Abstract
WRKY transcription factors play crucial roles in regulation mechanism leading to the adaption of plants to the complex environment. In this study, AhWRKY family was comprehensively analyzed using bioinformatic approaches in combination with transcriptome sequencing data of the drought-tolerant peanut variety ‘L422’. A total of 158 AhWRKY genes were identified and named according to their distribution on the chromosomes. Based on the structural features and phylogenetic analysis of AhWRKY proteins, the AhWRKY family members were classified into three (3) groups, of which group II included five (5) subgroups. Results of structure and conserved motifs analysis for the AhWRKY genes confirmed the accuracy of the clustering analysis. In addition, 12 tandem and 136 segmental duplication genes were identified. The results indicated that segmental duplication events were the main driving force in the evolution of AhWRKY family. Collinearity analysis found that 32 gene pairs existed between Arachis hypogaea and two diploid wild ancestors (Arachis duranensis and Arachis ipaensis), which provided valuable clues for phylogenetic characteristics of AhWRKY family. Furthermore, 19 stress-related cis-acting elements were found in the promoter regions. During the study of gene expression level of AhWRKY family members in response to drought stress, 73 differentially expressed AhWRKY genes were obtained to have been influenced by drought stress. These results provide fundamental insights for further study of WRKY genes in peanut drought resistance.
Introduction
The WRKY transcription factors are one of the largest families of transcriptional regulators in plants and form an integral part of signaling networks that regulate many plant processes [1]. They are important to plant growth by regulating development and responding to stresses [1–7]. WRKY proteins contain at least one WRKY domain which consists of approximately 60 amino acids with a highly conserved WRKYGQK heptapeptide at the N-terminus, and include a C2H2 (C–X4–5–C–X22–23–H–X–H) or C2HC (C–X7–C–X23–H–X–C) zinc-finger motif in C-terminus. WRKY transcription factors regulate gene expression by exclusively binding to W-box, which is a cis-element in the promoter regions of target genes [2]. Based on the number and characteristics of WRKY domains and zinc finger structure, respectively, the WRKY proteins can be divided into three (3) main groups, namely group I, II and III. The group I contains two (2) WRKY domains and a zinc finger motif of C2H2 type (C-X4-C-X22–23-H-X1-H), group II has a single WRKY domain and a zinc finger motif of C2H2 type (C-X4–5-C-X23-H-X1-H) and group III consists of a single WRKY domain and a zinc-binding motif of C2HC type (C-X7-C-X23-H-X1-C). Group II is further partitioned into five (5) subgroups (IIa, IIb, IIc, IId, and IIe) based on the additional conserved structural motifs of the WRKY structure [8].
A large number of studies on a functional analysis of WRKY genes have indicated the involvement of WRKY transcription factors in plant defense regulatory networks and developmental processes. SPF1 was the first WRKY transcription factor to be reported in sweet potato [9]. And with the continuous development of genome sequencing and other high throughput technologies, members of the WRKY gene family are being identified and perform diverse roles in various species including Ipomoea batatas [9], Arabidopsis thaliana [10], Oryza sativa [11], Glycine max [12], Gossypium raimondii [13], Lotus japonicas [14], Triticum aestivum [15], Vitis vinifera [16], Ananas comosus [17], Arachis duranensis and Arachis ipaensis [18]. For instance, in Cucumis, the expression of CsWRKY50 could positively regulate resistance to Pseudoperonospora cubensis involving multiple signaling pathways [19]. In rice, it has been reported that OsWRKY71 was up-regulated by defense signaling molecules, such as salicylic acid (SA), methyl jasmonate (MeJA) and pathogen infection, indicating that this member of the WRKY gene family might be involved in biotic stress response [20]. OsWRKY53 has also been found to positively regulate the brassinosteroid (BR) signaling and mediate the cross-talk between the hormone and other signaling pathways [21]. And OsWRKY78 member played an important role in regulation of stem elongation and seed development [22]. In Arabidopsis, AtWRKY8 may participate in defensive response to the infection of crucifer-infecting tobacco mosaic virus (TMV-cg) as the virus inhibited its expression [23]. It has also been reported that AtWRKY75 is involved in the regulation of Phosphate (Pi) deficiency response and root development [24]. AtWRKY6 has also been found to be related to the triggering of senescence process and defense response [25]. In transgenic Arabidopsis thaliana, overexpression of AtWRKY13 resulted in decreased Cadmium (Cd) accumulation with increased tolerance to it [26]. In grape, the expression of VvWRKY30 could improve salt tolerance by regulating the elimination of reactive oxygen and the accumulation of osmoticum [27]. Overexpression of TaWRKY2 conferred strong drought tolerance to transgenic wheat [28]. And ScWRKY1 was highly and transiently expressed in fertilized ovules bearing late torpedo-staged embryos of potato, revealing a key role during embryogenesis [29].
Despite peanut (Arachis hypogaea L.) being one of the most important oil crops in the world and widely cultivated in the tropical and subtropical regions, its yields, particularly on farmers’ fields, are still low which is largely due to biotic and abiotic stresses. Drought is one of the major abiotic stress factors in limiting peanut production. It affects the physiology, biochemistry and molecular biology of plants [30–32]. And with peanut mainly cultivated under rainfed conditions by resource-poor farmers, breeding for drought-resistant peanut cultivars is the most economical and effective way to reduce the yield loss caused by menace. However, drought resistance in itself is a complex trait governed by a large number of genes [33, 34]. As a result, the mining and identification of potential genes associated with this trait will be of great importance for the development and subsequent cultivation of drought-resistant peanut varieties. Also, in view of the important roles played by WRKY transcription factors in stress response, a functional analysis of these transcription factors could offer an alternative means to improve peanut stress tolerance in general.
Information about the WRKY gene family has been characterized in the two wild ancestral species of peanut. In a previous study, 75 AdWRKYs from A. duranensis and 77 AiWRKYs from A. ipaensis were identified through bioinformatic approaches and Arachis WRKY proteins potentially controlling disease-resistance were deduced [18]. So far, the whole genome sequence of cultivated peanut (Arachis hypogaea cv. Tifrunner) has been completely sequenced [35] and released (http://www.peanutbase.org/). However, WRKY gene family involved in drought-responsive processes has not been systematically identified in cultivated peanut. In this study, the sequence features, chromosomal distribution, phylogenetic relationships, cis-elements, gene duplications, and synteny of AhWRKYs were comprehensively analyzed through bioinformatic methods. The gene expression level of WRKY gene family members in response to drought stress on the basis of the transcriptome sequencing data was also studied. The results will provide valuable information for further functional investigations of the AhWRKY gene family and preliminary knowledge of AhWRKYs potentially involved in drought resistance.
Materials and methods
Identification of WRKYs
Two approaches were used in the process of identifying WRKYs. In the first approach, the genome of Arachis hypogaea cv. Tifrunner was downloaded from the peanut genome database (PeanutBase) (https://www.peanutbase.org). Also, a Pfam file with multiple sequence alignments and a hidden Markov model (HMM) corresponding to the WRKY domain (PF03106) was downloaded from the Pfam database (http://pfam.xfam.org/). HMMER 3.0 was used to search for WRKY genes from the downloaded genome based on the default parameters at a probability value of 0.01. In the second approach, WRKY protein sequences for Arabidopsis (AtWRKY) were downloaded from the Arabidopsis Information Resource website (http://www.arabidopsis.org/), whiles WRKY sequences of A. duranensis (AdWRKY) and A. ipaensis (AiWRKY) were obtained from previous study [18]. The WRKYs from the second approach were used to validate those obtained from the first approach. After then, all validated candidate genes with probable WRKY domain were further validated using PFAM and SMART programs (http://smart.embl-heidelberg.de) to retain the only sequences with WRKY domain.
Additionally, the primary structure parameters of the genes (length of sequences, molecular weight, and isoelectric point) were analyzed using the ExPASy website (http://web.expasy.org/protparam/). The prediction of subcellular location of identified WRKY proteins was performed using Plant-mPLoc [36], ProtComp 9.0 and WoLF PSORT (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi; http://linux1.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc; https://wolfpsort.hgc.jp).
Sequence alignment and phylogenetic analysis
Multiple sequence alignment of Arachis hypogaea WRKYs (AhWRKYs) domain sequences was executed using the Clustal X 2.1 program with default parameters [37] and results were colored using GeneDoc. Newly identified AhWRKYs were clustered into different groups based on the classification schemes of AtWRKYs, AdWRKYs, and AiWRKYs [8, 18]. The phylogenetic tree was generated and viewed using MEGA 7.0 with Maximum Likelihood (ML) method and 1000 replicates [38]. Gene structures were generated via an online tool GSDS (http://gsds.cbi.pku.edu.cn/) by comparing CDS of AhWRKYs with their corresponding full-length sequences [39]. The 1500 bp upstream of the coding region of AhWRKYs were selected and submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) for predicting the cis-regulatory element of promoter [40]. The conserved motifs in the AhWRKYs were identified statistically using the MEME program (http://alternate.meme-suite.org/tools/meme) with default settings except the maximum number of motifs to be found which was set at 10 [41].
Chromosomal distribution and gene duplication of AhWRKYs
Positional information of all candidate AhWRKYs was retrieved from the PeanutBase, and the locations were drafted using MG2C (http://mg2c.iask.in/mg2c_v2.0). Duplication events of genes were analyzed by means of Multiple Collinearity Scan toolkit (MCScanX) with the default parameters [42]. In order to exhibit the orthologous WRKY genes between peanut and other selected species, syntenic analysis maps were constructed using Circos software [43].
Plant materials used and drought treatment
Two peanut cultivars, L422 (drought-tolerant) and L677 (drought-sensitive) were used for this study. Seeds were planted in rainout shelters under well-watered conditions in Baoding, China. The relative moisture content of the soil was maintained at 70–75% until the plants reached the reproductive phase (pegging stage) which coincided with 60 days after planting (DAP). At pegging, each plot of the two cultivars was split into two units to set up the control and treatment groups. The control group continued to be under well-watered conditions while the treatment group received no irrigation. Fully expanded leaves from the main stem (third nodal) were randomly selected after 20 days (80 DAP) of exposure. Fresh leaves of control and treatment plants were collected at 80, 85, 95, and 100 DAP, respectively. Three (3) independent samples were collected and used as biological replicates. The samples were immediately frozen in liquid nitrogen and stored at -80°C for subsequent analyses.
Physiological index measurements
Physiological parameters were measured on the L422 and L677 plants both under well-watered and drought-stress conditions. The level of lipid peroxidation (MDA content) in the leaves was determined by Thiobarbituric acid (TBA) method [44]. Additionally, leaf superoxide dismutase (SOD) activity was measured spectrophotometrically at 560 nm according to a previously reported method [45]. Soluble protein content of samples was also measured by Coomassie Brilliant Blue method [46]. Each parameter measurement on each cultivar under each treatment group was replicated three (3) times using independent but parallel approaches. Student’s t-test was performed to determine the significance of treatment differences using Origin 8.0 software version v6.1052 (B232) (OriginLab Corp, Northhampton, MA, USA). Differences were considered significant if p-value was less than 0.05.
Total RNA extraction, cDNA library construction, and RNA-Seq
The isolation of total RNA from non-stressed and stressed plant leaves was optimized according to the instruction manual of the Trizol Reagent (Invitrogen, Carlsbad, CA, USA). RNA degradation and contamination were monitored on 1% agarose gel and its quality was evaluated using NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). The cDNA library construction and RNA sequencing were carried out on the Illumina HiseqTM 2500 platform using the 125-bp paired-end sequencing protocol of Illumina by Biomarker Technology Co. (Beijing, China).
Sequencing reads processing and mapping
Raw reads in a FASTA format were first processed using in-house Perl scripts. In this step, clean reads (clean data) was obtained by removing reads with low quality and containing adapters, and ploy-N. Q20 (99% base call accuracy), Q30 (99.9% base call accuracy), GC-content and sequence duplication level of the clean data were calculated. All the downstream analyses were based on high-quality clean data. Qualified clean reads were then mapped to the peanut reference genome sequence (Tifrunner.gnm1.ann1.CCJH) using a spliced aligner Tophat 2.0 software [47]. Only reads with a perfect match or only one mismatch were further analyzed and annotated according to the reference genome. Quantification of gene expression was calculated as FPKM (fragments per kilobase of exon model per million mapped reads) and the FPKM values were log2 transformed [48]. The aligned reads were analyzed using StringTie [49], and after then the assembled transcripts were further filtered using the FPKM value > 1.
Gene expression analysis and functional annotation of AhWRKYs
To identify differential expression genes (DEGs) between two different samples, the expression level of each transcript was calculated according to the FPKM method. EBSeq R software package (4.0) was used to execute gene differential expression analysis of two or more biological conditions in an RNA-seq experiment based on the negative binomial distribution model [50]. In case of no biological replicates, the package would estimate the variances of RNA-seq data by pooling similar genes together. p-values obtained via EBseq were further corrected using the Benjamini-Hochberg procedure [51], and the corrected p-values were used to determine the false discovery rate (FDR). Genes were considered to be differentially expressed when the value of log2 Fold Change was >1 or <-1 with an FDR value less than 0.05. To analyze the potential function of genes, all the genes were functionally annotated using Gene Ontology (GO) (http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.kegg.jp/) databases. GO functional classification enrichment analysis was performed using the OmicShare tools, an online platform for data analysis (http://www.omicshare.com/tools). Firstly, all the differentially expressed WRKY genes were mapped to GO terms in the Gene Ontology database, and gene numbers were calculated for every term. And then the differentially expressed WRKYs with significantly enriched GO terms were compared to the genome background and defined by hypergeometric test. Similarly, for each KEGG pathway, the numbers of differentially expressed WRKYs were compared to the entire reference gene set by hypergeometric tests using the OmicShare tools to determine the pathways enriched for differentially regulated genes. The calculated p-value was corrected through FDR Correction, taking FDR ≤ 0.05 as a threshold. The heatmap of AhWRKYs expression was also created using the OmicShare tools. All transcriptome data have been deposited at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/sra/PRJNA544421).
Validation of gene expression
Eleven (11) AhWRKY genes obtained by Illumina RNA-seq that showed differentially expressed and contained cis-recognition elements involved in the abiotic/biotic stress were randomly selected for validation by quantitative real-time PCR (qRT-PCR). All gene-specific primers were designed by Wcgene Biotech (Shanghai, China) (S1 Table). The housekeeping gene EF1b [52] was used as an internal control gene for qRT-PCR normalization. The qRT-PCR was run on the LightCycler® 96 instrument using Fast Super EvaGreen® qPCR Master Mix (US Everbright®Inc., China) based on the manufacturer’s instructions. The amplification program was set as follows: 95°C for 2 min followed by 45 cycles of 95°C for 5 s and 60°C for 1 min. Three (3) and two (2) biological and technical repetitions, respectively, were used for each gene sample. All data from qRT-PCR amplification were calculated with 2−△△CT method [53]. All expression data were analyzed using the Origin 8.0 software version v6.1052 (B232) (OriginLab Corp, Northhampton, MA, USA).
Results
Identification of WRKY proteins in peanut cultivars used
A total of 158 members of WRKY family were identified using bioinformatic approaches, mapped onto chromosomes and were designated from AhWRKY1 to AhWRKY158 based on their locations on the respective chromosome (S2 Table). The particularizations including open reading frame lengths (ORF), amino acid numbers, molecular weights (MW), isoelectric point (pI), and the predicted subcellular locations are all shown in S2 Table. The lengths of the 158 identified AhWRKY proteins varied from 128 (AhWRKY29) to 1345 (AhWRKY54) amino acids. Additionally, the pI and MW ranged from 4.92 (AhWRKY43) to 9.92 (AhWRKY85) and from 14.9 kDa to 148.6 kDa, respectively. Results from the predicted subcellular localization showed 155 AhWRKY proteins were located in the nuclear region, whereas one protein was located in the cell membrane. In addition, two (2) proteins were located in both the cell membrane and the nuclear region, which suggests they might have special biological functions. The detailed information, including gene loci number, conserved motif, zinc finger domain pattern and gene grouping are also given in S2 Table.
Multiple sequence alignment, phylogenetic tree analysis and group identification
To investigate the evolutionary relationship and classification of AhWRKYs, a total of 158 WRKY domains spanning approximately 60 amino acids were executed. For different groups, seven (7) Arabidopsis proteins were selected randomly as representative sequences (AtWRKY33; 60; 47; 48; 11; 22; 55). Detailed structures about the WRKY domain and zinc finger type are displayed in S1 Fig. One hundred and forty-two (142) of the 158 AhWRKY proteins contained WRKYGQK sequence which is a highly conserved sequence. However, six (6) variants of this conserved WRKY motif including GRKYGQK, WRKYGEK, WRKYGKK, WRKYGRK, WRKYDKK and WHKYGKK were distributed in subgroup IIc and I, respectively. Strikingly, C2HY, the zinc finger motif form variation was identified in AhWRKY13. Also, the WRKY sequences that had lost the WRKYGQK motif or the zinc finger motif were distributed in subgroups IIa, IIb, IIc and I, respectively.
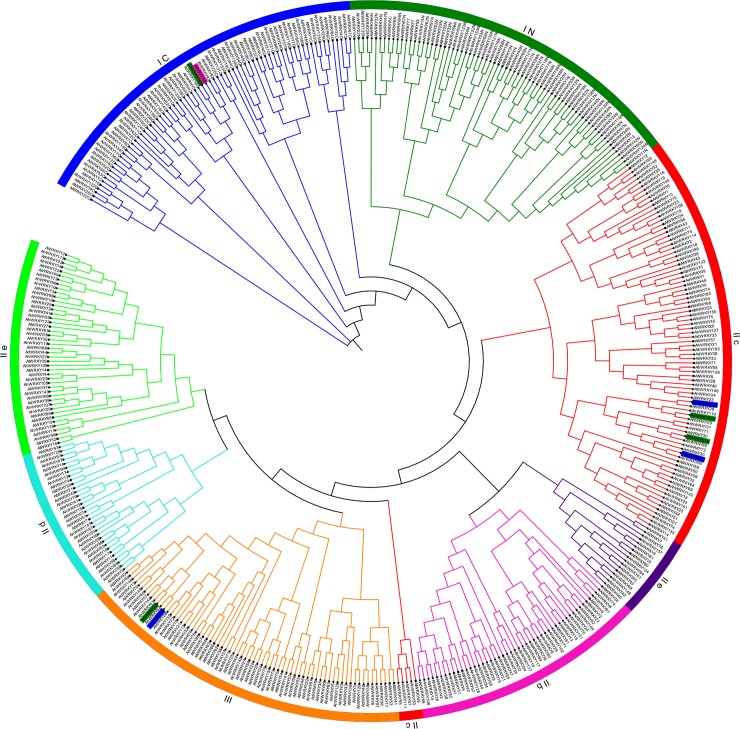
158 AhWRKYs, 72 AtWRKYs, 75 AdWRKYs, and 77 AiWRKYs were used to build the NJ phylogenetic tree (Fig 1). The phylogenetic relationship among AtWRKYs, AdWRKYs and AiWRKYs was consistent with the results of previous studies [8, 18]. Based on results of previous classification [8], the 158 AhWRKYs were grouped into three major clusters and five subgroups; I (33), IIa (6), IIb (22), IIc (38), IId (12), IIe (20), and III (27). However, I-N, I-C, IIb, IIc and III members were found to be nested in other groups or subgroups when the phylogenetic relationship between AdWRKYs and AiWRKYs was analyzed, which is similar to the results obtained in a previous study [18]. The result indicated that the origin of peanut WRKY proteins is still to be clarified. Additionally, the zinc finger motif (C-X4-C-X23-H-X1-C) was identified in AhWRKY71. Interestingly, three (3) WRKYs (AhWRKY71, AhWRKY72, and AhWRKY152) which belonged to group III possessed leucine-rich repeat (LRR) domain that was frequently involved in the formation of protein-protein interactions [10, 18, 54–56]. It is believed that these three (3) proteins may be associated with disease resistance in peanut.
Fig 1. Phylogenetic tree of WRKYs among Arachis hypogaea, Arabidopsis thaliana, Arachis duranensis and Arachis ipaensis.
The groups (I, II and III) and subgroups (IIa, b, c, d and e) were distinguished in different colors, and the different species were marked by different symbols. Stars represented WRKY genes from Arachis hypogaea, and squares indicated genes from Arabidopsis thaliana. Circles and right triangles indicated WRKY genes from Arachis ipaensis and Arachis duranensis, respectively.
Gene structure and motif composition of AhWRKYs family
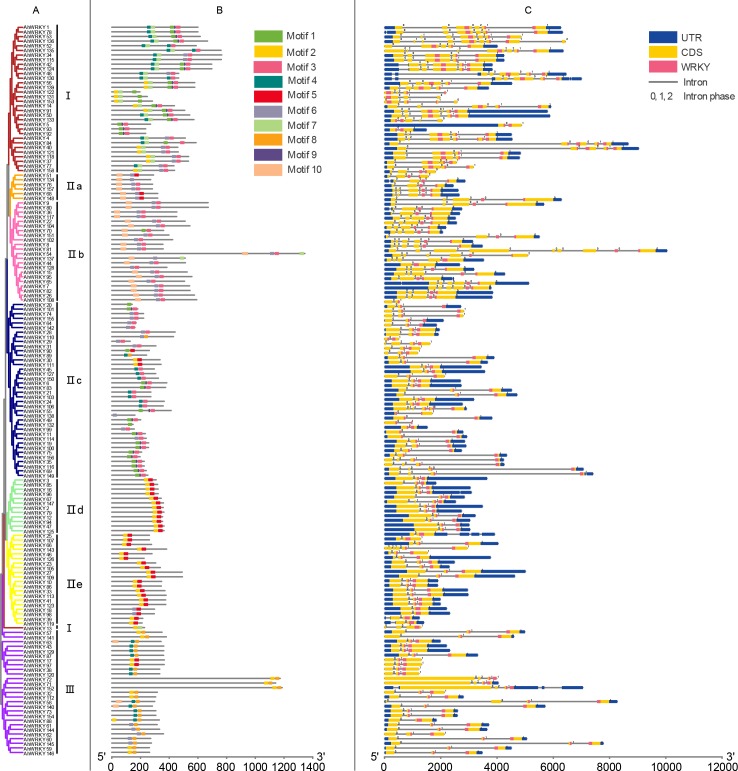
The exon-intron distribution and motif composition were analyzed to gain more insight into the AhWRKY genes based on phylogenetic relationships. Gene structure analysis showed that each of the AhWRKY consisted of exons separated by variable numbers of introns. The detailed exon-intron map is shown in Fig 2C. Seventy-one (71) AhWRKYs contained two introns and accounted for the maximum proportion (44.9%) of the genes. This was followed by 15 (9.5%), 20 (12.7%), 31 (19.6%), 11 (7.0%), 7 (4.4%) and 3 (1.9%) genes, possessing 1, 3, 4, 5, 6, and 8 introns, respectively. Among them, subgroup IIa genes contained either three (3) or four (4) introns, while most genes in subgroups IIc, IId and IId as well as group III had two (2) introns. The number of introns in group I and subgroup IIb were extremely variable, including 1, 2, 3, 4, 5, 6, 8 and 1, 3, 4, 5, 6, 8, respectively.
Fig 2. Structure characterization of AhWRKY gene family.
A Maximum Likelihood (ML) phylogenetic tree of AhWRKYs. The various groups were distinguished in different colors. B The conserved motifs of AhWRKYs. The 10 motifs were displayed in different-colored boxes. For details of motifs referred to S1 File. C The structures of AhWRKYs. The untranslated 5′- and 3′-regions were represented by blue boxes. The yellow boxes and gray lines indicated CDS and introns, respectively. The WRKY domains were highlighted by red boxes. The introns phases 0, 1 and 2 were indicated by numbers 0, 1 and 2, respectively.
Most members of AhWRKYs found in the same group had similar exon-intron structures. An intron was located between two complete codons named phase 0, and after the first or second nucleotides in the codon defined phase 1 and 2. Specifically, members of the subgroups IIa and IIb only found phase 0. The same results were observed in pineapple [17]. Phase 1 was widely distributed in other groups with phase 2 having the least distribution.
The conserved motifs were analyzed and assigned numbers from one (1) to ten (10). As shown in Fig 2B, motifs 1, 2, 4, and 6 contained a WRKYGQK sequence. Most AhWRKYs that have been observed to be in the same group or subgroup shared similar motif compositions. For instance, most members of groups IIb contained motifs 3, 6, and 10. Furthermore, subgroup IId and IIe shared motifs 2 and 5 except AhWRKY18 and AhWRKY98, indicating functional similarity among them. Overall, the similar exon-intron structures and conserved motif compositions of the WRKY members in the same group further validated the categorization of AhWRKYs as well as the phylogenetic relationships.
Chromosome location and gene duplication
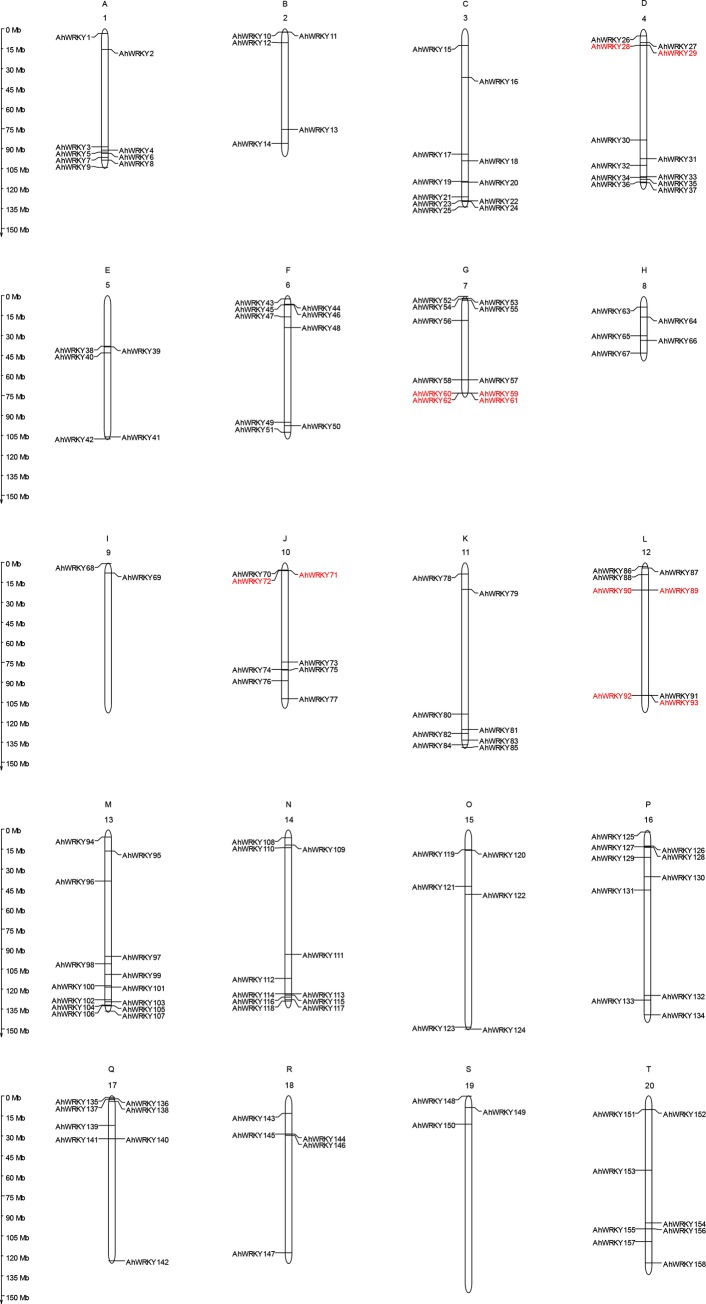
The 158 identified AhWRKYs were distributed across all the 20 peanut chromosomes (S2 Table; Fig 3) with chromosome 13 containing the highest number (14). On the other hand, chromosomes nine (9) had the least number (2) of AhWRKYs. Additionally, 5 chromosomes (10, 11, 12, 17, and 20) had eight (8) AhWRKYs whiles chromosomes 1, 3, 4, 6, 7, 14, and 16 harbored relatively more gene members than chromosomes 2, 5, 8, 15, 18, and 19. This is an indication that although the AhWRKYs were found on all 20 chromosomes, their distribution across the respective chromosomes was uneven.
Fig 3. Physical mapping of AhWRKY genes on chromosome.
The chromosome numbers were indicated at the top of each chromosome. The scale is in megabases (Mb). Tandem duplication genes were marked in red.
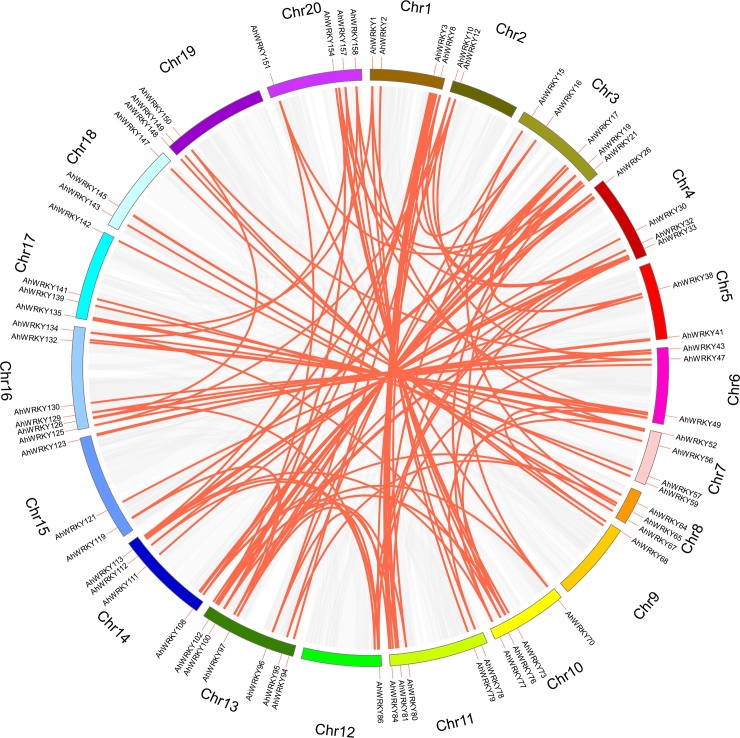
In the process of evolution, tandem duplication and segmental duplication contribute to the generation of gene families [57]. To explore these events, a chromosomal region within 200 kb containing two or more genes was defined as a gene cluster [58]. According to the defined criteria, 12 tandem duplication genes were found to form six gene clusters (AhWRKY92/AhWRKY93, AhWRKY59/AhWRKY60, AhWRKY89/AhWRKY90, AhWRKY28/AhWRKY29, AhWRKY61/AhWRKY62 and AhWRKY71/AhWRKY72). The chromosomes numbered four (4) and ten (10) had one (1) gene clusters, whereas chromosomes numbered seven (7) and twelve (12) contained two (2) gene clusters (Fig 3). On the other hand, 124 segmental duplication events (136 AhWRKYs) were found and were distributed on all 20 chromosomes (S3 Table, Fig 4). The results indicated that segmental duplication events were the main driving force in the evolution of AhWRKY gene family.
Fig 4. The segmental duplications of AhWRKY members.
The different color blocks indicated the part of peanut chromosomes. The gray lines indicated all segmental duplications in the peanut genome, and the red lines indicated segmentally duplicated WRKY gene pairs.
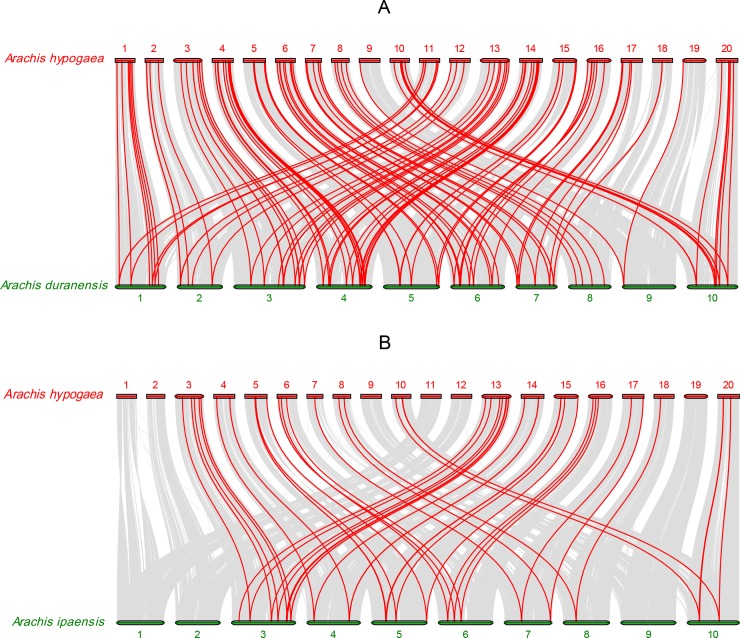
To further infer the phylogenetic mechanisms of peanut WRKY family, two comparative syntenic maps associated with two diploid wild ancestors (A. duranensis and A. ipaensis) that have been deduced as the donors of the A and B subgenomes, respectively, were constructed [59–61]. The numbers of collinear gene pairs between A. duranensis and A. ipaensis were 110 and 43, respectively (S4 Table). As shown in Fig 5, the collinear gene pairs between the Arachis hypogaea and A. duranensis were randomly distributed on all 20 chromosomes with chromosomes 9, 18 and 19 having the fewest collinear gene pairs. In addition, the collinear gene pairs between Arachis hypogaea and A. ipaensis were less than that of Arachis hypogaea and A. duranensis, and no collinear gene pairs were observed on chromosome 1, 2, 9, 11, 12 and 19. Notably, some collinear gene pairs (32 AhWRKYs) were found between Arachis hypogaea and A. duranensis/A. ipaensis.
Fig 5. The collinearity analysis of WRKY genes between Arachis hypogaea and two diploid wild ancestors (Arachis duranensis and Arachis ipaensis).
Gray lines and red lines indicated the all collinear blocks and while the syntenic WRKY gene pairs, respectively.
The cis-acting elements in promoter regions of AhWRKYs
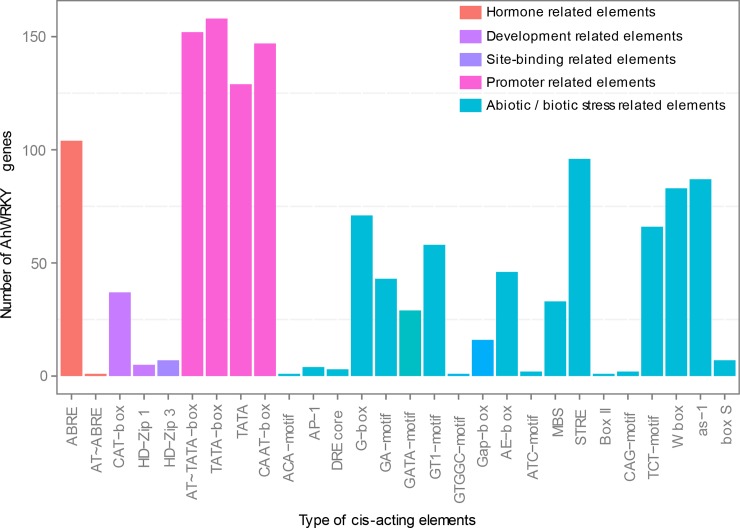
The cis-acting elements in the promoter are crucial to gene expression [62]. The 1,500-bp 5’-upstream promoter regions were searched using PlantCARE, and it was found out that the cis-acting elements were extremely diverse (S5 Table). The cis-elements were classified as stress-related elements, hormone-related elements, development-related elements, promoter related elements and site-binding related elements (Fig 6). The largest number of cis-acting elements (19) were related to abiotic/biotic stress responses (Fig 7), and mainly include the MYB binding site involving in drought-inducibility (MBS), the WRKY binding site regulating gene expression (W-box) and the light responsive elements (AE-box, GT1-motif, TCT-motif, GA-motif, G-box, etc.). Interestingly, 83 AhWRKYs contained W-box regulated gene expression by binding WRKY, indicating these genes may be auto-regulated or cross-regulated with others. These results indicated that most of AhWRKYs might play a vital role in various stress responses.
Fig 6. The number of AhWRKY genes containing various cis-acting elements.
x-axis and y-axis represented the type of cis-acting elements and the number of WRKY gene family members, respectively.
Fig 7. Predicted stress-related cis-elements in the AhWRKY promoters.
The upstream length to the translation start site can be inferred according to the scale at the bottom. Different cis-elements were represented by different shapes and colors.
Physiological changes in leaves of L422 and L677 under drought treatment
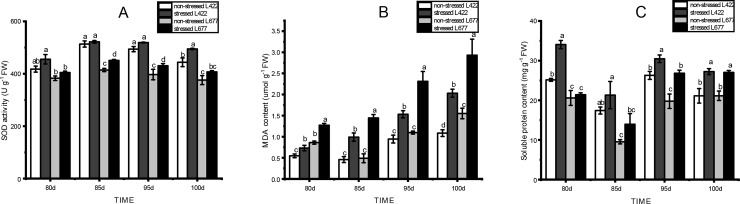
To investigate the peanut response to drought at the pod-setting stage, several phenotype responses induced by drought were studied. As expected, no significant phenotype differences were observed between the two lines under optimum water conditions. However, there were significant differences in the performances of the two lines when exposed to drought stress. The leaves of L677 began to shrivel up, while those of L422 cultivar displayed little or no phenotypic change. As shown in Fig 8A, there were significant differences in SOD activity in both L422 and L677 when the plants were upon exposure to drought stress. The SOD activity was generally higher in L422 than in L677 under stressed condition. The MDA content showed an increasing trend after drought stress in L677 and L422, while the MDA content of L677 was significantly (p < 0.05) higher than that of L422 (Fig 8B). The soluble protein content was significantly higher (p < 0.05) in L422 than that of in L677 under both stressed and non-stressed conditions (Fig 8C).
Fig 8.
The changes of SOD (A), MDA (B), and Soluble protein content (C) in leaves of drought-tolerant and drought-sensitive varieties under drought stress. Values are mean ± standard error (n = 3). Different letters (a, b, c and d) above line graphs refer to significant difference among treatments (p<0.05).
Expression analysis of AhWRKYs under drought using RNA-seq
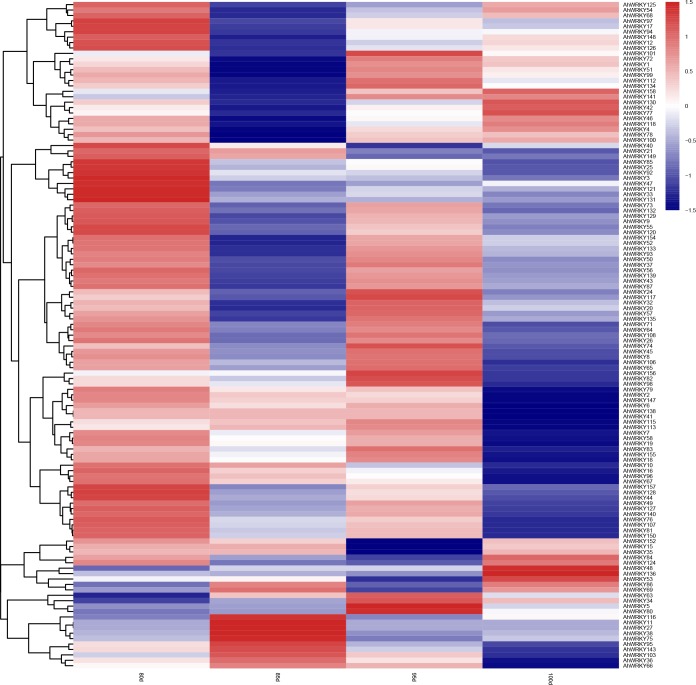
To investigate specifically the response of AhWRKYs to drought stress, expression analysis of the 158 AhWRKYs was carried based on transcriptome data obtained under drought (Fig 9 and S6 Table). The relative expression levels were represented as control vs treatment. Genes were considered to be differentially expressed when the value of log2 Fold Change was > 1 or < -1 and FDR was less than 0.05 threshold. Twenty (20) out of the 158 AhWRKYs were not expressed at all time points, which suggests they might be pseudogenes or may not have been expressed during this period. A total of 4780, 7130, 6240 and 3309 differentially expressed genes were identified at 80, 85, 95 and 100 DAP and include 43, 22, 52 and 14 differentially expressed WRKY genes, respectively. Seventy-three (73) differentially expressed WRKY genes were influenced by drought stress, of which forty-six (46) were identified in at least two (2) of the four (4) examined time points. Interestingly, two (2) of the genes (AhWRKY16 and 96) were initially up-regulated at 80 and 85 DAP while three (3) (AhWRKY17, 97 and 148) were up-regulated at 80 DAP and down-regulated at 85 DAP, respectively. These results suggest that the genes might have a significant role under drought stress at the early pod-setting stage (80–85 DAP). Also, three (3) genes (AhWRKY18, 45 and 106) were up-regulated at 95 DAP although down-regulation was later observed at 100 DAP, which inferred that these three (3) genes responded to drought stress at the latter stage of pod-setting. Moreover, twelve (12) genes (AhWRKY2, 6, 7, 24, 49, 58, 67, 68, 79, 81,132 and 155) were identified in three of the four examined time points. Overall, the results show AhWRKYs has an important role in peanut response to drought stress.
Fig 9. The transcriptomics analysis of the AhWRKY genes in response to drought stress.
Samples were collected 80, 85, 95 and 100 DAP from the control and drought stress treated peanut. Color scale represented the fold change of drought treatment to control of AhWRKYs with normalized log2-transformed count value. Red, blue and white indicated high expression, low expression and no expression, respectively.
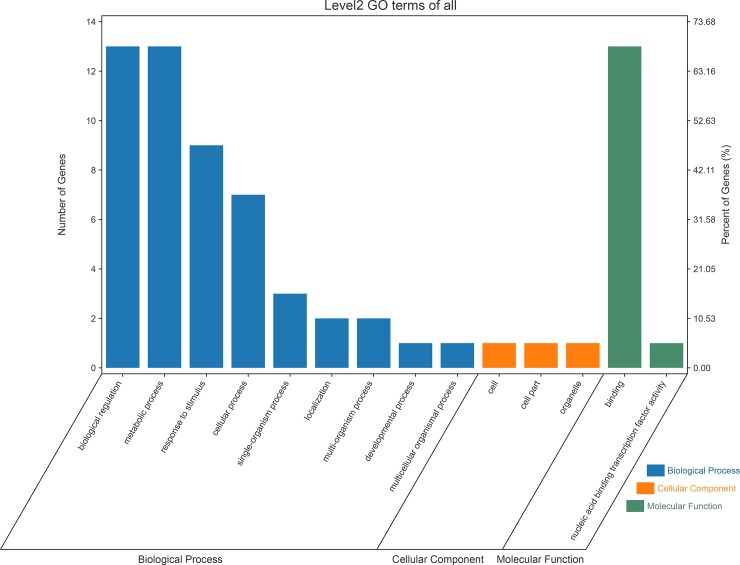
To functionally characterize the differentially expressed WRKY genes, annotation and pathway analysis were performed (S7 Table). The assigned functions of a total of nineteen (19) differentially expressed WRKYs covered a broad range of GO categories (Fig 10). The GO terms for differentially expressed WRKYs were mainly related to biological regulation (GO:0065007), metabolic process (GO:0008152), and response to stimulus (GO:0050896). For cellular components, only one (1) WRKY gene involved in the cell (GO:0005623), organelle (GO:0043226) and cell part (GO:0044464), respectively. Under molecular functions, the terms only were related to binding (GO:0005488) and nucleic acid binding transcription factor activity (GO:0001071). Genes usually interact with each other to play roles in certain biological functions. Pathway based GO analysis helped to further understand the biological functions of genes. According to the KEGG analysis, the seven (7) and six (6) differentially expressed WRKY genes were significantly enriched in plant-pathogen interaction (AhWRKY56, 12, 133, 50, 98, 152 and 139) and plant MAPK signaling pathway (AhWRKY56, 12, 133, 50, 98 and 139), respectively (S7 Table). According to the results of multiple sequence alignment, WRKY152 possessed leucine-rich repeat (LRR) domain, indicating that it might be associated with disease resistance in peanut. Moreover, WRKY152 was found to be involved in the pathway of plant-pathogen interaction via KEGG enrichment analysis. In conclusion, the results demonstrated that WRKY152 might play an important role in disease resistance processes. Overall, the results of GO and KEGG showed that AhWRKYs might not only participated in the drought stress process but also played an important role in other stress-resistant processes.
Fig 10. The distribution of GO terms for the differentially expressed WRKYs.
X-axis represents GO terms; Y-axis on the left is the percentage of differentially expressed WRKYs and on the right is the number of differentially expressed WRKYs.
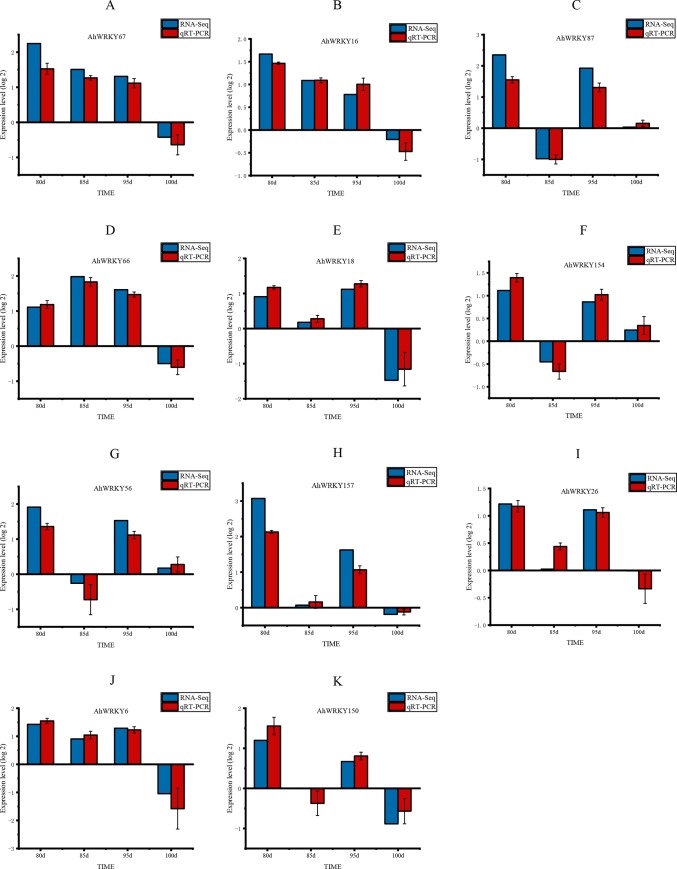
Validation of RNA sequencing using qRT-PCR
To validate the expression data obtained from RNA sequencing, 11 differentially expressed AhWRKY genes with cis-recognition elements noted to have been involved in the abiotic/biotic stress were randomly selected to perform qPCR. The PCR expressions were consistent with the RNA-seq data (Fig 11). A correlation coefficient of 0.89611 was obtained between the fold changes of qRT-PCR and RNA-seq (S2 Fig). The results of the qRT-PCR analysis validated the findings obtained from the RNA-seq data.
Fig 11. Confirmation of RNA-Seq results by quantitative real-time PCR (qRT-PCR).
All genes with negative values of expression level meant that they were down-regulated in response to drought stress.
Discussion
The cultivated peanut is an allotetraploid (amphidiploid with 2n = 4x = 40) and it’s mainly cultivated in areas characterized by unstable rainfall pattern. Drought seriously affects the productivity of peanut. The emergence of the genome sequences of Arachis duranensis and Arachis ipaensis has been an important resource for peanut research on drought resistance [59]. The complete genome sequence data of cultivated peanut has been officially released on PeanutBase (http://www.peanutbase.org/), and it provided the key data to explore functional diversity. WRKY family is a plant-specific transcription factor that participates in plant growth, development and stress responses [6] with 75 AdWRKYs and 77 AiWRKYs identified in A. duranensis and A. ipaensis, respectively [18]. The purpose of this study was to mine potential candidate AhWRKYs response to drought stress in peanut which will provide a valuable resource for genetic improvements of agronomically important traits. A total of 158 AhWRKYs were identified through genome-wide analysis of the cultivated peanut.
The sequence alignment and phylogenetic tree analysis based on the classification standards for the WRKY family showed the identified 158 AhWRKY proteins from peanut grouped into three main clusters (group I, II and III) with group II proteins further clustering into five subgroups (IIa, IIb, IIc, IId, and IIe) [8]. The subgroup IIc possessed the most members, which were similar to results obtained in many legume crops [12, 14, 18, 63, 64] and was more active in evolution which suggests an important function in legume crops. Majority of the AhWRKYs harbored the highly conserved WRKYGQK motif. However, variants were found in group I and subgroup IIc although most of them were distributed in subgroup IIc. Moreover, variants were mainly observed in Arachis duranensis and Arachis ipaensis [18]. The observation of variants in subgroup IIc has also been reported in soybean [12], Arabidopsis [8] and in pineapple [17]. By inference, WRKYGQK sequence was more likely to mutate in subgroup IIc and hence, members in this subgroup are likely to possess different binding specificities and biological functions by altering their DNA binding affinity [65]. Interestingly, some AhWRKYs were found to contain incomplete zinc finger motifs. However, C2HY was identified in AhWRKY13. These results were consistent with the transcription factor prediction based on the plant transcription factor database (http://planttfdb.cbi.pku.edu.cn/) [66], but the function of variations in zinc finger motif still needs to be further explored. Although AhWRKY5, AhWRKY13, AhWRKY92 and AhWRKY93 were classified into group I, they only possessed one WRKY domain while the loss of this domain was found in previous studies [11, 12].
The events of gene duplication have been considered as the main evolutionary driving force of new biological functions [67]. During evolutionary processes, tandem and segmental duplication contribute to the generation of gene family [57]. Among AdWRKYs and AiWRKYs, four (4) tandem duplication events with eight (8) genes and seven (7) segmental duplication events with 14 genes as well as four (4) tandem duplication events with nine (9) genes and 10 segmental duplication events with 17 genes were found, respectively [18]. In this study, six (6) tandem duplication events with 12 AhWRKYs and 124 segmental duplication events with 136 AhWRKYs were observed. The number of segmental duplications was significantly (p<0.05) more than tandem duplications, which was similar to the results of a previous study on A. duranensis and A. ipaënsis. Therefore, it could be deduced that segmental duplication was the main driver for the expansion of AhWRKYs family. Also, the comparative synteny mapping of phylogenetic mechanisms of AhWRKYs family with A. duranensis and A. ipaensis revealed the numbers of collinear gene pairs between A. duranensis and A. ipaensis were 110 and 43, respectively. However, only 32 collinear gene pairs were observed between Arachis hypogaea and the two diploid species. A. duranensis and A. ipaensis were deduced as the donors of the A and B subgenomes, respectively [59–61], and the divergence of the two diploid wild species was estimated to have occurred ~2.16 million years ago [68], which suggested that the 32 collinear gene pairs might have already existed before the divergence of two diploid wild species. In addition, the other AhWRKYs could not be mapped to any synteny blocks in this study, but it is not clear whether these genes appeared before or after the hybridization of two diploid wild species doubles.
Drought has a negative effect on physiological processes and hence reduces peanut yield [69]. However, when it is imposed under experimental conditions, it provides valuable clues which enable scientists to derive the function of AhWRKYs by comparing the phylogenetic and homologous relationships between AhWRKYs and other crop species. For example, AtWRKY46, the ortholog gene of AhWRKY43, was induced by drought, salt, and oxidative stresses [70]. This suggested that AhWRKY43 was involved not only in the process of peanut responding to drought stress but also in other physiological processes. AtWRKY53, the orthologous gene of AhWRKY17, was negatively regulated under drought tolerance by mediating stomatal movement [71]. AhWRKY43 and AhWRKY154 were induced by drought treatments, and have been described in their Arabidopsis sequence homologues (AtWRKY46, AtWRKY54 and AtWRKY70) as playing vital roles in drought responses and brassinosteroid-regulated plant growth [72], while AhWRKY154 showed no significant differential expression in the current study. The identification of differentially expressed genes under drought stress in Arachis duranensis and Arachis magna have been released in PeanutBase, which supplied valuable resources for study on drought-related genes in cultivated peanut [73]. Aradu.S7YD6, the ortholog to AhWRKY50, was significantly up-regulated 4.2- and 3.8-fold under drought treatment in Arachis duranensis and Arachis magna, respectively. Aradu.KG41H, the ortholog of AhWRKY43 and AhWRKY129, showed a high level of expression (3.9-fold increase) in Arachis magna. Based on the homologous relationships, it was deduced that the AhWRKYs may play a critical role in the responses to drought stress in cultivated peanut.
Conclusion
One hundred and fifty-eight AhWRKYs, distributing across 20 chromosomes of cultivated peanut, were identified and grouped into three main groups based on the sequence alignment and phylogenetic tree analysis. Gene duplication analysis indicated that segmental duplication events were the main driving force in the evolution of WRKY gene family in peanut. The synteny analysis of Arachis hypogaea and two diploid wild ancestors (A. duranensis and A. ipaensis) provided helpful information for the evolutionary characteristics of AhWRKYs. The functional annotations, cis-acting elements and expression analysis revealed that most AhWRKYs were involved in response to drought stress. These results will provide a valuable resource for further investigation of peanut drought tolerance characteristics.
Supporting information
The highly conserved WRKYGQK heptapeptide, and the amino acids forming the zinc-finger motif were covered in blue boxes, while the mutated amino acids were marked in red.
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors sincerely thank Dr. Zhengwen Sun (Hebei Agricultural University, China) and Dr. Yussif Baba Kassim (Hebei Agricultural University, China) for providing valuable suggestions.
Data Availability
All relevant data are within the paper, its Supporting Information files, and the RNA-Seq data have deposited in NCBI under the accession number PRJNA544421 (https://www.ncbi.nlm.nih.gov/sra/PRJNA544421).
Funding Statement
This work was financially sponsored by the National Natural Science Foundation of China (31801394, 31701459480 and 31771833); the Natural Science Foundation of Hebei Province (C2017204101); the China Agriculture Research System (CARS-13) and the Earmarked Fund for Hebei Oil Innovation Team of Modern Agro-industry Technology Research System (HBCT2018090202); the Science and Technology Supporting Plan Project of Hebei Province (16226301D); and the Innovation Fund Project for Graduate Student of Hebei Province (CXZZBS2019093).
References
- 1.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends in Plant Science. 2010;15(5):247–258. 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- 2.Agarwal P, Reddy M, Chikara J. WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Molecular Biology Reports. 2011;38(6):3883–3896. 10.1007/s11033-010-0504-5 [DOI] [PubMed] [Google Scholar]
- 3.Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology. 2007;10(4):366–371. 10.1016/j.pbi.2007.04.020 [DOI] [PubMed] [Google Scholar]
- 4.Ishihama N, Yoshioka H. Post-translational regulation of WRKY transcription factors in plant immunity. Current Opinion in Plant Biology. 2012;15(4):431–437. 10.1016/j.pbi.2012.02.003 [DOI] [PubMed] [Google Scholar]
- 5.Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J. WRKY transcription factors in plant responses to stresses. Journal of Integrative Plant Biology. 2017;59(2):86–101. 10.1111/jipb.12513 [DOI] [PubMed] [Google Scholar]
- 6.Ülker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Current Opinion in Plant Biology. 2004;7(5):491–498. 10.1016/j.pbi.2004.07.012 [DOI] [PubMed] [Google Scholar]
- 7.Viana VE, Busanello C, da Maia LC, Pegoraro C, Costa de Oliveira A. Activation of rice WRKY transcription factors: an army of stress fighting soldiers? Current Opinion in Plant Biology. 2018;45:268–275. 10.1016/j.pbi.2018.07.007 [DOI] [PubMed] [Google Scholar]
- 8.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends in Plant Science. 2000;5(5):199–206. 10.1016/s1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- 9.Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5' upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Molecular and General Genetics. 1994;244(6):563–571. 10.1007/bf00282746 [DOI] [PubMed] [Google Scholar]
- 10.Kalde M, Barth M, Somssich IE, Lippok B. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Molecular Plant-Microbe Interactions. 2003;16(4):295–305. 10.1094/MPMI.2003.16.4.295 [DOI] [PubMed] [Google Scholar]
- 11.Ross CA, Liu Y, Shen QJ. The WRKY gene family in rice (Oryza sativa). Journal of Integrative Plant Biology. 2007;49(6):827–842. 10.1111/j.1744-7909.2007.00504.x [DOI] [Google Scholar]
- 12.Yin G, Xu H, Xiao S, Qin Y, Li Y, Yan Y, et al. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biology. 2013;13:148 10.1186/1471-2229-13-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai C, Niu E, Du H, Zhao L, Feng Y, Guo W. Genome-wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton. Crop Journal. 2014;2(2–3):87–101. 10.1016/j.cj.2014.03.001 [DOI] [Google Scholar]
- 14.Song H, Wang P, Nan Z, Wang X. The WRKY transcription factor genes in Lotus japonicus. International Journal of Genomics. 2014;2014:420128 10.1155/2014/420128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okay S, Derelli E, Unver T. Transcriptome-wide identification of bread wheat WRKY transcription factors in response to drought stress. Molecular Genetics and Genomics. 2014;289(5):765–781. 10.1007/s00438-014-0849-x [DOI] [PubMed] [Google Scholar]
- 16.Wang M, Vannozzi A, Wang G, Liang Y-H, Tornielli GB, Zenoni S, et al. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Horticulture Research. 2014;1:14016 10.1038/hortres.2014.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie T, Chen C, Li C, Liu J, Liu C, He Y. Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics. 2018;19(1):490 10.1186/s12864-018-4880-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song H, Wang P, Lin JY, Zhao C, Bi Y, Wang X. Genome-wide identification and characterization of WRKY gene family in peanut. Frontiers in Plant Science. 2016;7:534 10.3389/fpls.2016.00534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luan Q, Chen C, Liu M, Li Q, Wang L, Ren Z. CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant Science. 2019;279:59–69. 10.1016/j.plantsci.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Bai X, Wang X, Chu C. OsWRKY71, a rice transcription factor, is involved in rice defense response. Journal of Plant Physiology. 2007;164(8):969–979. 10.1016/j.jplph.2006.07.006 [DOI] [PubMed] [Google Scholar]
- 21.Tian X, Li X, Zhou W, Ren Y, Wang Z, Liu Z, et al. Transcription factor OsWRKY53 positively regulates brassinosteroid signaling and plant architecture. Plant Physiology. 2017;175(3):1337–1349. 10.1104/pp.17.00946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C-Q, Xu Y, Lu Y, Yu H-X, Gu M-H, Liu Q-Q. The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta. 2011;234(3):541–554. 10.1007/s00425-011-1423-y [DOI] [PubMed] [Google Scholar]
- 23.Chen L, Zhang L, Li D, Wang F, Yu D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(21):E1963–E1971. 10.1073/pnas.1221347110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiology. 2007;143(4):1789–1801. 10.1104/pp.106.093971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant Journal. 2001;28(2):123–133. 10.1046/j.1365-313x.2001.01131.x [DOI] [PubMed] [Google Scholar]
- 26.Sheng Y, Yan X, Huang Y, Han Y, Zhang C, Ren Y, et al. The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant, Cell & Environment. 2019;42(3):891–903. 10.1111/pce.13457 [DOI] [PubMed] [Google Scholar]
- 27.Zhu D, Hou L, Xiao P, Guo Y, Deyholos MK, Liu X. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Science. 2019;280:132–142. 10.1016/j.plantsci.2018.03.018 [DOI] [PubMed] [Google Scholar]
- 28.Gao H, Wang Y, Xu P, Zhang Z. Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Frontiers in Plant Science. 2018;9:997 10.3389/fpls.2018.00997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagacé M, Matton DP. Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta. 2004;219(1):185–189. 10.1007/s00425-004-1253-2 [DOI] [PubMed] [Google Scholar]
- 30.Shao HB, Chu LY, Jaleel CA, Manivannan P, Panneerselvam R, Shao MA. Understanding water deficit stress-induced changes in the basic metabolism of higher plants—biotechnologically and sustainably improving agriculture and the ecoenvironment in arid regions of the globe. Critical Reviews in Biotechnology. 2009;29(2):131–151. 10.1080/07388550902869792 [DOI] [PubMed] [Google Scholar]
- 31.Ramachandra Reddy A, Chaitanya KV, Vivekanandan M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. Journal of Plant Physiology. 2004;161(11):1189–1202. 10.1016/j.jplph.2004.01.013 [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Liu ZH, Feng L, Zheng Y, Li DD, Li XB. Genome-wide functional analysis of cotton (Gossypium hirsutum) in response to drought. PloS One. 2013;8(11):e80879 10.1371/journal.pone.0080879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budak H, Hussain B, Khan Z, Ozturk NZ, Ullah N. From genetics to functional genomics: improvement in drought signaling and tolerance in wheat. Front Plant Sci. 2015;6:1012 10.3389/fpls.2015.01012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar P, Rouphael Y, Cardarelli M, Colla G. Vegetable Grafting as a Tool to Improve Drought Resistance and Water Use Efficiency. Front Plant Sci. 2017;8:1130 10.3389/fpls.2017.01130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertioli DJ, Jenkins J, Clevenger J, Dudchenko O, Gao D, Seijo G, et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nature Genetics. 2019;51(5):877–884. 10.1038/s41588-019-0405-z [DOI] [PubMed] [Google Scholar]
- 36.Chou KC, Shen HB. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One. 2010;5(6):e11335 10.1371/journal.pone.0011335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33(7):1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2014;31(8):1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30(1):325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Research. 2006;34:W369–W373. 10.1093/nar/gkl198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research. 2012;40(7):e49 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Research. 2009;19(9):1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu JJ, Wei Z, Li JH. Effects of copper on leaf membrane structure and root activity of maize seedling. Botanical Studies. 2014;55 10.1186/s40529-014-0047-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang MY, Zhang JH. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. Journal of Experimental Botany. 2002;53(379):2401–2410. 10.1093/jxb/erf090 [DOI] [PubMed] [Google Scholar]
- 46.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]
- 47.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14(4). 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 49.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology. 2015;33(3):290–+. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BMG, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments (vol 29, pg 1035, 2013). Bioinformatics. 2013;29(16):2073–2073. 10.1093/bioinformatics/btt337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Statistical Methodology. 1995;57(1):289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 52.Reddy DS, Bhatnagar-Mathur P, Cindhuri KS, Sharma KK. Evaluation and validation of reference genes for normalization of quantitative real-time PCR based gene expression studies in peanut. PloS One. 2013;8(10):e78555 10.1371/journal.pone.0078555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT method. Methods. 2001;25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 54.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Current Opinion in Structural Biology. 2001;11(6):725–732. 10.1016/s0959-440x(01)00266-4 [DOI] [PubMed] [Google Scholar]
- 55.Rinerson CI, Rabara RC, Tripathi P, Shen QJ, Rushton PJ. The evolution of WRKY transcription factors. BMC Plant Biology. 2015;15:66 10.1186/s12870-015-0456-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song H, Guo Z, Chen T, Sun J, Yang G. Genome-wide identification of LRR-containing sequences and the response of these sequences to nematode infection in Arachis duranensis. BMC Plant Biology. 2018;18(1):279 10.1186/s12870-018-1508-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology. 2004;4:10 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holub EB. The arms race is ancient history in Arabidopsis, the wildflower. Nature Reviews: Genetics. 2001;2(7):516–527. 10.1038/35080508 [DOI] [PubMed] [Google Scholar]
- 59.Bertioli DJ, Cannon SB, Froenicke L, Huang G, Farmer AD, Cannon EK, et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nature Genetics. 2016;48(4):438–446. 10.1038/ng.3517 [DOI] [PubMed] [Google Scholar]
- 60.Moretzsohn MC, Gouvea EG, Inglis PW, Leal-Bertioli SC, Valls JF, Bertioli DJ. A study of the relationships of cultivated peanut (Arachis hypogaea) and its most closely related wild species using intron sequences and microsatellite markers. Annals of Botany. 2013;111(1):113–126. 10.1093/aob/mcs237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seijo G, Lavia GI, Fernández A, Krapovickas A, Ducasse DA, Bertioli DJ, et al. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. American Journal of Botany. 2007;94(12):1963–1971. 10.3732/ajb.94.12.1963 [DOI] [PubMed] [Google Scholar]
- 62.Rombauts S, Déhais P, Van Montagu M, Rouzé P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Research. 1999;27(1):295–296. 10.1093/nar/27.1.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song H, Sun W, Yang G, Sun J. WRKY transcription factors in legumes. BMC Plant Biology. 2018;18(1):243 10.1186/s12870-018-1467-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srivastava R, Kumar S, Kobayashi Y, Kusunoki K, Tripathi P, Kobayashi Y, et al. Comparative genome-wide analysis of WRKY transcription factors in two Asian legume crops: Adzuki bean and Mung bean. Scientific Reports. 2018;8(1):16971 10.1038/s41598-018-34920-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, et al. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnology Journal. 2008;6(5):486–503. 10.1111/j.1467-7652.2008.00336.x [DOI] [PubMed] [Google Scholar]
- 66.Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Research. 2017;45(D1):D1040–D1045. 10.1093/nar/gkw982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biology. 2002;3(2):RESEARCH0008. 10.1186/gb-2002-3-2-research0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen X, Lu Q, Liu H, Zhang J, Hong Y, Lan H, et al. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Molecular Plant. 2019. 10.1016/j.molp.2019.03.005 [DOI] [PubMed] [Google Scholar]
- 69.Arruda IMa, Modacirino V, Buratto JS, Ferreira JM, Arruda IMa, Modacirino V, et al. Growth and yield of peanut cultivars and breeding lines under water deficit. Pesquisa Agropecuária Tropical. 2015;45:146–154. 10.1590/1983-40632015v4529652 [DOI] [Google Scholar]
- 70.Ding ZJ, Yan JY, Xu XY, Yu DQ, Li GX, Zhang SQ, et al. Transcription factor WRKY 46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant Journal. 2014;79(1):13–27. 10.1111/tpj.12538 [DOI] [PubMed] [Google Scholar]
- 71.Sun Y, Yu D. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Reports. 2015;34(8):1295–1306. 10.1007/s00299-015-1787-8 [DOI] [PubMed] [Google Scholar]
- 72.Chen J, Nolan TM, Ye H, Zhang M, Tong H, Xin P, et al. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell. 2017;29(6):1425–1439. 10.1105/tpc.17.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brasileiro AC, Morgante CV, Araujo AC, Leal-Bertioli SC, Silva AK, Martins AC, et al. Transcriptome profiling of wild arachis from water-limited environments uncovers drought tolerance candidate genes. Plant Molecular Biology Reporter. 2015;33:1876–1892. 10.1007/s11105-015-0882-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The highly conserved WRKYGQK heptapeptide, and the amino acids forming the zinc-finger motif were covered in blue boxes, while the mutated amino acids were marked in red.
(PDF)
(PDF)
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper, its Supporting Information files, and the RNA-Seq data have deposited in NCBI under the accession number PRJNA544421 (https://www.ncbi.nlm.nih.gov/sra/PRJNA544421).