Abstract
Prolonged hyperuricemia is a cause of gout and an independent risk factor for chronic health conditions including diabetes and chronic kidney diseases. Genome-wide association studies (GWASs) for serum uric acid (SUA) concentrations have repeatedly confirmed genetic loci including those encoding uric acid transporters such as ABCG2 and SLC9A2. However, many single nucleotide polymorphisms (SNPs) found in GWASs have been common variants with small effects and unknown functions. In addition, there is still much heritability to be explained. To identify the causative genetic variants for SUA concentrations in Korean subjects, we conducted a GWAS (1902 males) and validation study (2912 males and females) and found four genetic loci reaching genome-wide significance on chromosomes 4 (ABCG2) and 11 (FRMD8, EIF1AD and SLC22A12-NRXN2). Three loci on chromosome 11 were distributed within a distance of 1.3 megabases and they were in weak or moderate linkage disequilibrium (LD) states (r2 = 0.02–0.68). In a subsequent association analysis on the GWAS loci of chromosome 11 using closely positioned markers derived from whole genome sequencing data, the most significant variant to be linked with the nearby GWAS signal was rs121907892 (c.774G>A, p.W258*) of the SLC22A12 gene. This variant, and each of the three GWAS SNPs, were in LD (r2 = 0.06–0.32). The strength of association of SNPs with SUA concentration (negative logarithm of P-values) and the genetic distance (r2 of LD) between rs121907892 and the surrounding SNPs showed a quantitative correlation. This variant has been found only in Korean and Japanese subjects and is known to lower the SUA concentration in the general population. Thus, this low-frequency variant, rs121907892, known to regulate SUA concentrations in previous studies, is responsible for the nearby GWAS signals.
Introduction
Uric acid is the final product of purine metabolism produced from xanthine and hypoxanthine by the action of xanthine oxidase. Uric acid is mainly present as urate under normal physiological conditions. Homeostasis of serum uric acid (SUA) concentration is maintained through a balance between oral intake and renal/intestinal excretion. Prolonged elevation of SUA concentrations—hyperuricemia—accelerates the formation of monosodium urate crystals and causes gout. Hyperuricemia is independently associated with hypertension, diabetes, chronic kidney diseases and cardiovascular mortality [1–3]. In addition, recent clinical or experimental studies have suggested that hyperuria contributes to the development of metabolic syndrome [4].
The genetic contribution to SUA concentration is estimated at 40–70% [5]. To identify underlying genetic alterations involved in the regulation of SUA concentration, genome-wide association studies (GWASs) have been conducted in many populations, and genetic loci covering ABCG2 and SLC9A2 have been found most repeatedly and significantly [6–10]. Most of the genes identified in current GWASs seem plausible contributors to SUA concentrations because they encode for proteins that are responsible for uric acid excretion in the kidney or intestines, including the genes ABCG2 and SLC9A2 [11]. However, except for rs2231142 in ABCG2, most single nucleotide polymorphisms (SNPs) that have been reported to be significant so far are located in intronic or intergenic regions, and causative variants have not been identified for most loci. These collectively explained up to 7% of the inter-individual SUA variance in serum concentration, and the proportion explained by ABCG2 and SLC9A2 alone was about 2–3% [6,7,9]. Thus, much heritability is yet to be explained and many genetic variants remain to be identified.
While GWASs to date have provided valuable clues about the genetic background for SUA concentrations, additional GWASs using common variants with low effect are unlikely to be able to explain the residual heritability robustly. This “missing heritability” problem has also been raised in many other GWASs [12]. One hypothesis is that common variants with weak effects might have been found in GWASs because of their linkage disequilibrium (LD) relationships with causative variants with low-frequencies and large effects. To find such low-frequency causative variants responsible for GWAS signals, resequencing of target sites or whole genome sequencing (WGS) is required. Here, we identify such a low-frequency causative variant and its relationship with common variants in GWAS signals.
Results
GWAS and validation
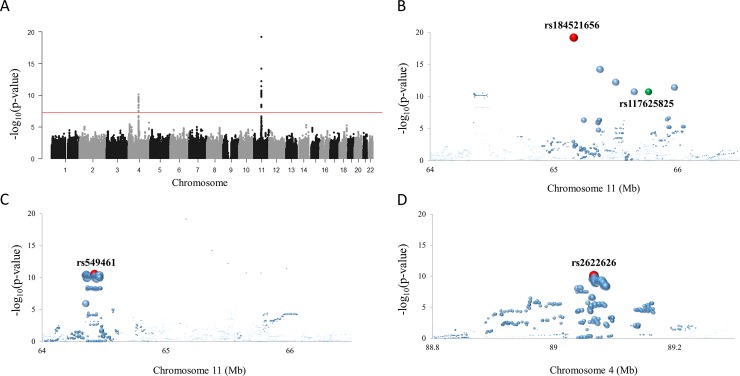
The baseline characteristics for study participants are described in S1 Table. The median SUA concentration was 6.4 mg/dL. As shown in the quantile–quantile (Q–Q) probability plot, the observed P-values of the GWAS were well fitted to expected values (S1 Fig). The genomic inflation factor was calculated as 1.0, demonstrating the homogeneity of the study population. The four association signals were found to reach genome-wide significance level (P < 5.0E–8; Table 1 and Fig 1). The most significant genetic association was found on chromosome 11, in a 65 Mb region (Fig 1B). The six SNPs with a low minor allele frequency (MAF) of 1.2–1.6% and p < 1E–10 were widely distributed over the region (Fig 1B). Because rs184521656 and the other five SNPs were in a moderate state of LD (r2 = 0.66–0.77) and those SNPs excluding rs184521656 showed strong LD relationships with each other (r2 > 0.95), these adjacent SNPs were considered as potentially separate signals (S2 Table). Rs184521656 is located on the intronic region of FRMD8. The other five SNPs are located on noncoding or intergenic regions of different genes and the areas in which they were located contain many genes. The third strongest signal was from chromosome 11, in a 64 Mb region. Rs549461 (P = 2.8E–11, MAF = 22%) and many significant SNPs with LD relationships (r2 > 0.83) are located on the SLC22A12-NTXN2 region (Fig 1C). Another signal (rs2622626, p = 7.5E–11, MAF = 30%) was located on the ABCG2 region of chromosome 4 (Fig 1D).
Table 1. Results of the genome-wide association and validation studies for serum uric acid concentrations.
| SNP | Chr | Position (bp) | Nearest genes | Minor allele | Major allele | GWAS (n = 1902) | Validation study (n = 2912) | Combined (n = 4814) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF (%) | Beta (SE) | P | MAF (%) | Beta (SE) | p | p | ||||||
| rs184521656 | 11 | 65161450 | FRMD8 (intron) | T | C | 1.2 | –1.70 (0.18) | 6.5E–20 | 1.3 | –0.99 (0.14) | 6.0E–12 | 1.1E–28 |
| rs117625825 | 11 | 65765725 | EIF1AD (3′ UTR) | A | G | 1.6 | –1.11 (0.16) | 1.9E–11 | 1.5 | –0.84 (0.13) | 1.2E–10 | 1.9E–20 |
| rs549461 | 11 | 64423032 | SLC22A12-NRXN2 (intron) | A | G | 22.4 | –0.33 (0.05) | 2.8E–11 | ||||
| rs2622626 | 4 | 89066715 | ABCG2 (intron) | C | A | 30.0 | –0.30 (0.05) | 7.5E–11 | ||||
GWAS, genome-wide association study; SNP, single nucleotide polymorphism; bp, base pair; Chr, chromosome; MAF, minor allele frequency; SE, standard error; UTR untranslated region
Fig 1.
(A) Manhattan plot of the GWAS results for SUA concentration. The horizontal red line represents the genome-wide significance level. Regional plots of the GWAS signals are shown for chromosome 11, 65–66 Mb (B), 64–65 Mb (C) and chromosome 4, 89 Mb (D). The red circle indicates the most significant SNP in the locus, and the circle size is proportional to the strength of LD (r2) with the most significant SNP. (B) Rs184521656 and rs117625825 are in a moderate LD relationship (r2 = 0.68).
Because the latter two loci in SLC22A12-NRXN2 and ABCG2 were found repeatedly in previous studies [11], we only validated the SNPs on the 65 Mb region of chromosome 11 by increasing the number of individuals sampled. Rs117625825 was chosen as the tag SNP, the best representative of the remaining five SNPs identified using Haploview [13] (S3 Table). Rs184521656 and rs117625825 were genotyped and analyzed as an independent group (Table 1); they were validated and had statistical significance. Because only males were included in the GWAS stage and both males and females were included in the validation study stage, we further tested the effect of these two SNPs (rs184521656 and rs117625825) on the SUA concentration of each male and female subject. In both males and females, these two SNPs were significantly associated with SUA concentration and the direction of their effect was also the same (S4 Table).
Association study using WGS data
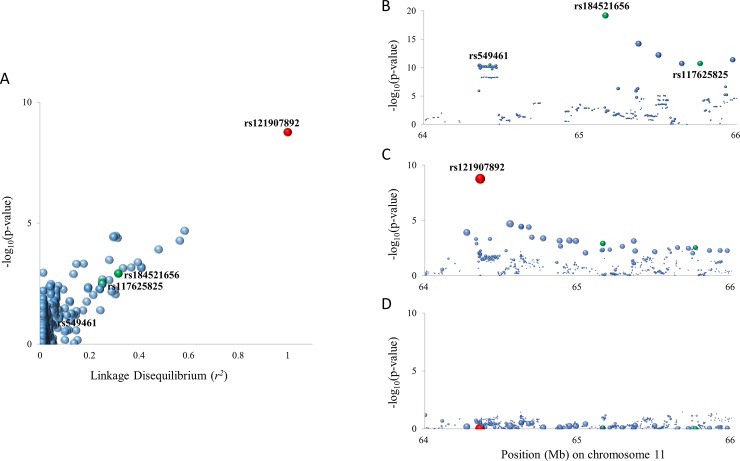
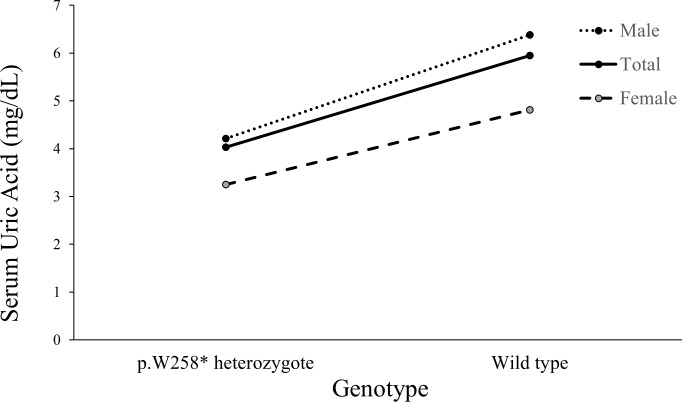
Although rs184521656 and rs117625825 have been validated successfully with statistical significance, it is difficult to regard these SNPs as causative variants because they are located in noncoding regions and the functions of their adjacent genes are insufficient to explain a causal relationship with SUA concentration. To identify more influential variants, we performed additional association analysis using closely spaced markers around the GWAS signal on chromosome 11, 63–67 Mb in extent, extracted from the 797 WGS data that formed part of the validation samples (S2 Fig). The most significant SNP, rs121907892 (P = 1.7E–09, MAF = 1.2%; Table 2), was positioned at the third nucleotide of amino acid codon #258 of SLC22A12 (NM_144585). The nucleotide change G>A of this SNP turns p.W258 into a stop codon (p.W258*). Interestingly, the degree of LD (estimated as r2) between rs121907892 and surrounding SNPs and the negative logarithm of the P-value (–log10 p) of their association showed positive correlation, which was also observed in the relationship with rs184521656, rs117625825 and rs549461 (Fig 2A). When associations of the SNPs with SUA concentration were reanalyzed after adjusting for rs121907892, their associations were weakened greatly (Fig 2B–2D and S5 Table). To confirm the association between rs121907892 and SUA concentration in larger samples, rs121907892 was genotyped and analyzed in an entire sample including discovery (GWAS) and validation samples. Among a total of 4708 individuals that were successfully genotyped, 130 had a heterozygous p.W258* variant and none had a homozygous variant (MAF = 1.4%). The strongest association was observed (P = 2.3E–88) and the SNP explained 8.1% of the inter-individual variance of SUA concentration in our study population (Table 2). The mean SUA concentrations of those with or without p.W258* were 4.03 and 5.95 mg/dL, respectively (Fig 3 and S6 Table).
Table 2. The association of rs121907892 with SUA concentration.
| SNP | Chr | Position (bp) | Nearest gene | Minor allele | Major allele | WGS (n = 797) | Validation study (n = 4708) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF (%) | Beta (SE) | p | MAF (%) | Beta (SE) | p | ||||||
| rs121907892 | 11 | 64361219 | SLC22A12 (p.W258*) | A | G | 1.2 | –1.68 (0.28) | 1.7E–09 | 1.4 | –2.10 (0.10) | 2.3E–88 |
Chr, chromosome; WGS, whole genome sequencing; MAF, minor allele frequency; SE, standard error
Fig 2. The role of rs121907892 in the GWAS signal on chromosome 11: 64–66 Mb region.
All dots in the figure are SNPs showing a genetic association with rs121907892, with r2 values > 0.01. Green circles are representative SNPs of each locus that reached genome-wide significance in the GWAS. The circle size is proportional to the strength of LD (r2) with rs121907892 (red circle). (A) Correlation between LD value (r2) with rs121907892 and the significance of association (–log10 p) of SNPs with serum uric acid (SUA) concentrations. (B) Regional plot for the results of the GWAS for SUA concentration. (C, D) Regional plots for the results of the association analysis for SUA concentration using SNPs derived from whole genome sequencing data before (C) and after (D) the conditional analysis incorporating rs121907892.
Fig 3. Effect of the p.W258* genotype on serum uric acid concentration in this study group.
Discussion
Using a stepwise approach, we found that a low-frequency SNP, rs121907892, was the lead variant of nearby GWAS signals for SUA concentrations. First, in our GWAS, we found four association signals reaching genome-wide significance, two previously established common SNPs (rs2622626 and rs549461) and two new low-frequency SNPs (rs184521656 and rs117625825). Although the latter two were validated using a subsequent study with increased sample size, because of their location in a noncoding region and weak cogency of their surrounding genes, we anticipated that there might be a causative variant nearby, and conducted further association analysis using more closely spaced markers derived from WGS data. The SNP rs121907892—the most significant variant found in this analysis—shows a LD relationship with surrounding variants including the three SNPs that reached genome-wide significance in the initial GWAS in chromosome 11 at 64–65 Mb (rs184521656, rs117625825 and rs549461). The degrees of LD between rs121907892 and each surrounding SNP and the significance of their associations with SUA concentrations were closely related. In addition, in the conditional analysis on rs121907892, the associations of these SNPs with SUA concentrations disappeared.
The SLC22A12 gene encodes a urate transporter in renal proximal tubules serving to reabsorb urate [14]. A SLC22A12 p.W258* variant was first found in a Japanese patient with idiopathic renal hypouricemia and exercise-induced acute renal failure [14], and has been reported further in Korean and Japanese patients with hereditary renal hypouricemia [15,16]. In addition to patients with this Mendelian inherited disorder, the effects of p.W258* on SUA concentrations or renal functions have been studied in the general population. The Suita study targeted a community-based general population in Japan and sequenced the SLC22A12 gene of 24 subjects with low SUA concentrations; it found that those with a homozygous or heterozygous p.W258* variant had low SUA concentrations but normal creatinine concentrations [17]. They suggested that loss-of-function variants of SLC22A12 might not be harmful in the general population. Another study examined the genotype distribution of p.W258* and SUA concentrations in 5023 Japanese subjects from the general population [18]. Five individuals with a homozygous p.W258* variant had SUA concentrations of < 1 mg/dL, and those with a heterozygous p.W258* variant had a significantly lower mean SUA concentration than did subjects with a wild-type allele, although they had a wider range of SUA concentrations (0.8–7.8 mg/dL) than did subjects with a homozygous p.W258* variant. The SLC22A12 p.W258* variant has been found only in Japanese and Koreans (http://www.1000genomes.org). Its allele frequency in Japanese and Korean general populations has been reported to be 0.02 and 0.01, respectively [18,19], consistent with our results. In our study, 130 subjects with the heterozygous p.W258* variant had slightly higher estimated glomerular filtration rate values than those without the variant (90.6 vs 88.1 mL/min/1.73 m2), but this was not statistically significant [20]. Thus, the role of SLC22A12 p.W258* in lowering the SUA concentration is relatively clear, but it does not seem to have adverse effects on renal function in the Japanese and Korean general populations.
Targeted sequencing of the SLC22A12 gene has identified low-frequency variants greatly affecting SUA concentrations in the general population. In addition to p.W258*, Iwai et al. identified additional rare variants (MAF < 0.01) in subjects with low SUA concentrations. Among these, p.R90H and p.R477H showed statistically significant associations with hypouricemia; a deletion of p.D313–P333 was identified in only one subject, but it was proved experimentally that it abolished the urate transport activity of SLC22A12 [17]. In addition, a subject with the p.R228E variant had a low SUA concentration, although this missense variant was not proved experimentally and was not statistically significant because it was found in only one subject. Direct sequencing of exons 7 and 9 of the SLC22A12 gene revealed a deletion (p.L415_G417del) and a missense variant (p.T467M) in 1.87 and 5.56% of the Czech and Slovak Roma populations, respectively [21]. Although that study did not directly address the association of these variants with SUA concentration, these variants were initially found in patients with primary renal hypouricemia in the same geographic area. We reviewed rare variants of the SLC22A12 gene in our WGS data and found three missense variants (p.R90H, p.R447H and p.Q382L) and one splicing site variant (c.661+1G>A). No subject had a homozygotic or a compound heterozygotic form of these variants; instead, all variant-positive subjects had only one of them in a heterozygous form and also showed low SUA concentrations (S7 Table). The p.Q382L variant was reported in a Japanese patient with renal hypouricemia as a compound heterozygotic form of p.W258*/p.Q382L [16]. The splicing site variant (c.661+1G>A) was found in only one person in our study. This was submitted to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/), and was classified as “Uncertain significance” under the “Familial renal hypouricemia” condition (accessed 31 July 2019) [22].
Population genetic models have demonstrated that a variant’s allele frequency is inversely related to the magnitude of its effect on a phenotype [23]. According to this, the common variants with MAF ≥ 0.05 that are mostly used in GWASs have inherently low “regression effects.” Therefore, there have been emerging attempts to find more powerful low-frequency variants. These variants inevitably show a population-specific distribution, because they are recent [24]. In addition to the 1000 genome projects involving 26 populations from all over the world, efforts are under way to create reference panels using WGS approaches in particular sample sets including the British (UK10K), Dutch (Genome of the Netherlands) [25], Sardinian [26] and Icelandic (deCODE Genetics) [27] populations. These have shown that population-based panels are particularly effective in discovering genotypes with a low population frequency. In these studies, some low-frequency variants strongly associated with specific phenotypes have been identified, as summarized recently [28]. The phenotypes included various complex traits or diseases such as blood lipid or glucose concentrations, bone mineral density, height, type 2 diabetes, and Alzheimer’s disease, but to our knowledge there have been no results for SUA concentrations.
The role of the common noncoding variant found in GWASs has been revealed through subsequent studies in some cases. Experiments have shown that T>C base changes in rs1421085, an intronic variant of the FTO gene found in GWASs on obesity, is involved in the regulation of adipocyte thermogenesis by increasing the expressions of nearby IRX3 and IRX5, but not FTO [29]. Rs7085104 upstream of AS3MT gene, a top GWAS-identified SNP associated with schizophrenia, is linked with a variable number tandem repeat sequence in exon 1 of AS3MT that is involved in the expression of the arsenite methyltransferase isoform (AS3MTd2d3) [30]. Surakka et al. showed that low-frequency functional variants that cause Mendelian-inherited dyslipidemia or have a known impact on lipids have larger average effects than the common variants found in GWASs, and contribute considerably to population variance in blood lipid concentrations [31]. Some of these low-frequency variants explain the association of common variants in the overlapping loci [31]. In our study, rs121907892 explained the association of surrounding common or low-frequency variants located over 1 Mb and at—least in Koreans—it might explain the signals found at this locus in previous GWASs for SUA concentration. This study linked the previously found GWAS signals and a variant that causes Mendelian-inherited disorders. It is also clearly shown that the degree of associations of SNPs with the phenotype correlates with the degree of LD between the low-frequency lead SNP and its surrounding SNPs.
The biggest weakness of this study is that rs121907892 is found only in Koreans and Japanese, so the results cannot be applied to other populations. Because this genetic locus has been repeatedly found in GWASs of other populations, there might be other low-frequency variants such as rs121907892. However, this study was conducted only in Koreans, so we could not find such variations in other ethnic groups. In addition, for many GWAS results for common variants lacking known molecular mechanisms, it will be necessary to use multifaceted approaches such as deep sequencing or functional experiments to discover their mechanism of action or the actual functioning variants surrounding them.
Methods
Study design and baseline measurements
The design of this study has been described previously [32]. A schema of this study is shown in S2 Fig. The subjects enrolled for this GWAS included unrelated adult male Koreans aged 20–65 years who visited the Seoul National University Hospital (SNUH, Seoul, Republic of Korea) for regular health checkups from May 2011 to December 2013. This study was originally designed to investigate the genetic background of abdominal obesity and metabolic syndrome and only male participants were recruited during that period. Participants with conditions that might influence body weight were excluded: individuals who had 1) been diagnosed with thyroid diseases or taken thyroid medication; 2) taken medication or undergone a procedure or operative treatment for obesity within 3 months of enrollment; 3) a medical history of stroke, cardiovascular diseases, diabetes mellitus treated with medication, cancer, or abdominal surgery. Repeated examinees and those with insufficient blood for testing were also excluded. In addition, individuals taking medications that could influence the SUA concentration, including allopurinol and antituberculotics, were excluded. After application of the exclusion criteria, 1,947 males were recruited for the GWAS. Since 2014, it has expanded to include both male and female participants. In this phase, 2,912 individuals (1,557 males and 1,355 females) were enrolled for the validation study. This study was approved by the institutional review board of SNUH (#C-1701-131-828) and all subjects gave written informed consent.
Anthropometric values including height and weight were measured by trained medical personnel, and the body mass index (BMI, in kg/cm2) was calculated subsequently. Blood tests were conducted after ≥ 12 h of fasting and the SUA concentration was measured by enzymatic colorimetric method using a COBAS 8000 c 702 analyzer (Roche Diagnostics, Japan) in mg/dL. Participants filled out a standardized questionnaire routinely used for health checkups that included items regarding previous diseases or medication history.
GWAS
Genome-wide SNP genotyping and imputation procedures have been described previously [32]. The Illumina HumanCore BeadChip kit (Illumina, San Diego, CA, USA) was used for genome-wide SNP genotyping of 1,947 subjects. Individuals with call rate < 0.88 were excluded. After SNPs with Hardy–Weinberg equilibrium (HWE) with P < 1.0E–6, call rate < 95%, or MAF < 1% had been excluded, the remaining 264,283 SNPs were used for genotype imputation. For the imputation, 1000 Genomes ASN Phase I integrated variant set release (v. 3) in NCBI build 37 (hg19) was used as the reference panel. We used PLINK (v. 1.07) for strand alignment, SHAPEIT2 for phasing, and IMPUTE2 for imputation [33–35]. Of the imputed SNPs, only those variants with Info Score ≥ 0.9 were selected. A total of 4,414,664 SNPs that were either genotyped or imputed with high confidence were used for the GWAS. One member of a pair of relatives was randomly selected based on genome-wide identity-by-descent estimates implemented in PLINK and the other member was excluded. In addition, after excluding those for whom phenotype information was not available, 1,902 males with both genotype and phenotype information were included in the GWAS. The distribution of SUA concentrations is described in S3 Fig. Genetic associations of SUA concentrations were tested using PLINK using a linear model adjusted for age and BMI. R software (v. 3.4.1) was used for drawing the SUA concentration distribution, Q–Q and Manhattan plots (https://www.R-project.org/). Integrative Genomics Viewer software (v2.3.6) was used for review of the association results [36]. SNPs were annotated to obtain the information including amino acid changes or population frequencies using ANNOVAR [37].
Validation study and meta-analysis
A validation study was conducted by increasing the number of subjects on two SNPs (rs184521656 and rs117625825) that were found to be the most significant in our GWAS (S2 Fig). Of the 2,912 subjects who were included for the validation study, 2,691 and 2,699 individuals were genotyped successfully using the Taqman method for rs184521656 and rs117625825, respectively. Detailed information about the probes used for Taqman genotyping is listed in S8 Table. ViiA 7 real-time quantitative polymerase chain reaction equipment and embedded software were used (Thermo Fisher Scientific, Waltham, MA, USA).
Association analysis between SUA concentration and these two SNPs were performed in the same method as GWAS using a linear model implemented in PLINK. Because the validation study included both male and female subjects, gender was added as another covariate in addition to age and BMI. The results of the GWAS and validation study for these two SNPs were combined to obtain an overall estimate of P-value through meta-analysis using inverse variance-based analysis assuming a fixed-effect model implemented in PLINK (Table 1).
Association analysis of target regions in chromosome 11 using WGS data
Of the 2,912 individuals included in the validation study, 850 were subjected to WGS with a mean depth of 15–30×. Paired-end sequencing reads generated on the Hiseq platform were aligned to the human reference sequence (hg19) and subsequent data processing including making BAM files and genetic variant call format files were performed using the Dynamic Read Analysis for GENomics (DRAGEN) platform by a commercial company (Macrogen Inc., Seoul, S. Korea). SNPs with a calling rate > 0.9 and HWE P > 1.0E–3 were selected for variants with local read depth ≥ 5 and genotype quality ≥ 20. Samples with heterozygous/homozygous ratios < 2 and call rate > 0.9 were selected; a total of 802/850 subjects passed the sample quality control (QC). Of these, 797 had phenotype data and were included in the final association analysis using WGS data. Of the 32,613 SNPs that passed the QC on the 63–67 Mb region of chromosome 11, only 6,773 SNPs with MAF > 1% were included for the association analysis. Associations of these SNPs with SUA concentrations were evaluated in the same way using PLINK.
Statistical significance
Associations with P < 5.0E–8 were considered genome-wide significant in the discovery GWAS stage [6,38]. In the following analysis, associations were considered significant if the P-value was lower than the Bonferroni correction level (0.05/the number of tests).
Supporting information
The genomic inflation factor was calculated as 1.
(PDF)
The numbers in the circles refer to the order in which the study was conducted.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by Basic Science Research Program through the National Research Foundation of Korea(NRF, https://www.nrf.re.kr/eng/index) funded by the Ministry of Education [NRF-2017R1A6A3A11031988 to S.I.]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jolly SE, Mete M, Wang H, Zhu J, Ebbesson SO, Voruganti VS, et al. Uric acid, hypertension, and chronic kidney disease among Alaska Eskimos: The Genetics of Coronary Artery Disease in Alaska Natives (GOCADAN) study. J Clin Hypertens (Greenwich). 2012;14(2):71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stack AG, Hanley A, Casserly LF, Cronin CJ, Abdalla AA, Kiernan TJ, et al. Independent and conjoint associations of gout and hyperuricaemia with total and cardiovascular mortality. QJM. 2013;106(7):647–658. 10.1093/qjmed/hct083 [DOI] [PubMed] [Google Scholar]
- 3.Sluijs I, Beulens JW, van der A DL, Spijkerman AM, Schulze MB, et al. Plasma uric acid is associated with increased risk of type 2 diabetes independent of diet and metabolic risk factors. J Nutr. 2013;143(1):80–85. 10.3945/jn.112.167221 [DOI] [PubMed] [Google Scholar]
- 4.Kim SK. Interrelationship of uric acid, gout, and metabolic syndrome: focus on hypertension, cardiovascular disease, and insulin resistance. J Rheum Dis. 2018;25(1):19–27. [Google Scholar]
- 5.Nath SD, Voruganti VS, Arar NH, Thameem F, Lopez-Alvarenga JC, Bauer R, et al. Genome scan for determinants of serum uric acid variability. J Am Soc Nephrol. 2007;18(12):3156–3163. 10.1681/ASN.2007040426 [DOI] [PubMed] [Google Scholar]
- 6.Kottgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45(2):145–154. 10.1038/ng.2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okada Y, Sim X, Go MJ, Wu JY, Gu D, Takeuchi F, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet. 2012;44(8):904–909. 10.1038/ng.2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giri AK, Banerjee P, Chakraborty S, Kauser Y, Undru A, Roy S, et al. Genome wide association study of uric acid in Indian population and interaction of identified variants with Type 2 diabetes. Sci Rep. 2016;6:21440 10.1038/srep21440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang B, Mo Z, Wu C, Yang H, Yang X, He Y, et al. A genome-wide association study identifies common variants influencing serum uric acid concentrations in a Chinese population. BMC Med Genomics. 2014;7:10 10.1186/1755-8794-7-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J, Lee Y, Park B, Won S, Han JS, Heo NJ. Genome-wide association analysis identifies multiple loci associated with kidney disease-related traits in Korean populations. PLoS One 2018;13:e0194044 10.1371/journal.pone.0194044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merriman TR. An update on the genetic architecture of hyperuricemia and gout. Arthritis Res Ther. 2015;17:98 10.1186/s13075-015-0609-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 14.Enomoto A, Kimura H, Chairoungdua A, Shigeta Y, Jutabha P, Cha SH, et al. Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature. 2002;417(6887):447–452. 10.1038/nature742 [DOI] [PubMed] [Google Scholar]
- 15.Kim HO, Ihm CG, Jeong KH, Kang HJ, Kim JM, Lim HS, et al. A case report of familial renal hypouricemia confirmed by genotyping of SLC22A12, and a literature review. Electrolyte Blood Press. 2015;13(2):52–57. 10.5049/EBP.2015.13.2.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichida K, Hosoyamada M, Hisatome I, Enomoto A, Hikita M, Endou H, et al. Clinical and molecular analysis of patients with renal hypouricemia in Japan-influence of URAT1 gene on urinary urate excretion. J Am Soc Nephrol. 2004;15(1):164–173. 10.1097/01.asn.0000105320.04395.d0 [DOI] [PubMed] [Google Scholar]
- 17.Iwai N, Mino Y, Hosoyamada M, Tago N, Kokubo Y, Endou H. A high prevalence of renal hypouricemia caused by inactive SLC22A12 in Japanese. Kidney Int. 2004;66(3):935–944. 10.1111/j.1523-1755.2004.00839.x [DOI] [PubMed] [Google Scholar]
- 18.Hamajima N, Naito M, Hishida A, Okada R, Asai Y, Wakai K. Serum uric acid distribution according to SLC22A12 W258X genotype in a cross-sectional study of a general Japanese population. BMC Med Genet. 2011;12:33 10.1186/1471-2350-12-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Choi HJ, Lee BH, Kang HK, Chin HJ, Yoon HJ, et al. Prevalence of hypouricaemia and SLC22A12 mutations in healthy Korean subjects. Nephrology (Carlton). 2008;13(8):661–666. [DOI] [PubMed] [Google Scholar]
- 20.Levey AS, Coresh J, Greene T, Marsh J, Stevens LA, Kusek JW, et al. Expressing the Modification of Diet in Renal Disease Study equation for estimating glomerular filtration rate with standardized serum creatinine values. Clin Chem. 2007;53(4):766–772. 10.1373/clinchem.2006.077180 [DOI] [PubMed] [Google Scholar]
- 21.Gabrikova D, Bernasovska J, Sokolova J, Stiburkova B. High frequency of SLC22A12 variants causing renal hypouricemia 1 in the Czech and Slovak Roma population; simple and rapid detection method by allele-specific polymerase chain reaction. Urolithiasis. 2015;43(5):441–445. 10.1007/s00240-015-0790-4 [DOI] [PubMed] [Google Scholar]
- 22.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–868. 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park JH, Gail MH, Weinberg CR, Carroll RJ, Chung CC, Wang Z, et al. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci U S A. 2011;108(44):18026–18031. 10.1073/pnas.1114759108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surakka I, Kristiansson K, Anttila V, Inouye M, Barnes C, Moutsianas L, et al. Founder population-specific HapMap panel increases power in GWA studies through improved imputation accuracy and CNV tagging. Genome Res. 2010;20(10):1344–1351. 10.1101/gr.106534.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46(8):818–825. 10.1038/ng.3021 [DOI] [PubMed] [Google Scholar]
- 26.Sidore C, Busonero F, Maschio A, Porcu E, Naitza S, Zoledziewska M, et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet. 2015;47(11):1272–1281. 10.1038/ng.3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47(5):435–444. 10.1038/ng.3247 [DOI] [PubMed] [Google Scholar]
- 28.Bomba L, Walter K, Soranzo N. The impact of rare and low-frequency genetic variants in common disease. Genome Biol. 2017;18(1):77 10.1186/s13059-017-1212-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373(10):895–907. 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Jaffe AE, Straub RE, Tao R, Shin JH, Wang Y, et al. A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nat Med. 2016;22(6):649–656. 10.1038/nm.4096 [DOI] [PubMed] [Google Scholar]
- 31.Surakka I, Horikoshi M, Magi R, Sarin AP, Mahajan A, Lagou V, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47(6):589–597. 10.1038/ng.3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bae YS, Im SW, Kang MS, Kim JH, Lee SH, Cho BL, et al. Genome-Wide Association Study of Bone Mineral Density in Korean Men. Genomics Inform. 2016;14(2):62–68. 10.5808/GI.2016.14.2.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–181. 10.1038/nmeth.1785 [DOI] [PubMed] [Google Scholar]
- 35.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. 10.1002/gepi.20303 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The genomic inflation factor was calculated as 1.
(PDF)
The numbers in the circles refer to the order in which the study was conducted.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.