment of gene editing efficiency
Genomic DNA was isolated from individual embryos (24–48 h post-fertilization [hpf]) using DirectPCR Lysis Reagent (Viagen, 102-T) supplemented with Proteinase K at 20 μg/ml (Qiagen, 158920). Samples were incubated at 55°C for 60 min followed by 85°C for 45 min. Genomic DNA was PCR amplified using specified primers (Supplementary Table S2). 8 μl of PCR product was mixed with 1.6 μl NEB Buffer 2 and 6.4 μl of water and was incubated at 95°C for 5 min followed by cooling from 95–85°C at −2°C/s and 85–25°C at −0.1°C/s. Following hybridization, the DNA was subject to T7 endonuclease (T7EI) digestion with 2 U of T7EI endonuclease (New England BioLabs [NEB], M0302) and 1 h incubation at 37°C. The reaction product was visualized on a 2% agarose gel.
For clonal analysis, the PCR product was cloned into pCRII-TOPO (Invitrogen, K460001) and Sanger sequenced (Genewiz). Sequences were compared to reference genome and non-targeting controls to identify indels using CrispRVariants (19) and MacVector (Version 15.5).
For CRISPR-STAT analysis, sperm samples from potential founders were collected with 10 μl capillary tubes as previously described, and then followed the HOTShot method with 25 μl alkaline lysis buffer to prepare DNA templates (20,21). Tail clips were prepared similarly. The CRISPR-STAT protocol was used to perform fluorescence-based genotyping (22). Primers sequences used for the CRISPR-STAT assay were slc6a15 FOR, tgtaaaacgacggccagtGGCCACGACCTACTACTGGTAT, which includes (lowercase) M13 tag for binding to a fluorescent-labeled M13 primer; and slc6a15 REV, gtgtcttTATAGATGCTGCGTCACGTTTC, which include the (lowercase) ‘PIG tail’ tag that helps ensure uniform product size, via Taq adenylation (23). Relative amounts of each uniquely sized PCR product (>100 bp and >100 peak height) were calculated with area under the curve measurements using Applied Biosystems Peak Scanner software.
For assessment of deletions, genomic DNA was PCR amplified using primers that flanked the deletion gRNAs. PCR products were purified (Qiagen, 28104), Sanger sequenced, and compared to the reference genome.
Analysis of HDR efficiency by next-generation sequencing
For analysis of HDR efficiency by next-generation DNA sequencing, 50 ηg of PCR amplicon was converted into blunt ends using T4 DNA polymerase (New England Biolabs, NEB) and Escherichia coli DNA polymerase I Klenow fragment (NEB). Libraries were prepared using the Kapa LTP Library Preparation Kit (Roche). Multiple indexing adapters were ligated to the ends of the DNA fragments. Ligation reaction products were purified by AMPure XP beads (NEB) to remove unligated adapters and quantified using Qubit (Thermo Fisher Scientific) and Bioanalyzer DNA chip (Agilent). Indexed sample libraries were normalized, pooled, and sequenced using the Illumina HiSeq4000 sequencer at 2 × 50 cycles. Reads were aligned to the zebrafish genome (GRCz10) using the Star aligner (24) and visualized using Integrative Genome Browser (IGV) (25,26).
Imaging
Micrographs of whole embryos and larval animals were taken with a Zeiss Discovery V8 stereomicroscope (Zeiss) equipped with epifluorescence and appropriate filters. Live imaging of fluorescent larvae was acquired with a Zeiss LSM800 laser scanning confocal microscope (Zeiss). For melanocyte counting, 48hpf embryos were visualized under the stereoscope and binned into four categories (WT, >101, 51–100 or 0–50 melanocytes). Individuals scoring embryos were blinded to the experimental conditions.
For analysis of chromosome condensation in h3f3a-EGFP F1 larvae, individual embryo was embedded in methylcellulose and confocal images were acquired with a Zeiss LSM800 laser scanning confocal microscope. A single 4hpf embryo was imaged under 20× objective and a mitotic cell was identified with an EGFP+ nuclear signal. 266 individual images were captured over 494 s and processed to a .avi file at 5 fps using Zeiss Zen software.
Hepatocyte ablation
HDR injection mixes were made as described above. Approximately 2 ηl of mix was injected into the cell of one-cell stage wild-type (Tu) embryos. At 4–6 days post fertilization (dpf), embryos were screened by confocal microscopy for mosaic mScarlet expression localized to the liver, and positive larvae were raised to adulthood. F0 adults were outcrossed with wild-type (TL) fish, and the F1 embryos were screened by confocal microscopy for mScarlet positive livers. Positive larvae were divided and treated with either 0.2% dimethylsufoxide (DMSO; Sigma Aldrich) or 10 μM metronidazole (Mtz; Sigma Aldrich 443-48-1) in egg water. After 24 h, the larvae were washed, anesthetized with 0.16 mg/ml tricaine-S (MS 222; Western Chemical, Inc.), and immobilized in 0.8% low melt point agarose (Invitrogen) on glass-bottom dishes (MatTek). Imaging was performed using a Nikon Ti2 inverted microscope equipped with a Yokogawa CSU-W1 spinning disk confocal unit and a Zyla 4.2 PLUS sCMOS camera (Andor), using CFI Plan Apochromat Lambda 10× NA 0.45 and 20× NA 0.8 objectives. Larvae were recovered from the agarose, allowed to recover in the absence of drug for 48 h, and imaged again.
Zebrafish husbandry
Zebrafish strains were maintained according to established guidelines (27). Wild-type animals were from the AB background except when indicated. Experiments were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All of the animals were handled according to approved institutional animal care and use committee (IACUC) protocols of the respective institutions.
RESULTS
Synthetic gRNAs lead to highly efficient induction of indels
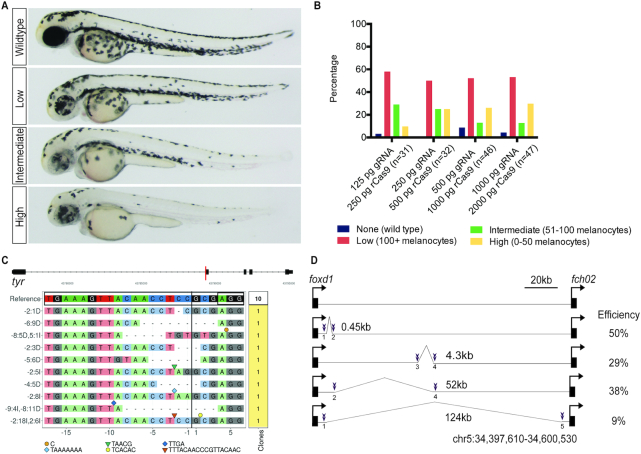
In order to evaluate the efficiency of synthetic gRNAs we targeted tyrosinase (tyr), a gene required for melanin synthesis. Wild-type zebrafish embryos were microinjected at the one-cell stage with recombinant Cas9 protein (rCas9) complexed with synthetic gRNA (IDT, Alt-R®). Embryos were scored for melanocyte number at 48 h post-fertilization (hpf) and divided into phenotypic categories representing the degree of gene editing (Figure 1A). We performed a dose response evaluation of editing efficiency as measured by phenotype and we find a modest increase in editing as the dose of rCas9 and gRNA increases (Figure 1B). In order to characterize the molecular alterations of synthetic gRNA injected embryos, we performed a clonal analysis. We find that 10/10 clones from a synthetic gRNA injected high-phenotype embryo contain a wide spectrum of indels (Figure 1C). We injected pairs of gRNAs to induce deletions in genomic DNA in a non-coding region 3′ of the foxd1 locus. We find that pairs of gRNAs are effective in inducing deletions in Fo injected embryos and that efficiency varies with deletion size (Figure 1D, Supplementary Figure S1). We targeted mlpha, a gene involved in melanosome trafficking, and observed highly penetrant phenotypes in F0 larvae that develop into adults phenocopying an established germline mutant (28), and transmitting an edited allele to F1 progeny (Supplementary Figure S2). Taken together, these data establish the efficacy of using synthetic gRNAs in combination with rCas9 to generate indels and deletions after microinjection in zebrafish embryos.
Figure 1.
Characterization of synthetic gRNAs in vivo. (A) Zebrafish embryos injected with rCas9 and a synthetic gRNA targeting tyr were examined by light microscopy. Embryos were binned into four categories based on the degree of phenotypic editing. (B) Dose–response of phenotype in embryos injected with rCas9 and synthetic gRNA targeting tyr. The percentage of embryos in each condition is plotted. (C) CrisprVariants (19) plot of indels in individual clones isolated from single embryos injected with synthetic gRNA targeting tyr. 10/10 clones contained indels. (D) Schematic of a 126kB region 3′ of foxd1 targeted for deletions using pairs of gRNAs. Guide locations are marked in purple and the deletion size is noted. Efficiency is calculated as the fraction of injected F0 embryos with the predicted allele-specific PCR product.
To monitor germline transmission of edited alleles, we applied CRISPR-STAT fluorescence-based genotyping (22) to sperm from F0 founders and their progeny. Sperm samples were collected from F0 founders injected with rCas9 and synthetic gRNA targeting slc6a15 at the one-cell stage. Results reveal mutant alleles containing indels as the major peaks, with additional minor peaks corresponding to wild-type and mutant alleles, indicating a high efficiency of CRISPR-induced mutagenesis in the germline (Supplementary Figure S3). We identified five distinct alleles that were transmitted to F1 progeny (Supplementary Figure S3B–F). The frequencies of observed CRISPR-induced indels in F1 progeny were similar to those found in the sperm of the F0 founder (Supplementary Figure S3G). These findings are consistent with high levels of somatic mosaicism and confirm that F0 sperm samples accurately predict the specific mutations transmitted to F1 progeny. These data demonstrate that CRISPR-induced indels created with synthetic gRNAs are found in the germline of founders and are efficiently transmitted to F1 progeny.
A genetic assay for homology-directed repair
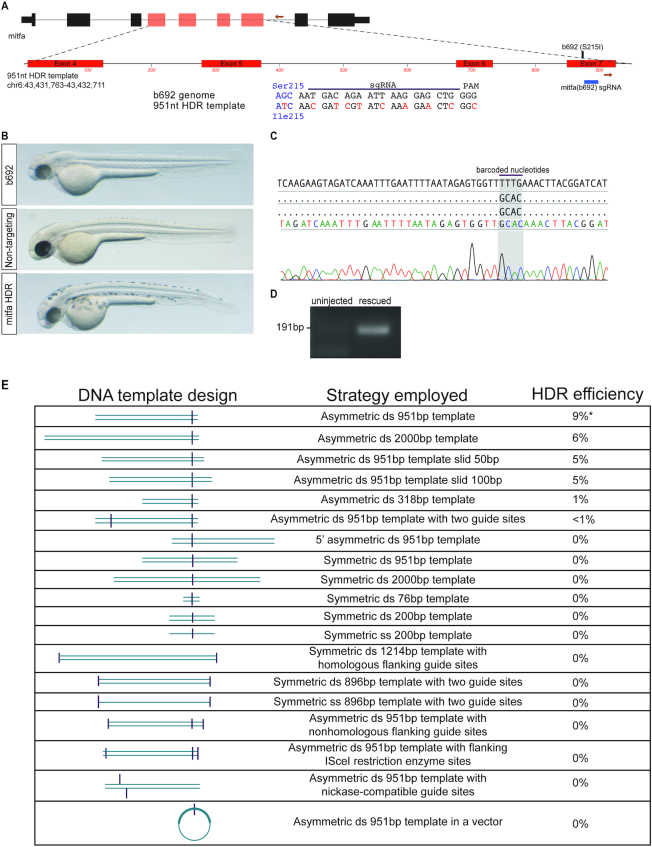
To develop a quantitative assay for optimizing HDR in zebrafish, we sought to target a locus that could provide phenotypic and molecular data on allele conversion. We initially examined the widely used mitfa(w2) mutant, which lacks melanocytes (29). Unexpectedly, injection of rCas9 complexed with a synthetic gRNA targeting the mutation site in w2 resulted in restoration of pigmented melanocytes (Supplementary Figure S4). We presume that indels caused by targeting the mitfa(w2) mutation site (Q113Stop) lead to in-frame deletions and restore protein function since this region of the protein is relatively unstructured. We next examined the mitfa(b692) mutant, which lacks melanocytes and harbors a mutation leading to a Ile215Ser substitution (30). We find that a synthetic gRNA targeting the mitfa(b692) mutation site does not restore pigmented melanocytes (Supplementary Figure S4). Therefore, we used mitfa(b692) to develop a quantitative phenotypic and molecular assay for optimizing HDR.
Using the b692-HDR assay we examined a range of synthetic templates to optimize HDR efficiency. mitfa(b692) embryos were microinjected with rCas9, synthetic gRNA, and synthetic DNA templates designed to restore the wild-type mitfa sequence and gene function. Microinjected embryos were scored at 48hpf for phenotypic evidence of melanocytes, indicative of allele conversion and mitfa function (Figure 2B). Melanocytes were not identified in uninjected embryos or in embryos injected with non-targeting gRNA. HDR templates were designed to feature multiple barcoded nucleotides and mutated gRNA recognition sequence, enabling unambiguous detection by sequencing and prevention of re-cleavage by rCas9. We found that a 951bp, double-stranded linear DNA template with an asymmetric gRNA site located at the 3′ end leads to a reproducible 9% rate of phenotypic rescue in mitfa(b692) embryos (Figure 2A). Melanocyte-positive embryos were subject to allele-specific PCR and Sanger sequencing to confirm precise HDR (Figure 2C and D). We examined DNA templates that differ in length, single vs. double stranded DNA, linear vs. circular templates, and the symmetry of the gRNA position within the template (Figure 2E). Unexpectedly we found that both longer (2kB) and shorter (318, 76 bp) DNA templates led to a decrease in HDR efficiency, as did relatively minor changes in template position relative to the gRNA site. Asymmetric positioning of gRNA site on the 951 bp template led to higher efficiency than symmetric positioning of the gRNA site on a template of identical length. These data indicate that HDR efficiency is sensitive to relatively minor changes in dsDNA template design, and they suggest that optimization of donor templates for specific loci may be necessary.
Figure 2.
A genetic assay for optimizing homology-directed repair using b692 mutant zebrafish. (A) Schematic depicting mitfa locus and 951 bp HDR template. The gRNA location in exon 7 is indicated. Primers used for allele-specific PCR are displayed as orange arrows. The HDR template encodes a wild-type Ile codon at position 215 and additional nucleotide changes to prevent re-cleavage at the gRNA recognition site (red). (B) mitfa(b692) embryos uninjected (top), injected with rCas9, 951bp DNA template, and non-targeting gRNA (middle) or mitfa gRNA and the 951 bp DNA template (bottom). All embryos shown at 48hpf. (C) Sanger sequencing results of allele-specific PCR products from a phenotypically rescued embryo. The sequence read spans the template-genome junction and includes template specific barcoded nucleotides. (D) Allele-specific PCR was performed on rescued embryos after microinjection with 951 bp template and compared with uninjected embryos. (E) Chart depicting attributes of DNA template, a description of the template, and observed rate of phenotypic rescue in the b692-HDR assay.
Using the b692 assay we examined multiple reagents and conditions reported to improve HDR efficiency in other model systems (Figure 2E). The addition of a second gRNA site in the optimal 951 bp template reduced HDR efficiency to <1%. We cloned the 951 bp template into a TopoTA plasmid vector and microinjected a circular plasmid into zebrafish. This plasmid-based template also failed to lead to measurable HDR in b692 embryos. In light of prior studies reporting increased HDR efficiency with the use of catalytically-mutated Cas9 (nickase Cas9) (31), we altered the 951 bp DNA template to allow us to test this approach. Microinjection of paired gRNAs and recombinant nickase Cas9 failed to lead to measurable HDR in b692 embryos. We did not, however, test ssODN donors as templates using nickase Cas9, which has been reported to be effective in some model systems. Based on reports that small molecule inhibitors of poly (ADP-ribose) polymerase (PARP) stimulate HDR efficiency (32), we performed microinjection of the 951 bp template and a PARP inhibitor, but did not observe any measurable HDR efficiency. Finally, we examined the effect of cleaving the ends of the linear 951 bp template with a restriction enzyme (ISce-I), but we did not observe measurable HDR under this condition.
Melanocyte restoration in b692 assay correlates with molecular efficiency
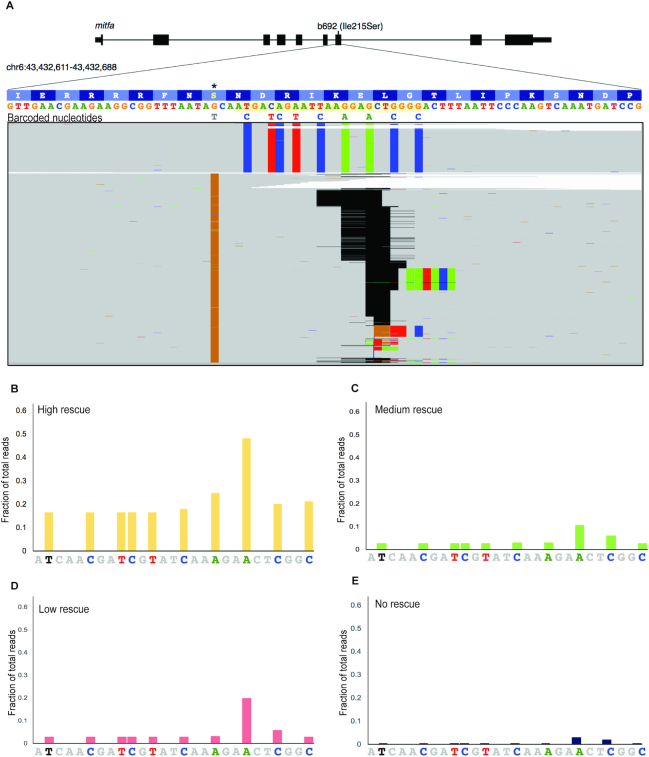
Using the b692-HDR assay we sought to measure the molecular efficiency of genome editing and correlate DNA template integration with melanocyte number. To this end, we performed single-cell injections in a large cohort of b692 embryos using the optimized reagents described: rCas9, synthetic b692 gRNA and 951 bp linear dsDNA template. Embryos were scored for the presence of melanocytes at 48hpf and binned by phenotype into four categories: high-, medium-, low- and no-editing. Uninjected embryos were included as a negative control. Genomic DNA was extracted from pools of five embryos in each category, and exon 7 was amplified by PCR. Amplicons from each group were subjected to next-generation sequencing. We find that that the efficiency of genome editing as determined by next-generation sequencing correlates with phenotype in b692 edited embryos. In high-rescued embryos, we find that 17% of reads (106 428 edited/641 281 total) harbor an edited codon restoring Ser215 to Ile, consistent with template integration. Analysis of aligned reads at exon 7 reveals the uniform presence of barcoded nucleotides from the HDR template across the gRNA target site (Figure 3A). Phenotypic rescue in medium and low categories revealed molecular efficiency of ∼3% (14 810/575 344 and 17 566/622 068 reads, respectively). Injected embryos with no phenotypic evidence of HDR had molecular efficiency of <0.3%. Alignment of reads demonstrates a mutually exclusive pattern of either HDR with template integration or indels at the gRNA site (Figure 3A), consistent with highly efficient dsDNA cleavage. We analyzed the molecular efficiency across all phenotypes and plotted the fraction of reads harboring barcoded nucleotides from the HDR template (Figure 3B–E). These data confirm the sensitivity of the b692-HDR assay to read out the molecular efficiency of HDR in vivo.
Figure 3.
Quantitative assessment of genome editing efficiency by next-generation sequencing. (A) Alignment of next generation sequencing reads from b692 genome edited embryos from the high rescue phenotype. Exon 7 of mitfa is displayed. The gRNA sequence is highlighted in yellow. b692 mutants have a T>G mutation leading to an isoleucine to serine substitution at codon 215(*). The 951bp HDR template restores the wild-type isoleucine codon (ATC) and encodes nine additional barcoded nucleotides as indicated. Indels in sequencing reads are represented as gapped regions in black. (B–E) The fraction of reads at each barcoded nucleotide in the HDR template is plotted for each phenotype category.
Genome editing using synthetic reagents leads to efficient fluorophore knock-in and germline transmission
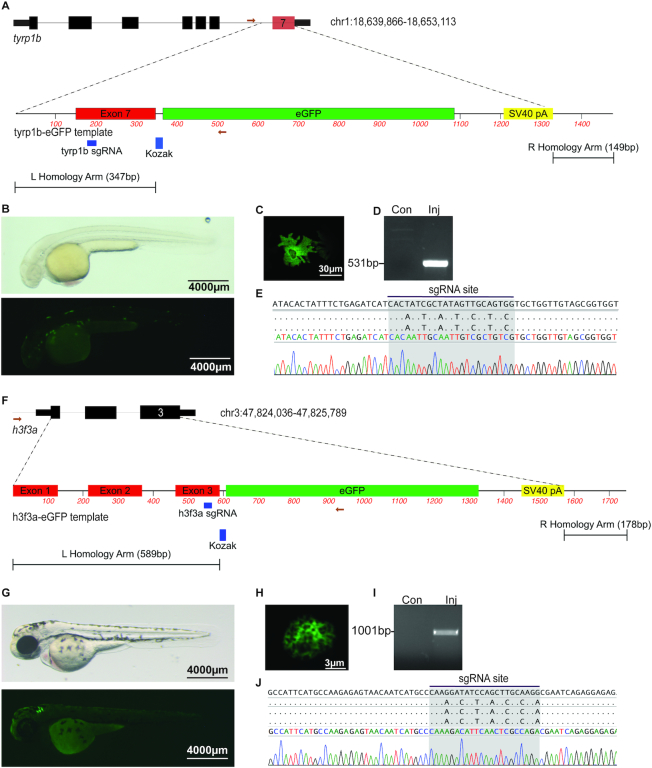
To determine the feasibility of using synthetic reagents to knock-in a fluorophore by HDR, we targeted tyrp1b, a melanocyte-specific gene. Using albino embryos (pigmentless due to a null mutation in slc45a2) we performed microinjection of rCas9 with a synthetic gRNA and a synthetic linear dsDNA template. The tyrp1b-EGFP template is a dsDNA template with the gRNA site mutated to prevent re-cleavage, removal of the terminal stop codon in tyrp1b, and inclusion of an EGFP cassette in frame with tyrp1b coding sequence (Figure 4A). We identified 7/153 (5%) microinjected embryos with EGFP-positive melanocytes at 48hpf (Figure 4B). Confocal imaging revealed the presence of EGFP-positive melanocytes with characteristic stellate morphology (Figure 4C). Allele-specific PCR was performed from injected embryos with EGFP-positive melanocytes (Figure 4D). Sanger sequencing revealed the presence of barcoded nucleotides encoded by the HDR template, consistent with template integration at the tyrp1b locus (Figure 4E).
Figure 4.
Genome editing leads to precise fluorophore knock-in. (A) Schematic depicting tyrp1b locus and tyrp1b-EGFP template. The gRNA recognition sequence in exon 7 is noted. (B) albino(b4) embryo injected with rCas9, tyrp1b synthetic gRNA, and the tyrp1b-EGFP template dsDNA template, light (top panel) or fluorescence (bottom panel) microscopy at 48hpf. (C) Confocal imaging reveals the presence of stellate EGFP-positive melanocyte. (D) Allele-specific PCR was performed on uninjected or injected/GFP+ embryos at 48hpf. (E) Sanger sequencing from allele-specific PCR detects template integration and modified nucleotides at the gRNA recognition site. (F) Schematic depicting h3f3a locus and h3f3a-eGFP template. The gRNA recognition sequence in exon 3 is noted. (G) Wild-type embryo injected with rCas9, h3f3a gRNA, and dsDNA template is photographed with light (top panel) or fluorescence (bottom panel) microscopy (48hpf). (H) Confocal imaging reveals the presence of an EGFP-positive nuclei. (I) Allele-specific PCR was performed on uninjected or HDR injected/GFP+ embryos at 48hpf. (J) Sanger sequencing was performed from allele-specific PCR to detect template integration. Barcoded nucleotides at the gRNA binding site are identified.
To determine the generalizability of this approach at other loci, we targeted h3f3a, one of several genes encoding histone H3.3. We designed a linear dsDNA template to knock EGFP in frame with h3f3a, creating a C-terminal fusion (Figure 4F). In this template, the entire coding sequence of h3f3a is replaced. We microinjected rCas9 with a synthetic gRNA targeting h3f3a and a synthetic dsDNA template into wild type embryos. We identified 12/152 (8%) embryos with EGFP-positive cells at 48hpf (Figure 4G). Confocal imaging revealed the presence of cells with a nuclear EGFP-positive signal characteristic of chromatin (Figure 4H). Allele-specific PCR and Sanger sequencing performed on EGFP positive embryos revealed precise template integration (Figure 4I, J). We raised mosaic F0 embryos to adulthood and assayed the rate of germline transmission. Of the 14 adult fish that mated, 2 (14%) produced EGFP+ F1 embryos (Supplementary Figure S5A, B). We characterized the integration site in F1 adult animals and we find precise template integration (Supplementary Figure S5C). We performed confocal microscopy on F1 embryos and were able to visualize chromosome condensation and mitosis in individual cells imaged at 4 h post-fertilization (Supplementary File). Taken together, these data demonstrate the ability of synthetic gRNAs and dsDNA templates to knock-in allele-specific fluorophores at several genomic loci to transmit edited alleles through the germline.
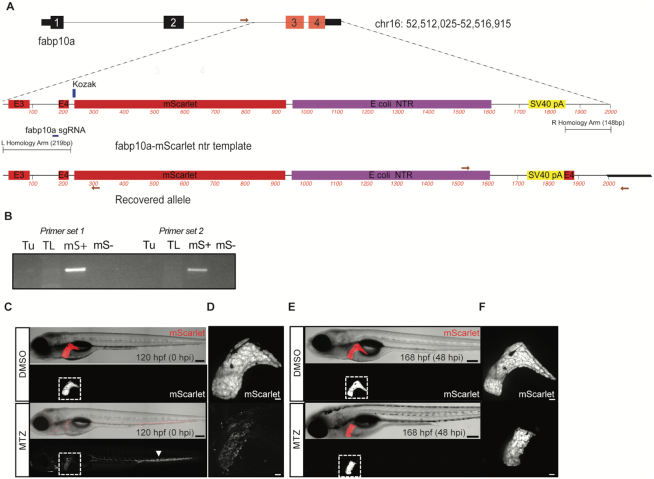
To expand the applications of genome editing using synthetic dsDNA templates, we targeted the liver-specific gene fabp10a using a template that knocks in mScarlet and bacterial nitroreductase (ntr) to facilitate hepatocyte ablation studies (Figure 5A). The zebrafish liver is capable of regeneration after injury (33), and expression of ntr has been used in combination with metronidazole (Mtz) to cause cellular injury (34). We injected embryos with rCas9, gRNA targeting fabp10a, and the HDR template and raised mScarlet+ F0 embryos to adulthood. Of adult fish that mated, 2/8 (25%) produced mScarlet+ F1 embryos. We characterized a recovered fabp10a allele in F1 animals using allele-specific PCR and confirmed integration of the template in mScarlet+ embryos (Figure 5B). Molecular analysis indicated that the 5′ end of the HDR template resulted in an in-frame fusion between exon 4 and mScarlet. At the 3′ end of the recovered allele, we identified a fragment of exon 4 distal to the gRNA recognition sequence, likely resulting from ligation to the template. We treated transgenic F1 embryos with Mtz to examine whether an fab10a-ntr fusion was functional and could lead to liver-specific cytotoxicity. Treatment of fabp10a-mScarlet-NTR larvae with Mtz from 96 to 120 hpf resulted in a dramatic reduction in the mScarlet fluorescent signal, signifying hepatocyte ablation without obvious damage to other tissues (Figure 5C, D). Remnants of the ablated hepatocytes were observed circulating in the vasculature. After a recovery period of 48 h in the absence of Mtz, mScarlet signal was observed in the liver of the Mtz-treated larvae, consistent with liver regeneration (Figure 5E, F).
Figure 5.
Knock-in of a synthetic dsDNA template encoding a fluorophore and bacterial nitroreductase (ntr) enables tissue-specific cellular ablation. (A) Schematic depicting fabp10a locus and fabp10a-mScarlet ntr template for HDR. A schematic of the recovered allele from F1 progeny confirms integration of mScarlet after the last coding exon of fabp10a and includes a duplicated exon 4. Primers used for PCR are displayed as red arrows. (B) Allele-specific PCR was performed to confirm integration of the template in mScarlet+ (mS+) embryos. (C–F) Mtz exposure induced hepatocyte injury in fabp10a-mScarlet-NTR larvae. (C) Confocal imaging revealed loss of fluorescence in the liver (outlined) of Mtz-treated but not DMSO-treated larvae. Remnants of ablated hepatocytes were found distributed throughout the vasculature (arrowhead). (D) Confocal imaging of liver in DMSO- and Mtz-treated larvae after injury. (E) At 48 h post injury (hpi), mScarlet signal was observed in the liver (outlined) of the Mtz-treated larvae, demonstrating liver regeneration. (F) Confocal imaging of liver in DMSO and Mtz treated larvae at 48 hpi. Scale bars are 200 μm (C, E) and 30 μm (D, F).
DISCUSSION
Here, we describe the development and application of genome editing in zebrafish using synthetic gRNAs in combination with linear dsDNA template and rCas9 protein. In order to optimize the efficiency of homology-directed repair we developed a genetic assay using mitfa(b692) mutant zebrafish. Our data demonstrate that the b692-HDR assay permits correlation between phenotype and HDR allele conversion (genotype). Using this assay, we systematically tested multiple template designs to determine which parameters are optimal for efficient knock-in by HDR. Based on these design principles we were successful in performing HDR knock-in of fluorophores at tyrp1b, h3f3a and fabp10a, achieving germline transmission rates of 14–25% at two loci, among the highest reported in zebrafish. At fabp10a integration of the HDR template led to liver specific mScarlet expression and included a functional ntr cassette for lineage ablation. At h3f3a integration of the HDR template led to a histone-EGFP fusion that permits visualization of chromosome condensation during mitosis in early embryogenesis. These results reveal the flexibility of approaches possible with synthetic dsDNA templates.
Unexpectedly, we found that small alterations in template design are associated with significant differences in HDR efficiency in the b692 HDR assay. These observations suggest that empiric optimization and testing of donor templates may be required to achieve knock-in at some loci. Intriguingly a prior report of donor optimization at a different locus (albino) using a genetic assay reached similar conclusions (5). In that report, the authors also found differences between templates and repair efficiency. Interestingly, they found the highest rate of repair with high concentration of a circular plasmid based template, though toxicity was also significant. Germline transmission rates in that study were slightly lower (∼11%) as compared with ours, and linear DNA templates had minimal efficacy. In our hands a plasmid containing the 951 bp sequence did not lead to any HDR, though we did not include flanking gRNA sites as the investigators in Irion et al. did in their design. These comparisons suggest that design optimization for HDR may differ between loci or depend on the specific reagents used.
Synthetic reagents hold an advantage in that both gRNA and dsDNA templates can be designed in silico and commercially manufactured in a short time frame. The efficiency of commercial gRNAs may result from chemical modifications that protect gRNAs from degradation. The use of gRNAs produced by commercial solid phase synthesis raises the possibility of incorporating degeneracy in the gRNA sequence to permit a single gRNA to target multiple related genes. Unlike in vitro synthesized gRNAs, synthetic gRNAs are not constrained by a requirement for a 5′ GG dinucleotide necessary for synthesis by RNA polymerase. The target sites we identified for HDR did not contain a 5′ GG. Had we used in vitro synthesized gRNAs we would have had to either incorporate a 5′ GG overhang in the template used for gRNA synthesis or introduce a 2 bp mismatch into the target recognition sequence. We did not perform a comparison of these approaches, but it seems reasonable to assume that mismatches may reduce cleavage efficiency at some target sites (35). Synthetic reagents also pose unique challenges for HDR projects. Our template designs were partially constrained by repetitive DNA sequences, particularly in non-coding DNA, that precluded commercial synthesis and limited the length of some homology arms. Commercial synthesis may also be associated with errors depending on template complexity, though the precise error rate is unknown. Sequence confirmation of in silico designed templates should therefore be considered.
The use of CRISPR/Cas9 to generate loss of function mutations in zebrafish and other model organisms makes it a uniquely valuable resource for genetics, developmental biology, and disease models. Our studies offer insight into the relative efficiency of HDR and provide investigators with a workflow for generating knock-in alleles. Our approach to genome editing should allow investigators to pursue a broad range of in vivo applications, including tissue-restricted lineage ablation, fluorophore and epitope knock-in, and generation of conditional alleles. Future improvements in HDR efficiency may occur with new variations in Cas9 protein and molecular and chemical tools to further enhance the efficiency of homology directed repair.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to members of the Houvras laboratory for critical discussion and manuscript review. We thank the Weill Cornell Genomics Resources Core Facility for next generation sequencing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Surgery, Weill Cornell Medical College (to Y.H.); National Institutes of Health/National Cancer Institute pre-doctoral fellowship [F31CA192813 to S.E.D.]; National Institutes of Health/National Cancer Institute pre-doctoral fellowship [F31CA213997 to R.M.]; Medical Scientist Training Program of General Medical Sciences of the NIH [T32GM007739] to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program (RM); National Center for Advancing Translational Sciences of the NIH [TL1-TR000459 to C.K.G.]; American Liver Foundation (to P.J.W.); National Institutes of Health post-doctoral fellowship [F32AA025271 to P.J.W.]; National Institutes of Health [R01DK090311 to W.G., R01DK105198 to W.G., R24OD017870 to W.G.]; Claudia Adams Barr Program in Innovative Basic Cancer Research (to W.G.); Wolfram Goessling is a Pew Scholar in the Biomedical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding for open access charge: Departmental funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jao L.E., Wente S.R., Chen W.. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shah A.N., Davey C.F., Whitebirch A.C., Miller A.C., Moens C.B.. Rapid reverse genetic screening using CRISPR in zebrafish. Nat. Methods. 2015; 12:535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hruscha A., Krawitz P., Rechenberg A., Heinrich V., Hecht J., Haass C., Schmid B.. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013; 140:4982–4987. [DOI] [PubMed] [Google Scholar]
- 4. Burg L., Palmer N., Kikhi K., Miroshnik E.S., Rueckert H., Gaddy E., MacPherson Cunningham C., Mattonet K., Lai S.L., Marin-Juez R. et al.. Conditional mutagenesis by oligonucleotide-mediated integration of loxP sites in zebrafish. PLos Genet. 2018; 14:e1007754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Irion U., Krauss J., Nusslein-Volhard C.. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development. 2014; 141:4827–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Kaini P., Sander J.D., Joung J.K., Peterson R.T., Yeh J.R.. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One. 2013; 8:e68708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Auer T.O., Duroure K., De Cian A., Concordet J.P., Del Bene F.. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014; 24:142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hisano Y., Sakuma T., Nakade S., Ohga R., Ota S., Okamoto H., Yamamoto T., Kawahara A.. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci. Rep. 2015; 5:8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ota S., Taimatsu K., Yanagi K., Namiki T., Ohga R., Higashijima S.I., Kawahara A.. Functional visualization and disruption of targeted genes using CRISPR/Cas9-mediated eGFP reporter integration in zebrafish. Sci. Rep. 2016; 6:34991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kimura Y., Hisano Y., Kawahara A., Higashijima S.. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep. 2014; 4:6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B. et al.. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015; 33:985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rahdar M., McMahon M.A., Prakash T.P., Swayze E.E., Bennett C.F., Cleveland D.W.. Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E7110–E7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gutierrez-Triana J.A., Tavhelidse T., Thumberger T., Thomas I., Wittbrodt B., Kellner T., Anlas K., Tsingos E., Wittbrodt J.. Efficient single-copy HDR by 5′ modified long dsDNA donors. Elife. 2018; 7:e39468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoshijima K., Jurynec M.J., Grunwald D.J.. Precise editing of the zebrafish genome made simple and efficient. Dev. Cell. 2016; 36:654–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chari R., Mali P., Moosburner M., Church G.M.. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods. 2015; 12:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chari R., Yeo N.C., Chavez A., Church G.M.. sgRNA Scorer 2.0: a species-Independent model to predict CRISPR/Cas9 activity. ACS Synth. Biol. 2017; 6:902–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perez A.R., Pritykin Y., Vidigal J.A., Chhangawala S., Zamparo L., Leslie C.S., Ventura A.. GuideScan software for improved single and paired CRISPR guide RNA design. Nat. Biotechnol. 2017; 35:347–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Varshney G.K., Zhang S., Pei W., Adomako-Ankomah A., Fohtung J., Schaffer K., Carrington B., Maskeri A., Slevin C., Wolfsberg T. et al.. CRISPRz: a database of zebrafish validated sgRNAs. Nucleic Acids Res. 2016; 44:D822–D826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindsay H., Burger A., Biyong B., Felker A., Hess C., Zaugg J., Chiavacci E., Anders C., Jinek M., Mosimann C. et al.. CrispRVariants charts the mutation spectrum of genome engineering experiments. Nat. Biotechnol. 2016; 34:701–702. [DOI] [PubMed] [Google Scholar]
- 20. Draper B.W., Moens C.B.. A high-throughput method for zebrafish sperm cryopreservation and in vitro fertilization. J. Vis. Exp. 2009; doi:10.3791/1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meeker N.D., Hutchinson S.A., Ho L., Trede N.S.. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. BioTechniques. 2007; 43:610.612, 614. [DOI] [PubMed] [Google Scholar]
- 22. Carrington B., Varshney G.K., Burgess S.M., Sood R.. CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Res. 2015; 43:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holleley C.E., Geerts P.G.. Multiplex Manager 1.0: a cross-platform computer program that plans and optimizes multiplex PCR. BioTechniques. 2009; 46:511–517. [DOI] [PubMed] [Google Scholar]
- 24. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thorvaldsdottir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Westerfield M. The Zebrafish Book, 5th Edition: A Guide for the Laboratory Use of Zebrafish (Danio rerio). 2007; Eugene: University of Oregon Press. [Google Scholar]
- 28. Sheets L., Ransom D.G., Mellgren E.M., Johnson S.L., Schnapp B.J.. Zebrafish melanophilin facilitates melanosome dispersion by regulating dynein. Curr. Biol. 2007; 17:1721–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lister J.A., Robertson C.P., Lepage T., Johnson S.L., Raible D.W.. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999; 126:3757–3767. [DOI] [PubMed] [Google Scholar]
- 30. Lister J.A., Close J., Raible D.W.. Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Dev. Biol. 2001; 237:333–344. [DOI] [PubMed] [Google Scholar]
- 31. Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E.. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016; 34:339–344. [DOI] [PubMed] [Google Scholar]
- 32. Anantha R.W., Simhadri S., Foo T.K., Miao S., Liu J., Shen Z., Ganesan S., Xia B.. Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance. Elife. 2017; 6:e21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cox A.G., Goessling W.. The lure of zebrafish in liver research: regulation of hepatic growth in development and regeneration. Curr. Opin. Genet. Dev. 2015; 32:153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Curado S., Stainier D.Y., Anderson R.M.. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat. Protoc. 2008; 3:948–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al.. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.