OBJECTIVES:
Immune activation and intestinal microbial dysbiosis could induce diarrhea-predominant irritable bowel syndrome (IBS-D). We examined the roles of ileal immunoglobulin A (IgA) and IgA-coated bacteria in IBS-D pathogenesis.
METHODS:
Peripheral blood, fecal samples, and ileal and cecal biopsies were collected from 32 healthy volunteers and 44 patients with IBS-D. Quantitative reverse transcriptase polymerase chain reaction was used to assess differential gene expression. IgA levels in the blood and fecal samples were quantified by an enzyme-linked immunosorbent assay. IgA+ cells were assessed by immunofluorescence imaging. Flow-cytometry-based IgA+ bacterial cell sorting and 16S rRNA gene sequencing (IgA-SEQ) was used to isolate and identify fecal IgA+ bacteria.
RESULTS:
Fecal IgA, particularly IgA1, was upregulated in patients with IBS-D. IgA class switch and B cell–activating factor-receptor were increased in the terminal ileum of patients. The intestinal microbiota composition was altered in patients compared with that in controls. IgA-SEQ showed that the proportion of fecal IgA-coated bacteria was increased significantly in patients with IBS-D. IgA+ bacteria in patients with IBS-D showed higher abundances of Escherichia–Shigella, Granulicatella, and Haemophilus compared with healthy controls and IgA− bacteria in patients with IBS-D. The Escherichia–Shigella IgA coating index was positively correlated with anxiety and depression. The Escherichia–Shigella relative abundance, luminal IgA activity, and some altered IgA-coated bacteria were positively associated with the clinical manifestations of IBS-D.
DISCUSSION:
Microbial dysbiosis may promote the terminal ileal mucosa to produce higher levels of IgA, increasing the proportion of IgA-coated bacteria by activating IgA class switching, which might regulate local inflammation and clinical manifestations in IBS-D. IgA may mediate the effects of microbial dysbiosis on the pathogenesis of IBS-D.
INTRODUCTION
Irritable bowel syndrome (IBS) is a common digestive tract dysfunction. According to recent statistics, the global prevalence of IBS is as high as 11% (1). The Rome IV criteria indicate that patients with IBS primarily suffer from recurrent abdominal pain, which is related to defecation or altered bowel habits (2). Patients with IBS tend to suffer from low-grade inflammation in the intestinal mucosa, although the underlying mechanisms remain unclear (3). Recent studies revealed enhanced humoral immunity in diarrhea-predominant IBS (IBS-D) (4,5). However, how humoral responses to microbiota regulate the pathogenesis of IBS-D is unclear.
Immunoglobulin A (IgA) is a critical molecule in mucosal immunity and is exposed to both commensal and potential pathogens (6). IgA is the main type of antibody produced on the mucosal surface and protects against pathogen infection and mucosal penetration by the indigenous bacteria (7). Commensal intestinal microbial members can also induce IgA secretion and transform to show an IgA-coated pattern (7,8). The role of fecal IgA in regulating intestinal homeostasis and pathophysiology of IBD has been widely examined (9–13). Increasing studies demonstrated upregulated fecal IgA levels in patients with IBD (9–13) and an indispensable function of IgA-coated bacteria in the pathogenesis of IBD (12). Nevertheless, the roles of mucosal IgA and IgA-coated bacteria in the pathogenesis of IBS-D are unclear. Large amounts of mucosa-associated lymphoid tissue aggregate at the ileocecum, indicating that this region not only influences the flow rate and prevents reflux (14) but also is an important area of mucosal immunity in the gastrointestinal tract (15). However, whether ileocecal IgA and IgA-coated bacteria are altered, which may affect the pathophysiology of IBS-D, are unknown.
Class switch recombination (CSR), which alters the existing immunoglobulin heavy chain (IGH) with the α heavy chain without modifying antigen specificity, confers mature B cells with the ability to express IgA (6). This process requires stimulation of B cells, followed by the germline transcription of specific gene regions of the IGH and expression of the activation-induced cytidine deaminase (AID) (16). Thus, the expression of AID during the recombination process is considered as the most important marker for detecting the sites of CSR in vivo (7). A proliferation-inducing ligand (APRIL) and B cell–activating factor (BAFF), which are tumor necrosis factor family ligands, contribute to B lymphocyte and plasma cell homeostasis and survival and regulate CSR and antibody production (17,18). The process is mediated by surface receptors, particularly transmembrane activator, calcium modulator, and cyclophilin ligand interactor (TACI), B-cell maturation antigen (BCMA), and BAFF receptor (BAFF-R) (4,18). In addition, promoters upstream of the IgA switch regions can be activated by transforming growth factor β1 (TGF-β1) to induce IgA production (19,20). In this study, we investigated whether alterations occur in ileal IgA and IgA-coated bacteria and whether these changes were correlated with the clinical manifestations of IBS-D.
MATERIALS AND METHODS
Participants
Thirty-two healthy controls and 44 patients with IBS-D were prospectively recruited. All patients met the Rome IV criteria (2). Before inclusion in the study, all potential participants underwent a thorough medical history and physical examination and were requested to complete a structured clinical questionnaire to ensure that the specific symptoms were present only in the patient group, but not in the healthy group. This study was approved by the Ethics Committee of the Qilu Hospital of Shandong University. All subjects signed informed consent. Inclusion and exclusion criteria are listed in Table 1 (Supplementary Digital Content 1, http://links.lww.com/CTG/A218).
Clinical assessment
All participants completed clinical questionnaires on (i) pain frequency (number of days with pain per 14 days), (ii) pain severity (by a visual analogue scale) (21), (iii) bowel movements (maximum number of stool frequencies), and (iv) stool form (following the Bristol Stool Form Scale) (22). The IBS symptom severity scale (IBS-SSS) was used to assess the symptoms of patients (21). Subjects enrolled in the study also completed the Hospital Anxiety and Depression Scale (HADS) to evaluate their psychological symptoms (23).
Biological samples
Peripheral blood.
Peripheral ethylenediaminetetraacetic acid blood samples were obtained from all subjects. The plasma obtained by centrifugation was cryopreserved (−80°C) until quantification of immunoglobulin. Blood cells were collected in a plastic tube, mixed with DNA/ribonucleic acid (RNA) Shield (Zymo Research, Irvine, CA), and cryopreserved (−80°C) until RNA extraction.
Fecal samples.
Fresh fecal samples collected before bowel preparation were flash-frozen in liquid nitrogen and preserved in a cryogenic refrigerator (−80°C) until further analysis.
Ileal and cecal biopsies.
Colonoscopy with intubation of the terminal ileum was performed for terminal ileal and cecal biopsies using traditional forceps. Two biopsies were obtained at each site. One fragment was formalin-fixed and paraffin-embedded for further microscopic observation, and the other one was flash-frozen (−196°C) and cryopreserved (−80°C) in RNase-free tubes until RNA extraction.
Study design
Fecal sample preparation.
A fecal sample (40 mg) was placed in a 15-mL Eppendorf tube containing 2 mL of ice-cold collection medium, which was composed of phosphate-buffered saline containing ethylenediaminetetraacetic acid and soybean trypsin inhibitor. The sample was vortexed to disrupt the pellets with a metal stirrer inserted in the tube. After incubation on ice for 10 minutes, the samples were vortexed again and centrifuged (360g for 10 minutes at 4°C). The supernatant (1 mL) was pipetted into a clean microtube and then centrifuged (14,000g for 15 minutes at 4°C). Next, 0.8 mL aliquots of the supernatant were transferred into a new microtube, and 0.2 mL of glycerol and 0.02 mL of 100 mM phenylmethanesulfonyl fluoride were added to the tube and vortexed. The samples were stored at −80°C until analysis.
IgA quantification.
IgA was quantified in the plasma samples and prepared fecal samples by an enzyme-linked immunosorbent assay (ELISA; Thermo Fisher Scientific, Waltham, MA). The total amount of protein in the fecal samples was determined with a Pierce bicinchoninic acid Protein Assay Kit (Thermo Fisher Scientific), and the amount of IgA was expressed as the percentage of IgA among the total protein.
Immunofluorescence.
Following the standard procedures, tissue sections (4 μm) were incubated with antihuman IgA (Abcam, Cambridge, UK) for IgA+ cell identification after incubation with a secondary antibody (Goat Anti-Mouse IgG H&L Alexa Fluor 488 preabsorbed, Abcam). The number of positive cells per high-power field (hpf, 400×) was calculated using CellSens software (Olympus, Tokyo, Japan) in at least 5 fields without overlap.
RNA quantitative analysis
Ileal and cecal messenger RNAs (mRNAs) were isolated using the RNAprep Pure Tissue Kit (Tiangen Biotech, Beijing, China) and blood mRNA using the Quick-RNA Whole Blood kit (Zymo Research). cDNA was synthesized using the All-in-One First-Strand cDNA Synthesis Kit (GeneCopoeia, Rockville, MD). Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) was performed to quantify the expression of AID, IGHA1 (heavy chain of the immunoglobulin A1), IGHA2 (heavy chain of the immunoglobulin A2), TGF-β1, BAFF-R, BCMA, and TACI on an Applied Biosystems StepOnePlus Real-Time PCR System (Foster City, CA) using validated SYBR Green Gene Expression Assays. Relative expression was assessed using the 2−ΔΔCt method, and the expression of these genes was normalized to the housekeeping gene, glyceraldehyde phosphate dehydrogenase (Sangon Biotech, Shanghai, China). Primers for AID, TGF-β1, BAFF-R, BCMA, and TACI were purchased from GeneCopoeia. IGHA1 and IGHA2 were amplified using previously reported primer sets (24,25).
IgA-coated bacteria flow cytometry
We used a validated method for detecting IgA-coated bacteria (12). Briefly, Mpbio Fastprep Lysing Matrix D Tubes (MP Biomedicals, Santa Ana, CA) were used to homogenize the feces, and anti-IgA-phycoerythrin (1:5; clone IS11-8E10; Miltenyi Biotec, Bergisch Gladbach, Germany) was used to stain the samples before flow cytometry (ACEA NovoCyte, San Diego, CA). Anti-phycoerythrin MicroBeads (Miltenyi Biotec) and a Quadro magnetic activated cell sorting (MACS) Starting Kit (LS; Miltenyi Biotec) were used to sort fecal bacteria. After MACS separation, the negative and positive portions were assembled for 16S rRNA gene sequencing analysis.
Microbiota analysis.
The fecal samples were shipped to Majorbio (Shanghai, China) for Illumina MiSeq sequencing using an Illumina MiSeq platform (San Diego, CA).
Statistical analysis
All the samples were masked to our statistician. Data are represented as the median (range) or mean (SD). The normality of data distribution was tested by the Kolmogorov–Smirnov test. An unpaired Student t test (2 tailed) was used to analyze normally distributed parametric data, whereas the Mann–Whitney U test was used to compare data with non-Gaussian distributions. Results were considered statistically significant when P < 0.05. Spearman rank correlation was adopted to analyze the relationships between biological and clinical variables. Statistical analyses were performed using SPSS version 24.0 software (SPSS, Chicago, IL) and GraphPad Prism 5.0 software (GraphPad, La Jolla, CA).
Microbiota composition analysis. All the samples passed quality control. Microbiota composition analysis was conducted using the I-Sanger bioanalysis platform from Majorbio (www.i-sanger.com). To quantify the different bacteria in the IgA+ or IgA− fraction, the average IgA-coating index (ICI) was determined (12,13,26).
RESULTS
General characteristics
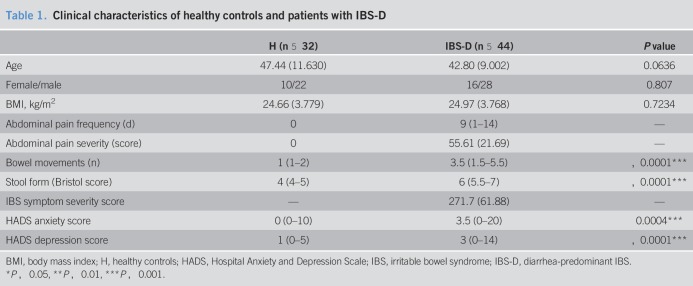
Thirty-two healthy controls and 44 patients with IBS-D were evaluated in this study. No differences were found in age, gender, or body mass index among participants (Table 1). Bowel movements and stool scores were higher in the IBS-D group than in the control group (Table 1). In addition, patients with IBS-D exhibited significantly higher anxiety and depression scores than healthy controls (Table 1).
Table 1.
Clinical characteristics of healthy controls and patients with IBS-D
Patients with IBS-D show higher IgA production in the terminal ileum
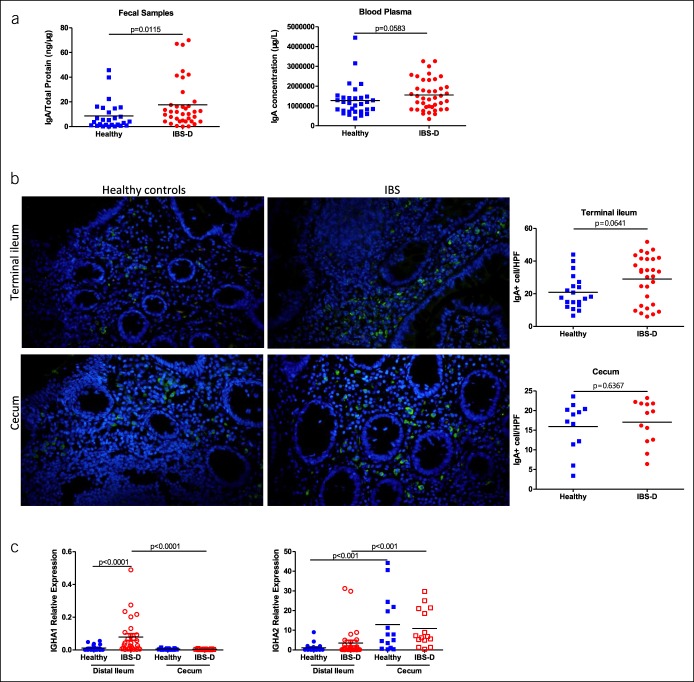
Although there was no difference in plasma IgA levels between the control and IBS-D groups, the fecal IgA level significantly increased in the IBS-D group (Figure 1a). Because fecal IgA was produced in both the small and large intestines, we next investigated whether IgA+ cells were increased in the terminal ileum or cecum by immunofluorescence. Interestingly, the number of IgA+ cells in the terminal ileum tended to be higher in patients with IBS-D (28.97 ± 14.20 cells/hpf) compared with that in healthy controls (20.93 ± 10.27 cells/hpf, P = 0.0641; Figure 1b), although no difference was detected in the cecal mucosa between the 2 groups (Figure 1b), indicating increased IgA production in the ileum but not in the cecum. There are 2 IgA subtypes, IgA1 and IgA2. To determine which IgA subtype was increased in IBS-D, we analyzed the transcription of IGHA1 and IGHA2 by qRT-PCR and identified significantly enhanced expression of IGHA1 in the terminal ileal mucosa of patients with IBS-D, although no differences in IGHA1 were detected in the blood or colonic mucosa between the 2 groups (Figure 1c and see Figure 1A, Supplementary Digital Content 1, http://links.lww.com/CTG/A217).
Figure 1.
(a) Immunoglobulin A concentration in the fecal samples and blood of healthy subjects and subjects with IBS-D. (b) Representative images and statistical graphs of immunoglobulin A+ cells in the ileal and cecal mucosa of healthy participants and participants with IBS-D. (c) Quantitative gene expression of IGHA in the mucosal samples in the IBS-D and healthy control groups. Glyceraldehyde phosphate dehydrogenase is used as a housekeeping gene. In the statistical graph, blue colors represent the healthy controls and red colors represent patients with IBS-D, the same as below. IBS-D, diarrhea-predominant irritable bowel syndrome.
IgA class switch and BAFF-R are increased in the terminal ileum of patients with IBS-D
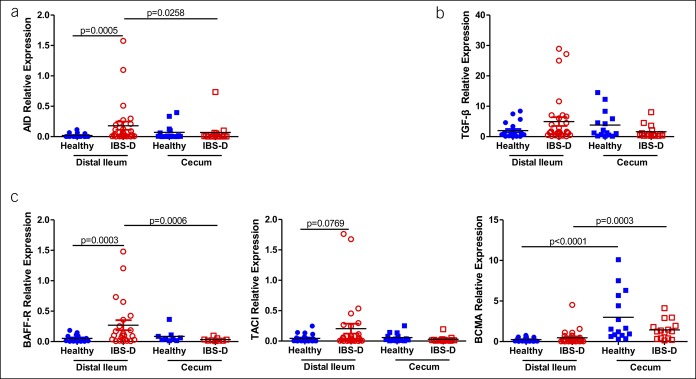
Next, we assessed CSR by measuring AID expression by qRT-PCR. As expected, in the distal ileal mucosa, AID expression was significantly increased in IBS-D compared with that in healthy subjects. However, no difference was detected in the cecal mucosa and blood between groups (Figure 2a and see Figure 1A, Supplementary Digital Content 1 http://links.lww.com/CTG/A217). Because the BAFF–APRIL pathway is essential for the IgA class switch via BAFF-R, TACI, and BCMA, we then quantified these 3 key receptors. BAFF-R expression was significantly upregulated in the distal ileal mucosa of patients with IBS-D (P = 0.0003). TACI showed an increasing trend in IBS-D, although the increase was not significant (P = 0.0769) (Figure 2c). There was no difference in the expression of BCMA or TGF-β1, a potent inducer of IgA production, between the 2 groups (Figure 2b,c). Importantly, there was no difference in AID, BAFF-R, TACI, BCMA, or TGF-β between the 2 groups in the cecal mucosa and blood samples (Figure 2 and Figure 1B, Supplementary Digital Content 1, http://links.lww.com/CTG/A217).
Figure 2.
(a) Quantitative gene expression of activation-induced cytidine deaminase in the mucosal samples in the IBS-D and healthy control groups. (b–c) Quantitative gene expression of class switch recombination–regulating genes, including transforming growth factor β1, B cell–activating factor-receptor, transmembrane activator, calcium modulator, and cyclophilin ligand interactor, and B-cell maturation antigen in the mucosal samples in the IBS-D and healthy control groups. Glyceraldehyde phosphate dehydrogenase is used as a housekeeping gene. IBS-D, diarrhea-predominant irritable bowel syndrome
Intestinal microbiota composition is altered in patients with IBS-D
It has been shown that the interactions between the intestinal microbiota and antibody responses regulate intestinal inflammation (4,10,13,27,28). We next investigated whether the intestinal microbiota are altered in IBS-D compared with those in controls. To this end, the feces were obtained from 36 patients with IBS-D and 26 healthy volunteers to analyze their microbial compositions. The bacterial compositions were compared by the Wilcoxon rank sum tests, which revealed that bacteria in patients with IBS-D were distinct from those in healthy controls. As shown in Figure 3, at the phylum level, patients exhibited a lower relative abundance of Bacteroidetes and Tenericutes (Figure 3a). They also had a lower relative abundance of Bacteroidia and Mollicutes and a higher relative abundance of Erysipelotrichia at the class level (Figure 3b). At the order level, patients with IBS-D showed a decreased proportion of Bacteroidales and Mollicutes_RF39 and an increased proportion of Erysipelotrichales and Micrococcales (Figure 3c). At the family level, patients with IBS-D exhibited a lower relative abundance of Bacteroidaceae, Rikenellaceae, Acidaminococcaceae, Christensenellaceae, norank_o_Mollicutes_RF39, and unclassified_o_Bacteroidales and a higher relative abundance of Erysipelotrichaceae and Carnobacteriaceae (Figure 3d). Moreover, at the genus level, patients with IBS-D showed an increase in Escherichia–Shigella, Ruminococcus gnavus_group, Dorea, norank_f__Lachnospiraceae, Haemophilus, and Granulicatella and a decrease in the relative abundance of Bacteroides, Alistipes, Phascolarctobacterium, Eubacterium]_ventriosum_group, Ruminococcaceae_UCG-002, Christensenellaceae_R-7_group, Sutterella, norank_o__Mollicutes_RF39, Lachnospiraceae_UCG-003, Ruminiclostridium_9, and Family_XIII_UCG-001 compared with that in healthy controls (Figure 3e).
Figure 3.
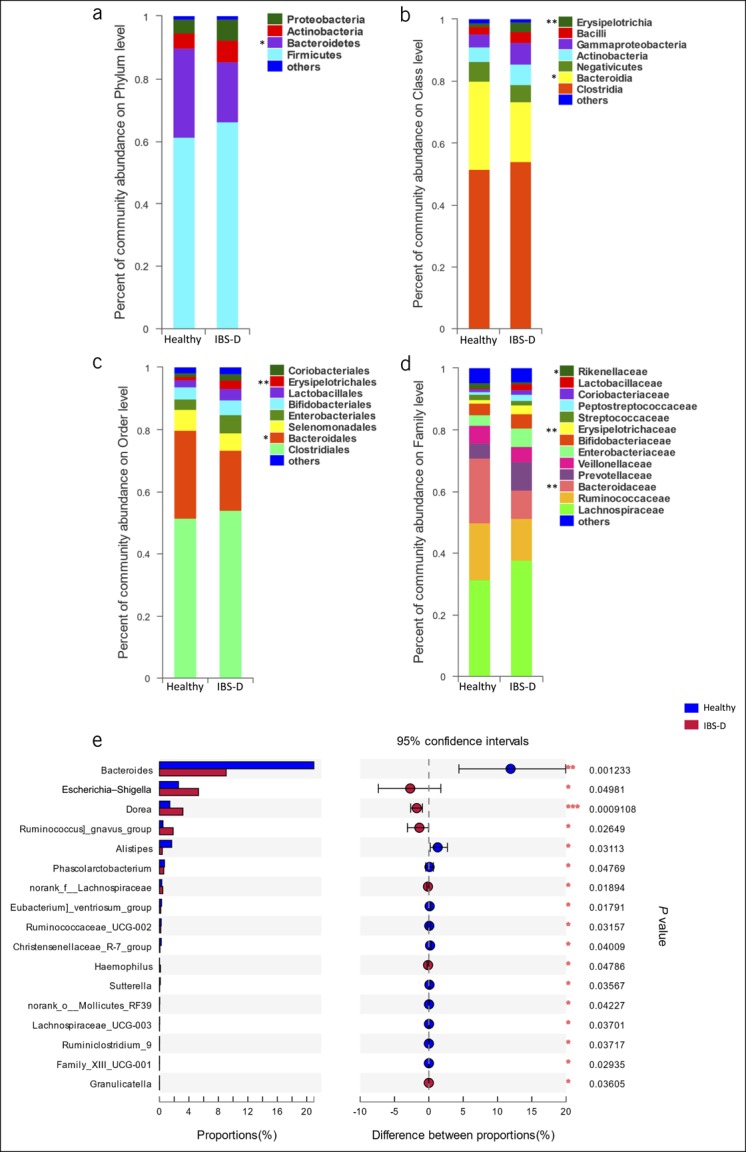
(a–d) The average relative abundance of fecal bacteria (>1%) in the IBS-D and healthy groups on the phylum, class, order, and family levels, respectively. (e) The Wilcoxon rank sum tests bar plot on the genus level, which shows 17 distinct bacterial genera in patients with IBS-D compared with those in healthy subjects. *P < 0.05, **P < 0.01, ***P < 0.001. IBS-D, diarrhea-predominant IBS.
Highly IgA-coated fecal microbiota are recognized in patients with IBS-D
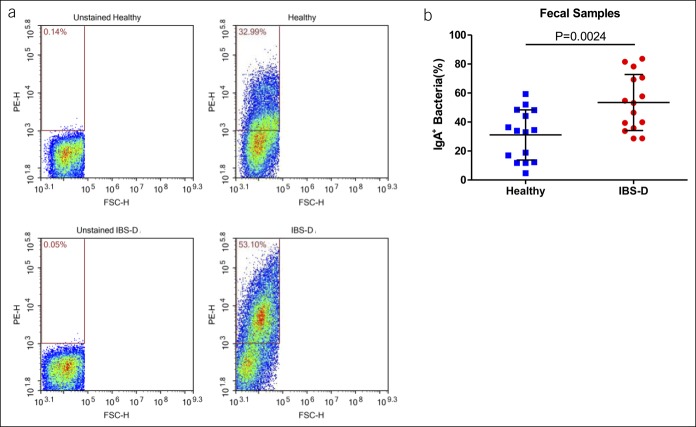
Because IgA-coated (IgA+) bacteria can induce IBD, we investigated whether the level and composition of IgA+ bacteria were altered in patients with IBS-D. As shown in Figure 4, the patients showed a significantly higher percentage of fecal IgA+ bacteria compared with healthy controls (IBS-D: 53.44 ± 19.35 vs controls: 31.09% ± 17.31%, P = 0.0024).
Figure 4.
(a) Representative images of fecal immunoglobulin A+ bacteria in healthy participants and participants with IBS-D. (b) Proportion of fecal immunoglobulin A+ bacteria in healthy participants and participants with IBS-D. FSC-H, forward scatter-height; H, healthy controls; IBS-D, diarrhea-predominant irritable bowel syndrome; PE-H, phycoerythrin (mark IgA)-height.
We first stained fecal bacteria with phycoerythrin-labeled anti-IgA antibody and analyzed by flow cytometry. Some of these bacteria were coated with IgA (12,29–31). We used MACS to isolate IgA+ (highly coated) and IgA− (noncoated) bacteria. Flow cytometry was adopted to confirm the efficiency of sorting (Figure 5A). IgA+ bacteria in patients with IBS-D differed from IgA− bacteria. In patients with IBS-D, we observed higher relative abundances of Escherichia–Shigella, Romboutsia, Streptococcus, Tyzzerella_4, Klebsiella, Intestinibacter, Granulicatella, and Haemophilus in IgA+ bacteria compared with those in IgA− bacteria (Figures 5b,c). IgA+ bacteria in patients with IBS-D showed a higher abundance of Escherichia–Shigella compared with those in healthy controls (IBS-D IgA+: 6.346% ± 12.01% vs H: 0.4814 ± 0.6319%, P = 0.003199) and IgA− bacteria in patients with IBS-D (IBS-D IgA+: 6.346% ± 12.01% vs IBS-D IgA−: 1.831% ± 4.854%, P = 0.007347).
Figure 5.
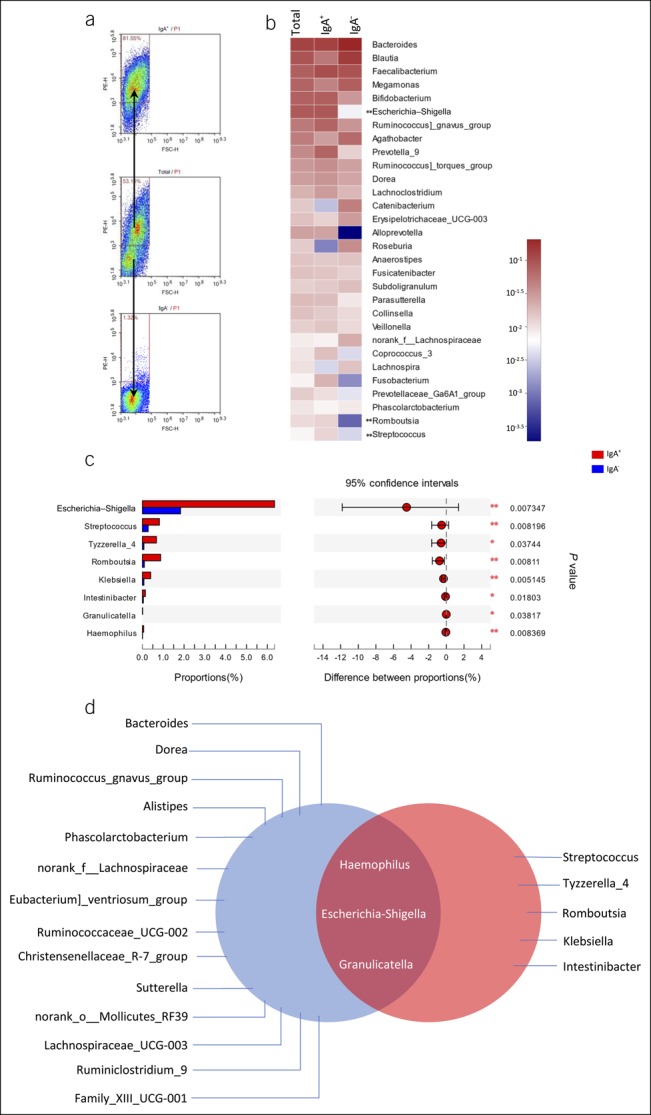
(a) Representative results of fecal bacterial cell sorting from participants. (b) Heat map describing the average relative abundance of fecal bacterial genera from patients with IBS-D on a logarithmic scale. IgA+ and IgA− fractions are compared by the linear discriminant analysis effect size. Genera with a significant increase in the IgA+ fraction are marked with asterisks. (c) The Wilcoxon rank sum test bar plot on the genus level between IgA+ and IgA− fractions of fecal bacteria from patients with IBS-D. (d) Intersection of highly IgA-coated bacteria (red) and 17 distinct bacteria (blue) in patients with IBS-D. *P < 0.05, **P < 0.01, ***P < 0.001. FSC-H, forward scatter-height; IBS-D, diarrhea-predominant irritable bowel syndrome; IgA, immunoglobulin A; PE-H, phycoerythrin (mark IgA)-height.
It has been well established that dysbiosis is one of the important causes of IBS. In this study, we found 17 distinct bacteria in patients with IBS-D compared with those in healthy controls (Figure 3e). We speculated that bacteria related to IBS-D might exist in these 17 genera. The proportion of fecal IgA+ bacteria was significantly increased in patients with IBS-D. We further analyzed the microbiome variation between IgA+ bacteria and IgA− bacteria in patients with IBS-D and found that there were 8 highly IgA-coated genera (Figure 5c). When we overlapped the 2 ranges, combining the 2 factors of disease and IgA encapsulation, 3 taxa emerged, i.e., Escherichia–Shigella, Granulicatella, and Haemophilus (Figure 5d). These data indicate that these 3 taxa are strongly associated with the pathogenesis of IBS-D.
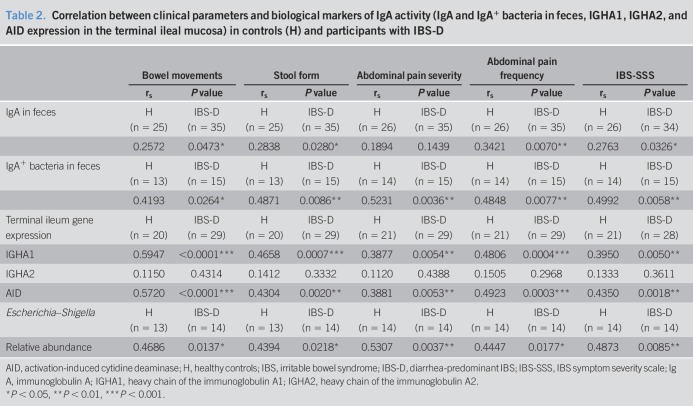
Clinical correlations
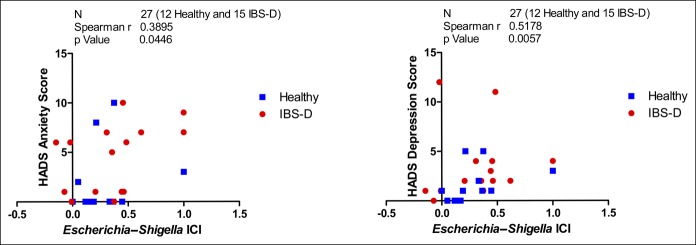
To gain insight into the mechanisms of intestinal dysfunction regulated by ileal IgA and IgA-coated bacteria, we analyzed the association between the clinical manifestations and biological variables of these bacteria (Table 2 and see Table 2, Supplementary Digital Content 1, http://links.lww.com/CTG/A219). The number of bowel movements, stool consistency, intensity and frequency of abdominal pain, and IBS-SSS were positively correlated with IgA and IgA+ bacteria in the feces and with the terminal ileum mRNA expression of IGHA1 and AID, although no association was found with IGHA2 expression (Table 2). No biological variables were associated with the HADS anxiety score or depression score (data not shown). Furthermore, the relative abundance of Escherichia–Shigella was positively correlated with bowel movements, stool form, pain intensity and frequency, and IBS-SSS (Table 2), and the Escherichia–Shigella ICI score was positively associated with the HADS anxiety score and HADS depression score (P = 0.0446 and 0.0057, respectively) (Figure 6).
Table 2.
Correlation between clinical parameters and biological markers of IgA activity (IgA and IgA+ bacteria in feces, IGHA1, IGHA2, and AID expression in the terminal ileal mucosa) in controls (H) and participants with IBS-D
Figure 6.
Correlation between Escherichia–Shigella ICI and HADS score. ICI, immunoglobulin A-coating index score; HADS, Hospital Anxiety and Depression score.
DISCUSSION
In this study, we demonstrated increased ileal IgA production and IgA-coated bacteria in IBS-D, indicating that abnormalities in the intestinal microbiota promote resident IgA class switching in the terminal ileal mucosa, leading to abnormal elevation of IgA, which may be the cause and involved in the pathogenesis of IBS-D. Although the mechanism of intestinal IgA in regulating intestinal homeostasis and IBD has been well established, how it regulates the pathogenesis of IBS-D remains unclear. Patients with IBS-D exhibited activated IgA class switching in the terminal ileum, as evidenced by increased AID gene expression. Interestingly, these patients showed higher mRNA expression of IGHA1, but not IGHA2, indicating that IgA1 is differentially regulated in IBS-D. Selective IgA CSR is a process regulated by multiple complex mechanisms, in which the core step is the innate mucosa immune recognition of the gut microbiota (6). Patients with IBS-D tended to show higher BAFF-R and TACI expression in the terminal ileal mucosa. APRIL and BAFF participate in regulating lymphocyte survival and activation (17,32). BAFF can interact with 3 receptors in the tumor necrosis factor receptor family, BAFF-R, TACI, and BCMA, whereas APRIL binds to TACI and BCMA (17,33). Of the 3 receptors, TACI is the major inducing receptor of IgA CSR. Notably, in B cells, APRIL and BAFF require the co-operation of microbial toll-like receptor (TLR) ligands to effectively induce IgA class switching (6). Activation of TLR-mediated signals participates in upregulating TACI expression in B cells (20). MyD88, a conserved activating adaptor of transcription factor nuclear factor κB, binds the cytoplasmic domain of TACI (28). Thus, TACI can trigger CSR through the AID by activating nuclear factor κB through a TLR–MyD88 pathway (28). In accordance with these results, our data indicated that increased IgA in the feces was associated with upregulation of TACI in the terminal ileum and that patients with IBS-D tend to express higher levels of TACI. In addition, BAFF-R combines with BAFF to deliver CSR-inducing signals and survival signals to peripheral B cells (34,35). By contrast, activation of BCMA merely transmits survival signals to B cells without initiating CSR (6,36). In our study, BAFF-R was upregulated in IBS-D, whereas BCMA showed no difference, indicating that the BAFF–BAFF-R interaction is the cause of increased ileal IgA production.
IgA-coated bacteria have been implicated in the regulation of IBD. Interestingly, IgA-coated bacteria were also more abundant in patients with IBS-D. We used the IgA-SEQ method to investigate the immune response of the intestinal microbiota. Accordingly, the pattern of IgA coating varied among different individuals. Among these differences, we found 3 genera in common: Escherichia–Shigella, Granulicatella, and Haemophilus. However, Escherichia–Shigella not only showed the highest order of magnitude but also changed significantly and thus showed the greatest potential to be involved in IBS-D. It has been reported that Escherichia–Shigella promotes an inflammatory status (37), and chronic infection of adherent-invasive Escherichia may cause persistent peripheral inflammation (38). In our study, patients with IBS-D showed a higher relative abundance of Escherichia–Shigella compared with healthy subjects, mainly because of IgA-coated Escherichia–Shigella. Moreover, the Escherichia–Shigella ICI score was positively corelated with participants' anxiety and depression intensity. In a recent report, Escherichia–Shigella was found to be associated with a peripheral inflammatory state in patients with Alzheimer disease (39). Escherichia–Shigella has also been reported to be related to autism spectrum disorders (40). These results support that Escherichia–Shigella is associated with the central nervous system disorders, which is consistent with our research. We also observed a significant reduction in Bacteroides in patients with IBS-D, which agrees with the results of previous studies (41–44).
In conclusion, our study revealed that patients with IBS-D exhibit higher IgA levels and increased proportions of IgA-coated bacteria by activating IgA class switching, which are associated with the clinical manifestations. Thus, IgA may mediate the effects of microbial dysbiosis on the pathophysiology of IBS-D. Given the importance of the mucosal antibody response to microbiota and its increase in IBS-D, further studies of the underlying mechanisms are needed.
CONFLICTS OF INTEREST
Guarantor of the article: Yanqing Li, MD, PhD.
Specific author contributions: Y.L. and X.Y.Y. are joint first authors. Y.Q.L. obtained funding. Y.L., Y.Z.C., and Y.Q.L. designed the study. Y.L., L.X.L., L.L., and X.L.Z. collected the data. Y.L. and X.Y.Y. analyzed the data and drafted the manuscript. Y.Q.L. and Y.Z.C. contributed to the interpretation of the results and critical revision of the manuscript for important intellectual content and approved the final version of the manuscript. All authors have read and approved the final manuscript.
Financial support: This study was supported by grants from the National Natural Science Foundation of China (81873550 and 81670489).
Potential competing interests: None to report.
Ethical approval: This study was approved by the Ethics Committee of the Qilu Hospital of Shandong University, Jinan, Shandong, China (2018–126). All patients signed informed consent.
Study Highlights.
WHAT IS KNOWN
✓ Low-grade inflammation occurs in the intestinal mucosa of patients with IBS, and increased humoral immunity occurs in diarrhea-predominant IBS.
✓ Fecal IgA is involved in regulating intestinal homeostasis and the pathogenesis of IBD.
✓ How humoral responses to microbiota regulate the pathogenesis of IBS-D and the roles of mucosal IgA and IgA-coated bacteria in the pathogenesis of IBS-D are unclear.
WHAT IS NEW HERE
✓ Patients with IBS-D exhibited activated IgA class switching and higher IgA levels in the terminal ileum.
✓ Patients with IBS-D presented an increased proportion of IgA-coated bacteria, which was related to the strength of the inflammatory response triggered by intestinal bacteria.
✓ Ileal IgA and IgA-coated bacteria were associated with the major clinical manifestations of IBS-D.
✓ Escherichia–Shigella, particularly IgA-coated Escherichia–Shigella, which was anomalously enriched and positively correlated with anxiety and depression scores, was associated with the clinical manifestations of IBS-D.
TRANSLATIONAL IMPACT
✓ Our study revealed that patients with IBS-D exhibit higher levels of IgA and IgA class switching and an increased proportion of IgA-coated bacteria, which are associated with the clinical manifestations. IgA may mediate the effects of microbial dysbiosis on the pathophysiology of IBS-D. Understanding the role of IgA in the pathogenesis of IBS-D will provide a foundation for potential therapeutic interventions to prevent or ameliorate inflammatory reactions of IBS-D.
Supplementary Material
ACKNOWLEDGMENTS
We thank all participants in this study. Special thanks to the Laboratory of Translational Gastroenterology, Qilu Hospital of Shandong University (Jinan, Shandong, China), and Robot Engineering Laboratory for precise diagnosis and therapy of GI tumor, Qilu Hospital of Shandong University (Jinan, Shandong, China) for technical support and professional guidance. Besides, we thank Editage (www.editage.cn) for English language editing.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A218, http://links.lww.com/CTG/A217, http://links.lww.com/CTG/A219
References
- 1.Lovell RM, Ford AC. Global prevalence of and risk factors for irritable bowel syndrome: A meta-analysis. Clin Gastroenterol Hepatol 2012;10:712–21.e4. [DOI] [PubMed] [Google Scholar]
- 2.Mearin F, Lacy BE, Chang L, et al. Bowel disorders. Gastroenterology 2016;150:1393–407. [DOI] [PubMed] [Google Scholar]
- 3.Barbara G, De Giorgio R, Stanghellini V, et al. A role for inflammation in irritable bowel syndrome? Gut 2002;51(Suppl 1):i41–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vicario M, González-Castro AM, Martínez C, et al. Increased humoral immunity in the jejunum of diarrhoea-predominant irritable bowel syndrome associated with clinical manifestations. Gut 2015;64:1379–88. [DOI] [PubMed] [Google Scholar]
- 5.Forshammar J, Isaksson S, Strid H, et al. A pilot study of colonic B cell pattern in irritable bowel syndrome. Scand J Gastroenterol 2008;43:1461–6. [DOI] [PubMed] [Google Scholar]
- 6.Cerutti A. The regulation of IgA class switching. Nat Rev Immunol 2008;8:421–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol 2012;12:821–32. [DOI] [PubMed] [Google Scholar]
- 8.Slack E, Balmer ML, Fritz JH, et al. Functional flexibility of intestinal IgA—broadening the fine line. Front Immunol 2012;3:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin R, Chen H, Shu W, et al. Clinical significance of soluble immunoglobulins A and G and their coated bacteria in feces of patients with inflammatory bowel disease. J Transl Med 2018;16:359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okai S, Usui F, Ohta M, et al. Intestinal IgA as a modulator of the gut microbiota. Gut Microbes 2017;8:486–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang T, Ding C, Zhao M, et al. Sodium butyrate reduces colitogenic immunoglobulin A-coated bacteria and modifies the composition of microbiota in IL-10 deficient mice. Nutrients 2016;8:E728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014;158:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viladomiu M, Kivolowitz C, Abdulhamid A, et al. IgA-coated E. coli enriched in Crohn's disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med 2017;9:eaaf9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahai P, Mandiga P, Lobo S. Anatomy, abdomen and pelvis, large intestine. In StatPearls. StatPearls Publishing StatPearls Publishing LLC.: Treasure Island, FL: 2019. [PubMed] [Google Scholar]
- 15.Chhabra P, Spano AJ, Bowers D, et al. Evidence for the role of the cecal microbiome in maintenance of immune regulation and homeostasis. Ann Surg 2018;268:541–9. [DOI] [PubMed] [Google Scholar]
- 16.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol 2002;20:165–96. [DOI] [PubMed] [Google Scholar]
- 17.Schneider P. The role of APRIL and BAFF in lymphocyte activation. Curr Opin Immunol 2005;17:282–9. [DOI] [PubMed] [Google Scholar]
- 18.Moisini I, Davidson A. BAFF: A local and systemic target in autoimmune diseases. Clin Exp Immunol 2009;158:155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pardali E, Xie XQ, Tsapogas P, et al. Smad and AML proteins synergistically confer transforming growth factor beta1 responsiveness to human germ-line IgA genes. J Biol Chem 2000;275:3552–60. [DOI] [PubMed] [Google Scholar]
- 20.Puga I, Cols M, Cerutti A. Innate signals in mucosal immunoglobulin class switching. J Allergy Clin Immunol 2010;126:889–95; quiz 96–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francis CY, Morris J, Whorwell PJ. The irritable bowel severity scoring system: A simple method of monitoring irritable bowel syndrome and its progress. Aliment Pharmacol Ther 1997;11:395–402. [DOI] [PubMed] [Google Scholar]
- 22.Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol 1997;32:920–4. [DOI] [PubMed] [Google Scholar]
- 23.Zigmond AS, Snaith RP. The hospital anxiety and depression scale. Acta Psychiatr Scand 1983;67:361–70. [DOI] [PubMed] [Google Scholar]
- 24.Feldman S, Kasjanski R, Poposki J, et al. Chronic airway inflammation provides a unique environment for B cell activation and antibody production. Clin Exp Allergy 2017;47:457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chitnis N, Clark PM, Kamoun M, et al. An expanded role for HLA genes: HLA-B encodes a microRNA that regulates IgA and other immune response transcripts. Front Immunol 2017;8:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kau AL, Planer JD, Liu J, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med 2015;7:276ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maes M, Twisk FN, Kubera M, et al. Increased IgA responses to the LPS of commensal bacteria is associated with inflammation and activation of cell-mediated immunity in chronic fatigue syndrome. J Affect Disord 2012;136:909–17. [DOI] [PubMed] [Google Scholar]
- 28.He B, Santamaria R, Xu W, et al. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat Immunol 2010;11:836–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawamoto S, Tran TH, Maruya M, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science 2012;336:485–9. [DOI] [PubMed] [Google Scholar]
- 30.Bunker JJ, Flynn TM, Koval JC, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 2015;43:541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Waaij LA, Mesander G, Limburg PC, et al. Direct flow cytometry of anaerobic bacteria in human feces. Cytometry 1994;16:270–9. [DOI] [PubMed] [Google Scholar]
- 32.Dillon SR, Gross JA, Ansell SM, et al. An APRIL to remember: Novel TNF ligands as therapeutic targets. Nat Rev Drug Discov 2006;5:235–46. [DOI] [PubMed] [Google Scholar]
- 33.Waldschmidt TJ, Noelle RJ. Immunology. Long live the mature B cell--a baffling mystery resolved. Science 2001;293:2012–3. [DOI] [PubMed] [Google Scholar]
- 34.Thompson JS, Bixler SA, Qian F, et al. BAFF-R, a newly identified TNF receptor that specifically interacts with BAFF. Science 2001;293:2108–11. [DOI] [PubMed] [Google Scholar]
- 35.Castigli E, Wilson SA, Scott S, et al. TACI and BAFF-R mediate isotype switching in B cells. J Exp Med 2005;201:35–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connor BP, Raman VS, Erickson LD, et al. BCMA is essential for the survival of long-lived bone marrow plasma cells. J Exp Med 2004;199:91–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soares DM, Figueiredo MJ, Martins JM, et al. A crucial role for IL-6 in the CNS of rats during fever induced by the injection of live E. coli. Med Microbiol Immunol 2012;201:47–60. [DOI] [PubMed] [Google Scholar]
- 38.Small CL, Reid-Yu SA, McPhee JB, et al. Persistent infection with Crohn's disease-associated adherent-invasive Escherichia coli leads to chronic inflammation and intestinal fibrosis. Nat Commun 2013;4:1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cattaneo A, Cattane N, Galluzzi S, et al. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging 2017;49:60–8. [DOI] [PubMed] [Google Scholar]
- 40.Strati F, Cavalieri D, Albanese D, et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collins SM. A role for the gut microbiota in IBS. Nat Rev Gastroenterol Hepatol 2014;11:497–505. [DOI] [PubMed] [Google Scholar]
- 42.Salonen A, de Vos WM, Palva A. Gastrointestinal microbiota in irritable bowel syndrome: Present state and perspectives. Microbiology 2010;156:3205–15. [DOI] [PubMed] [Google Scholar]
- 43.Jeffery IB, O'Toole PW, Ohman L, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 2012;61:997–1006. [DOI] [PubMed] [Google Scholar]
- 44.Rajilic-Stojanovic M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 2011;141:1792–801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.