Abstract
In experimental autoimmune encephalomyelitis (EAE), a model for multiple sclerosis (MS), induction generates successive waves of clonally expanded CD4+, CD8+, and γ+ T cells in the blood and central nervous system, similar to gluten challenge studies of Celiac patients. In MS patients, we also observe major expansions of CD8+ T cells. In EAE, we find that most expanded CD4+ T cells are specific for the inducing myelin peptide MOG35–55 but in contrast, surrogate peptides derived from a yeast peptide-MHC display library for some of the clonally expanded CD8+ T cells inhibit disease by suppressing the proliferation of MOG-specific CD4+ T cells. These results suggest the induction of autoreactive CD4+ T cells triggers an opposing mobilization of regulatory CD8+ T cells.
Susceptibility to Multiple sclerosis (MS) and many other autoimmune diseases correlates strongly with specific major histocompatibility complex (MHC) class II alleles1–3, and CD4+ T cell involvement in MS and experimental autoimmune encephalomyelitis (EAE) is well established1,4,5. Additionally, the presence of CD8+ T and γδ+ T cells in EAE and MS brain lesions has also been described, but their role in the disease, if any, is not understood6–8. Previously, we have shown that celiac patients exposed to gluten mobilize not only gluten-specific CD4+ T cells in the blood, as expected, but also gut homing CD8+ and γδ+ T cells as well9.
Here, we asked whether the coordinated T cell response that we saw in the Celiac study9 might also occur in EAE, and found that it does, both in the blood and in the central nervous system (CNS). While the expanded CD4+ T cells are largely specific for the MOG35–55 peptide, clonally expanded CD8+ T cells were non-responsive to myelin peptides or proteins. To identify the target antigens, we screened six CD8+ TCRs on a class I MHC molecule H2-Db yeast p-MHC display library10,11 and obtained surrogate peptides (SP) for two of these TCRs and these SP greatly reduced severity. Further analyses show that these T cells represent a unique subset of regulatory CD8+ T cells which suppress MOG35–55 specific CD4+ T cell proliferation, suggesting that the induction of autoreactive CD4+ T cells in EAE triggers a counteracting wave of regulatory CD8+ T cells. In newly diagnosed MS patients, TCR analysis showed modest CD4+ clonal expansions, very pronounced CD8+ T cell clonal expansions, and similar skewing towards an IL-17 phenotype in the γδ+ T cells. Most importantly, given the similarities described above in the dynamics of T cell responses in celiac disease, EAE, and MS, it seems likely that pathogenic CD4+ and γδ+ Τ cell responses opposed by regulatory CD8+ T cell responses may be a common phenomenon across autoimmune diseases.
Results
Mobilization of three distinct T cell types following EAE induction
Here, we performed a broad survey of T cell dynamics post EAE induction5 (Fig. 1a and Extended Data Fig. 1a). We observed a gradual and significant increase in the frequency of total CD4+ T cells in the blood from day (D)0-D10 post-immunization (PI), peaking around D10, declining to below baseline level at D15, with an eventual recovery on D17. A similar drop and recovery in the frequency of total CD4+ T cells was again observed on D19 and D21, respectively (Fig. 1b). We also observed a similar significant increase in the frequency of total CD8+ and γδ+ T cells in the blood at D10 PI, and the kinetics and the magnitude of response among these T cells precisely matched that of CD4+ T cells (Fig. 1b). This pattern of synchronous behavior was also observed in the CNS (Fig. 1c).
Fig. 1. Concomitant activation of all T cells following EAE immunization.
(a) C57BL/6J mice were immunized for EAE induction and at different days post-immunization (PI) blood, CNS, spleen, and draining lymph nodes (LN) cells were harvested and analyzed for total frequency of T cells. (b, c, d, and e) Total frequency of CD4+, CD8+, and γδ+ T cells in the blood, CNS, spleen, and in the LN different days PI [(D0 (Unimmunized) (n = 5), D3 (n = 4), D5 (n = 4), D7 (n = 4), D10 (n = 4), D12 (n = 4), D15 (n = 5), D17 (n = 5), D19 (n = 5), D21 (n = 5), D23 (n = 5), and D30 (n = 3))]. Significance of differences observed in changes in frequency of T cells between days was determined using one-way ANOVA followed by Dunnets post hoc multiple comparison test. Data are shown as mean ± SEM. Representative data from two independent experiments. *p = 0.05; **p = 0.0097; ***p = 0.0008; ****p < 0.0001.
Splenic and lymph node (LN) T cells exhibited a different pattern, with a gradual decline in the frequency of total CD4+, CD8+, and γδ+ T cells from D0-D7, a rise in frequency until D17, and another dip between D17 and D30 (Fig. 1d and 1e). Parallel to this, there was also corresponding changes in the frequency of effector cells (Extended Data Fig. 2a, 2c, and 2e) and naïve T cells PI (Extended Data Fig. 2b, 2d, and 2f).
CD4+, CD8+, and γδ+ T cells clonally expand following EAE induction
To determine whether these waves of T cells constitute a focused immune response, we performed single-cell paired TCR sequencing9,12 (Fig. 2a) of effector T cells (Fig. 2b and Supplementary Table. 1). All three types of T cells showed increased clonal expansion starting at D7 (Fig. 2c and Extended Data Fig. 1b-e). Among γδ+ T cells, we found that nearly all the clonally expanded and some non-clonal γδ+ T cells in the blood and CNS are enriched for natural γδ+ 17 T cells (nTγδ17)13 TCRs (Extended Data Fig. 1f and 1g).
Fig. 2. CD4+, CD8+, and γδ+ T cells are clonally expanded following EAE.
(a) C57BL/6J mice were immunized for EAE induction and on different days PI [(D0 (unimmunized), D7, D10, D15, and D19)] blood and CNS infiltrating CD4+, CD8+, and γδ+ T cells were single cell sorted based on (b) activation markers (CD44hiCD62Llow) and their TCRs sequenced. Pie chart depicting clonal expansion of CD4+, CD8+, and γδ+ T cells different days PI in (c) blood and (d) CNS. Each pie chart is an aggregate of the number of TCR sequences from 3 individual mice pooled together per time point per tissue. The number of cells with β or both γ and δ chains successfully identified is shown above its pie chart. For each TCR clone expressed by two or more cells (clonally expanded), the absolute number of cells expressing that clone is shown with a distinct colored section. Sequencing data is from one experiment constituting 3 individual animal per time point.
Clonally expanded CD8+ T cells are not responsive to myelin antigens
To determine whether the expanded CD4+ T cell clones are MOG responsive, we expressed four of the these CD4+ TCRs (Supplementary Table. 2) in SKWαβ−/− cells and stained them with a MOG35–55 I-Ab peptide-MHC tetramer, which showed robust tetramer staining (Fig. 3a-d), reinforcing the primacy of MOG specific CD4+ T cells in this disease14–17 and validating our strategy of sequencing activated/effector cells as a way to enrich for clonally expanded T cells important in an immune response.
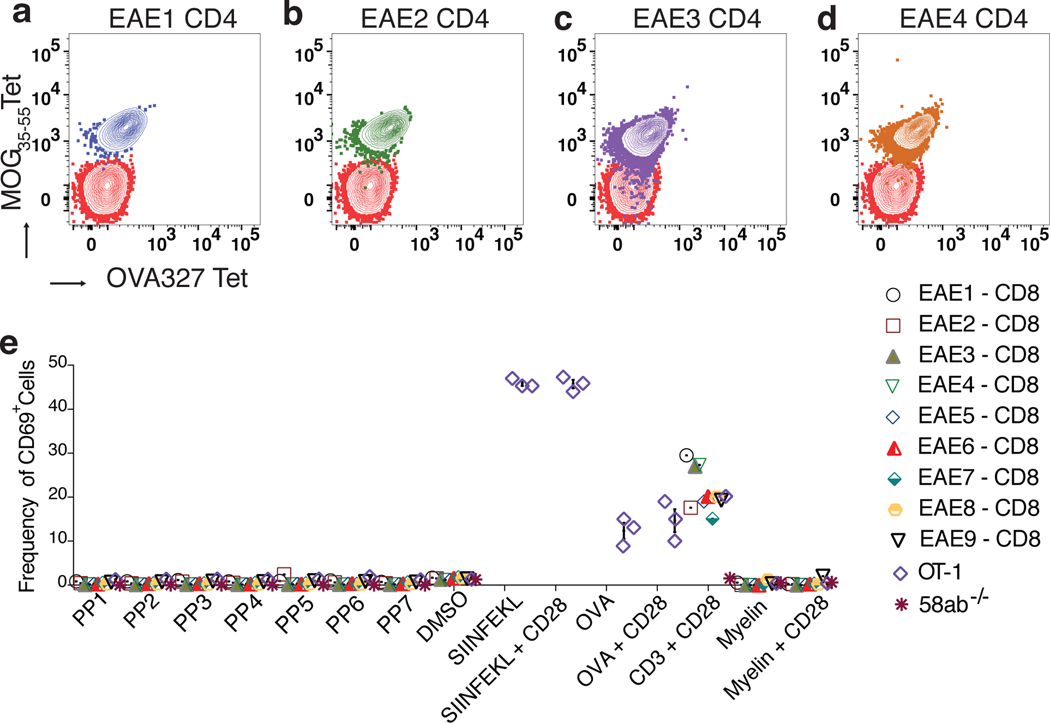
Fig. 3. Clonally expanded CD8 TCRs are not responsive to myelin.
(a-d) 4 clonally expanded CD4 TCRs (EAE1-CD4 to EAE4-CD4) were expressed on SKWαβ−/− cells and stained with MOG35–55 and OVA327 I-Ab pMHC tetramer. 9 clonally expanded CD8 TCRs (EAE1-CD8 to EAE9-CD8) were expressed on 58αβ−/− cells. (e) Cells were stimulated with pools of myelin peptides from myelin oligodendrocyte glycoprotein (MOG), myelin basic protein (MBP), proteolipid protein (PLP), myelin associated glycoprotein (MAG), SIINFEKL, myelin or ovalbumin (OVA) protein, and anti-CD3 + anti-CD28 for 12–16 hr, and examined for activation marker CD69. Each peptide pool (PP)1 – PP7 comprised of variable length peptides (8–12MERs), and each PP contained 50 peptides. (e) 58αβ−/− cells expressing OT-1 TCR were stimulated with either SIINFEKL peptide or OVA protein. Representative data from three independent experiments.
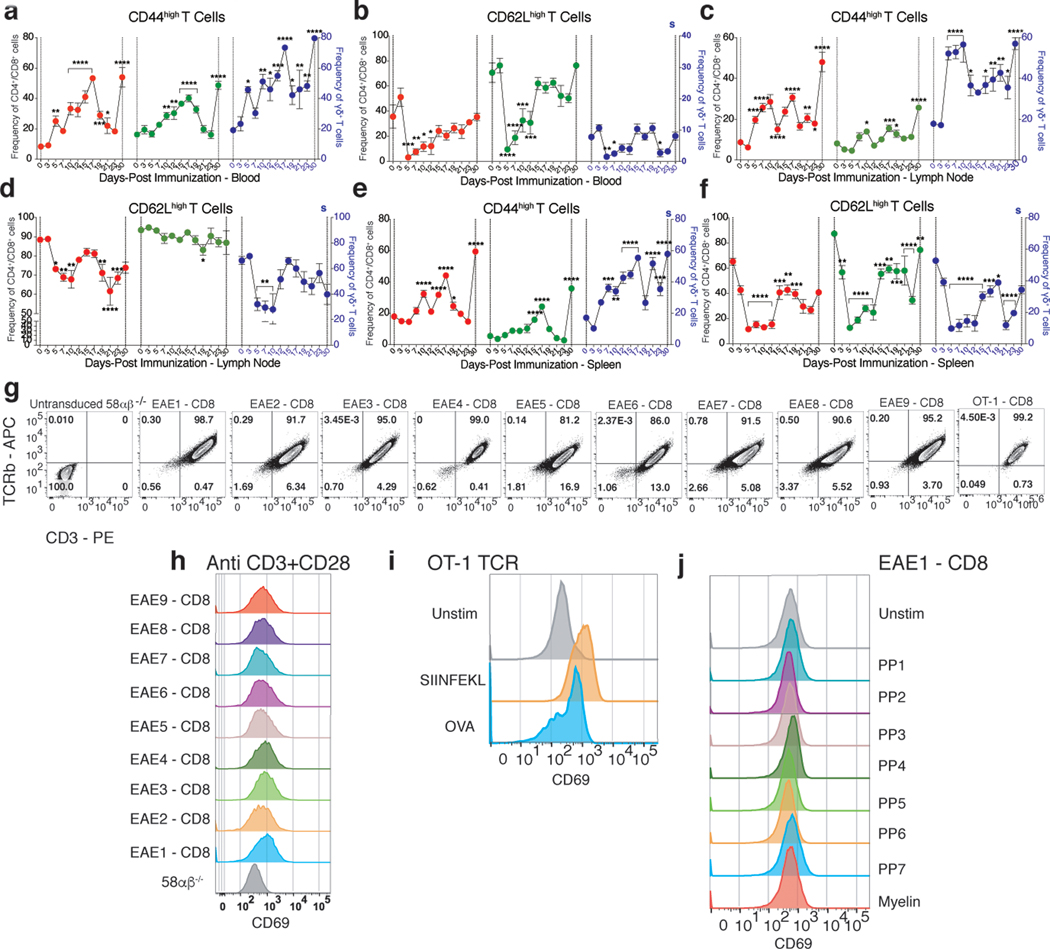
To investigate the antigen(s) specificity of the CD8+ T cells in EAE, 58αβ−/− cells expressing nine of the clonally expanded and common CD8+ TCRs (EAE1-CD8 to EAE9-CD8 (Extended Data Fig. 2g and Supplementary Table. 3) were co-cultured with bone marrow derived dendritic cells (BMDCs) pulsed with myelin protein derived peptide (Supplementary Table. 4). Surprisingly, none of the 350-myelin peptides stimulated any of the CD8+ TCR cell lines (Fig. 3e). Cells expressing ovalbumin specific TCR (OT-1) stimulated with the SIINFEKL peptide, gave a robust activation (Fig. 3e and Extended Data Fig. 2i) as well as anti-CD3 + anti-CD28 stimulation (Extended Data Fig. 2h). The myelin proteins also did not stimulate any of the CD8+ TCR cell lines, whereas OT-1 T cells were robustly stimulated with ovalbumin protein (Fig. 3e and Extended Data Fig. 2i and 2j) suggesting that very few of the activated and clonally expanded CD8+ T cells during EAE are specific for myelin antigens.
Generation of a H2-Db yeast peptide-MHC library
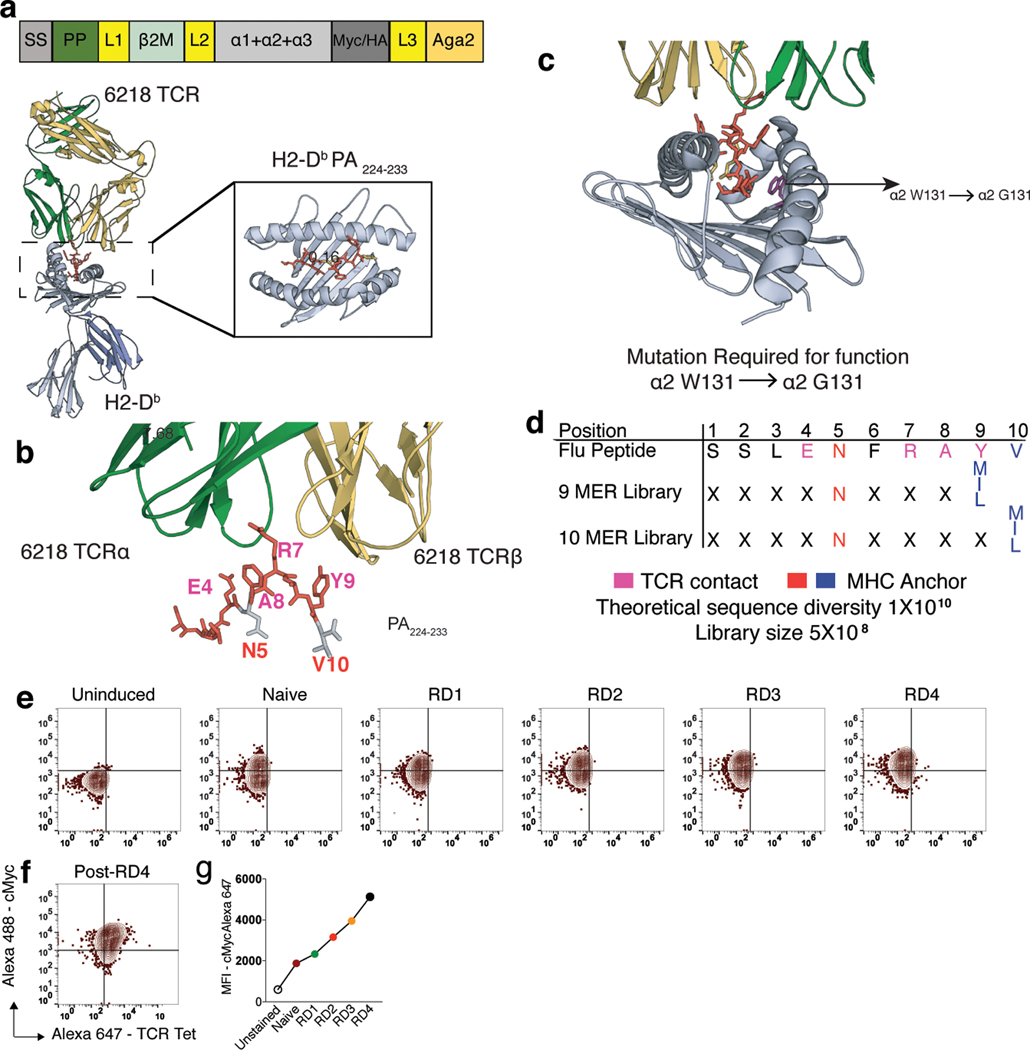
To discover the peptide antigens for these EAE-CD8 TCRs, we designed yeast peptide-H2-Kb and H2-Db library constructs with SIINFEKL or SSLENFRAYV peptides10,11,18,19 (Extended Data Fig. 3a and 3b). Initial constructs did not bind to their cognate TCRs, indicating incorrect folding. To rescue proper folding, we subjected both the H2-Kb and H2-Db full length construct to error-prone mutagenesis10. While this was unsuccessful with H2-Kb, we found that a single H2-Db mutation restored TCR recognition (Extended Data Fig. 3c and 3e-g) and thus we generated two different peptide libraries with this mutation and mutagenized the 9 and 10 aa inserts with limited diversity at the MHC anchor positions (Extended Data Fig. 3a-d). The estimated diversity for both peptide libraries were 5X108.
Class I peptides Immunization protects mice from EAE
Six of the clonally expanded EAE-CD8 TCRs (Supplementary Table. 3) were used to screen the H2-Db yeast-pMHC libraries10,11. After four rounds of selection, the flu specific control (6218), EAE6, and EAE7 TCRs showed robust tetramer staining (Fig. 4a-c and Supplementary Table. 5). While we obtained a perfect match with the flu TCR and its known peptide (Fig. 4d), we did not find any matches in the mouse genome for the two EAE TCR peptides (Fig. 4e and 4f). Nevertheless, they can still serve as important surrogates when complexed with H2-Db.
Fig. 4. Clonally expanded EAE CD8 TCRs bind to novel peptides.
Tetramer staining of 9 MER and 10 MER H2-Db yeast-pMHC library with (a) 6218 TCR, (b) EAE6-CD8 TCR, and (c) EAE7-CD8 TCR at the end of 3rd round (RD) of selection. Heatmaps of amino acid preference by position for (d) 6218 (left) (e) EAE6-CD8 (center) and (f) EAE7-CD8 (right) TCRs after three RDs of selection. Sequences of top 7 peptides post RD3 selection for each TCRs shown below with its amino acid preference. MHC anchor residues are colored in red (P5, N) or blue (P9/10, M/I/L). Each TCR was screened on the yeast library once.
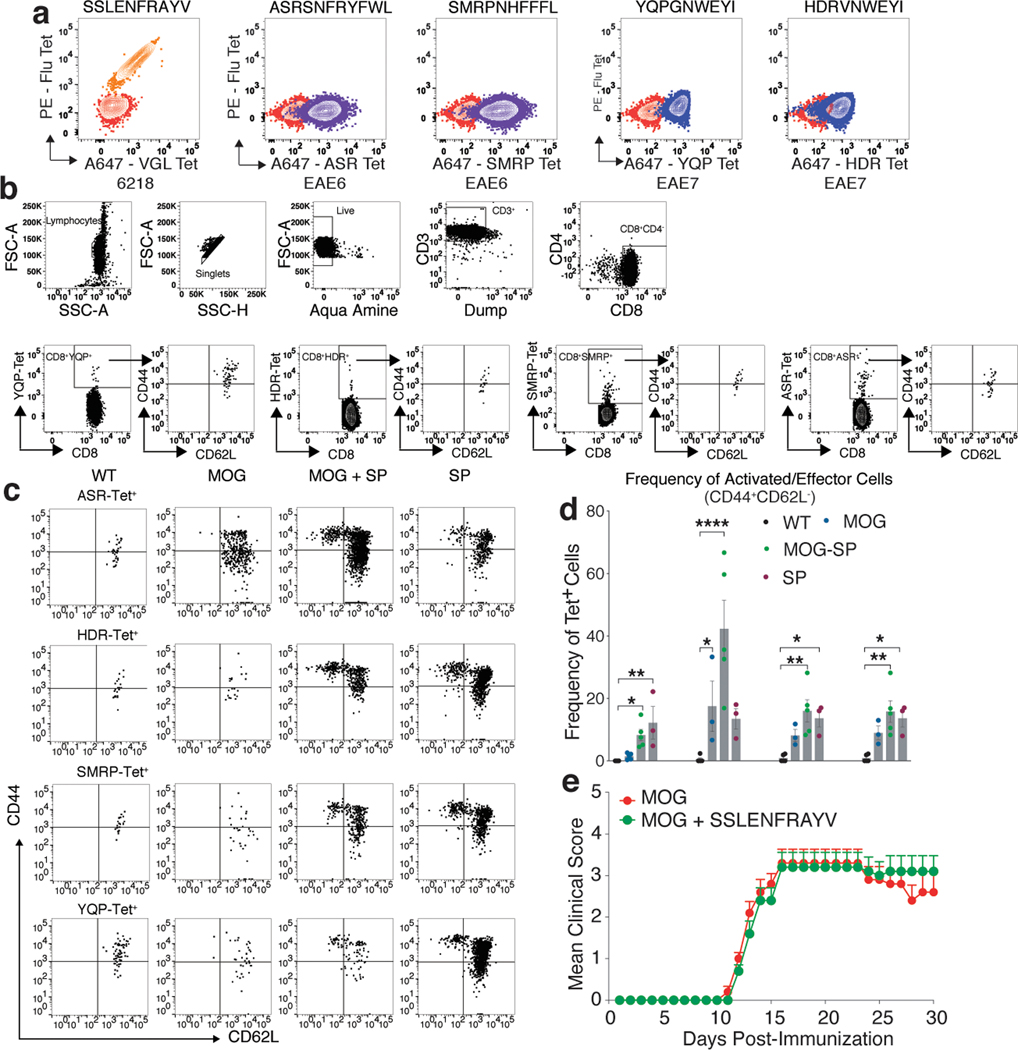
To characterize these CD8 SP, Jurkat αβ−/− cells expressing 6218, EAE6, and EAE7-CD8 TCRs were stained with corresponding yeast library enriched pMHC tetramers (SSLENFRAYV for 6218, ASRSNRYFWL and SMRPNHFFFL for EAE6-CD8, and YQPGNWEYI for EAE7-CD8), which showed robust staining (Extended Data Fig. 4a). Moreover, upon analyzing the enriched peptide sequences for EAE7-CD8 TCR, we noticed that the 36th enriched peptide (HDRVNWEYI) was very similar to the top enriched YQPGNWEYI peptide. EAE7-CD8 TCR cell line was stained with HDRVNWEYI H2-Db pMHC tetramers which showed robust tetramer staining (Extended Data Fig. 4a).
To determine the immune response elicited by these SP, we immunized mice either with adjuvant, MOG35–55, all four peptides together [YQPGNWEYI (YQP), HDRVNWEYI (HDR), ASRSNRYFWL (ASR), and SMRPNHFFFL (SMRP) – SP immunization], or MOG35–55 with all four peptides together (MOG + SP immunization). 10D PI, spleen and LN CD8+ T cells were enriched separately with SP-H2-Db tetramers (Extended Data Fig. 4b). We also tetramer enriched MOG specific CD4+ T cells from mice immunized with MOG35–55. EAE immunization elicited MOG specific CD4+ T cells20 (Fig. 5a and 5c). We detected very few SP specific CD8+ T cells in WT, MOG35–55, or adjuvant immunized mice. However, with MOG + SP immunization, the frequency of CD8+ T cells specific for ASR, HDR, and SMRP increased (Fig. 5b and 5d). In comparison to WT mice, SP immunization, elicited a higher frequency of CD8+ T cells specific for HDR and SMRP, with no change in the number of ASR and YQP specific CD8+ T cells (Fig. 5b and 5d) and these cells exhibited activated and effecter phenotypes after immunization (Extended Data Fig. 4b-d). Thus, these SP’s identify a pre-existing pool of specific CD8+ T cells in mice that can be activated, and some proliferate upon immunization.
Fig. 5. CD8+ T cell specific surrogate peptides immunization abrogates EAE.
From unimmunized and D10 PI mice, spleen and LN cells were harvested and enriched for (a) CD4+ T cells specific for I-Ab MOG35–55 or (b) CD8+ T cells specific for SP (ASRSNRYFWL, SMRPNHFFFL YQPGNWEYI, and HDRVNWEYI). Representative dot-plots are shown for unimmunized and immunized mice. Representative data from two independent experiments. (c) Frequency of CD4+ T cells specific for MOG35–55 among WT (n = 4) and immunized mice (n = 5) are shown as a bar graph. Data are shown as mean ± SEM. (d) Frequency of SP specific CD8+ T cells among WT (n = 4) and different immunization groups are shown as a bar graph (n = 5 mice per group). Data are shown as mean ± SEM. (e) EAE clinical scores among C57BL/6J mice immunized with an emulsion containing MOG35–55 + CFA + PTX (n =10), or with MOG35–55 + SP + CFA + PTX (n = 10), or SP + CFA + PTX (n =10). (f) C57BL/6J mice were immunized for EAE with an emulsion containing MOG35–55 + CFA + PTX (n =10) and seven days post-MOG35–55 immunization, mice were challenged with SP + ICFA + PTX (n = 10). (g) C57BL/6J mice were immunized for EAE with an emulsion containing SP + CFA + PTX and seven days post primary immunization, mice were challenged with MOG35–55 + ICFA + PTX (n =10). (e-g) The clinical scores following immunization were recorded, and the significance of differences between clinical courses was calculated by regression analysis with the two-way ANOVA followed by Bonferroni post hoc multiple comparison test. Data are shown as mean ± SEM. Representative data from two independent experiments. ** p = 0.0040, **** p < 0.0001.
To test the effect of these peptides on EAE, we induced EAE with or without the SPs. While the MOG immunization induced severe disease in 100% of the mice, the addition of the SP with MOG resulted in much less severe or no disease, with only a 30% incidence of very mild disease, with most mice exhibiting no symptoms at all (Fig. 5e and Supplementary Table. 6). Immunization of mice with MOG35–55 + a flu peptide resulted in no significant difference on EAE severity (Extended Data Fig. 4e). We tested the prophylactic or therapeutic effect of these peptides in EAE by immunizing the mice with SP a week prior or after MOG immunization (MOG and SP Challenge, respectively) and with both of these challenges, mice had less severe disease (Fig. 5f and 5g and Supplementary Table. 7). Overall, MOG + SP immunization significantly ameliorates EAE.
Class I peptides immunization suppresses MOG35–55 specific CD4 T cells
To investigate whether this decreased severity with the SP addition was directly affecting MOG specific CD4+ T cells, we analyzed the frequency of MOG35–55 specific CD4+ T cells in spleen and LN with a MOG35–55 I-Ab tetramer upon MOG or MOG + SP immunizations. As expected, MOG immunization resulted in an increase in the number of MOG35–55 CD4+ T cells (Fig. 6a and 6b and Extended Data Fig. 5a). Interestingly, there was a significant reduction in the frequency of these T cells among MOG + SP immunized mice (Fig. 6a and Extended data Fig. 5b and 5c). To examine this in vitro, we CellTrace™ Violet Dye (CTV) labelled CD4+ T cells from MOG immunized mice and that were co-cultured and stimulated with MOG35–55 in the presence or absence of CD8+ T cells derived from MOG + SP or SP immunized mice and the magnitude of CD4+ T cell proliferation was quantified. CD4+ T cells proliferated robustly to MOG35–55 in the absence of CD8+ T cells (Fig. 6c). However, the addition of CD8+ T cells from either MOG + SP or SP immunized mice resulted in a significant decline in the proliferative capacity of CD4+ T cells (Fig. 6d) indicating that either condition elicits CD8+ T cells which actively suppresses MOG35–55 specific CD4+ T cells proliferation. However, CD8+ T cells from either the WT, MOG + flu peptide immunized or from the adjuvant immunized mice did not suppress MOG35–55 stimulated CD4+ T cells (Extended data Fig. 6a-e). Furthermore, MOG + SP induced CD8+ T cells did not suppress the proliferative capacity of ovalbumin specific CD4+ T cells, suggesting that suppression is antigen specific (Extended data Fig. 6h).
Fig. 6. CD8+ T cell specific surrogate peptides immunization suppresses MOG35–55 specific CD4+ T cells.
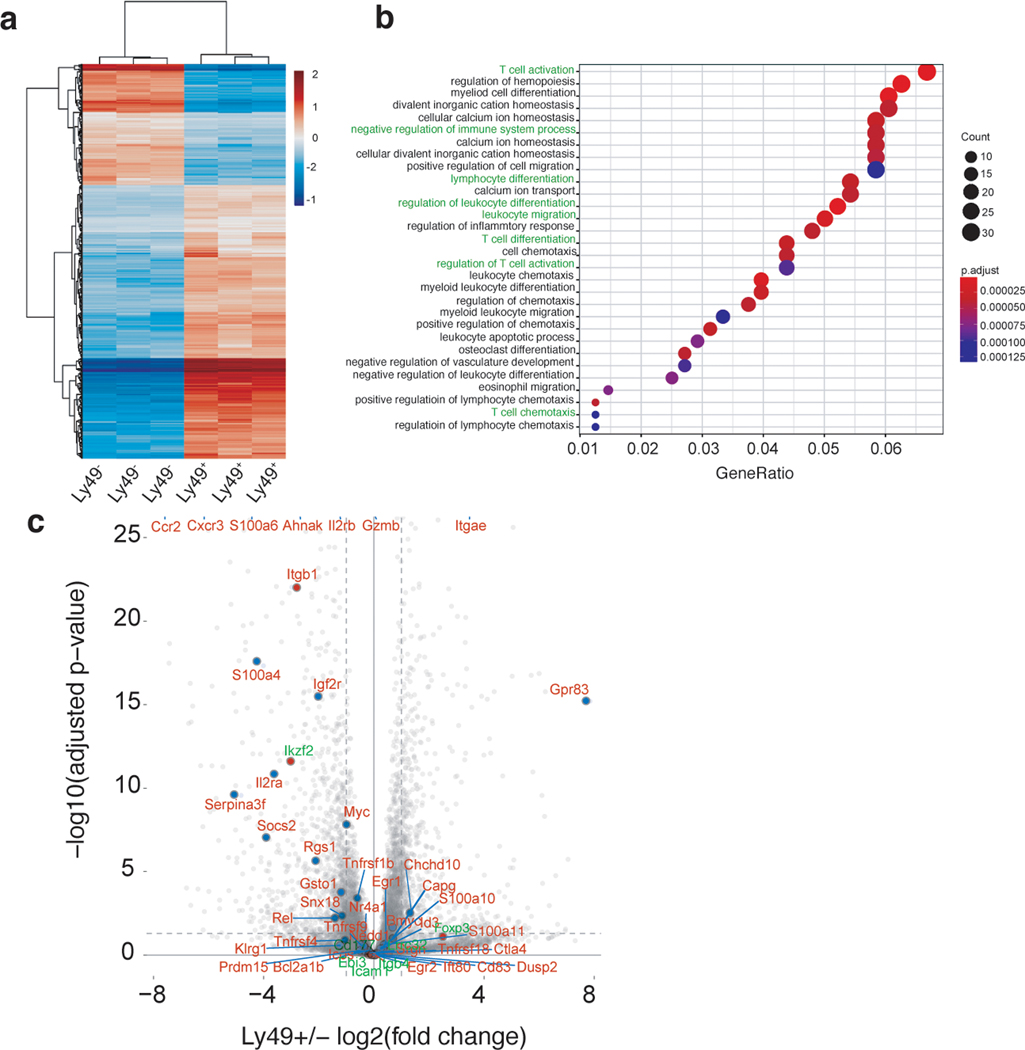
C57BL/6J mice were immunized with an emulsion containing MOG35–55 + CFA + PTX (n = 5), or MOG35–55 + SP + CFA + PTX (n = 4). From (a) unimmunized (n = 4) and D10 PI mice (n = 5 mice/group), LN and spleen cells were harvested and enriched for MOG35–55 I-Ab pMHC specific CD4+ T cells. FACS dot-plots from representative animal from different groups are shown (see extended data Fig. 8a-c for data from additional mice per group). (b) Frequency of MOG35–55 specific CD4+ T cells from each group are shown as a bar graph. Representative data from two independent experiments. Data are shown as mean ± SEM. C57BL/6J mice were immunized with MOG35–55 + CFA + PTX (n = 2 mice), or with MOG35–55 + SP + CFA + PTX (n = 2 mice), or SP + CFA + PTX (n = 2 mice). From unimmunized (n = 2) and D10 PI mice, LN and spleen cells were harvested, enriched for either CD4+, CD8+ T cells, or antigen presenting cells (APC) using Miltenyi cell enrichment kits followed by FACS. Dye labelled CD4+ T cells from MOG immunized mice were co-cultured with APC from MOG immunized mice either in the (c) absence or (d) presence of CD8+ T cells from MOG and MOG + SP or SP immunized mice. 7 days post co-culture, cells were analyzed for proliferation. Representative data from two independent experiments. (e) Frequency of CD8+ T cells with a regulatory phenotype (CD44+CD122+Ly49+) are shown among WT (n = 4) and different immunization groups (n = 5 mice/group). The significance of differences among the frequency of CD122+CD44+Ly49+ cells between different immunization group was determined using one-way ANOVA followed by Tukey’s post hoc multiple comparison test. Representative data from two independent experiments. Data are shown as mean ± SEM. *p = 0.0382; ***p = 0.001. CD4+ T cells from MOG immunized were co-cultured either (f) without or with (g) total CD8+ T cells or (h) purified CD8+CD44+CD122+Ly49+[Ly49+] or (i) CD8+CD44+CD122+Ly49−[Ly49−] T cells from MOG + SP immunized mice (n = 2 mice/group). Representative data from two independent experiments. (j) A heatmap of gene expression in RNA-seq samples. Genes were selected on the basis that they are differentially expressed (log2 (fold change) > 0.75 and the adjusted p value < 0.005) and the two-tailed Benjamini Hochberg adjusted p value < 0.005) as defined by DESeq2 in both Ly49+/Ly49− and MOG/MOG + SP comparisons. Columns show samples. Rows and columns are ordered based on hierarchical clustering. Normalized gene expression values are centered for each gene by subtracting the average value of all samples from each sample value. Representative data from two independent experiments.
Class I peptides immunization elicits a unique subset of regulatory CD8+ T cells
Interestingly, we find that MOG + SP and SP immunization elicit significantly higher frequency of CD8+ T cells and individual SP specific CD8+ T cells expressing CD44, CD122, and Ly49, as shown by Cantor and colleagues to be the markers for Qa-1b restricted regulatory CD8+ T cells21 (Fig. 6e and Extended Data Fig. 5d-h). We find that the EAE CD8-TCRs described here are not Qa-1b restricted, as we did not see any effect of the anti-Qa-1b antibody22 on the CD8 suppression of MOG specific CD4+ T cells (Extended Data Fig. 6g). To determine whether CD8+ T cells with this phenotype can actively suppress MOG35–55 specific CD4+ T cells in vitro, CD4+ T cells from MOG immunized were co-cultured either with total CD8+ T cells or purified CD8+CD44+CD122+Ly49+(Ly49+) or CD8+CD44+CD122+Ly49−(Ly49−) T cells from MOG + SP immunized mice. We found that total CD8+ T cells as well as Ly49+CD8+ T cells from MOG + SP immunized mice suppressed MOG35–55 specific CD4+ T cell proliferation (Fig. 6f-g, and 6i) whereas no suppression is observed with Ly49− T cells (Fig. 6f and 6h). Additionally, we adoptively transferred Ly49+ and Ly49− cells into mice prior to EAE induction23 and found Ly49+ cells from MOG + SP immunized mice significantly reduced EAE with no effect upon transferring Ly49− cells (Extended data Fig. 6i). This suggests that not only Ly49+ cells can suppress CD4+ T cells in vitro but that they also suppress EAE in vivo.
We also tested the effects of SP on another autoimmune disease model, Experimental autoimmune uveitis (EAU). We induced EAU in C57BL6/J24 and compared EAU pathology to mice immunized with human interphotoreceptor binding protein (IRBP) + SP. IRBP immunization mice produced a mild inflammatory response in 40% of the mice (Extended Data Fig. 6j and 6k). Interestingly, IRBP + SP immunization resulted in a more severe inflammatory response in a much greater fraction of the mice (80%) (Extended Data Fig. 6l). We also performed in vitro suppression assays in which we found that CD4+ T cells robustly proliferated in the absence of CD8+ T cells from IRBP + SP immunized mice (Extended Data Fig. 3m) and neither Ly49+ or the Ly49− CD8+ T cells from IRBP + SP mice were able to suppress the proliferation of IRBP specific CD4+ T cells (Extended Data Fig. 6n and 6o). Thus, the EAE SPs seem specific to that disease.
It has been shown that Qa-1b restricted regulatory cells mediate their effect through Perforin23. To test this possible mechanism in our system, we co-cultured CD4+ T cells from MOG35–55 immunized mice with CD8+ T cells derived from Perforin knockout mice immunized with MOG + SP and found that the this completely abolished their suppressive capacity (Extended Data Fig. 6f). Additionally, we performed RNA-seq analysis of SP specific CD8+ T cells from MOG and MOG + SP immunized mice as well as CD8+ (Ly49+ versus Ly49−) T cells from MOG + SP immunized mice. Gene ontology enrichment analysis of the differentially expressed genes between Ly49+ vs Ly49− showed genes involved in various T cell functions (Extended data Fig. 7a and 7b and Supplementary Table. 8). Interestingly, SP specific CD8+ T cells showed a marked upregulation of Ly49 genes, most of which are inhibitory, in addition to NK cell receptor genes, and genes associated with CD8+ T cell effector and memory functions (Fig. 6j). In addition, Ly49+ and SP specific CD8+ T cells also express many genes associated with regulatory CD4+ T cells25 (Extended Data Fig. 7c). Overall, our results strongly suggest that the SP immunization elicits CD8+ T cells with a regulatory phenotype which suppresses pathogenic MOG35–55 specific CD4+ T cells through cytotoxicity, ultimately resulting in resistance to EAE.
Parallels with Multiple Sclerosis
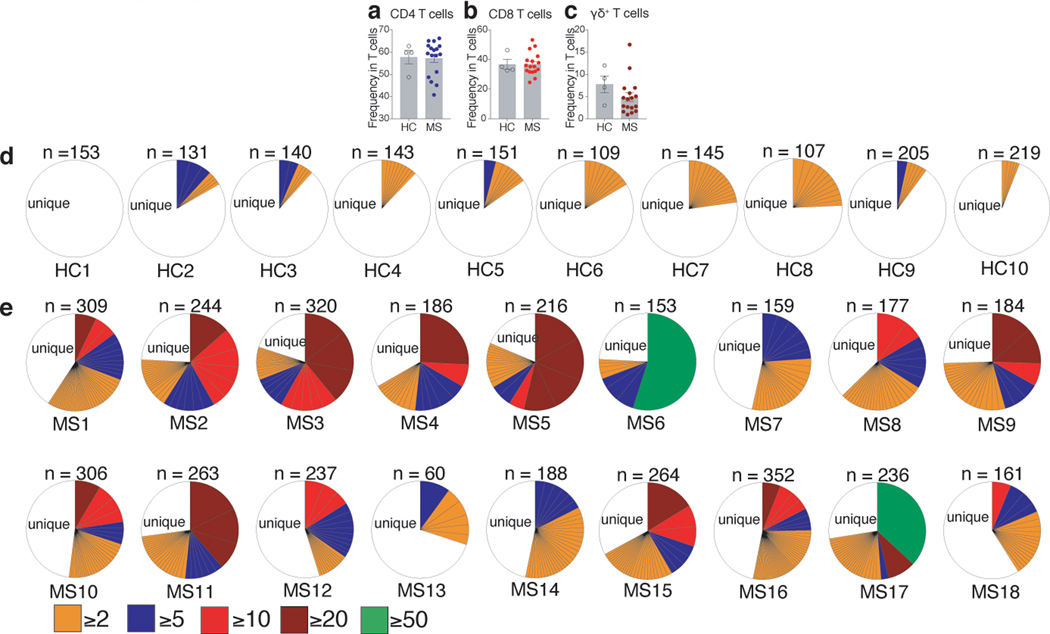
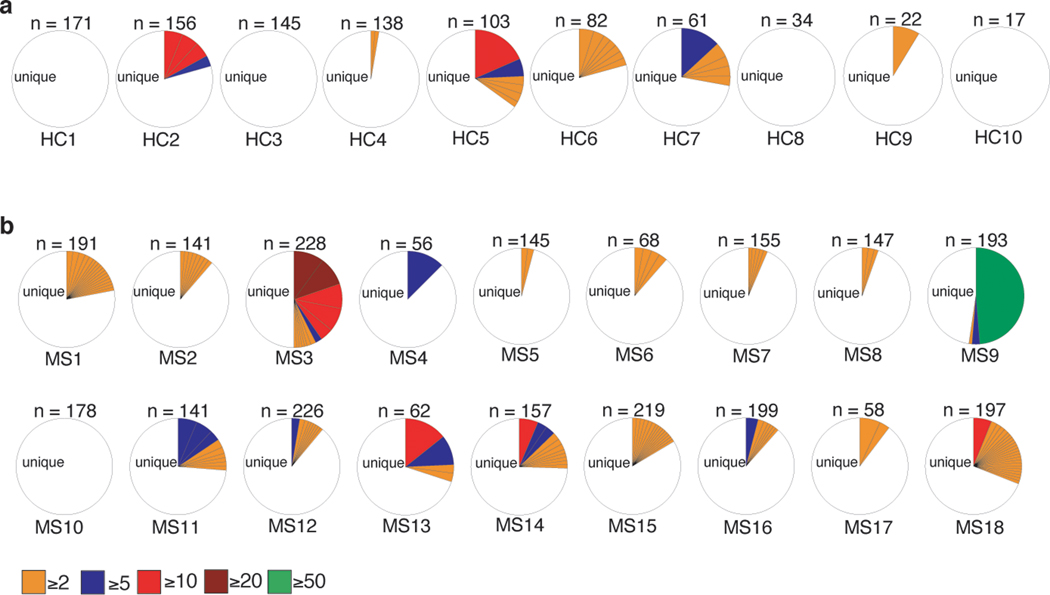
To determine are there any similarities with MS, we first determined the frequency of CD4+, CD8+, and γδ+ T cells directly in recently diagnosed MS patients and found no differences in the frequency of total T cells in the peripheral blood compared to healthy controls (HC) (Extended Data Fig. 8a-c). However, when we performed single cell TCR sequencing of activated brain homing (CD38+HLA-DR+CD49d+CD29+) CD4+, CD8+, and γδ+ T cells from PBMCs of newly diagnosed MS patients and HCs (Supplementary Table. 9), we observed a massive oligoclonal expansion of CD8+ T cells in MS patients compared to HCs (Extended Data Fig. 8d and 8e), which is similar to that observed in EAE and celiac disease9. We also observed only a few oligoclonal expansions of CD4+ T cells in MS patients (Extended Data Fig. 9a and 9b).
Additionally, we find that γδ+ T cells are clonally expanded in MS patients and also in HCs (Extended data Fig. 10a and 10b). The oligoclonal expansions of γδ+ T cells in MS has been noted before26,27,28. Unlike mouse nTγδ17 cells, the differentiation of human γδ17 T cells is poorly understood and it has been shown that in vitro activation of Vγ9+δ2+ T cells induces ROR-related orphan receptor gamma (RORγ) expression and IL-17 production29,30. In fact, we detected a significant increase in the frequency of RORC transcript positive γδ+ T cells in MS patients (Extended data Fig. 10c). Therefore, it is possible that in chronic autoinflammatory setting like MS some of these expanded γδ+ T cells potentially produce IL-17 and contribute to disease pathogenesis as like EAE.
Discussion
We show here that the simultaneous mobilization of oligoclonal T cells, seen earlier in celiac patients9, has a parallel not only in EAE, but also to some extent in newly diagnosed MS patients. When we systematically characterized each of these cell types, we found two of the three are likely pathogenic: specifically, the γδ+ T cells that dominate the response are well known to be producers of a pro-inflammatory cytokine, IL-1713,31,32,33, while the CD4+ T cell response is predominantly MOG-specific14,15,17, a key driver of EAE pathology4. In contrast to these two cell types, the clonally expanded CD8+ T cells exhibit a regulatory function. These T cells have a distinct phenotype and extend observations centered on Qa-1b restricted regulatory CD8+ T cells21,23,34–37,36,43,44 to include peptides presented by the classical class I MHC molecule H2-Db.
More importantly, our study shows the value of studying T cell specificity and activity from “the ground up”, that is, identifying the T cells that are most active in a given response by single cell paired TCR sequencing, using both activation markers and clonal expansion as key indicators, and ligand identification either with a yeast display library or candidate antigens and reporter cells transfected with the relevant TCR pairs14. This is in contrast to traditional methods38 which typically involve knowing (or guessing) what the relevant antigens are.
In summary, the work presented here indicates that there is a subset of CD8+ T cells that can suppress pathogenic CD4+ T cells in mice and likely in humans, and that this gives rise to the dynamic co-mobilization of T cells upon disease induction that we see in both Celiac disease and EAE. Determining the ligands for these regulatory CD8+ T cells in autoimmune diseases could thus be of significant value therapeutically.
Methods
Laboratory animals
Female C57BL/6J mice (referred to as either B6 or WT) and female Perforin knockout mice (Stock No: 002407) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). The experimental procedures used in this study were approved by the Animal Care and Use Committee of the Stanford University.
Human Samples
Peripheral blood mononuclear cells (PBMCs) were obtained from healthy blood donations from Stanford Blood Center. Healthy human subjects were male and female, ages 22–47 yrs. PBMCs from Multiple sclerosis patients were obtained from the Multiple Sclerosis Center at the University of California, San Francisco (UCSF). The committee on Human Research at UCSF approved the protocol, and informed consent was obtained from all participants. Detailed information on the patient population included in the study is provided as Supplementary Table. 7.
Generation of soluble TCRs
Soluble TCRs were generated as previously described10. TCR variable mouse-constant human (VmCh) chimeras containing an engineered C domain disulfide were cloned into the pAcGP67a insect expression vector (BD Biosciences, 554756) encoding either a C-terminal acidic GCN4-zipper-Biotin acceptor peptide (BAP)-6xHis tag (for α chain) or a C-terminal basic GCN4 zipper-6xHis tag (for β chain)39. Each chain also encoded a 3C protease site between the C terminus of the TCR ectodomains and the GCN4 zippers to allow for cleavage of zippers. Baculoviruses for each TCR construct were created in SF9 cells via co-transfection of BD Baculogold linearized baculovirus DNA (BD Biosciences, 554739) with Cellfectin II (Life Technologies, 10362–100). TCRα and β chain viruses were coinfected in a small volume (2 ml) of High Five cells in various ratios to find a ratio to ensure 1:1 α:β stoichiometry. To prepare soluble TCRs, 1L of High Five cells were infected with the appropriate ratio of TCRα and TCRβ viruses for 48 hr at 28C. Collected culture media were conditioned with 100 mM Tris-HCl (pH 8.0), 1 mM NiCl2, 5 mM CaCl2, and the subsequent precipitation was cleared via centrifugation. The media were then incubated with Ni-NTA resin (QIAGEN 30250) at room temperature for 3 hr and eluted in 1×HBS+200 mM imidazole (pH 7.2). TCRs were then site-specifically biotinylated by adding recombinant BirA ligase, 100 μM biotin, 50 mM Bicine pH 8.3, 10 mM ATP, and 10 mM Magnesium Acetate and incubating 4C overnight. The reaction was then purified via size-exclusion chromatography using an AKTAPurifier (GE Healthcare) on a Superdex 200 column (GE Healthcare). Peak fractions were pooled and then tested for biotinylation using an SDS-PAGE gel shift assay. Proteins were typically 100% biotinylated.
Generation of a mouse yeast displayed H2-Db peptide library, Tag enrichment, Staining, and selection
The single chain trimer (SCT) H2-Db yeast constructs were synthesized as N-terminal fusions to the yeast surface protein Aga2p. Full length SCT H2-Db constructs were cloned into the vector pYAL. These constructs contained an Aga2p leader sequence followed by the 9–10MER peptide sequence, a Gly-Ser (GGGGS)3 linker, the murine β2-microglobulin (β2M) sequence, a second glycine linker (GGGGS)4, the mouse H2-Db heavy chain sequence, either a Myc or HA epitope tag, a third glycine linker (GGGGS)3, and the Aga2 protein. SCT H2-Db MHC constructs were then electroporated into EBY-100 yeast as previously described40,41 and induced for expression in SGCAA pH 4.5 media at 20C 24–72 hr until maximum epitope tag staining was observed (typically 40%–70% of total population). The full length H2-Db yeast construct was mutagenized as described previously40. Briefly, the construct was mutagenized via error prone PCR (Genemorph II kit, Agilent 200550), with final error rate of ~ 4–5 nucleotide substitutions per kbp as judged by ligating error prone constructs into the pYAL vector and sequencing the clones. Yeast libraries were created by electroporation of competent EBY-100 cells via homologous recombination of linearized pYAL-cMyc/HA vector. Final libraries contained approximately 5×108 yeast transformants. Peptide libraries were created in the same manner as the error prone libraries, except pMHC constructs were instead randomized along the peptide by using mutagenic primers allowing all 20 amino acids via an NNK codon as previously described. The libraries allowed only limited diversity at the known MHC anchor residues to maximize the number of correctly folded and displayed pMHC clones in the library. For H2-Db, P5 and P9 anchors were limited to Asn (N) and Met/Ile/Leu (M/I/L) using AAC and MTS codons, respectively. The resulting PCR product was used as template for a second PCR reaction in which 50 nucleotides of sequence homologous to the vector was added to both ends of the PCR product. Then, 50 ug of this second PCR product and ~10 ug of linearized vector were purified and used for electroporating yeast to create each library. Before selecting on the H2-Db 9MER and 10MER pMHC libraries, each library was enriched for its respective epitope tag to maximize the percentage of yeast in the initial pool with correctly folded and displayed pMHC molecules presented on their surface. To achieve this, each of the libraries was induced separately in 500mL SGCAA at 20C for 24–72 hr with a starting density of 1×107 cells/mL. When maximum epitope tag staining was observed, approximately 1.4×109 induced yeast cells were washed once in PBS + 0.5% BSA and 1mM EDTA (PBE buffer) and resuspended in 5mL PBE with 200 uL of Miltenyi streptavidin microbeads (Miltenyi, 130–048-101). The cell and bead mixture were incubated at 4C with rotation for 1 hr, washed again in PBE, resuspended in 5mL PBE, and passed through a cell strainer onto a prewetted MACS LD column (Miltenyi 130–042-901). After allowing the column to fully empty, it was washed twice with 2 mL PBE, and the flow-through was collected. Cells were isolated from the flow-through by centrifugation and resuspended in 5 mL PBE with 80 uL of anti-cMyc AlexaFluor647 or anti-HA AlexFluor647 antibody (Cell signaling, 2233 and 3444), respectively incubated at 4C with rotation for one hour. The cells were washed and resuspended in 5 ml PBE, with 220 uL of Miltenyi anti-AlexaFluor647 microbeads added (Miltenyi, 130–091-395). This mixture was incubated at 4C for 30 minutes with rotation and protected from light. The cells were then washed, resuspended in 6mL PBE, and split evenly between two pre-wet MACS LS columns (Miltenyi, 130–042-401). After allowing the columns to fully empty, each column was washed two times with 3 mL of PBE, and the flow-through was set aside. The cells were eluted from the columns with 5mL PBE per column. A small fraction of the eluate (5–20 uL) was reserved to compare AlexaFluor647 staining to that of the flow-through for a quantification of tag enrichment. The rest of the eluted cells were pooled, collected by centrifugation, resuspended in a total of 40 mL SDCAA media, and the cell density was measured by spectrophotometer at 600 nm. The cell density was then adjusted to an OD of 1 or less with the addition of SDCAA, and the yeast were cultured at 30C overnight. The cells were the passaged for another round of overnight growth in SDCAA. For induction of the tag enriched library for the first round of selection, 10 times the number of cells retrieved in the eluate were taken for culture at 20C in 500 mL of SGCAA. To stain pMHC with TCR tetramers, biotinylated TCR was incubated with streptavidin coupled to AlexaFluor647, AlexaFluor488, or Phycoerythrin in a 5:1 ratio for 5 min on ice to ensure complete tetramer formation. Yeast cells were then stained with 250 nM tetramer + anti-Myc-AlexaFluor488 or anti-HA-AlexaFluor488 antibodies (Cell Signaling, 2279 or 2350, respectively) for 3 hr on ice and washed twice with ice cold PBE buffer before analysis via flow cytometry (Accuri C6 flow cytometer). All the yeast selections and sequencing of yeast libraries were done as previously described10,11.
List of primers used for H2-Db Libraries
For generating H2-Db error-prone libraries:
Forward primer: −5’TGCAGTTACTTCGCTGTTTTTCAATATTTTCTGTTATTGCTAGCGT TTTAGCAAGCAGCCTGGAGAACTTCAGAGCCTACGTGG-3’
Reverse: 5’-GAACAAAAGCTTATCTCCGAAGAAGACTTG-3’.
For the random H2-Db library:
Forward primer for 9MER HA library (initial randomization PCR): 5’-TCAATATTTTCTGTT ATTGCTAGCGTTTTAGCANNKNNKNNKNNKAACNNKNNKNNKMTSGGTGGAGGAGGTTCTG-3’.
Reverse primer for 9MER HA library (initial randomization PCR): 5’-TCCACCACCACCAGC GTAGTCTGGAACGTCGTATGGGTAGGATCCCTCCCA-3’.
To add overlap for homologous recombination with linearized pYAL vector:
Forward primer:5’-ATTTTCAATTAAGATGCAGTTACTTCGCTGTTTTTCAATATTTTCTG TTATTGCTAGCGTTTTAGCA-3’.
Reverse primer:5’-TCCACCACCACCAGCGTAGTCTGGAACGTCGTATG GGTAGGATCC CTCCCA-3’.
Class I and Class II peptide monomer production, tetramerization, and tetramer enrichment
Peptide-I-Ab monomer production: The peptide-I-Ab monomer was generated as previously described20,42. Briefly, the extracellular portion of I-Ab α chain was linked to acidic zipper on the C-terminus, followed by AviTag (GLNDIFEAQKIEWHE) and 6X Histidine tag. The peptides-myelin oligodendrocyte glycoprotein (MOG) 38–48 (GWYRSPFSRVV) or ovalbumin (OVA) 327–337 (VHAAHAEINEA) were tethered to the N-terminus of I-Ab β chain, followed by basic zipper and 6X Histidine tag. The “disulfide trap” is introduced through the oxidation of the cysteine at p+2 position and the cysteine at position 72 of I-Ab α chain mutated from valine to ensure the proper peptide binding register20,43. The α chain and the peptide-β chain were cloned separately into the pAcGP67A vectors by Gibson Assembly (New England Biosciences, E2611S). Baculoviruses for each construct were created in SF9 cells via co-transfection of BD baculogold linearized baculovirus DNA (BD Biosciences, 554739) with Cellfectin II (Life Technologies, 10362–100). The α and β chain viruses were coinfected in a small volume (2 ml) of High Five cells in various ratios to find a ratio to ensure 1:1 α:β stoichiometry. To prepare soluble monomers, 1L of High Five cells were infected with the appropriate ratio of α and β viruses for 48 hr at 28C. Collected culture media was conditioned with 100 mM Tris-HCl (pH 8.0), 1 mM NiCl2, 5 mM CaCl2 and the subsequent precipitation was cleared via centrifugation. The media is then incubated with Ni-NTA resin (QIAGEN, 30250) at room temperature for 3 hr and eluted in 1×HBS+200 mM imidazole (pH 7.2). TCRs were then site-specifically biotinylated by adding recombinant BirA ligase, 100 μM biotin, 50 mM Bicine pH 8.3, 10 mM ATP, and 10 mM Magnesium Acetate and incubating 4C overnight. The reaction was then purified via size-exclusion chromatography using an AKTAPurifier (GE Healthcare) on a Superdex 200 column (GE Healthcare). Peak fractions were pooled and then tested for biotinylation using an SDS-PAGE gel shift assay. Proteins were typically 100% biotinylated. Peptide-H2-Db monomer production: The peptide-H2-Db monomers were refolded with the appropriate peptide and human β2-microglobulin as previously described44. Briefly, H2-Db and human β2-microglobulin were separately expressed in BL21DE3 (ThermoFisher, C600003) in the form of inclusion bodies. In the H2-Db construct, H2-Db α chain was linked to AviTag and 6X Histidine tag. The refolding was carried out using rapid dilution. Following biotinylation by BirA, protein was purified by size-exclusion chromatography (Superdex 200 10/300 GL) and stored in −80C. For YQPGNWEYI (YQP), HDRVNWEYI (HDR), ASRSNRYFWL (ASR) and SMRPNHFFFL (SMRP), we individually refolded the monomers and purified them. For peptide-H2-Db monomer of 6218 flu peptides (QGLSNMRVRL, VGLENMRVRL, VSLRNMRSYL and SSLENFRAYV), we refolded H2-Db with a photo-cleavable peptide (FAPGNY-Anp-AL) and exchanged the target peptides into H2-Db upon UV cleavage of FAPGNY-Anp-AL45. Peptide-MHC tetramer formation: All tetramers were freshly prepared as previously described44. Briefly, for tetramerization, the amount of fluorophore-conjugated streptavidin and pMHC monomer were mixed with 4:1 molar ratio. One fifth amount of the fluorophore-conjugated streptavidin was added to the monomer solution every 10 minutes in room temperature. Enrichment of tetramer-positive T cells in mice and cell lines: Single cell suspensions of spleen and LN cells were prepared from unimmunized or immunized mice, resuspended in 200 μL FACS buffer (Ca2+/Mg2+-free sterile PBS with 0.5 % BSA, 0.5 mM EDTA) with Fc block (1:100) and 10 μM biotin. Following tetramer concentrations were used for staining cells: I-Ab-MOG38–48 tetramer (15 nM), I-Ab-OVA327–337 tetramer (15 nM), H2-Db-ASR tetramer (25 nM), H2-Db-SMRP tetramer (10 nM), H2-Db-YQP tetramer (25 nM) and H2-Db-HDR tetramer (25 nM). The cells were tetramer stained for an hour at room temperature and washed with FACS buffer. For I-Ab-MOG38–48 and I-Ab-OVA327–337 tetramers, the enrichment of tetramer-positive cells was done using the EasySep™ PE Positive Selection Kit (STEMCELL Technologies, 18557). If the cells were stained with both I-Ab tetramers and H2-Db tetramers, the enrichment of tetramer-positive cells were done using anti-PE MicroBeads (Miltenyi Biotec, 130–048-801) and anti-His MicroBeads (Miltenyi Biotec, 130–094-258) according to manufacturer instructions. Following tetramer enrichment, cells were surface stained with an antibody cocktail for 20 minutes at 4C. Stained cells were washed using FACS buffer and analyzed on LSR II (Becton Dickinson) or single cell sorted/bulk sorted on FACS Aria Fusion SORP (Becton Dickinson). Lentivirally transduced Jurkat TCR cell lines were stained with tetramers at 20 nM concentration in FACS buffer with 10 μM biotin at room temperate for 1 hr and followed by surface staining with appropriate antibodies for 20 minutes 4C. Following surface staining the cells were washed with FACS buffer and analyzed on LSR II (Becton Dickinson).
Single cell mouse and human TCR sequencing and data analysis
All human TCR primers used were previously published12. All mouse TCR primer sequences are provided in Supplementary Table. 1. TCR sequencing was done according to previously established protocols9,12.
Induction and evaluation of EAE
EAE was actively induced in C57BL/6J mice according to previously established protocol. Briefly, for the induction of EAE mice were injected subcutaneously in the posterior right and left flank with an emulsion containing 200 μg of MOG35–55 or CD8 specific SP derived from yeast library ASRSNRYFWL, SMRPNHFFFL YQPGNWEYI, and HDRVNWEYI, and an equal volume of complete Freund’s adjuvant (CFA; Sigma-Aldrich, F5881) supplemented with 200 μg of Mycobacterium tuberculosis H37Ra (Difco Laboratories, 231141). On the day of immunization and 2 days post-immunization, each mouse received 200 ng of PTX (List Biological Laboratories, 180) by intraperitoneal injection. Mice were scored daily for clinical signs of EAE beginning on day 5 after injection as follows: 0, no clinical expression of disease; 1, flaccid tail without hind-limb weakness; 2, hind limb weakness; 3, complete hind-limb paralysis and floppy tail; 4, hind-limb paralysis accompanied by a floppy tail and urinary or fecal incontinence; and 5, moribund. Clinical quantitative trait variables were assessed as described previously46.
CNS-infiltrating mononuclear cell isolation
CNS infiltrating cells were isolated according to previously established protocol46. Briefly, at different days post-immunization, animals were perfused with saline, and brains and spinal cords were removed. A single-cell suspension was obtained and passed through a 70-μm strainer. Mononuclear cells were obtained by Percoll gradient (37%/70%) centrifugation and collected from the interphase. Cells were washed, labeled with antibody conjugated to fluorochrome dyes, and analyzed by flow cytometry.
Expression of TCRs, H2-Db, and Qa-1b by lentiviral transduction
TCRα, β, H2-Db, and β2M constructs were cloned into a lentiviral construct. For TCR expression, alpha and beta TCR lentiviral constructs were transfected into 293X cells separately. The virus was harvested after 72 hr of transfection and transduced into Jurkat αβ−/− or SKW αβ−/− cells. SKW or Jurkat cells were enriched for highest expression of TCRαβ by using a Miltenyi anti-APC selection (Miltenyi 130–090-855). Similar strategy was used for the expression of H2-Db and β2M, except that T2 cells were used for transduction and expression.
T cell stimulation assays
T cell stimulation assays were performed as previously described11. All the T cell peptide stimulation experiments were done in 96 well round bottom plates with a 200 ul total volume. T2, K562 cells or BMDCs were pulsed with 10–100ug of the peptides for 45 minutes, washed once and plated (10, 000 cells/well). Cell lines expressing TCRs (100, 000 cells/well) were co-cultured with APCs for 18 hr. At the end of stimulation, cells were harvested, washed, and stained with TCRβ, human CD3, and CD69 and analyzed on LSR II (Becton Dickinson) for activation.
In vitro proliferation/suppression assay
Spleen and LN cells were harvested from WT or immunized mice. Single cell suspensions were prepared, and RBCs were lysed using Ack lysis buffer (ThermoFisher scientific A1049201). Total CD4+ (Miltenyi 130–049-201) and CD8+ (Miltenyi 130–049-401) T cells were positively purified using Miltenyi kits following established manufacturer protocol followed by FACS sorting. Similarly, antigen presenting cells were isolated using Miltenyi Pan Dendritic Cell (DC) Isolation Kit (Miltenyi 130–100-875). Post CD4+ T cell enrichment, cells were counted and labelled with CellTrace™ Violet Dye (ThermoFisher scientific C34557) according to manufacturer instructions. In vitro proliferation/suppression assays were set up according to previously published protocol47. Briefly, labelled CD4+ T cells were co-cultured either with CD8+ T cells (1:1 ratio, 0.25X106 cells/well) or without CD8+ T cells in the presence of pan DC’s (0.75X106 cells/well). In some of the suppression experiments cells were pre-included with 10ug/ml of anti-Qa-1b neutralizing antibody (6A8.6F10.1A16, BD Biosciences)22,48. Cells were cultured in a total volume of 200 ul in a 96-well round bottom plate. The CD4+ T cells were stimulated with either MOG35–55 or not. On day 7, the cells were washed and stained with surface antibodies and analyzed on LSR II (Becton Dickinson).
Adoptive transfer
EAE was actively induced in C57BL/6J mice were actively immunized with an emulsion containing 200 μg of MOG35–55 + CD8 specific PPTs derived from yeast library (ASRSNRYFWL, SMRPNHFFFL YQPGNWEYI, and HDRVNWEYI), and an equal volume of complete Freund’s adjuvant (CFA; Sigma-Aldrich, F5881) supplemented with 200 μg of Mycobacterium tuberculosis H37Ra (Difco Laboratories, 231141). On the day of immunization and 2 days post-immunization, each mouse received 200 ng of PTX (List Biological Laboratories, 180) by intraperitoneal injection. Day 10 post-immunization, spleen and lymph nodes were harvested, CD8+ cells were obtained using a CD8 enrichment kit and sorted for CD44+CD122+Ly49+ (Ly49+) and CD44+CD122+Ly49− (Ly49−) cells23. FACS purified Ly49+ and Ly49− cells (8 million cells/mice) were adoptively transferred at the time of active MOG immunization and mice were scored daily for clinical signs of EAE beginning on day 5 after as described previously46.
Induction and evaluation of EAU
Experimental autoimmune uveitis (EAU) was induced in mice as described previously24. Briefly, mice were injected subcutaneously in the posterior right and left flank with an emulsion containing 300 ug of human interphotoreceptor binding protein (IRBP) peptide 1–20 in CFA (1:1 v/v), 0.2 ug of PTX on day 0 and 0.2 ug again on day 2. Mice were euthanized on day 21 post-immunization. Mouse eyes were enucleated, fixed, and pupil-optic nerve sections were examined by histology as previously described49.
Whole transcriptome sequencing and data analysis
Whole transcriptome sequencing was done as previously described50. T cells were bulk sorted directly into Trizol (Qaigen). RNA was extracted with a RNeasy Plus Micro Kit (Qiagen). After analysis on the 2100 Bioanalyzer, the resulting library was sequenced on the HiSeq 4000 platform (Illumina). For each sample in the whole transcriptome sequencing library, 75-base pair paired-end reads were acquired from the sequencer. Each sample condition was completed in triplicate, except for the WT sample for which one sample was generated. Read quality was determined with FastQC 0.11.4. Using TopHat v2.0.13, we aligned the reads to the mouse reference genome (NCBI/ assembly GRCm38). On average, 90% of reads were aligned to the reference genome. One MOG + SP sample was removed from downstream analysis as an identified outlier. Differential gene expression analysis and read count normalization used as input for heatmaps were determined via DESeq251. TPM values were calculated with RSEM v1.3.052. Heatmaps were generated with the R package “pheatmap” 53. Gene ontology analysis plots were generated with the R package “enrichplot” 54.
Extended Data
Extended Data Fig. 1. Massive clonal expansion of all T cells following EAE immunization.
C57BL/6J mice were immunized for EAE induction and on different days post-immunization (PI) [D0 (Unimmunized), D3, D5, D7, D10, D12, D15, D17, D19, D21, D23, and D30], blood, draining lymph nodes (LN), spleen, and CNS infiltrating cells were harvested and stained with a cocktail of cell surface antibodies. (a) Stained cells were analyzed for total frequency of different T cell types and their activation status was determined with the gating strategy as shown in the figure. C57BL/6J mice were immunized for EAE induction and on different days post-immunization (PI) [D0 (unimmunized), D7, D10, D15, and D19], blood and CNS infiltrating CD4+, CD8+, and γδ+ T cells were single cell sorted. The cells underwent single-cell paired TCR sequencing. (n = 3 mice per group/time point). In total, we sequenced 1302 (CD4+), 1660 (CD8+), and 1451 (γδ+) paired TCR sequences. (b, c, and d) Average percent clonal expansion of CD4+, CD8+, and γδ+ T cells among unimmunized and immunized mice in all days and tissues combined together. Data are shown as mean ± SEM. (e) Percent of identical CD4+, CD8+, and γδ+ TCR sequences shared between blood and the CNS within each day PI. (f) Frequency of major groups (Group 1–4) of natural γδ+ T17 (nTγδ17) cells in blood and CNS different days and tissue PI. (g) Corresponding paired TCRγ and TCRδ sequences which define each major group (Group 1–4) of nTγδ17 cells.
Extended Data Fig. 2. Concomitant activation of all T cells following EAE immunization and clonally expanded CD8 TCRs are not specific to myelin peptides or proteins.
(a, c, and e) C57BL/6J mice were immunized for EAE induction and on different days PI [D0 (Unimmunized) (n = 5), D3 (n = 4), D5 (n = 4), D7 (n = 4), D10 (n = 4), D12 (n = 4), D15 (n = 5), D17 (n = 5), D19 (n = 5), D21 (n = 5), D23 (n = 5), and D30 (n = 3)], blood, draining LN, and spleen cells were harvested and analyzed for total frequency of activated (CD44high) and (b, d, and f) naïve (CD62Lhigh) CD4+, CD8+, and γδ+ T cells in the blood, draining LN, and spleen. Significance of differences observed in changes in frequency of naïve and activated T cells between days was determined using one-way ANOVA followed by Dunnets post hoc multiple comparison test. Data are shown as mean ± SEM. Representative data from two independent experiments. *p = 0.046; **p = 0.0023; ***p = 0.0002; ****p < 0.0001. 9 clonally expanded CD8 TCRs (EAE1-CD8 to EAE9-CD8) were retrovirally transduced to express on 58αβ−/− cells. (g) Un-transduced and transduced cell lines were stained with fluorochrome labelled anti-TCRβ and anti-CD3 to determine the surface expression of TCRs. (h) Un-transduced and transduced EAE-CD8 TCR cell lines were stimulated with plate bound anti-CD3 and soluble anti-CD28 for 12–16 hr and surface stained with activation marker CD69. (i) Un-transduced 58αβ−/− or OT-1 TCR transduced cell lines were stimulated with BMDCs pulsed with SIINFEKL peptide or whole Ovalbumin (OVA) protein for 12–16 hr, washed, and stained with activation marker CD69. (j) Unstimulated 58αβ−/− or EAE-CD8 TCR transduced cell lines were stimulated with pool of peptides (PP1–7) from MOG, MBP, PLP, MAG and SIINFEKL peptide and examined for activation marker CD69 (CD69 expression shown in figure for EAE1-CD8 TCR). Peptides are of variable lengths (8–12MERs). Each peptide pool contained 50 peptides. Representative data from three independent experiments.
Extended Data Fig. 3. Generation and functional validation of a H2-Db yeast peptide-MHC library.
(a) Schematic of the murine Class I MHC H2-Db displayed on yeast as β2m, α1, α2, and α3 with peptide covalently linked to the MHC N-terminus. (b) Design of the peptide library displayed by H2-Db. Design is based on the structure of the 6218 TCR bound to H2-Db-restricted acid polymerase peptide 224–233 (SSLENFRAYV, DbPA224) (Day E B et al., 2011, PDB, 3PQY). (c) Mutation required for proper folding of the H2-Db displayed on yeast (α2-W131 to α2-G131). Mutations were derived from error prone mutagenesis. (d) Design for two different lengths of H2-Db libraries. For nine amino acid (9 MER) library, residues from P1 to P9 were randomized, with limited diversity at MHC anchor positions P5 (Asparagine, N) and P9 (Methionine, Isoleucine, and Leucine, M/I/L). For ten amino acid (10 MER) library, residues from P1 to P10 were randomized, with limited diversity at MHC anchor positions P5 (Asparagine, N) and P10 (Methionine, Isoleucine, and Leucine, M/I/L). TCR contact residues are colored pink and MHC anchor residues are colored red or blue. (e and g) Selection of PA224-H2-Db error prone library with 6218 soluble TCR. Increased cMyc expression among induced yeast peptide-H2-Db error prone library at different rounds (RD1-RD4) of selection and (f) 6218 soluble TCR tetramer staining on the post-RD4 error prone H2-Db library. Each TCR was screened on the yeast library once.
Extended Data Fig. 4. In vitro and In vivo characterization of CD8+ T cells specific for surrogate peptides following EAE.
(a) EAE-CD8 TCR transduced Jurkat αβ−/− T cells were pMHC tetramer stained with 6218, EAE6, and EAE7-CD8 TCR specific SP (SSLENFRAYV, ASRSNRYFWL, SMRPNHFFFL YQPGNWEYI, and HDRVNWEYI), respectively. (b) From unimmunized (n = 4), MOG (n = 5), MOG + SP (n = 5), or SP immunized mice (n = 5), spleen and LN cells were harvested, and enriched for SP specific CD8+ T cells with pMHC tetramers. Representative flow cytometry gating strategy is shown for different cell surface markers and tetramer specific cells. (c) Representative flow cytometry data is shown for activation status (defined as CD44+CD62L−) on CD8+ T cells specific for SP (ASR, HDR, SMRP, and YQP-tet+) from WT and different immunization groups (MOG, MOG + SP, and SP). (d) Activated/effector phenotype of CD8+ T cells specific for SP (ASR, HDR, SMRP, and YQP-tet+) from WT (n = 5) and different immunization groups [MOG (n = 3), MOG + SP (n = 4), and SP (n = 3)] is quantified and shown as a bar graph (n = 5 mice per group). The significance of differences among the frequency of activated/effector cells between different immunization group was determined using one-way ANOVA followed by Tukey’s post hoc multiple comparison test. *p = 0.0169; **p = 0.0020; ****p < 0.0001. Data are shown as mean ± SEM. (e) C57BL/6J mice were immunized for EAE with an emulsion containing MOG35–55 + CFA + PTX (n =10) and Flu peptide (SSLENFRAYV) + MOG35–55 + CFA + PTX (n =10). The clinical scores following immunization were recorded. Data are shown as mean ± SEM. Representative data from two independent experiments.
Extended Data Fig. 5. CD8+ T cell specific surrogate peptide immunization suppresses MOG35–55 specific CD4+ T cells and induces CD8+ T cells with a regulatory phenotype.
C57BL/6J mice were immunized with an emulsion containing MOG35–55 + CFA + PTX (n = 5), or with MOG35–55 + SP + CFA + PTX (n = 5). From (a) unimmunized and (b and c) D10 PI mice, spleen and LN cells were isolated, stained, and enriched for MOG35–55 I-Ab pMHC specific CD4+ T cells and an irrelevant tetramer. Representative FACS plots for different groups are shown. (d-g) From unimmunized (n = 5) and D10 PI MOG (n = 5), MOG + SP (n = 5), and SP (n = 4) immunized mice, spleen and LN cells were harvested stained, and enriched for SP specific CD8+ T cells using pMHC tetramer. Representative FACS dot-plots for CD8+ T cell with a regulatory phenotype (CD44+CD122+Ly49+) from each group is shown. (h) Each tetramer+ (ASR, HDR, SMRP, and YQP-tet+) CD8+ T cells were sub-gated for CD122, CD44, and Ly49 and the frequency of CD122+CD44+Ly49+ cells among each SP specific cells are shown among different immunization groups. The significance of differences among the frequency of CD122+CD44+Ly49+ cells between different immunization group was determined using one-way ANOVA followed by Tukey’s post hoc multiple comparison test. ***p = 0.0002. Data are shown as mean ± SEM. Representative data from two independent experiments.
Extended Data Fig. 6. CD8+ T cells elicited upon MOG + SP immunization are specific, their suppression is mediated by Perforin, adoptive transfer of CD122+CD44+Ly49+ abrogates EAE, and SP triggers a more severe, inflammatory retinal uveitis than IRBP peptide alone.
C57BL/6J mice were immunized with an emulsion containing MOG35–55 + CFA + PTX, MOG35–55 + SP + CFA + PTX, or MOG35–55 + Flu peptide + CFA + PTX. Spleen and LN cells were harvested from D10 PI mice, enriched for either CD4+, CD8+ T cells, or antigen presenting cells (APC) using Miltenyi cell enrichment kits followed by FACS. CellTraceTM Violet (CTV) Dye labelled CD4+ T cells from MOG immunized mice were co-cultured with APC from MOG immunized mice either in the (a) absence of CD8+ T cells or in the presence of CD8+ T cells from (b) MOG + SP, (b) WT, (d) CFA + PTX, (e) MOG + flu, or (f) CD8+ T cells from Perforin knockout mice immunized with MOG + SP. (g) CTV labelled CD4+ T cells from MOG35–55 + CFA + PTX immunized mice were co-cultured with CD8+ T cells from MOG35–55 + SP + CFA + PTX in the presence of anti-Qa-1b antibody (10 ug/ml). In addition (h) CTV labelled CD4+ T cells from OVA329–337 CFA + PTX were co-cultured with CD8+ T cells from MOG35–55 + SP + CFA + PTX immunized mice. 7 days post co-culture, cells were washed and stained with surface markers and analyzed for CD4+ T cell proliferation (CTV Dye dilution). Representative data from two independent experiments. C57BL/6J mice were immunized with an emulsion containing MOG35–55 + SP + CFA + PTX (n = 10) and 10 days post-immunization spleen and lymph node cells were isolated, stained, enriched for CD8+ T cells with Miltenyi cell enrichment kit followed by FACS to CD44+CD122+Ly49+ (Ly49+) and CD44+CD122+Ly49− (Ly49−) cells. Sorted Ly49+ and Ly49− cells were adoptively transferred (8 million cells/animal) to C57BL/6J mice (n = 5 mice/group) at the time of MOG35–55 + CFA + PTX immunization. (i) The clinical scores following adoptive transfer of Ly49+ and Ly49− cells and MOG35–55 + SP + CFA + PTX immunization were recorded, and the significance of differences between clinical courses was calculated by regression analysis with the two-way ANOVA followed by Bonferroni post hoc multiple comparison test. **** p < 0.0001. Data are shown as mean ± SEM. Representative data from two independent experiments. (j) In the wild-type, untreated mouse eye, the retina shows a normal laminar pattern and there are no leukocytes in the vitreous. (k) After subcutaneous injection of IRBP (interphotoreceptor binding protein) peptide antigen, there was only a mild inflammatory response in 40% of eyes with activated leukocyte invasion of the vitreous (red arrow) and mild disruption of the and retina outer nuclear layer photoreceptors (black arrow). (l) Following subcutaneous injection of both IRBP + SP there was a severe inflammatory response in 80% of eyes with activated leukocyte invasion of the vitreous (red arrows) and severe disruption of the retina photoreceptors (black arrows). (RGC, retinal ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; RPE, retinal plexiform layer). Five C57BL/6J mice were examined for each condition. EAU was induced in mice and mice were euthanized on day 21 post-immunization. Mouse eyes were enucleated, fixed, and pupil-optic nerve sections were examined by histology. C57BL/6J mice were immunized with an emulsion containing IRBP + CFA + PTX or IRBP + SP + CFA + PTX. Spleen and LN cells were harvested from Day 10 post-immunized mice, enriched for either CD4+, CD8+ T cells, or antigen presenting cells (APC) using Miltenyi cell enrichment kits followed by FACS. CellTraceTM Violet (CTV) Dye labelled CD4+ T cells from IRBP immunized mice were co-cultured with APC from IRBP immunized mice with purified (n) CD8+CD44+CD122+Ly49+[Ly49+], (o) CD8+CD44+CD122+Ly49−[Ly49−] or (m) without CD8+ T cells from IRBP + SP immunized mice 7 days post co-culture, cells were washed and stained with surface markers and analyzed for CD4+ T cell proliferation (CTV Dye dilution).
Extended Data Fig. 7. Transcriptional profiling of Ly49+ Vs Ly49− cells.
C57BL/6J mice were immunized with an emulsion containing SP + CFA + PTX (n = 3). (a) Spleen and LN cells were harvested from D10 mice, enriched for either CD8+ T cells using a Miltenyi CD8+ T cell enrichment kit followed by sorting for CD122+CD44+Ly49− [Ly49−] and CD122+CD44+Ly49+ [Ly49+] cells and then performed bulk RNA-seq on these cells. (a) A heatmap of differentially expressed genes (log2(fold change) > 2 and the adjusted p value < 0.005) in Ly49+/Ly49− RNA-seq samples. Columns show samples. Rows and columns are ordered based on hierarchical clustering. Normalized gene expression values are centered for each gene by subtracting the average value of all samples from each sample value (2–3 mice/group). (b) Gene Ontology enrichment analysis of genes differentially expressed (log2(fold change) > 2 and the two-tailed Benjamini Hochberg adjusted p value < 0.005 from DESeq2) between Ly49+/Ly49− RNA-seq samples. The y-axis represents the top 30 enriched Gene Ontologies (genes from gene ontologies highlighted in green are in Supplementary Data Table. 6). The x-axis value is the fraction of genes in that ontology that are differentially expressed. The color of the dot represents that significance of the Gene Ontology enrichment (one-tailed Fisher’s exact test), and the size of the dot represents the number of genes differentially expressed. The plot was made with the R package “clusterProfiler.” (c) Volcano plot representing gene expression differences between the Ly49+ vs Ly49− samples (3 mice/group). Each point is a gene. List of genes specifically expressed in CD4+ Tregs (Zemmour D et al., 2018) are colored red if they are expressed in both MOG + SP RNA seq samples, and green if not. The horizontal dotted line is made at −log10(0.05), and the two vertical dotted lines represent a fold change of log2(2). Genes with a negative fold change are highly expressed in Ly49+ cells.
Extended Data Fig. 8. Major clonal expansion of CD8+ T cells in recent onset MS patients.
PBMCs from healthy controls (HC) (n = 4) and MS (n = 18) patients were stained and analyzed by flow cytometry to determine the frequency of T cells. Frequency of (a) CD4+, (b) CD8+, and (c) γδ+ T cells in T cells is shown, data are shown as mean ± SEM. Brain homing and activated (CD49d+CD29+HLA-DR+CD38+) CD8+ T cells were single cell sorted from PBMCs of (d) healthy controls (HCs) and (e) newly diagnosed MS patients. The cells underwent single-cell paired TCR sequencing. Pie charts depicting clonal expansion of CD8+ T cells among HC (n = 10) and MS (n =18) patients. The number of cells with β chain successfully identified is shown above its pie chart. For each TCR clone expressed by two or more cells (clonally expanded), the absolute number of cells expressing that clone (≥2, ≥5, ≥10, ≥20, and ≥50) is shown with a distinct colored section.
Extended Data Fig. 9. TCR repertoire of brain homing activated CD4+ T cells in recent onset MS patients.
Brain homing and activated (CD49d+CD29+HLA-DR+CD38+) CD4+ T cells were single cell sorted from PBMCs of (a) healthy controls (HCs) and (b) recent onset MS patients. The cells underwent single-cell paired TCR sequencing. Pie charts depicting clonal expansion of CD4+ T cells among HC (n = 10) and MS (n = 18) patients. The number of cells with β chain successfully identified is shown above its pie chart. For each TCR clone expressed by two or more cells (clonally expanded), the absolute number of cells expressing that clone (≥2, ≥5, ≥10, ≥20, and ≥50) is shown with a distinct colored section.
Extended Data Fig. 10. TCR repertoire of brain homing activated γδ+ T cells in recent onset MS patients.
Brain homing and activated (CD49d+CD29+HLA-DR+CD38+) γδ+ T cells were single cell sorted from PBMCs of (a) healthy controls (HCs) and (b) recent onset MS patients. The cells underwent single-cell paired TCR sequencing. Pie charts depicting clonal expansion of γδ+ T cells among HC and MS patients. The number of cells with δ chain successfully identified is shown above its pie chart. For each TCR clone expressed by two or more cells (clonally expanded), the absolute number of cells expressing that clone (≥2, ≥5, ≥10, ≥20, and ≥50) is shown with a distinct colored section. From the single cell sorted γδ+ T cells, RAR-related orphan receptor (ROR) transcripts were amplified using gene specific primers and sequenced simultaneously with γ and δ chains. (c) The number of γδ+ T cells positive for the RORC transcript is shown among HC (n = 10) and MS (n =18) patients. Significance of differences observed in the number of positive cells for RORC transcript was determined using paired t test. *p = 0.0301. Data are shown as mean ± SEM.
Supplementary Material
Acknowledgments
We thank members of Davis, Chien, Garcia, and Steinman laboratories for helpful discussions. We particularly thank Arnold Han and Murad M Mamedov in help designing mouse TCR primers, Vamsee Mallajyosulla for help with the H2-Db library constructs. We thank the Stanford Functional Genomics Facility for DNA and RNA sequencing (NIH shared facility equipment grant, S10OD018220). We thank Catherine Carswell, Bianca Gomez, and the Stanford Shared FACS facility for assistance in sorting cells and providing access to the equipment. We also thank Allison Nau, Drs. Lawrence Steinman, and Cristina Tato for critical reading of the manuscript and helpful discussions. We thank Christopher Bohlen from the lab of the late Ben Barres for myelin proteins. N.S. was supported by a Postdoctoral Fellowship from National Multiple Sclerosis Society (NMSS) and also a Career Transition Grant from NMSS. F.Z. was supported by the Ovarian Cancer Research Fund Alliances. Human samples collection was supported by a grant from NMSS RG-1611–26299 (JO and SH). M.M.D. and K.C.G. are funded by the US National Institutes of Health (U19 AI057229) and the Howard Hughes Medical Institute. Additional early support also came from the Simons Foundation (to MMD).
Footnotes
Data availability
RNA-seq data and Yeast p-MHC selection data are deposited to the Gene Expression Omnibus (GEO) data repository with accession number GSE130975. Source data for each figure are provided. Other data that support the findings of this study are available from the corresponding author upon reasonable request.
All authors declare no competing interests.
Supplementary Information
Available online
References
- 1.International Multiple Sclerosis Genetics Consortium et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fallang L-E et al. Differences in the risk of celiac disease associated with HLA-DQ2.5 or HLA-DQ2.2 are related to sustained gluten antigen presentation. Nat. Immunol. 10, 1096–1101 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Sollid LM, Qiao S-W, Anderson RP, Gianfrani C & Koning F Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 64, 455–460 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zamvil S et al. T-cell clones specific for myelin basic protein induce chronic relapsing paralysis and demyelination. Nature 317, 355–358 (1985). [DOI] [PubMed] [Google Scholar]
- 5.Blankenhorn EP et al. Genetics of experimental allergic encephalomyelitis supports the role of T helper cells in multiple sclerosis pathogenesis. Ann. Neurol. 70, 887–896 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skulina C et al. Multiple sclerosis: brain-infiltrating CD8+ T cells persist as clonal expansions in the cerebrospinal fluid and blood. PNAS 101, 2428–2433 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babbe H et al. Clonal Expansions of Cd8+ T Cells Dominate the T Cell Infiltrate in Active Multiple Sclerosis Lesions as Shown by Micromanipulation and Single Cell Polymerase Chain Reaction. J. Exp. Med. 192, 393–404 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blink SE & Miller SD The contribution of gammadelta T cells to the pathogenesis of EAE and MS. Curr. Mol. Med. 9, 15–22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han A et al. Dietary gluten triggers concomitant activation of CD4+ and CD8+ αβ T cells and γδ T cells in celiac disease. Proc. Natl. Acad. Sci. U.S.A. 110, 13073–13078 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birnbaum ME et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell 157, 1073–1087 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gee MH et al. Antigen Identification for Orphan T Cell Receptors Expressed on Tumor-Infiltrating Lymphocytes. Cell 172, 549–556.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han A, Glanville J, Hansmann L & Davis MM Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 32, 684–692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Y-L et al. A Highly Focused Antigen Receptor Repertoire Characterizes γδ T Cells That are Poised to Make IL-17 Rapidly in Naive Animals. Front Immunol 6, 118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langrish CL et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201, 233–240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroenke MA, Carlson TJ, Andjelkovic AV & Segal BM IL-12– and IL-23–modulated T cells induce distinct types of EAE based on histology, CNS chemokine profile, and response to cytokine inhibition. J. Exp. Med. 205, 1535–1541 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben Nun A, Wekerle H & Cohen IR The rapid isolation of clonable antigen‐specific T lymphocyte lines capable of mediating autoimmune encephalomyelitis. European Journal of Immunology 11, 195–199 (1981). [DOI] [PubMed] [Google Scholar]
- 17.Jäger A, Dardalhon V, Sobel RA, Bettelli E & Kuchroo VK Th1, Th17, and Th9 Effector Cells Induce Experimental Autoimmune Encephalomyelitis with Different Pathological Phenotypes. The Journal of Immunology 183, 7169–7177 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denton AE et al. Affinity Thresholds for Naive CD8+ CTL Activation by Peptides and Engineered Influenza A Viruses. The Journal of Immunology 187, 5733–5744 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Day EB et al. Structural basis for enabling T-cell receptor diversity within biased virus-specific CD8+ T-cell responses. PNAS 108, 9536–9541 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon JJ et al. Naive CD4+ T Cell Frequency Varies for Different Epitopes and Predicts Repertoire Diversity and Response Magnitude. Immunity 27, 203–213 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H-J & Cantor H Regulation of self-tolerance by Qa-1-restricted CD8(+) regulatory T cells. Semin. Immunol. 23, 446–452 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu L, Kim H-J, Werneck MBF & Cantor H Regulation of CD8+ regulatory T cells: Interruption of the NKG2A-Qa-1 interaction allows robust suppressive activity and resolution of autoimmune disease. Proc. Natl. Acad. Sci. U.S.A. 105, 19420–19425 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H-J et al. CD8+ T regulatory cells express the Ly49 Class I MHC receptor and are defective in autoimmune prone B6-Yaa mice. Proc. Natl. Acad. Sci. U.S.A. 108, 2010–2015 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agarwal RK & Caspi RR Rodent models of experimental autoimmune uveitis. Methods Mol. Med. 102, 395–419 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Zemmour D et al. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol. 19, 291–301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hvas J, Oksenberg JR, Fernando R, Steinman L & Bernard CC Gamma delta T cell receptor repertoire in brain lesions of patients with multiple sclerosis. J. Neuroimmunol. 46, 225–234 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Wucherpfennig KW et al. Gamma delta T-cell receptor repertoire in acute multiple sclerosis lesions. PNAS 89, 4588–4592 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gandhi R, Laroni A & Weiner HL Role of the innate immune system in the pathogenesis of multiple sclerosis. J. Neuroimmunol. 221, 7–14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caccamo N et al. Differentiation, phenotype, and function of interleukin-17-producing human Vγ9Vδ2 T cells. Blood 118, 129–138 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Moens E et al. IL-23R and TCR signaling drives the generation of neonatal Vgamma9Vdelta2 T cells expressing high levels of cytotoxic mediators and producing IFN-gamma and IL-17. J. Leukoc. Biol. 89, 743–752 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Sutton CE et al. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity 31, 331–341 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Price AE, Reinhardt RL, Liang H-E & Locksley RM Marking and quantifying IL-17A-producing cells in vivo. PLoS ONE 7, e39750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrington LE et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6, 1123–1132 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Elias D, Tikochinski Y, Frankel G & Cohen IR Regulation of NOD mouse autoimmune diabetes by T cells that recognize a TCR CDR3 peptide. Int Immunol 11, 957–966 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Kumar V, Stellrecht K & Sercarz E Inactivation of T cell receptor peptide-specific CD4 regulatory T cells induces chronic experimental autoimmune encephalomyelitis (EAE). J. Exp. Med. 184, 1609–1617 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu D et al. Analysis of regulatory CD8 T cells in Qa-1-deficient mice. Nat. Immunol. 5, 516–523 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Panoutsakopoulou V et al. Suppression of autoimmune disease after vaccination with autoreactive T cells that express Qa-1 peptide complexes. J. Clin. Invest. 113, 1218–1224 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis MM & Brodin P Rebooting Human Immunology. Annual Review of Immunology 36, 843–864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Shea EK, Lumb KJ & Kim PS Peptide ‘Velcro’: Design of a heterodimeric coiled coil. Current Biology 3, 658–667 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Birnbaum ME et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell 157, 1073–1087 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams JJ et al. T Cell Receptor Signaling Is Limited by Docking Geometry to Peptide-Major Histocompatibility Complex. Immunity 35, 681–693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson RW et al. T Cell Receptor Cross-Reactivity between Similar Foreign and Self Peptides Influences Naive Cell Population Size and Autoimmunity. Immunity 42, 95–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stadinski BD et al. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. PNAS 107, 10978–10983 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altman JD et al. Phenotypic Analysis of Antigen-Specific T Lymphocytes. Science 274, 94–96 (1996).8810254 [Google Scholar]
- 45.Grotenbreg GM et al. Discovery of CD8+ T cell epitopes in Chlamydia trachomatis infection through use of caged class I MHC tetramers. PNAS 105, 3831–3836 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krementsov DN et al. Sex‐specific control of central nervous system autoimmunity by p38 mitogen‐activated protein kinase signaling in myeloid cells. Ann. Neurol. 75, 50–66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tennakoon DK et al. Therapeutic induction of regulatory, cytotoxic CD8+ T cells in multiple sclerosis. J. Immunol. 176, 7119–7129 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Bian Y et al. MHC Ib molecule Qa-1 presents Mycobacterium tuberculosis peptide antigens to CD8+ T cells and contributes to protection against infection. PLOS Pathogens 13, e1006384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahajan VB, Skeie JM, Assefnia AH, Mahajan M & Tsang SH Mouse eye enucleation for remote high-throughput phenotyping. J Vis Exp e3184–e3184 (2011). doi: 10.3791/3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mamedov MR et al. A Macrophage Colony-Stimulating-Factor-Producing γδ T Cell Subset Prevents Malarial Parasitemic Recurrence. Immunity 48, 350–363.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolde R pheatmap: Pretty Heatmaps. R package version 1.0. 8. (2015). [Google Scholar]
- 54.Yu G clusterProfiler: An universal enrichment tool for functional and comparative study. doi: 10.1101/256784 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.