ABSTRACT
Circular RNAs (circRNAs) are new types of endogenous non-coding RNAs, which are identified to have critical regulatory roles in cancer biology. In this study, we aimed to find the abnormally expressed circRNAs in hepatocellular carcinoma (HCC) and investigate the function and mechanism of circRNAs in HCC progression. Upregulation of hsa_circ_0091581 was identified by RNA-sequencing and validated by quantitative real-time polymerase chain reaction (qRT-PCR). Hsa_circ_0091581 expression was correlated with tumor size, disease-free survial and overall survival of HCC patients. Functionally, hsa_circ_0091581 could promote the proliferation of HCC cells in vitro. Mechanism research showed that hsa_circ_0091581 promoted cell proliferation via hsa_circ_0091581/miR-526b/c-Myc axis in HCC cells. Also, the expression of hsa_circ_0091581 in HCC could be regulated by c-Myc. These results revealed that hsa_circ_0091581/miR-526b/c-Myc/hsa_circ_0091581 positive feedback loop plays a vital role in HCC progression and hsa_circ_0091581 may be a potential prognostic biomarker and therapeutic target for HCC.
KEYWORDS: Circular RNA, hepatocellular carcinoma, miR-526b, c-Myc
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide which rank as the third leading cause of cancer-related mortalities [1,2]. Despite the wide use of advanced therapies such as resection, transplantation and ablation, the prognosis of HCC patients is still poor due to high recurrence and metastasis [3–5]. HCC develops through a multistep transformation process which involves many genetic and epigenetic changes. Recently, exploring potential biomarkers and investigating the molecular mechanisms of HCC carcinogenesis are hot topics.
More and more non-coding RNAs, including microRNA, circular RNA, long non-coding RNA, were found to be abnormally expressed in HCC and identified as diagnostic biomarkers and therapeutic targets [6]. Circular RNAs (circRNAs) are a class of transcripts constructed with a closed loop, which have limited protein-coding capacity [7]. CircRNA was found to play important roles in HCC progression such as cell growth, invasion, migration and apoptosis [8–11]. These findings indicate that circRNAs may act as potential biomarkers and therapeutic targets in HCC and investigation of circRNAs in HCC may have great significance.
In this study, we found some abnormally expressed circRNAs in HCC using circRNA sequencing. Hsa_circ_0091581 which derived from back-splicing of Glypican 3 (GPC3) transcript was found to be upregulated in HCC and verified using quantitative real-time polymerase chain reaction (qRT-PCR). We then estimated the clinical value of hsa_circ_0091581 in HCC and further investigated the functions and mechanisms of hsa_circ_0091581 in HCC cell proliferation.
Materials and methods
Ethic and patient samples
This study was approved by the Ethics Committee of Shenzhen University General Hospital and Southwest Hospital of Third Military Medical University. Written informed consent was obtained from each patient. HCC and adjacent normal tissues were obtained from surgical resections of patients at Shenzhen University General Hospital and Southwest Hospital of Third Military Medical University. The clinicopathological data are shown in Table 1.
Table 1.
Correlation between the clinicopathologic variables and hsa_circ_0091581 expression in HCC tissues.
| Variables | Case | hsa_circ_0091581 expression low/high | P value |
|---|---|---|---|
| Gender | |||
| Male | 41 | 19/22 | |
| Female | 5 | 4/1 | 0.173 |
| Age (y) | |||
| ≤60 | 37 | 18/19 | |
| >60 | 9 | 5/4 | 0.336 |
| TNM stage | |||
| I | 39 | 21/18 | |
| II–III | 7 | 2/5 | 0.207 |
| Tumor size (cm) | |||
| ≤5 | 32 | 21/11 | |
| >5 | 14 | 2/14 | 0.002 |
| Tumor number | |||
| 1 | 33 | 19/14 | |
| ≥2 | 13 | 4/9 | 0.095 |
| AFP (ng/mL) | |||
| ≤20 | 21 | 13/8 | |
| >20 | 25 | 10/15 | 0.118 |
| ALT(U/L) | |||
| ≤40 | 40 | 20/20 | |
| >40 | 6 | 3/3 | 0.667 |
| Liver cirrhosis | |||
| Yes | 6 | 2/4 | |
| No | 40 | 21/19 | 0.333 |
| HBV | |||
| Negative | 5 | 2/3 | |
| Positive | 41 | 21/20 | 0.500 |
hsa_circ_0091581 expression low/high: the expression of hsa_circ_0091581 in HCC tissues was lower/higher than the median; AFP: alpha-fetoprotein; ALT, alanine aminotransferase; HBV: hepatitis B virus.
RNA isolation, RNase R treatment and RNA-sequencing
Total RNA was extracted using Trizol (Invitrogen) from three HCC tissues and paired adjacent normal tissues (ANT). RNase R (Epicentre, Madison, WI) was used to remove linear RNA before the construction of RNA-seq libraries. High-throughput sequencing was then performed by HiSeq 3000 platform (Illumina, San Diego, CA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from tissues or cells was extracted using Trizol (Invitrogen). The cDNA for qPCR analyses was synthesized using M-MLV Reverse Transcriptase (Promega, Madison, USA). Real Master Mix (SYBR Green) (Tiangen, Beijing, China) was used for real-time PCR. The primers for hsa_circ_0091581 were, F: 5′-TGGAGAACGTACTGCTTGGTC-3′, R:5′-TGGAGTCAGGCTTGGGTAGT-3′; GAPDH was used as an internal reference for hsa_circ_0091581 and the primers were, F: 5′-TCGACAGTCAGCCGCATCTTCTTT-3′, R: 5′-ACCAAATCCGTTGACTCCGACCTT-3′. MiR-526b expression was detected using Hairpin-itTM Quantitation PCR Kit (GenePharma, Shanghai, China) and U6 were used as reference for miR-526b. The primers for U6 were, F: 5′- TCGCTTCGGCAGCACATA-3′, R: 5′- TTTGCGTGTCATCCTTGC-3′.
Cell culture and transfection
Human HCC cell lines (MHCC97-L, SMMC-7721, HuH-7 and HepG2) and human normal hepatic cell line HL-7702 were cultured with DMEM (Gibco, CA, USA) containing 10% fetal bovine serum in a humidified atmosphere of 5% CO2. Plasmids or siRNAs and the corresponding negative controls were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The siRNA sequences were, for hsa_circ_0091581 siRNA, sense: 5ʹ- ACCACCACUAGGCCUUUGAAATT-3ʹ, antisense: 5ʹ-UUUCAAAGGCCUAGUGGTGGUTT-3ʹ. c-Myc siRNA, sense: 5ʹ- CAGAAAUGUCCUGAGCAAUTT-3ʹ, antisense: 5ʹ- AUUGCUCAGGACAUUUCUGTT-3ʹ.
CCK8 cell proliferation assay
CCK-8 assay was used to study cell proliferation. Cells were seeded in 96-well plates at a density of 5 × 103 cells per well and transfected with oligoribonucleotides. CCK-8 kit (Dojindo Molecular Technologies, Inc, Japan) was added to each well 24, 48, 72 h after transfection. The optical density was measured at the wavelength of 450 nm according to the manufacturer’s protocol.
Colony formation assay
Cells were seeded in six-well plates at a density of 500 cells per well and transfected with oligoribonucleotides. After culturing for 2 weeks, formed colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet.
Western blot assay
Cells were collected and lysed with cell RIPA (Beyotime, China) to extract proteins. The proteins were separated on SDS-polyacrylamide gels and transferred on a polyvinylidene fluoride membrane (Millipore). Immunoblotting of the membrane was performed using the following primary antibodies: anti-c-Myc (Cell Signaling Technology, Danvers, MA, USA), anti-GAPDH (Cell Signaling Technology).
Dual-luciferase assay
The hsa_circ_0091581 sequences containing potential wild-type or mutant binding sites of miR-526b were constructed into pmirGLO vectors (Promega Corporation, Fitchburg, WI, USA). The luciferase vectors and miR-526b mimics were transfected into SMMC-7721 cells along with pRL-TK vector (Promega). The Dual-Luciferase Reporter Assay System (Promega) was used to detect luciferase activity and Renilla luciferase activity was normalized to Firefly luciferase activity.
Statistical analysis
Statistical analyses were performed using SPSS version 19.0 software (SPSS, Inc. Chicago, IL). Chi-squared test, Student’s t-test and log-rank test were performed for comparisons, as appropriate. P < 0.05 was considered as statistical significance.
Results
Hsa_circ_0091581 expression was upregulated in HCC tissues and correlated with clinical features and prognosis of HCC patients
To screen the abnormally expressed circRNAs, circular RNA sequencing was performed in three pairs of HCC tissues and adjacent normal tissues and hsa_circ_0091581 was found to be significantly upregulated in HCC tissues compared with adjacent normal tissues (Figure 1(a)). Then, qRT-PCR assay was used to detect hsa_circ_0091581 expression in 46 pairs of HCC tissue and adjacent normal tissues. The result suggested that hsa_circ_0091581 expression was relatively higher in HCC tissues than adjacent normal tissues (Figure 1(b)). We also detected hsa_circ_0091581 expression using qRT-PCR in four human HCC cell lines (MHCC97L, SMMC-7721, HuH-7 and HepG2) and one human normal hepatic cell line (HL-7702). The hsa_circ_0091581 expression was relatively lower in HL-7702 cell lines than human HCC cell lines (Figure 1(c)). So, we chose hsa_circ_0091581 to study the function in HCC.
Figure 1.
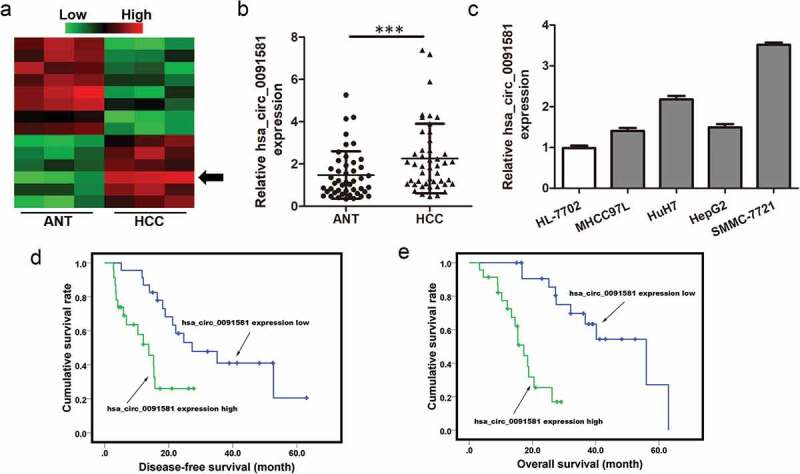
Hsa_circ_0091581 expression was upregulated in HCC tissues and correlated with prognosis of HCC patients. (a) Heatmap showing the circRNA expression profiles of HCC tissues and ANTs. Red and green indicate high and low expression, respectively. (b) Relative expression of hsa_circ_0091581 in 46 pairs of HCC tissues and ANTs was detected using qRT-PCR. (c) Relative expression of hsa_circ_0091581 in four human HCC cell lines and one human normal hepatic cell line was detected using qRT-PCR. The data were analyzed using the delta Ct method and presented as mean ± SD. ***P < 0.001. (d) Disease-free survival of patients with high-level hsa_circ_0091581 expression and low-level hsa_circ_0091581 expression; P = 0.004. (e) Overall survival of patients with high-level hsa_circ_0091581 expression and low-level hsa_circ_0091581 expression; P < 0.001. Hsa_circ_0091581 expression low/high: the expression of hsa_circ_0091581 in HCC tissues was lower/higher than the median.
To explore whether hsa_circ_0091581 expression was associated with clinical characteristics of HCC patients, we divided hsa_circ_0091581 expression into two groups according to the median. The clinicopathological data of the two groups were compared. Hsa_circ_0091581 expression was relatively higher in HCC tissues of tumor size >5 cm (Table 1). Kaplan–Meier analysis showed that higher hsa_circ_0091581 expression in HCC tissues was associated with low disease-free survival rate (DFS) and overall survival (OS) rate of HCC patients (Figure 1(d,e)). These results illustrated that hsa_circ_0091581 might act as an effective prognostic biomarker for HCC.
Downregulation of hsa_circ_0091581 inhibited the proliferation of HCC cells
Since hsa_circ_0091581 expression in HCC tissues was associated with HCC tumor size, we further studied the function and mechanism of hsa_circ_0091581 in cell proliferation in vitro. Because SMMC-7721 cells had the highest hsa_circ_0091581 level, we downregulated hsa_circ_0091581 in SMMC-7721 cells with siRNA. The siRNA only downregulated the expression of hsa_circ_0091581 but not the expression of the GPC3 mRNA (Figure 2(a,b)). Then, CCK8 assay and colony formation assay showed that hsa_circ_0091581 knockdown significantly suppressed the proliferation of SMMC-7721 cells (Figure 2(c,d)). These results suggested that downregulation of hsa_circ_0091581 inhibited the proliferation of HCC cells.
Figure 2.
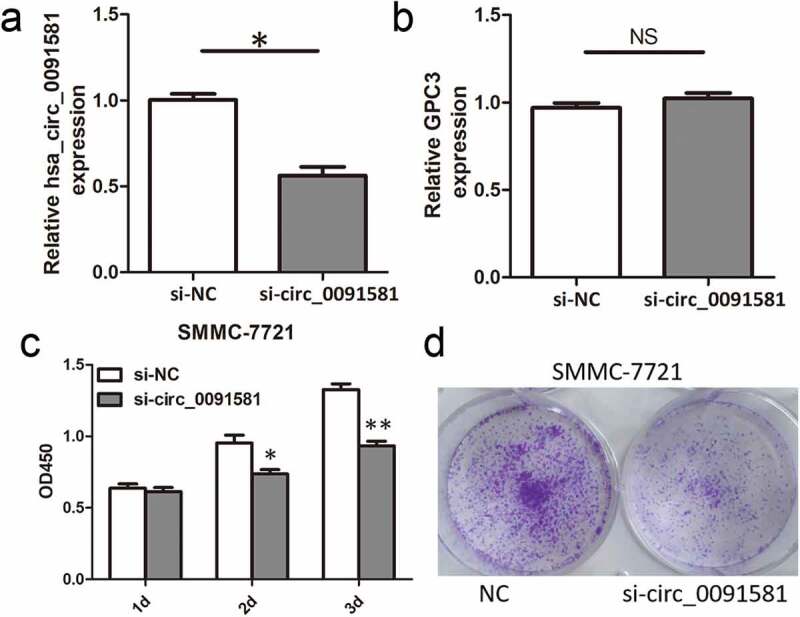
Downregulation of hsa_circ_0091581 inhibited the proliferation of HCC cells. (a, b) Relative hsa_circ_0091581 and GPC3 mRNA expression in SMMC-7721 cells transfected with hsa_circ_0091581 siRNA. (c, d) CCK8 and colony formation proliferation assays of SMMC-7721 cells transfected with hsa_circ_0091581 siRNA. Results are presented as mean ± SD. NS, not significant, *P < 0.05, **P < 0.01.
Hsa_circ_0091581 could act as a sponge of miR-526b in HCC cells
Emerging studies proved that circRNAs may act as a sponge to repress miRNA availability. We then investigate whether hsa_circ_0091581 could bind to some miRNAs. Using bioinformatics prediction tool based on TargetScan and MiRanda, we found that miR-526b was a potential target of hsa_circ_0091581. Thus, we performed luciferase reporter assays to determine whether hsa_circ_0091581 functioned as an miR-526b sponge. We constructed luciferase reporter plasmid containing wild-type (wt) or mutant (mut) binding site of miR-526b (Figure 3(a)). We found that co-transfection of miR-526b mimics and wt luciferase reporter plasmid of hsa_circ_0091581 into SMMC-7721 cells significantly decreased the relative luciferase activity. And, co-transfection of miR-526b mimics and mut luciferase reporter plasmid of hsa_circ_0091581 into SMMC-7721 cells did not affect the relative luciferase activity (Figure 3(b)). Moreover, downregulation of hsa_circ_0091581 could increase the expression of miR-526b in SMMC-7721 cells (Figure 3(c)). We also detected the expression of miR-526b in HCC tissues and adjacent normal tissues and found that miR-526b was significantly downregulated in HCC tissues (Figure 3(d)). What is more, the miR-526b expression was negatively correlated with hsa_circ_0091581 expression in HCC tissues (Figure 3(e)). Taken together, these results demonstrated that hsa_circ_0091581 could act as a sponge of miR-526b in HCC cells.
Figure 3.
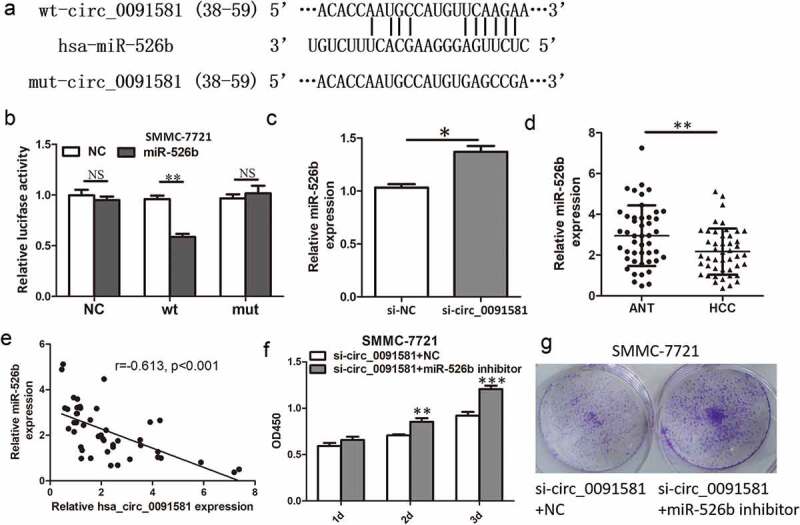
Hsa_circ_0091581 could act as a sponge of miR-526b in HCC cells. (a) Predicted miR-526b binding site and corresponding mutant sites in the hsa_circ_0091581 sequence (wt, wild-type; mut, mutant). (b) Relative luciferase activity in SMMC-7721 cells co-transfected with wt or mut luciferase reporters and miR-526b mimics or corresponding negative control. (c) Relative miR-526b expression in SMMC-7721 cells transfected with hsa_circ_0091581 siRNA. (d) Relative miR-526b expression in 46 pairs of HCC tissues and ANTs was detected using qRT-PCR. (e) Correlation between relative hsa_circ_0091581 expression and relative miR-526b expression analyzed with Spearman’s analysis. (g, f) CCK8 and colony formation proliferation assays of SMMC-7721 cells co-transfected with hsa_circ_0091581 siRNA and miR-526b inhibitor or corresponding negative controls. Results are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
For miR-526b was proved to act as tumor suppressor gene and inhibit cell proliferation in HCC [11], we wonder whether hsa_circ_0091581 promoted proliferation by repressing miR-526b availability in HCC cells. We found that miR-526b inhibitor could reverse the proliferation inhibition induced by hsa_circ_0091581 knockdown (Figure 3(f,g)). These results demonstrated that hsa_circ_0091581 promoted proliferation by sponging miR-526b in HCC cells.
C-Myc was a target of miR-526b and could upregulate hsa_circ_0091581 expression in HCC
Through microRNA.org, we found there was a potential binding site of miR-526b in c-Myc 3ʹUTR (Figure 4(a)). Then, we performed luciferase reporter assay and found that miR-526b could reduce the luciferase activity of SMMC-7721 cells transfected with the reporter containing wild-type c-Myc 3ʹUTR but not the reporter vector containing the mutant binding sites of c-Myc 3ʹUTR (Figure 4(b)). Moreover, reduced c-Myc protein level was found in SMMC-7721 cells transfected with miR-526b mimics (Figure 4(c)). Hsa_circ_0091581 knockdown increased the c-Myc protein level and this effect could be reversed by miR-526b inhibitor (Figure 4(d)). Because c-Myc is a well-known oncogene in HCC and promotes proliferation, hsa_circ_0091581 might promote proliferation by upregulating c-Myc via miR-526b in HCC cells.
Figure 4.
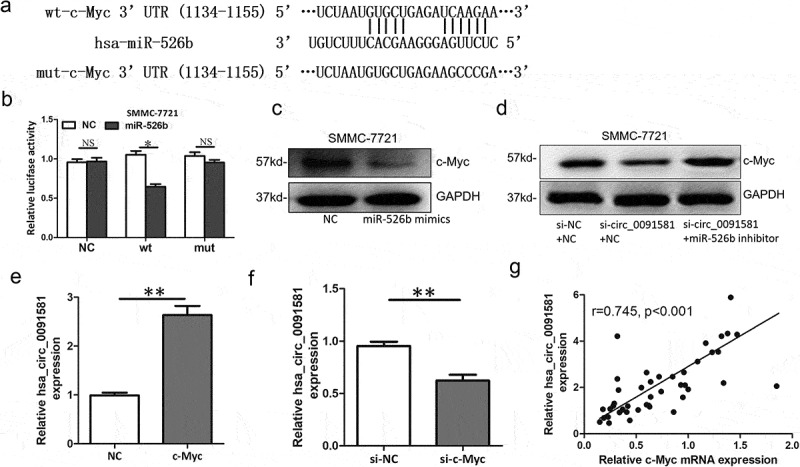
C-Myc was a target of miR-526b and could upregulate hsa_circ_0091581 expression in HCC. (a) Predicted miR-526b binding site and corresponding mutant sites in the c-Myc 3ʹUTR (wt, wild-type; mut, mutant). (b) Relative luciferase activity in SMMC-7721 cells co-transfected with wt or mut luciferase reporters and miR-526b mimics or corresponding negative control. (c) Relative c-Myc protein level in SMMC-7721 cells transfected with miR-526b mimics. (d) Relative c-Myc protein level in SMMC-7721 cells co-transfected with hsa_circ_0091581 siRNA and miR-526b inhibitor or corresponding negative controls. (e, f) Relative hsa_circ_0091581 expression in SMMC-7721 cells transfected with c-Myc overexpression plasmids or siRNA. (g) Correlation between relative hsa_circ_0091581 expression and relative c-Myc mRNA expression analyzed with Spearman’s analysis. Results are presented as mean ± SD. *P < 0.05, **P < 0.01.
Hsa_circ_0091581 is derived from the GPC3 gene locus and a study had revealed that c-Myc could bind to the endogenous GPC3 promoter and directly transcriptionally activate GPC3 [12]. So, theoretically, c-Myc could upregulate hsa_circ_0091581 expression in HCC. Then, we overexpressed c-Myc with overexpression plasmids and knockdown c-Myc with siRNA, respectively. The expression of hsa_circ_0091581 mRNA was detected by qRT-PCR. The results showed that hsa_circ_0091581 increased after c-Myc overexpression and decreased after c-Myc knockdown (Figure 4(e,f)). Moreover, we found a positive correlation between c-Myc mRNA expression and hsa_circ_0091581 expression in HCC tissues (Figure 4(g)). These results suggested that c-Myc could upregulate hsa_circ_0091581 expression in HCC.
Discussion
Increasing numbers of studies have focused on the role of circRNA in various diseases especially cancer. Studies have proved that circRNAs contain miRNA-binding sites and act as miRNA sponges; thus, circRNAs play an important regulatory role in cancer via interacting with cancer-associated miRNAs [13].
In this study, we identified a new circRNA, hsa_circ_0091581, that was significantly upregulated in HCC tissues. Hsa_circ_0091581 is derived from the GPC3 gene locus. GPC3 expression increases at both the gene and protein levels in HCC patients, and its high expression associates with poor prognosis. Mechanistic studies found GPC3 plays important roles in HCC progression by binding to molecules such as Wnt signaling proteins and growth factors. GPC3 has also been used as a target for molecular imaging and therapeutic intervention in HCC [14]. We also found that the expression of hsa_circ_0091581 correlated with poor clinical outcomes of patients with HCC. So, hsa_circ_0091581 may be a potential prognostic biomarker for HCC.
Functionally, hsa_circ_0091581 could promote the proliferation of HCC cells and this could explain the association between hsa_circ_0091581 expression and HCC tumor size. Through mechanism research, we found hsa_circ_0091581 promoted HCC cells proliferation by sponging miR-526b. MiR-526b has already been found to be downregulated in HCC and act as a tumor suppressor gene in HCC [11]. We also found that miR-526b could suppress the expression of c-Myc through directly targeting it, and this could explain why hsa_circ_0091581 upregulated the expression of c-Myc. c-Myc is a cell cycle regulation-related proteins and could promote cell proliferation [15], so, hsa_circ_0091581/miR-526b/c-Myc axis plays a vital role in HCC cells proliferation.
Hsa_circ_0091581 is derived from the GPC3 gene locus and a study had revealed that c-Myc could bind to the endogenous GPC3 promoter and directly transcriptionally activate GPC3 [12]. We also demonstrated that c-Myc could upregulate hsa_circ_0091581 expression in HCC. So, we can say that c-Myc could upregulate hsa_circ_0091581 expression via binding to the endogenous GPC3 promoter and directly transcriptionally activate hsa_circ_0091581 in HCC. Thus, a hsa_circ_0091581/miR-526b/c-Myc/hsa_circ_0091581 positive feedback loop forms and it plays a vital role in HCC cells proliferation (Figure 5). In this positive feedback loop, hsa_circ_0091581 may be a potential therapeutic target for HCC.
Figure 5.
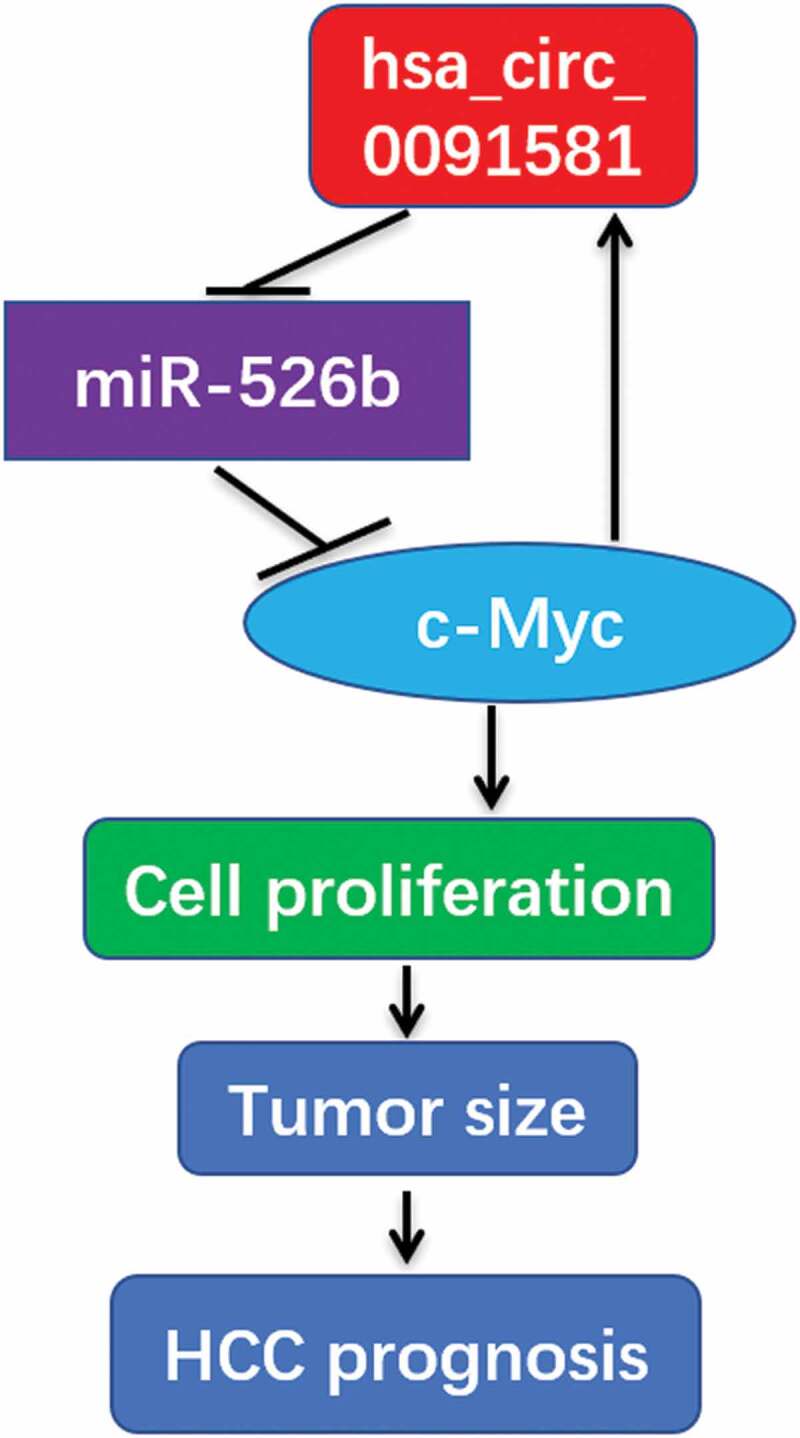
Diagram showing roles of hsa_circ_0091581/miR-526b/c-Myc/hsa_circ_0091581 positive feedback loop in HCC.
In summary, we for the first time prove that hsa_circ_0091581 expression is upregulated in HCC tissues and down-regulation of hsa_circ_0091581 inhibits the proliferation of HCC cells. Hsa_circ_0091581/miR-526b/c-Myc/hsa_circ_0091581 positive feedback loop plays a vital role in HCC cells proliferation. Hsa_circ_0091581 may be a potential prognostic biomarker and therapeutic target for HCC.
Funding Statement
This work was supported by the National Natural Science Foundation of China [81430063] and Guangdong Provincial Science and Technology Program [2019B030301009].
Disclosure statement
All the authors declare that they have no conflict of interest.
Ethical approval
All the procedures performed in studies involving human participants were in accordance with the declaration of Helsinki and its later amendments or comparable ethical standards. This study was approved by the Ethics Committee of Shenzhen University General Hospital and Southwest Hospital of Third Military Medical University.
Informed consent
Informed consent was obtained from each patient included in the study.
References
- [1].Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. [DOI] [PubMed] [Google Scholar]
- [2].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [3].Sun HZ, Song YL, Wang XY.. Effects of different anesthetic methods on cellular immune and neuroendocrine functions in patients with hepatocellular carcinoma before and after surgery. J Clin Lab Anal. 2016;30:1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Takayama T. Surgical treatment for hepatocellular carcinoma. Jpn J Clin Oncol. 2011;41:447–454. [DOI] [PubMed] [Google Scholar]
- [5].Poon RTP. Differentiating early and late recurrences after resection of HCC in cirrhotic patients: implications on surveillance, prevention, and treatment strategies. Ann Surg Oncol. 2009;16:792. [DOI] [PubMed] [Google Scholar]
- [6].Wang M, Yu F, Li P. Circular RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers (Basel). 2018;10:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li MF, Li YH, He YH, et al. Emerging roles of hsa_circ_0005075 targeting miR-431 in the progress of HCC. Biomed Pharmacother. 2018;99:848–858. [DOI] [PubMed] [Google Scholar]
- [9].Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214–1227. [DOI] [PubMed] [Google Scholar]
- [10].Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun. 2018;499:1044–1049. [DOI] [PubMed] [Google Scholar]
- [11].Liu X, Yang L, Tu J, et al. microRNA-526b servers as a prognostic factor and exhibits tumor suppressive property by targeting Sirtuin 7 in hepatocellular carcinoma. Oncotarget. 2017;8:87737–87749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li L, Jin R, Zhang X, et al. Oncogenic activation of glypican-3 by c-Myc in human hepatocellular carcinoma. Hepatology. 2012;56:1380–1390. [DOI] [PubMed] [Google Scholar]
- [13].Qu S, Liu Z, Yang X, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–309. [DOI] [PubMed] [Google Scholar]
- [14].Zhou F, Shang W, Yu X. Glypican-3: A promising biomarker for hepatocellular carcinoma diagnosis and treatment. Med Res Rev. 2018;38:741–767. [DOI] [PubMed] [Google Scholar]
- [15].Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–5610. [DOI] [PubMed] [Google Scholar]


