Abstract

Label-free high-throughput screening using mass spectrometry has the potential to provide rapid large-scale sample analysis at a speed of more than one sample per second. Such speed is important for compound library, assay and future clinical screening of millions of samples within a reasonable time frame. Herein, we present a liquid atmospheric pressure matrix-assisted laser desorption/ionization (AP-MALDI) setup for high-throughput large-scale sample analysis (>5 samples per second) for three substance classes (peptides, antibiotics, and lipids). Liquid support matrices (LSM) were used for the analysis of standard substances as well as complex biological fluids (milk). Throughput and analytical robustness were mainly dependent on the complexity of the sample composition and the current limitations of the commercial hardware. However, the ultimate limits of liquid AP-MALDI in sample throughput can be conservatively estimated to be beyond 10–20 samples per second. This level of analytical speed is highly competitive compared with other label-free MS methods, including electrospray ionization and solid state MALDI, as well as MS methods using multiplexing by labeling, which in principle can also be used in combination with liquid AP-MALDI MS.
Label-free high-throughput screening (HTS) of large sample sets using mass spectrometry (MS) has gained increased attention in recent years.1−5 Especially, the reduced numbers of false positive or negative results is a particular advantage in contrast to the non-MS label-based screening approaches such as fluorescence-based assays.6,7 The latter also require elaborate sample preparations using costly labels such as dyes.
So far the main focus for mass spectrometric HTS has been on electrospray ionization (ESI)8 and solid-state matrix-assisted laser desorption/ionization (MALDI)9 as ionization techniques, and a critical review has been published recently.10
ESI is a versatile and well-studied platform for the ionization of a broad range of pharmaceutically interesting compounds.11−16 However, a major drawback of ESI is a lack of speed in the supply of samples, which is a prerequisite for HTS applications. The fastest commercially available system using ESI is the Agilent RapidFire for which a maximum throughput of 2.5 s per sample was reported without using the supplied solid-phase extraction.17
Solid-state MALDI achieved an analytical speed of up to 2.5 samples per second (0.4 s per sample) for certain analytes using the Bruker RapifleX Pharma Pulse.9 However, the time for spotting and the actual biochemical assay were considerably longer. Acoustic Mist Ionization (AMI) and Desorption Electrospray Ionization (DESI) yielded comparable throughput with 0.45 s18 and 0.4 s19 per sample, respectively.
For the different MS ionization methods, biochemical matrices or necessary assay components can be challenging due to their imparted ion signal suppression,18 impeding crystallization in the case of MALDI or being generally incompatible with the necessary requirements regarding the sample environment or mass spectrometry (nonvolatile salts). However, the suitability of commonly used buffers for MS analysis was investigated20 and it was shown that label-based non-MS assays can be readily adapted for MALDI MS analysis.2 The implementation of an additional MALDI spot washing step offers the possibility to reduce buffer concentrations and hence make more assays accessible for analysis with MALDI MS.9
Liquid atmospheric pressure (AP) MALDI combines the advantages of both the analysis speed of conventional solid-state MALDI under AP and the versatility of ESI. Different types of biomolecules21−23 can be analyzed over a wide range of pH values24 and in a complex biological matrix,25 illustrating the general suitability of liquid AP-MALDI for biochemical screening assays. Additionally, the predominant formation of multiply charged analyte ions offers the possibility for further target characterization by highly informative MS/MS.26
Experimental Section
AP-MALDI MS
A detailed description of the in-house developed AP-MALDI setup can be found in a previous publication.27 Briefly, a heated transfer tube (1 mm internal diameter, 6 cm length) was placed at the inlet of a Synapt G2-Si HDMS instrument (Waters, Wilmslow, U.K.). Ions were generated using a pulsed 337 nm nitrogen laser (3 ns pulse duration; 30 Hz pulse repetition rate) and extracted from a target plate across a gap of approximately 3 mm to the ion transfer tube with a potential difference of 3.5 kV. A counter N2 gas flow of 180 L/h was applied to the ion transfer tube. Target plate movement was achieved using a Waters Research Enabled Software (WREnS)-controlled xy-stage and its start was synchronized with the start of the MS data acquisition. Data acquisition was set to TOF, sensitivity and positive ion mode with an m/z range of 100–2000. Manual calibration was performed by AP-LDI using sodium iodide and an acquisition time of 3 min with an m/z range of 100–2000 using Intellistart (MassLynx; Waters).
Materials
Ethylene glycol, propylene glycol, glycerol, water, tris base (trizma), acetonitrile (MeCN), trifluoroactic acid (TFA), bradykinin, α-cyano-4-hydroxycinnamic acid (CHCA), 2,5-dihydroxybenzoic acid (DHB), ampicillin sodium salt (AMP), and penicillinase from Bacillus cereus were purchased from Sigma-Aldrich (Gillingham, U.K.).
Solid MALDI Sample Preparation
Solid MALDI samples were prepared by mixing matrix solution with analyte solution at a ratio of 1:1 (v/v), spotting 1 μL of the mixture onto the target plate and leaving it to dry at room temperature. The CHCA matrix solution was prepared by dissolving CHCA in 0.1% TFA/MeCN (50:50; v/v) to yield a final concentration of 10 mg/mL. Similarly, a 20 mg/mL DHB matrix solution was prepared in 0.1% TFA/MeCN (70:30; v/v).
Liquid MALDI Sample Preparation
Liquid MALDI samples were prepared by mixing a liquid support matrix (LSM) with analyte solution at a ratio of 1:1 (v/v). The LSM consisted of a CHCA solution (5–30 mg/mL, 50:50 or 70:30 H2O/MeCN; v/v) with ethylene or propylene glycol added equal to 60–70% of the solution volume. For milk lipid extracts glycerol-based LSM was used to enhance droplet stability.
For the lactamase assay, an aqueous solution of AMP was prepared at a concentration of 50 μg/mL. The penicillinase was dissolved in 0.1 M tris buffer around pH 8 to a final concentration of 1800–3600 units/mL. Tris buffer or enzyme solution were respectively added to AMP and incubated in a block heater for 2 h at 35 °C.
Milk for lipid analysis was obtained from the dairy cow herd at the Centre for Dairy Research (CEDAR) at the University of Reading (UoR). A total of 100 milk samples were collected from 100 healthy cows through a Dairymaster 50 rotary parlor (Dairymaster Ltd., Bromsgrove, U.K.). All milk samples were pooled and stored in 2 mL cryotubes at −80 °C.
Prior to the analysis, one cryotube of milk was defrosted at room temperature for 5 min and 50 μL of pooled milk was aliquoted in 1.5 mL tubes. The aliquots (50 μL) were mixed with 450 μL of hexane/isopropanol (3:2; v/v) and vortexed for 5 s. No centrifugation was required, and the supernatant (lipids fraction) was directly used as analyte solution for analysis.
Results and Discussion
Subsecond sample analyses require data acquisition with significantly faster scan rates in order to obtain an adequate number of sampling points. This is critical for accurate recording of each sample’s ion signal as well as the separation of individual subsequent samples in high-throughput applications. Thus, using the liquid AP-MALDI source, analyses were carried out in TOF mode without ion mobility measurement. The MS scan time was set between 0.1 and 0.03 s and the interscan delay time to its shortest value (0.01 s), which resulted in an actual interscan delay time of 50 ms due to data transfer restrictions and automated software-driven delay time adjustment.
Bradykinin was selected as a peptidic analyte standard for the proof of principle of high-throughput analysis using liquid AP-MALDI MS. A commercial 96-well MALDI sample plate (Waters) was manually spotted with a mixture of the LSM and analyte. Figure 1 shows the results that can be obtained by moving the plate at a speed of approximately 5 mm/s. At this speed 96 samples spotted in 8 rows were analyzed in approximately 76 s, that is, 1.3 samples per second. As the plate moved at a constant speed across each row, the MALDI samples were irradiated by the laser only for a fraction of the time. In addition, as the laser pulse repetition rate was not synchronized with the sample presentation rate, the laser randomly irradiated different areas of the sample droplets, arguably leading to some fluctuation in sample-to-sample ion signal intensities due to the dome shape of the sample droplets and the laser beam’s angle of incidence on the MALDI sample plate of approximately 60°. Additionally, the effect of deceleration and acceleration of the stage at the turning points caused some peak broadening at these positions. Yet, all sample spots yielded intense analyte ion signals with low sample-to-sample variation and subsequent samples were easily distinguished. It should be noted here that liquid MALDI can produce “ESI-like” multiply charged ions and therefore the base peak in the MALDI sample spectra is in all cases the doubly protonated bradykinin ion signal (see Figure 1b,c).
Figure 1.
(a) Liquid AP-MALDI extracted ion chromatogram (m/z 530.79) of 96 sample wells with bradykinin as analyte (1 μL total sample volume spotted, 25 pmol analyte on target) at 5 mm/s stage movement speed. (b) Mass spectrum of all scans acquired for the first sample. (c) Mass spectrum of all scans acquired for the last sample.
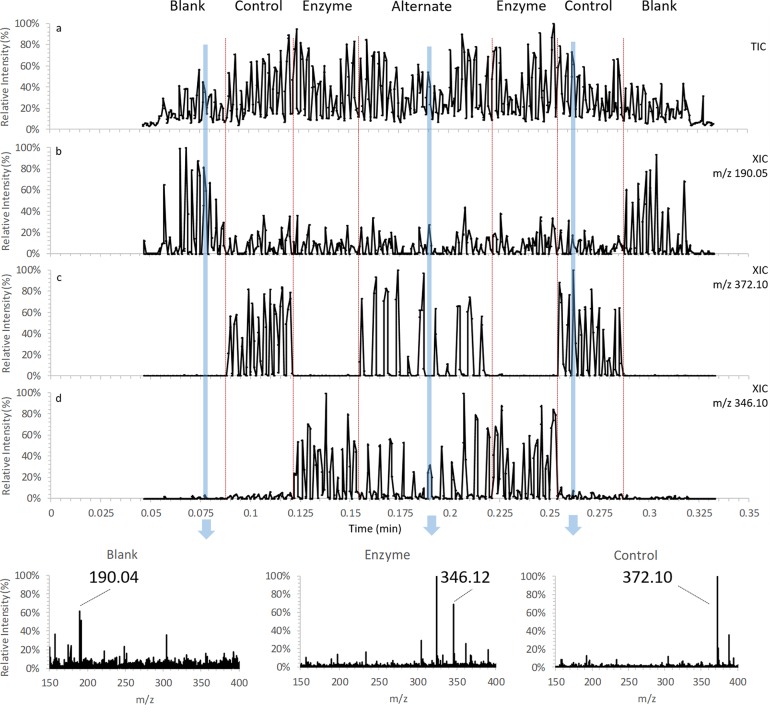
As the next step, a slightly faster stage for the MALDI plate was installed and a simple biochemical assay to detect lactamase activity was chosen to demonstrate the potential of liquid AP-MALDI MS for screening assays. The β-lactam antibiotic ampicillin was incubated for 2 h with penicillinase in the appropriate buffer as well as with buffer only (control). A target plate was then spotted with MALDI samples consisting of LSM with water as analyte solution (blank), LSM with ampicillin in buffer only (control), and LSM with lactamase-treated ampicillin in buffer. Because of the geometrical constraints of the setup with the new stage (due to the larger footprint of the new stage and space restrictions of the current setup), only 11 of the 12 columns could be used and a total of 88 MALDI samples were spotted and irradiated. Figure 2 displays the total ion chromatogram (TIC) and selected extracted ion chromatograms (XICs) obtained from analyzing all 88 samples with a stage movement speed of 26 mm/s.
Figure 2.
Liquid AP-MALDI MS ion chromatograms of 88 samples consisting of LSM with water only (“blank”), LSM with ampicillin incubated with penicillinase (64.1 amol on target; “enzyme”) and without penicillinase (“control”) acquired at 0.18 s/sample (26 mm/s) speed. Some of the “enzyme” and “control” samples were alternatingly spotted in the two middle rows of the sample plate (“alternate”). (a) Total ion chromatogram and extracted ion chromatogram of (b) protonated CHCA, (c) sodiated intact ampicillin, and (d) sodiated decarboxylated hydrolyzed ampicillin. (e) Mass spectra of all scans for each respective sample as indicated in the chromatograms (m/z 190.04 protonated CHCA, m/z 346.12 sodiated decarboxylated hydrolyzed ampicillin, m/z 372.10 sodiated intact ampicillin).
The XIC for CHCA (m/z 190.05; Figure 2b) shows 11 peaks at higher intensity for the spots where matrix was only spotted with water (blank). The sodiated ampicillin ion signal at m/z 372.1 was only detected when the MALDI samples with ampicillin but without lactamase were irradiated (Figure 2c). Strong ion signals at m/z 346 were only detected from the samples with the penicillinase-incubated ampicillin (Figure 2d). These ion signals can be assigned to the sodium adduct ions of the decarboxylated hydrolyzed ampicillin, a typical product of lactamase treatment. As seen in Figure 2, unambiguous identification of treated and untreated ampicillin samples can be made. Virtually no carryover between the samples was observed.
Importantly, the analytical speed demonstrated by the analysis of these assay samples is >5.5 samples per second, which is twice as fast as the latest published data for HTS analysis of large sample sets by mass spectrometry.9,18 However, the current maximum laser pulse repetition rate and the instrument’s scan and interscan delay times arguably result in some limitations with respect to the ion signal intensity and stability, explaining some of the ion signal fluctuations in Figure 2. At a laser pulse repetition rate of 30 Hz and a stage speed of 26 mm/s each sample will be irradiated by only three laser shots, since the sample diameter is ∼2.5 mm. Thus, as indicated earlier some samples will be irradiated with two laser shots hitting the laser beam-facing side of the dome-shaped sample droplet and one laser shot hitting the shadow side of the droplet while others will have the opposite irradiation pattern as there is no synchronization between laser pulse repetition rate and sample presentation rate. Thus, higher laser pulse repetition rates (1 kHz or more) and a reduction of the instrument’s scan and interscan delay times should allow for analytical speeds well beyond 10 samples per second with much-improved ion signal abundance and stability.
With the above-mentioned restrictions of the faster stage and speed, 88 samples of bradykinin were also analyzed using this new stage. For further comparison, both liquid and solid AP-MALDI MS data were acquired. As expected, the liquid AP-MALDI MS data of this experiment (see Figure 3a) show a similar ion signal stability compared to the penicillinase assay data but a slightly worse ion signal stability compared to the earlier bradykinin data due to the lower number of laser shots (≤3 vs approximately 15) and scans per sample and their associated limitations on ion signal stability as discussed above. However, the comparison to the solid AP-MALDI MS data (see Figure 3) clearly shows that the predominant analyte ion signal in solid AP-MALDI MS is the singly, not doubly, charged protonated bradykinin ion (see Figure 3c–e), and more importantly, that the ion signal intensity and stability is significantly worse with many samples (scans), showing only poor or no analyte ion signal (see Figure 3b).
Figure 3.
(a) Liquid AP-MALDI extracted ion chromatogram of 88 sample wells (1 μL total sample volume spotted, 25 pmol analyte on target) at 26 mm/s stage movement speed for doubly charged bradykinin. (b) Solid AP-MALDI extracted ion chromatogram of 88 sample wells (1 μL total sample volume spotted, 25 pmol analyte on target) at 26 mm/s stage movement speed for singly charged bradykinin using DHB as matrix. (c) Mass spectrum obtained under the above high-throughput screening conditions from all (3) scans of one liquid AP-MALDI sample. (d) Mass spectrum obtained under the above high-throughput screening conditions from all (3) scans of one solid AP-MALDI sample prepared with DHB. (e) Mass spectrum obtained from one solid AP-MALDI sample prepared with CHCA but manually acquired over 100 scans (>100 laser shots). For solid AP-MALDI samples prepared with CHCA, bradykinin was not detected under the above automated high-throughput screening conditions using similar laser energies needed for DHB to produce bradykinin ion signals.
Finally, this method was applied to the analysis of heterogeneous biofluid. Bovine milk was collected at the University of Reading’s research farm at CEDAR, and after a short, one-pot sample preparation, samples were spotted as described before. To achieve a good signal-to-noise ratio for analyte detection, somewhat longer acquisition times, that is, approximately 1 s per sample, were chosen with the stage moving fast from sample to sample while spending more time on each sample.
For all sample spots, mass spectra with good signal-to-noise ratios were obtained (see Figure 4), highlighting the suitability of liquid AP-MALDI for the fast analysis of complex biological samples. As the stage resides longer on each sample and more importantly on a specific similar spot on each sample, the ion signal for each spot becomes more rectangularly shaped, again indicating that similarly well-defined ion signals should be obtained with higher laser pulse repetition rates at faster stage movement (and faster scan times and less interscan delay). For selected peaks MS/MS data were acquired and characteristic fragment peaks at m/z 184.07 were found, indicating the loss of the headgroup of glycerophospholipids. Similar peaks for bovine milk were reported earlier.28
Figure 4.
Liquid AP-MALDI MS analysis of 88 sample wells of bovine milk at 1.0 s/sample. Top panel: TIC with an enlargement of a part of the TIC as inset. Bottom panel: a representative mass spectrum for one sample (13 scans). Labeled peaks can be assigned to glycerophospholipids.
Conclusion
The data presented demonstrate that liquid AP-MALDI is suitable for high-throughput sample screening with a speed of greater than 5 samples per second for the analysis of peptides, lipids, and antibiotics. A simple biochemical assay was conducted and a clear distinction between converted and nonconverted substrate for each sample was achieved. The technique was successfully applied to complex raw milk extracts, although a somewhat lower analysis speed was chosen to obtain quality spectra with a multitude of lipid species. The sample throughput obtained with liquid AP-MALDI is higher compared to those recently reported for the RapifleX and RapidFire techniques. For the more complex milk samples, currently lower speeds are applied, which are still highly competitive with ESI-based systems.
Further advancement toward higher throughput can be achieved using MALDI sample plate formats with smaller volumes and sample spacing. Preliminary experiments with 384-well plates using 0.4 μL sample volume have shown the importance of spotting reproducibility and the use of higher laser pulse repetition rates. For the 96-well plate (2.5 mm sample diameter) in this study, up to three laser pulses are used for one sample at 30 Hz laser pulse repetition rate and 26 mm/s sample movement speed. Thus, TIC variations may be due to only two laser shots hitting the sample and different laser fluences caused by the angle of the sample droplet. For lower volume plates, these effects will become even more pronounced. Hence, higher laser pulse repetition rates combined with less interscan delay and faster scan times should further enhance performance of the technique, especially regarding reproducibility.
Acknowledgments
This research was conducted as part of a studentship funded by Waters Corporation and the University of Reading.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
References
- Haslam C.; Hellicar J.; Dunn A.; Fuetterer A.; Hardy N.; Marshall P.; Paape R.; Pemberton M.; Resemannand A.; Leveridge M. The Evolution of MALDI-TOF Mass Spectrometry toward Ultra-High-Throughput Screening: 1536-Well Format and Beyond. J. Biomol. Screening 2016, 21 (2), 176–186. 10.1177/1087057115608605. [DOI] [PubMed] [Google Scholar]
- Beeman K.; Baumgärtner J.; Laubenheimer M.; Hergesell K.; Hoffmann M.; Pehl U.; Fischer F.; Pieck J.-C. Integration of an In Situ MALDI-Based High-Throughput Screening Process: A Case Study with Receptor Tyrosine Kinase c-MET. SLAS Discovery 2017, 22 (10), 1203–1210. 10.1177/2472555217727701. [DOI] [PubMed] [Google Scholar]
- Sun S.; Kennedy R. T. Droplet Electrospray Ionization Mass Spectrometry for High Throughput Screening for Enzyme Inhibitors. Anal. Chem. 2014, 86 (18), 9309–9314. 10.1021/ac502542z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roddy T. P.; Horvath C. R.; Stout S. J.; Kenney K. L.; Ho P.-I.; Zhang J.-H.; Vickers C.; Kaushik V.; Hubbard B.; Wang Y. K. Mass Spectrometric Techniques for Label-free High-Throughput Screening in Drug Discovery. Anal. Chem. 2007, 79 (21), 8207–8213. 10.1021/ac062421q. [DOI] [PubMed] [Google Scholar]
- Haarhoff Z.; Wagner A.; Picard P.; Drexler D. M.; Zvyaga T.; Shou W. Coupling Laser Diode Thermal Desorption with Acoustic Sample Deposition to Improve Throughput of Mass Spectrometry–Based Screening. J. Biomol. Screening 2016, 21 (2), 165–175. 10.1177/1087057115607184. [DOI] [PubMed] [Google Scholar]
- Fang X.; Zheng Y.; Duan Y.; Liu Y.; Zhong W. Recent Advances in Design of Fluorescence-Based Assays for High-Throughput Screening. Anal. Chem. 2019, 91 (1), 482–504. 10.1021/acs.analchem.8b05303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese J.; Johnson R. L.; Simeonov A.; Xia M.; Zheng W.; Austin C. P.; Auld D. S. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 2007, 3 (8), 466–79. 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- Covey T.; Kovarik P.; Liu C.. High Speed System for Analysis of Biological Samples that Corrects for ESI Ionization Suppression in Real Time. In Poster at 67th ASMS Conference; ASMS, 2019.
- Winter M.; Ries R.; Kleiner C.; Bischoff D.; Luippold A. H.; Bretschneider T.; Büttner F. H. Automated MALDI Target Preparation Concept: Providing Ultra-High-Throughput Mass Spectrometry–Based Screening for Drug Discovery. SLAS Technol. 2019, 24 (2), 209–221. 10.1177/2472630318791981. [DOI] [PubMed] [Google Scholar]
- Kempa E. E.; Hollywood K. A.; Smith C. A.; Barran P. E. High throughput screening of complex biological samples with mass spectrometry – from bulk measurements to single cell analysis. Analyst 2019, 144 (3), 872–891. 10.1039/C8AN01448E. [DOI] [PubMed] [Google Scholar]
- Wang C.; Wang M.; Han X. Applications of mass spectrometry for cellular lipid analysis. Mol. BioSyst. 2015, 11 (3), 698–713. 10.1039/C4MB00586D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth W. F.; McClean S.; Massaro C. F.; Smyth T. J.; Brooks P.; Robledo V. R. Characterization of Synthetic and Natural Product Pharmaceuticals by Functional Group Analysis using Electrospray Ionization-Ion Trap Mass Spectrometry: A Mini-Review. Anal. Lett. 2015, 48 (17), 2661–2675. 10.1080/00032719.2015.1045590. [DOI] [Google Scholar]
- Hilton G. R.; Benesch J. L. P. Two decades of studying non-covalent biomolecular assemblies by means of electrospray ionization mass spectrometry. J. R. Soc., Interface 2012, 9 (70), 801–816. 10.1098/rsif.2011.0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltashov I. A.; Bobst C. E.; Abzalimov R. R.; Wang G.; Baykal B.; Wang S. Advances and challenges in analytical characterization of biotechnology products: Mass spectrometry-based approaches to study properties and behavior of protein therapeutics. Biotechnol. Adv. 2012, 30 (1), 210–222. 10.1016/j.biotechadv.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladwig P. M.; Barnidge D. R.; Willrich M. A. V. Mass Spectrometry Approaches for Identification and Quantitation of Therapeutic Monoclonal Antibodies in the Clinical Laboratory. Clin. Vaccine Immunol. 2017, 24 (5), e00545–16. 10.1128/CVI.00545-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehender H.; Mayr L. M. Application of mass spectrometry technologies for the discovery of low-molecular weight modulators of enzymes and protein–protein interactions. Curr. Opin. Chem. Biol. 2007, 11 (5), 511–517. 10.1016/j.cbpa.2007.08.031. [DOI] [PubMed] [Google Scholar]
- Bretschneider T.; Ozbal C.; Holstein M.; Winter M.; Buettner F. H.; Thamm S.; Bischoff D.; Luippold A. H. RapidFire BLAZE-Mode Is Boosting ESI-MS Toward High-Throughput-Screening. SLAS Technol. 2019, 24 (4), 386–393. 10.1177/2472630318822449. [DOI] [PubMed] [Google Scholar]
- Sinclair I.; Bachman M.; Addison D.; Rohman M.; Murray D. C.; Davies G.; Mouchet E.; Tonge M. E.; Stearns R. G.; Ghislain L.; Datwani S. S.; Majlof L.; Hall E.; Jones G. R.; Hoyes E.; Olechno J.; Ellson R. N.; Barran P. E.; Pringle S. D.; Morris M. R.; Wingfield J. Acoustic Mist Ionization Platform for Direct and Contactless Ultrahigh-Throughput Mass Spectrometry Analysis of Liquid Samples. Anal. Chem. 2019, 91 (6), 3790–3794. 10.1021/acs.analchem.9b00142. [DOI] [PubMed] [Google Scholar]
- Wleklinski M.; Loren B. P.; Ferreira C. R.; Jaman Z.; Avramova L.; Sobreira T. J. P.; Thompson D. H.; Cooks R. G. High throughput reaction screening using desorption electrospray ionization mass spectrometry. Chem. Sci. 2018, 9 (6), 1647–1653. 10.1039/C7SC04606E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J.; Haslam C.; Hardy N.; Leveridge M.; Marshall P. A Systematic Investigation of the Best Buffers for Use in Screening by MALDI–Mass Spectrometry. SLAS Discovery 2017, 22 (10), 1262–1269. 10.1177/1087057116681726. [DOI] [PubMed] [Google Scholar]
- Hale O. J.; Cramer R. Collision-induced dissociation of doubly-charged barium-cationized lipids generated from liquid samples by atmospheric pressure matrix-assisted laser desorption/ionization provides structurally diagnostic product ions. Anal. Bioanal. Chem. 2018, 410 (5), 1435–1444. 10.1007/s00216-017-0788-6. [DOI] [PubMed] [Google Scholar]
- Ryumin P.; Brown J.; Morris M.; Cramer R. Protein identification using a nanoUHPLC-AP-MALDI MS/MS workflow with CID of multiply charged proteolytic peptides. Int. J. Mass Spectrom. 2017, 416, 20–28. 10.1016/j.ijms.2016.12.006. [DOI] [Google Scholar]
- Ryumin P.; Cramer R. The composition of liquid atmospheric pressure matrix-assisted laser desorption/ionization matrices and its effect on ionization in mass spectrometry. Anal. Chim. Acta 2018, 1013, 43–53. 10.1016/j.aca.2018.01.070. [DOI] [PubMed] [Google Scholar]
- Towers M.; Cramer R. Liquid matrices for analyses by UV-MALDI mass spectrometry. Spectroscopy 2007, 22 (11), 29–32. [Google Scholar]
- Hale O. J.; Morris M.; Jones B.; Reynolds C. K.; Cramer R. Liquid Atmospheric Pressure Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Adds Enhanced Functionalities to MALDI MS Profiling for Disease Diagnostics. ACS Omega 2019, 4 (7), 12759–12765. 10.1021/acsomega.9b01476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer R.; Pirkl A.; Hillenkamp F.; Dreisewerd K. Liquid AP-UV-MALDI Enables Stable Ion Yields of Multiply Charged Peptide and Protein Ions for Sensitive Analysis by Mass Spectrometry. Angew. Chem., Int. Ed. 2013, 52 (8), 2364–2367. 10.1002/anie.201208628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryumin P.; Brown J.; Morris M.; Cramer R. Investigation and optimization of parameters affecting the multiply charged ion yield in AP-MALDI MS. Methods 2016, 104, 11–20. 10.1016/j.ymeth.2016.01.015. [DOI] [PubMed] [Google Scholar]
- Calvano C. D.; De Ceglie C.; Aresta A.; Facchini L. A.; Zambonin C. G. MALDI-TOF mass spectrometric determination of intact phospholipids as markers of illegal bovine milk adulteration of high-quality milk. Anal. Bioanal. Chem. 2013, 405 (5), 1641–1649. 10.1007/s00216-012-6597-z. [DOI] [PubMed] [Google Scholar]