Abstract
A protein delivery tool based on bacterial type III secretion system (T3SS) has been broadly applied in biomedical researches. In this review, we summarize various applications of the T3SS-mediate protein delivery which enables translocation of proteins directly into mammalian cells without protein purification. Some of the remarkable advancements include delivery of antigens for therapeutic vaccines, nucleases for genome editing, transcription factors for cellular reprogramming and stem cells differentiation, and signaling molecules for post-translational proteomics studies. With continued improvement of the T3SS-mediated protein delivery tools, even wider application of the technology is anticipated.
Keywords: Protein delivery, Type III secretion system, Vaccine, Cellular, Reprogramming, Genome editing, Stem cell differentiation
1. Introduction
Intracellular delivery of desired proteins, including gene editing enzymes, transcription factors, signaling molecules, antigens, and antibodies, into various target cells serves diverse purposes in biomedical researches, such as cellular and molecular biology as well as medical genetics and immunology studies. Currently, transgene techniques, such as transfection of plasmid DNA or mRNA and viral infections, are widely used for the ectopic expression of target proteins in eukaryotic cells (Woltjen et al., 2009; Zhou and Freed, 2009). These approaches, however, often result in massive overproduction of the target proteins, altering endogenous gene expression patterns, making functional dynamics studies and omics approaches difficult to interpret. Moreover, introduction of foreign genetic materials into the host cells poses the potential danger of insertion/recombination mediated mutagenesis, making it difficult to meet the strict safety requirements for biomedical applications.
Transient protein delivery offers a safe alternative, without the concern for genome disruption or incomplete silencing of the exogenously introduced genes. The challenge of delivering proteins into mammalian cells is the limited capability of proteins to penetrate the cell membranes. It had been reported that target proteins can be delivered into mammalian cells by proteofection which involves conjugating the proteins with the cell-penetrating peptides (CPPs) to guide their translocation. CPPs are comprised of short cationic or amphipathic oligopeptides that have the ability to translocate across the plasma membrane of eukaryotic cells (Rádis-Baptista et al., 2017; Seo et al., 2017), such as the 11 amino acid peptide (TATp, YGRKKRRQRRR) from the human immunodeficiency virus (HIV) transcriptional activator TAT protein, which is rich in basic amino acids arginine and lysine (Elsayed et al., 2009; Inoue et al., 2006; Ziegler et al., 2005). When the TATp is synthesized as recombinant fusion proteins or covalently cross-linked to full-length proteins, they mediate membrane penetration for the target protein delivery into host cells. This process appears to follow an endocytotic or phagocytic pathway and does not directly transfer the cargo into the cytosol, limiting their ability to target subcellular organelles (Mackay and Szoka Jr, 2003). Although these methods are promising, the extremely low efficiency and obtaining sufficient amount of soluble pure proteins hinder their use as general methods for cellular protein delivery (Higuchi et al., 2015).
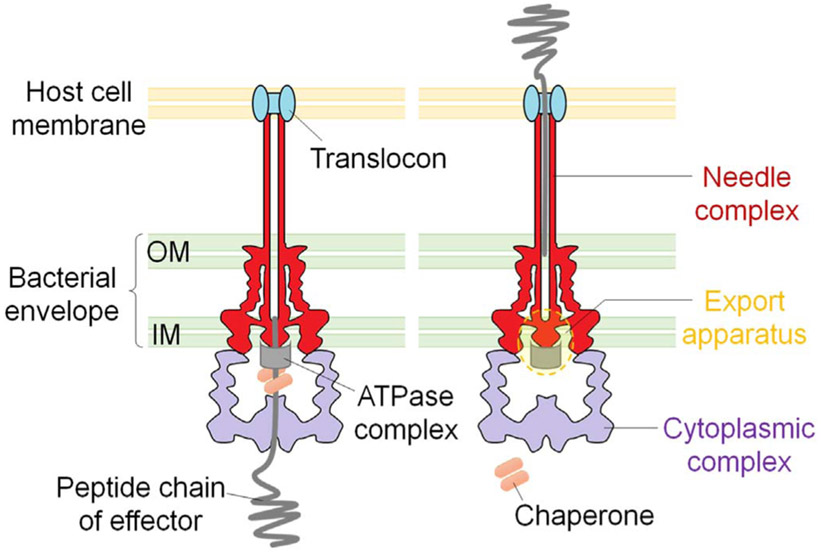
Some pathogenic bacteria have evolved complex nanomachines that allow them to deliver specific effector proteins into the cytosols of eukaryotic cells. Type III secretion system (T3SS) is a highly coordinated multiprotein system which consists of structural, regulatory and secreted proteins. The structure of the type III secretion nanomachine (or injectisome) is highly conserved among Gram-negative bacteria, such as Salmonella, Shigella, pathogenic E. coli, Pseudomonas and Yersinia (Cornelis and Wolfwatz, 1997; Hauser, 2009). Recently, a high-resolution in situ structure of Salmonella T3SS injectisome has been described following cryo-electron tomography (Hu et al., 2017). As shown in Fig. 1, the T3SS injectisome is composed of a needle complex, an inner membrane export apparatus, and a cytoplasmic platform that energizes the secretion process and selectively sorts substrates for their orderly delivery to the secretion machine (Hu et al., 2017; Lara-Tejero et al., 2011). The needle complex is composed of a multi-ring cylindrical base with ~26 nm in diameter that is anchored on the bacterial envelope and a needle-like structure that projects ~60 nm from the bacterial surface (Kubori et al., 1998; Marlovits et al., 2004). The entire structure is traversed by a channel ~2 nm in diameter that serves as a conduit for the passage of proteins injected through the type III secretion machinery (Radics et al., 2014). Protein export through the injectisome is fueled by an ATPase at the cytoplasmic sorting platform (Woestyn et al., 1994).
Fig. 1.
Schematic of type III secretion injectisome.
The T3SSs can be induced via cell-cell contact between bacteria and hosts (Rosqvist et al., 1994). In Pseudomonas and Yersinia species, when a host cell contact has been sensed by the bacteria, a set of pore-forming proteins are transported through the needle and are inserted into the eukaryotic cell membrane to form a pore of approximately 3–6 nm in diameter, called translocon. Following the pore formation, type III secretion regulatory protein (repressor) is secreted, resulting in transcriptional activation of the whole T3SS regulon genes (Rietsch et al., 2005; Urbanowski et al., 2007; Urbanowski et al., 2005). This activation results in rapid production and specific injection of the type III effectors into the target cells, without wasting the effector proteins into the culture supernatant, and thus called “polar translocation” (Sundin et al., 2004). Secretion of the type III effectors can also be triggered in the absence of host cells, requiring low calcium environment, such as in the presence of calcium chelator EGTA (Heesemann et al., 1986). The release of the type III effectors into culture medium under low calcium growth condition is T3SS-dependent but does not require the formation of translocon, thus defined as “non-polar translocation” (Vallis et al., 1999; Yahr and Wolfgang, 2006). All of the type III effectors harbor a short N-terminal secretion signal (Michiels et al., 1990), which binds to respective chaperones that stabilize them inside the bacterial cells, preventing them from premature folding while facilitating interaction with the secretion machinery, thus favor secretion (Gauthier and Finlay, 2003; Rodgers et al., 2008; Wattiau and Cornelis, 1993). The effector proteins pass through the needle complex in an unfolded or only partially folded form, and subsequently refold within the cytosol of the host cells, where they display their biological activities against various host targets (Fig.1). (Feldman et al., 2002).
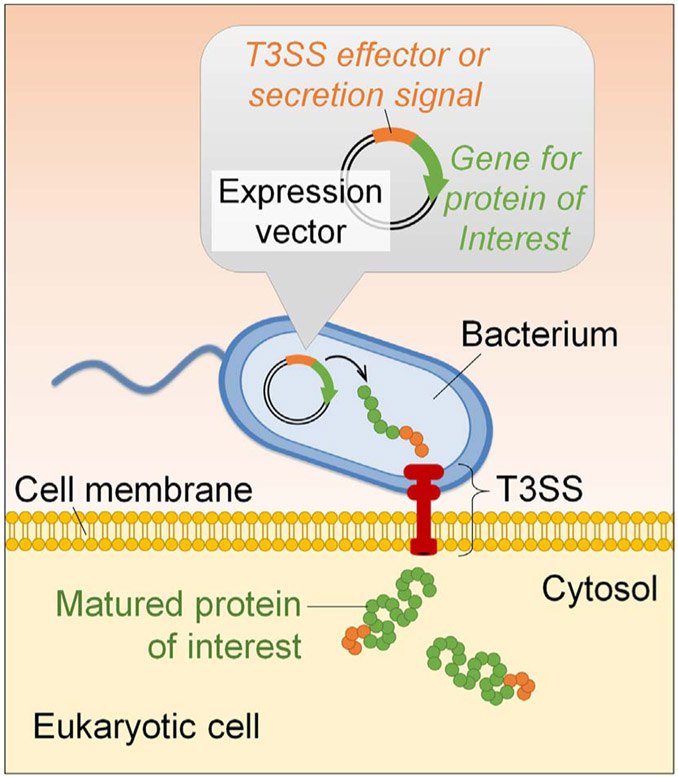
As we gain more knowledge on the biogenesis and molecular regulation of the T3SS, it has emerged as a promising tool for protein delivery directly into the target cells. When proteins of interests are fused with the type III effectors or secretion signals, the recombinant fusion proteins can be efficiently injected into the cytosols of host cells (Fig. 2). The ease of bacterial genetic and physiological manipulations has made them extremely attractive for use in biomedical applications. Below, we will focus on the applications of the bacterial T3SS as a protein delivery tool, describe successful examples of protein delivery into mammalian cells thus far for various purposes (Table 1) and discuss future works that lie ahead.
Fig. 2.
Schematic representation of bacterial T3SS-based protein delivery tool.
Table 1.
Proteins that have been delivered by the bacterial T3SSs.
| Target protein | Host cell | Delivery strain | T3SS effector fusion | Application | Ref. | |
|---|---|---|---|---|---|---|
| Antigen or epitope | Pathogens' antigen | |||||
| Influenza virus nucleoprotein (IVNP) | Antigen-presenting cells (APCs) | Salmonella enterica serovar Typhimurium (ΔaroA/sptP) | SptP-IVNP366–374 | Antiviral vaccine | Rüssmann et al., 1998 | |
| p60 antigen of Listeria monocytogenes | APCs | Yersinia enterocolitica (wild-type) | YopE1–138-p60 | Anti-bacterial vaccine | Rüssmann et al., 2000 | |
| Listeriolysin O and p60 of L. monocytogenes | APCs | S. typhimurium (ΔaroA/sptP) |
YopE1–138-listeriolysin O; YopE1–138-p60 |
Anti-bacterial vaccine | Rüssmann et al., 2001 | |
| Listeriolysin O of L. monocytogenes | Dendritic cells (DCs) | Y. enterocolitica (pYV) | YopE1–138-listeriolysin O 51–363 | Vaccination of mice against Listeria infection | Trülzsch et al., 2005 | |
| Gag proteins of simian and human immunodeficiency viruses (SIV and HIV) | APCs | S. typhimurium (ΔphoP/phoQ) |
SopE1–104-SIV-Gag4–284; SopE1–104-HIV-GagΔ392–426 |
Antiviral vaccine | Chen et al., 2006 | |
| p60 of L. monocytogenes Tumor-associated antigen (TAA) or model protein | Macrophages | S. typhimurium (ΔaroA/sptP) | SspH2-p60130–484 | Anti-bacterial vaccine | Panthel et al., 2008 | |
| MAGE | DCs | Y. enterocolitica (ΔyopHOPEM) | YopE1–130-MAGE-A1 | Stimulate specific cytolytic T lymphocytes (CTL) | Duffour, 1999 | |
| p60 from L. monocytogenes | APCs | S. typhimurium (ΔaroA/sptP) | YopE1–138-p60217–225 | Anti-tumor immunotherapy (fibrosarcoma) | Panthel et al., 2006 | |
| NY-ESO-1 | APCs | S. typhimurium (ΔphoP/phoQ) | SopE1–100-NY-ESO-1 | Anti-tumor immunotherapy (sarcoma) | Nishikawa et al., 2006 | |
| Ovalbumin (Ova) | DCs | Pseudomonas aeruginosa (ΔexoS/exoT) | ExoS1–54-Ova | Anti-tumor immunotherapy | Epaulard et al., 2006 | |
| Hepatitis B virus x (HBx) | Macrophages | S. typhimurium (ΔaroA) | SspH2-HBx | Anti-tumor immunotherapy (hepatocellular carcinoma) | Wang et al., 2008 | |
| Survivin | APCs, macrophages | S. typhimurium (ΔpurD/htrA) | SseF-Survivin | Anti-tumor immunotherapy (colon carcinoma and glioblastoma) | Xiong et al., 2010 | |
| Tyrosinase-related protein 2 (TRP2) | APCs | S. typhimurium (ΔaroA) | SopE1–100-Hsp70-TRP2153–417 | Anti-tumor immunotherapy (melanoma) | Zhu et al., 2010 | |
| TRP2 | APCs | P. aeruginosa (ΔexoS/exoT) | ExoS1–54-TRP2L125–376 | Anti-tumor immunotherapy (glioma tumor) | Derouazi et al., 2010 | |
| Pan-HLA-DR-binding epitope (PADRE); TRP2; GP100 | APCs | P. aeruginosa (ΔexoS/exoT/aroA/lasI) |
ExoS1–54-PADRE-TRP2L125–376 ExoS1–54-PADRE-GP10021–150 |
Anti-tumor immunotherapy | Wang et al., 2012 | |
| Vascular endothelial growth factor receptor 2 (VEGFR2) | APCs | S. typhimurium (ΔaroA/sptP) | YopE1–138-VEGFR2352–411 | Anti-tumor immunotherapy (melanoma) | Jellbauer, 2012 | |
| Ovalbumin (Ova) | APCs | P. aeruginosa (ΔexoS/exoT/uvrA/uvrB) | ExoS1–54-Ova | Anti-tumor immunotherapy (melanoma) | Le Gouëllec et al., 2013;Chauchet et al., 2016 | |
| Antibody | Heavy-chain antibodies (Nanobody or VHH) recognizing amylase (Vamy) and GFP (Vgfp) Nanobody recognizing EGFP (VHHGFP4) | HeLa cells | Enteropathogenic E. coli (EPEC) (Δeae/tir/map/espF) | EspF1–20-Vamy EspF1–20-Vgfp | Target specific intracellular proteins | Blancotoribio et al., 2010 |
| Nanobody recognizing EGFP (VHHGFP4) | HeLa cells | Y. enterocolitica (ΔyopHOPEMT/asd) | YopE1–138-VHHGFP4 | Nanobody-dependent subcellular localization | Ittig et al., 2015 | |
| Enzyme | Cytosolic enzymes | |||||
| Adenylate cyclase (Cya) | HeLa cells | Y. enterocolitica (wild-type) | YopE1–130-Cya | Study the translocation mechanism | Sory and Cornelis, 1994 | |
| Dihydrofolate reductase (DHFR) | – | Y. enterocolitica (ΔyopHOPEMT) | YopE1–52-DHFR | Study the type III secretion mechanism | Feldman et al., 2002 | |
| Glycogen synthase kinase (GSK) | HeLa cells | Yersinia pestis (ΔyopE) | YopE1–129-GSK | Study the translocation of T3S and T4S substrates | Garcia et al., 2006 | |
| Nucleases | ||||||
| Cre recombinase | TE26 cells; mouse ESCs and iPSCs | P. aeruginosa (ΔexoSTY) | ExoS1–54-NLS-Cre (Fig. 3) | Gene editing | Bichsel et al., 2011 | |
| TALEN | HeLa cells; mouse ESCs; human ESCSs and iPSCs | P. aeruginosa (ΔexoSTY/ndk/xcpQ/lasI/rhlI/popN) | ExoS1–54-NLS-TALEN (Fig. 3) | Gene editing | Jia et al., 2014; Jia et al., 2015 | |
| Transcription factor | MyoD | Mouse embryonic fibroblasts (MEF) | P. aeruginosa (ΔexoSTY) | ExoS1–54-MyoD (Fig. 3) | Muscle cell reprogramming | Bichsel et al., 2013 |
| Gata4, Mef2c and Tbx5 | Mouse ESCs | P. aeruginosa (ΔexoSTY/ndk/xcpQ/lasI/rhlI/popN) | ExoS1–54-Gata4 (Fig. 3) ExoS1–54-Mef2c (Fig. 3) ExoS1–54-Tbx5 (Fig. 3) | Cardiomyocytes diffferentiation | Bai et al., 2015 | |
| Oct4, Sox2 and Nanog | human fibroblasts and hematopoietic human stem cells | P. aeruginosa (ΔexoST) | ExoS1–54-Oct4 | Induction of the pluripotency; cell reprogramming | Berthoin et al., 2016 | |
| Fluorescent protein | GFP | HEp-2 cells | Y. enterocolitica (wild-type) | YopE1–138–GFP | Study of protein translocation via T3SS | Jacobi et al., 1998 |
| The 11th strand of the GFP β-barrel (GFP11) | HeLa cells | S. typhimurium (ΔpipB2) | PipB2-GFP11; SteA-GFP11 | Tracking intracellular localization of effectors | Van Engelenburg and Palmer, 2010 | |
| EGFP and mCherry | HeLa cells | Y. enterocolitica (ΔyopHOPEMT/asd) | YopE1–138-EGFP; YopE1–138-EGFP-NLS; YopE1–138-mCherry; YopE1–138-NLS-mCherry | Tracking intracellular localization of translocated proteins | Ittig et al., 2015 | |
| Others | Neutrophil cytosolic factor p67-phox | p67phox-deficient lymphocytes | P. aeruginosa (wild-type) | ExoS1–129-p67phox | Cellular therapy | Polack, 2000 |
| Anti-inflammatory cytokines IL-10 and IL-1 receptor antagonist (IL-1ra) | Lung tissue | Shigella flexneri (wild-type) | YopE1–50-IL-10; IpaH1–60-IL-10; IpaH1–60-IL-1ra | Immunomodulation | Chamekh, 2008 | |
| Bacterial T3SS effectors | HeLa cells | Y. enterocolitica (ΔyopHOPEMT) | YopE1–138-IpgB1 YopE1–138-IpgB2 YopE1–138-Map | Study the cell biological effects of bacterial effectors | Wölke et al., 2011 | |
| Pro-apoptotic protein: BH3 interacting-domain death agonist (BID) and truncated BID (tBID) | HeLa cells | Y. enterocolitica (ΔyopHOPEMT/asd) | YopE1–138-BID YopE1–138-tBID | Study of mechanism of apoptosis/intracellular signal transduction | Ittig et al., 2015 | |
2. Engineering bacteria for heterologous protein delivery
Many pathogenic bacteria encode T3SS as a virulence factor, such as species of Shigella, Salmonella, Enteropathogenic Escherichia coli (EPEC), Yersinia, Pseudomonas and phytopathogenic bacteria Xanthomonas campestris that cause bacillary dysentery, typhoid fever, diarrhea, plague and various animal and plant infections, respectively. T3SSs of these pathogens induce high cytotoxicity via type III effectors that tamper with the host cell signal transduction and in some cases induce apoptosis (Deslandes and Rivas, 2012; Gong et al., 2010; Hilbi et al., 1998). To make use of the bacterial T3SS in the protein delivery into eukaryotic cells, they must be attenuated first to minimize their pathogenic capacity. Three most widely studied delivery strains, Salmonella spp., Yersinia spp. and Pseudomonas aeruginosa, have all been subjected to extensive genetic alterations for attenuation (Table 1).
Recombinant attenuated Salmonella typhimurium strains have been widely investigated as delivery systems for heterologous antigens because of their ability to induce humoral, mucosal, and cellular immune responses after oral administration (Panthel et al., 2008). Several modulating strategies have been developed in order to attenuate virulence while retaining its adjuvant capacity. Modifications of virulence-associated factors, such as lipopolysaccharide synthesis (ΔmsbB), type III effector (ΔsptP) and a master two-component regulatory system (ΔphoP/phoQ), were employed to attenuate the Salmonella (Crull et al., 2011; Frahm et al., 2015). In addition, metabolic mutations (auxotroph) affecting the synthesis of cell wall (Δasd), nucleotides (ΔpurI, ΔpurD), or amino acids (ΔaroA, resulting in blockade of chorismate biosynthesis which is the precursor for aromatic amino acids and siderophores) were also used to attenuate the delivery strain (Arrach et al., 2010; Clairmont et al., 2000; Xin et al., 2012). The combination of auxotrophic mutations and defect in virulence factors enabled the Salmonella to maintain the wild-type phenotype in vitro but became self-limiting in vivo during host infection (Juárez-Rodríguez et al., 2012).
In Salmonella, there are two types of T3SSs, encoded by pathogenicity island-1 (SPI-1) and pathogenicity island-2 (SPI-2) (Hapfelmeier et al., 2005). The SPI-1 T3SS is expressed and assembled in the early stages of infection, during which host cell invasion by bacteria is facilitated by the SPI-1 T3SS (Crull et al., 2011). In contrast, the SPI-2 T3SS, first discovered in 1996 by Holden's group (Shea et al., 1996), is assembled in systemic infection stage, during which bacteria reside within phagocytic vacuoles of the host cells, thus the SPI-2 T3SS ensures the necessary communication between bacteria and host cells. Both SPI-1 and SPI-2 T3SS have their unique effectors, many of them have been utilized as the type III secretion signal for heterologous protein delivery (Table 1).
Yersinia species (Y. pestis, Y. enterocolitica and Y. pseudotuberculosis) encode T3SS on a virulence plasmid (pYV) which enables them to overcome the host immune defense and survive in the lymphoid tissues (Trülzsch et al., 2003). This virulence apparatus allows bacteria to deliver toxic effectors, called Yops (Yersinia outer proteins), inside the host cells to disrupt normal intracellular activities (Cornelis and Wolfwatz, 1997). With the identification of Yersinia T3SS effectors, a variety of attenuated strains have been constructed, such as strain ΔYopHOPEMT, in which all known Yop effectors have been deleted or truncated (Boland and Cornelis, 1998; Iriarte and Cornelis, 1998). This strain was further made deficient of the asd gene, making the bacteria unable to proliferate in the absence of exogenous meso-diaminopimelic acid (DAP) (Hoang et al., 1997; Kudryashev et al., 2013). This multi-gene mutant strain maintains its ability to translocate effector proteins into the cytosols of eukaryotic cells but does not elicit cytotoxicity, and thus can be used as a protein delivery vector (Ittig et al., 2015).
Another attenuated bacterium, P. aeruginosa, has also been widely investigated as T3SS-based protein delivery tool. P. aeruginosa is an extracellular pathogen whose T3SS function prevents bacteria from getting internalized by the phagocytic cells. Approximately forty genes, encoded in five operons that are clustered together on the P. aeruginosa chromosome, are involved in the biogenesis and regulation of the T3SS. Once T3SS is activated by direct contact with the host cells, P. aeruginosa rapidly injects proteinous exotoxins, ExoS, ExoT, ExoY, ExoU, and NDK, into host cells via the T3SS injectisome. These toxins possess various enzymatic activities to influence the physiology of host cells. For instance, ExoS possesses both ADP ribosyltransferase and GTPase activating activities, triggering rapid apoptosis in various host cells upon injection (Kaufman et al., 2000). To utilize P. aeruginosa T3SS as a tool for protein delivery, various attenuated strains have been generated by genetic deletion of genes implicated in the bacterial virulence, including the exotoxin genes (exoS, exoT, exoY and ndk) (Bichsel et al., 2011; Epaulard et al., 2006; Neeld et al., 2014), the quorum-sensing genes (lasI and rhlI) which coordinate the expression of virulence genes and biofilm formation (Hentzer et al., 2003), and the type II secretion gene (xcpQ) which is responsible for the secretion of various exotoxins (such as exotoxin A, elastase etc.) (Sandkvist, 2001). An auxotroph ΔaroA has also been used as a metabolic mutation to attenuate P. aeruginosa (Epaulard et al., 2008). Furthermore, a “killed but metabolically active” (KBMA) P. aeruginosa strain was constructed and investigated as an in vivo antigen delivery vector. Besides disarmament by deleting virulence genes, the KBMA strain was also deleted of two additional genes, uvrA and uvrB, which encode an exonuclease that is involved in the nucleotide excision repair mechanism. Following photo-inactivation, long-wave UV light radiation in the presence of DNA crosslinking agent amotosalen, the uvrAB double mutant bacteria are doomed to die due to their inability to replicate, although their T3SS remains active (Chauchet et al., 2016; Le Gouëllec et al., 2013).
Recently, a D-glutamate auxotroph has been applied as an effective strategy for virulence attenuation and self-limited growth of various bacterial pathogens. A P. aeruginosa ΔmurI mutant, defective of D-glutamate synthesis that is essential for bacterial cell wall synthesis (Fisher, 2008), showed a requirement for exogenous D-glutamate for growth. As no D-amino acids are available in mammals, the P. aeruginosa ΔmurI mutant can be eliminated as quickly as 10 h after intravenous administration in a mouse model, much shorter persistence time than that of ΔaroA auxotrophic strain (3–4 days) (Cabral et al., 2017). For T3SS-mediated protein delivery, an important issue is how to quickly remove bacterial vectors upon completion of the protein delivery. The D-glutamate auxotrophy allows scientists to rapidly eliminate bacteria both in vitro and in vivo.
It should be noted that wild-type Yersinia and Pseudomonas are extracellularly located because of the cell-paralyzing effect of injected effectors on phagocytic cells. Defect in the effector injection, however, results in rapid uptake of Yersinia and Pseudomonas by the phagocytic cells. Salmonella on the other hand, are rapidly internalized into non-phagocytic epithelial cells by SP-1 T3SS-injected effectors. (Galán et al., 2014; Hauser, 2009).
3. Antigen delivery — vaccination
The bacterial T3SS has been exploited to deliver antigenic peptides and proteins into various target cells. Type III effectors were shown efficiently injected into a wide range of host cells, including professional antigen presenting cells (APC), such as macrophages and dendritic cells (DCs) (Toussaint et al., 2013). The premise behind the use of this system for vaccine delivery is that the bacteria can inject the antigen directly into the cytoplasm of the APCs for processing and presentation to activate T cell response against the specific antigen. Viral and bacterial epitopes, as well as peptides from human tumors, have been delivered by the bacterial T3SS with the aim to elicit immune response (vaccination) or cancer immunotherapy (Panthel et al., 2006).
Russmann et al. have first demonstrated the utility of S. typhimurium T3SS as a tool to deliver heterologous antigenic protein inside host cells to induce class I-restricted immune responses (Rüssmann et al., 1998). The epitope of influenza virus nucleoprotein (IVNP366–374) was fused to a Salmonella type III secreted effector SptP and was successfully delivered into the cytosol of mouse thymoma cells in a T3SS-dependent manner. When mice were orally vaccinated with an attenuated S. typhimurium strain expressing the SptP-IVNP366–374, specific cytotoxic T lymphocyte (CTL) responses were induced, resulting in protection of vaccinated animals against lethal infections by the influenza virus (Rüssmann et al., 1998). Since then, several groups have utilized attenuated Salmonella species for antigen delivery by fusing the antigenic protein epitopes to various type III effectors (Chen et al., 2006; Nishikawa et al., 2006; Panthel et al., 2008; Rüssmann et al., 2001; Wang et al., 2008; Xiong et al., 2010). Through the T3SS mediated delivery of various pathogen-associated antigens, CD4 and CD8 T-cell responses have been elicited against specific antigens, including Listeria monocytogenes p60 protein as well as simian and human immunodeficiency virus Gag proteins, providing robust protection against challenges by the respective wild-type pathogens (Chen et al., 2006; Panthel et al., 2008; Rüssmann et al., 2001). In addition to Salmonella, the Yersinia T3SS has also been used to deliver the model antigens listeriolysin O and p60 of L. monocytogenes into APCs. Successful vaccination of mice against Listeria infection was correlated with protective CD8 T-cell induction. (Rüssmann et al., 2000; Trülzsch et al., 2005).
The live-attenuated Salmonella vectors for antigen delivery have also attracted growing interests in the field of tumor immunotherapy. Although tumors express tumor-associated antigens (TAAs), cancer vaccines often fail due to inadequate antigen delivery and/or insufficient activation of innate immunity. Engineering non-pathogenic bacterial vectors to deliver TAAs of choice provided an efficient way of presenting TAAs in an immunogenic form (Toussaint et al., 2013). The effectiveness of Salmonella T3SS-based antitumor vaccines had been tested and confirmed in various tumor models. TAA survivin was fused to a T3SS effector SseF and the fusion protein was expressed and translocated into the cytosols of murine macrophages by an attenuated strain of S. typhimurium, MvP728 (deleted of purD/htrA). In vivo coadministration of the MvP728 expressing SseF-survivin with a ligand for CD1d-reactive NKT cells, α-glucuronosylceramide (GSL1), enhanced effector-memory CTL responses. Furthermore, a combined use of MvP728/survivin with GSL1 produced antitumor activity in mouse models of CT26 colon carcinoma and orthotopic DBT glioblastoma (Xiong et al., 2010). Other antitumor applications include the Salmonella T3SS-meditated tyrosinase-related protein 2 (TRP2) epitope delivery for melanoma (Zhu et al., 2010), hepatitis B virus x (HBx) delivery for hepatocellular carcinoma (Wang et al., 2008) and NY-ESO-1 tumor antigen delivery for sarcoma (Nishikawa et al., 2006). Moreover, the Salmonella genus itself possesses inherent tumor targeting capacity coupled to antitumor activity. A genetically attenuated S. typhimurium strain VNP20009 (purI/msbB double mutant) was observed to colonize in high tumor-to-normal tissue ratios (range from 250: 1 to 25,000: 1 compared to the liver) and inhibit tumor growth in mice (Nemunaitis et al., 2003). However, in a clinical trial of intravenous VNP20009 administration to a heterogeneous group of cancer patients did not result in significant antitumor effect. (Toso et al., 2002). In another clinical study, intratumoral injection of the VNP20009 expressing an E. coli cytosine deaminase showed certain extent of antitumor effect without significant adverse reactions (King et al., 2009; Nemunaitis et al., 2003). However, it appears that this approach is no longer under active development (Toussaint et al., 2013).
P. aeruginosa T3SS has also been applied for antigen delivery studies. ExoS is one of the best studied T3SS effectors in P. aeruginosa, and its N-terminal 54 amino acids are sufficient to serve as a secretion signal (ExoS1–54) for T3SS-dependent secretion or injection (Bichsel et al., 2011; Epaulard et al., 2006). Heterologous antigens can be translocated into various APCs by fusing with the ExoS1–54 (Derouazi et al., 2010; Wang et al., 2012). Tumor antigens of various length and epitope composition, including TRP-2, gp100, and MUC18, were evaluated against glioma tumor cells. CTL immunity and T-cell receptor (TCR) repertoire diversity were investigated following the vaccination (Derouazi et al., 2010). This work demonstrated that P. aeruginosa T3SS is suitable for rapid screen and evaluation of tumor antigens of varying length and epitope composition. In addition, the ‘killed but metabolically active’ (KBMA) P. aeruginosa strain showed promising effects in antitumor immunotherapy as a safe antigen delivery vector. A model protein OVA257–264 (SIINFEKL) epitope from chicken ovalbumin was co-expressed with ExoS1–54 and shown able to translocate into antigen-presenting cells in vitro via T3SS with considerably attenuated cytotoxicity as compared to the wild-type vector. In a mouse model of cancer, the KBMA strain, which cannot replicate in its host, efficiently disseminates into lymphoid organs and triggers antigen-specific CD8 T cell priming, resulting in protection of the mice against aggressive B16-OVA melanoma development (Epaulard et al., 2006; Le Gouëllec et al., 2013; Wang et al., 2012). In addition, multiple rounds of KBMA immunization triggered a long-lasting immune response, resulting in a pool of predominantly effector memory cells that protected the mice from tumor challenge (Chauchet et al., 2016).
4. Nuclease delivery — genome editing
Recently developed genome editing tools for mammalian cells, TALEN and CRISPR, revolutionized the whole biomedical research field (Kim et al., 2017; Komor et al., 2016; Li et al., 2015). The most promising field of their applications is genome editing of pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), for disease modeling, drug screening and replacement therapy (Santostefano et al., 2015). In the past few years, our laboratory has been exploring the application of P. aeruginosa T3SS in the delivery of various nucleases, such as Cre recombinase and TA-LENs, into mammalian cells, including mouse and human PSCs (Bichsel et al., 2011; Jia et al., 2015; Jia et al., 2014).
4.1. A P. aeruginosa–based delivery toolbox
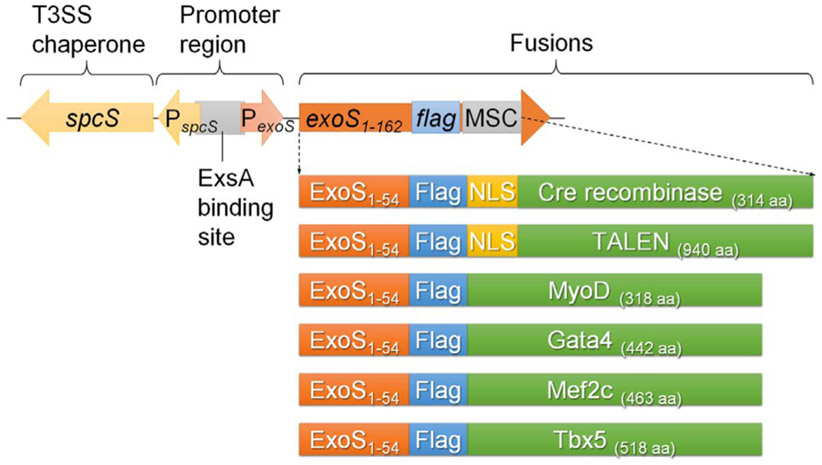
An attenuated P. aeruginosa strain Δ8, deleted of 7 virulence-related genes (exoS/T/Y, ndk, xcpQ, lasI, rhlI) and one T3S suppressor gene (popN), was employed as the protein delivery vector (Bai et al., 2015; Jia et al., 2015; Neeld et al., 2014). Proteins of interest were cloned and expressed on an Escherichia-Pseudomonas shuttle expression plasmid (Fig. 3), which encodes the T3S effector ExoS promoter with N-terminal ExoS1–54 signal sequence, followed by a FLAG tag and a multiple cloning site (MCS). Also on the vector, an intact spcS gene encoding the chaperone for the ExoS (Fig. 3) (Shen et al., 2008). Proteins of interest can be fused in-frame utilizing the MCS and the fusion proteins can be detected by following the FLAG tag. Under the guidance of ExoS1–54 secretion signal and the assistance of ExoS chaperone (SpcS), the target proteins can be efficiently injected into mammalian cells via the T3SS. Moreover, by incorporation of a nuclear localization sequence (NLS), the target proteins can be translocated into the nucleus of mammalian cells (Bichsel et al., 2011).
Fig. 3.
Expression vector of a P. aeruginosa T3SS-based protein delivery toolbox and the fusion proteins that had been effectively translocated inside host cells by using this toolbox. ExsA is the master regulator for P. aeruginosa T3SS.
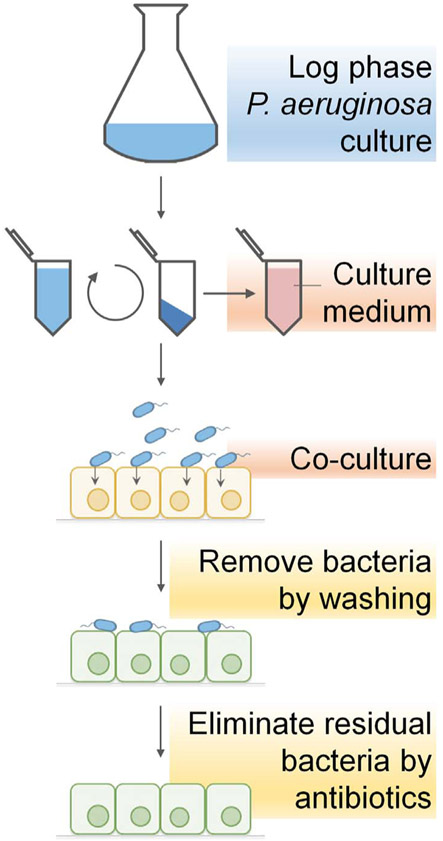
The manipulation of protein delivery involves a simple bacterial infection procedure (Fig. 4), which includes growing attenuated P. aeruginosa strain that contains ExoS1–54-protein fusion expression vector and co-culturing with the target mammalian cells. During the co-culture, T3SS of P. aeruginosa is fully activated upon contact with the host cells, the ExoS1–54 fusions are then rapidly expressed and injected into the host cells. Infection is terminated by washing to remove the floating bacterial cells (remove > 90% bacteria cells), and then further culturing the cells in medium containing antibiotics to eliminate residual bacterial cells. Complete elimination of the residual bacterial cells can be achieved with an overnight treatment of a membrane permeable antibiotic, ciprofloxacin (Bai et al., 2015).
Fig. 4.
Schematic representation of the infection procedure of P. aeruginosa T3SS-based protein delivery.
4.2. Cre recombinase delivery
The Cre-LoxP system was used to demonstrate the utility of this T3SS mediated protein delivery toolbox (Bichsel et al., 2011). An ExoS1–54-Cre fusion was delivered into TE26 cells by the T3SS of an attenuated P. aeruginosa. The TE26 is a human sarcoma cell line with a lacZ reporter gene preceded by a floxed SV40 transcriptional terminator that prevents the downstream lacZ expression. Following 3 h of co-culture with the bacteria at multiplicity of infection (MOI) of 100, nearly 100% of the TE26 cells were detected positive for the injected Cre fusion protein. As an NLS was fused to the Cre, the T3SS-delivered Cre was able to translocate into the nucleus of the target cells. Upon Cre mediated recombination, the DNA between loxP sites is removed, allowing lacZ expression which can be evaluated by β-galactosidase activity. Bacterially delivered Cre induced as high as 42% of the TE26 cells to express β-galactosidase, indicating not every Cre injected cells underwent loxP mediated recombination. Interestingly, the proportion of LacZ positive cells was similar to the cell population in S phase of the cell-cycle. When the TE26 cells were synchronized, obtaining 78% of the cell population in S phase, the Cre delivery resulted in 75% β-galactosidase positive cells, suggesting that Cre-mediated DNA recombination mainly occurs among cells in the S phase of cell-cycle, presumably when the loxP sites are freely accessible to the Cre recombinase (Bichsel et al., 2011). By using the same T3SS-Cre delivery tool, a floxed reprogramming cassette, containing four transcription factors Oct4-Sox2-c-Myc-Klf4, has also been efficiently excised from a mouse embryonic fibroblast-derived iPSC (Bichsel et al., 2011). This work also demonstrated for the first time that functional nuclear proteins can be delivered through the bacterial T3SS.
4.3. TALEN protein delivery
Transcription activator-like effector nuclease (TALEN) is a genome editing tool that specifically recognizes target sequence as a dimer and introduces a double-strand DNA break (DSB) on the target site. The DSBs can then be repaired by either homologous recombination (HR) or non-homologous end joining (NHEJ) (Wright et al., 2014). TALEN is a protein fusion of a DNA-binding domain from transcription activator-like effector (TALE) and a DNA cleavage domain from FokI endonuclease. The TALE can be engineered to bind to any desired DNA sequence, so when combined with a nuclease activity, DNA can be cut at intended locations (Cermak et al., 2011). In nature, TALEs are secreted by Xanthomonas bacteria via their T3SS when they infect plants (Boch and Bonas, 2010; Drehkopf et al., 2017). In 2014, our laboratory demonstrated that TALEN can also serve as substrate for the T3SS of P. aeruginosa (Jia et al., 2014). By fusing with P. aeruginosa type III secretion signal ExoS1–54, a pair of gfp-targeting TALEN was efficiently delivered into HeLa cells and introducing DSBs in the target site of venus gene that had been integrated into the HeLa cell genome. Despite a large sized protein (~110-kD), P. aeruginosa T3SS is able to efficiently inject the ExoS1–54-TALEN fusion protein into host cells, reaching almost 100% efficiency with MOI > 50 in a short infection time (1 ~ 3h). With the integration of an NLS to the N-terminal of TALENs, the T3SS-delivered TALENs were efficiently translocated into the nucleus of target cells where they remain intact for ~10 h before being degraded. Bacterial delivery of the TALENs resulted in ~20% cells lost of their GFP which was further confirmed of DNA alterations at the intended target site, presumably resulted from NHEJ (Jia et al., 2014).
The same strategy was also shown functional in mouse embryonic stem cells (mESCs) expressing a gfp transgene. Following 3h of co-culture at MOI of 100, the bacterial T3SS efficiently delivered the TALEN proteins into the mESC nucleus where they remained detectable for 8 h. The T3SS mediated TALEN delivery resulted in at least two-fold higher GFP-targeting efficiency than that by transfection of the TALEN-coding plasmids. Further, in combination with a single-stranded oligonucleotide DNA template, the T3SS injected TALENs effectively introduced a single nucleotide change in the GFP gene via homologous recombination, converting a “GAG” to a “TAG” stop codon, thus switching off the GFP. This approach resulted in a higher efficiency of target site modification (~3 folds) than the conventional transfection method (Jia et al., 2015).
The T3SS mediated TALEN delivery was further demonstrated effective in human ESC as well as human iPSCs. In a human ESC that expresses GFP, T3SS mediated delivery of GFP-targeting TALENs induced GFP silencing at a greater efficiency than that by plasmid transfection. Also, T3SS mediated delivery of TALENs that target HPRT1, a gene associated with human neurological disease (Cristini et al., 2010), efficiently mediated DSBs at the HPRT1 site, and the introduction of a single-stranded oligonucleotide, designed for homologous recombination, resulted in high-efficiency modification of the target gene (Jia et al., 2015). These studies demonstrated the utility of T3SS in the delivery of genome editing nucleases to achieve a high efficiency target gene modification.
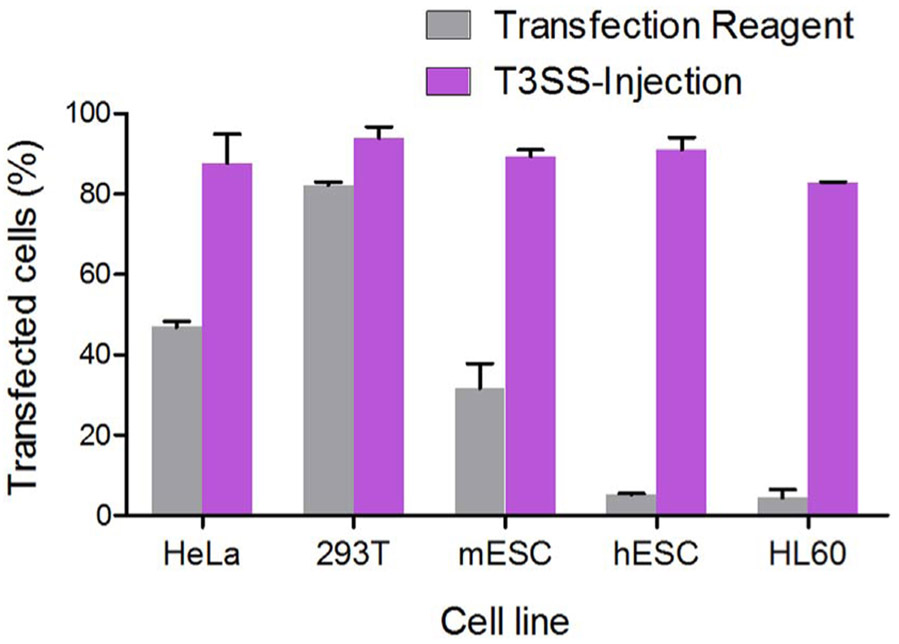
For precise genome editing in PSC, T3SS-based DNA editing enzyme delivery has two major advantages: First, nucleases are introduced in the form of protein rather than nucleic acids, thus allows us to control not only the amount but also duration of the nuclease inside the target cells. For the DNA based transfection or transduction, persistent gene expression results in excessive nuclease activity which increases the chance of stochastic insertions and deletions (Kim et al., 2017). Second, T3SS-mediated protein injection is highly efficient for pluripotent stem cells (PSCs). Human PSCs, in particular, are recalcitrant to DNA transfection or viral transduction, therefore currently available techniques yield extremely low genome editing efficiency (Komor et al., 2016; Li et al., 2015). As shown in Fig. 5, transfection rates for plasmid DNA are high in 293T and HeLa cells but very low for the human ESCs (H9) and promyelocytic leukemia cells (HL60). However, the T3SS mediated protein delivery is high in all four cell types, reaching close to 90% delivery efficiencies.
Fig. 5.
Efficiency comparison of transfection and T3SS-mediated protein delivery. For transfection, different cells were transfected with plasmid (pFLAG-CMV2-iExoS) encoding the FLAG-tagged inactive ExoS (E381A, iExoS for short) by a commercially available transfection reagent. For T3SS-injection, cells were infected by attenuated P. aeruginosa Δ8 expressing FLAG-iExoS fusion. The number of transfected cells was assessed 24 h posttransfection (grey bars) or 4 h post-injection (purple bars) by labeling the cells with anti-FLAG antibody and then flow cytometric analysis. Error bars represent standard deviation of triplicate FACS assays. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The T3SS mediated nuclease delivery method does have limitations. The most significant one relates to the editing efficiency being restricted by the cell cycle status. As the DNA recombination mediated by Cre or TALEN mainly occurs during the S phase of cell-cycle, only a portion of the whole target cell population can undergo recombination. Considering generation times of the target cells, ~15 h for human PSCs and ~25 h for somatic cells (Hindley and Philpott, 2013), the ~6 h half-life of the translocated nuclease (TALEN) inside host cells is not long enough to edit the whole cell population, despite > 90% cells being injected of the nuclease. Engineered TALENs with higher intracellular stability should resolve the issue. Alternatively, multiple rounds of nuclease delivery may also overcome the limitation.
Another genome editing tool, CRISPR-Cas9, has aroused significant interests of the academic community. The ease with which the reagents work in different hands, the wide variety of proven model systems, and the ease with which guide RNAs can be designed for the endonuclease Cas9, drove the rapid uptake of CRISPR in academic laboratories all over the world (Komor et al., 2016). Our laboratory has attempted to deliver Cas9 by P. aeruginosa T3SS. Unfortunately, the fusion of ExoS1–54-Cas9 caused significant toxicity to the P. aeruginosa. Recent studies demonstrated that Cas9 can be split into two distinct fragments, the N- and C-terminal fragments, which spontaneously form a functional Cas9 nuclease when brought together (Truong et al., 2015; Zetche et al., 2015). Future attempts should include T3SS mediated delivery of the N- and C-terminal Cas9 fragments separately, allowing functional Cas9 to form inside the host cells. Along the line, efficient delivery of gRNA will be another significant hurdle that needs to be overcome.
5. Transcription factors delivery—directing cell fate
With the advent of iPS cells generation a decade ago by forced expression of four transcription factors (Takahashi et al., 2007; Takahashi and Yamanaka, 2006), it has been well documented that transcription factor (TF) delivery is an effective way to direct cellular reprogramming as well as stem cell differentiation (Chambers and Studer, 2011; Cherry and Daley, 2012; Yamanaka and Blau, 2010). In 2013, our group demonstrated that the P. aeruginosa T3SS-based protein delivery method could be used to deliver single transcription factor into somatic cells to induce cell type conversion, or cellular reprogramming (Bichsel et al., 2013). MyoD is a key muscle regulatory factor, overexpression of which is able to induce transdifferentiation of numerous cell types into functional myocytes (Puri and Mercola, 2012). Using a genetically attenuated strain of P. aeruginosa, MyoD was delivered into mouse embryonic fibroblasts using the T3SS (Fig. 3). Translocated MyoD protein effectively activated muscle-specific gene expression, resulting in 30% of the fibroblasts converted into myocytes one week later (Bichsel et al., 2013). Recently, by using Pseudomonas T3SS and ExoS1–54 secretion signal, the core embryonic transcription factor Oct4, Sox2 and Nanog have been delivered efficiently into the nucleus of primary human fibroblasts and cord blood CD34+ hematopoietic stem cells, inducing a strong activation of the pluripotency gene expression (Berthoin et al., 2016).
Besides reprogramming, T3SS-base TF delivery has also been used to direct de novo differentiation from pluripotent stem cells. Using the P. aeruginosa T3SS-based protein delivery method, three transcriptional factors (GATA4, MEF2c, and TBX5) had been translocated into murine embryonic stem cells (mESCs), effectively targeting into nucleus, with an average intracellular half-life of 5.5 h. Exogenous TFs delivery activated the endogenous cardiac gene expression, and multiple rounds of TFs injection significantly improved the efficiency of de novo cardiomyocytes differentiation. Interestingly, the T3SS-mediated TFs delivery showed an additive effect with activin A which is a growth factor with the ability to induce mesodermal fate at a proper stoichiometry (Bai et al., 2015). More recently, five human transcriptional factors, namely GATA4, MEF2C, TBX5, ESRRG, and MESP1, were efficiently translocated into human ESCs as well as human iPSCs by using the same strategy, resulting in a significant enhancement in the cardiomyocyte differentiation (unpublished data).
The PSC-derived cardiomyocytes described to date exhibit cardiac subtype heterogeneity and rather immature compared to adult cardiomyocytes (Rajala et al., 2011), including embryonic atrial-, ventricular- and nodal-like cardiomyocytes as defined by intracellular electrophysiological measurements of action potentials (APs) (Zhang et al., 2011). Swine transplantation studies have shown that implanted hESC derived cardiomyocytes have pacemaking activities, which is a potential cause of ventricular arrhythmias (Kehat et al., 2004). Future challenges lie on the derivation of specific subtypes and maturation of PSC-derived cardiomyocytes, which will be crucial for their clinical use (David and Franz, 2012). Recently, several studies demonstrated the reprogramming effect of various transcription factors on cardiomyocyte subtypes commitment (Ionta et al., 2015; Jung et al., 2014; Kapoor et al., 2013). In the future, the T3SS-based protein delivery system can be used to deliver specific factors into cardiac progenitor cells to drive cell type-specific cardiomyocytes, such as ventricular, atrial or pacemaker cells.
6. Signaling protein delivery—study of intracellular signal transduction
Intracellular protein function can be investigated by cDNA transfection, microinjection, or protein transduction (or proteofection) of purified proteins. However, they often result in the overrepresentation of the target protein and/or highly heterogeneous cell populations, making downstream studies difficult to interpret. In addition, protein purification leads to high cost and technical difficulties for certain types of proteins. Many intracellular signal transduction processes involve posttranslational modifications, such as phosphorylation, methylation, acetylation or cleavage (Hynes et al., 2013; Olsen and Mann, 2013). Due to the transient nature of such signaling events, the influence of a particular signaling protein on the global cellular network is difficult to dissect using conventional methods. Fortunately, using T3SS mediated direct injection of “active” signaling proteins, researchers were able to dissect the influence of particular signaling molecule on a global cellular network. A Y. enterocolitica T3SS-based protein delivery toolbox was developed to investigate the intracellular function of effectors from pathogens and transient signaling protein from host cell itself (Ittig et al., 2015; Wölke et al., 2011).
Phagocytosis, cell migration and regulation of epithelial cell monolayer integrity are central cellular aspects that are controlled by Rho GTPase family members (Jaffe and Hall, 2005). Bacterial toxins and regulatory proteins targeting these cellular switches are very effective pathogenic factors. However, research on the intracellular function of the GTPase-targeting effectors is hindered by their high cytotoxicity, unable to obtain cell clones that successfully express the toxins after transfection. This difficulty was overcome by employing a Yersinia T3SS toolbox (Trülzsch et al., 2003; Wölke and Heesemann, 2011). Using this toolbox, GTPase-targeting effectors IpgB1 and IpgB2 of Shigella and Map of Escherichia coli were fused with the N-terminal secretion domain of YopE and delivered into HeLa cells. This strategy lead to the studies of cellular functions of the effectors from diverse pathogens in the context of bacterial-host interaction, avoiding the artificial approach of ectopically expressing the effectors.
In addition, the above Yersinia toolbox has been used to investigate the mechanism of apoptosis. BH3 interacting-domain (BID) is a proapoptotic member of the Bcl-2 protein family that can be cleaved by caspase-8 (Sarosiek et al., 2013). The truncated BID (tBID) translocates into mitochondria and triggers apoptosis via cytochrome c release and caspase 3 activation (Li et al., 1998). The systematic impact of tBID on cellular signaling was investigated by fusing with the N-terminal 138-amino-acid fragment of Yersinia T3S effector YopE and translocating into HeLa cells. The translocated tBID triggered massive phosphorylation events that affect multiple cellular functions, confirming the central role of caspase 3 in this process (Ittig et al., 2015). Unlike plasmid transfection mediated expression of the tBID, T3SS mediated direct injection of the tBID protein enabled them to analyze the real-time changes in target cell signal transduction pathways, assessing the cellular networks influenced by the tBID.
Analyzing protein functions is a key focus in the post-genomic era. The protein injection machinery allows researchers to investigate the functions of proteins by directly manipulating their intracellular levels. This fast change occurs directly at the protein level, rather than at the mRNA or promoter induction, thus enables us to address protein functions independent of their gene transcription or translation. This is especially important in cases where the gene product is essential for viability, as gene knockout is lethal while knockdown causes severe growth defect.
7. Reporter protein delivery
To investigate the subcellular localization and molecular interaction of a protein of interest, fusion expression with reporter proteins (such as GFP, β-galactosidase or β-lactamase) is an effective strategy. T3SS-based reporter protein delivery can be used to study the mechanism of translocation itself as well as the cellular function of the effector proteins secreted by diverse bacterial pathogens. Adenylate cyclase (Sory and Cornelis, 1994), dihydrofolate reductase (DHFR) (Feldman et al., 2002), and glycogen synthase kinase (GSK) tag (Garcia et al., 2006) were used as reporters of translocation to identify the secretion signal for T3SS.
It is also possible for real-time tracking of the movement trajectory of T3SS effectors within the host cells throughout the course of infection. Fluorescent protein fusion is a widely used technique for tracking the dynamical movement of the protein of interest in living cells. Among a large number of fluorescent proteins, GFP is the most commonly used one. However, GFP is a notoriously poor substrate for the T3SS. The reason may well be the GFP's inability to maintain unfolded form to fit through the needle complex of the T3SS (Jacobi et al., 1998). Interestingly, a split-GFP method was successfully utilized (Van Engelenburg and Palmer, 2010). The 11th strand of the GFP β-barrel, which is composed of 13 amino acids, was fused to Salmonella SPI-2 T3SS effectors (PipB2, SteA, or SteC, respectively) and the complementary fragment corresponding to the first 10 strands of GFP was expressed in the host cells. Upon T3SS effector translocation, complementation of the two fragments result in fluorescent tagging of the effector population in the host cell. The PipB2 displayed a highly dynamic behavior on tubules emanating from the Salmonella containing vacuole (SCV) during late stages of infection. SteA displayed fluorescent signal on the SCV and tubules, enriched in Golgi of the host cell. While SteC localized predominantly to the SCV-associated actin nests. However, the split-GFP method is not suitable to track effectors whose actions are instantaneous after translocation, as reconstitution of the fluorescence requires mature time (Van Engelenburg and Palmer, 2010).
A single-domain antibody (sdAb), also known as a Nanobody or VHH, is an antibody fragment consisting of a single monomeric variable antibody domain (Hamers-Casterman et al., 1993). As a protein molecule, sdAb can also be injected into mammalian cells by the bacterial T3SS. Nanobodies that target GFP and amylase have successfully been delivered inside HeLa cells by the T3SS of enteropathogenic E. coli (EPEC) as well as Y. enterocolitica (Blancotoribio et al., 2010; Ittig et al., 2015), constituting an alternative way to target specific intracellular proteins.
8. Current limitations and problems
First, cytotoxicity of the delivery strain. The bacterial residual cytotoxicity prevents us from a prolonged co-incubation of the delivery strain with target cells, limiting the delivery time to around 5 h in the case of P. aeruginosa. Pathogen-associated molecular pattern molecules (PAMPs) are known to activate host inflammatory responses (Tang et al., 2012), and many of them are essential components for bacterial viability, such as cell wall component peptidoglycan and outer membrane component lipid A, thus unable to remove from the delivery strains, significantly impeding many of the in vivo applications of the T3SS mediated protein delivery technology.
Interestingly, however, the PAMPs seem do not influence the viability of cultured host cells in the case of P. aeruginosa, as T3SS defective mutant strains almost completely lost of their cytotoxic effect on the cultured mammalian cells. These suggested that the attenuated Pseudomonas strain is still capable of secreting unknown cytotoxins into the host cells. Indeed, a number of new effector proteins have recently been found to get injected into the host cells via the T3SS (Burstein et al., 2015; Neeld et al., 2014). Efforts are underway to delete all of these new type III effectors. It should also be pointed out that the translocon pores formed on the host cells membrane upon infection have previously been shown to cause cytotoxic effect (Galle et al., 2008), although the relative contribution of such pores to the cytotoxicity, compared to all other type III injected factors, remains to be determined.
Second, low level expression of certain foreign genes. In the P. aeruginosa delivery strain, the expression levels of foreign target genes vary greatly when placed under the control of exoS promoter, some are high while others dismal, although transcribed at comparable levels. Codon usage seems not the cause of such difference, as codon optimization did not improve the expression (unpublished results). We are addressing the possible involvement of mRNA secondary structures that hinder efficient translation, as this was shown to be the case in E.coli (Kudla et al., 2009).
Third, low translocation efficiencies of certain foreign proteins. Once expressed in the delivery strain, certain proteins (such as GFP) are expressed at reasonable level, yet very low amount was translocated into the target cells via the T3SS (Ittig et al., 2015; Jacobi et al., 1998). Due to the limited size of the internal conduit of the type III needle, it is believed that the proteins are translocated in an unfolded state. The T3SS may not be able to translocate proteins that are stably folded or those difficult to unfold. Efforts are underway to study the relevance of protein structure to the efficiency of protein translocation, with the hope to understand the dynamic relationship between protein structures and the translocation efficiency.
9. Conclusions
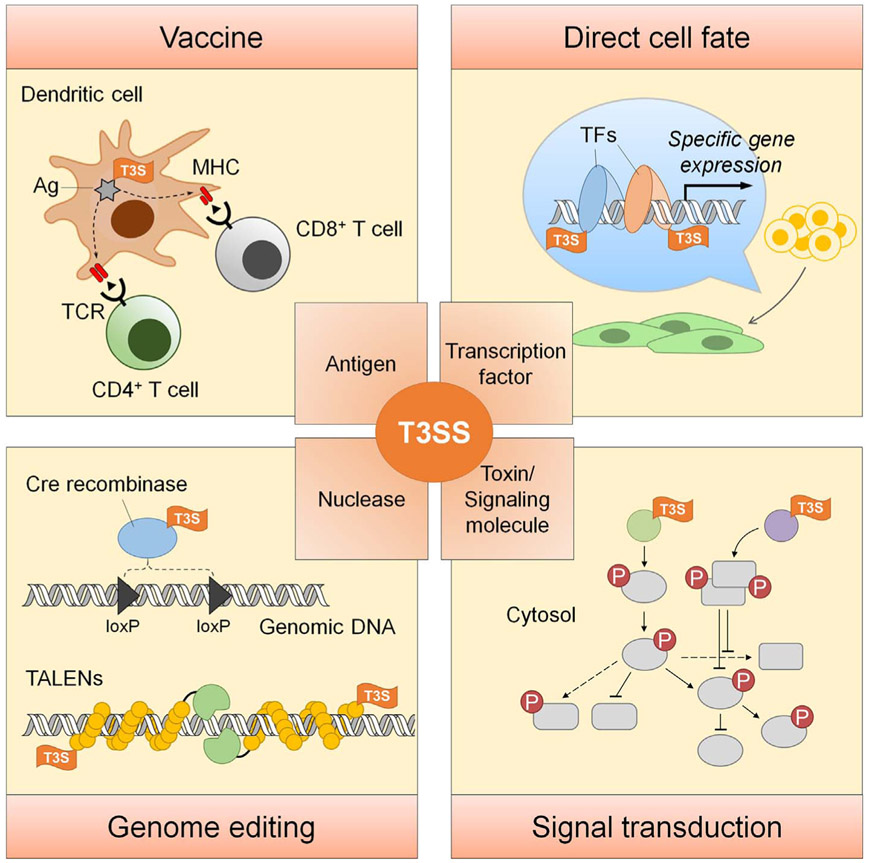
In-depth knowledge of the mechanisms of T3SS by which bacteria combat with their natural hosts, along with a firm understanding of bacterial engineering, enabled the development of innovative protein delivery tool using live bacterial T3SS. With this, we can achieve controlled delivery of target molecules into a large number of mammalian cells without the need for in vitro purification of the protein. Also, multiple components can be delivered simultaneously or sequentially at desired time points for desired amounts (by adjusting MOI), truly realizing temporal and spatial control of the delivery. So far, this protein delivery method has been applied to several biomedical research works, including antigen delivery for therapeutic vaccines, nuclease delivery for gene editing, transcription factor delivery for cells differentiation, and signaling molecule delivery for phosphoproteomics study (Fig. 6). Nevertheless, wider application of this tool needs to further address existing challenges such as substrate selectivity, stability of the fusion protein, post-translational modification of the target proteins, cytotoxicity of the delivery strain toward target cells, and quick elimination of the bacteria after protein delivery. Active investigation and improvements on these and many other remaining questions will provide easy to use protein delivery toolkits for biomedical researchers in the years to come. We expect that this efficient and transgene-free method of protein delivery will be adopted widely by a broader research community.
Fig. 6.
Summary of the applications of T3SS-mediated protein delivery. Ag, antigen; MHC, main histocompatibility complex; TCR, T-cell receptor; TF, transcription factor. T3S, type III secretion signal.
Acknowledgments
This work was supported by the National Institutes of Health/National Center for Advancing Translational Sciences Clinical and Translational Science Award to the University of Florida [Grant UL1 TR000064], Florida Department of Health Biomedical Research Programs [3KB04], International Science & Technology Cooperation Program of China [2015DFG32500], National Science Foundation of China [31670130, 31370168, 31370167 and 31600110] and Tianjin Municipal Science and Technology Commission, China [15JCYBJC53900 and 15JCZDJC33000].
References
- Arrach N, Cheng P, Zhao M, Santiviago CA, Hoffman RM, McClelland M, 2010. High-throughput screening for Salmonella avirulent mutants that retain targeting of solid tumors. Cancer Res. 70 (6), 2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Lim CH, Jia J, Santostefano K, Simmons C, Kasahara H, Wu W, Terada N, Jin S, 2015. Directed differentiation of embryonic stem cells into Cardiomyocytes by bacterial injection of defined transcription factors. Sci. Rep 5 (2), 15014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoin L, Toussaint B, Garban F, Le GA, Caulier B, Polack B, Laurin D, 2016. Targeted release of transcription factors for cell reprogramming by a natural microsyringe. Int. J. Pharm 513 (1–2), 678. [DOI] [PubMed] [Google Scholar]
- Bichsel C, Neeld DK, Hamazaki T, Wu D, Chang LJ, Yang L, Terada N, Jin S, 2011. Bacterial delivery of nuclear proteins into pluripotent and differentiated cells. PLoS One 6 (1), 477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichsel C, Neeld D, Hamazaki T, Chang LJ, Yang LJ, Terada N, Jin S, 2013. Direct reprogramming of fibroblasts to myocytes via bacterial injection of MyoD protein. Cell. Reprogramming 15 (2), 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancotoribio A, Muyldermans S, Frankel G, Fernández LÁ, 2010. Direct injection of functional single-domain antibodies from E. coli into human cells. PLoS One 5 (12), e15227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Bonas U, 2010. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol 48 (48), 419. [DOI] [PubMed] [Google Scholar]
- Boland A, Cornelis GR, 1998. Role of YopP in suppression of tumor necrosis factor alpha release by macrophages during Yersinia infection. Infect. Immun 66 (5), 1878–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Satanower S, Simovitch M, Belnik Y, Zehavi M, Yerushalmi G, Ben-Aroya S, Pupko T, Banin E, 2015. Novel type III effectors in Pseudomonas aeruginosa. MBio 6 (2), e00161–00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral MP, García P, Beceiro A, Rumbo C, Pérez A, Moscoso M, Bou G, 2017. Design of live attenuated bacterial vaccines based on D-glutamate auxotrophy. Nat. Commun 8, 15480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Bailer JA, Somia NV, Bogdanove AJ, Voytas DF, 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39 (12), e82–e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Studer L, 2011. Cell fate plug and play: direct reprogramming and induced pluripotency. Cell 145 (6), 827–830. [DOI] [PubMed] [Google Scholar]
- Chauchet X, Hannani D, Djebali S, Laurin D, Polack B, Marvel J, Buffat L, Toussaint B, Gouëllec AL, 2016. Poly-functional and long-lasting anticancer immune response elicited by a safe attenuated Pseudomonas Aeruginosa vector for antigens delivery. Mol. Ther. Oncolytics 3, 16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamekh M, Phalipon A, Quertainmont R, Salmon I, Sansonetti P, Allaoui A, 2008. Delivery of biologically active anti-inflammatory cytokines IL-10 and IL-1ra in vivo by the Shigella type III secretion apparatus. J. Immunol 180 (6), 4292–4298. [DOI] [PubMed] [Google Scholar]
- Chen LM, Briones G, Donis RO, Galán JE, 2006. Optimization of the delivery of heterologous proteins by the Salmonella enterica Serovar Typhimurium Type III secretion system for vaccine development. Infect. Immun 74 (10), 5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry AB, Daley GQ, 2012. Reprogramming cellular identity for regenerative medicine. Cell 148 (6), 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clairmont C, Lee KC, Pike J, Ittensohn M, Low KB, Pawelek J, Bermudes D, Brecher SM, Margitich D, Turnier J, 2000. Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimurium. J. Infect. Dis 181 (6), 1996–2002. [DOI] [PubMed] [Google Scholar]
- Cornelis GR, Wolfwatz H, 1997. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol 23 (5), 861–867. [DOI] [PubMed] [Google Scholar]
- Cristini S, Navone S, Canzi L, Acerbi F, Ciusani E, Hladnik U, De GP, Alessandri G, Colombo A, Parati E, 2010. Human neural stem cells: a model system for the study of Lesch-Nyhan disease neurological aspects. Hum. Mol. Genet 19 (10), 1939. [DOI] [PubMed] [Google Scholar]
- Crull K, Bumann D, Weiss S, 2011. Influence of infection route and virulence factors on colonization of solid tumors by Salmonella enterica serovar Typhimurium. Fems Immunol. Med. Microbiol 62 (1), 75. [DOI] [PubMed] [Google Scholar]
- David R, Franz WM, 2012. From pluripotency to distinct cardiomyocyte subtypes. Physiology 27 (3), 119–129. [DOI] [PubMed] [Google Scholar]
- Derouazi M, Wang Y, Marlu R, Epaulard O, Mayol JF, Pasqual N, Le GA, Polack B, Toussaint B, 2010. Optimal epitope composition after antigen screening using a live bacterial delivery vector: application to TRP-2. Bioeng. Bugs 1 (1), 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Rivas S, 2012. Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17 (11), 644–655. [DOI] [PubMed] [Google Scholar]
- Drehkopf S, Hausner J, Jordan M, Scheibner F, Bonas U, Büttner D, 2017. A TAL-based reporter assay for monitoring type III-dependent protein translocation in Xanthomonas. Methods Mol. Biol 121–139. [DOI] [PubMed] [Google Scholar]
- Duffour MT, Chaux P, Lurquin C, Cornelis G, Boon T, van der Bruggen P, 1999. A MAGE-A4 peptide presented by HLA-A2 is recognized by cytolytic T lymphocytes. Eur. J. Immunol 29 (10), 3329–3337. [DOI] [PubMed] [Google Scholar]
- Elsayed A, Futaki S, Harashima H, 2009. Delivery of macromolecules using arginine-rich cell-penetrating peptides: ways to overcome Endosomal entrapment. AAPS J. 11 (1), 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epaulard O, Toussaint B, Quenee L, Derouazi M, Bosco N, Villiers C, Le Berre R, Guery B, Filopon D, Crombez L, 2006. Anti-tumor immunotherapy via antigen delivery from a live attenuated genetically engineered Pseudomonas aeruginosa type III secretion system-based vector. Mol. Ther 14 (5), 656–661. [DOI] [PubMed] [Google Scholar]
- Epaulard O, Derouazi M, Margerit C, Marlu R, Filopon D, Polack B, Toussaint B, 2008. Optimization of a type III secretion system-based Pseudomonas aeruginosa live vector for antigen delivery. Clin. Vaccine Immunol 15 (2), 308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MF, Muller S, Wuest E, Cornelis GR, 2002. SycE allows secretion of YopE-DHFR hybrids by the Yersinia enterocolitica type III Ysc system. Mol. Microbiol 46 (4), 1183–1197. [DOI] [PubMed] [Google Scholar]
- Fisher SL, 2008. Glutamate racemase as a target for drug discovery. Microb. Biotechnol 1 (5), 345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm M, Felgner S, Kocijancic D, Rohde M, Hensel M, Rd CR, Erhardt M, Weiss S, 2015. Efficiency of conditionally attenuated Salmonella enterica serovar Typhimurium in bacterium-mediated tumor therapy. MBio 6 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE, Lara-Tejero M, Marlovits TC, Wagner S, 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol 68 (68), 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galle M, Schotte P, Haegman M, Wullaert A, Yang HJ, Jin S, Beyaert R, 2008. The Pseudomonas aeruginosa Type III secretion system plays a dual role in the regulation of caspase-1 mediated IL-1beta maturation. J. Cell. Mol. Med 12 (5A), 1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JT, Ferracci F, Jackson MW, Joseph SS, Pattis I, Plano LR, Fischer W, Plano GV, 2006. Measurement of effector protein injection by type III and type IV secretion systems by using a 13-residue phosphorylatable glycogen synthase kinase tag. Infect. Immun 74 (10), 5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier A, Finlay BB, 2003. Translocated intimin receptor and its chaperone interact with ATPase of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol 185 (23), 6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Vu GP, Bai Y, Yang E, Liu F, Lu S, 2010. Differential expression of Salmonella type III secretion system factors InvJ, PrgJ, SipC, SipD, SopA and SopB in cultures and in mice. Microbiology 156 (Pt 1), 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R, 1993. Naturally occurring antibodies devoid of light chains. Nature 363 (6428), 446–448. [DOI] [PubMed] [Google Scholar]
- Hapfelmeier S, Stecher B, Barthel M, Kremer M, Müller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, 2005. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J. Immunol 174 (3), 1675. [DOI] [PubMed] [Google Scholar]
- Hauser AR, 2009. The type III secretion system of Pseudomonas Aeruginosa: infection by injection. Nat. Rev. Microbiol 7 (7), 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesemann J, Gross U, Schmidt N, Laufs R, 1986. Immunochemical analysis of plasmid-encoded proteins released by enteropathogenic Yersinia sp. grown in calcium-deficient media. Infect. Immun 54 (2), 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J 22 (15), 3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi A, Ling QD, Kumar SS, Munusamy MA, Alarfaj AA, Chang Y, Kao SH, Lin KC, Wang HC, Umezawa A, 2015. Generation of pluripotent stem cells without the use of genetic material. Lab. Investig 95 (1), 26–42. [DOI] [PubMed] [Google Scholar]
- Hilbi H, Moss JE, Hersh D, Chen Y, Arondel J, Banerjee S, Flavell RA, Yuan J, Sansonetti PJ, Zychlinsky A, 1998. Shigella-induced apoptosis is dependent on Caspase-1 which binds to IpaB. J. Biol. Chem 273 (49), 32895–32900. [DOI] [PubMed] [Google Scholar]
- Hindley C, Philpott A, 2013. The cell cycle and pluripotency. Biochem. J 451 (2), 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Williams S, Schweizer HP, Lam JS, 1997. Molecular genetic analysis of the region containing the essential Pseudomonas aeruginosa asd gene encoding aspartate-beta-semialdehyde dehydrogenase. Microbiology 143 (Pt 3), 899–907. [DOI] [PubMed] [Google Scholar]
- Hu B, Laratejero M, Kong Q, Galán JE, Liu J, 2017. In situ molecular architecture of the Salmonella Type III secretion machine. Cell 168 (6), 1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes NE, Ingham PW, Lim WA, Marshall CJ, Massagué J, Pawson T, 2013. Signalling change: signal transduction through the decades. Nat. Rev. Mol. Cell Biol 14 (6), 393–398. [DOI] [PubMed] [Google Scholar]
- Inoue M, Tomizawa K, Matsushita M, Lu YF, Yokoyama T, Yanai H, Takashima A, Kumon H, Matsui H, 2006. p53 protein transduction therapy: successful targeting and inhibition of the growth of the bladder cancer cells. Eur. Urol 49 (1), 161–168. [DOI] [PubMed] [Google Scholar]
- Ionta V, Liang W, Kim EH, Rafie R, Giacomello A, Marbán E, Cho HC, 2015. SHOX2 overexpression favors differentiation of embryonic stem cells into cardiac pacemaker cells, improving biological pacing ability. Stem Cell Rep. 4 (1), 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriarte M, Cornelis GR, 1998. YopT, a new Yersinia Yop effector protein, affects the cytoskeleton of host cells. Mol. Microbiol 29 (3), 915–929. [DOI] [PubMed] [Google Scholar]
- Ittig SJ, Schmutz C, Kasper CA, Amstutz M, Schmidt A, Sauteur L, Vigano MA, Low SH, Affolter M, Cornelis GR, 2015. A bacterial type III secretion-based protein delivery tool for broad applications in cell biology. J. Cell Biol 211 (4), 913–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi CA, Roggenkamp A, Rakin A, Zumbihl R, Leitritz L, Heesemann J, 1998. In vitro and in vivo expression studies of yopE from Yersinia enterocolitica using the gfp reporter gene. Mol. Microbiol 30 (4), 865–882. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A, 2005. Rho GTPases: biochemistry and biology. Ann. Rev. Cell Dev. Biol 21 (1), 247. [DOI] [PubMed] [Google Scholar]
- Jellbauer S, Panthel K, Hetrodt JH, Rüssmann H, 2012. CD8 T-cell induction against vascular endothelial growth factor receptor 2 by Salmonella for vaccination purposes against a murine melanoma. PLoS One 7 (4), e34214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Jin Y, Bian T, Wu D, Yang L, Terada N, Wu W, Jin S, 2014. Bacterial delivery of TALEN proteins for human genome editing. PLoS One 9 (3), e91547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Bai F, Jin Y, Santostefano KE, Ha UH, Wu D, Wu W, Terada N, Jin S, 2015. Efficient gene editing in pluripotent stem cells by bacterial injection of transcription activator-like effector nuclease proteins. Stem Cells Transl. Med 4 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juárez-Rodríguez MD, Yang J, Kader R, Alamuri P, Roy Curtiss I, Clark-Curtiss JE, 2012. Live attenuated Salmonella vaccines displaying regulated delayed lysis and delayed antigen synthesis to confer protection against mycobacterium tuberculosis. Infect. Immun 80 (2), 815–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JJ, Husse B, Rimmbach C, Krebs S, Stieber J, Steinhoff G, Dendorfer A, Franz WM, David R, 2014. Programming and isolation of highly pure physiologically and pharmacologically functional sinus-nodal bodies from pluripotent stem cells. Stem Cell Rep. 2 (5), 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N, Liang W, Marban E, Cho HC, 2013. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol 31 (1), 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman M, Jia J, Zeng L, Ha U, Chow M, Jin S, 2000. Pseudomonas aeruginosa mediated apoptosis requires ADP-ribosylating activity of the ExoS. Microbiology 146 (10), 2531–2541. [DOI] [PubMed] [Google Scholar]
- Kehat I, Khimovich L, Caspi O, Gepstein A, Shofti R, Arbel G, Huber I, Satin J, Itskovitz-Eldor J, Gepstein L, 2004. Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat. Biotechnol 22 (10), 1282–1289. [DOI] [PubMed] [Google Scholar]
- Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR, 2017. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol 35 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I, Itterson M, Bermudes D, 2009. Tumor-targeted Salmonella typhimurium overexpressing cytosine deaminase: a novel, tumor-selective therapy. Methods Mol. Biol 542, 649. [DOI] [PubMed] [Google Scholar]
- Komor AC, Badran AH, Liu DR, 2016. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168 (1–2), 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Matsushima Y, Nakamura D, Uralil J, Laratejero M, Sukhan A, Galán JE, Aizawa SI, 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280 (5363), 602–605. [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB, 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324 (5924), 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashev M, Stenta M, Schmelz S, Amstutz M, Wiesand U, Castaño-Díez D, Degiacomi MT, Münnich S, Bleck CK, Kowal J, 2013. In situ structural analysis of the Yersinia enterocolitica injectisome. elife 2, e00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Tejero M, Kato J, Wagner S, Liu X, Galán JE, 2011. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331 (6021), 1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gouëllec A, Chauchet X, Laurin D, Aspord C, Verove J, Wang Y, Genestet C, Trocme C, Ahmadi M, Martin S, 2013. A safe bacterial microsyringe for in vivo antigen delivery and immunotherapy. Mol. Ther 21 (5), 1076–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Hongmei, Fujimoto Lisa, Sasakawa Naoko, Shirai Noriko, Ohkame Saya, 2015. Precise correction of the dystrophin gene in Duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem cell reports 4 (1), 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J, 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94 (4), 491. [DOI] [PubMed] [Google Scholar]
- Mackay JA, Szoka FC Jr., 2003. HIV TAT protein transduction domain mediated cell binding and intracellular delivery of nanoparticles. J. Dispers. Sci. Technol 24 (3), 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galán JE, Unger VM, 2004. Structural insights into the assembly of the Type III secretion needle complex. Science 306 (5698), 1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels T, Wattiau P, Brasseur R, Ruysschaert JM, Cornelis G, 1990. Secretion of Yop proteins by Yersiniae. Infect. Immun 58 (9), 2840–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeld D, Jin Y, Bichsel C, Jia J, Guo J, Bai F, Wu W, Ha UH, Terada N, Jin S, 2014. Pseudomonas aeruginosa injects NDK into host cells through a type III secretion system. Microbiology 160 (Pt 7), 1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemunaitis J, Cunningham C, Senzer N, Kuhn J, Cramm J, Litz C, Cavagnolo R, Cahill A, Clairmont C, Sznol M, 2003. Pilot trial of genetically modified, attenuated Salmonella expressing the E. coli cytosine deaminase gene in refractory cancer patients. Cancer Gene Ther. 10 (10), 737. [DOI] [PubMed] [Google Scholar]
- Nishikawa H, Sato E, Briones G, Chen LM, Matsuo M, Nagata Y, Ritter G, Jäger E, Nomura H, Kondo S, 2006. In vivo antigen delivery by a Salmonella typhimurium type III secretion system for therapeutic cancer vaccines. J. Clin. Investig 116 (7), 1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Mann M, 2013. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 12 (12), 3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panthel K, Meinel KM, Domçnech VES, Geginat G, Linkemann K, Busch DH, Rüssmann H, 2006. Prophylactic anti-tumor immunity against a murine fibrosarcoma triggered by the Salmonella type III secretion system. Microbes Infect. 8 (10), 2539–2546. [DOI] [PubMed] [Google Scholar]
- Panthel K, Meinel KM, Sevil Domènech VE, Trülzsch K, Rüssmann H, 2008. Salmonella type III-mediated heterologous antigen delivery: a versatile oral vaccination strategy to induce cellular immunity against infectious agents and tumors. Int. J. Med. Microbiol 298 (1–2), 99–103. [DOI] [PubMed] [Google Scholar]
- Polack B, Vergnaud S, Paclet MH, Lamontte D, Toussaint B, Morel F, 2000. Protein delivery by Pseudomonas type III secretion system: ex vivo complementation of p67(phox)-deficient chronic granulomatous disease. Biochem. Biophys. Res. Commun 275 (3), 854–858. [DOI] [PubMed] [Google Scholar]
- Puri PL, Mercola M, 2012. BAF60 A, B, and Cs of muscle determination and renewal. Genes Dev. 26 (24), 2673–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radics J, Königsmaier L, Marlovits TC, 2014. Structure of a pathogenic type 3 secretion system in action. Nat. Struct. Mol. Biol 21 (1), 82–87. [DOI] [PubMed] [Google Scholar]
- Rádis-Baptista G, Campelo IS, Jrl M, Melo LM, Vjf F, 2017. Cell-penetrating peptides (CPPs): from delivery of nucleic acids and antigens to transduction of engineered nucleases for application in transgenesis. J. Biotechnol 252, 15. [DOI] [PubMed] [Google Scholar]
- Rajala K, Pekkanen-Mattila M, Aalto-Setala K, 2011. Cardiac differentiation of pluripotent stem cells. Stem Cells Int. 2011, 383709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietsch A, Valletgely I, Dove SL, Mekalanos JJ, 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas Aeruginosa. Proc. Natl. Acad. Sci. U. S. A 102 (22), 8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers L, Gamez A, Riek R, Ghosh P, 2008. The type III secretion chaperone SycE promotes a localized disorder-to-order transition in the natively unfolded effector YopE. J. Biol. Chem 283 (30), 20857–20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosqvist R, Magnusson KE, Wolf-Watz H, 1994. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13 (4), 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüssmann H, Shams H, Poblete F, Fu Y, Galán JE, Donis RO, 1998. Delivery of epitopes by the Salmonella type III secretion system for vaccine development. Science 281 (5376), 565. [DOI] [PubMed] [Google Scholar]
- Rüssmann H, Weissmüller A, Geginat G, Igwe EI, Roggenkamp A, Bubert A, Goebel W, Hof H, Heesemann J, 2000. Yersinia enterocolitica-mediated translocation of defined fusion proteins to the cytosol of mammalian cells results in peptide-specific MHC class I-restricted antigen presentation. Eur. J. Immunol 30 (5), 1375. [DOI] [PubMed] [Google Scholar]
- Rüssmann H, Igwe EI, Sauer J, Hardt WD, Bubert A, Geginat G, 2001. Protection against murine listeriosis by oral vaccination with recombinant Salmonella expressing hybrid Yersinia type III proteins. J. Immunol 167 (1), 357–365. [DOI] [PubMed] [Google Scholar]
- Sandkvist M, 2001. Biology of type II secretion. Mol. Microbiol 40 (2), 271. [DOI] [PubMed] [Google Scholar]
- Santostefano KE, Hamazaki T, Biel NM, Jin S, Umezawa A, Terada N, 2015. A practical guide to induced pluripotent stem cell research using patient samples. Lab. Investig. 95 (1), 4–13. [DOI] [PubMed] [Google Scholar]
- Sarosiek KA, Chi X, Bachman JA, Sims JJ, Montero J, Patel L, Flanagan A, Andrews DW, Sorger P, Letai A, 2013. BID preferentially activates BAK while BIM preferentially activates BAX, affecting chemotherapy response. Mol. Cell 51 (6), 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo BJ, Hong YJ, Do JT, 2017. Cellular reprogramming using protein and cell-penetrating peptides. Int. J. Mol. Sci (3), 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea JE, Hensel M, Gleeson C, Holden DW, 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A 93 (6), 2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen DK, Quenee L, Bonnet M, Kuhn L, Derouazi M, Lamotte D, Toussaint B, Polack B, 2008. Orf1/SpcS chaperones ExoS for type three secretion by Pseudomonas aeruginosa. 生物医学与环境科学 21 (2), 103–109. [DOI] [PubMed] [Google Scholar]
- Sory MP, Cornelis GR, 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol 14 (3), 583–594. [DOI] [PubMed] [Google Scholar]
- Sundin C, Thelaus J, Bröms JE, Forsberg A, 2004. Polarisation of type III translocation by Pseudomonas aeruginosa requires PcrG, PcrV and PopN. Microb. Pathog 37 (6), 313. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S, 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 (4), 663–676. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S, 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 (5), 861–872. [DOI] [PubMed] [Google Scholar]
- Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT, 2012. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol. Rev 249 (1), 158–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, Sherry RM, Topalian SL, Yang JC, Stock F, 2002. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J. Clin. Oncol 20 (1), 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint B, Chauchet X, Wang Y, Polack B, Le GA, 2013. Live-attenuated bacteria as a cancer vaccine vector. Exp. Rev. Vaccines 12 (10), 1139. [DOI] [PubMed] [Google Scholar]
- Trülzsch K, Roggenkamp A, Aepfelbacher M, Wilharm G, Ruckdeschel K, Heesemann J, 2003. Analysis of chaperone-dependent Yop secretion/translocation and effector function using a mini-virulence plasmid of Yersinia enterocolitica. Int. J. Med. Microbiol 293 (2), 167–177. [DOI] [PubMed] [Google Scholar]
- Trülzsch K, Geginat G, Sporleder T, Ruckdeschel K, Hoffmann R, Heesemann J, Rüssmann H, 2005. Yersinia outer protein P inhibits CD8 T cell priming in the mouse infection model. J. Immunol 174 (7), 4244–4251. [DOI] [PubMed] [Google Scholar]
- Truong DJ, Kühner K, Kühn R, Werfel S, Engelhardt S, Wurst W, Ortiz O, 2015. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 43 (13), 6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Lykken GL, Yahr TL, Wickner WT, 2005. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa Type III secretion system. Proc. Natl. Acad. Sci. U. S. A 102 (28), 9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Brutinel ED, Yahr TL, 2007. Translocation of ExsE into Chinese hamster ovary cells is required for transcriptional induction of the Pseudomonas aeruginosa type III secretion system. Infect. Immun 75 (9), 4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallis AJ, Yahr TL, Barbieri JT, Frank DW, 1999. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect. Immun 67 (2), 914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelenburg SB, Palmer AE, 2010. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat. Methods 7 (4), 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Hou Y, Huang H, Liu GR, White AP, Liu SL, 2008. Two oral HBx vaccines delivered by live attenuated Salmonella: both eliciting effective anti-tumor immunity. Cancer Lett. 263 (1), 67–76. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gouëllec AL, Chaker H, Asrih H, Polack B, Toussaint B, 2012. Optimization of antitumor immunotherapy mediated by type III secretion system-based live attenuated bacterial vectors. J. Immunother 35 (3), 223. [DOI] [PubMed] [Google Scholar]
- Wattiau P, Cornelis GR, 1993. SycE, a chaperone-like protein of Yersinia enterocolitica involved in Ohe secretion of YopE. Mol. Microbiol 8 (1), 123–131. [DOI] [PubMed] [Google Scholar]
- Woestyn S, Allaoui A, Wattiau P, Cornelis GR, 1994. YscN, the putative energizer of the Yersinia Yop secretion machinery. J. Bacteriol 176 (6), 1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölke S, Heesemann J, 2012. Probing the cellular effects of bacterial effector proteins with the Yersinia toolbox. Future Microbiol 7 (4), 449–456. [DOI] [PubMed] [Google Scholar]
- Wölke S, Ackermann N, Heesemann J, 2011. The Yersinia enterocolitica type 3 secretion system (T3SS) as toolbox for studying the cell biological effects of bacterial Rho GTPase modulating T3SS effector proteins. Cell. Microbiol 13 (9), 1339–1357. [DOI] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A, 2009. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458 (7239), 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DA, Li T, Yang B, Spalding MH, 2014. TALEN-mediated genome editing: prospects and perspectives. Biochem. J 462 (1), 15–24. [DOI] [PubMed] [Google Scholar]
- Xin W, Wanda SY, Zhang X, Santander J, Scarpellini G, Ellis K, Alamuri P, Curtiss R, 2012. The Asd + −DadB+ dual-plasmid system offers a novel means to deliver multiple protective antigens by a recombinant attenuated Salmonella vaccine. Infect. Immun 80 (10), 3621–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G, Husseiny MI, Song L, Erdreich-Epstein A, Shackleford GM, Seeger RC, Jäckel D, Hensel M, Metelitsa LS, 2010. Novel cancer vaccine based on genes of Salmonella pathogenicity island 2. Int. J. Cancer 126 (11), 2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahr TL, Wolfgang MC, 2006. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol 62 (3), 631–640. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Blau HM, 2010. Nuclear reprogramming to a pluripotent state by three approaches. Nature 465 (7299), 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetche B, Volz SE, Zhang F, 2015. A split Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol 33 (2), 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Jiang J, Han P, Yuan Q, Zhang J, Zhang X, Xu Y, Cao H, Meng Q, Chen L, Tian T, Wang X, Li P, Hescheler J, Ji G, Ma Y, 2011. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 21 (4), 579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Freed CR, 2009. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells 27 (11), 2667–2674. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhou P, Cai J, Yang G, Liang S, Ren D, 2010. Tumor antigen delivered by Salmonella III secretion protein fused with heat shock protein 70 induces protection and eradication against murine melanoma. Cancer Sci. 101 (12), 2621–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Nervi P, Dürrenberger M, Seelig J, 2005. The cationic cell-penetrating peptide CPP(TAT) derived from the HIV-1 protein TAT is rapidly transported into living fibroblasts: optical, biophysical, and metabolic evidence. Biochemistry 44 (1), 138. [DOI] [PubMed] [Google Scholar]